Abstract
Background
The right ventricle (RV) is not designed to sustain high pressure leading to failure. There are no current medications to help RV contraction, so further information is required on adaption of the RV to such hypertension.
Methods
The Right Ventricle in Children (RVENCH) study assessed infants with congenital heart disease undergoing cardiac surgery with hypertensive RV. Clinical and echocardiographic data were recorded, and samples of RV were taken from matched infants, analysed for proteomics and compared between pathologies and with clinical and echocardiographic outcome data.
Results
Those with tetralogy of Fallot (TOF) were significantly more cyanosed than those with ventricular septal defect (median oxygen saturation 83% vs 98%, P=0.0038), had significantly stiffer RV (tricuspid E wave/A wave ratio 1.95 vs 0.84, P=0.009) and had most had restrictive physiology. Gene ontology in TOF, with enrichment analysis, demonstrated significant increase in proteins of contractile mechanisms and those of calmodulin, actin binding and others associated with contractility than inventricular septal defect. Structural proteins were also found to be higher in association with sarcomeric function: Z-disc, M-Band and thin-filament proteins. Remaining proteins associated with actin binding, calcium signalling and myocyte cytoskeletal development. Phosphopeptide enrichment led to higher levels of calcium signalling proteins in TOF.
Conclusion
This is the first demonstration that those with an RV, which is stiff and hypertensive in TOF, have a range of altered proteins, often in calcium signalling pathways. Information about these alterations might guide treatment options both in terms of individualised therapy or inotropic support for the Right ventricle when hypertensive due to pulmoanry hypertension or congenital heart disease.
Keywords: congenital heart disease, right ventricle, ventricular hypertrophy, proteomics
Key questions.
What is already known about this subject?
Proteomic changes in myocardium have been studied in adult patients with or without ischaemic arrest, and our laboratory has identified genetic changes in the myocardium of children undergoing cardiac surgery.
What does this study add?
To date, this is a unique study that identifies the proteomic differences in right ventricular myocardium from infants undergoing cardiac surgery for cyanotic and acyanotic heart diseases with a hypertensive right ventricle.
How might this impact on clinical practice?
It is of clinical concern how to manage such children, as to the timing of surgery, the type of inotrope that might benefit the child with each cardiac condition and individual mechanisms that might be accessed in precision medicine to determine the best therapy for each child. This study is the first step in that process to identify the mechanisms involved and the best treatment for the individual child.
Background
It is estimated that about 1% of all live-born babies are affected by cardiac congenital malformations. Intracardiac surgical repair in these patients is associated with a low mortality in infants and children (<5%). More importantly, a growing number of patients survive into adulthood with hypertensive right ventricles (RV). Heart failure due to RV dysfunction is a major cause of morbidity in this population, and a significant proportion of them will die as a result. There is recent evidence that the mortality is increasing in the teenage population who are dying of the complications of cardiac surgery performed over a decade ago, and it is our responsibility to improve this for the next generation.1 The gravity of the problem is well explained by the 2009 WHO annual report that puts the disability-adjusted life-year for congenital heart disease higher than diseases such as diabetes or hypertension.2 At present, pharmacotherapy of heart failure may improve clinical symptoms, but there is no targeted pharmacotherapy to improve RV systolic and diastolic function or to prevent or even reverse the remodelling of the RV. Significant morbidity secondary to RV dysfunction leads to greater length of time on the paediatric intensive care unit (PICU) postoperatively and more expensive repeat interventions and reoperations. Since each day on the PICU can cost up to £2000, and each reoperation costs £25000, it will be beneficial to the healthcare economy to improve the quality of individualised care we deliver to these children.
One drawback in this area is the fact that patients with RV dysfunction are grouped together despite the fact that there are different congenital cardiac disorders that will trigger different functional, molecular and cellular remodelling in the RV. It is therefore vital to dissect potential differences in cell signalling and remodelling in patients with different causes of RV dysfunction. Consequently, a greater understanding of the underlying mechanisms of RV function in different congenital cardiac disorder groups will help to formulate strategies targeting individual cardiac disorders. The National Heart, Lung, and Blood Institute (Bethesda, Maryland, USA) has been awarded top priority to research on pathophysiology of the RV. RV dysfunction is caused by changes in the cell fate and tissue composition. These changes occur due to cell signalling that may be initiated by various factors including oxidative stress and lead to alterations in cell survival, structural (cytoskeletal) and extracellular proteins, protease activity and calcium handling properties.3 From our recent studies, we have shown that chronic hypoxaemia in cyanotic children with tetralogy of Fallot (TOF) induced the expression of genes associated with apoptosis and reduced the expression of genes associated with normal myocyte contractility and function.3
It is well established that convergence of the signalling pathways activated by tumour growth factor-β, Wnts and cadherin are critical for development and its importance re-emerge in disease. It plays a key role in cell fate (growth and survival) and in matrix synthesis and remodelling, which are all pertinent processes in RV dysfunction. We already have extensive evidence for their important role in blood vessel remodelling and propose that modulation of these signalling pathways contribute to RV dysfunction. Although right ventricular failure (RVF) is the hallmark of pulmonary arterial hypertension (PAH), the mechanism of RVF is unclear. Development of PAH-induced RVF is associated with an increased reactive oxygen species (ROS) production. Increases in oxidative stress lead to generation of nitro-tyrosine residues in tissue inhibitor of metalloproteinase and liberate active matrix metalloproteinase.4
The purpose of this study is to investigate the cellular signalling changes associated with RV hypertension in the setting of congenital heart disease. This would lead to a better understanding of the determinants of RV function in patients with hypertrophy associated with different congenital cardiac malformations. It was proposed that:
Changes in key myocardial cellular signalling pathways triggered by hypertension-induced RV dysfunction in congenital cardiac disorders depend on the type of malformation and systemic oxygen saturation.
The pathology-induced cellular and molecular changes will result in differences in postoperative stay within the hospital.
Identifying differences in the cellular signalling pathways between those with large septal defects and those with TOF might enable us to tailor therapies for each pathology to improve outcome.
The clinical outcome in our patients often depends on the ability of the RV to cope with the anatomical substrate and the effect of cardiopulmonary bypass. We hoped to be able to understand better the effect of these and hence to modify our treatment accordingly. This might lead to better and perhaps individualised care for our patients undergoing cardiopulmonary bypass for congenital heart diseases.
Patients and methods
Inclusion criteria
We compared patients with RV hypertension who underwent cardiopulmonary bypass and consented for inclusion between 1 July 2015 and 31 March 2017. We report on two groups of children with right ventricular pressures at systemic level: group 1: pulmonary hypertension due to large septal defects, such as ventricular septal defect (VSD) and group 2: obstructed RV (as in TOF). In group 1, left to right shunt was the cause of RV hypertension. In group 2, the children were cyanosed, and the RV was functioning under a significant obstructive afterload.
Exclusion criteria
Children with additional syndromes were excluded (apart from Trisomy 21) due to the unknown effect on ventricular function and protein analysis. Parents who were not able to consider the information at least 1 week before operation were excluded, hence excluding any children undergoing emergency surgery. In addition, we excluded those with sepsis or coexisting morbidity, which might alter the protein analysis.
Fully informed consent was obtained from parents prior to admission for operation. Clinical data were obtained from patient’s clinical records and Heartsuite Database at the time of cardiac surgery. Demographic data were recorded, including details of cardiac anatomy and systemic oxygen saturations. At operation, tissue samples were obtained from right ventricular myocardium as soon as possible after instituting cardiopulmonary bypass to minimise the effect of this on protein analysis.
Protein analysis technique is online supplement.
openhrt-2017-000716supp001.pdf (50.5KB, pdf)
Clinical data
The patient clinical outcome data that was recorded were standard operative and postoperative data. This included bypass time, cross clamp time, recorded pressure in the superior vena cava at 4 hours postoperative, time in intensive care, time on mechanical ventilation, time of pleural effusion drainage, requirement for postoperative oxygen therapy and time to discharge. In addition, we recorded the requirement for inotropic support both for length of time this was needed but also which inotrope was used.
