Abstract
Synj2 (synaptojanin 2) encodes an inositol polyphosphate phosphatase that functions in recycling neurotransmitter vesicles and is implicated in spermatogenesis. Transcription of Synj2 is thought to occur from one of two promoters based on analysis of a variable 5′ untranslated region. Clustering all known mouse Synj2 transcripts led us to uncover a novel subset of transcripts that appears to derive from a region located within intron 7. We identified two alternate splice variants emanating from use of this promoter. These alternate splice variants manifest developmental stage specificity and somatic versus gametic differences in expression.
Introduction
Synaptojanins belong to a family of inositol polyphosphate phosphatases characterized by the presence of an N-terminal phosphatidylinositol polyphosphate 3′ and 4′ phosphatase domain similar to the yeast Sac1 domain and a central inositol polyphosphate 5′ phosphatase (IPP5Pase) domain [1, 2]. Mammals possess two paralogs, Synj1 and Synj2, encoding proteins that are approximately 55% identical over their Sac1-like and IPP5Pase domains but with divergent carboxyl termini [3, 4]. Synj1 was identified in rat as p145, a Grb2-binding brain protein enriched in presynaptic nerve terminals and also present in cultures of neuronal but not glial cells [5]. It accounts for the majority of phosphatidylinositol 5-phosphatase activity in adult rat brain lysates [6]. Synj1 functions in clathrin-mediated endocytosis via interactions with dynamin 1 [5], amphiphysin [7], endophilin [8, 9] and syndapin [10]. Synj1 knockout mice die perinatally and exhibit neurological defects including increased neuronal levels of phosphatidylinositol-4,5-bisphosphate and an accumulation of presynaptic clathrin-coated secretory vesicles. Synj2 knockout mice have not been reported. However, in vitro small interfering RNA experiments implicate human SYNJ2 in clathrin-coated receptor internalization and coated vesicle formation, both of which are drastically diminished in its absence [11]. Interestingly, overexpressing SYNJ2 rescues these defects, whereas overexpressing SYNJ1 does not [11], suggesting that the paralogs have different functions in receptor-mediated endocytosis.
In contrast to Synj1, rat Synj2 is ubiquitously expressed, albeit more so in brain and testis than in other tissues [3]. Synj2 undergoes extensive alternative splicing, mostly within the 3′ region. In mice, 7 splice variants (α – ζ) are reported [12, 13], whereas rats have three major splice variants (A, B1 and B2) [4]. In rats, these variants encode proteins with marked differences in subcellular localization and protein-protein interactions. For example, Synj2A (but not B1 or B2) is recruited to mitochondria by association with outer mitochondrial protein 25 where it functions in intracellular distribution of the organelle [14], whereas Synj2B1/B2 localize to the sperm manchette [4]. The manchette is a transient microtubule-derived structure implicated in nuclear elongation and chromatin organization during sperm maturation [15] and consistent with a role in spermatogenesis, our laboratory discovered multiple non-synonymous substitutions in the t haplotype allele of mouse Synj2 and linked these mutations with the phenomenon of t haplotype-specific male sterility [16]. Lastly, constitutive activation of Rac1, a small GTPase, causes translocation of human Synj2 from the cytoplasm to the plasma membrane via the C-terminus of Synj2 resulting in inhibition of receptor-mediated endocytosis [4, 17].
In mouse, transcription of Synj2 occurs from one of two promoters. The resulting transcripts differ in their 5′ untranslated region (UTR) but translation is initiated from the same start codon [12]. The potential for a third, downstream promoter was suggested by the identification of a novel transcript in the RIKEN collection [18] with a 5′ UTR mapping within the 3′ end of intron 7. A more comprehensive clustering of all known Synj2 transcripts onto the genomic locus identified a small subset of transcripts that originate within this intron. The resulting transcripts, including the RIKEN transcript AK019694, truncate most of the Sac1 homology domain yet leave intact the catalytic site encoded by this domain. In this report, we describe the promoter activity resident in intron 7. In addition, we demonstrate stage-specific and somatic versus gametic alternate splicing within the 5′ UTR derived from use of this promoter in the developing mouse testis. Our analysis identifies a novel promoter that does not conform to known canonical promoter sequences and may define a new class of RNA polymerase II promoter.
