Version Changes
Revised. Amendments from Version 2
In response to the referees, some spelling mistakes in chemical compounds were modified throughout the paper.
Abstract
Background: Diagnoses of respiratory tract infections usually happen in the late phase of the disease and usually result in reduction of the pathogen load after broad-spectrum antibiotic therapy, but not in eradication of the pathogen. The development of a non-invasive, fast, and accurate method to detect pathogens has always been of interest to researchers and clinicians alike. Previous studies have shown that bacteria produce organic gases. The current study aimed to identify the volatile organic compounds (VOCs) produced by three respiratory tract pathogens, including Staphylococcus aureus, Escherichia coli and Candida albicans.
Methods: The VOCs produced were identified by gas chromatography–mass spectrometry (GC-MS), with prior collection of microbial volatile compounds using solid phase microextraction (SPME) fiber. The volatile compounds were collected by obtaining bacterial headspace samples.
Results: Results showed that these three organisms have various VOCs, which were analyzed under different conditions. By ignoring common VOCs, some species-specific VOCs could be detected. The most important VOC of E. coli was indole, also some important VOCs produced by S. aureus were 2,3-pentandione, cis-dihydro-α-terpinyl acetate, 1-decyne, 1,3-heptadiene, 2,5-dimethyl pyrazine, ethyl butanoate and cyclohexene,4-ethenyl. Furthermore, most of the identified compounds by C. albicans are alcohols.
Conclusions: The detection of VOCs produced by infectious agents maybe the key to make a rapid and precise diagnosis of infection, but more comprehensive studies must be conducted in this regard.
Keywords: Candida albicans, Escherichia coli, gas chromatography-mass spectrometry, Staphylococcus aureus, volatile organic compounds
Introduction
Infectious diseases are the main reason for morbidity and mortality in developing countries, especially among children 1. Staphylococcus aureus is a common inhabitant of the upper respiratory tract in children, and the causative agent for many infections. It is believed that people under 20 are more likely to have these bacteria. There is a greater possibility that S. aureus exists in the respiratory tract of infants aged 3 months or younger than in people of other ages 2. Moreover, S. aureus is colonized in the nasopharynx in 10–35% of children, and in almost 35% of the adult population 3.
Escherichia coli is one of the most significant pathogens affecting preterm infants 4. Some studies in developing countries have suggested that gram-negative rods (such as E. coli) are the major causes of infection in premature infants (0–6 days) 5– 7. Furthermore, infections caused by E. coli are one of the most important causes of death in the early neonatal period 5. Candida albicans is an opportunistic pathogen and an agent of nosocomial infection 8.
Generally, the causative agents of respiratory tract infections are diagnosed in late phases of the disease 7. Such infections need broad-spectrum antibiotic therapy, the consequences of which are a reduction in the pathogen load, but not eradication. Moreover, such therapies increase the probability of drug-resistant infections spreading 9. Accurate and rapid detection of pathogens is a critical step for adequate treatment of infection 10. and a non-invasive diagnostic method that has a high degree of accuracy needs to be developed 11.
It has been shown that bacteria produce organic gases. Different types of microorganisms have a distinct metabolism, and they produce various types of volatile organic compounds (VOCs) 12– 14. Attempts have been made to identify the VOCs of pathogenic organisms 15– 20. There are several sophisticated methods available that have been used for recognizing VOCs; these include gas chromatography-mass spectrometry (GC-MS) 21, selected ion flow tube mass spectrometry (SIFT-MS) 22, electronic noses (eNoses) 23, and ion-molecule reaction mass spectrometry (IMRMS) 24. Previous studies suggest that GC-MS is the most appropriate and reliable technique for the isolation and identification of VOCs 25– 27.
The current study aimed to identify the volatile organic compounds (VOCs) produced by three respiratory tract pathogens, including Staphylococcus aureus, Escherichia coli and Candida albicans, to determine if these could be used as biomarkers.
Materials and methods
Model organisms, medium and growth conditions
The bacterial strains used in this study were E. coli (ATCC 25922) and S. aureus (ATCC 25923), as gram-negative and gram-positive model organisms, and C. albicans (ATCC 10231) was used as a human pathogenic fungi model. These organisms model were obtained from the Microbiology Laboratory of Medicinal plants and Drugs Research Institute, Shahid Beheshti University. Monocultures of all strains were cultured 24 hours in nutrient agar, and then sub-cultured aerobically at 37°C in 30 ml of two different types of broth medium, Mueller Hinton broth (MB) and tryptic soy broth (TSB), in 100 ml sterilized glass bottles. For a more careful assessment of VOCs produced by each microorganism, the headspace was extracted from both media at three different time points: 2, 4 and 24 hours. To increase the possibility of VOC production, bottles containing cultured microorganism were shaken at 150 rpm during incubation time 28. A suspension of microorganisms with approximately OD 600 ~0.5 in culture media was used during the headspace extraction 10, and the corresponding sterile broth mediums were used as the blank samples 29.
Headspace extraction
A solid phase microextraction (SPME) fiber holder (57330-U, Sigma-Aldrich) containing fiber coated with divinyl benzene/carboxen/poly dimethyl siloxane 50/30 µm (DVB/CAR/PDMS) (57328-U, Sigma-Aldrich) was used for absorption of volatile compounds from the headspace of pathogens. To provide conditions that increase the rate of VOC absorption, after incubation time, 2ml of NaCl 36% was added to each culture. Then the DVB/CAR/PDMS fiber was suspended from the top of the bottle containing the culture and placed on a magnetic stirrer hotplate at 70°C for 30 minutes 30. After that, the fiber was placed at the injection site of GC-MS and all the absorbed VOCs entered the device. Eventually each VOC is represented as a chromatogram peak in the monitor that is connected to the GC-MS. For thermal desorption, the SPME fiber remained in the injector for 2 minutes before it was exposed to the headspace of the pathogen samples 31. To avoid possible false discoveries each state was tested at least three times.
GC-MS
To study the bacterial VOCs, a Thermo-Finnigan Trace GC-MS system (Thermo Quest-Finnigan Co) equipped with a DB-5 column (60 m length, 0.25 mm inner diameter, and 0.25 μm film thickness) with helium carrier gas at a flow rate of 1.1 ml/min was used. The starting temperature was 50°C, increasing at a rate of 10°C/minute up to 250°C. The GC-MS was set in splitless mode and a quadrupole ion trap with ionization energy of 70 eV was used in the filament.