Tissue studies
Biopsies were taken from the free wall of the RV of patients with VSD (n=6) or with TOF (n=6) immediately after starting cardiopulmonary bypass. Tissue was either snap frozen in liquid nitrogen or placed in Allprotect tissue reagent (Qiagen, Manchester, UK) before being stored at −80°C.
Echocardiography
Echocardiography was performed using a Vivid 9 or IE33 echocardiographic machines using standard assessment of preoperative and postoperative characteristics, including cardiac anatomy in addition to function. Left heart function assessment included fractional shortening in long axis view, mitral annular plane systolic excursion (not indexed), mitral E wave/A wave (E/A wave) ratio and volume time integral of left ventricular outflow tract Doppler. Right heart function similarly included tricuspid annular plane systolic excursion (not indexed), fractional area length change and lateral wall tissue Doppler. An assessment was made of diastolic function by Doppler interrogation of inflow velocity through the mitral and tricuspid valve and also the presence or absence of diastolic A wave velocity in the pulmonary artery.
Statistical comparison
For the data that were parametric, mean and SD are presented with paired t-test values (P<0.05 being significant). For the data that were non-parametric, median (range) is presented with Wilcoxon values (P<0.05 being significant). Significance of proteomic data was determined as described above.
Results
Clinical data
The demographic and clinical data are displayed in table 1; children in each group had similar age, sex ratio and weight. On preoperative echocardiogram, all children had systemic level right ventricular pressure, with an unrestrictive VSD. Those with TOF were significantly cyanosed, with severe infundibular pulmonary stenosis. All had good systolic function preoperatively, and there was no difference in bypass time or cross-clamp time at operation.
Table 1.
Patient demographics, preoperative and intraoperative characteristics
| Group 1 VSD | Group 2 TOF | P value | |
| Age (years) | 0.38 (0.13) | 0.37 (0.13) | ns |
| Weight (kg) | 4.81 (0.41) | 5.89 (1.01) | ns |
| Oxygen saturations (%) | 97.5 (2.6) | 82.8 (6.2) | 0.0038 |
| Male:female | 3:3 | 3:3 | ns |
| Systolic blood pressure (mm Hg) | 83.8 (10.9) | 88.5 (13.6) | ns |
| Fractional shortening (%) | 34.7 (2.5) | 34.0 (8.5) | ns |
| Bypass time (minutes) | 117 (127) | 140 (28) | ns |
| Cross-clamp time (minutes) | 86 (95) | 87(51) | ns |
Data displayed as mean (SD).
P<0.05 was significant.
TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Postoperatively, there were some differences in the RV function, as seen in table 2. Those from the TOF group having good systolic function and good cardiac output. However, there was evidence of increased stiffness in the RV with a significantly higher tricuspid E/A ratio (VSD group 0.84 (0.3) compared with TOF group 1.95 (0.4) (P=0.009) and high incidence of pulmonary diastolic forward flow (A wave) (0/6 in the VSD group and 5/6 in the TOF group (P=0.001)).
Table 2.
Echocardiographic assessment postoperatively, comparison of VSD group and TOF group
| Group 1 VSD | Group 2 TOF | P value | |
| Heart rate | 125 (14) | 141 (14) | 0.044 |
| Fractional shortening (%) | 30.43 (2) | 32.5 (4) | ns |
| MAPSE (mm) | 7 (1) | 6.8 (1) | ns |
| Aortic VTI (mL/s) | 12.2 (3) | 13 (3) | ns |
| TAPSE (mm) | 6 (1) | 7.43 (2) | ns |
| Tricuspid E/A ratio | 0.84 (0.3) | 1.95 (0.4) | 0.009 |
| Tricuspid S′ (cm/s) | 6 (1) | 4.7 (1) | ns |
| Pulmonary A wave | 0/6 | 5/6 | 0.001 |
Data displayed as mean (SD).
P<0.05 is significant.
E/A ratio, E wave/A wave ratio; MAPSE, mitral annular plane systolic excursion; S′, tissue Doppler assessment of free wall movement; TAPSE, tricuspid annular plane systolic excursion; TOF, tetralogy of Fallot; VSD, ventricular septal defect; VTI, velocity time integral.
Proteomics analysis
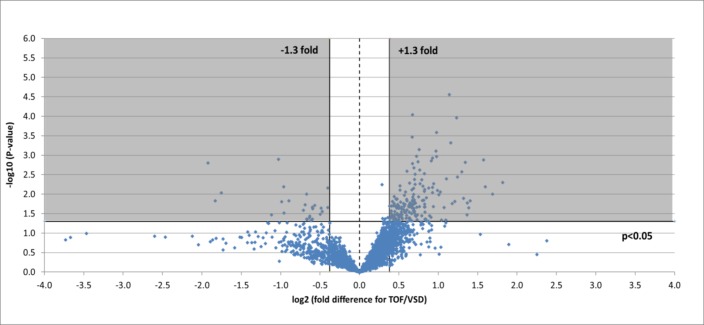
A total of 3610 protein accession numbers (representing 3342 gene names; note that this number differs in table 3 (2357 gene names) due to the PantherDB database version used) were identified that were found in RV samples from both heart pathologies and that fulfilled all criteria outlined above. Of these, 200 were found to significantly change (±1.3-fold, P<0.05) between pathologies: 26 were lower in TOF samples and 174 were higher in TOF (figure 1). Gene ontology analysis was performed using these proteins found in both TOF and VSD samples and which significantly changed or not (table 4).
Table 3.
