Abstract
Porcine faecal waste is a serious environmental pollutant. Carriage of antimicrobial-resistance genes (ARGs) and virulence-associated genes (VAGs), and the zoonotic potential of commensal Escherichia coli from swine are largely unknown. Furthermore, little is known about the role of commensal E. coli as contributors to the mobilization of ARGs between food animals and the environment. Here, we report whole-genome sequence analysis of 103 class 1 integron-positive E. coli from the faeces of healthy pigs from two commercial production facilities in New South Wales, Australia. Most strains belonged to phylogroups A and B1, and carried VAGs linked with extraintestinal infection in humans. The 103 strains belonged to 37 multilocus sequence types and clonal complex 10 featured prominently. Seventeen ARGs were detected and 97 % (100/103) of strains carried three or more ARGs. Heavy-metal-resistance genes merA, cusA and terA were also common. IS26 was observed in 98 % (101/103) of strains and was often physically associated with structurally diverse class 1 integrons that carried unique genetic features, which may be tracked. This study provides, to our knowledge, the first detailed genomic analysis and point of reference for commensal E. coli of porcine origin in Australia, facilitating tracking of specific lineages and the mobile resistance genes they carry.
Keywords: antimicrobial resistance, commensal E. coli, virulence, IS26, animal E. coli, microbial genomic epidemiology
Abbreviations
APEC, avian pathogenic Escherichia coli; ARG, antimicrobial-resistance gene; EHEC, enterohaemolytic Escherichia coli; EMAI, Elizabeth MacArthur Agricultural Institute; EPEC, enteropathogenic Escherichia coli; ETEC, enterotoxigenic Escherichia coli; ExPEC, extraintesintal pathogenic Escherichia coli; IPEC, intestinal pathogenic Escherichia coli; IS, insertion sequence; MDR, multidrug resistant; MLST, multilocus sequence typing; VAG, virulence-associated gene.
Data Summary
One hundred and forty-three whole-genome sequences of porcine faecal Escherichia coli sequenced in this project have been deposited at the European Molecular Biology Laboratory (EMBL) European Nucleotide Archive under study accession number PRJEB21464 [https://www.ebi.ac.uk/ena/data/view/PRJEB21464]. For individual sample accession numbers, please refer to Table S1 (available in the online version of this article). Further strain data is available in Tables S2–S6.
Impact Statement
The data presented in this manuscript describes for the first time, to our knowledge, a genomic analysis of commensal Escherichia coli from commercial swine-production facilities in Australia. Pig production routinely involves antibiotic use for disease treatment and prophylaxis, and feed additives containing zinc and heavy metals to control infectious disease. The study is significant because it reports phylogenetically diverse E. coli that are multidrug resistant (MDR; resistant to three or more classes of antimicrobials) and carry class 1 integrons altered by IS26. This initial descriptive work is important as a basis for the analysis of porcine faecal E. coli in all countries that produce swine commercially, due to the scale of global pork production and the vast quantities of faecal waste that are used as manure. The contamination of this waste with MDR bacteria, antimicrobial-resistance genes and unmetabolized antimicrobial residues is a concern. It is necessary to characterize food-chain-associated micro-organisms, such as E. coli, with zoonotic potential and multiple resistance genes, as they may pose a threat to public health.
Introduction
Escherichia coli is the most frequently isolated Gram-negative pathogen affecting human health [1]. Isolates are frequently resistant to multiple antibiotics and modelling studies forecast that multidrug resistant (MDR; resistant to three or more classes of antimicrobials) E. coli infections will account for 30 % of 10 million fatal MDR infections annually by 2050 [2]. In addition to the pathogenic variants, commensal E. coli comprise an important component of the gut microbiota. E. coli are shed into the environment in high numbers. For example, each gram of faeces from commercially reared pigs contains between 104 and 108 E. coli [3]. It is important to understand the characteristics of these E. coli given the huge quantities of faeces generated and disseminated by intensive pig production. China, the world’s largest producer of swine, produces an estimated 0.618 billion to 1.29 billion metric tonnes of swine faeces each year [4, 5].
Pathogenic E. coli are broadly divided into intestinal pathogenic E. coli (IPEC) and extraintestinal pathogenic E. coli (ExPEC). ExPEC have a faecal origin, having persisted asymptomatically in the gut before opportunistically colonizing extraintestinal sites where they cause a diverse range of diseases, including urinary-tract infections (UTI), pyelonephritis, wound infections, sepsis and meningitis [6]. ExPEC are thought to have foodborne reservoirs and may enter the food chain via a number of sources [7–11]. The zoonotic potential of commensal porcine E. coli as a source of ExPEC that cause disease in humans is unknown. ExPEC cannot be reliably detected in a diagnostic test as they are yet to be shown to possess unique identifying features relative to other pathotypes of E. coli [12]. Instead, as we aim to do here, whole-genome sequencing can be used to discriminate strains indistinguishable by other methods, and identify any genetic relationships between E. coli strains isolated from pigs and humans.
Horizontal gene transfer, mediated by mobile genetic elements, plays an important role in the evolution of E. coli. Commensal or pathogenic bacteria may, in a single horizontal gene transfer event, acquire a mobile genetic element carrying multiple antimicrobial-resistance genes (ARGs), virulence-associated genes (VAGs) and other genetic cargo that encode traits that offer a niche advantage [13–17]. The release of MDR commensal E. coli into the environment, such as when pig faeces are used as manure, facilitates horizontal transfer of resistance and virulence genes into other microbial communities in a manner that is poorly understood. ARGs cluster on mobile genetic elements and form complex resistance regions that are often independently mobile. Indirect selection pressure can, in the absence of antibiotic use, lead to the persistence of transferred genes. For example, heavy metals such as copper and zinc in feed formulations for food animals select for ARGs that co-localize with metal-resistance genes [18, 19]. Selection pressure afforded by any one of a number of antibiotics and heavy metals (zinc, cadmium, mercury) that contaminate faecal waste or those used in food-producing and hospital environments is sufficient to select for the retention and spread of complex resistance regions [20, 21]. Understanding of how ARGs assemble on mobile genetic elements, and the extent to which these then traffic through human, food animal and environmental reservoirs, remains limited.
Class 1 integrons are a reliable proxy for the presence of multiple ARGs within bacteria in clinical and veterinary settings [22]. They are gene capture and expression elements that can integrate AMR gene cassettes from the environmental resistome and express them via a promoter residing in the class 1 integrase gene. They are often mobilized by mercury-resistance transposons belonging to the Tn21 family, which have been disseminated globally on a wide variety of conjugative plasmid backbones [23]. Resistance genes can also be acquired, lost and rearranged in bacteria by genetic events that involve insertion sequences (ISs) such as IS26, ISEcp1and ISCR1 [24–27]. IS26 is prominent in this regard due to its unique mechanisms of transposition (conservative and replicative), ability to recognize itself, lack of copy number control and ability to mobilize a wide range of ARGs [15, 26, 28–30]. Furthermore, IS26 is recognized to play a key role in: (i) the evolution of plasmids and genomic islands that carry combinations of VAGs and ARGs [16, 17, 31, 32]; (ii) driving the formation of cointegrate plasmids encoding VAGs and ARGs [33]; and (iii) initiating deletions in large multidrug-resistance plasmids that enhance plasmid stability and expand host range [34].
Infectious-disease management relies on the surveillance of antimicrobial resistance and emerging pathogens using a One Health approach. There is currently no published data available that records whole-genome-sequence-based phylogeny, or ARG or VAG carriage in commensal E. coli from Australian pigs, and only one comparable study is available from overseas [35]. Here, for the first time, to our knowledge, we present whole-genome sequence analysis of 103 class 1 integron-positive commensal E. coli from pigs commercially reared in Australia. We present data characterizing their phylogenetic diversity, carriage of VAGs and ARGs, and an analysis of the class 1 integrons they carry.
