Abstract
Introduction:
Successful implementation of whole slide imaging (WSI) for routine clinical practice has been accomplished in only a few pathology laboratories worldwide. We report the transition to an effective and complete digital surgical pathology workflow in the pathology laboratory at Cannizzaro Hospital in Catania, Italy.
Methods:
All (100%) permanent histopathology glass slides were digitized at ×20 using Aperio AT2 scanners. Compatible stain and scanning slide racks were employed to streamline operations. eSlide Manager software was bidirectionally interfaced with the anatomic pathology laboratory information system. Virtual slide trays connected to the two-dimensional (2D) barcode tracking system allowed pathologists to confirm that they were correctly assigned slides and that all tissues on these glass slides were scanned.
Results:
Over 115,000 glass slides were digitized with a scan fail rate of around 1%. Drying glass slides before scanning minimized them sticking to scanner racks. Implementation required introduction of a 2D barcode tracking system and modification of histology workflow processes.
Conclusion:
Our experience indicates that effective adoption of WSI for primary diagnostic use was more dependent on optimizing preimaging variables and integration with the laboratory information system than on information technology infrastructure and ensuring pathologist buy-in. Implementation of digital pathology for routine practice not only leveraged the benefits of digital imaging but also creates an opportunity for establishing standardization of workflow processes in the pathology laboratory.
Keywords: Digital pathology, informatics, pathology, whole slide imaging, workflow
INTRODUCTION
Digital pathology has matured considerably since the advent of whole slide imaging (WSI) almost 20 years ago. Although diagnoses based on examining glass slides using a traditional optical microscope still remain the gold standard, an increasing number of papers have been published showing the validity of a “digital” diagnosis using WSI.[1,2,3,4,5,6,7,8,9,10] Recent progress achieved with WSI technology (e.g., improved image resolution, throughput of scanners, image management software, teleconsultation tools, and image analysis algorithms) and declining storage costs have made the proposition of investing in digital pathology more attractive for laboratories. Accordingly, many pathology laboratories have considered large-scale implementation of WSI for routine clinical histopathology practice.[10,11,12,13,14,15,16,17,18,19,20,21,22] However, the number of pathology departments that have actually gone “fully digital” is relatively low despite the well-documented advantages of adopting WSI (e.g., workload balancing, telepathology, and enhanced multidisciplinary tumor boards). In 2015, the Department of Pathology in Cannizzaro Hospital in Catania, Italy, embarked on digitizing all of their histology cases using WSI.
The aim of this paper is to report the Pathology laboratory's experience at Cannizzaro Hospital in Catania following their digital transition and to highlight effective solutions implemented and instructive problems encountered in developing a fully “digital workflow.”
METHODS
In 2015 at Cannizzaro Hospital in Catania (Italy), the department of pathology decided to start using digital slides for routine surgical pathology practice. The intent was to digitize all (100%) histopathology glass slides. Only slides obtained from formalin-fixed paraffin-embedded tissue (FFPE) were digitized. This included hematoxylin and eosin (H& E), special histochemical, and immunohistochemical stained slides, but not immunofluorescence. Scanning of all frozen section slides was abandoned because of the high scan failure rate experienced with these handmade glass slides. Digitizing cytology slides was also not undertaken due to the need for Z-stack image acquisition which increases scan time and file size.[19] For cytology cases, snapshots were instead directly taken from the cytology glass slides using a digital camera mounted to a microscope (Leica DM4000) and these static images were saved directly to the laboratory information system (Pathox 13.0.0). The digital pathology system employed was initially validated by comparing glass slides and digital images. The scanning system was operated by technologists who were trained to use these devices to support routine daily work. The Canizzaro Hospital is not a university-based hospital, so there are no pathology trainees. The digital pathology system was deployed primarily to support clinical diagnostic work. However, the system was also adopted to assist with showing select cases as whole slide images in multidisciplinary team meetings or tumor boards.
