Abstract
The three-dimensional (3D) cultivation of intestinal cells and tissues in dynamic bioreactor systems to represent in vivo intestinal microenvironments is essential for developing regenerative medicine treatments for intestinal diseases. We have previously developed in vitro human intestinal tissue systems using a 3D porous silk scaffold system with intestinal architectures and topographical features for the adhesion, growth, and differentiation of intestinal cells under static culture conditions. In this study, we designed and fabricated a multifunctional bioreactor system that incorporates pre-epithelialized 3D silk scaffolds in a dynamic culture environment for in vitro engineering of human intestine tissues. The bioreactor system allows for control of oxygen levels in perfusion fluids (aerobic simulated intestinal fluid (SIF), microaerobic SIF, and anaerobic SIF), while ensuring control over the mechanical and chemical microenvironments present in native human intestines. The bioreactor system also enables 3D cell culture with spatial separation and cultivation of cocultured epithelial and stromal cells. Preliminary functional analysis of tissues housed in the bioreactor demonstrated that the 3D tissue constructs survived and maintained typical phenotypes of intestinal epithelium, including epithelial tight junction formation, intestinal biomarker expression, microvilli formation, and mucus secretion. The unique combination of a dynamic bioreactor and 3D intestinal constructs offers utility for engineering human intestinal tissues for the study of intestinal diseases and discovery options for new treatments.
Keywords: bioreactor, intestine, perfusion, oxygen, peristalsis, silk, three-dimensional
Introduction
The human intestine performs vital functions for human health and physiology. The intestines secrete fluids, hormones, and digestive enzymes in addition to digesting food, absorbing nutrients, and supporting the immune system via metabolic homeostasis.1,2 In the past decade, diseases of the human intestine, such as inflammatory bowel disease (IBD) or infections by organisms such as Clostridium difficile, have affected a substantial proportion of the human population and have become an increasing health and socio-economic burden in the U.S. and other developed countries.3 Thus, there is a clear need for improved treatment options to minimize or avoid such diseases across the globe, yet progress in understanding mechanisms of infection or intestinal malfunctions remains poorly defined, and new therapeutic treatments remain to be discovered.
Current research into intestinal disease biology and drug development is mostly conducted either in 2D cell culture with intestinal epithelial cells or in animal models. The 2D cell culture models provide limited utility due to the nature of the substrate and architecture utilized. Although animal models allow researchers to develop novel therapies for intestinal diseases, they are costly and often do not reflect the corresponding human disease, as many new drugs that have successfully passed animal trials fail during the human clinical trial phase.4 Tissue engineering offers an alternative to animal models for quantitative studies of disease modeling and tests of new drugs in a verifiable, cost-efficient manner through a biologically driven approach by which the artificial tissues are regenerated by combining human cells, biosignals, materials, and bioreactors in vitro.5
The intestines are highly heterogeneous hollow organs with biological, mechanical and chemical differences between lumen and stroma. All segments of the human intestine are comprised of four layers: intestinal epithelium, subepithelium, muscle layer, and serosa.1 Under physiological conditions, the intestines experience rhythmic peristaltic motions characterized by wave-like muscle contractions traveling along the bowel wall as well as intralumenal intestinal fluid flow regulated by vasodilatation and vasoconstriction. The intestines also exhibit a proximal-to-distal, intraluminal oxygen gradient due to the oxygen consumption of intestinal microbiota.6 All the above-mentioned elements in intestine microenvironments are involved in intestinal development, microbiome functions, and bowel diseases. For example, ineffective intestinal propulsion results in intestinal pseudo-obstruction, yet intestinal inflammation may cause the inhibition of peristaltic contractions.7 Intestinal epithelial cells function within an oxygen tension level with dynamic and rapid fluctuation; however, in inflammatory bowel disease (IBD), the physiological oxygen pattern is dramatically dysregulated.8 Therefore, when engineering human intestine models for disease research and drug discovery, it is important to recreate the in vivo dynamic nature of the tissue microenvironments.
Traditional engineering of in vitro human intestinal tissues is based on the culture of intestinal epithelial cells on 2D substrates, such as rigid plastics and transwell inserts, under static conditions.9,10 However, 2D static culture cannot accurately recreate the microenvironments of native intestines. As a result, important mechanisms of infectious diseases cannot be accurately recapitulated in 2D static culture, hindering the development of drugs for the treatment of intestine diseases. As a result, significant efforts have been made to mimic the dynamics of the original intestinal microenvironment by integrating perfusion-based bioreactor systems into static cultures. For instance, a perfusion bioreactor was designed and fabricated for intestinal tissue engineering where intestinal epithelial organoid units were dynamically seeded onto tubular polymer scaffolds and survived under flow conditions for 2 days.11 Recently, human-gut-on-a-chip models utilized fluid flow as well as cyclic mechanical strain to intestinal epithelial cells grown on a membrane at levels similar to those experienced in the living intestine in vivo. This system supported a longer term cell culture than the static conditions and enabled the colonization of some aerobic probiotics.12,13 More recently, a mechanically driven bioreactor mimicking the cyclic contraction and relaxation of the intestine tissue using an electro-responsive elastomeric membrane was designed for the in vitro remodeling of human intestine.14 While these bioreactor-based intestinal systems are useful in vitro tools to study intestinal infections and drug treatments,15,16 they do not reproduce an in vivo-like, oxygen-restricted luminal microenvironment for the growth of bacterial communities of obligate anaerobes prevalent in human intestines. Moreover, the native intestinal epithelium is a monolayer lining the 3D tubular tissue architecture, and the apical surface of the epithelium is exposed to the low-oxygen-tension environment in the lumen, while the basal surface receives a constant supply of nutrients and oxygen through arterial blood supply. However, current systems do not address the sophisticated spatial organization and distinct routes for oxygen and nutrient supply of the epithelial cells, and therefore the cells may not behave physiologically, even with the support of dynamic devices.