VOCs were identified using the National Institute of Standards and Technology (NIST) reference library. To analyze the GC-MS data, Xcalibur 3.0 with Foundation 3.0 SP2 software (Thermo Fisher Scientific) was used, and the kovats retention index (RI) was calculated for each chromatographic peak.
When calculating the RI, a series of standards were used: n-alkanes were injected into the GC-MS the day before starting experiments, using the same temperature profile that would be used for the analysis of VOCs. The NIST17 Mass Spectral Library (NIST7/2017/EPA/NIH) was used to identify each compound according to its RI. Since there may be several types of volatile compounds have similar RI, to validate the final results extensive studies were also performed by a phytochemist to determine if the compounds were organic. The common VOCs released from the sterile environment (Blank samples) and tests were not considered.
Results
The VOCs produced by S. aureus, E. coli and C. albicans were assessed under six different conditions (using two types of media and taking measurements at three time points). The Xcalibur raw files for these three pathogens are available at https://doi.org/10.6084/m9.figshare.5178004.v1 32.
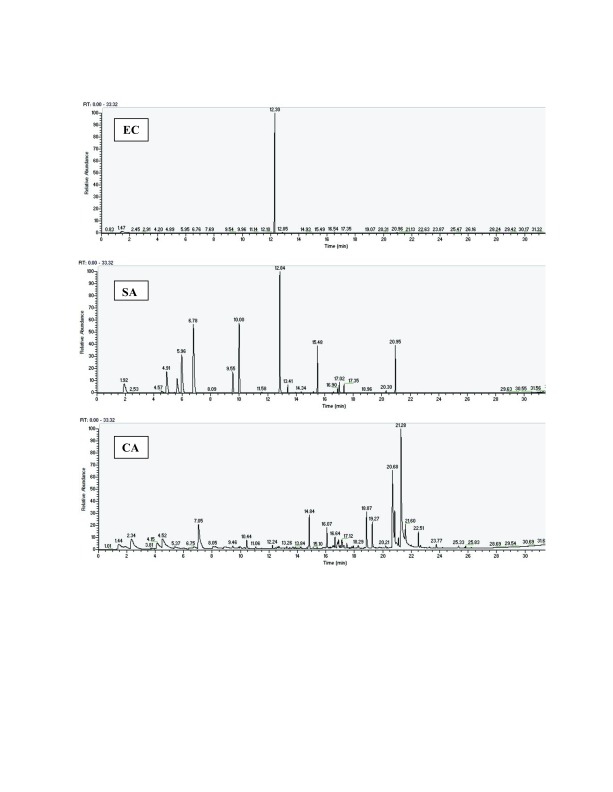
One chromatogram of the six chromatograms obtained is displayed in Figure 1, showing the chromatogram obtained 4 hours after culture in TSB medium, for each pathogen. The five other chromatograms are also available, as Supplementary File S1, Supplementary File S2, Supplementary File S3, Supplementary File S4 and Supplementary File S5.
Figure 1. Three chromatograms, for samples taken 4 hours after culture in TSB media.
EC: E. coli, SA: S. aureus and CA: C. albicans. The other chromatograms are available in the Supplementary material.
The processed GC-MS data obtained in the current study is available in a total of 18 tables as supplementary GC-MS data. It shows the details of the VOCs detected for each of the three pathogens, each analyzed under different conditions (using two types of media and taking measurements at three time points, as explained above).
For a better overview the detected VOCs are shown in three tables (at the 2 hour time point in Table 1, at the 4 hour time point in Table 2 and at the 24 hour time point in Table 3), alongside the percentage of the total area that the average peak of the detected VOC covered. In other words it is proportional to amount of the compound that is present.
Table 1. The identified VOCs for E. coli, S. aureus and C. albicans, and the percentage of the total area that their average peak covered (peak area %), after 2hours in MB and TSB media.
In total, 25 types of VOCs by E. coli, 33 types by S. aureus and 28 types by C. albicans were generated in this period.
| Compound |
E. coli
in MB |
E. coli
in TSB |
S. aureus
in MB |
S. aureus
in TSB |
C. albicans
in MB |
C. albicans
in TSB |
|---|---|---|---|---|---|---|
| (e)-2-hexyl ester-butanoic acid | 1.84 | 0.79 | - | - | 6.75 | 3.78 |
| 1-(1,5-dimethyl-4-hexyl-4-methyl-benzene | 3.19 | 0.41 | - | - | - | - |
| 1,2-benzenedicarboxylic acid | - | - | 0.39 | - | - | 0.2 |
| 1,2-butadiene | - | - | - | 1.73 | - | - |
| 1,3-butadiene | - | - | - | - | - | 26.68 |
| 1,3-heptadiene | - | - | - | 4.88 | - | 4.55 |
| 1,5-decadiene | - | - | - | - | - | 0.86 |
| 1,9-decadiene | 0.05 | 0.05 | - | - | - | 0.39 |
| 1-decyne | - | 0.07 | 0.85 | - | 1.55 | 1.55 |
| 1-penten-3-ol | - | 0.02 | - | 5.14 | - | - |
| 2,3-pentandione | - | 1.33 | - | - | - | - |
| 2,5-(1,1-dimethylethyl)-phenol | 0.13 | 0.1 | - | 0.11 | - | 0.43 |
| 2,5-dimethyl pyrazine | - | - | - | 20.19 | - | 3.07 |
| 2,6-bis(1,1-dimethylethyl)-4-methyl-phenol | - | 0.04 | 0.5 | - | - | 0.64 |
| 2,6-dibutyl-2,5-cyclohexadiene-1,4-dione | 0.03 | - | - | - | - | - |
| 2-ethenyl-6-methyl-pyrazine | 1.1 | 0.63 | 6.58 | 6.63 | - | 3.63 |
| 2-ethyl hexanol | - | - | - | 2.32 | - | - |
| 2-heptanone | 0.05 | - | - | 2.31 | - | - |
| 2-hexan-1-ol | - | - | - | - | - | 0.22 |
| 2-methyl-2-undecanethiol | 0.24 | 0.13 | 1.98 | 1.09 | - | - |
| 3-methyl-1,5-heptadiene | - | - | - | - | 3.77 | 1.03 |
| 3-propionyloxypentadecane | 0.57 | 0.18 | 7.36 | 1.3 | 2 | 0.61 |
| 4-t-butyl-2-(1-methyl-2-