Imaging technology
The laboratory employed the Leica digital pathology platform. Aperio AT2 whole slide scanners (400 slide capacity) were used to scan slides. The image management system was eSlide Manager (Version 12.1.0.5029 Aperio, Leica). A total of 300 slides were continuously digitized daily. Before the end of each work day, the scanner was loaded with glass slides and an overnight batch scanning session initiated. This workflow resulted in a 12 h delay in the delivery of slides to pathologists; however, thus far this has not caused any major impact to clinical practice. Given that potential downtime of an overnight scanning session (e.g., caused by glass slides sticking to scanner racks) could significantly impede workflow in the laboratory, two AT2 scanners were run simultaneously. Implementation of continuous workflow within the laboratory (i.e., cutting, staining, and then immediate scanning before signout activity) allowed the laboratory to achieve complete slide creation and digitization of 300 slides within the same day. Regular maintenance required white/color balance and line adjustment of the scanner to be made once a month.
Information technology infrastructure
eSlide Manager was bidirectionally integrated with the anatomic pathology laboratory information system (APLIS) (Pathox version 13.0.0, Tesi Elettronica e Sistemi Informativi S.P.A., Milan, Italy). Interface exchanges were handled through HL7 version 2.5 messages. The integration took 1 week of work. This is in contrast with other reported similar implementations that required more time to deploy.[22,23] The workstation used for pathologists to review digital slides included a HP computer with an Intel Core i5-4590 CPU @ 3.30GHz 3.30 GHz processor, installed memory of 8 GB, and a 64 bit operating system. The graphic interface was NVIDIA NVS315. Digital images were displayed on a 32’’ LED monitor of 4K resolution (3840 pixels × 2160 pixels).
e-Slides were directly accessed from the APLIS. Specifically, a virtual slide tray was created and incorporated within the APLIS [Figure 1]. Accessioning of cases and real-time tracking of digital slides occurred directly in the APLIS. The creation of a single slide tray within the APLIS Pathox that displays the macroimage (thumbnail) of several slides permitted incorporation of digital slides that could be acquired from different scanners (e.g., Aperio AT2, Hamamatsu NanoZoomer XR, Roche iscan Coreo) without disrupting end-user workflow [Figure 2]. All images were saved with network-attached storage (96TB Qnap NAS TVS-EC1280U-SAS-RP) using a 100 Mbit/s network connection. A decision was made not to delete any images so that archived whole slide images were always available when they were opened from the APLIS. Any rescanned images replaced previous images so that pathologists always had access to the last saved images.
Figure 1.
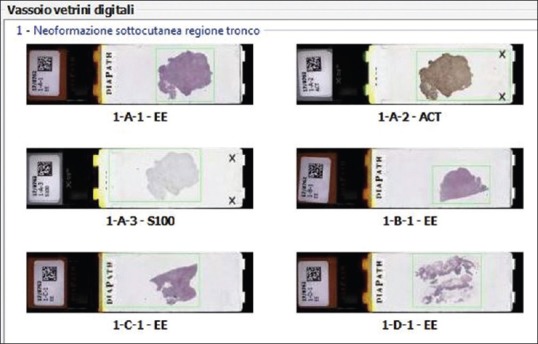
Image of a virtual slide tray created and incorporated within the anatomical pathology laboratory information system
Figure 2.