Methods
Management of farms and animals
The study was conducted using E. coli sourced separately from two pig-production farm systems located approximately 250 km apart. Farms were designated descriptors F1 and F2. Isolate numbers consist of farm number, a pig number and a letter designating a single isolate from that pig (i.e. F1_404D indicates farm 1, isolate D from pig 404). At both farms, pigs were intensively housed and kept in total confinement. Both farms have used neomycin in the past for the treatment of diarrhoeal disease. No antibiotics were being used during the first sampling time at F1; however, the pigs sampled at the second sampling time had received a course of neomycin (see below). No antibiotics had been administered to the pigs at F2 prior to sampling.
E. coli strains used in the study
E. coli isolates were collected via rectal-swab sampling of pigs between 19 and 30 days of age. At farm 1, rectal swabs were collected in May 2007 from pigs during an outbreak of diarrhoeal disease, but prior to treatment with neomycin. These pigs were subsequently removed from the shed. The causative agent of the outbreak was unknown. A new batch of healthy sows and their piglets were transferred to this shed and the sows were given neomycin in-feed. Once the piglets were weaned they also received neomycin in-feed for 7–10 days. The second sampling occurred on these piglets in June 2007 after the course of antibiotics. At farm 2, rectal swabs were performed on healthy weaners that were not treated with antibiotics.
E. coli were isolated at the Elizabeth MacArthur Agricultural Institute (EMAI), Australia. Up to ten E. coli colonies were selected from individual pigs using MacConkey agar. The total collection from farm 1 was 164 isolates from 33 pigs, whilst from farm 2 was 171 isolates from 23 pigs. All strains were screened by PCR for the class 1 integrase gene intI1. This screening indicated that 117/164 (71 %) E. coli from farm 1 and 168/171 (98 %) from farm 2 carried intI1. Initially, 50 intI1-positive isolates from F1 and 100 intI1-positive isolates from F2 were selected for whole-genome sequencing. Two enterotoxigenic E. coli (ETEC) strains, M10 and ETEC286_3, which were submitted to the EMAI from Australian veterinary services, as clinical, pig-derived strains, were also sequenced and included in the phylogenetic analysis as reference strains.
Storage
All strains were freshly cultured in LB medium and frozen as glycerol stocks made using 500 µl M9 salts solution and 500 µl 50 % (v/v) glycerol and stored at −80 °C. All strains were cultured in LB medium prior to isolation of total cellular DNA used for sequencing.
DNA extraction, whole-genome sequencing and assembly
Total DNA was extracted using the ISOLATE II genomic DNA kit (Bioline) following the manufacturers standard protocol for bacterial cells and stored at −20 °C. Whole-genome sequencing libraries were prepared from separate aliquots of sample DNA using the Illumina Nextera DNA kit with modifications. In brief, the DNA was first quantified using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific). All sample DNA concentrations were standardized to equal concentration to achieve uniform reaction efficiency in the tagmentation step. Standard Illumina Nextera adaptors were used for sample tagmentation. The PCR-mediated adapter addition and library amplification was carried out using customized indexed i5 and i7 adaptor primers (IDT), which were developed based on the standard Nextera XT indexed i5 and i7 adapters (e.g. N701–N729 and S502–S522). Libraries were then pooled and size selected using SPRI-Select magnetic beads (Beckman Coulter). Finally, the pooled library was quality checked and quantified on an Agilent Bioanalyzer 2100 using the DNA HS kit (Agilent). Whole-genome sequencing for the majority of F1 strains and ETEC strains was performed as previously reported [36], using an Illumina MiSeq sequencer and MiSeq V3 chemistry. Whole-genome sequencing of the remaining F1 and F2 strains was performed using an Illumina HiSeq 2500 v4 sequencer in rapid PE150 mode. Sequence read quality was initially assessed using FastQC version 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Illumina raw reads passing quality control were assembled into draft genome sequences using the A5 assembly pipeline, version A5-miseq 20140604 [37]. Genome sequences have been deposited in the European Molecular Biology Laboratory European Nucleotide Archive with study accession number PRJEB21464. Accession numbers for each sample are listed in Table S1.
Strain selection
Sequence data was successfully generated for 141 strains and these were screened by blast for intI1, ARGs and VAGs, and subjected to Phylosift analysis as described below. These analyses indicated 12 strains were negative for intI1 and that a number of clones were isolated from individual pigs. We, therefore, excluded intI1-negative strains and selected representatives of the clonal isolates, thereby excluding a further 26 strains. The subset of strains that were sequenced were identified as F1+F2 (n=103 from 42 pigs). This subset consisted of 35 strains from 21 pigs sampled at farm 1 and 68 strains from 21 pigs sampled at farm 2; among the F1 strains, 17 were disease-associated strains from 12 pigs (isolate numbers 1–30, designated ‘disease’ in Tables S1–S5) and 18 were isolated from 11 healthy pigs (isolate numbers 365–409, designated ‘healthy’ in Tables S1–S5). Only 11 isolates in the collection carried toxin genes (eltA, n=2; eltB, n=2; stA, n=0; stB, n=11) associated with porcine ETEC and no ETEC adhesins were detected. Notably, only five of these were from diseased pigs, whilst six were from healthy pigs. This highlights the role that host factors, such as stress and immune health, play in the manifestation of pre- and post-weaning diarrhoea in pigs and we, therefore, argue that this collection should be considered commensal.
Assembly statistics
Comprehensive assembly statistics for 143 sequenced porcine-derived E. coli, (141+2 ETEC) are available in Table S1. Isolates not included in this study are highlighted grey. The number of scaffolds per genome ranged from 29 to 1571, with a mean of 235. Each genome sequence had a median sequencing coverage of at least 20 ×, with a maximum of 94× and mean of 54×.
Phenotypic resistance testing
F1 strains were tested at the EMAI using the calibrated dichotomous susceptibility (CDS) test for resistance to 12 antibiotics [38]. The following were tested: ampicillin (25 µg), cefoxitin (30 µg), nalidixic acid (30 µg), ciprofloxacin (2.5 µg), imipenem (10 µg), sulphafurazole (300 µg), trimethoprim (5 µg), tetracycline (10 µg), neomycin (30 µg), gentamicin (10 µg), azithromycin (15 µg) and chloramphenicol (30 µg). F2 strains were tested for resistance to antibiotics at the i3 Institute, University of Technology Sydney, Australia, using the same method and panel of antibiotics as the F1 collection. F2 strains were also tested with streptomycin (25 µg) and kanamycin (50 µg) (Table S2).
Gene identification and serotyping
Resistance, virulence and plasmid-associated genes were identified using local blastn v2.2.30+ [39] searches with an E value of 1.0×10−3 (Tables S3–S5). The gene databases used were ResFinder, PlasmidFinder, ISFinder, SerotypeFinder and VirulenceFinder [Data references 1–5] [40–44]. Our virulence database was supplemented with additional virulence genes from GenBank, available in Table S6. Genes were considered present if the subject nucleotide sequence was >90 % identical over 100 % of the length of the query sequence. blast hits with >90 % identity but covering less than 100 % of the query were considered positive if they were truncated by a scaffold break or insertion. Integrons were characterized in SnapGene (GSL Biotech) using blastn output. The collection was then retroactively screened for characterized integrons using blastn. Where strains carry two intI1 genes, de novo assembly software is unable to assemble the two complete integrons with Illumina short read data, as it cannot determine which cassette array belongs to which intI1 copy. The presence of two integrons in strains in this collection was, therefore, initially inferred by blast identification of their cassette arrays and downstream regions (e.g. IS26 deletion signatures), and then confirmed by read-mapping using Bowtie2 and Tablet [45, 46].