nitroethyl)cyclohexane |
0.93 | 0.73 | 12.17 | 5.4 | 10.98 | 2.45 |
| 5.5-dodecadinyl-1, 12-diol | - | - | 0.48 | 0.5 | 1.19 | 6.38 |
| allyl butylhydroquinone | - | - | - | 0.31 | - | - |
| anisol | - | 0.05 | - | 1.19 | - | - |
| benzaldehyde | 2.13 | 1.34 | 3.22 | 8.98 | - | 0.64 |
| benzene acetaldehyde | - | - | 8.74 | 7.04 | - | - |
| benzophenone | 0.03 | - | - | - | - | - |
| bisabolene | 1.21 | 0.03 | - | - | - | - |
| butyl cyclohexyl acetate | - | - | - | 0.4 | - | - |
| butyraldehyde | - | - | - | - | - | 0.67 |
| cadinene | - | - | - | - | - | 1.79 |
| carbamic acid | - | - | - | - | 48.49 | 0.5 |
| caryophyllene | - | - | - | 0.09 | - | - |
| cedran-1,8-diol | 0.14 | 0.09 | 2.63 | 0.39 | - | 0.48 |
| cedrol | - | - | 0.71 | 0.23 | - | - |
| copaene | 0.01 | - | - | - | - | - |
| cyclohexene, 4-ethenyl- | - | - | 29.64 | 2.18 | - | 0.47 |
| decanol | - | 0.93 | - | - | - | - |
| decene | - | - | - | 2.85 | - | - |
| dimethyl octenal | - | - | - | 1.39 | - | - |
| dimethyl ethyl cyclohexanol | - | - | - | 1.04 | - | 0.59 |
| dodecane | 0.06 | - | - | 0.96 | - | - |
| dodecanol | - | 0.27 | - | - | - | - |
| dodecenol | - | - | - | - | - | 0.5 |
| eicosane | - | - | - | - | - | 0.12 |
| ethyl butanoate | - | - | - | - | 5.31 | 4.64 |
| heptadecane | - | - | 12.35 | 5.33 | - | - |
| humulen | - | - | 0.71 | - | - | - |
| indole | 82.61 | 90.97 | - | 0.48 | - | - |
| limonene | 0.68 | - | - | - | - | - |
| longifolene | - | - | - | - | 4.96 | 0.43 |
| longifolene | - | - | - | 0.52 | - | - |
| methone | - | - | - | 7.49 | - | 1.4 |
| muurola-4,5-diene | 0.47 | - | - | - | - | - |
| naphthalenol | - | 0.84 | - | 0.21 | - | 0.44 |
| neryl acetate | 0.06 | 0.03 | - | - | - | - |
| nonadecanone | - | - | 0.62 | - | - | - |
| ocimene | - | - | - | - | - | 2.24 |
| octacosane | 0.41 | 0.06 | 1.37 | 1.22 | 1.2 | 1.02 |
| octyl acetate | - | - | - | - | - | 0.4 |
| pentadecane | 0.03 | - | 0.86 | 0.68 | - | 4.1 |
| phthalic acid, butyl ester | 0.19 | 0.13 | 0.97 | - | - | 0.22 |
| phenyl ethyl pyrrole | - | 0.06 | - | - | - | - |
| sesquiphellandrene | 1.1 | - | - | - | - | 4.79 |
| tetra butyl cyclohexyl acetate | - | - | 1.7 | - | - | - |
| tetradecane | 0.01 | - | - | - | - | - |
| tetradecanol | - | - | - | - | - | 0.34 |
| zingiberene | 1.63 | - | 1.03 | - | 2.87 | 7.91 |
| α-acetoxydihydrocoumarin | - | 0.52 | 1.88 | 0.25 | - | - |
| β-santalol | - | - | - | - | 4.87 | 1.34 |
Table 2. The identified VOCs for E. coli, S. aureus and C. albicans, and the percentage of the total area that their average peak covered (peak area %), after 4 hours in MB and TSB media.
In total, 9 types of VOCs by E. coli, 19 types by S. aureus and 42 types by C. albicans were generated in this period.
| Compound |
E. coli
in MB |
E. coli
in TSB |
S. aureus
in MB |
S. aureus
in TSB |
C. albicans
in MB |
C. albicans
in TSB |
|---|---|---|---|---|---|---|
| (e)-2-hexyl ester-butanoic acid | - | - | - | - | 6.64 | 4.24 |
| (z)-2-octene-1-ol | - | - | - | - | 0.69 | 0.54 |
| (z)-4-decan-1-ol | - | - | - | - | 0.70 | 0.46 |
| 1,2-benzenedicarboxylic acid | - | - | - | - | 0.29 | 0.31 |
| 1,2-butadiene | - | - | 0.37 | 4.02 | - | - |
| 1,3-butadiene | - | - | - | - | 1.40 | - |
| 1,3-heptadiene | - | - | 81.33 | - | 0.23 | 0.47 |
| 1,5-decadiene | - | - | - | - | 1.01 | 3.00 |
| 1,9-decadiene | - | 0.01 | - | - | 0.20 | 3.59 |
| 1-decyne | 2.36 | - | 0.59 | 16.47 | 2.81 | 1.40 |
| 1-methoxy-2-propanol | - | - | 0.20 | - | - | - |
| 2-(phenylmethylene)-octanal | - | - | - | - | 1.55 | 1.63 |
| 2,3-pentandione | - | - | 7.07 | 21.67 | - | - |
| 2,5-(1,1-dimethylethyl)-phenol | - | - | - | - | 0.69 | 0.43 |
| 2,5-dimethyl pyrazine | - | - | - | - | - | 3.48 |
| 2-acetyl-1-pyrroline | - | 0.07 | - | - | - | - |
| 2-ethenyl-6-methyl-pyrazine | 0.01 | - | 0.80 | - | 7.33 | - |
| 2-ethyl hexanol | - | - | - | 1.05 | - | - |
| 2-heptanone | 0.02 | 0.33 | - | - | - | - |
| 2-hexan-1-ol | - | - | - | - | 0.33 | 0.23 |
| 2h-tetrazole-5-carboxylicacid, 2-phenyl | - | - | 0.48 | - | 1.44 | 1.59 |
| 2-methyl-2-undecanethiol | - | - | 0.24 | - | - | - |
| 3-methyl-1,5-heptadiene | - | - | 0.60 | - | 0.76 | 0.53 |
| 4-t-butyl-2-(1-methyl-2-
nitroethyl)cyclohexane |
- | - | - | - | 6.05 | 5.81 |