Materials and Methods
Mice
All experiments were conducted using B6SJL F1 male mice obtained from The Jackson Laboratory (Bar Harbor, ME). The day of birth was designated as postpartum day (dpp) 0. Mice were maintained under normal conditions. All procedures were approved by the Animal Care and Use Committee.
Northern Analysis and Probe Synthesis
A multiple tissue Northern blot of Balb/c-derived polyA mRNA (Clontech) was probed following the manufacturer’s protocol. The probe consisted of a 623bp fragment containing 273bp of intron 7, all of exons 8 and 9 and part of exon 10 (NM_011523) radiolabeled using RediPrime II (GE Healthcare) as per manufacturer’s protocol. Washes were performed at 60 °C in increasingly stringent solutions of SSC supplemented with 0.5% v/v SDS. Probe signal was detected by autoradiography.
RNA extraction and reverse transcription
Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer’s protocol. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). First-strand cDNA was synthesized from 2 μg of total RNA using MMLV reverse transcriptase (RT) from a RETROscript kit (Ambion) according to the manufacturer’s protocol. Negative controls were included.
Polymerase Chain Reaction (PCR) and Primer Extension
Standard PCR conditions were used (95 °C for one minute followed by 30 cycles of 95 °C for 30 sec., 55 °C for 30 sec., 72 °C for 30 sec.). Taq polymerase, buffer and dNTPs were purchased from Eppendorf. First-strand cDNA for primer extension was synthesized from total RNA primed with an exon 10-specific primer. 5′ rapid amplification of cDNA ends (RACE) was performed with the SMART RACE kit (Clontech) following the recommended protocol using a nested Synj2 exon 10-specific primer and the anchor primer provided with the kit.
Protein Extraction
All procedures were performed on ice except where noted. Mouse brain, liver and testis tissues from a wild-type male were harvested immediately after euthanasia, minced in chilled buffer AB (25 mM Tris pH 7.9, 12.5% glycerol, 2.5 mM MgCl2, 0.1 mM EDTA, 12 mM KCl, 10 mM 2-mercaptoethanol supplemented with protease inhibitor cocktail (SIGMA)) and homogenized using a rotor-stator. Prior to high-speed centrifugation to obtain the cytoplasmic fraction, polyethylenimine (PEI) was added to the homogenate in order to precipitate nucleic acid and nucleic acid-binding proteins. Cytoplasmic protein was isolated by centrifugation at 100,000x g for 1 hour at 4 °C (S100). The pellet was resuspended in chilled buffer C (50 mM Tris pH 7.9, 12.5% glycerol, 0.1 mM EDTA, 25 mM KCl, 10 mM 2-ME supplemented with protease inhibitor cocktail) and rocked at 4 °C for 30 minutes in order to extract soluble proteins precipitated by PEI. The PEI-insoluble fraction was separated from the particulate fraction by centrifugation at 100,000x g for 1 hour at 4 °C (PEI100). The remaining pellet was extracted with 1.5% v/v Triton X-100 in buffer C overnight at 4 °C followed by centrifugation at 100,000x g for 1 hour at 4 °C (P100). The S100, PEI100 and P100 fractions were flash-frozen in liquid N2 and stored at −80 °C until needed.
SDS-PAGE and Western Blotting
Proteins were resolved on 8.5% SDS-PAGE as previously described [19]. Electroblotting was performed as previously described [20] with the following modifications: proteins were blotted onto Immobilon-P (Millipore) in a transfer solution consisting of 15.6 mM Tris pH 8.2, 120 mM glycine, 20% v/v methanol and 0.1% v/v SDS. Analysis of Synj2 expression was performed with anti-Synj2 antibody (1:500 dilution; gift from P. De Camilli) recognizing a C-terminal epitope (amino acid 1255 to 1391) of rat Synj2 [4] followed by detection using an alkaline phosphatase-conjugated secondary antibody supplemented with NBT and BCIP (SIGMA).