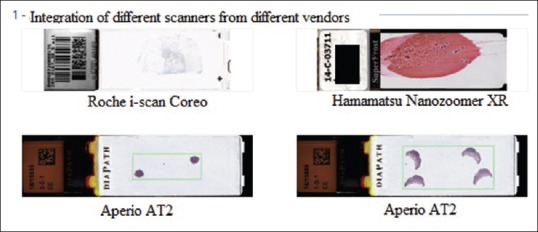
The creation of a single slide tray within the anatomic pathology laboratory information system Pathox is shown that displays the macroimage (thumbnail) of several slides which permits incorporation of digital slides acquired from different scanners
Digital workflow
Before being scanned, all glass slides received a two-dimensional (2D) tracking barcode on their label that was read by the WSI scanner. By reading the 2D barcode, each eSlide was automatically assigned to the correct case within the APLIS. The scanning station was situated in the same room used to house the staining and coverslipping machine (Sakura Tissue-Tek Prisma and Coverslipper). Stained and coverslipped glass slides were loaded into scanner slide racks after they were dried at 60°C in an oven for an hour. Special attention was dedicated to keep glass slides clean. Compatibility between the staining and scanning racks streamlined operations [Figure 3a and b]. The choice of scanner took into account the need for such compatibility making sure that technicians were able to easily load slides from staining racks directly into the Aperio AT2. Virtual slide trays offered pathologists a double check to verify (i) that e-slides were correctly displayed [Figure 4a] and (ii) whether the tissue present on the glass slide had been correctly recognized by the “tissue finder” of the scanning system and then scanned [Figure 4b]. After scanning, glass slides were still assembled onto trays and physically delivered to the assigned pathologist. Each pathologist could also access their assigned cases directly from the APLIS, thus supporting the hybrid possibility to access and consult digital slides from the virtual tray on a monitor or to render a diagnosis with glass slides using a conventional microscope.
Figure 3.
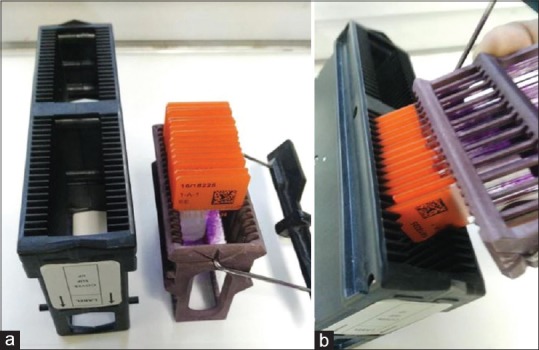
(a) Compatible scanner and staining slide racks (b) allow slides to be easily loaded for scanning with minimal laborious human intervention
Figure 4.
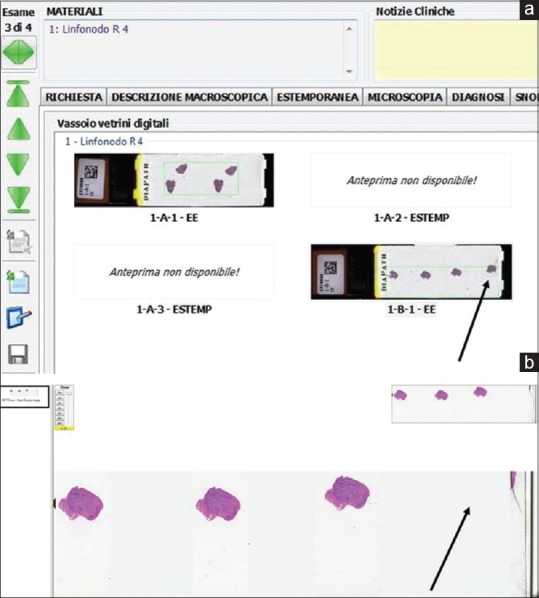
Screenshot of the virtual slide tray incorporated within the anatomic pathology laboratory information system showing how whole slide images are connected with the tracking system. The virtual slide tray displays macroimages of scanned slides that allow pathologists to check whether tissue present on the glass slide was correctly recognized by the “tissue finder” tool of the scanning system. (a) Two slides of a case are shown that were missing, due to a failure to read the barcode during scanning. (b) In this case, the tissue finder tool was unable to recognize the entire tissue sample
RESULTS
Following go live of the production system >115,000 histology glass slides were digitized. The scan fail rate experienced was around 1%. The reasons for cases failing to scan were due to overly thick slides and excess glue present on the coverslip. Excess mounting medium not only lead to air bubbles, but when it obscured the slide labels, this affected the automatic registration of scanned slides in some instances. Only a few slide breakages were documented. Drying glass slides before scanning reduced the likelihood of them sticking to the scanner racks.