Phylogrouping and multilocus sequence typing (MLST)
E. coli phylogroups were determined using the scheme published by Clermont et al. [47]. The genes chuA (GenBank accession no. U67920.1), yjaA (GenBank accession no. NC_000913.3) and the DNA fragment TspE4.C2 (GenBank accession no. AF222188.1) were sourced from GenBank and identified in silico using blastn. MLST was performed in silico using the PubMLST database (http://pubmlst.org/) and the Achtman E. coli MLST scheme (http://mlst.warwick.ac.uk/mlst/).
Phylogenetic analyses
Maximum-likelihood phylogenetic distances between genomes were analysed using the PhyloSift pipeline [48], and a tree was generated using FastTree2 [49]. The tree was visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and iTOL (https://itol.embl.de/). The FastTree2 protocol was modified to resolve short branches, as described previously [50].
Results
Our study collection consisted of 103/335 (31 %) strains of E. coli isolated from rectal swabs of pigs from two farms in New South Wales, Australia, that were PCR-positive for the class 1 integron integrase gene, intI1. Initial screening indicated that 117/164 (71 %) E. coli from farm 1 and 168/171 (98 %) from farm 2 carried intI1.
Population structure of E. coli isolated from porcine rectal swabs
Strains in our study collection were classified by phylogrouping, in silico MLST and in silico serotyping. The majority of the strains in our study collection 74/103 (72 %) belonged to phylogroup A, while the remainder belonged to phylogroup B1 (18; 17 %), phylogroup B2 (5; 5 %) and phylogroup D (6; 6 %).
We identified 37 distinct sequence types, 21 of which were previously isolated from swine, as reported by the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli; accessed June 2017). Only seven sequence types were common to both F1 and F2. The most prominent sequence types were ST10, ST361, ST641, ST542, ST48 and ST218. Twenty-five sequence types were represented by a single isolate. Six strains with a single SNP in a reference allele were assigned putative sequence types (Fig. 1, Table S3; denoted by an asterisk). A designation of non-typable was given to the five remaining strains for which one or more alleles could not be determined.
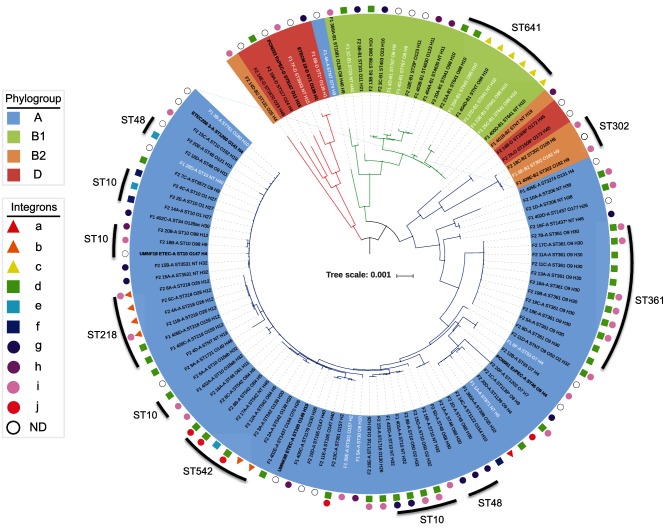
Fig. 1.
A mid-point rooted, maximum-likelihood phylogenetic tree inferred using PhyloSift v1.0.1, FastTree2, FigTree v1.4.2 and iTOL. The tree contains all 103 pig E. coli isolates sequenced in this study, 2 porcine ETEC strains and 4 reference pig-sourced sequences. The labels of strains isolated from pigs with diarrhoea are in white, and of ETEC and reference strains are in bold. Branches are coloured by clade (clade 1, red; clade 2, green; clade 3, blue). Shading over tip labels indicates phylogroup (A, blue; B1, green; B2, orange; D, red). Tip labels also contain multilocus sequence type and serotype. Asterisks indicate single-locus variants of a given sequence type. The tree scale shows the distance for 1 amino acid substitution per 1000 sites in the analysis. Clusters of the seven most common sequence types have been marked with an outer line. Integrons shown in Fig. 3 are annotated by shapes indicating the presence of sul1 (triangles), IS26-truncated 3′-CS (squares) and sul3 (circles). Integrons (a–j) are coloured red, orange, yellow, green, aqua, blue, purple, magenta, pink and crimson. Strains that were intI1 positive, but were not characterized are annotated with a white circle. Integrons were not determined for reference genomes used in the analysis.
In silico O:H typing using SerotypeFinder predicted 47 serotypes for 85 strains. The remaining 18 strains were O-non-typable with 10 different H types (Fig. 1, Table S3). In general, strains of any given sequence type carried the same O:H alleles, though intra-sequence type variability was observed among eight sequence types (ST10, ST48, ST218, ST542, ST641, ST302, ST4630 and ST1437).
Phylogenetic analysis
To determine genetic relatedness, we used PhyloSift, FastTree2, FigTree v1.4.2, and iTOL to generate and visualize a mid-point rooted, maximum-likelihood phylogenetic tree containing the F1+F2 pig E. coli draft whole-genome sequences, two ETEC strains (ETEC286_3 and ETECM_10) and four pig-pathogenic E. coli complete genome sequences [E. coli UMNK88 (NC_017641.1), UMNF18 (NZ_AGT D01000001.1), PCN033 (NZ_CP006632.1) and PCN061 (NZ_CP006636.1)] (Fig. 1). Tree topology was highly congruent with Achtman MLST and in silico serotyping, grouping strains by sequence type, and then further by serotype. Clade structure was generally congruent with phylogroup analyses; however, seven strains belonging to phylogroups B2 and D formed a separate clade. We identified three major clades, with the seven B2/D phylogroup strains forming clade 1. Clade 2 consisted almost exclusively of phylogroup B1 strains, ST641 was the dominant sequence type; however, one phylogroup A strain (F1_4A) was an unexpected member of this clade. Clade 3 was composed of two separate sub-clades, one consisted of six B2 and D strains (three ST302, two ST1508 and a non-typable) and the other exclusively containing phylogroup A strains (ST10 and sequence types within CC10, as well as ST361 and ST542, strains that were common in our study collection).
ARGs and heavy-metal-resistance genes
We identified a total of 17 ARGs in the collection and strains carried between 1 and 15 ARGs each. A total of 100/103 (97 %) strains carried three or more resistance genes. Surprisingly, strains belonging to phylogroup A carried the highest mean number of ARGs (10 per strain). Strains belonging to phylogroup B1, B2 and D each carried a mean of eight ARGs per strain. The most common ARGs among the strains in our collection were: the penicillin-resistance gene blaTEM-1, (84; 82 %); aphA1, encoding resistance to kanamycin and neomycin (76; 74 %); the co-linked streptomycin-resistance genes, strA and strB (73; 71 %); and the tetracycline-resistance gene tetA (73; 71 %). Quinolone-resistance genes oqxAB, which typically localize on plasmids, were less frequently identified (27; 26 %). Genes encoding extended-spectrum β-lactamases, extended-spectrum carbapenemases and resistance to macrolides were not detected. Heavy-metal-resistance genes, including the copper-resistance gene cusA (103; 100 %), the Tn21 mercury-resistance gene merA (71; 69 %) and the tellurite-resistance gene terA (40; 39 %), were identified frequently (Fig. 2).
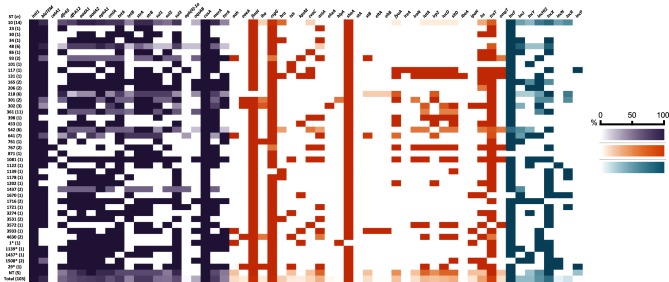
Fig. 2.