Cell Culture and Luciferase Assay
All assays were performed using NIH-3T3 cells grown in disposable 16-well tissue-culture trays (Costar) to approximately 75–80% confluency. Cells were grown in enriched DMEM supplemented with 10% calf serum (Gibco) at 37 °C in 5% CO2. Candidate promoter fragments from intron 7 were PCR amplified and subcloned into pCR2.1 using the TOPO-TA kit (Invitrogen) per manufacturer’s instructions. Promoter fragments were removed by digesting with XhoI and HindIII and cloned into XhoI/HindIII-linearized pGL3-Basic (Promega). Each fragment was cloned into pGL3-Basic in both orientations. Six constructs were tested: 213 (+), 213 (−), 278 (+), 278 (−), 365 (+) and 365 (−). Stoichiometrically equal amounts of each construct were transfected in triplicate using Lipofectamine (Invitrogen) per manufacturer’s protocol. Cells were cultured for 24 hours followed by lysis in luciferase reagent (Bright-Glo; Promega). Luciferase activity was measured in an HT Synergy plate reader (Bio-Tek) with a gain settingof 200. Data were analyzed in Excel (Microsoft Corp.)
Results
Truncated Synj2 transcripts map to intron 7
Synj2 transcripts derive from one of two promoters based on the finding of two distinct 5′ UTRs in full-length transcripts [12]. While attempting to estimate the relative preference for these promoters based on the number of times each 5′ UTR recurs in the EST dataset at NCBI, we identified three out of 204 variants (UniGene Mm.236068, UGID: 324106) that have 5′ UTRs mapping to intron 7. Of these, transcript AK019694 [18] had the longest 5′ UTR whereas transcripts AK161467 and AY823997 were shorter variants of AK019694 (Supplemental Fig. 1). We interpreted this to mean that intron 7 harbors a promoter.
To determine if these transcripts originate from a promoter within intron 7, we performed RT-PCR with primers specific to the 5′ UTR of AK019694. Whereas primers designed to the 5′ most region of AK019694 consistently failed to amplify a product, primers designed to more distal regions of the 5′ UTR produced an amplicon (Figure 1A). Next, we isolated an ~530bp fragment by 5′ RACE (Figure 1B). Based on sequence analysis, the 5′ end of this transcript maps within intron 7 at nucleotide 2181 (the length of intron 7 is 2518bp). We refer to this transcript as Synj2DSac1.
Figure 1.
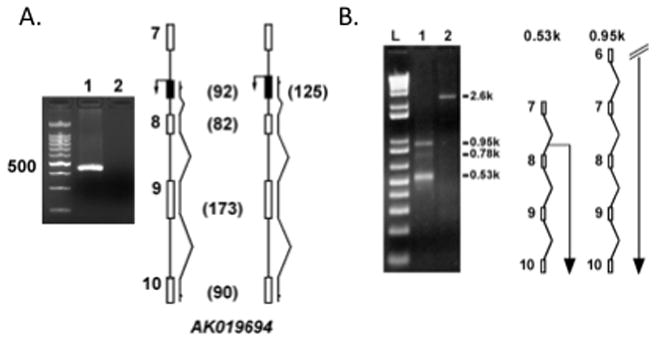
5′ UTR mapping analysis. A: Staggered primers to the 5′ UTR of AK019694 were used in combination with an exon 10 primer to amplify cDNA transcripts derived from poly dT-primed reverse transcription reactions. Primers within the first 30 nucleotides of AK019694 consistently failed (lane 2 and right diagram) whereas internal primers produced an amplicon of the expected size (lane 1 and left diagram). B: 5′ RACE produced a ~530bp product (Lane 1) whose 5′ end mapped to intron 7. Larger fragments were observed, corresponding to transcripts most likely derived from the far-upstream promoters. Lane 2 is the human transferrin receptor positive control.