Digital slides were on average 340 MB in size when scanned at ×20. Scan time was around 2 min per slide. Of note, the entire 1st year of implementation required extensive LEAN modifications to be made to workflow processes within the laboratory including the adoption of standardized protocols and the introduction of a 2D barcode tracking system. Incorrectly printed barcodes were encountered in 0.5% of cases that resulted in reading errors. Focal focus problems were present in about 0.5%–1% of scanned slides; specifically, we found that focus problems sometimes affected either a part or the entire surface of one or multiple slides per case. Slide focus issues seemed to correlate with the number of focus points produced at the time of scanning. The scanner was subsequently configured to minimize such errors. Overall, this did not preclude the ability to render a final diagnosis. Pathologists were responsible for checking the quality of scanned images.
DISCUSSION
Our experience indicates that WSI was successfully adopted for the purpose of performing primary diagnoses in surgical pathology. All diagnoses were interchangeably performed on either a computer monitor or light microscope. Scanning glass slides also permitted pathologists to easily and immediately retrieve archival slides within the APLIS by simply clicking on available whole slide images from prior cases. This approach significantly reduced the time required to retrieve slides. Digital slides were also easier to share with colleagues. The laboratory is in the process of implementing a dedicated tool within the APLIS (“Telepathox”) to further facilitate teleconsultation using secure HTTPS protocols. Evaluating and discussing pathological findings present in certain cases (e.g., the margin status of a tumor) at tumor boards on a computer monitor or conference room screen proved to be preferable and was well received by clinician colleagues.
All histopathology slides prepared from FFPE tissues were digitized at Cannizzaro Hospital in Catania. However, although our laboratory had gone “fully digital,” handmade frozen section slides and cytology slides were not scanned. Scanning all frozen sections was omitted because of logistic reasons and technical problems. Scanners were not located in close proximity to the cryostats and staining machines used for frozen sections, requiring technicians to move between different rooms to prepare frozen sections and digitize slides. The resulting delay was exacerbated when pathologists requested additional frozen section levels. Furthermore, hand-prepared frozen section slides were out of focus in many cases, and tissue sections were sometimes not properly recognized by the scanner's tissue finder tool. Our laboratory is accordingly evaluating hybrid robotic and WSI instruments (e.g., LV1 by Aperio) as a possible solution for digitizing frozen section slides.
The validation process employed followed the guidelines published by the College of American Pathologists.[11] Glass slides with H&E-stained FFPE tissue sections, histochemical stains, and immunohistochemical slides were all validated. Even though digital slides were subsequently validated for primary diagnosis, our laboratory supported a hybrid workflow for 1 year, as in addition to using digital slides, pathologists were subsequently still provided with glass slides. At the beginning of our journey, this hybrid approach allowed pathologists to become more confident and overcome their learning curve with WSI. However, this hybrid approach did not allow users (pathologists and technicians) to immediately benefit from all of the advantages of digitization, with a reduction in workload related to not delivering glass slides being the most evident advantage of going fully digital. Moreover, although we did not perform an accurate timed workflow analysis, a decrease in the turnaround time of histological diagnoses was apparent when just a digital workflow was applied. The average turnaround time for our cases measured from complete “stained status” of a glass slide to “digitized status” when the WSI was available was around 3 h. This led to a gain of 8 h in our laboratory due to skipping the prior lengthy manual case assembly and delivery of glass slides to pathologists. Immediate selection of slides for running additional stains or for molecular analysis, and immediate access to review previous slides belonging to patients, strongly favored abandoning the hybrid approach involving glass slides. The LEAN approach embraced to facilitate digital workflow, and the integrated tracking system with complete interface to the APLIS, was a prerequisite to the successful shift toward primary digital histological diagnosis. This is in line with the findings reported by recent similar publications.[24,25,26,27]
We started scanning slides using both ×20 and ×40 magnifications. The ×40 scanning time (4 min) took twice as long compared to the ×20 scanning procedure (2 min). Generated image files were also four times larger (1.2 GB/slide) from ×40 than ×20 scans. Although some authors have suggested that scans at ×40 are more favorable for viewing certain cellular and/or nuclear details,[14] we found that routinely scanning our surgical pathology slides at ×20 was satisfactory for the majority of cases. Setting the viewer (Imagescope) to maximum magnification (200%) allowed pathologists to achieve a “virtual (digital) ×40” with only minimal blurring of the slide. However, resolution issues were encountered when evaluating breast carcinoma, melanoma, cases for mitotic figures, and Helicobacter pylori in gastric biopsies. In these cases, pathologist deferred their examination to the glass slides.