We have previously engineered intestinal tissues using tubular 3D silk scaffolds with compartments to separately accommodate different cells.17,18 We have also developed bioreactors for the optimal mechanical condition of various engineered constructs in vitro.19−21 In this paper, we present a bioreactor system which combines 3D intestine-like tissues17 and a benchtop bioreactor design to impart many dynamic aspects of the actual in vivo environment of human intestine, including intralumenal perfusion of simulated intestine fluid (SIF), local oxygen microenvironment, and rhythmic peristaltic movement. This custom-designed, pulsatile perfusion bioreactor, with programmable manipulation, allows adjustment of luminal flow rate, oxygen control of perfusion fluids, and pneumatic stimulation pressure. In addition, this system is also capable of achieving the intestine-like nutrition/oxygen supply to the cells by the luminal perfusion of the hollow channel compartment of the 3D tissue construct with oxygen deficient SIFs and the nutrition/oxygen diffusions in the scaffold bulk in support of epithelial and subepithelial cells. This dynamic platform can mimic the human intestine with increased accuracy to overcome many of the limitations with the previously described in vitro intestinal models, providing a more representative proof-of-concept model of the human intestine.
Materials and Methods
Materials
The Caco-2 (CRL-2102) cell line was obtained from ATCC (Rockville, MD). The HT29-MTX cell line was obtained from the Public Health England Culture Collections (Salisbury, Great Britain). Human Intestinal Myofibroblasts (H-InMyoFib) were purchased from Lonza. Both Caco-2 and HT29-MTX cells were grown in DMEM supplemented with 10% fetal bovine serum, 10 μg/mL human transferrin (Gibco), and 1% antibiotics and antimycotics. Myofibroblasts were cultured in SMGM-2 BulletKit medium (Lonza). Polydimethylsiloxane (PDMS, Sylgard 184) was purchased from Dow Corning, and Ecoflex 00–30 was purchased from Smooth-On. A Teflon cylinder, Teflon tubes, adaptor, and silicone rubber tubes were purchased from McMaster-Carr.
Cell Seeding and Preperfusion Culture
Human intestinal epithelial cells (Caco-2 and HT29-MTX) and myofibroblasts (H-InMyoFib) were seeded onto the silk scaffolds as previously described.17 Briefly, human intestinal epithelial cells (Caco-2 and HT29-MTX cells) were seeded on the luminal surface of silk scaffolds, and primary human intestinal myofibroblasts were delivered into the scaffold bulk space. After seeding, the constructs were precultured for 2 weeks under static, normoxic conditions (37 °C, 5% CO2, 21% O2) in 12 well plates to form a confluent layer of intestinal epithelium, lining the interior wall of the scaffolds.
Design and Assembly of the Perfusion Cartridge and the Entire Bioreactor
The design and assembly of the perfusion cartridge and the entire bioreactor is described in detail in the Results and Discussion section.
Preparation of Oxygen-Deficient SIF
A mixture of 5% CO2, 10% O2, and 85% N2 gas was used to pre-equilibrate the Simulated Intestinal Fluid (SIF;22,23 Ricca; 0.57% potassium dihydrogen phosphate (Sigma), 0.39% potassium chloride (Sigma), and 0.1% sodium hydroxide (Sigma), pH = 6.7–6.9). A total of 500 mL of SIF was then placed in a PTFE stoppered Pyrex reaction kettle (Sigma), and a glass bubbler was fitted to allow the SIF to be bubbled with the gas mixture of 5% CO2, 10% O2, and 85% N2 at room temperature for up to 30 min.
Bioreactor Pressure Response Measurements
The pressure response of the bioreactors was measured by a custom-made device. Briefly, a pressure monitoring system was connected to the bioreactor and an air injection system. Then, the deformation of the bioreactor caused by a cyclic pumping process was recorded with a video camera. The values of the changes in diameter and pressure were extracted from each frame of recorded video with a time resolution of ∼0.3 s.
Perfusion Culture of Human Intestinal Tissue
The pre-equilibrated luminal and subepithelial media were pumped, respectively, from the perfusion liquid reservoirs into the inner channel (epithelial lumen) and through the bulk (subepithelial myofibroblasts) of the bioengineered intestinal scaffold using an NE-1600 six channel programmable syringe pump (New Era Pump Systems, Inc.; Farmingdale, NY, USA). The luminal channel was perfused at a constant flow rate of 30 μL/min, corresponding to a shear stress of 0.02 dyn/cm2. The medium in the chamber to support the myofibroblasts was changed every other day. For the generation of the intestinal equivalent, the generated matrix-cell constructs were perfused for up to 4 weeks with cell culture medium, followed by histological as well as immunofluorescence analysis of the growth behavior of the cells. As a control, matrix-cell constructs were cultivated statically. The medium was changed every other day.
Confocal Imaging
At specific time points after tissue perfusion, the scaffolds with intestinal cells were fixed and imaged by confocal microscope as previously described.17 Briefly, the scaffolds were fixed with 4% paraformaldehyde (PFA, Santa Cruz). Silk scaffolds were cut in half along the longitudinal axis in order to better expose the lumen to the blocking solutions and antibodies during the following incubation steps. All specimens were then permeabilized using 0.1% Triton x-100 in phosphate-buffered saline (PBS, Gibco), then blocked with 5% bovine serum albumin (BSA, Sigma) for 2 h. These specimens were incubated overnight at 4 °C with antihuman ZO-1 (BD Transduction Laboratorie, 1:50), anti-e-cadherin (abcam, 1:50), antihuman-MUC-2 (Santa Cruz Biotech, 1:50), and anti-villin (abcam, 1:100), then immersed in Alexa Fluor 488 donkey antimouse and Alexa Fluor 546 goat–antirabbit secondary antibodies (Invitrogen) at a dilution of 1:100. Scaffolds were then counterstained with dihydrochloride (DAPI; Invitrogen) before being mounted using Vectashield mounting medium (Vector Laboratories). These 3D scaffolds were scanned using a Leica SP2 confocal microscope (Leica Microsystems) and Nikon A1R (Nikon Instruments Inc.) with Z-series capability. Scaffolds were observed under a confocal microscope with a filter set for DAPI (Ex/Em: 350/470 nm), Texas Red (Ex/Em: 540/605 nm), and GFP/FITC (Ex/Em: 488/514 nm). The 3D rendering images and confocal 3D maximum projection images were assembled with Leica confocal software (ver 2.61, Leica), NIS-Elements ARsoftware package (ver 4.20.01, Nikon), and ImageJ. The measured fields in the 3D lumen were randomly selected. Twenty randomly selected fields were analyzed for each scaffold sample using NIS-Elements ARsoftware. To obtain an average mucus thickness over the epithelial surface in each 3D picture, five points were measured and the average thickness calculated.