Tissue-Specific Synj2 Expression
Rat Synj2 is expressed in several adult tissues, with the exception of liver [3]. In testis, the two most prominent transcripts are 5.2 and 3.5kb and, to a much lesser extent, a 7.5kb transcript that is prevalent in other tissues is faintly observed [3]. To determine transcript sizes and tissue expression profiles and to identify alternate splice variants of mouse Synj2, we probed a multiple tissue Northern blot (Clontech). The probe consisted of a fragment conserved in all reported alternate splice variants, with the exception that it included the 3′-most 273bp of intron 7. In contrast to rat [3], Synj2 expression was observed in all tissues, including liver, except for skeletal muscle (Supplemental Fig. 2). A 1.7kb transcript occurred most frequently although alternatively spliced variants were observed in heart and testis, and to a lesser extent in brain and liver. In testis, four transcript sizes were observed, corresponding to 0.9, 1.7, 3.3 and 3.8kb, whereas significantly larger transcript variants (≤ 7.5kb) were observed in heart and brain
Synj2 Partitions into Soluble and Particulate Fractions
Prior studies provided evidence that rat Synj2 partitions into the particulate fraction [3] whereas others have shown that human SYNJ2 cycles between the cytoplasm and plasma membrane via interaction with Rac1 [17]. In order to determine the partitioning properties of mouse Synj2, we fractionated proteins into cytoplasmic and membrane-bound fractions. In addition, because Synj2 contains an RNA-binding motif (RP1; IPR000504), we co-precipitated nuclear components with the membrane fraction by addition of polyethylenimine, a positively charged polyelectrolyte that precipitates nucleic acids and nucleic acid-binding proteins [21]. Thereafter, we investigated the partitioning of Synj2 into cytoplasmic, nuclear or particulate fractions by Western blot analysis using an antibody to a carboxyl terminus 137 amino acid epitope of rat Synj2B [4]. Two isoforms of Synj2 were observed in brain: one with a molecular mass of 160 kDa in the S100 and PEI100 fractions, and a doublet centered at approximately 116 kDa in the pellet fraction (Figure 2). The 116-kDa isoform corresponds to the predicted size encoded by Synj2 DSac1. Synj2 was not detected in either testis or liver (data not shown) even though transcription is observed by Northern blot (Supplemental Fig. 2). The most plausible explanation is that the epitope detected by the antibody is absent from the testis variant of mouse Synj2 because exons encoding the epitope are targets of alternative splicing.
Figure 2.
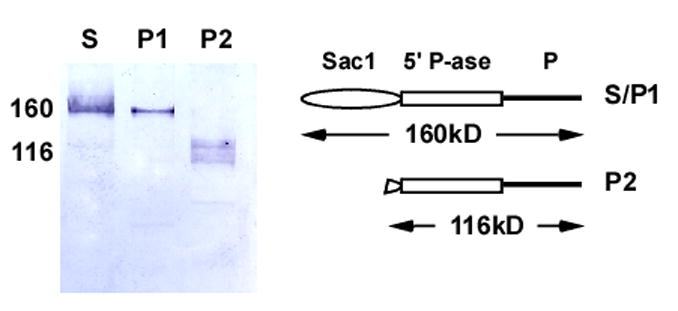
Western analysis of soluble and particulate fractions from brain lysates. The soluble and PEI-precipitated isoforms of Synj2 are indistinguishable (S, P1) whereas the membrane-associated isoform (P2) migrates at ~120 kDa as a doublet. A model representing the two isoforms is shown to the right.
Differential expression of Synj2 in somatic versus gametic cells
Synj2 is thought to be required during spermatogenesis or for normal sperm function [4, 16]. The mitotic phase of mouse spermatogenesis begins at ~7 dpp. Dividing spermatogonia transition into meiosis at ~11 dpp and spermatids are observed by ~21.5 dpp whereas mice are reproductively mature at ~35 dpp. During these stages, changes in the number and architecture of the somatic cells (Sertoli cells) that nourish the developing sperm are also occurring. To assess expression of Synj2 DSac1 during spermatogenesis, we performed RT-PCR on 7 dpp (mitosis), 14 dpp (zygotene), 18 dpp (pachytene), 21 dpp (meiosis II), 28 dpp (post-meiotic spermatid), 35 dpp (maturation) and adult testis-derived mRNA. We also assayed expression of Synj2 DSac1 in Sertoli cells derived from spermatogonia-depleted adult Kitw/wv mice [22]. As shown in Figure 3, Synj2 DSac1 is expressed at the earliest stage (7 dpp). At 14 dpp, a smaller amplicon coamplifies to a lesser extent with the larger amplicon seen exclusively at 7 dpp and by 21 dpp becomes the prominent transcript. In the testis of wild type mice, only the smaller amplicon is expressed at 28 and 35 dpp, and in adults; whereas in Kitw/wv testes, which are spermatogonia depleted, only the larger amplicon is expressed. The simplest interpretation of this result is that this variant of Synj2 DSac1 is somatic cell-specific whereas the smaller variant is germ cell-specific. The difference between the two variants is due to alternative splicing of the intron 7-derived 5′ UTR.