Number of proteins in proteomics analysis, per ontology category and per pathology
| Proteins (n) | Percentage of proteins | ||||
| Ontology | Category | TOF RV | VSD RV | TOF RV | VSD RV |
| Total number of accession numbers | 5848 | 3654 | |||
| Total number of unique proteins | 3190 | 2374 | |||
| Molecular function | Antioxidant activity | 14 | 13 | 0.4 | 0.5 |
| Binding | 841 | 649 | 26.4 | 27.3 | |
| Catalytic activity | 1051 | 809 | 32.9 | 34.1 | |
| Channel regulator activity | 5 | 4 | 0.2 | 0.2 | |
| Receptor activity | 64 | 37 | 2.0 | 1.6 | |
| Signal transducer activity | 21 | 15 | 0.7 | 0.6 | |
| Structural molecule activity | 285 | 235 | 8.9 | 9.9 | |
| Translation regulator activity | 26 | 21 | 0.8 | 0.9 | |
| Transporter activity | 146 | 102 | 4.6 | 4.3 | |
| Biological process | Biological adhesion | 77 | 55 | 2.4 | 2.3 |
| Biological regulation | 232 | 165 | 7.3 | 7.0 | |
| Cellular component organisation or biogenesis | 414 | 339 | 13.0 | 14.3 | |
| Cellular process | 1338 | 1023 | 41.9 | 43.1 | |
| Developmental process | 217 | 162 | 6.8 | 6.8 | |
| Growth | 1 | 1 | 0.0 | 0.0 | |
| Immune system process | 139 | 83 | 4.4 | 3.5 | |
| Localisation | 409 | 311 | 12.8 | 13.1 | |
| Locomotion | 19 | 16 | 0.6 | 0.7 | |
| Metabolic process | 1235 | 943 | 38.7 | 39.7 | |
| Multicellular organismal process | 193 | 140 | 6.1 | 5.9 | |
| Reproduction | 27 | 24 | 0.8 | 1.0 | |
| Response to stimulus | 248 | 171 | 7.8 | 7.2 | |
| Cell component | Cell junction | 20 | 15 | 0.6 | 0.6 |
| Cell part | 914 | 717 | 28.7 | 30.2 | |
| Extracellular matrix | 31 | 21 | 1.0 | 0.9 | |
| Extracellular region | 97 | 64 | 3.0 | 2.7 | |
| Macromolecular complex | 359 | 286 | 11.3 | 12.0 | |
| Membrane | 207 | 152 | 6.5 | 6.4 | |
| Organelle | 566 | 446 | 17.7 | 18.8 | |
| Synapse | 8 | 5 | 0.3 | 0.2 | |
| Protein class | Calcium-binding protein | 85 | 64 | 2.7 | 2.7 |
| Cell adhesion molecule | 52 | 38 | 1.6 | 1.6 | |
| Cell junction protein | 21 | 18 | 0.7 | 0.8 | |
| Chaperone | 63 | 49 | 2.0 | 2.1 | |
| Cytoskeletal protein | 210 | 167 | 6.6 | 7.0 | |
| Defense/immunity protein | 40 | 31 | 1.3 | 1.3 | |
| Enzyme modulator | 268 | 200 | 8.4 | 8.4 | |
| Extracellular matrix protein | 55 | 40 | 1.7 | 1.7 | |
| Hydrolase | 255 | 189 | 8.0 | 8.0 | |
| Isomerase | 54 | 42 | 1.7 | 1.8 | |
| Ligase | 79 | 56 | 2.5 | 2.4 | |
| Lyase | 43 | 38 | 1.3 | 1.6 | |
| Membrane traffic protein | 93 | 69 | 2.9 | 2.9 | |
| Nucleic acid binding | 412 | 323 | 12.9 | 13.6 | |
| Oxidoreductase | 198 | 165 | 6.2 | 7.0 | |
| Receptor | 86 | 54 | 2.7 | 2.3 | |
| Signalling molecule | 107 | 80 | 3.4 | 3.4 | |
| Storage protein | 1 | 1 | 0.0 | 0.0 | |
| Structural protein | 27 | 20 | 0.8 | 0.8 | |
| Surfactant | 1 | 1 | 0.0 | 0.0 | |
| Transcription factor | 110 | 73 | 3.4 | 3.1 | |
| Transfer/carrier protein | 83 | 64 | 2.6 | 2.7 | |
| Transferase | 227 | 171 | 7.1 | 7.2 | |
| Transmembrane receptor regulatory/adaptor protein | 13 | 10 | 0.4 | 0.4 | |
| Transporter | 123 | 81 | 3.9 | 3.4 | |
| Viral protein | 1 | 0 | 0.0 | 0.0 | |
Proteins are also shown as a percentage of the number of unique proteins per pathology. The ontology categories with the 10 highest percentage of proteins per pathology are given in bold.
RV, right ventricle; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Figure 1.
Volcano plot of entire set of proteins quantified in the right ventricle of TOF versus VSD patients (positive log2 (fold change) indicates higher expression in TOF samples). Each point represents the difference in expression (log2 fold difference) between the groups, and the associated significance of this change (independent unpaired samples t-test). Proteins significantly altered (±1.3-fold, P<0.05) are found within the grey shaded boxes. TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Table 4.
Number of proteins in proteomics analysis, per ontology category, statistically compared between TOF and VSD RV samples
| Proteins (n) | Percentage of total unique proteins (2357) | ||||||||
| Ontology | Category | Total | No change | Higher TOF | Lower TOF | Total | No change | Higher TOF | Lower TOF |
| Total number of accession numbers | 3610 | 3410 | 174 | 26 | |||||
| Total number of unique proteins | 2357 | 2244 | 100 | 13 | 100 | 95.2 | 4.2 | 0.6 | |
| Molecular function | Antioxidant activity | 13 | 13 | 0 | 0 | 0.6 | 0.6 | 0.0 | 0.0 |
| Binding | 643 | 607 | 34 | 2 | 27.3 | 25.8 | 1.4 | 0.1 | |
| Catalytic activity | 806 | 776 | 25 | 5 | 34.2 | 32.9 | 1.1 | 0.2 | |
| Channel regulator activity | 4 | 4 | 0 | 0 | 0.2 | 0.2 | 0.0 | 0.0 | |
| Receptor activity | 36 | 32 | 4 | 0 | 1.5 | 1.4 | 0.2 | 0.0 | |
| Signal transducer activity | 14 | 13 | 0 | 1 | 0.6 | 0.6 | 0.0 | 0.0 | |
| Structural molecule activity | 234 | 217 | 15 | 2 | 9.9 | 9.2 | 0.6 | 0.1 | |
| Translation regulator activity | 21 | 20 | 1 | 0 | 0.9 | 0.8 | 0.0 | 0.0 | |
| Transporter activity | 101 | 95 | 5 | 1 | 4.3 | 4.0 | 0.2 | 0.0 | |
| Biological process | Biological adhesion | 54 | 53 | 1 | 0 | 2.3 | 2.2 | 0.0 | 0.0 |
| Biological regulation | 162 | 156 | 4 | 2 | 6.9 | 6.6 | 0.2 | 0.1 | |
| Cellular component organisation or biogenesis | 335 | 319 | 12 | 4 | 14.2 | 13.5 | 0.5 | 0.2 | |
| Cellular process | 1014 | 965 | 42 | 7 | 43.0 | 40.9 | 1.8 | 0.3 | |
| Developmental process | 158 | 149 | 7 | 2 | 6.7 | 6.3 | 0.3 | 0.1 | |