| 5.5-dodecadinyl-1, 12-diol | - | - | - | - | 5.16 | 0.51 |
| 6-methyl-5-hepten-2-one | - | - | - | - | 0.31 | 0.20 |
| benzaldehyde | - | - | - | - | 0.83 | 0.68 |
| butyraldehyde | - | - | 0.38 | - | - | - |
| cadinene | - | - | - | - | 0.79 | 0.34 |
| carbamic acid | - | - | - | - | 18.10 | 8.19 |
| caryophyllene | - | 0.02 | - | 4.88 | - | - |
| cedrol | - | - | - | - | 0.36 | 0.36 |
| cis-dihydro-α-terpinyl acetate | - | - | - | 17.90 | - | - |
| cyclohexene, 4-ethenyl- | - | - | 0.08 | 6.77 | - | - |
| dimethyl octenal | - | - | - | - | 0.58 | - |
| dimethylethyl cyclohexanol | - | - | - | - | 0.79 | 0.22 |
| dodecenal | - | - | - | - | 0.52 | 0.39 |
| dodecenol | - | - | - | - | 0.61 | 2.54 |
| eicosane | - | - | - | - | - | 0.29 |
| ethyl butanoate | 0.01 | - | 7.57 | 12.21 | 6.63 | 0.73 |
| indole | 97.05 | 99.46 | - | 0.82 | 0.34 | - |
| levomenthol | - | - | - | 3.74 | - | - |
| longifolene | - | - | - | - | 0.31 | 0.26 |
| longifolol | - | - | - | - | 5.66 | 22.07 |
| methyl isopropyl hexenal | - | - | - | - | 1.11 | 0.59 |
| naphthalenol | - | - | - | - | - | 0.37 |
| octacosane | - | - | - | - | 1.74 | 1.60 |
| octyl acetate | - | - | - | - | 0.34 | - |
| pentadecane | - | - | - | 0.87 | 1.80 | 1.38 |
| phthalic acid, butyl ester | - | - | - | - | 0.60 | 0.36 |
| tetradecanol | - | - | - | - | 0.61 | 0.26 |
| tridecanol | - | - | - | - | 0.34 | 0.34 |
| zingiberene | - | - | - | - | 1.27 | 1.12 |
| β-santalol | - | 0.03 | - | 5.16 | 6.52 | 14.15 |
| sesquiphellandrene | - | - | - | - | 2.00 | 0.70 |
Table 3. The identified VOCs for E. coli, S. aureus and C. albicans, and the percentage of the total area that their average peak covered (peak area %), after 24 hours in MB and TSB media.
In total, 16 types of VOCs by E. coli, 26 types by S. aureus and 27 types by C. albicans were generated in this period.
| Compound |
E. coli
in MB |
E. coli
in TSB |
S. aureus
in MB |
S. aureus
in TSB |
C. albicans
in MB |
C. albicans
in TSB |
|---|---|---|---|---|---|---|
| (e)-2-hexyl ester-butanoic acid | 0.04 | 0.03 | 0.21 | 0.04 | 0.50 | - |
| (z)-2-octene-1-ol | - | - | - | - | - | 0.18 |
| (z)-4-decan-1-ol | - | - | - | - | - | 0.60 |
| 1,2-benzenedicarboxulic acid | - | - | - | - | 0.13 | 0.28 |
| 1,2-butadiene | - | - | 3.71 | 0.10 | - | - |
| 1,3-heptadiene | - | - | 21.75 | - | - | - |
| 1,5-decadiene | - | - | - | - | 0.10 | 0.26 |
| 1,9-decadiene | 0.02 | 0.03 | - | - | - | - |
| 1-decyne | - | 0.02 | 0.81 | 59.78 | - | 0.75 |
| 1-methoxy-2-propanol | - | - | 6.74 | 0.02 | - | - |
| 2-(phenylmethylene)-octanal | - | - | - | - | 0.71 | 0.86 |
| 2,3-pentandione | 0.03 | 0.48 | 15.53 | 0.66 | - | - |
| 2,5-dimethyl pyrazine | - | - | 0.62 | - | - | 0.55 |
| 2-acetyl-1-pyrroline | - | 6.37 | - | 1.11 | - | - |
| 2-decenal | - | - | 0.06 | 0.01 | - | - |
| 2-ethenyl-6-methyl-pyrazine | 0.06 | - | 1.35 | - | 0.39 | - |
| 2-ethyl hexanol | - | - | 0.13 | 0.02 | - | 0.51 |
| 2-heptanone | - | 0.21 | - | - | - | - |
| 2h-tetrazole-5-carboxylicacid,
2-phenyl |
- | - | 3.21 | - | - | - |
| 2-methyl tetradecane | 0.05 | 0.03 | - | - | - | - |
| 2-methyl-1-propanol | - | - | 0.15 | 0.04 | 5.89 | 16.03 |
| 2-methyl-2-undecanethiol | 0.10 | - | 2.54 | - | - | 61.65 |
| 2-octyl-1-ol | - | - | - | - | 0.15 | - |
| 2-octyne | - | - | - | - | - | 0.27 |
| 3-methy-4-pentene-3-ol | - | - | - | - | - | 0.13 |
| 3-methyl-1,5-heptadiene | - | - | 0.19 | - | - | - |
| 3-methyl-1-pentene | - | - | - | - | - | 0.41 |
| -4-t-butyl-2-(1-methyl-2-
nitroethyl)cyclohexane |
- | - | - | - | 80.87 | |
| 5.5-dodecadinyl-1, 12-diol | 0.01 | 0.03 | 0.77 | 0.05 | - | - |
| butyraldehyde | - | - | 1.41 | 0.09 | 0.14 | 0.24 |
| carbamic acid | - | - | - | - | 0.48 | - |
| caryophyllene | - | 0.21 | - | 0.14 | - | 1.25 |
| cedrol | - | - | - | - | 1.93 | 2.68 |
| cis-dihydro-α-terpinyl acetate | - | 0.64 | - | 34.77 | - | - |
| cyclohexene, 4-ethenyl- | - | - | 3.54 | 0.15 | - | - |
| ethyl acetoacetate | - | - | - | - | - | 0.46 |
| ethyl butanoate | 0.06 | 0.31 | 28.72 | 0.46 | - | 1.64 |
| indole | 99.61 | 88.86 | 0.07 | 0.02 | - | - |
| levomenthol | - | - | - | 1.66 | - | - |
| longifolol | - | - | - | - | 0.33 | 0.24 |
| octacosane | - | - | - | - | 0.33 | 0.44 |
| pentadecane | - | 0.35 | 0.17 | 0.08 | - | - |