Figure 3.
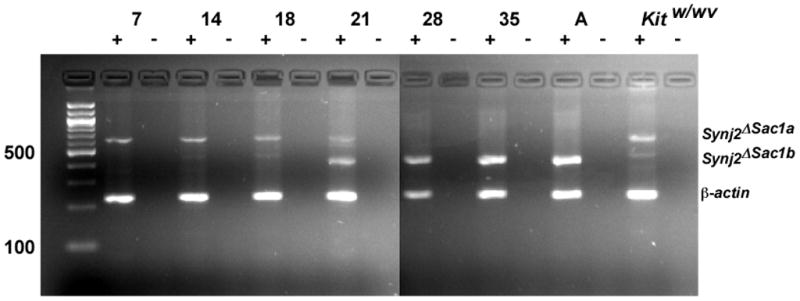
Developmental and cell-type differential expression of Synj2 DSac1. Testis at different stages of development were assayed for Synj2 DSac1 expression. A switch in alternate transcripts is seen at ~21 dpp. The larger transcript is somatic cell-specific whereas the smaller transcript appears to be germ cell-specific.
Promoter activity resides within intron 7
We tested the promoter activity of various intron 7 fragments. First, we cloned a 365bp fragment of intron 7, beginning at nucleotide 2154 and extending to the junction with exon 8, into pGL3-Basic (Promega) in both orientations. Successively smaller fragments (273 and 213bp) were also cloned in in both orientations. In all, 8 constructs were tested – 2 for each of the 3 promoter fragments in the plus and minus orientations, the empty vector (pGL3-Basic) and a positive-control (pGL3-Control). All constructs were transfected in stoichiometrically equal amounts by lipofection into NIH-3T3 cells in triplicate. After 24 hours, the cells were lysed in luciferase assay solution and luminosity measured for each construct. As seen in Figure 4, the larger construct (365bp) expressed luciferase at a level approximately 20 percent of the constitutively active control, pGL3-Control. Approximately 3–4 fold less activity was observed from the smaller constructs (273 and 213bp). Importantly, luciferase activity was observed only for the promoter fragments in the plus orientation whereas luminosity from the minus orientation did not differ from the empty vector control. Computational analysis of the 365bp promoter fragment failed to identify canonical promoter sequences although several transcription factor binding sites were identified (Table 1 and Supplemental Fig. 3), including GC-box binding sites for SP1 and members of the Zfx and Zfy transcription factor family implicated in mammalian sex determination. These results demonstrate that intron 7 harbors a promoter.
Figure 4.