Scanning Electron Microscopy (SEM) and Quantification of Microvilli
The silk scaffolds with cells were cross-linked with 2.5% glutaraldehyde, followed by progressive dehydration in a graded series of ethanols (30%, 50%, 75%, 95%, and twice in 100%, 30 minutes at each concentration). The samples were subsequently dried by critical point drying with a liquid CO2 dryer (AutoSamdri-815, Tousimis Research Corp., Rockville, MD). Prior to imaging using a scanning electron microscope (Zeiss UltraPlus SEM or Zeiss Supra 55 VP SEM, Carl Zeiss SMT Inc., Peabody, MA) at a voltage of 2–3 kV, the samples were coated with a thin layer (10 nm thick) of Pt/Pd using a sputter coater (208HR, Cressington Scientific Instruments Inc., Cranberry Twp., PA).
Quantitative RT-PCR
Intestinal epithelial cells on the luminal surface of scaffolds were detached with 0.25% trypsin-EDTA and a cell scraper. Total RNA was isolated using the Qiagen Mini mRNA Extraction kit. RNA was reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Six nanograms of cDNA were used for real-time PCR amplification for each well. The primer sequences were as follows: (1) GAPDH Forward: GAAGGTGAAGGTCGGAGTC, Reverse: GAAGATGGTGATGGGATTTC; (2) ZO-1, Forward: CTGGTGAAATCCCGGAAAAATGA, Reverse: TTGCTGCCAAACTATCTTGTGA; (3) E-cadherin, Forward: ATCGGTTGTTCAATGCGTCC, Reverse: CCTTCAGGATTTGGTACATGACA; (4) Villin, Forward: CGGAAAGCACCCGTATGGAG, Reverse: CGTCCACCACGCCTACATAG. For each gene tested, we performed three experimental replicates and four biological replicates. Gene expression levels were normalized to the GAPDH mRNA level.
Results and Discussion
Bioreactor Design and Functionality
Design and Assembly of the Perfusion Cartridge
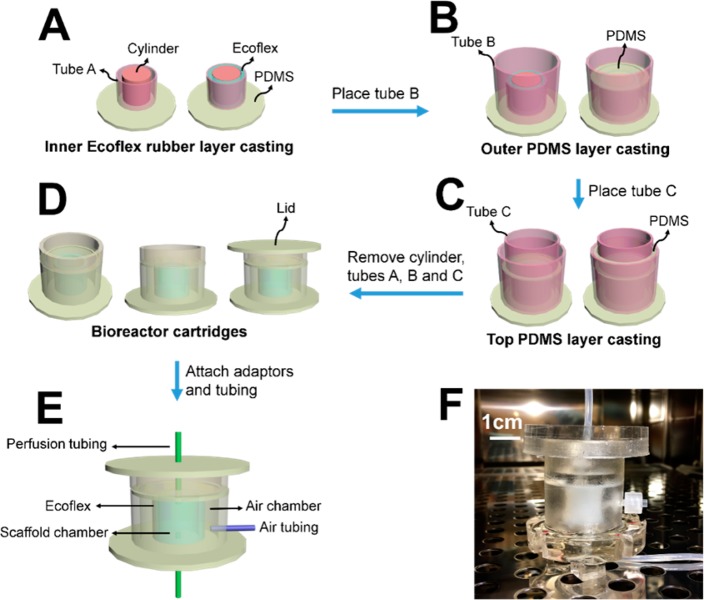
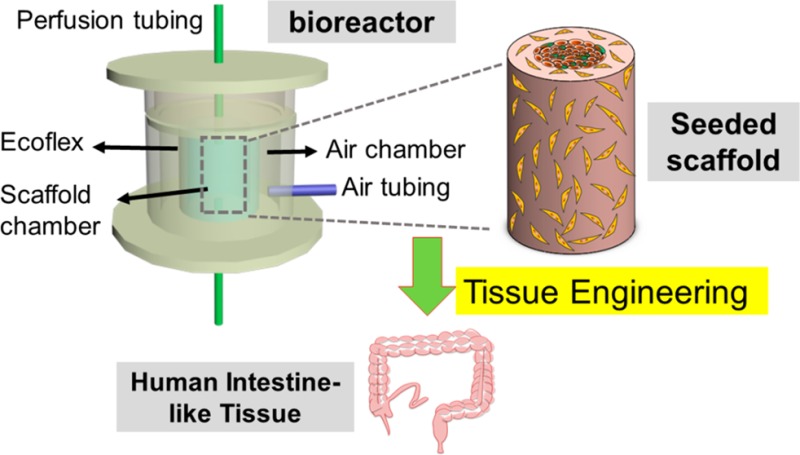
As illustrated in Figure 1, the perfusion cartridge bioreactors were prepared from custom-cast polydimethylsiloxane (PDMS) by a three-step replica molding process: casting of the inner Ecoflex rubber layer (Figure 1A), casting of the outer PDMS layer (Figure 1B), and casting of the top PDMS layer (Figure 1C). Each device consists of an inner wall, inside which the 3D intestinal scaffold is mounted, and an outer wall, forming an air chamber surrounding the inner wall. The inner wall is composed of highly stretchable Ecoflex (blue) and less deformable conventional PDMS (white), while the outer wall is composed of entirely PDMS (white). Access holes were punched in the center of the top and bottom layers for the perfusion tubing to connect to the scaffolds in the chamber (Figure 1E). A photograph of a perfusion cartridge is shown in Figure 1F. A precultured intestinal scaffold (Figure 2A–C) is mounted in the middle of the scaffold chamber by connecting the inner luminal channel with the perfusion tubing through the adapters. Medical glue was applied on the connection locations on the top and bottom to prevent leakage. Dimensions of each compartment are shown in Table 1.