The ability to rapidly create and scan around 300 slides daily was attainable because the laboratory utilized loading of compatible slide staining racks directly into the Aperio AT-2 scanner which required minimal human intervention. In fact, Sakura's staining racks are also compatible with several other scanners including the iScan Coreo by Roche, the Ultra Fast Scanner by Philips and Pannoramic 1000 by 3DHistech. In the foreseeable future, it is likely that more vendors will support “plug and play” of their devices and that the process of slide creation, staining, coverslipping, and scanning will become streamlined with fewer steps and reliance on less instruments that can perform more or all of these functions. Apart from the scanner used, our experience showed that scan time varied with the type of tissue present on the slide (i.e., larger tissue pieces took longer to scan than small biopsy specimens) and was influenced by the number of focus points used when digitizing slides, as well as on the performance of the network when transferring data to the server. Limiting the number of focus points during scanning to accelerate the scanning operation seems feasible given that pathologists were able to still satisfactorily interpret digital slides even though a small proportion of them had focal areas that were out of focus. Other measures that ensured the successful implementation of WSI were running two AT2 scanners simultaneously to expedite scanning and afford redundancy. With the everyday use of scanners, the laboratory realized that running several scanners with fewer slides, but with high throughput could be more efficient and practical than having few scanners with high slide capacity. In an ideal workflow, high slide capacity scanners should be used only for overnight scanning sessions. Our scanning approach combined with the automatic assignment of cases to pathologists allowed whole slide images to be made available to pathologists on the same day as slide staining.
Successful implementation of WSI was also ensured by drying of glass slides before scanning to prevent scan failures as a result of sticky material, as well as making sure that the digital pathology system eSlide manager was integrated with the APLIS. Employing a bidirectional interface with the APLIS simplified workflow and leveraged the laboratory barcode tracking system and thereby promoted the willingness of pathologists to adopt digital pathology. As reported by others, our experience similarly indicated that the transition to digital pathology was more dependent on optimizing preimaging variables and supporting an integrated, standardized workflow than on information technology problems.[24] Indeed, Cheng et al. from Singapore who routinely use WSI in their clinical practice specified that suboptimal glass slide preparation (e.g., dirty slide surfaces and entrapped air bubbles) can limit the quality of digital slides and hence their ability to be used to make diagnoses.[1]
To date, lack of interoperability between proprietary images and different image viewers has been one of the major barriers to the widespread adoption of WSI.[24] By creating a virtual slide tray within the APLIS, which may integrate different scanners from different vendors, allowed us to have a continuous undisrupted end-user workflow. This was possible either due to compatibility of the image management system with scanners (in our case eSlide manager was compatible with Hamamatsu scanners) or through integrating different image management systems from different vendors with the APLIS (i.e., eSlide manager and the Virtuoso image management system from Roche). This helps overcome the lack of interoperability between proprietary images. We believe that, in the near future, more open platforms that permit integrating different scanners will be more readily available.