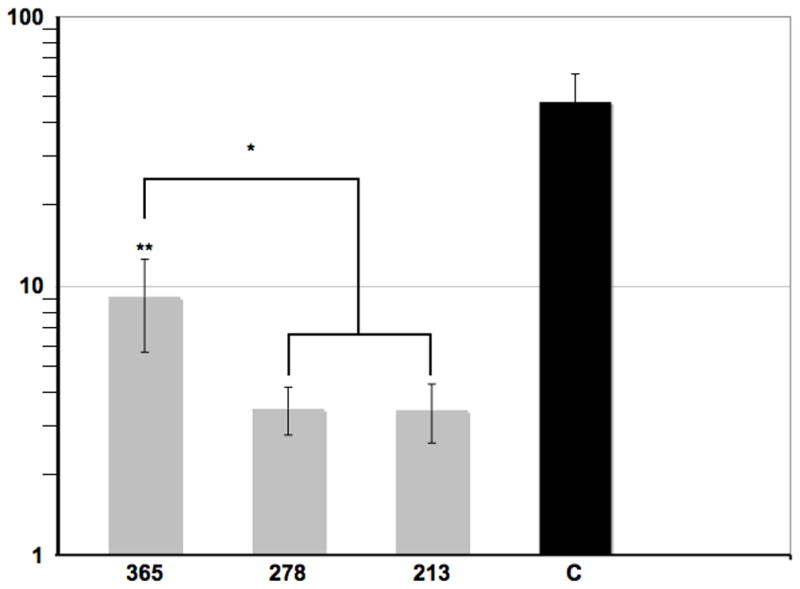
Luciferase assay. Three overlapping fragments (365bp, 273bp and 213bp) of intron 7 were cloned separately into a luciferase reporter construct and transfected into NIH-3T3 cells. Luminosity was measured and compared to a control luciferase (C) driven by an SV40 promoter. All assays were performed in triplicate and error bars are SD. Asterisks indicate significance (*, p ≤ 0.05; **, p ≤ 0.01).
Table 1.
Distribution of transcription factor binding sites in the Intron 7 promoter
| Factor Name | Anchor1 | Strand2 |
|---|---|---|
| Calsenilin, presenilin binding protein, EF hand transcription factor | 241 | + |
| Cellular and viral Myb-like transcriptional regulators | 7 | − |
| E-box binding factors | 26 | + |
| EGR/nerve growth factor induced protein C & related factors | 191, 193 | − |
| EVI1-myleoid transforming protein | 348 | − |
| GC-Box factors SP1/GC | 188, 194, 323 | − |
| Human and murine ETS1 factors | 310, 348 | +/− |
| Krueppel like transcription factors | 190, 192, 194,231 | − |
| Myc associated zinc fingers | 193 | − |
| Myeloid zinc finger 1 factors | 195 | − |
| Myoblast determining factors | 35, 36 | +/− |
| MYT1 C2HC zinc finger protein | 314 | − |
| NeuroD, Beta2, HLH domain | 154 | + |
| Nuclear factor kappa B/c-rel | 94, 95 | +/− |
| Pur-α | 190 | − |
| RXR heterodimer binding sites | 58, 244 | −/+ |
| v-ERB and RAR-related orphan receptor alpha | 239 | + |
| Vertebrate homologues of enhancer of split complex | 27 | + |
| Vertebrate SMAD family of transcription factors | 204, 209, 334 | +/−/+ |
| Zfx and Zfy-transcription factors implicated in mammalian sex determination | 248 | − |
| Zinc binding protein factors | 190, 193 | + |
MatInspector software from Genomatix (www.genomatix.de) was used to analyze the 365bp promoter fragment. Only matches with a Matrix Similarity Score ≥ 0.9 are shown.
Anchor corresponds to the center nucleotide position of a binding site; coordinates are from the 365bp promoter fragment.
Strand corresponds to orientation within the gene: + 5′ → 3′, − 3′ → 5′.
Synj2DSac1 encodes a truncated isoform of the full-length protein
We analyzed the 5′ sequence of Synj2DSac1 to identify the putative translation start site and determined that the first in-frame start codon is located at nucleotide 40 of exon 8 (Supplemental Fig. 3). The predicted AUG, surrounded by an imperfect Kozak consensus sequence (ACCUAUGA) [23], is in-frame with the remaining Synj2 coding sequence and predicts an ~120-kDa protein consistent with the size observed by Western analysis (Figure 2). This newly predicted N-terminus is 54 amino acid residues upstream of the catalytic site of the Sac1 homology domain [1].