Figure 1.
Design and assembly of the perfusion cartridge. (A) Casting of the inner Ecoflex rugger layer of the device. (B) Casting of the outer PDMS layer of the device. (C) Casting of the top PDMS layer of the device. (D) The complete perfusion cartridges. (E) The attachment of adaptors and tubing. (F) A photograph of a perfusion cartridge.
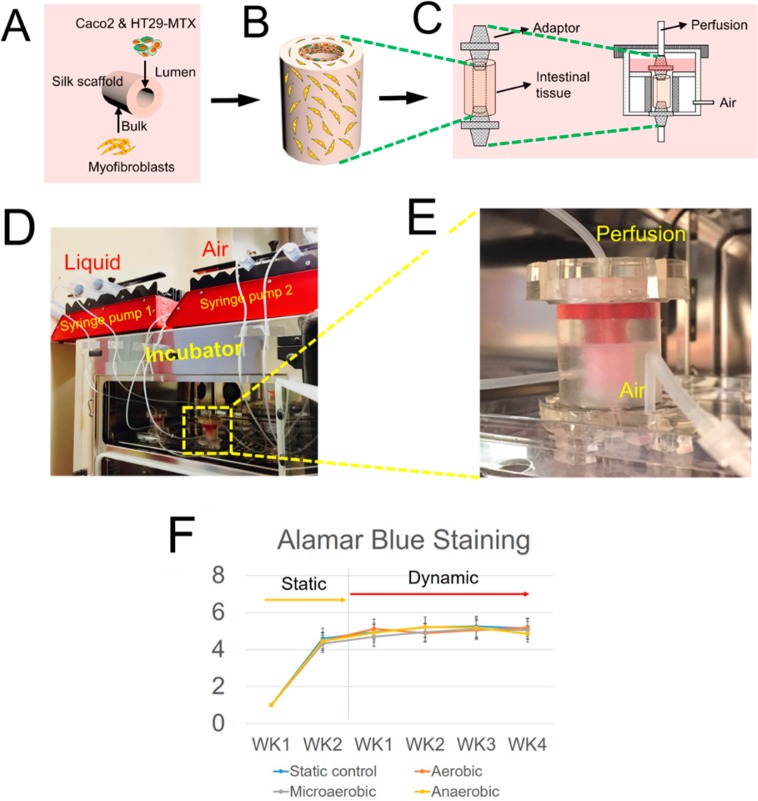
Figure 2.
Assembled perfusion cartridge. (A–C) Schematics showed the seeding of intestinal cells on the 3D porous silk scaffold and the placement of the scaffolds in the dynamic bioreactor. (D) Overview of the entire bioreactor. (E) A photograph of a working perfusion cartridge. (F) AlamarBlue Proliferation Assay showed tissue viability from all experimental groups over time.
Table 1. Dimension of Each Compartment and Teflon Modes.
| parts | outer diameters | inner diameters | thickness of wall | height |
|---|---|---|---|---|
| Teflon cylinder | 1/2″ | N/A | N/A | 1″ |
| Teflon tube A | 1″ | 3/4″ | 1/8″ | 1″ |
| Teflon tube B | 1.5″ | 1″ | 0.25″ | 2″ |
| Teflon tube C | 1″ | 3/4″ | 1/8″ | 1″ |
| silicon tube | 1/8″ | 1/16” | ||
| adaptors | 1/16″ |
Design and Assembly of the Bioreactor System
Figure 2 displays the schema and the photographs of the entire assembled bioreactor system. The major components of the dynamic bioreactor system include a perfusion cartridge (described above), a syringe pump (pump 1) controlling fluid circulation, a syringe pump (pump 2) controlling the deformation of the ECOFLEX membrane in the perfusion cartridge, and a pregassing unit. The top and bottom of the chamber housing the bioengineered scaffold sealed in the PDMS perfusion cartridge were connected, respectively, by the perfusion liquid (syringe pump 1) and the waste collection unit with tubing, while the air chamber of the cartridge was connected to the “syringe air pump 2” with tubing. The details of the system setup of perfusion and pneumatic stimulation of the device were described as two aspects: (i) flow perfusion with oxygen deficient SIFs and (ii) pneumatic stimulation.
(i) For flow perfusion with oxygen deficient SIFs. The bioreactor perfuses the intestinal tissue, which was fit between two adaptors with the oxygen deficient SIF circulating through the lumen of the intestinal tissue at a constant rate of 30 μL/h. The rest of the bioreactor was filled with nutrient medium at normal oxygen tension to support the stromal cells in the scaffold bulk and the epithelial cells on the luminal surface by providing nutrients and oxygen to the basal surface (Figure 2C). To achieve the oxygen control of the perfusion SIFs, a gas mixture of 5% CO2, 10% O2, and 85% N2 was used to pre-equilibrate the fluids before perfusion into the cartridge (Figure 3A,B). Using an oxygen probe, the oxygen concentration of the SIF was measured at ∼15.7% (aerobic SIF) without any gas purging. The oxygen concentration of the SIF decreased to ∼4.7% (microaerobic SIF) after 10 min of purging, and the oxygen concentration of the SIF decreased to 0.3% (anaerobic SIF) after 30 min of purging. By using the oxygen deficient SIFs as luminal perfusion fluids in the bioreactor system, we were able to precisely control the oxygen level on the tissue lumen. This feature of the bioreactor is critical as the intestinal lumen is largely devoid of oxygen. A proximal-to-distal, intraluminal oxygen gradient with a marked decrease in pO2 along the gastrointestinal tract, e.g., 3–7% in the midstomach, 2–4% in the mid-duodenum, ∼1% in the midsmall intestine, and <0.4% in the distal colon, exists in a living mouse.24 Similar to the in vivo case, our previous 3D models of human intestinal tissue17 exhibited depth-graded oxygen profiles in the luminal direction under static cultures. To generate a more controlled system, here, we implemented a bioreactor and oxygen control system that houses the tissue and allows for a differential control of oxygen levels in the tissue. In addition, the bioreactor system also incorporates SIFs to further mimic intestinal microenvironments. SIFs allowed for better selection of peptide candidates for oral delivery,25 and therefore, this system can provide a suitable platform for intestinal drug discovery research.