| thiophene | - | - | 0.18 | - | - | - |
| zingiberene | - | - | - | - | 0.20 | - |
| β-santalol | - | 0.49 | - | 0.21 | - | 1.21 |
Some VOCs were common among organisms and were generated by two or three organisms at an approximately equal rate, including 1,2-benzenedicarboxylic acid, 1,9-decadiene, 2,5-(1,1-dimethylethyl)-phenol, 2,6-bis(1,1-dimethylethyl)-4-methyl-phenol, 3-propionyl oxy pentadecane and anisol ( Table 1). Some common VOCs were produced at a greater rate between one organism and another. It can be concluded that these VOCs could also be more important in the organism that produces greater quantities. 1-penten-3-ol was produced from E . coli in TSB medium after 2 hours (0.02%); under identical conditions, more of it was produced by S. aureus (5.14%) than by E. coli. Furthermore, indole was produced from E. coli after 2 hours of culture in two types of medium (82.61% for MB and 90.97% for TSB) and was also produced by S. aureus after 2 hours in TSB medium, although at a much lower rate (0.48%) ( Table 1).
Uncommon VOCs of E. coli detected 2 hours after culture included 1-(1,5-dimethyl)-4-hexyl-4-methyl-benzene, 2,3-pentandione, 2,6-dibutyl-2,5-cyclohexadiene-1,4-dione, benzophenone, bisabolene, copaene, decanol, dodecanol, indole, limonene, muurola-4,5-diene, neryl acetate, phenyl ethyl pyrrole and tetradecane ( Table 1).
Uncommon VOCs of S. aureus detected 2 hours after culture included 1,2-butadiene, 1-penten-3-ol, 2,5-dimethyl pyrazine, 2-ethyl hexanol, allyl butyl hydroquinone, benzene acetaldehyde, butyl cyclohexyl acetate, caryophyllene, cedrol, cyclohexene, 4-ethenyl-, decene, dimethyl octenal, heptadecane, humulen, longifolene, methone, nonadecanone and tetrabutyl cyclohexyl acetate ( Table 1).
Uncommon VOCs of C. albicans detected 2 hours after culture included 1,3-butadiene, 1,5-decadiene, 2-hexan-1-ol, 3-methyl-1,5-heptadiene, butyraldehyde, cadinene, carbamic acid, dodecenol, eicosane, ethyl butanoate, longifolene, ocimene, octyl acetate, tetradecanol and β-santaloland ( Table 1).
Uncommon VOCs of E. coli identified 4 hours after culture included 1,9-decadiene, 2-acetyl-1-pyrroline, 2-heptanone and indole ( Table 2).
Uncommon VOCs of S. aureus identified 4 hours after culture included 1,2-butadiene, 1,3-heptadiene, 1-decyne, 1-methoxy-2-propanol, 2,3-pentandione, 2-ethyl hexanol, 2-methyl-2-undecanethiol, butyraldehyde, cis-dihydro-α-terpinyl acetate, cyclohexene,4-ethenyl- and levomenthol ( Table 2).
Uncommon VOCs of C. albicans identified 4 hours after culture included (e)-2-hexyl ester- butanoic acid, (z)-2-octene-1-ol, (z)-4-decan-1-ol, 1,2-benzenedicarboxulic acid, 1,3-butadiene, 1,5-decadiene, 2-(phenyl methylene)-octanal, 2,5-(1,1-dimethylethyl)-phenol, 2,5-dimethyl pyrazine, 2-ethenyl-6-methyl-pyrazine, 2-hexan-1-ol, 4-t-butyl-2-(1-methyl-2-nitroethyl) cyclohexane, 5.5-dodecadinyl-1, 12-diol, 6-methyl-5-hepten-2-one, benzaldehyde, cadinene, carbamic acid, cedrol, , dimethyl octenal, dimethyl ethyl cyclohexanol, dodecenal, dodecenol, eicosane, longifolene, longifolol, methyl isopropyl hexenal, naphthalenol, octacosane, octyl acetate, phthalic acid butyl ester, tetradecanol, tridecanol, zingiberene and sesquiphellandrene ( Table 2).
Uncommon VOCs of E. coli identified 24 hours after culture included 1,9-decadiene, 2-acetyl-1-pyrroline, 2-heptanone, 2-methyl tetradecane and indole ( Table 3).
Uncommon VOCs of S. aureus identified in 24 hours after culture were included; 1,2-butadiene, 1,3-heptadiene, 1-decyne, 1-methoxy-2-propanol, 2,3-pentandione, 2,5-dimethyl pyrazine, 2-decenal, 2h-tetrazole-5-carboxylicacid, 2-phenyl, 3-methyl-1,5-heptadiene, caryophyllene, cis-dihydro-α-terpinyl acetate, cyclohexene,4-ethenyl-, ethyl butanoate, levomenthol and thiophene ( Table 3).
Uncommon VOCs of C. albicans identified 24 hours after culture included (z)-2-octene-1-ol, (z)-4-decan-1-ol, 1,2-benzenedicarboxylic acid, 1,5-decadiene, 2-(phenyl methylene)-octanal, 2,5-dimethyl pyrazine, 2-methyl-1-propanol, 2-methyl-2-undecanethiol, 2-octyl-1-ol, 2-octyne, 3-methy-4-pentene-3-ol, 3-methyl-1-pentene, 4-t-butyl-2-(1-methyl-2-nitroethyl) cyclohexane, carbamic acid, cedrol, , ethyl acetoacetate, longifolol, octacosane and zingiberene ( Table 3).