Discussion
We demonstrate the feasibility of a straightforward strategy designed on the premise that clustering EST and mRNA sequences onto the genomic locus of origin can identify novel promoters. It is accepted that enhancer and repressor elements are frequently found within introns. Alternative or cryptic promoters are often located in the first or second intron of a gene and when used in place of the upstream promoter, produce variation in the amino terminus or result in novel 5′ UTRs. The previously identified transcription start sites in Synj2 illustrate the latter case [12]. However, promoters located in distal introns that produce mRNA transcripts encoding truncated isoforms of full-length proteins are not as common, but occur [24–27]. Use of these promoters might lead to novel ways of controlling protein function. For example, they may affect allosteric control by the omission of regulatory domains, or change substrate specificity in the case of enzymes; modify protein complex formation by disrupting protein-protein interactions; or alter the spatial activity of the protein by changes in subcellular targeting.
We have shown that the fragment of intron 7 of Synj2 harbors a promoter located >65kb downstream of the canonical promoter. Computational analysis did not identify hallmark eukaryotic promoter sequences, such as TATA box or Inr [28], although a number of transcription factor binding sites were strongly predicted, including sites for GC-box binding proteins, NfκB and Smad. The significance of these sites remains unknown. The promoter is moderately active relative to a constitutively active, strong promoter such as SV40.
Synj2 is a member of a rare class of enzymes having distinct but related catalytic activities located within separate domains. The N-terminal domain is a Sac1 homology domain homologous to the yeast Sac1p protein [29]. It encodes a phosphatidylinositol polyphosphate phosphatase that converts phosphatidylinositoI-3-phosphate (PI3P), PI4P and PI4,5P2 to PI [1]. The catalytic site of this domain belongs to a family of phosphatases that includes the Yersinia pestis tyrosine phosphatase (Yop51) [30], and the bovine acid phosphatase 1 (Acp1; LMW-PTP) [31]. It is characterized by the presence of consensus catalytic residues, CX5RS/T, located in the PTP-loop. Our studies that Synj2DSac1 truncates a significant fraction of the Sac1 homology domain without deleting the catalytic site residues. Whether or not this catalytic site is functional remains to be determined.
The alternate splice variants of Synj2DSac1 produced by germ cells versus somatic cells may indicate cell-type specific differential expression of splice factors or accessory proteins. This phenomenon could lead to synthesis of germ cell-specific and somatic cell-specific protein isoforms that perform distinct functions. It is revealing that the switch from high to low transcript sizes for Synj2DSac1 occurs at the tail-end of prophase I of meiosis as seen in Figure 3, possibly reflecting the need to transition from meiosis to a transcriptionally arrested spermatid. The absence of detectable Synj2 and any of its isoforms in mouse testis by Western blot was unexpected, although its absence may be due to alternative splicing in the 3′ region of the gene resulting in a loss of the epitope, consistent with the large transcript variation observed at this locus.
Supplementary Material
We characterized a novel promoter within intron 7 of synj2
We delineate the region of this promoter having the highest transcriptional activity
We demonstrate that transcripts from this promoter are differentially expressed in the testis
We correlate these transcripts to differences in protein isoforms
Acknowledgments
I thank Pietro De Camilli for the anti-Synj2 antibody, Chris Smith for DNA sequencing and Andrew Christie for insightful comments on the manuscript. I also thank Jeffrey Calhoun, William Frels, Nataliya Ilyashenko and Ashley Wolf for technical assistance. This work was supported by a grant from the INBRE program of the National Institutes of Health (P20 RR016463).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guo S, et al. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–5. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 2.McPherson PS, et al. A presynaptic inositol-5-phosphatase. Nature. 1996;379:353–7. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- 3.Nemoto Y, et al. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J Biol Chem. 1997;272:30817–21. doi: 10.1074/jbc.272.49.30817. [DOI] [PubMed] [Google Scholar]
- 4.Nemoto Y, et al. Identification and characterization of a synaptojanin 2 splice isoform predominantly expressed in nerve terminals. J Biol Chem. 2001;276:41133–42. doi: 10.1074/jbc.M106404200. [DOI] [PubMed] [Google Scholar]