Routinely scanning our surgical pathology slides at ×20 was sufficient for the majority of cases even though some authors such as Al-Janabi et al. concluded that the use of ×40 was more favorable for viewing certain cellular and/or nuclear details.[14] While some institutions have developed an entire “digital pathology service” to manage and sustain their digital pathology operation that includes a team composed of pathologists, technicians, and laboratory managers,[1] it is important to highlight that, as in our laboratory, pathologists often bear the ultimate responsibility in assuring that digital slides are of diagnostic quality.
CONCLUSION
Implementing a digital pathology system indirectly created an opportunity for standardization of procedures, thereby introducing a LEAN process within the laboratory from grossing of specimens to scanning slides. Now that the pathology laboratory at Cannizzaro Hospital in Catania has successfully transitioned to a digital pathology platform we are enabled to start taking advantage of our ability to use digital slides more frequently for interdepartmental meetings, more easily share whole slide images (e.g., for teleconsultation with experts), and embrace image analysis. The next phase of our digital pathology journey will explore scanning of cytology slides and also adoption of the DICOM standard which will offer the opportunity to integrate images with those already present in the picture archiving and communication system of the hospital.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2017/8/1/51/221132
REFERENCES
- 1.Cheng CL, Tan PH. Digital pathology in the diagnostic setting: Beyond technology into best practice and service management. J Clin Pathol. 2017;70:454–7. doi: 10.1136/jclinpath-2016-204272. [DOI] [PubMed] [Google Scholar]
- 2.Ho J, Parwani AV, Jukic DM, Yagi Y, Anthony L, Gilbertson JR, et al. Use of whole slide imaging in surgical pathology quality assurance: Design and pilot validation studies. Hum Pathol. 2006;37:322–31. doi: 10.1016/j.humpath.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Boyce BF. Whole slide imaging: Uses and limitations for surgical pathology and teaching. Biotech Histochem. 2015;90:321–30. doi: 10.3109/10520295.2015.1033463. [DOI] [PubMed] [Google Scholar]
- 4.Cheng CL, Azhar R, Sng SH, Chua YQ, Hwang JS, Chin JP, et al. Enabling digital pathology in the diagnostic setting: Navigating through the implementation journey in an academic medical centre. J Clin Pathol. 2016;69:784–92. doi: 10.1136/jclinpath-2015-203600. [DOI] [PubMed] [Google Scholar]
- 5.Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: A Systematic review. Arch Pathol Lab Med. 2017;141:151–61. doi: 10.5858/arpa.2016-0025-RA. [DOI] [PubMed] [Google Scholar]
- 6.Snead DR, Tsang YW, Meskiri A, Kimani PK, Crossman R, Rajpoot NM, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016;68:1063–72. doi: 10.1111/his.12879. [DOI] [PubMed] [Google Scholar]
- 7.Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: A systematic review. Arch Pathol Lab Med. 2017;141:151–61. doi: 10.5858/arpa.2016-0025-RA. [DOI] [PubMed] [Google Scholar]
- 8.Evans AJ, Kiehl TR, Croul S. Frequently asked questions concerning the use of whole-slide imaging telepathology for neuropathology frozen sections. Semin Diagn Pathol. 2010;27:160–6. doi: 10.1053/j.semdp.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Thorstenson S, Molin J, Lundström C. Implementation of large-scale routine diagnostics using whole slide imaging in Sweden: Digital pathology experiences 2006-2013. J Pathol Inform. 2014;5:14. doi: 10.4103/2153-3539.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vodovnik A. Distance reporting in digital pathology: A study on 950 cases. J Pathol Inform. 2015;6:18. doi: 10.4103/2153-3539.156168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantanowitz L, Sinard JH, Henricks WH, Fatheree LA, Carter AB, Contis L, et al. Validating whole slide imaging for diagnostic purposes in pathology: Guideline from the college of American pathologists pathology and laboratory quality center. Arch Pathol Lab Med. 2013;137:1710–22. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantanowitz L, Dickinson K, Evans AJ, Hassell LA, Henricks WH, Lennerz JK, et al. American telemedicine association clinical guidelines for telepathology. J Pathol Inform. 2014;5:39. doi: 10.4103/2153-3539.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans AJ, Krupinski EA, Weinstein RS, Pantanowitz L. 2014 American telemedicine association clinical guidelines for telepathology: Another important step in support of increased adoption of telepathology for patient care. J Pathol Inform. 2015;6:13. doi: 10.4103/2153-3539.153906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Janabi S, Huisman A, Nap M, Clarijs R, van Diest PJ. Whole slide images as a platform for initial diagnostics in histopathology in a medium-sized routine laboratory. J Clin Pathol. 2012;65:1107–11. doi: 10.1136/jclinpath-2012-200878. [DOI] [PubMed] [Google Scholar]
- 15.Riber-Hansen R, Vainer B, Steiniche T. Digital image analysis: A review of reproducibility, stability and basic requirements for optimal results. APMIS. 2012;120:276–89. doi: 10.1111/j.1600-0463.2011.02854.x. [DOI] [PubMed] [Google Scholar]
- 16.Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: Issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017;141:944–59. doi: 10.5858/arpa.2016-0074-OA. [DOI] [PubMed] [Google Scholar]
- 17.Parwani AV, Hassell L, Glassy E, Pantanowitz L. Regulatory barriers surrounding the use of whole slide imaging in the United States of America. J Pathol Inform. 2014;5:38. doi: 10.4103/2153-3539.143325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Têtu B, Evans A. Canadian licensure for the use of digital pathology for routine diagnoses: One more step toward a new era of pathology practice without borders. Arch Pathol Lab Med. 2014;138:302–4. doi: 10.5858/arpa.2013-0289-ED. [DOI] [PubMed] [Google Scholar]
- 19.Hanna MG, Monaco SE, Cuda J, Xing J, Ahmed I, Pantanowitz L, et al. Comparison of glass slides and various digital-slide modalities for cytopathology screening and interpretation. Cancer. 2017;125:701–9. doi: 10.1002/cncy.21880. [DOI] [PubMed] [Google Scholar]
- 20.Abels E, Pantanowitz L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J Pathol Inform. 2017;8:23. doi: 10.4103/jpi.jpi_11_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stathonikos N, Veta M, Huisman A, van Diest PJ. Going fully digital: Perspective of a Dutch academic pathology lab. J Pathol Inform. 2013;4:15. doi: 10.4103/2153-3539.114206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaacs M, Lennerz JK, Yates S, Clermont W, Rossi J, Pfeifer JD, et al. Implementation of whole slide imaging in surgical pathology: A value added approach. J Pathol Inform. 2011;2:39. doi: 10.4103/2153-3539.84232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H, Birsa J, Farahani N, Hartman DJ, Piccoli A, O’Leary M, et al. Digital pathology and anatomic pathology laboratory information system integration to support digital pathology sign-out. J Pathol Inform. 2016;7:23. doi: 10.4103/2153-3539.181767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin J, Treanor D. Digital pathology in clinical use: Where are we now and what is holding us back? Histopathology. 2017;70:134–45. doi: 10.1111/his.12993. [DOI] [PubMed] [Google Scholar]
- 25.Ho J, Ahlers SM, Stratman C, Aridor O, Pantanowitz L, Fine JL, et al. Can digital pathology result in cost savings? A financial projection for digital pathology implementation at a large integrated health care organization. J Pathol Inform. 2014;5:33. doi: 10.4103/2153-3539.139714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanna MG, Pantanowitz L. Bar coding and tracking in pathology. Clin Lab Med. 2016;36:13–30. doi: 10.1016/j.cll.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Hartman DJ, Pantanowitz L, McHugh JS, Piccoli AL, OLeary MJ, Lauro GR, et al. Enterprise implementation of digital pathology: Feasibility, challenges, and opportunities. J Digit Imaging. 2017;30:555–60. doi: 10.1007/s10278-017-9946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