| Growth | 1 | 1 | 0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Immune system process | 81 | 75 | 6 | 0 | 3.4 | 3.2 | 0.3 | 0.0 | |
| Localisation | 310 | 295 | 13 | 2 | 13.2 | 12.5 | 0.6 | 0.1 | |
| Locomotion | 15 | 14 | 1 | 0 | 0.6 | 0.6 | 0.0 | 0.0 | |
| Metabolic process | 939 | 900 | 34 | 5 | 39.8 | 38.2 | 1.4 | 0.2 | |
| Multicellular organismal process | 137 | 130 | 5 | 2 | 5.8 | 5.5 | 0.2 | 0.1 | |
| Reproduction | 24 | 22 | 1 | 1 | 1.0 | 0.9 | 0.0 | 0.0 | |
| Response to stimulus | 169 | 158 | 9 | 2 | 7.2 | 6.7 | 0.4 | 0.1 | |
| Cell component | Cell junction | 14 | 1 | 0 | 0.6 | 0.6 | 0.0 | 0.0 | |
| Cell part | 709 | 675 | 30 | 4 | 30.1 | 28.6 | 1.3 | 0.2 | |
| Extracellular matrix | 20 | 20 | 0 | 0 | 0.8 | 0.8 | 0.0 | 0.0 | |
| Extracellular region | 64 | 63 | 1 | 0 | 2.7 | 2.7 | 0.0 | 0.0 | |
| Macromolecular complex | 282 | 268 | 12 | 2 | 12.0 | 11.4 | 0.5 | 0.1 | |
| Membrane | 149 | 144 | 5 | 0 | 6.3 | 6.1 | 0.2 | 0.0 | |
| Organelle | 440 | 417 | 20 | 3 | 18.7 | 17.7 | 0.8 | 0.1 | |
| Synapse | 5 | 5 | 0 | 0 | 0.2 | 0.2 | 0.0 | 0.0 | |
| Protein class | Calcium-binding protein | 62 | 60 | 2 | 0 | 2.6 | 2.5 | 0.1 | 0.0 |
| Cell adhesion molecule | 38 | 35 | 3 | 0 | 1.6 | 1.5 | 0.1 | 0.0 | |
| Cell junction protein | 18 | 18 | 0 | 0 | 0.8 | 0.8 | 0.0 | 0.0 | |
| Chaperone | 49 | 49 | 0 | 0 | 2.1 | 2.1 | 0.0 | 0.0 | |
| Cytoskeletal protein | 167 | 153 | 12 | 2 | 7.1 | 6.5 | 0.5 | 0.1 | |
| Defense/immunity protein | 31 | 29 | 2 | 0 | 1.3 | 1.2 | 0.1 | 0.0 | |
| Enzyme modulator | 200 | 195 | 4 | 1 | 8.5 | 8.3 | 0.2 | 0.0 | |
| Extracellular matrix protein | 39 | 38 | 1 | 0 | 1.7 | 1.6 | 0.0 | 0.0 | |
| Hydrolase | 189 | 180 | 9 | 0 | 8.0 | 7.6 | 0.4 | 0.0 | |
| Isomerase | 41 | 39 | 2 | 0 | 1.7 | 1.7 | 0.1 | 0.0 | |
| Ligase | 56 | 56 | 0 | 0 | 2.4 | 2.4 | 0.0 | 0.0 | |
| Lyase | 38 | 36 | 2 | 0 | 1.6 | 1.5 | 0.1 | 0.0 | |
| Membrane traffic protein | 69 | 67 | 2 | 0 | 2.9 | 2.8 | 0.1 | 0.0 | |
| Nucleic acid binding | 320 | 306 | 12 | 2 | 13.6 | 13.0 | 0.5 | 0.1 | |
| Oxidoreductase | 165 | 164 | 1 | 0 | 7.0 | 7.0 | 0.0 | 0.0 | |
| Receptor | 52 | 48 | 4 | 0 | 2.2 | 2.0 | 0.2 | 0.0 | |
| Signalling molecule | 79 | 76 | 3 | 0 | 3.4 | 3.2 | 0.1 | 0.0 | |
| Storage protein | 1 | 1 | 0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Structural protein | 20 | 17 | 1 | 2 | 0.8 | 0.7 | 0.0 | 0.1 | |
| Surfactant | 1 | 1 | 0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Transcription factor | 72 | 67 | 5 | 0 | 3.1 | 2.8 | 0.2 | 0.0 | |
| Transfer/carrier protein | 64 | 61 | 3 | 0 | 2.7 | 2.6 | 0.1 | 0.0 | |
| Transferase | 170 | 161 | 8 | 1 | 7.2 | 6.8 | 0.3 | 0.0 | |
| Transmembrane receptor regulatory/adaptor protein | 10 | 10 | 0 | 0 | 0.4 | 0.4 | 0.0 | 0.0 | |
| Transporter | 81 | 77 | 3 | 1 | 3.4 | 3.3 | 0.1 | 0.0 | |
| Viral protein | 0 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | |
The total number of proteins, and number significantly changed, or not, between pathologies are shown. Proteins are also shown as a percentage of the number of unique proteins. The ontology categories with the five highest percentage of proteins per pathology are given in bold.
RV, right ventricle; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
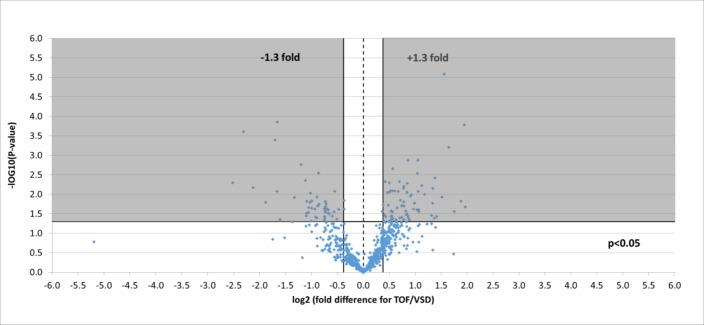
Four hundred and fourteen phosphorylated proteins were also successfully identified, each containing at least one translation modification at serine, threonine or tyrosine (resulting in a total of 700 phosphorylation site matches). Of the phosphorylation sites identified, 117 significantly changed representing 97 unique proteins: 46 were lower in TOF and 71 were higher (figure 2).
Figure 2.
Volcano plot of entire set of phosphorylated proteins quantified in the right ventricle of TOF versus VSD patients (positive log 2 (fold change) indicates higher expression in TOF samples). Each point represents the difference in expression (log 2 fold difference) between the groups, and the associated significance of this change (independent unpaired samples t-test). Proteins significantly altered (±1.3 fold, P<0.05) are found within the grey shaded boxes. TOF, tetralogy of Fallot; VSD, ventricular septal defect.
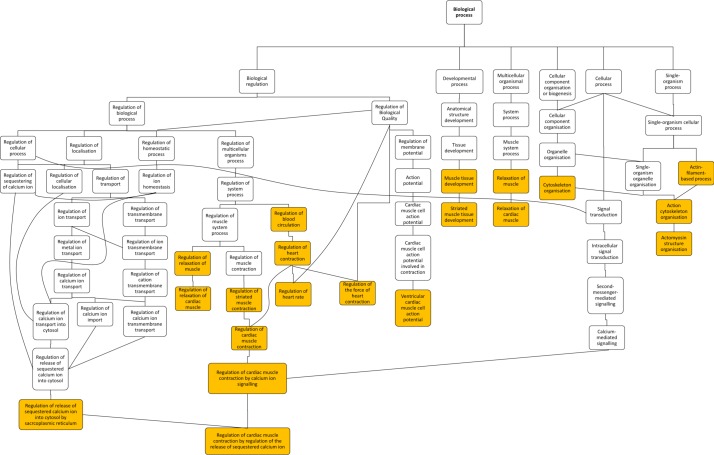
Gene ontology enrichment analysis showed that cellular component proteins associated with the muscle/contractile fibre part (11 proteins) and specifically the Z-disc (eight proteins) were significantly enriched in those proteins that were higher in patients with TOF. For individual proteins associated with Gene Ontology (GO) terms, see table 5. Proteins and associated gene names are displayed in table 6. Molecular function proteins associated with α-catenin, calmodulin and cytoskeletal actin binding (3, 7 and 14 proteins, respectively) were enriched, as well as biological processes predominantly associated with heart and muscle contractility and regulation (figure 3). There was no enrichment of proteins higher in VSD patients. Phosphorylated proteins associated with the biological processes actomyosin structure organisation, regulation of calcium-mediated signalling and regulation of calcium ion transmembrane transporter activity (5, 3 and 3 phosphoproteins, respectively), and the cellular components somatodendritic compartment, Z-disc and actomyosin (2, 4 and 5 phosphoproteins, respectively) were significantly enriched.
Table 5.