Discussion
As previous studies have shown, organisms are able to produce either common or specific VOCs 33– 35. In the current study, GC-MS was used to detect VOCs generated by three pathogenic organisms in the human respiratory tract. The VOCs of E. coli, S. aureus and C. albicans were analyzed at three different time points, using two different types of media ( Figure 1).
Results of the current study suggest that VOCs exclusively produced by E. coli are 1-(1,5-dimethyl)-4-hexyl-4-methyl-benzene, 2,6-dibutyl-2,5-cyclohexadiene-1,4-dione, benzophenone, bisabolene, copaene, decanol, dodecanol, indole, limonene, muurola-4,5-diene, nerylacetate, phenyl ethyl pyrrole, sesquiphellandrene, tetradecane, 2-acetyl-1-pyrroline and 2-methyl tetradecane. The most important compound among these is Indole, because it is generated at the three time points and also it was the most produced VOC by E. coli (at least 82%). Other studies have confirmed this finding 28, 29, 35. E. coli produced tryptophanase and this enzyme degrades tryptophan to indole and the other compounds 36. In future studies, it is advisable to measure the amount of indole in the exhaled air of infected patients with E. coli and compare it with the current results. This is because in the patient’s lungs the level of tryptophan is not the same as culture medium. It is also suggested that the amount of released indole from this bacterium should be evaluated under at in-vitro conditions and with using the simplest culture medium (relative to TSB and MB). In this way, we will have a more detailed thought of the importance of the Indole production by E. coli.
The current study has shown that the specific VOCs produced by S. aureus are 1,2-butadiene, 1-penten-3-ol, 2,5-dimethyl pyrazine, 2-ethyl hexanol, allyl butyl hydroquinone, benzene acetaldehyde, butylcyclohexyl acetate, caryophyllene, cyclohexene, 4-ethenyl-, decene, heptadecane, humulen, longifolene, methone, nonadecanone, tetrabutylcyclohexyl acetate, 1,3-heptadiene, 1-decyne, 1-methoxy-2-propanol, 2,3-pentandione, cis-dihydro-α-terpinyl acetate, levomenthol, 2-decenal, ethyl butanoate and thiophene. Moreover, 1,2-butadiene, 2,5-dimethyl pyrazine, 2-ethyl hexanol, caryophyllene, cyclohexene, 4-ethenyl, 1,3-heptadiene, 1-decyne, 1-methoxy-2-propanol, 2,3-pentandione, cis-dihydro-α-terpinyl acetate, and levomenthol were detected under more than one of the six conditions that were tested, so they are significant. Another important point is that the percentage of the total area that the average peaks for 2,3-pentandione, cis-dihydro-α-terpinyl acetate, 1-decyne, 1,3-heptadiene, 2,5-dimethyl pyrazine, ethyl butanoate and cyclohexene,4-ethenyl covered were at least 15%; thus, they are remarkable VOCs for S. aureus. Some of the VOCs produced by S. aureus in the current study have been reported in other studies 34, 37 but some of them have not 28, 33. The origin of all produced VOCs is not exactly known. However it is believed some released VOCs by this bacterium is because of the ability to degrade amino acids in its growth environment 11.
This study suggested that the specific VOCs produced by C. albicans include 1,3-butadiene, 1,5-decadiene, 2-hexan-1-ol, cadinene, carbamic acid, dodecenol, eicosane, longifolene, ocimene, octyl acetate, tetradecanol, β-sesquiphellandrene, (z)-2-octene-1-ol, (z)-4-decan-1-ol, 2-(phenyl methylene)-octanal, 4-t-butyl-2-(1-methyl-2-nitroethyl) cyclohexane, longifolol, 6-methyl-5-hepten-2-one, dodecenal, methyl isopropyl hexenal, tridecanol, 2-methyl-2-undecanethiol, 2-octyl-1-ol, 2-octyne, 3-methy-4-pentene-3-ol, 2-methyl-1-propanol and 3-methyl-1-pentene. also, 1,3-butadiene, 1,5-decadiene, 2-hexan-1-ol, cadinene, carbamic acid, dodecenol, eicosane, longifolene, octyl acetate, tetradecanol, β-sesquiphellandrene, (z)-2-octene-1-ol, (z)-4-decan-1-ol, 2-(phenyl methylene)-octanal, 4-t-butyl-2-(1-methyl-2-nitroethyl) cyclohexane, longifolol, octyl acetate, β-sesquiphellandreneand 2-methyl-2-undecanethiol were detected under more than one of the six conditions that were tested, so they are significant. Furthermore, 1,3-butadiene, carbamic acid, longifolol, β-santalol, 2-methyl-1-propanol, 2-methyl-2-undecanethiol and 4-t-butyl-2-(1-methyl-2-nitroethyl) cyclohexane were produced in greater quantities . Several studies have analyzed the VOCs of C. albicans and have noted that most of these identified compounds are alcohols 38– 40. That is because if favorable growth conditions are available for this bacterium (a sufficient level of oxygen, aromatic amino acids, and an alkaline pH) will produce large amounts of alcohol that results from its metabolism 41.
It is suggested that the findings of future studies on the exhaust air of respiratory infections patients with these three pathogens should be compared with the identified VOCs in this study. Although there may be some differences between the results of in-vitro and in-vivo studies there seems to be significant similarities over the dominant detected VOCs.
Finding a non-invasive and rapid method for diagnosis of infectious agents is a subject of interest, so it has been investigated in several studies 33, 42– 45. The current study showed that using SPME fiber and GC-MS for extraction and detection of VOCs allowed detection of more specific VOCs for the three pathogenic respiratory tract organisms, E. coli, S. aureus and C. albicans, which could be used as biomarkers for their identification. It is essential that more comprehensive studies be conducted to create a more complete profile of VOCs for these organisms, and so that the methods can be developed further.
Data availability
The Xcalibur raw files for the three studied pathogens are available at https://doi.org/10.6084/m9.figshare.5178004.v1 32.