Heat map depicting carriage of ARGs (aqua), VAGs (orange) and plasmid incompatibility groups (purple) by sequence type. A darker colour indicates high carriage amongst a given sequence type, a lighter colour indicates lower carriage and white indicates no carriage. For full screening data see Tables S3–S5.
Five ARGs were identified as gene cassettes carried by class 1 integrons (Fig. 3). Cassettes carried by the majority of strains included those conferring: aminoglycoside resistance, aadA1 (69; 67 %) and aadA2 (72; 70 %); chloramphenicol resistance, cmlA (60; 58 %); and trimethoprim resistance, dfrA12 (62; 60 %) and dfrA5 (51; 50 %). Among sulphonamide-resistance genes, sul3 was identified in more strains (62; 60 %) than sul1 (48; 47 %) or sul2 (46; 45 %) (Fig. 2, Table S3). sul1 and sul3 were associated with integrons (Fig. 3).
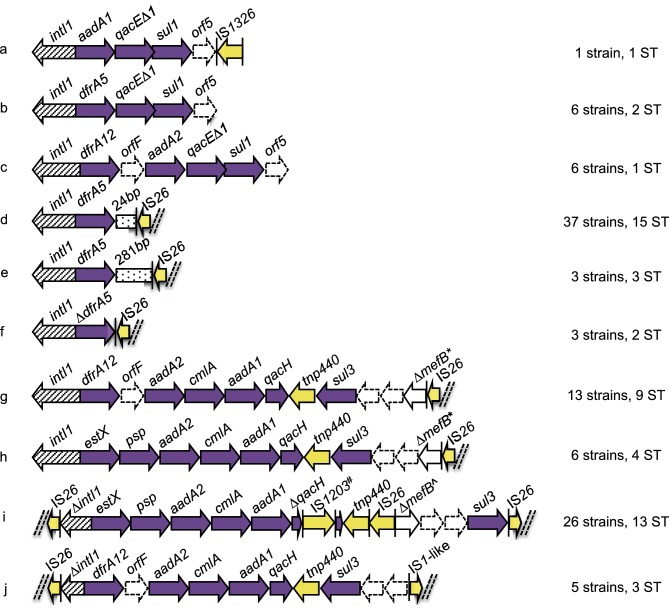
Fig. 3.
Schematic diagram (not to scale) of integrons within porcine strains that were sequenced. Arrows represent ORFs. Arrows with broken lines indicate hypothetical proteins. Vertical bars represent inverted repeats. Dashed double diagonal lines represent sequence scaffold breaks. Intergenic sequences are not shown. ARGs (purple) and IS/transposable elements (yellow) are colour coded. *, 260 bp of mefB remaining; ^, 111 bp of mefB remaining; #, IS1203-like.
Structurally diverse class 1 integrons
Among our study collection, we sought to characterize the diversity of class 1 integrons present. It is challenging to assemble complete sequences for such regions using Illumina sequence data, because of the presence of repeated elements. However, we identified numerous structurally diverse class 1 integrons, hereafter referred to as integrons (a–j) (Fig. 1 and 3). Notably IS26 altered the 3′ region in six of the most common structures (d–i).
Four different class 1 integrons (g–j) carried a sul3 gene. The first time sul3 was linked with E. coli from a food-animal source in Australia was in 2015 in a highly virulent porcine ST4245 ExPEC strain [50]. Moreover, sul3 was first reported in a human in Australia in 2017 in a commensal E. coli ST95 [51]. In integrons (g) and (h), the sul3 module, which comprises a putative transposase tnp440, sul3, two hypothetical proteins (orfA and orfB) and 260 bp of the macrolide efflux gene mefB truncated by IS26, was the same. Integrons (g) and (h) differed from each other in their respective cassette arrays. Integron (i) differed from (g) and (h) both in its sul3 module, which carried an additional copy of IS26, length of the mefB gene fragment (111 bp) and an insertion of an IS1203-like element in qacH. In integron (j), an IS26 insertion leaves only 197 bp of intI1 remaining, mefB is absent and an IS1-like element is adjacent to orfB. Only three of the integrons (a–c) among our strain collection carried a sul1 gene. Screening indicated that at least 22 strains carry two integron structures. The most common co-carriage pattern was (d, i) (14/22), though (b, i) (2/22), (d, j) (4/22) and (d, g) (2/22) also occurred (Fig. 1, Table S3). Eight sequence types carried more than one integron, including predominant types ST10, ST361 and ST542 (Table S3).
VAGs
To assess the virulence potential of commensal pig E. coli strains in our collection, we screened for a total of 94 genes that have been associated with either intestinal disease or extraintestinal disease caused by E. coli pathotypes. Twenty-nine of these genes were present in at least one strain (Fig. 2, Table S4). All strains possessed between 3 and 16 VAGs. The mean number of VAGs for each phylogroup was: A, 5; B1, 9; B2, 11; D, 9. The VAGs were present in diverse gene combinations between and within sequence types. Most VAGs were typical of extraintestinal E. coli pathotypes (ExPEC), whilst ETEC toxin gene (eltA, eltB, stA, stB) carriage was only observed in 11 strains and no ETEC adhesins were present.
Plasmid incompatibility groups
We screened the collection for plasmid replication-associated genes from nine plasmid incompatibility groups that are commonly associated with carriage and mobility of ARGs. IncF was the most common replicon (89; 86 %), followed by IncX (61; 59 %) and IncHI2 (43; 42 %). All replicons were present across multiple sequence types (Fig. 2, Table S5).
Discussion
Globally, there is a poor representation of genomic sequences for commensal E. coli isolated from the faeces of pigs, and none in Australia. Here, for the first time, to our knowledge, we have sequenced the genomes of E. coli isolated from the faeces of predominantly healthy pigs and determined their Clermont phylogroup, multilocus sequence type (Achtman) and serotype, as well as carriage of ARGs and VAGs. The phylogenetic relationships shared by the 103 strains, the types of resistance genes that reside within the class 1 integrons and the structures of class 1 integrons were also investigated. Despite sampling only two commercial piggeries, we identified a wide variety of multilocus sequence types. The diversity of isolates differed to previous studies on E. coli in pigs [35, 52] and this may be due to our selection of intI1-positive strains or simply reflect geographical differences. Our findings suggest that commensal E. coli populations residing within the faeces of pigs are often resistant to multiple antimicrobial agents and carry numerous VAGs. Notably, we also identified genetic epidemiological markers for tracking antimicrobial-resistance loci residing on mobile genetic elements in commensal E. coli.
Commensal E. coli lineages are associated with disease
The dominant lineages in our collection were phylogroup A E. coli belonging to sequence types residing within CC10, particularly ST10, ST48 and ST218. ST10 has previously been reported as the dominant sequence type from pigs in Germany, Denmark, Ireland and Spain [3, 35, 52–54]. Our data and the observations of others suggest E. coli of CC10 sequence type may be opportunistic, MDR pathogens with a broad animal host range. E. coli CC10 can colonize humans, swine, poultry, dogs, migratory birds, rodents, camels and cattle [9, 12, 55–61]. E. coli CC10 can also be isolated from raw and treated wastewater, and from urban streams [8]. E. coli CC10 is increasingly associated with intestinal disease in humans [62, 63], and extraintestinal infections in pigs [64, 65], dogs [57] and humans, including UTI, pyelonephritis and sepsis [9, 66–68]. E. coli CC10 are often MDR, and the resistance genes they carry can encode resistance to extended-spectrum β-lactams [69, 70]. ST10 is a noted ExPEC sequence type in humans and has been identified in food animals, retail meats and the environment [58, 71–74]. The core attributes of ST10 that enable it to colonize diverse niches remain unknown. The phylogenetic diversity we observed within porcine faecal ST10 suggests that such attributes may vary between strains. Whole-genome sequence analysis of E. coli ST10 genomes from different regions of the world and from different hosts is needed to understand the full diversity and success of this sequence type.