Enriched gene ontology terms for significantly higher total and phosphorylated protein levels in the right ventricle of patients with tetralogy of Fallot compared with ventricular septal defect
| Ontology | Protein type | GO term | Description | P value | Proteins (n) | Protein symbol |
| Cellular component | Total | GO:0044449 | Contractile fibre part | 4E-05 | 11 | CFL2, CMYA5, CSRP3, FHOD3, FLNC, MYOM1, MYOZ2, NEBL, OBSCN, RYR2, SVIL |
| GO:0030018 | Z-disc | 9E-05 | 8 | CFL2, CSRP3, FHOD3, FLNC, MYOZ2, NEBL, OBSCN, RYR2 | ||
| Phosphorylated | GO:0042641 | Actomyosin | 6E-05 | 4 | CDC42BPB, MYO18A, NEBL, SORBS1 | |
| GO:0036477 | Somatodendritic compartment | 1E-04 | 2 | MAPT, STX4 | ||
| GO:0044449 | Contractile fibre part | 7E-04 | 5 | AHNAK, BAG3, JPH2, NEBL, OBSCN | ||
| GO:0030018 | Z-disc | 8E-04 | 4 | BAG3, JPH2, NEBL, OBSCN | ||
| Molecular function | Total | GO:0045294 | Alpha-catenin binding | 1E-05 | 3 | AJUBA, CDH2, PKP2 |
| GO:0003779 | Actin binding | 4E-05 | 14 | AJUBA, CFL2, CSRP3, FHOD3, FLII, FLNC, LIMCH1, MYOZ2, NEBL, SNTA1, SNTB2, SORBS1, SVIL, TWF2 | ||
| GO:0005516 | Calmodulin binding | 9E-04 | 7 | CAMK2D, OBSCN, PPP3CC, RYR2, SNTA1, SNTB2, STRN | ||
| Biological process | GO:0010881 | Regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 3E-05 | 4 | CAMK2D, GSTM2, PLN, RYR2 | |
| GO:0031032 | Actomyosin structure organisation | 9E-05 | 7 | CFL2, CSRP3, FHOD3, LIMCH1, MYOZ2, OBSCN, SORBS1 | ||
| GO:0010882 | Regulation of cardiac muscle contraction by calcium ion signalling | 9E-05 | 4 | CAMK2D, GSTM2, PLN, RYR2 | ||
| GO:0014706 | Striated muscle tissue development | 1E-04 | 6 | CFL2, CSRP3, MYOM1, PLN, SEMA3C, SVIL, | ||
| GO:1903522 | Regulation of blood circulation | 1E-04 | 9 | ADCY6, CAMK2D, CSRP3, GSTM2, PKP2, PLN, RYR2, SNTA1, TMEM65 | ||
| GO:0060537 | Muscle tissue development | 1E-04 | 6 | CFL2, CSRP3, MYOM1, PLN, SEMA3C, SVIL | ||
| GO:0010880 | Regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 2E-04 | 4 | CAMK2D, GSTM2, PLN, RYR2 | ||
| GO:0008016 | Regulation of heart contraction | 2E-04 | 8 | CAMK2D, CSRP3, GSTM2, PKP2, PLN, RYR2, SNTA1, TMEM65 | ||
| GO:0007010 | Cytoskeleton organisation | 3E-04 | 16 | AJUBA, CFL2, CSRP3, ERBB2IP, FHOD3, FLII, LIMCH1, MYOZ2, NTMT1, OBSCN, PKP2, PRKCDBP, SHANK1, SORBS1, SVIL, TUBA4A | ||
| GO:0055119 | Relaxation of cardiac muscle | 4E-04 | 3 | CAMK2D, GSTM2, PLN | ||
| GO:0086005 | Ventricular cardiac muscle cell action potential | 4E-04 | 3 | PKP2, RYR2, SNTA1 | ||
| GO:0055117 | Regulation of cardiac muscle contraction | 4E-04 | 5 | CAMK2D, GSTM2, PKP2, PLN, RYR2 | ||
| GO:0002026 | Regulation of the force of heart contraction | 4E-04 | 4 | CAMK2D, CSRP3, PLN, RYR2 | ||
| GO:1901077 | Regulation of relaxation of muscle | 6E-04 | 2 | CAMK2D, PLN | ||
| GO:1901897 | Regulation of relaxation of cardiac muscle | 6E-04 | 2 | CAMK2D, PLN | ||
| GO:0002027 | Regulation of heart rate | 8E-04 | 5 | CAMK2D, PKP2, PLN, RYR2, SNTA1 | ||
| GO:0006942 | Regulation of striated muscle contraction | 1E-03 | 5 | CAMK2D, GSTM2, PKP2, PLN, RYR2 | ||
| GO:0090075 | Relaxation of muscle | 1E-03 | 3 | CAMK2D, GSTM2, PLN | ||
| Phosphorylated | GO:0031032 | Actomyosin structure organisation | 1E-05 | 5 | CDC42BPB, MYLK3, MYO18A, OBSCN, SORBS1 | |
| GO:0050789 | Regulation of biological process | 3E-04 | 25 | AHNAK, BAG3, BVES, CDC42BPB, JPH2, KANK2, KPNA4, LRRFIP2, MAPT, MYLK3, MYO18A, NUCKS1, OBSCN, OSBP, PLCL1, PLN, PSIP1, RBM20, RSF1, SCN7A, SGTA, SORBS1, SPEG, STX4, ZBTB7A | ||
| GO:0050848 | Regulation of calcium-mediated signalling | 4E-04 | 3 | JPH2, MAPT, PLN | ||
| GO:1901019 | Regulation of calcium ion transmembrane transporter activity | 4E-04 | 3 | AHNAK, JPH2, PLN |
Table 6.
List of proteins and associated gene name abbreviations
| Gene symbol | Protein name |
| ADCY6 | Adenylate cyclase type 6 |
| AHNAK | Neuroblast differentiation-associated protein AHNAK |
| AJUBA | LIM domain-containing protein ajuba |
| BAG3 | BAG family molecular chaperone regulator 3 |
| BCAT2 | Branched-chain-amino-acid aminotransferase, mitochondrial |
| BCKDHA | 2-Oxoisovalerate dehydrogenase subunit alpha, mitochondrial |
| BCKDK | (3-methyl-2-oxobutanoate dehydrogenase (lipoamide)) kinase, mitochondrial |
| BVES | Blood vessel epicardial substance |
| C6orf57 | Chromosome six open reading frame 57 |
| CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta |
| CDC42BPB | Serine/threonine-protein kinase MRCK beta |
| CDH2 | Cadherin-2 |
| CFL2 | Cofilin-2 |
| CMYA5 | Cardiomyopathy-associated protein 5 |
| COA5 | Cytochrome c oxidase assembly factor 5 |
| COBL | Protein cordon-bleu |
| COL6A5 | Collagen alpha-5(VI) chain |
| COX1 | Cytochrome c oxidase subunit 1 |
| CPT2 | Carnitine O-palmitoyltransferase 2, mitochondrial |
| CSRP3 | Cysteine and glycine-rich protein 3 |
| DMD | Dystrophin |
| ERBB2IP | Protein LAP2 |
| FDX1 | Adrenodoxin, mitochondrial |
| FHOD3 | FH1/FH2 domain-containing protein 3 |
| FLII | Protein flightless-1 homolog |
| FLNC | Filamin-C |
| GSTM2 | Glutathione S-transferase Mu 2 |
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D0 |
| HRC | Sarcoplasmic reticulum histidine-rich calcium-binding protein |
| JPH2 | Junctophilin-2 |
| KANK2 | KN motif and ankyrin repeat domain-containing protein 2 |
| KPNA4 | Importin subunit alpha-3 |
| KRT10 | Keratin, type I cytoskeletal 10 |
| KRT14 | Keratin, type I cytoskeletal 14 |
| LCN2 | Neutrophil gelatinase-associated lipocalin |
| LDB3 | LIM domain binding 3 isoform 1 |
| LIMCH1 | LIM and calponin homology domains-containing protein 1 |
| LRRFIP2 | Leucine-rich repeat flightless-interacting protein 2 |
| MAPT | Microtubule-associated protein |
| MRPL12 | 39S Ribosomal protein L12, mitochondrial |
| MYLK3 | Myosin light chain kinase 3 |
| MYO18A | Unconventional myosin-XVIIIa |
| MYOM1 | Myomesin-1 |
| MYOM2 | Myomesin (M-protein) 2, 165 kDa, isoform CRA_c |
| MYOZ2 | Myozenin-2 |
| ND1 | NADH-ubiquinone oxidoreductase chain 1 |
| NEBL | Nebulette |
| NTMT1 | N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| NUCKS1 | Nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 |
| OBSCN | Obscurin |
| OGDH | 2-Oxoglutarate dehydrogenase, mitochondrial |
| OSBP | Oxysterol-binding protein 1 |
| PKP2 | Plakophilin-2 |
| PLCL1 | Inactive phospholipase C-like protein 1 |
| PLN | Cardiac phospholamban |
| PPM1K | Protein phosphatase 1K, mitochondrial |
| PPP3CC | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform |
| PRKAR2A | Protein kinase cAMP-dependent regulatory type II alpha isoform 1 |
| PRKCDBP | Protein kinase C delta-binding protein |
| PSIP1 | PC4 and SFRS1-interacting protein |
| RBM20 | RNA-binding protein 20 |
| RSF1 | Remodelling and spacing factor 1 |
| RYR2 | Ryanodine receptor 2 |
| SCN7A | Sodium channel protein type 7 subunit alpha |
| SDHD | Succinate dehydrogenase (ubiquinone) cytochrome b small subunit, mitochondrial |
| SDHAF1 | Succinate dehydrogenase assembly factor 1, mitochondrial |
| SEMA3C | Semaphorin-3C |
| SGTA | Small glutamine-rich tetratricopeptide repeat-containing protein alpha (Alpha-SGT) (Vpu-binding protein) (UBP) |
| SHANK1 | SH3 and multiple ankyrin repeat domains protein 1 |
| SNTA1 | Alpha-1-syntrophin |
| SNTB2 | Beta-2-syntrophin |
| SORBS1 | Sorbin and SH3 domain-containing protein 1 |
| SPEG | Striated muscle preferentially expressed protein kinase |
| SRL | Sarcalumenin |
| STRN | Striatin |
| STX4 | Syntaxin-4 |
| SVIL | Supervillin |
| SYNPO2L | Synaptopodin 2-like, isoform CRA_a |
| TIMM22 | Mitochondrial import inner membrane translocase subunit Tim22 |
| TMEM65 | Transmembrane protein 65 |
| TNNI3 | Mutant cardiac troponin I |
| TPM1 | Tropomyosin 1 (Alpha), isoform CRA_a |
| TSFM | Elongation factor Ts, mitochondrial |
| TUBA4A | Tubulin alpha-4A chain |
| TUBB4A | Tubulin beta-4A chain |
| TWF2 | Twinfilin-2 |
| XIRP1 | Xin actin-binding repeat-containing protein 1 |
| ZBTB7A | Zinc finger and BTB domain-containing protein 7A |
Figure 3.