Acknowledgements
This paper is part of Najmeh Karami’s PhD thesis, named: "Identification the volatile metabolic profiling of six respiratory pathogenic organisms". We would like to express our specific thanks to Shahid Beheshti University of Medical Sciences for the financial support. We also appreciate all colleagues that helped us in the Pediatric Infections Research Center, Mofid Children’s Hospital, Shahid Beheshti University of Medical Sciences and Medicinal Plant and Drug Research Institute, Shahid Beheshti.
Funding Statement
This work was supported by the Shahid Beheshti University of Medical Sciences
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 3; referees: 3 approved]
Supplementary material
Supplementary File S1: Three chromatograms, for samples taken 2 hours after culture in MB media. EC: E. coli, SA: S. aureus and CA: C. albicans.
Supplementary File S2: Three chromatograms, for samples taken 4 hours after culture in MB media. EC: E. coli, SA: S. aureus and CA: C. albicans.
Supplementary File S3: Three chromatograms, for samples taken 24 hours after culture in MB media. EC: E. coli, SA: S. aureus and CA: C. albicans.
Supplementary File S4: Three chromatograms, for samples taken 2 hours after culture in TSB media. EC: E. coli, SA: S. aureus and CA: C. albicans.
Supplementary File S5: Three chromatograms, for samples taken 24 hours after culture in TSB media. EC: E. coli, SA: S. aureus and CA: C. albicans.
Supplementary File S6: GC-MS data analysis, showing the details of the detected VOCs of three pathogens in 6 modes.
References
- 1. Rodríguez L, Cervantes E, Ortiz R: Malnutrition and gastrointestinal and respiratory infections in children: a public health problem. Int J Environ Res Public Health. 2011;8(4):1174–205. 10.3390/ijerph8041174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang YC, Chen CJ: Nasal carriage of methicillin-resistant Staphylococcus aureus during the first 2 years of life in children in northern Taiwan. Pediatr Infect Dis J. 2015;34(2):131–5. 10.1097/INF.0000000000000517 [DOI] [PubMed] [Google Scholar]
- 3. Regev-Yochay G, Dagan R, Raz M, et al. : Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. JAMA. 2004;292(6):716–20. 10.1001/jama.292.6.716 [DOI] [PubMed] [Google Scholar]
- 4. Stoll BJ, Hansen NI, Sánchez PJ, et al. : Early onset neonatal sepsis: the burden of group B Streptococcal and E. coli disease continues. Pediatrics. 2011;127(5):817–26. 10.1542/peds.2010-2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walker CL, Rudan I, Liu L, et al. : Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381(9875):1405–16. 10.1016/S0140-6736(13)60222-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Russo TA, Johnson JR: Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 2003;5(5):449–56. 10.1016/S1286-4579(03)00049-2 [DOI] [PubMed] [Google Scholar]
- 7. Zaidi AK, Huskins WC, Thaver D, et al. : Hospital-acquired neonatal infections in developing countries. Lancet. 2005;365(9465):1175–88. 10.1016/S0140-6736(05)71881-X [DOI] [PubMed] [Google Scholar]
- 8. Pierce GE: Pseudomonas aeruginosa, Candida albicans, and device-related nosocomial infections: implications, trends, and potential approaches for control. J Ind Microbiol Biotechnol. 2005;32(7):309–18. 10.1007/s10295-005-0225-2 [DOI] [PubMed] [Google Scholar]
- 9. Filipiak W, Sponring A, Baur MM, et al. : Characterization of volatile metabolites taken up by or released from Streptococcus pneumoniae and Haemophilus influenzae by using GC-MS. Microbiology. 2012;158(Pt 12):3044–53. 10.1099/mic.0.062687-0 [DOI] [PubMed] [Google Scholar]
- 10. Pauwels RA, Buist AS, Calverley PM, et al. : Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine. 2012. [DOI] [PubMed] [Google Scholar]
- 11. Filipiak W, Sponring A, Baur MM, et al. : Molecular analysis of volatile metabolites released specifically by staphylococcus aureus and pseudomonas aeruginosa. BMC Microbiol. 2012;12(1):113. 10.1186/1471-2180-12-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schulz S, Dickschat JS: Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24(4):814–42. 10.1039/b507392h [DOI] [PubMed] [Google Scholar]
- 13. Thorn RM, Reynolds DM, Greenman J: Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J Microbiol Methods. 2011;84(2):258–64. 10.1016/j.mimet.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 14. Wilson AD, Baietto M: Advances in electronic-nose technologies developed for biomedical applications. Sensors (Basel). 2011;11(1):1105–76. 10.3390/s110101105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barker M, Hengst M, Schmid J, et al. : Volatile organic compounds in the exhaled breath of young patients with cystic fibrosis. Eur Respir J. 2006;27(5):929–36. 10.1183/09031936.06.00085105 [DOI] [PubMed] [Google Scholar]
- 16. Buszewski B, Kesy M, Ligor T, et al. : Human exhaled air analytics: biomarkers of diseases. Biomed Chromatogr. 2007;21(6):553–66. 10.1002/bmc.835 [DOI] [PubMed] [Google Scholar]
- 17. Hamilton-Kemp T, Newman M, Collins R, et al. : Production of the long-chain alcohols octanol, decanol, and dodecanol by Escherichia coli. Curr Microbiol. 2005;51(2):82–6. 10.1007/s00284-005-4469-x [DOI] [PubMed] [Google Scholar]
- 18. Julák J, Stránská E, Procházková-Francisci E, et al. : Blood cultures evaluation by gas chromatography of volatile fatty acids. Med Sci Monit. 2000;6(3):605–10. [PubMed] [Google Scholar]
- 19. Friedrich MJ: Scientists seek to sniff out diseases: electronic "noses" may someday be diagnostic tools. JAMA. 2009;301(6):585–6. 10.1001/jama.2009.90 [DOI] [PubMed] [Google Scholar]
- 20. Röck F, Barsan N, Weimar U: Electronic nose: current status and future trends. Chem Rev. 2008;108(2):705–25. 10.1021/cr068121q [DOI] [PubMed] [Google Scholar]
- 21. Farag MA, Ryu CM, Sumner LW, et al. : GC–MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry. 2006;67(20):2262–8. 10.1016/j.phytochem.2006.07.021 [DOI] [PubMed] [Google Scholar]