- 5.McPherson PS, et al. p145, a major Grb2-binding protein in brain, is co-localized with dynamin in nerve terminals where it undergoes activity-dependent dephosphorylation. J Biol Chem. 1994;269:30132–9. [PubMed] [Google Scholar]
- 6.Woscholski R, Waterfield MD, Parker PJ. Purification and biochemical characterization of a mammalian phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase. J Biol Chem. 1995;270:31001–7. doi: 10.1074/jbc.270.52.31001. [DOI] [PubMed] [Google Scholar]
- 7.David C, et al. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci U S A. 1996;93:331–5. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Heuvel E, et al. Identification of the major synaptojanin-binding proteins in brain. J Biol Chem. 1997;272:8710–6. doi: 10.1074/jbc.272.13.8710. [DOI] [PubMed] [Google Scholar]
- 9.Ringstad N, Nemoto Y, De Camilli P. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc Natl Acad Sci U S A. 1997;94:8569–74. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qualmann B, et al. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–13. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rusk N, et al. Synaptojanin 2 functions at an early step of clathrin-mediated endocytosis. Curr Biol. 2003;13:659–63. doi: 10.1016/s0960-9822(03)00241-0. [DOI] [PubMed] [Google Scholar]
- 12.Seet LF, et al. Molecular cloning of multiple isoforms of synaptojanin 2 and assignment of the gene to mouse chromosome 17A2–3.1. Biochem Biophys Res Commun. 1998;247:116–22. doi: 10.1006/bbrc.1998.8564. [DOI] [PubMed] [Google Scholar]
- 13.Khvotchev M, Sudhof TC. Developmentally regulated alternative splicing in a novel synaptojanin. J Biol Chem. 1998;273:2306–11. doi: 10.1074/jbc.273.4.2306. [DOI] [PubMed] [Google Scholar]
- 14.Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. Embo J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange BM, Gull K. A molecular marker for centriole maturation in the mammalian cell cycle. J Cell Biol. 1995;130:919–27. doi: 10.1083/jcb.130.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schimenti JC, Reynolds JL, Planchart A. Mutations in Serac1 or Synj2 cause proximal t haplotype-mediated male mouse sterility but not transmission ratio distortion. Proc Natl Acad Sci U S A. 2005;102:3342–7. doi: 10.1073/pnas.0407970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malecz N, et al. Synaptojanin 2, a novel Rac1 effector that regulates clathrin-mediated endocytosis. Curr Biol. 2000;10:1383–6. doi: 10.1016/s0960-9822(00)00778-8. [DOI] [PubMed] [Google Scholar]
- 18.Kawai J, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–90. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dissing U, Mattiasson B. Polyelectrolyte complexes as vehicles for affinity precipitation of proteins. J Biotechnol. 1996;52:1–10. doi: 10.1016/s0168-1656(96)01594-5. [DOI] [PubMed] [Google Scholar]
- 22.Nocka K, et al. Molecular bases of dominant negative and loss of function mutations at the murine c-kit/white spotting locus: W37, Wv, W41 and W. Embo J. 1990;9:1805–13. doi: 10.1002/j.1460-2075.1990.tb08305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990;87:8301–5. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abuladze N, et al. Structural organization of the human NBC1 gene: kNBC1 is transcribed from an alternative promoter in intron 3. Gene. 2000;251:109–22. doi: 10.1016/s0378-1119(00)00204-3. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Lunec J. Characterisation of a novel p53 down-regulated promoter in intron 3 of the human MDM2 oncogene. Gene. 2005;361:112–8. doi: 10.1016/j.gene.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Rohrschneider LR, et al. The intron 5/6 promoter region of the ship1 gene regulates expression in stem/progenitor cells of the mouse embryo. Dev Biol. 2005;283:503–21. doi: 10.1016/j.ydbio.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 27.Yu S, et al. Pituitary tumor AP-2alpha recognizes a cryptic promoter in intron 4 of fibroblast growth factor receptor 4. J Biol Chem. 2003;278:19597–602. doi: 10.1074/jbc.M212432200. [DOI] [PubMed] [Google Scholar]
- 28.Fickett JW, Hatzigeorgiou AG. Eukaryotic promoter recognition. Genome Res. 1997;7:861–78. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- 29.Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–50. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuckey JA, et al. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature. 1994;370:571–5. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 31.Su XD, et al. The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase. Nature. 1994;370:575–8. doi: 10.1038/370575a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