Hierarchy of enriched gene ontology biological processes for expression of proteins that are higher in patients with TOF compared with VSD. Highlighted terms correspond to table 5. Figure exported from GOrilla analysis. TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Structural and extracellular matrix protein and calcium signalling comparisons
Structural proteins in human skeletal muscle were cross-referenced against protein lists found in our different samples. One hundred and nine accession numbers (representing 97 proteins) were detected, and 12 (12.4%) of these were significantly higher in TOF. Six of these were associated with sarcomeric function that included Z-disc proteins (muscle LIM protein (CSRP3), filamin-C, ZASP), M-band and giant muscle proteins (myomesin-1 and obscurin) and the thin filament protein, tropomyosin-1. The remaining six proteins were associated with actin-binding (supervillin, cardiomyopathy-associated protein 5, synaptopodin 2-like protein and cordon-bleu protein), calcium signalling (phospholamban) and myocyte cytoskeletal development (striated muscle preferentially expressed protein kinase; by similarity with members of myosin light chain kinase family). Additionally, adenylate cyclase six was higher in TOF samples. Phosphopeptide enrichment led to higher levels of 9 proteins in TOF samples compared with VSD, some with multiple phosphorylation sites. These were calcium signalling proteins (neuroblast differentiation-associated protein AHNAK–Ser4425 and AHNA K–Ser5110, sarcoplasmic reticulum histidine-rich calcium-binding protein –Ser145 and –Ser457, junctophilin-2 –Threonine (Thr)490, phospholamban-Ser16 and phospholamban-Ser16/Thr17), Z-disk proteins (obscurin-Ser5563 and dystrophin-Ser432), BAG family molecular chaperone regulator 3-Ser289 (which stimulates expression of cytoskeleton), striated muscle preferentially expressed protein kinase-Ser2448 and protein kinase-Ser2014/Ser2015, and nuclear ubiquitous casein and cyclin-dependent kinase substrate-Ser113 (highly expressed in cardiac tissue, but with unknown function). Six phosphoproteins had lower levels in TOF samples: AHNAK-Ser1068 and AHNAK-Ser5369 (calcium signalling), myomesin-1-Ser36 and myomesin-2-Ser39, myomesin-2-Ser76, myomesin-2-Ser78, myomesin-2-Ser1461 (M-band proteins), Xin actin-binding repeat-containing protein 1-Ser208 (actin-binding) and protein kinase cAMP-dependent regulatory type II alpha-Ser78 (protein kinase activity).
Of the list of extracellular matrix proteins searched, four were higher in VSD (collagen VI, keratin-10 and keratin-14 and tubulin-β4 A), and five were higher in TOF samples (cofilin-2, cadherin-2, myomesin-1, tropomyosin-1 and tubulin-α4 A). Additionally, three phosphorylated proteins were lower in TOF samples: myomesin 1-Ser36 (also in structural proteins list), myomesin 2 at phosphosites-Ser39, phosphosites-Ser76 and phosphosites-Ser1461 and troponin I at Ser199.
Clinical outcome data
All children survived without any major complications. However, as expected there were significant differences in outcome data (table 7). The number of days of mechanical ventilation, the pressure in the superior vena cava at 4 hours, the peak rate of dopamine, the length of time of required dopamine infusion, the peak rate of milrinone, the length of time of milrinone requirement and the number of days of pleural drains in situ were all not significantly different in the two groups. One child with TOF had longer time in intensive care, longer time on milrinone, longer hospital stay and longer time in oxygen than the others. As a result, there was a mildly significant difference in the length of time that supplemental oxygen was required between the VSD group and the TOF group. However, this all resolved by discharge. Two children had arrhythmias in the VSD group and one with supraventricular tachycardia and with complete heart block but both resolved by the time of leaving the intensive care. Three children with TOF had junctional ectopic tachycardia but resolved with standard approach of cooling and short-term amiodarone usage. None required long-term medication. All children were discharged home on furosemide and spironolactone. Two children with VSD repair were discharged home on captopril. One child with TOF required captopril at discharge and one child had propranolol for stiff RV.
Table 7.
Clinical outcome data
| Group 1 VSD n=6 | Group 2 TOF n=6 | P value | |
| Ventilation time (median days) | 0.833 | 5.2 | ns |
| SVC pressure (median mm Hg) | 11.2 | 10.7 | ns |
| Dopamine (median peak rate, µg/kg/min) | 4.2 | 7.5 | ns |
| Dopamine (median hours) | 12.5 | 59 | ns |
| Milrinone (median peak rate, µg/kg/min) | 0.5 | 0.57 | ns |
| Milrinone (median hours) | 39.7 | 88 | ns |
| Pleural drains (median days) | 2.3 | 2.3 | ns |
| Arrhythmias (number of children) | 2 | 3 | ns |
| Oxygen duration (median, days) | 2 | 5.1 | 0.043 |
| Intensive care stay (median, days) | 2.7 | 4.5 | ns |
| Hospital stay (median days) | 6.5 | 9.5 | ns |
SVC, superior vena cava; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Discussion
The present study is the first to attempt to correlate changes in myocardial proteins with clinical and echocardiographic data in order to try to understand the adaptive, maladaptive and remodelling pathways in the two groups of patients with hypertensive RVs. These two groups were chosen since they both have RVs that were functioning at systemic pressure. Both groups of infants were of the same age at the time of operation, and there was the same sex distribution. Hence, confounders relating to these diseases have been removed. The only difference between the two groups was the physiology—one being volume loaded and the other being after loaded—in addition to the systemic oxygen saturation being significantly lower in the group with TOF.
We confirmed previous studies that have shown the RV to be stiff in those children who have TOF,5 and our gene ontology protein enrichment data lends further support to this, with proteins found to be higher in patients with TOF in the contractile part of the cell and in particular the Z-disc. Enrichment of differentially expressed proteins within processes regulating heart and muscle contractility also support this.