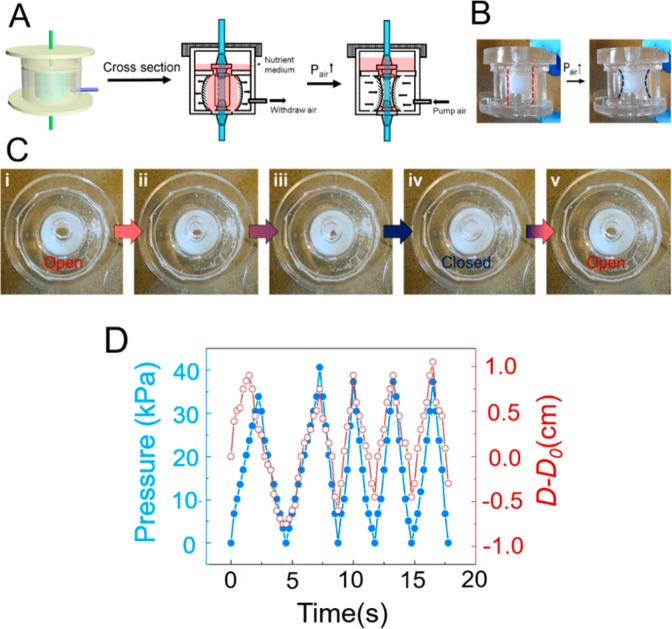
Figure 3.
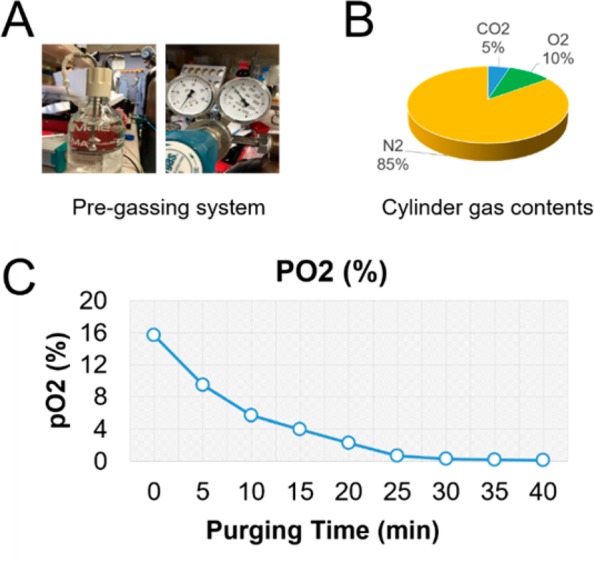
Preparation of oxygen-deficient SIFs. (A) Photographs of the pregassing system of the bioreactor. A mixture of 5% CO2, 10% O2, and 85% N2 gas (B) was used to pre-equilibrate the simulated intestinal fluids (SIF). (C) Real-time measurements of average oxygen levels during the gas purging.
(ii) Pneumatic stimulation. Propulsive peristalsis is characterized by a circular muscle contraction wave traveling along the digestive tract, from the esophagus into the stomach and the small intestine. The frequency range of contraction of the gastrointestinal tract is 3/min in the stomach, 11–12/min in the duodenum, and 9/min in the ileum and is determined by the frequency of the basic electrical rhythm.26 To mimic in vivo peristalsis, the bioreactor was designed to provide control over peristaltic motion. To achieve this, a tube was used to connect the socket and a syringe pump and air was pumped into the chamber to create controlled air pressure differentials between the inside and outside of the chamber, causing the highly stretchable ECOFLEX membrane to contract and expand. The resultant deformation of the ECOFLEX membrane simulates intestinal peristalsis and provides mechanical stimulation to the tissue inside the inner wall (Figure 3A–C). In vivo, the frequencies of intestinal contractions are about 7–20 per minute, with slower frequencies closer to the distal ileum and higher frequencies toward the duodenum.7 Therefore, the frequency of the deformation in the system was set to 12 contractions per minute to mimic physiological intestinal peristalsis. The real-time pressure of the luminal fluid was monitored by a pressure sensor. Performance tests showed that pulsatile perfusion driven by the syringe pump was stable and was recorded at a range of 0–40 mmHg (Figure 4D). In healthy adults, the average pressure in the jejunum is around 20 mmHg, while under some pathologic conditions such as chronic inflammatory states, intraluminal pressures are often increased.7 The present bioreactor system, therefore, is capable of imparting the physiological mechanical stimuli that are present in the native intestine to the cultured tissue constructs.
Figure 4.
Schematic illustration of perfusion bioreactors that employ a mechanism of pneumatic stimulation. (A) Schematic cross-sectional view of a bioreactor cartridge design. Each device consists of an inner wall, inside which the 3D intestinal scaffold (orange) is mounted, and an outer wall, forming an air chamber surrounding. The inner wall is composed of highly stretchable Ecoflex (blue) and less deformable conventional PDMS (red), while the outer one is composed entirely of PDMS. Operation of a prototype bioreactor cartridge, (B) side view, and (C) top view. (D) Real-time measurements of average pressures generated from the pneumatic stimulation.
The Phenotypes of the Dynamic Bioreactor System-Produced Intestine Constructs
Three-dimensional hollow scaffold systems17 were used to investigate the phenotypes of the dynamic bioreactor system-produced intestine constructs. The 3D intestine model was bioengineered by cultivating intestinal epithelial cells (Caco-2 and HT-29-MTX), on the luminal surface of silk scaffolds and primary human intestinal myofibroblasts (H-InMyoFibs) within the scaffold bulk as feeder cells (Figure 2). After cell seeding, the 3D tissue constructs were maintained in static culture for 2 weeks for the formation of confluent and differentiated intestinal epithelium in the scaffolds. The tissues were then placed into the multifunctional bioreactor that we described above (perfusion with oxygen deficient SIF and peristaltic motion) and maintained in the dynamic culture for up to 4 weeks. According to the oxygen level of the perfusion fluids, tissues were divided into aerobic, microaerobic, and anaerobic groups. The same tissue constructs cultured in static plates served as controls. Cell/tissue viability over time was determined by AlamarBlue Proliferation Assay (Figure 2F). The assays demonstrated that tissue viability was sustainable in all groups from week 1 to week 4. As the number of H-InMyoFibs within the bulk was much higher than the monolayer of epithelium on the luminal surface, the results mostly reflect the viability of the H-InMyoFibs, which are not significantly affected by oxygen or SIFs.