MDR porcine E. coli carry structurally diverse class 1 integrons
Notably, sul3 was the most frequently identified sul gene in our collection and three different sul3-containing integron structures were identified. Carriage of class 1 integrons possessing sul3 has been observed in disease-associated and commensal E. coli isolates from animals and humans, as well as in bacterial species other than E. coli from different countries [75–77]. In Australia, the carriage of sul3 by E. coli has been reported infrequently, although it has been identified in several uropathogenic E. coli isolates [78], in a highly virulent porcine ST4245 ExPEC strain [50], and in a human commensal ST95 E. coli on a virulence plasmid that carries multiple ARGs and VAGs [14]. In Europe, class 1 integrons containing sul3 have been observed in commensal E. coli from both humans and animals, indicating they are widely disseminated in a variety of E. coli lineages [14, 79–82]. Structures similar to ours have also been reported in different Salmonella enterica serovars, suggesting inter-species transfer of class 1 integrons carrying sul3 may have occurred [75].
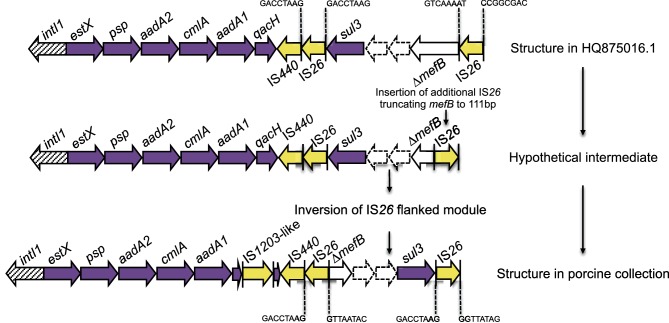
The potential role for sul3 integrons in intraspecies and interspecies exchange of antibiotic resistance makes it desirable both to understand their evolution and to track their movement through bacterial populations. In Fig. 4, we have provided a model that could explain the micro-evolutionary events that created the novel sul3 integron depicted in structure (i). This integron likely evolved from a progenitor similar to one described by Curiao et al. in a human-derived extended-spectrum β-lactamase positive E. coli on an IncI1 plasmid from Spain (GenBank accession no. HQ875016.1), as this is the only report to describe IS26 adjacent to sul3 [76]. Conceivably, the novel structure (i) emerged from insertion of a second copy of IS26, which further truncated mefB, followed by an inversion event. To our knowledge, this is the first study to identify a 111 bp mefB variant. Integron (i) was observed within the collection in 26 E. coli strains of different sequence types, suggesting horizontal transfer of a mobile element(s) carrying the integron, though we were unable to determine which mobile elements were responsible for this. Further work is needed to examine this hypothesis.
Fig. 4.
Schematic diagram (not to scale) of proposed evolutionary pathway to the sul3-∆mefB arrangement shown in Fig. 3(i). IS26 8 bp direct repeats are annotated.
IS26-mediated deletions of mefB can be used to track sul3-containing integrons and additional resistance genes they may acquire due to the unique ability of IS26 to target itself [26]. A number of different truncated variants of the mefB gene are carried by sul3 integrons found in human- and animal-derived E. coli [14, 75–77]. Our data suggests the class 1 integrase upstream of the sul3 module is likely to be functional based on the presence of different antibiotic cassette arrays associated with a 260 bp mefB deletion (g, h). blastn analysis identified sul3 integrons carrying ∆mefB with an identical 260 bp deletion in porcine isolates P328.10.99.C2 (GenBank accession no. FJ196386.1) and P528.10.99.C4 (GenBank accession no. FJ196388.1) from Great Britain, though the associated cassette arrays were not completely characterized [77]. Furthermore, plasmid pCAZ590 (GenBank accession no. LT669764.1) isolated from poultry in Germany carried an identical integron (estX-psp-aadA2-cmlA-aadA1-qacI-tnp440-sul3-orf1-orf2-∆mefB:260bp-IS26) to 4(h) with an additional blaSHV-12 gene 73 bp upstream of IS26 [83]. Although the evolutionary events that lead to this derivative structure are not known, this plasmid illustrates how IS26 augmented integrons continue to evolve and acquire genes that confer resistance to critically important human antibiotics.
The deletion event in the 3′-CS of the integron depicted in (d) (dfrA5-IS26) may serve as another genetic signature for tracking resistance genes, and bacteria that carry them, through different hosts and environments [15, 84–86]. Previously, we observed the integron structure (d) on plasmids carrying VAGs in atypical EPEC strains isolated from cattle with gastrointestinal disease and E. coli strains linked to EHEC O26:H− isolated from a human patient with haemorrhagic colitis [16, 17]. In each of these earlier cases, the IS26 that interrupted the 3′-CS of the integron formed part of the left boundary of Tn6026, an IS26-flanked, globally disseminated transposon that harbours multiple ARGs [15–17, 87, 88]. Twenty-seven strains carrying integron (d) possess the resistance genes present in Tn6026 (blaTEM, sul2, strAB, aphA1) suggesting this transposon is also carried in our collection, though further studies are necessary to confirm this. This again highlights that tracking IS26 deletions is useful for tracking not only the integrons they interrupt, but also additional resistance genes that may be acquired in association with the IS26.
The carriage of more than one integron in a number of prominent sequence types in the collection suggests that plasmid or transposon-mediated horizontal transfer of resistance determinants may occur within the microbiota of the porcine gut. This transfer is likely mediated by plasmids present in the collection, though transposons and IS elements may be involved. Long-read sequencing is required to test this hypothesis.
Zoonotic potential of commensal. E. coli from swine
In considering the zoonotic potential of pig faecal E. coli, we determined the proportion of strains in our collection that carried IPEC and ExPEC VAGs. A limitation of investigating zoonotic potential for extraintestinal disease is the genetic redundancy identified in the virulence attributes from ExPEC. A recent study suggested that the number of virulence factors carried by an ExPEC strain is the only independent factor that can explain extraintestinal virulence in a mouse model of sepsis [89]. Our collection contained two strains possessing large numbers of VAGs, belonging to ST131 and ST117, representative of pandemic ExPEC clones that cause hospital- and community-acquired infections in humans worldwide [58, 90, 91]. They have both been linked with poultry and have only rarely been isolated from porcine sources [9, 58]. The single ST131 strain in our porcine collection carried 10 ARGs and 16 VAGs. The ST117 strain carried 8 ARGs and 16 VAGs, including the full array of iron-acquisition genes fyuA, irp2, ireA, iroN, iutA, iucD and sitD. Several of these genes are typically encoded on virulence plasmids circulating in APEC [92] and this profile is similar to ST117-O111:H4 strains from poultry reported by Mora et al. [93]. The presence of ST117 and ST131 in our collection is intriguing, and warrants further investigation.
Most of the VAGs identified in our collection were those associated with the ability to cause extraintestinal disease in humans, as well as intestinal persistence [6, 94]. Carriage of genes that are under positive selection in uropathogenic E. coli [95], such as heat-stable agglutinin gene hra [96], murine uroepithelial cell adhesin gene iha [97], iron-acquisition genes fyuA, iutA, iucD and sitD, and the serum survival genes iss and traT, suggest that some strains may be capable of causing extraintestinal disease in humans. Conversely, it also highlights how many ExPEC VAGs can be considered important intestinal fitness factors. The most intriguing IPEC VAG was intimin gene eaeA, found in eight strains, that is characteristic of several intestinal E. coli pathotypes, including EHEC, EPEC and atypical EPEC [98]. These strains also carried ExPEC VAGs and may represent hybrid pathotypes.