The contractile units of myocytes are the sarcomeres, and within these, the thin actin filaments and thick myosin filaments generate the contractile force. Many other proteins surround and support these to make up the sarcomeric scaffold, which affects the efficiency and transmission of the generated force.6 In our patients, there were no significant differences in expression between actin or myosin; however in the RV of hearts with high after load, as in TOF, there were increases in the amounts of supporting proteins, for example, muscle LIM protein, filamin-C, obscurin, myomesin-1 and ZASP, which have roles in cross-linking and connecting filaments, Z-disc assembly, mechano-sensing and myocyte signalling.7–13 The evidence that MYOM1 is present in the myocardium of neonatal dogs might have relevance to our patients, if we later found it to be reduced in amount in older children with the same condition.12 It is possible that overexpression of these proteins is linked with decreased ability to relax and that there is failure of the RV to develop past the phenotype seen in the neonate. Further studies would be needed to determine whether this disappears in older children with TOF once repair had been performed. The giant protein titin stretches between the Z-disc and M-line of the sarcomere and is thought to be a key player in passive myocardial stiffness and contraction.14 15 Our data showed a non-significant increase (1.4-fold) in titin content of TOF samples; further interrogation showed no significant correlation between titin expression and E/A wave ratio as a stiffness indicator. However, previous work has shown that it may be the ratio of cardiac titin isoforms that is important in heart failure and stiffness (which was not looked at here) and not total expression.15 We have shown that in the hearts of infants with TOF, which have evidence of increased stiffness and pressure afterload, there is an increase in some of the contractile proteins. It should however be noted that in this study, due to unavailability of healthy patient control tissue, we cannot definitively say that proteins are abnormally altered in either pathology, only that there are differences between the pathologies. It is unclear whether the predominantly enhanced expression of proteins in TOF is due to right ventricular remodelling of the myocytes and the onset of hypertrophy or a compensatory mechanism. Studies on aortas from rats with type 2 diabetes have shown that increased ROS caused by oxidative stress upregulates contractile protein expression resulting in vascular wall remodelling.16
Perhaps of greater importance is the differences in phosphorylation of proteins involved in the ventricle contractile processes. Phosphorylation of proteins indicates a change in protein function (either activation or deactivation) and enrichment was seen in proteins at various phosphorylation sites with higher levels in TOF, associated with calcium signalling and contractility pathways. Of the phosphorylation sites we detected in our proteomics analysis, many of them have not been detected previously in heart tissue (PhosphositePlus, accessed March 201717), the exceptions being the following (R denotes regulatory protein, and K denotes associated kinases where known): phospholamban-ser16 (R: protein phosphatase 1 regulatory inhibitor subunit 1A,18 K: cAMP-dependent protein kinase catalytic subunit alpha19) and -thr17 (R: tumour necrosis factor-α receptor subtypes 1 and 2,20 K: cAMP-dependent protein kinase catalytic subunit alpha19), myomesin 2-ser76 (K: cAMP-dependent protein kinase catalytic subunit alpha,21 protein kinase cAMP-dependent regulatory type II-α-ser78 and cardiac troponin I-ser199) (K: cAMP-dependent protein kinase catalytic subunit alpha).22 The interaction of cardiac troponin I-Ser199 with its putative kinase has been shown to be inhibited by tropomyosin I,23 which was significantly higher in TOF samples, resulting in altered force of contraction.
Previous studies have shown some changes in calcium signalling in congenital heart disease, but it has not been possible to determine whether this is a generic response to RV hypertension or to specific cardiac physiologies.24 Proteins associated with calcium signalling were detected with adenylate cyclase 6, calcium/calmodulin-dependent protein kinase, phospholamban, ryanodine receptor 2 and sarcalumenin all being significantly higher in TOF. Phosphosites for phospholamban at Ser16, sarcoplasmic reticulum histidine-rich calcium-binding protein at Ser145 and junctophilin-2 at Thr490 were higher in TOF. However, cardiac troponin I at Ser199 was lower. The finding for phospholamban-Ser16 is similar to our previous studies and was linked to hypoxaemia.25 26
Although children with TOF clearly are more cyanosed, it is not clear how this would affect the RV. The biopsies are taken from the cavity of the RV that would normally see the venous (desaturated) blood. Systemic desaturation would not be expected to affect the endocardial surface of the RV.27 28 It should be noted that hypoxia-inducible factor-1-alpha (HIF-1α) was not detected in our proteomics analysis, despite oxygen saturation levels indicating hypoxia, especially in the TOF patients. It would be expected that HIF-1α be present in all our samples as has previously been described for CHD,29 suggesting the abundance was too low to detect with our protocol. However, we know that the RV is stiff after definitive cardiac surgery for TOF, which might relate to abnormalities of contractile proteins, secondary to an obstructed RV in comparison with the volume loaded RV of children with large post-tricuspid left to right shunt (VSD).30 For the first time, we have made a direct comparison between the hypertensive RV of children with VSD and the RV of children with TOF.
The novel findings for the rest of the proteins/phosphoproteins provide further evidence for maladaptation of the RV in cyanotic TOF patients with implications for increased stiffness after open-heart surgery. The lack of functional measurements, for instance, single permeabilised cardiomyocyte measurements to assess systolic and diastolic properties, calcium sensitivity and cross-bridge kinetics, make it difficult to understand the functional relevance of the observed differences in cellular signalling between the two patient groups. It would be helpful to understand this in future studies, but the amount of tissue available from such small children limits the possibility under the current approvals for this study. A large piece of RV tissue would be required, and there is some doubt that the digestion process required to achieve single cells might render the sample unrepresentative.
Comparison with known cardiac disease
Genes known to be affected in myocardial disorders with ventricular dysfunction (hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM) and arrhythmogenic right ventricular cardiomyopathy (ARVC) (KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathways map0510, map0514 and map05412 respectively31) were compared with our proteomics data. The majority of the affected proteins in these diseases were also present in our samples (11/15 HCM, 9/13 DCM, 5/6 ARVC); however, only tropomyosin-1 (normally affected in HCM and DCM, but not ARVC) was significantly higher in TOF samples. This suggests there was no downregulation of contractility as seen in maladaptation and right ventricular cardiomyopathy but more likely the upregulation due to increased myocardial wall stress.32
Conclusion
For the first time, it is now possible to suggest a cellular mechanism for the stiff RV in TOF, in that we have shown abnormalities in the calcium signalling pathways of the right ventricular myocardium. We have shown increased expression of myocardial contractile and extracellular proteins in TOF as compared with VSD, showing an adaptive physiology that is unrelated to age. We have shown that there is also parallel abnormality of clinical, echocardiographic and outcome data that might allow us, in future, to customise the treatment of the individual child in relation to their post-translational proteomics. It is too early to relate one finding directly to the other, but further work needs to be performed to determine the relationship between these important changes in RV hypertension in congenital heart disease.
Footnotes
Contributors: RMT, M-SG, SJG and MG designed the study and protocol. MC collected RV samples. CG performed and analysed the clinical and echocardiographic data. ARB, DI, SA-G and KH performed the laboratory analysis. RMT and AJB wrote the first draft, and all authors provided input to and approved the final version of the manuscript.
Funding: Sparks, the Childrens charity * 13BTL01. This study was supported by the NIHR Biomedical Centre at the University Hospitals Bristol NHS Foundation Trust and the University of Bristol. University Hospitals Bristol NHS Foundation Trust department of research and innovation number CH/2014/4571 provided sponsorship.
Disclaimer: The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.
Competing interests: None declared.
Patient consent: Detail has been removed from this case description/these case descriptions to ensure anonymity. The editors and reviewers have seen the detailed information available and are satisfied that the information backs up the case the authors are making.