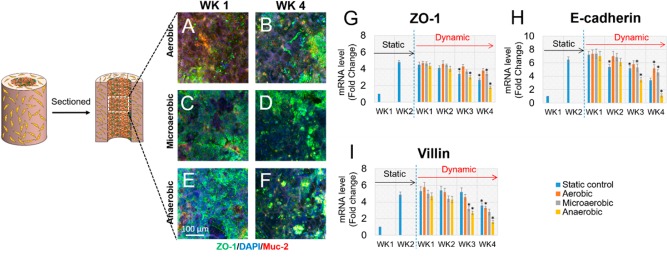
To further evaluate the phenotype of tissue constructs in the bioreactor, immunofluorescence microscopy was used to detect the expression of some differentiation markers in intestinal epithelium, including Zonula occludens-1 (ZO-1), an epithelial tight junction biomarker, and MUC2, a major component of mucus (Figure 5A–F). Both ZO-1 and MUC2 were observed in all groups through week 1 to week 4 in the dynamic bioreactors, suggesting that the 3D bioreactor systems were suitable for differentiation of the intestinal tissues. Additionally, qRT-PCR was performed to qualitatively detect gene expression of intestinal epithelial biomarkers, including ZO-1, E-cadherin, and villin (Figure 5G–I). qRT-PCR data revealed that, in addition to ZO-1, the gene expression levels of other biomarkers of human intestinal epithelium, such as E-cadherin and villi, were not significantly changed after switching from a static to dynamic culture in the first 2 weeks. Compared to the static group, aerobic perfusion sustained relatively stable gene expression levels of biomarkers up to 4 weeks. However, at week 3 postperfusion, gene expression of markers under microaerobic and anaerobic perfusion conditions was significantly lower than week 1 postperfusion. Similar results were found when microvilli and the mucus layer of tissue constructs under static and dynamic cultures were imaged and quantified separately (Figures 6 and 7). The microvilli number and the thickness of the mucus layer under aerobic conditions were stably maintained at least for 4 weeks post bioreactor incubation, while under microaerobic and anaerobic perfusion conditions, the microvilli number and mucus layer tended to decrease. In general, the bioreactor system incorporating a 3D intestinal construct and a dynamic system design with control over perfusion fluid, oxygen tension, and mechanical stimulation supported more stable growth and differentiation of the intestinal epithelium when compared to static culture. Tissue constructs in the bioreactor with microaerobic and anaerobic SIFs yielded comparable phenotypes to static cultures in the first 2 weeks; however, tissue functions in the static cultures decreased gradually from week 3. This can be explained by the lack of oxygen supply to the epithelium. In the bioreactors, cells in the bulk space were maintained in a static medium. Under this situation, cells with oxygen deficient SIFs do not get enough oxygen, and as a result their functions diminished with time. Future bioreactor designs will include a perfusion circuit to the subepithelial tissue to promote oxygen transport in the bulk space and to support the basal surface of the epithelium. In addition, the decline in gene expression levels of biomarkers occurred in the system with aerobic flow from week 1 to week 4. This may indicate that the SIFs perfused through the lumens may lack nutrients that the apical surface of the epithelium would need. To address this issue, the composition of SIFs may be modified to be more compatible with Caco-2/HT29-MTX cells.27
Figure 5.
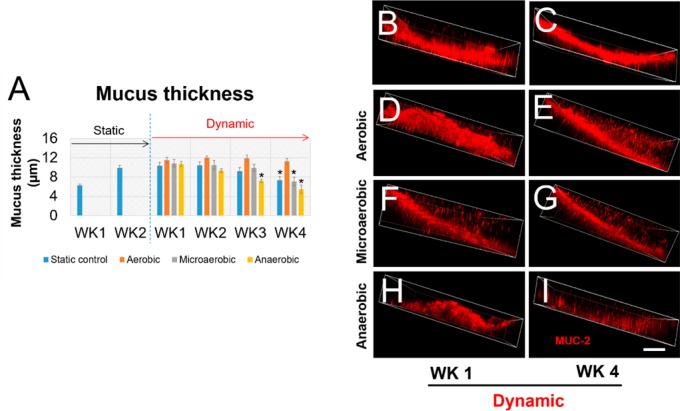
(A–F) Scaffolds with intestinal cells collected from different time points were sectioned in half along the longitudinal axis for immunostaining. Immunostaining of MUC-2 and ZO-1of Caco-2/HT29-MTX cultured on scaffolds 1 week and 4 weeks post-bioreactor culture were imaged by confocal microscopy. MUC2 was visualized as red, ZO-1 as green, and DAPI as blue. (G–I) Intestinal epithelial cells on the luminal surface of scaffolds were detached, and total RNA was isolated for evaluation of gene expression levels of different biomarkers. Gene expression levels of intestinal epithelium biomarkers, ZO-1 (G), E-cadherin (H), and villin (I), were evaluated by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) overtime in cultures. Data are presented as mean ± SEM, n = 3 in each group, *p < 0.001.
Figure 6.