The frequency of VAGs in phylogroups A and B1, a mean of 5 and 9 VAGs per isolate, respectively, was unexpected because E. coli belonging to phylogroups A and B1 are considered to have low virulence potential [99, 100]. The carriage of multiple VAGs in pig E. coli is consistent with earlier studies [101, 102]. In China, ExPEC have been isolated from a variety of tissues and bodily fluids of pigs with septicaemia, meningitis and respiratory disease with increasing frequency since 2004 [64, 65]. It is notable that 35 % of 81 isolates in one of these studies belonged to phylogroup A, clonal complex 10 [64]. In European wild boars, which are assumed to be ancestors of domestic pigs in Europe [103], E. coli strains carry, on average, 7 or more VAGs, with some strains carrying up to 16 VAGs [104]. Collectively, these observations suggest that E. coli phylogroup A and B1, at least those sourced from swine, carry multiple VAGs.
Contribution of food-production animals to the evolution of pathogens and antimicrobial resistance
MDR E. coli carrying ARGs associated with mobile genetic elements and VAGs are released into the environment by food-production animals via faecal effluent. In Australia, the capacity for pig production to contribute to the evolution and dissemination of pathogens and ARGs is restricted compared to that of pig-production systems in many other countries, due to a range of factors. Firstly, Australia has a large landmass that is surrounded by ocean, preventing the movement of animals from neighbouring countries. Secondly, importation of food animals into Australia has been restricted since the 1970s [105]. Thirdly, antibiotics such as fluoroquinolones cannot legally be administered to food animals and many others are restricted from use in food-animal production [106, 107]. However, even in the restricted environment in Australia, phenotypic resistance to clinically important antibiotics, including extended-spectrum cephalosporins and fluoroquinolones, has been observed in E. coli that belong to globally disseminated E. coli lineages ST744, ST100 and ST1 [108]. Globally, genomic surveillance is needed to understand the relative contribution of food-production animals to the complex web of interactions between microbiota and the mobile resistome, to provide baseline carriage rates for antimicrobial genes and VAGs, and to monitor the emergence of novel drug-resistant pathogens [84, 109].
In summary, we report, to our knowledge, the first genomic study of commensal E. coli isolated from commercial pigs used for food consumption and provide data to inform assessment of potential risks pig commensal E. coli may pose to human health. Our results show that swine are a reservoir: (i) for phylogroup A and B1 E. coli that carry VAGs, (ii) the sul3 gene, (iii) class 1 integrons associated with IS26, and (iv) E. coli lineages belonging to CC10. Our study has identified several new genetic signatures that may be used in tracking mobile ARGs.
Data bibliography
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67, 2640–2644 (2012).
Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34, D32–D36 (2006).
Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53, 2410–2426 (2015).
Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52, 1501–1510 (2014).
Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58, 3895–3903 (2014).
Funding information
This work was supported by the Australian Research Council, linkage grant LP150100912. This project was partly funded by the Australian Centre for Genomic Epidemiological Microbiology (Ausgem), a collaborative partnership between the NSW Department of Primary Industries and the University of Technology Sydney. C.J.R. and E.R.W. are recipients of Australian Government Research Training Program Scholarships.
Acknowledgements
We thank Fiona MacIver for assistance with preparing the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
References
- 1.Poolman JT, Wacker M. Extraintestinal pathogenic Escherichia coli, a common human pathogen: challenges for vaccine development and progress in the field. J Infect Dis. 2016;213:6–13. doi: 10.1093/infdis/jiv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London: Wellcome Trust, UK Government; 2016. [Google Scholar]
- 3.Herrero-Fresno A, Larsen I, Olsen JE. Genetic relatedness of commensal Escherichia coli from nursery pigs in intensive pig production in Denmark and molecular characterization of genetically different strains. J Appl Microbiol. 2015;119:342–353. doi: 10.1111/jam.12840. [DOI] [PubMed] [Google Scholar]
- 4.Han S. Environmental Impacts of China's Pork Industry. Washington, DC:: Wilson Center; 2014. [Google Scholar]
- 5.Wang F-H, Ma W-Q, Dou Z-X, Ma L, Liu X-L, et al. The estimation of the production amount of animal manure and its environmental effect in China. China Environmental Science. 2006;26:614–617. [Google Scholar]
- 6.Johnson JR, Russo TA. Molecular epidemiology of extraintestinal pathogenic (uropathogenic) Escherichia coli. Int J Med Microbiol. 2005;295:383–404. doi: 10.1016/j.ijmm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JR, Kuskowski MA, Smith K, O'Bryan TT, Tatini S. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis. 2005;191:1040–1049. doi: 10.1086/428451. [DOI] [PubMed] [Google Scholar]
- 8.Varela AR, Manageiro V, Ferreira E, Guimarães MA, da Costa PM, et al. Molecular evidence of the close relatedness of clinical, gull and wastewater isolates of quinolone-resistant Escherichia coli. J Glob Antimicrob Resist. 2015;3:286–289. doi: 10.1016/j.jgar.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Manges AR. Escherichia coli and urinary tract infections: the role of poultry-meat. Clin Microbiol Infect. 2016;22:122–129. doi: 10.1016/j.cmi.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Nordstrom L, Liu CM, Price LB. Foodborne urinary tract infections: a new paradigm for antimicrobial-resistant foodborne illness. Front Microbiol. 2013;4:29. doi: 10.3389/fmicb.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis. 2010;16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordoni G, Woodward MJ, Wu H, Alanazi M, Wallis T, et al. Comparative genomics of European avian pathogenic E. coli (APEC) BMC Genomics. 2016;17:960. doi: 10.1186/s12864-016-3289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levings RS, Djordjevic SP, Hall RM. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob Agents Chemother. 2008;52:2529–2537. doi: 10.1128/AAC.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran RA, Holt KE, Hall RM. pCERC3 from a commensal ST95 Escherichia coli: a ColV virulence-multiresistance plasmid carrying a sul3-associated class 1 integron. Plasmid. 2016;84-85:11–19. doi: 10.1016/j.plasmid.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury PR, Charles IG, Djordjevic SP. A role for Tn6029 in the evolution of the complex antibiotic resistance gene loci in genomic island 3 in enteroaggregative hemorrhagic Escherichia coli O104:H4. PLoS One. 2015;10:e0115781. doi: 10.1371/journal.pone.0115781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 2010;24:1160–1166. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]
- 17.Venturini C, Hassan KA, Chowdhury PR, Paulsen IT, Walker MJ, et al. Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS One. 2013;8:e78862. doi: 10.1371/journal.pone.0078862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Li LG, Xia Y, Zhang T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. Isme J. 2017;11:651–662. doi: 10.1038/ismej.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben W, Wang J, Pan X, Qiang Z. Dissemination of antibiotic resistance genes and their potential removal by on-farm treatment processes in nine swine feedlots in Shandong Province, China. Chemosphere. 2017;167:262–268. doi: 10.1016/j.chemosphere.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Qiao M, Wang FH, Zhu YG. Use of commercial organic fertilizer increases the abundance of antibiotic resistance genes and antibiotics in soil. Environ Sci Pollut Res Int. 2017;24:701–710. doi: 10.1007/s11356-016-7854-z. [DOI] [PubMed] [Google Scholar]