- 22. Allardyce RA, Langford VS, Hill AL, et al. : Detection of volatile metabolites produced by bacterial growth in blood culture media by selected ion flow tube mass spectrometry (SIFT-MS). J Microbiol Methods. 2006;65(2):361–5. 10.1016/j.mimet.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 23. de Heer K, van der Schee MP, Zwinderman K, et al. : Electronic nose technology for detection of invasive pulmonary aspergillosis in prolonged chemotherapy-induced neutropenia: a proof-of-principle study. J Clin Microbiol. 2013;51(5):1490–5. 10.1128/JCM.02838-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dolch ME, Frey L, Hornuss C, et al. : Molecular breath-gas analysis by online mass spectrometry in mechanically ventilated patients: a new software-based method of CO 2-controlled alveolar gas monitoring. J Breath Res. 2008;2(3):037010. 10.1088/1752-7155/2/3/037010 [DOI] [PubMed] [Google Scholar]
- 25. Bos LD, Sterk PJ, Schultz MJ: Volatile metabolites of pathogens: a systematic review. PLoS Pathog. 2013;9(5):e1003311. 10.1371/journal.ppat.1003311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reade S, Mayor A, Aggio R, et al. : Optimisation of sample preparation for direct SPME-GC-MS analysis of murine and human faecal volatile organic compounds for metabolomic Studies. J Anal Bioanal Tech. 2014;5(2):184 10.4172/2155-9872.1000184 [DOI] [Google Scholar]
- 27. Karami N, Karimi A, Aliahmadi A, et al. : Identification of bacteria using volatile organic compounds. Cell Mol Biol (Noisy-le-grand). 2017;63(2):112–21. 10.14715/cmb/2017.63.2.18 [DOI] [PubMed] [Google Scholar]
- 28. Zhu J, Bean HD, Kuo YM, et al. : Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization-mass spectrometry. J Clin Microbiol. 2010;48(12):4426–31. 10.1128/JCM.00392-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tait E, Perry JD, Stanforth SP, et al. : Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. J Chromatogr Sci. 2014;52(4):363–73. 10.1093/chromsci/bmt042 [DOI] [PubMed] [Google Scholar]
- 30. Schulz S, Fuhlendorff J, Reichenbach H: Identification and synthesis of volatiles released by the myxobacterium Chondromyces crocatus. Tetrahedron. 2004;60(17):3863–72. 10.1016/j.tet.2004.03.005 [DOI] [Google Scholar]
- 31. Tait E, Hill KA, Perry JD, et al. : Development of a novel method for detection of Clostridium difficile using HS-SPME-GC-MS. J Appl Microbiol. 2014;116(4):1010–9. 10.1111/jam.12418 [DOI] [PubMed] [Google Scholar]
- 32. Karami N, Mirzajani F, Rezadoost H, et al. : Diagnosis of Three Different Pathogenic Microorganisms by Gas chromatography–mass spectrometry. figshare. 2017. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boots AW, Smolinska A, van Berkel JJ, et al. : Identification of microorganisms based on headspace analysis of volatile organic compounds by gas chromatography-mass spectrometry. J Breath Res. 2014;8(2):027106. 10.1088/1752-7155/8/2/027106 [DOI] [PubMed] [Google Scholar]
- 34. Effmert U, Kalderás J, Warnke R, et al. : Volatile mediated interactions between bacteria and fungi in the soil. J Chem Ecol. 2012;38(6):665–703. 10.1007/s10886-012-0135-5 [DOI] [PubMed] [Google Scholar]
- 35. Garner CE, Smith S, de Lacy Costello B, et al. : Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007;21(8):1675–88. 10.1096/fj.06-6927com [DOI] [PubMed] [Google Scholar]
- 36. Martino PD, Fursy R, Bret L, et al. : Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003;49(7):443–9. 10.1139/W03-056 [DOI] [PubMed] [Google Scholar]
- 37. Nizio KD, Perrault KA, Troobnikoff AN, et al. : In vitro volatile organic compound profiling using GC×GC-TOFMS to differentiate bacteria associated with lung infections: a proof-of-concept study. J Breath Res. 2016;10(2):026008. 10.1088/1752-7155/10/2/026008 [DOI] [PubMed] [Google Scholar]
- 38. Hertel M, Hartwig S, Schütte E, et al. : Identification of signature volatiles to discriminate Candida albicans, glabrata, krusei and tropicalis using gas chromatography and mass spectrometry. Mycoses. 2016;59(2):117–26. 10.1111/myc.12442 [DOI] [PubMed] [Google Scholar]
- 39. Perl T, Jünger M, Vautz W, et al. : Detection of characteristic metabolites of Aspergillus fumigatus and Candida species using ion mobility spectrometry-metabolic profiling by volatile organic compounds. Mycoses. 2011;54(6):e828–e37. 10.1111/j.1439-0507.2011.02037.x [DOI] [PubMed] [Google Scholar]
- 40. Zehm S, Schweinitz S, Würzner R, et al. : Detection of Candida albicans by mass spectrometric fingerprinting. Curr Microbiol. 2012;64(3):271–5. 10.1007/s00284-011-0064-5 [DOI] [PubMed] [Google Scholar]
- 41. Ghosh S, Kebaara BW, Atkin AL, et al. : Regulation of aromatic alcohol production in Candida albicans. Appl Environ Microbiol. 2008;74(23):7211–8. 10.1128/AEM.01614-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Basanta M, Jarvis RM, Xu Y, et al. : Non-invasive metabolomic analysis of breath using differential mobility spectrometry in patients with chronic obstructive pulmonary disease and healthy smokers. Analyst. 2010;135(2):315–20. 10.1039/b916374c [DOI] [PubMed] [Google Scholar]
- 43. Bean HD, Dimandja JM, Hill JE: Bacterial volatile discovery using solid phase microextraction and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;901:41–6. 10.1016/j.jchromb.2012.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bean HD, Zhu J, Hill JE: Characterizing bacterial volatiles using secondary electrospray ionization mass spectrometry (SESI-MS). J Vis Exp. 2011; (52): pii: 2664. 10.3791/2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bunge M, Araghipour N, Mikoviny T, et al. : On-line monitoring of microbial volatile metabolites by proton transfer reaction-mass spectrometry. Appl Environ Microbiol. 2008;74(7):2179–86. 10.1128/AEM.02069-07 [DOI] [PMC free article] [PubMed] [Google Scholar]