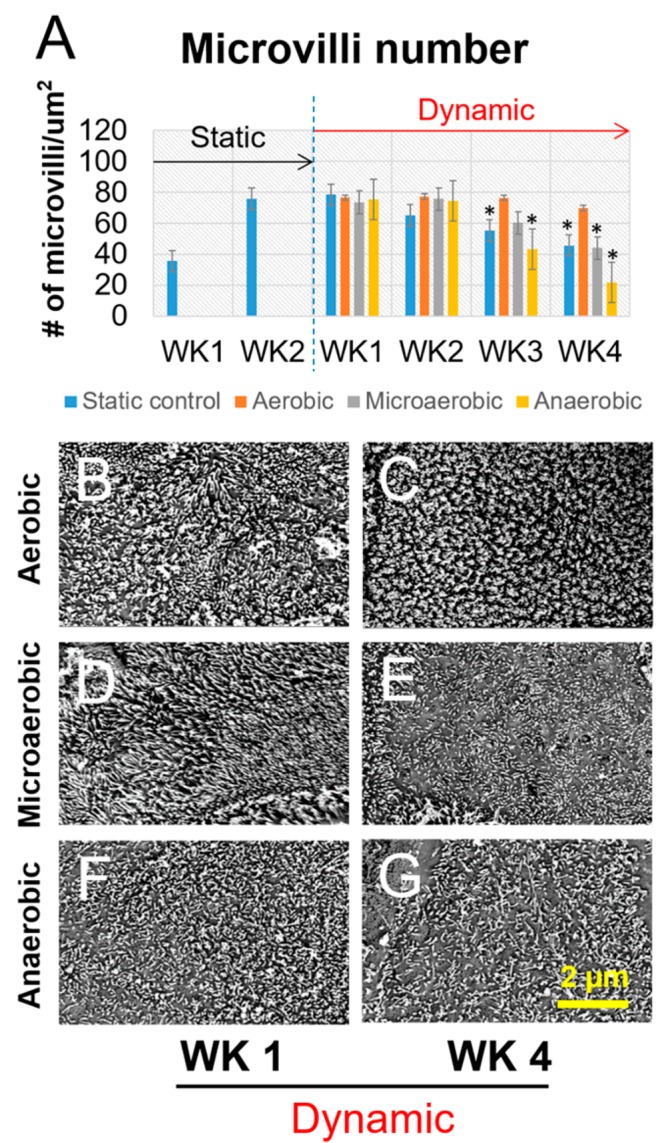
Visualization and quantification of epithelial microvilli. Data are presented as mean ± SEM, n = 5 in each group, *p < 0.001.
Figure 7.
Analysis of mucus thickness generated in the static culture and bioreactor cultures with oxygen control. (A) The quantification of mucus thickness. (B–G) Confocal images of MUC-2 staining on samples from different culture conditions. Scale bar = 10 μm. Data are presented as mean ± SEM, n = 5 in each group, *p < 0.001.
Research on bioreactors has led to tailorable systems integrating environmental factors such as perfusion in addition to biomechanical, biochemical, and electrical stimulation.28 Moreover, current efforts addressing issues of scale and morphology have further advanced the field.29,30 In this study, we have presented a bioreactor design built for the study of engineered intestinal tissues. The overall aim of the intestinal bioreactor was to support the growth and differentiation of intestinal epithelial cells on 3D scaffolds in vitro by mimicking the in vivo physiological environment, using a pulsatile perfusion system with intraluminal SIF perfusion and oxygen control. The bioreactor system can generate physiologically relevant peristaltic motion frequencies (12 contractions/min) and intraluminal pressure to the epithelium (0–40 mmHg). Additionally, the pregassing unit was used to deplete the oxygen from SIFs to achieve oxygen control of the luminal fluid (0–15.7%). We demonstrated that this bioreactor technology provided appropriate biochemical and physiological regulatory signals by guiding the differentiation and by maintaining the genotypes of intestinal epithelial cells on 3D intestinal tissue constructs. The epithelial cells continued to express key intestinal biomarkers after switching to the bioreactor. We report for the first time the recapitulation of functional human intestinal epithelium on a multifunctional 3D bioreactor system (3D tissue, oxygen deficient SIF perfusion, and peristaltic motion) that could be maintained for long time frames (>2 weeks). Overall, the design of tissue deformation modes (cyclic stretch/flexure) and SIF perfusion with an oxygen control in combination with 3D intestinal tissue make this novel bioreactor a useful tool in investigating intestinal function and drugs for infections and infectious diseases.
Conclusions
In summary, we designed a new bioreactor system, which supports the extended cultivation of 3D human intestinal tissues, a key feature for longer term investigation of mechanisms of intestinal functions and dysfunctions in normal and disease states. We further demonstrated the successful design and implementation of this new device with human epithelial and fibroblast cells, to establish proof of concept. Our results suggest that such a system is invaluable in studies of microbiome–intestinal interactions and signaling and encourage us to further study the refinements to support organoid cultures as well as robust screening studies for new therapeutic discovery.
Author Contributions
∥ These authors contributed equally to this work.
Author Contributions
W.Z., Y.C., Y.L., and D.L.K. conceived of the experiments. W.Z., Y.C., and S.L. performed the experiments, designed research, and analyzed data with assistance from T.R., S.Z., D.M.C., N.K., E.M., and D.L.K. Y.C., W.Z., and D.L.K. wrote the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
We thank the National Institutes of Health (U19A1131126) and the Bill and Melinda Gates Foundation for support of this work.
The authors declare no competing financial interest.