- 22.Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. Isme J. 2015;9:1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng C, Sun J, Zheng F, Lu W, Yang Q, et al. New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol. 2016;16:71. doi: 10.1186/s12866-016-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmer CJ, Hall RM. IS26-Mediated formation of transposons carrying antibiotic resistance genes. mSphere. 2016;1:e00038-16. doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harmer CJ, Moran RA, Hall RM. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio. 2014;5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cantón R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 29.Dionisi AM, Lucarelli C, Owczarek S, Luzzi I, Villa L. Characterization of the plasmid-borne quinolone resistance gene qnrB19 in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2009;53:4019–4021. doi: 10.1128/AAC.00294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toleman MA, Walsh TR. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 31.Doublet B, Praud K, Weill FX, Cloeckaert A. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J Antimicrob Chemother. 2009;63:282–289. doi: 10.1093/jac/dkn500. [DOI] [PubMed] [Google Scholar]
- 32.Siebor E, Neuwirth C. Emergence of Salmonella genomic island 1 (SGI1) among Proteus mirabilis clinical isolates in Dijon, France. J Antimicrob Chemother. 2013;68:1750–1756. doi: 10.1093/jac/dkt100. [DOI] [PubMed] [Google Scholar]
- 33.Mangat CS, Bekal S, Irwin RJ, Mulvey MR. A novel hybrid plasmid carrying multiple antimicrobial resistance and virulence genes in Salmonella enterica serovar Dublin. Antimicrob Agents Chemother. 2017;61:e02601-16. doi: 10.1128/AAC.02601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porse A, Schønning K, Munck C, Sommer MO. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol. 2016;33:2860–2873. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed S, Olsen JE, Herrero-Fresno A. The genetic diversity of commensal Escherichia coli strains isolated from non-antimicrobial treated pigs varies according to age group. PLoS One. 2017;12:e0178623. doi: 10.1371/journal.pone.0178623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darling AE, Worden P, Chapman TA, Chowdhury PR, Charles IG, et al. The genome of Clostridium difficile 5.3. Gut Pathog. 2014;6:4. doi: 10.1186/1757-4749-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 38.Bell SM, Newton P, Nguyen TT. Antibiotic Susceptibility Testing by the CDS Method: a Manual for Medical and Veterinary Laboratories. Randwick, NSW, Australia: South Eastern Area Laboratory Services; 2013. p. 6. [Google Scholar]
- 39.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. 2015;53::2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, et al. Tablet-next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, et al. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyrsch E, Chowdhury PR, Abraham S, Santos J, Darling AE, et al. Comparative genomic analysis of a multiple antimicrobial resistant enterotoxigenic E. coli O157 lineage from Australian pigs. BMC Genomics. 2015;16:165. doi: 10.1186/s12864-015-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moran RA, Hall RM. Evolution of regions containing antibiotic resistance genes in FII-2-FIB-1 ColV-Colla virulence plasmids. Microb Drug Resist. 2017 [Epub ahead of print] doi: 10.1089/mdr.2017.0177. [DOI] [PubMed] [Google Scholar]
- 52.Bednorz C, Oelgeschläger K, Kinnemann B, Hartmann S, Neumann K, et al. The broader context of antibiotic resistance: zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. Int J Med Microbiol. 2013;303:396–403. doi: 10.1016/j.ijmm.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Cortés P, Blanc V, Mora A, Dahbi G, Blanco JE, et al. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl Environ Microbiol. 2010;76:2799–2805. doi: 10.1128/AEM.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Gibbons JF, McGrath K, Bai L, Li F, et al. Molecular characterization of blaESBL-producing Escherichia coli cultured from pig farms in Ireland. J Antimicrob Chemother. 2016;71:3062–3065. doi: 10.1093/jac/dkw278. [DOI] [PubMed] [Google Scholar]
- 55.Alcalá L, Alonso CA, Simón C, González-Esteban C, Orós J, et al. Wild birds, frequent carriers of extended-spectrum β-Lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb Ecol. 2016;72:861–869. doi: 10.1007/s00248-015-0718-0. [DOI] [PubMed] [Google Scholar]
- 56.Ho PL, Lo WU, Lai EL, Law PY, Leung SM, et al. Clonal diversity of CTX-M-producing, multidrug-resistant Escherichia coli from rodents. J Med Microbiol. 2015;64:185–190. doi: 10.1099/jmm.0.000001-0. [DOI] [PubMed] [Google Scholar]
- 57.Liu X, Liu H, Li Y, Hao C. High prevalence of β-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in Shaanxi, China. Front Microbiol. 2016;7:1843. doi: 10.3389/fmicb.2016.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manges AR, Harel J, Masson L, Edens TJ, Portt A, et al. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog Dis. 2015;12:302–310. doi: 10.1089/fpd.2014.1860. [DOI] [PubMed] [Google Scholar]
- 59.Rodrigues C, Machado E, Peixe L, Novais A. IncI1/ST3 and IncN/ST1 plasmids drive the spread of blaTEM-52 and blaCTX-M-1/-32 in diverse Escherichia coli clones from different piggeries. J Antimicrob Chemother. 2013;68:2245–2248. doi: 10.1093/jac/dkt187. [DOI] [PubMed] [Google Scholar]
- 60.Shabana II, Zaraket H, Suzuki H. Molecular studies on diarrhea-associated Escherichia coli isolated from humans and animals in Egypt. Vet Microbiol. 2013;167:532–539. doi: 10.1016/j.vetmic.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 61.Trobos M, Christensen H, Sunde M, Nordentoft S, Agersø Y, et al. Characterization of sulphonamide-resistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology. 2009;155:831–836. doi: 10.1099/mic.0.024190-0. [DOI] [PubMed] [Google Scholar]
- 62.Guiral E, Mendez-Arancibia E, Soto SM, Salvador P, Fabrega A, et al. CTX-M-15-producing enteroaggregative Escherichia coli as cause of travelers' diarrhea. Emerg Infect Dis. 2011;17:1950–1953. doi: 10.3201/eid1710.110022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reuland EA, Overdevest IT, Al Naiemi N, Kalpoe JS, Rijnsburger MC, et al. High prevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin Microbiol Infect. 2013;19:542–549. doi: 10.1111/j.1469-0691.2012.03947.x. [DOI] [PubMed] [Google Scholar]
- 64.Ding Y, Tang X, Lu P, Wu B, Xu Z, et al. Clonal analysis and virulent traits of pathogenic extraintestinal Escherichia coli isolates from swine in China. BMC Vet Res. 2012;8:140. doi: 10.1186/1746-6148-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan C, Tang X, Zhang X, Ding Y, Zhao Z, et al. Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China. Vet J. 2012;192:483–488. doi: 10.1016/j.tvjl.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 66.Giufrè M, Graziani C, Accogli M, Luzzi I, Busani L, et al. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother. 2012;67:860–867. doi: 10.1093/jac/dkr565. [DOI] [PubMed] [Google Scholar]
- 67.Salvador E, Wagenlehner F, Köhler CD, Mellmann A, Hacker J, et al. Comparison of asymptomatic bacteriuria Escherichia coli isolates from healthy individuals versus those from hospital patients shows that long-term bladder colonization selects for attenuated virulence phenotypes. Infect Immun. 2012;80:668–678. doi: 10.1128/IAI.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Usein CR, Papagheorghe R, Oprea M, Condei M, Strãuţ M. Molecular characterization of bacteremic Escherichia coli isolates in Romania. Folia Microbiol. 2016;61:221–226. doi: 10.1007/s12223-015-0427-6. [DOI] [PubMed] [Google Scholar]
- 69.Pires J, Kuenzli E, Kasraian S, Tinguely R, Furrer H, et al. Polyclonal intestinal colonization with extended-spectrum cephalosporin-resistant enterobacteriaceae upon traveling to India. Front Microbiol. 2016;7:1069. doi: 10.3389/fmicb.2016.01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tagg KA, Ginn AN, Partridge SR, Iredell JR. MALDI-TOF mass spectrometry for multilocus sequence typing of Escherichia coli reveals diversity among isolates carrying blaCMY-2-like genes. PLoS One. 2015;10:e0143446. doi: 10.1371/journal.pone.0143446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, et al. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J Antimicrob Chemother. 2015;70:2757–2762. doi: 10.1093/jac/dkv188. [DOI] [PubMed] [Google Scholar]