Ethics approval: Ethical approval was granted by the National Research Ethics Service number 14/NW/1256, IRAS 143683.
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1. Knowles RL, Bull C, Wren C, et al. . Mortality with congenital heart defects in England and Wales, 1959-2009: exploring technological change through period and birth cohort analysis. Arch Dis Child 2012;97:861–5. 10.1136/archdischild-2012-301662 [DOI] [PubMed] [Google Scholar]
- 2. Murray CJ, Richards MA, Newton JN, et al. . UK health performance: findings of the global burden of disease study 2010. Lancet 2013;381:997–1020. 10.1016/S0140-6736(13)60355-4 [DOI] [PubMed] [Google Scholar]
- 3. Ghorbel MT, Cherif M, Jenkins E, et al. . Transcriptomic analysis of patients with tetralogy of Fallot reveals the effect of chronic hypoxia on myocardial gene expression. J Thorac Cardiovasc Surg 2010;140:337–45. 10.1016/j.jtcvs.2009.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qipshidze N, Tyagi N, Metreveli N, et al. . Autophagy mechanism of right ventricular remodeling in murine model of pulmonary artery constriction. Am J Physiol Heart Circ Physiol 2012;302:H688–96. 10.1152/ajpheart.00777.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaturvedi RR, Shore DF, Lincoln C, et al. . Acute right ventricular restrictive physiology after repair of tetralogy of Fallot: association with myocardial injury and oxidative stress. Circulation 1999;100:1540–7. 10.1161/01.CIR.100.14.1540 [DOI] [PubMed] [Google Scholar]
- 6. Agarkova I, Perriard JC. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol 2005;15:477–85. 10.1016/j.tcb.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 7. van der Ven PF, Obermann WM, Lemke B, et al. . Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton 2000;45:149–62. [DOI] [PubMed] [Google Scholar]
- 8. Lin C, Guo X, Lange S, et al. . Cypher/ZASP is a novel A-kinase anchoring protein. J Biol Chem 2013;288:29403–13. 10.1074/jbc.M113.470708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fürst DO, Goldfarb LG, Kley RA, et al. . Filamin C-related myopathies: pathology and mechanisms. Acta Neuropathol 2013;125:33–46. 10.1007/s00401-012-1054-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffmann C, Moreau F, Moes M, et al. . Human muscle LIM protein dimerizes along the actin cytoskeleton and cross-links actin filaments. Mol Cell Biol 2014;34:3053–65. 10.1128/MCB.00651-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buyandelger B, Ng KE, Miocic S, et al. . MLP (muscle LIM protein) as a stress sensor in the heart. Pflugers Arch 2011;462:135–42. 10.1007/s00424-011-0961-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reddy KB, Fox JE, Price MG, et al. . Nuclear localization of Myomesin-1: possible functions. J Muscle Res Cell Motil 2008;29:1–8. 10.1007/s10974-008-9137-x [DOI] [PubMed] [Google Scholar]
- 13. Lange S, Agarkova I, Perriard JC, et al. . The sarcomeric M-band during development and in disease. J Muscle Res Cell Motil 2005;26:375–9. 10.1007/s10974-005-9019-4 [DOI] [PubMed] [Google Scholar]
- 14. Granzier H, Labeit S. Cardiac titin: an adjustable multi-functional spring. J Physiol 2002;541(Pt 2):335–42. 10.1113/jphysiol.2001.014381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagueh SF, Shah G, Wu Y, et al. . Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation 2004;110:155–62. 10.1161/01.CIR.0000135591.37759.AF [DOI] [PubMed] [Google Scholar]
- 16. Chettimada S, Ata H, Rawat DK, et al. . Contractile protein expression is upregulated by reactive oxygen species in aorta of Goto-Kakizaki rat. Am J Physiol Heart Circ Physiol 2014;306:H214–24. 10.1152/ajpheart.00310.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hornbeck PV, Zhang B, Murray B, et al. . PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 2015;43:D512–20. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodriguez P, Mitton B, Nicolaou P, et al. . Phosphorylation of human inhibitor-1 at Ser67 and/or Thr75 attenuates stimulatory effects of protein kinase a signaling in cardiac myocytes. Am J Physiol Heart Circ Physiol 2007;293:H762–9. 10.1152/ajpheart.00104.2007 [DOI] [PubMed] [Google Scholar]
- 19. Valverde CA, Mundiña-Weilenmann C, Said M, et al. . Frequency-dependent acceleration of relaxation in mammalian heart: a property not relying on phospholamban and SERCA2a phosphorylation. J Physiol 2005;562(Pt 3):801–13. 10.1113/jphysiol.2004.075432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Defer N, Azroyan A, Pecker F, et al. . TNFR1 and TNFR2 signaling interplay in cardiac myocytes. J Biol Chem 2007;282:35564–73. 10.1074/jbc.M704003200 [DOI] [PubMed] [Google Scholar]
- 21. Obermann WM, van der Ven PF, Steiner F, et al. . Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band. Mol Biol Cell 1998;9:829–40. 10.1091/mbc.9.4.829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kooij V, Zhang P, Piersma SR, et al. . PKCα-specific phosphorylation of the troponin complex in human myocardium: a functional and proteomics analysis. PLoS One 2013;8:e74847 10.1371/journal.pone.0074847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wijnker PJ, Li Y, Zhang P, et al. . A novel phosphorylation site, Serine 199, in the C-terminus of cardiac troponin I regulates calcium sensitivity and susceptibility to calpain-induced proteolysis. J Mol Cell Cardiol 2015;82:93–103. 10.1016/j.yjmcc.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Y, Feng W, Zhang H, et al. . Ca²+-regulatory proteins in cardiomyocytes from the right ventricle in children with congenital heart disease. J Transl Med 2012;10:67 10.1186/1479-5876-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imura H, Caputo M, Parry A, et al. . Age-dependent and hypoxia-related differences in myocardial protection during pediatric open heart surgery. Circulation 2001;103:1551–6. 10.1161/01.CIR.103.11.1551 [DOI] [PubMed] [Google Scholar]
- 26. Modi P, Imura H, Caputo M, et al. . Cardiopulmonary bypass-induced myocardial reoxygenation injury in pediatric patients with cyanosis. J Thorac Cardiovasc Surg 2002;124:1035–6. 10.1067/mtc.2002.122536 [DOI] [PubMed] [Google Scholar]
- 27. Jeewa A, Manickaraj AK, Mertens L, et al. . Genetic determinants of right-ventricular remodeling after tetralogy of Fallot repair. Pediatr Res 2012;72:407–13. 10.1038/pr.2012.95 [DOI] [PubMed] [Google Scholar]
- 28. Lee W, Yoo SJ, Roche SL, et al. . Determinants and functional impact of restrictive physiology after repair of tetralogy of fallot: new insights from magnetic resonance imaging. Int J Cardiol 2013;167:1347–53. 10.1016/j.ijcard.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 29. Yin HL, Luo CW, Dai ZK, et al. . Hypoxia-inducible factor-1α, vascular endothelial growth factor, inducible nitric oxide synthase, and endothelin-1 expression correlates with angiogenesis in congenital heart disease. Kaohsiung J Med Sci 2016;32:348–55. 10.1016/j.kjms.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 30. Norgård G, Gatzoulis MA, Moraes F, et al. . Relationship between type of outflow tract repair and postoperative right ventricular diastolic physiology in tetralogy of Fallot. Implications for long-term outcome. Circulation 1996;94:3276–80. 10.1161/01.CIR.94.12.3276 [DOI] [PubMed] [Google Scholar]
- 31. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zagorski J, Sanapareddy N, Gellar MA, et al. . Transcriptional profile of right ventricular tissue during acute pulmonary embolism in rats. Physiol Genomics 2008;34:101–11. 10.1152/physiolgenomics.00261.2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
openhrt-2017-000716supp001.pdf (50.5KB, pdf)