References
- Turner J. R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9 (11), 799–809. 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Peterson L. W.; Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14 (3), 141–53. 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- Burnham C. A.; Carroll K. C. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev. 2013, 26 (3), 604–30. 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak I. W.; Evaniew N.; Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl Res. 2014, 6 (2), 114–118. [PMC free article] [PubMed] [Google Scholar]
- Marx V. Tissue engineering: Organs from the lab. Nature 2015, 522 (7556), 373–7. 10.1038/522373a. [DOI] [PubMed] [Google Scholar]
- Barbara G.; Feinle-Bisset C.; Ghoshal U. C.; Quigley E. M.; Santos J.; Vanner S.; Vergnolle N.; Zoetendal E. G. The Intestinal Microenvironment and Functional Gastrointestinal Disorders. Gastroenterology 2016, 150, 1305. 10.1053/j.gastro.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Gayer C. P.; Basson M. D. The effects of mechanical forces on intestinal physiology and pathology. Cell. Signalling 2009, 21 (8), 1237–44. 10.1016/j.cellsig.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 2013, 7 (7), 1256–61. 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larregieu C. A.; Benet L. Z. Drug Discovery and Regulatory Considerations for Improving In Silico and In Vitro Predictions that Use Caco-2 as a Surrogate for Human Intestinal Permeability Measurements. AAPS J. 2013, 15 (2), 483–497. 10.1208/s12248-013-9456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers A. R.; Conradi R. A.; Burton P. S. Caco-2 Cell Monolayers as a Model for Drug Transport across the Intestinal-Mucosa. Pharm. Res. 1990, 7 (9), 902–910. 10.1023/A:1015937605100. [DOI] [PubMed] [Google Scholar]
- Kim S. S.; Penkala R.; Abrahimi P. A perfusion bioreactor for intestinal tissue engineering. J. Surg. Res. 2007, 142 (2), 327–31. 10.1016/j.jss.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Ingber D. E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol. (Camb) 2013, 5 (9), 1130–40. 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Huh D.; Hamilton G.; Ingber D. E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12 (12), 2165–74. 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Cei D.; Costa J.; Gori G.; Frediani G.; Domenici C.; Carpi F.; Ahluwalia A. A bioreactor with an electro-responsive elastomeric membrane for mimicking intestinal peristalsis. Bioinspir Biomim 2017, 12 (1), 016001. 10.1088/1748-3190/12/1/016001. [DOI] [PubMed] [Google Scholar]
- Maurer M.; Wollny T.; Berlinghof F.; Raasch M.; Huber O.; Mosig A. Development of a human gut-on-chip model to study macrophage dependent inflammatory response. Infection 2017, 45, S10–S10. [Google Scholar]
- Lee J.; Choi J. H.; Kim H. J. Human gut-on-a-chip technology: will this revolutionize our understanding of IBD and future treatments?. Expert Rev. Gastroenterol. Hepatol. 2016, 10 (8), 883–885. 10.1080/17474124.2016.1200466. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Lin Y.; Davis K. M.; Wang Q.; Rnjak-Kovacina J.; Li C.; Isberg R. R.; Kumamoto C. A.; Mecsas J.; Kaplan D. L. Robust bioengineered 3D functional human intestinal epithelium. Sci. Rep. 2015, 5, 13708. 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCicco RePass M. A.; Chen Y.; Lin Y.; Zhou W.; Kaplan D. L.; Ward H. D. Novel Bioengineered Three-Dimensional Human Intestinal Model for Long-Term Infection of Cryptosporidium parvum. Infect. Immun. 2017, 85 (3), e00731-16. 10.1128/IAI.00731-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.; Quinn K. P.; Bellas E.; Georgakoudi I.; Kaplan D. L. Noninvasive metabolic imaging of engineered 3D human adipose tissue in a perfusion bioreactor. PLoS One 2013, 8 (2), e55696. 10.1371/journal.pone.0055696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian B.; Rudym D.; Cannizzaro C.; Perrone R.; Zhou J.; Kaplan D. L. Tissue-engineered three-dimensional in vitro models for normal and diseased kidney. Tissue Eng., Part A 2010, 16 (9), 2821–31. 10.1089/ten.tea.2009.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Buduo C. A.; Wray L. S.; Tozzi L.; Malara A.; Chen Y.; Ghezzi C. E.; Smoot D.; Sfara C.; Antonelli A.; Spedden E.; Bruni G.; Staii C.; De Marco L.; Magnani M.; Kaplan D. L.; Balduini A. Programmable 3D silk bone marrow niche for platelet generation ex vivo and modeling of megakaryopoiesis pathologies. Blood 2015, 125 (14), 2254–64. 10.1182/blood-2014-08-595561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingels F.; Deferme S.; Destexhe E.; Oth M.; Van den Mooter G.; Augustijns P. Simulated intestinal fluid as transport medium in the Caco-2 cell culture model. Int. J. Pharm. 2002, 232 (1–2), 183–92. 10.1016/S0378-5173(01)00897-3. [DOI] [PubMed] [Google Scholar]
- Lind M. L.; Jacobsen J.; Holm R.; Mullertz A. Development of simulated intestinal fluids containing nutrients as transport media in the Caco-2 cell culture model: assessment of cell viability, monolayer integrity and transport of a poorly aqueous soluble drug and a substrate of efflux mechanisms. Eur. J. Pharm. Sci. 2007, 32 (4–5), 261–70. 10.1016/j.ejps.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Marra A.; Isberg R. R. Invasin-dependent and invasin-independent pathways for translocation of Yersinia pseudotuberculosis across the Peyer’s patch intestinal epithelium. Infect. Immun. 1997, 65 (8), 3412–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muheem A.; Shakeel F.; Jahangir M. A.; Anwar M.; Mallick N.; Jain G. K.; Warsi M. H.; Ahmad F. J. A review on the strategies for oral delivery of proteins and peptides and their clinical perspectives. Saudi Pharm. J. 2016, 24 (4), 413–28. 10.1016/j.jsps.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudie D. M.; Murray K.; Hoad C. L.; Pritchard S. E.; Garnett M. C.; Amidon G. L.; Gowland P. A.; Spiller R. C.; Amidon G. E.; Marciani L. Quantification of Gastrointestinal Liquid Volumes and Distribution Following a 240 mL Dose of Water in the Fasted State. Mol. Pharmaceutics 2014, 11 (9), 3039–3047. 10.1021/mp500210c. [DOI] [PubMed] [Google Scholar]
- Patel N.; Forbes B.; Eskola S.; Murray J. Use of simulated intestinal fluids with Caco-2 cells and rat ileum. Drug Dev. Ind. Pharm. 2006, 32 (2), 151–61. 10.1080/03639040500465991. [DOI] [PubMed] [Google Scholar]
- Martin I.; Wendt D.; Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004, 22 (2), 80–6. 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Nokhbatolfoghahaei H.; Rad M. R.; Khani M. M.; Shahriari S.; Nadjmi N.; Khojasteh A., Application of bioreactors to improve functionality of bone tissue engineering constructs: A systematic review. Curr. Stem Cell Res. Ther. 2017, DOI: 10.2174/1574888X12666170822100105. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A.; Langer R. A decade of progress in tissue engineering. Nat. Protoc. 2016, 11 (10), 1775–81. 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]