- 72.Toval F, Köhler CD, Vogel U, Wagenlehner F, Mellmann A, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol. 2014;52:407–418. doi: 10.1128/JCM.02069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 74.Moran RA, Anantham S, Pinyon JL, Hall RM. Plasmids in antibiotic susceptible and antibiotic resistant commensal Escherichia coli from healthy Australian adults. Plasmid. 2015;80:24–31. doi: 10.1016/j.plasmid.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Antunes P, Machado J, Peixe L. Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3' conserved sequence region among Salmonella isolates. Antimicrob Agents Chemother. 2007;51:1545–1548. doi: 10.1128/AAC.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Curiao T, Cantón R, Garcillán-Barcia MP, de La Cruz F, Baquero F, et al. Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-beta-lactamase-producing Escherichia coli clones from humans. Antimicrob Agents Chemother. 2011;55:2451–2457. doi: 10.1128/AAC.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J, Keelan P, Bennett PM, Enne VI. Characterization of a novel macrolide efflux gene, mef(B), found linked to sul3 in porcine Escherichia coli. J Antimicrob Chemother. 2009;63:423–426. doi: 10.1093/jac/dkn523. [DOI] [PubMed] [Google Scholar]
- 78.Gündoğdu A, Long YB, Vollmerhausen TL, Katouli M. Antimicrobial resistance and distribution of sul genes and integron-associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J Med Microbiol. 2011;60:1633–1642. doi: 10.1099/jmm.0.034140-0. [DOI] [PubMed] [Google Scholar]
- 79.Bischoff KM, White DG, Hume ME, Poole TL, Nisbet DJ. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol Lett. 2005;243:285–291. doi: 10.1016/j.femsle.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 80.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 81.Sáenz Y, Vinué L, Ruiz E, Somalo S, Martínez S, et al. Class 1 integrons lacking qacEDelta1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet Microbiol. 2010;144:493–497. doi: 10.1016/j.vetmic.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 82.Sunde M, Solheim H, Slettemeås JS. Genetic linkage between class 1 integrons with the dfrA12-orfF-aadA2 cassette array and sul3 in Escherichia coli. Vet Microbiol. 2008;130:422–425. doi: 10.1016/j.vetmic.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 83.Alonso CA, Michael GB, Li J, Somalo S, Simón C, et al. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J Antimicrob Chemother. 2017;72:1589–1596. doi: 10.1093/jac/dkx024. [DOI] [PubMed] [Google Scholar]
- 84.Djordjevic SP, Stokes HW, Chowdhury PR. Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front Microbiol. 2013;4:86. doi: 10.3389/fmicb.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dawes FE, Kuzevski A, Bettelheim KA, Hornitzky MA, Djordjevic SP, et al. Distribution of class 1 integrons with IS26-mediated deletions in their 3'-conserved segments in Escherichia coli of human and animal origin. PLoS One. 2010;5:e12754. doi: 10.1371/journal.pone.0012754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levings RS, Lightfoot D, Partridge SR, Hall RM, Djordjevic SP. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J Bacteriol. 2005;187:4401–4409. doi: 10.1128/JB.187.13.4401-4409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cain AK, Liu X, Djordjevic SP, Hall RM. Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb Drug Resist. 2010;16:197–202. doi: 10.1089/mdr.2010.0042. [DOI] [PubMed] [Google Scholar]
- 88.Reid CJ, Chowdhury PR, Djordjevic SP. Tn6026 and Tn6029 are found in complex resistance regions mobilised by diverse plasmids and chromosomal islands in multiple antibiotic resistant Enterobacteriaceae. Plasmid. 2015;80:127–137. doi: 10.1016/j.plasmid.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Bleibtreu A, Gros PA, Laouénan C, Clermont O, Le Nagard H, et al. Fitness, stress resistance, and extraintestinal virulence in Escherichia coli. Infect Immun. 2013;81:2733–2742. doi: 10.1128/IAI.01329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis. 2013;207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stoesser N, Sheppard AE, Pankhurst L, de Maio N, Moore CE, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio. 2016;7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu Ge X, Jiang J, Pan Z, Hu L, Wang S, et al. Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2:K1:H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1:K1 and human ExPEC O18:K1 strains. PLoS One. 2014;9:e112048. doi: 10.1371/journal.pone.0112048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mora A, López C, Herrera A, Viso S, Mamani R, et al. Emerging avian pathogenic Escherichia coli strains belonging to clonal groups O111:H4-D-ST2085 and O111:H4-D-ST117 with high virulence-gene content and zoonotic potential. Vet Microbiol. 2012;156:347–352. doi: 10.1016/j.vetmic.2011.10.033. [DOI] [PubMed] [Google Scholar]
- 94.Schierack P, Walk N, Ewers C, Wilking H, Steinrück H, et al. ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ Microbiol. 2008;10:1742–1751. doi: 10.1111/j.1462-2920.2008.01595.x. [DOI] [PubMed] [Google Scholar]
- 95.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Srinivasan U, Foxman B, Marrs CF. Identification of a gene encoding heat-resistant agglutinin in Escherichia coli as a putative virulence factor in urinary tract infection. J Clin Microbiol. 2003;41:285–289. doi: 10.1128/JCM.41.1.285-289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, et al. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 99.Ewers C, Li G, Wilking H, Kiessling S, Alt K, et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol. 2007;297:163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 100.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 101.Dixit SM, Gordon DM, Wu XY, Chapman T, Kailasapathy K, et al. Diversity analysis of commensal porcine Escherichia coli - associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology. 2004;150:1735–1740. doi: 10.1099/mic.0.26733-0. [DOI] [PubMed] [Google Scholar]
- 102.Schierack P, Walk N, Reiter K, Weyrauch KD, Wieler LH. Composition of intestinal Enterobacteriaceae populations of healthy domestic pigs. Microbiology. 2007;153:3830–3837. doi: 10.1099/mic.0.2007/010173-0. [DOI] [PubMed] [Google Scholar]
- 103.Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 104.Römer A, Wieler LH, Schierack P. Analyses of intestinal commensal Escherichia coli strains from wild boars suggest adaptation to conventional pig production conditions. Vet Microbiol. 2012;161:122–129. doi: 10.1016/j.vetmic.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 105.Turner A. Quarantine, exports and animal disease in Australia 1901-2010. Aust Vet J. 2011;89:366–371. doi: 10.1111/j.1751-0813.2011.00822.x. [DOI] [PubMed] [Google Scholar]
- 106.Cheng AC, Turnidge J, Collignon P, Looke D, Barton M, et al. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg Infect Dis. 2012;18:1453–1460. doi: 10.3201/eid1809.111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jordan D, Chin JJ, Fahy VA, Barton MD, Smith MG, et al. Antimicrobial use in the Australian pig industry: results of a national survey. Aust Vet J. 2009;87:222–229. doi: 10.1111/j.1751-0813.2009.00430.x. [DOI] [PubMed] [Google Scholar]
- 108.Abraham S, Jordan D, Wong HS, Johnson JR, Toleman MA, et al. First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J Glob Antimicrob Resist. 2015;3:273–277. doi: 10.1016/j.jgar.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 109.Wyrsch ER, Chowdhury PR, Chapman TA, Charles IG, Hammond JM, et al. Genomic Microbial epidemiology is needed to comprehend the global problem of antibiotic resistance and to improve pathogen diagnosis. Front Microbiol. 2016;7:843. doi: 10.3389/fmicb.2016.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.