Abstract
Multiple sclerosis (MS) is a debilitating neurological disease whose onset and progression are influenced by the interplay of genetic and environmental factors. Epigenetic modifications, which include post-translational modification of the histones and DNA, are considered mediators of gene-environment interactions and a growing body of evidence suggests that they play an important role in MS pathology and could be potential therapeutic targets. Since epigenetic events regulate transcription of different genes in a cell type-specific fashion, we caution on the distinct functional consequences that targeting the same epigenetic modifications might have in distinct cell types. In this review, we primarily focus on the role of histone acetylation and DNA methylation on oligodendrocyte and T cell function and its implications for MS. We find that decreased histone acetylation and increased DNA methylation in oligodendrocyte lineage (OL) cells enhance myelin repair, which is beneficial for MS, while the same epigenetic processes in T cells augment their pro-inflammatory phenotype, which can exacerbate disease severity. In conclusion, epigenetic-based therapies for MS may have great value but only when cellular specificity is taken into consideration.
Introduction
Increasing evidence supports the role of environmental factors on Multiple Sclerosis (MS) disease onset and course [1], yet the underlying mechanisms remain poorly understood. Epigenetics defines the study of histone and DNA modifications that affect gene expression after environmental exposure [2]. Because epigenetics is regulated in a strictly cell-specific manner, the same modification might bear distinct functional outcomes in different cell types. MS is characterized by immune dysregulation and demyelination [3]. This review will discuss epigenetics studies on immune and oligodendrocyte lineage cells, in the context of the disease. Due to space limitations we refer the readers to several excellent reviews on the general concepts of epigenetics [2],[4],[5]. Nuclear DNA in cells is wrapped around histone proteins into nucleosomal structures, which allow critical amino acids in the histone tails to be accessible to enzymes catalyzing post-translational modifications, with distinctive functional outcomes [2].
Histone deacetylation and myelin repair
Histone acetylation is regulated by two opposing enzymatic activities: histone acetyltransferases (HATs) are the “writers” as they deposit acetyl groups onto lysine residues, while histone deacetylases (HDACs) are the “erasers” as they remove them. HATs include: KAT2A, KAT2B, KAT6–8, CREBBP, and EP300 [2]. HDACs can be grouped into 4 classes: class I, II, and IV HDACs catalyze deacetylation in a zinc-dependent manner while class III includes NAD+-dependent HDACs [2].
In oligodendrocyte progenitors, the removal of acetyl marks by HDACs is essential for proper differentiation and myelin formation [6] during developmental myelination [7] and remyelination [8]. In the developing brain, the differentiation of progenitors into oligodendrocytes is initially characterized by the occurrence of global histone deacetylation [7] due to HDAC1 and HDAC2 activity [8],[9]. Histone deacetylation was shown to be critical during the first two postnatal weeks and repress progenitor genes to initiate the expression of differentiation markers. Administration of class I HDAC pharmacological inhibitors during the neonatal period was characterized by impaired developmental myelination, characterized by down-regulated expression of myelin genes, reduced number of myelinated fibers and accumulation of progenitor cells compared to untreated controls. The arrested differentiation of progenitors could be recovered by allowing a recovery period after suspension of the treatment with the pharmacological inhibitors. Genetic ablation of Hdac1 and Hdac2 during early embryonic development had even more dramatic consequences, resulting in severe hypomyelination [10]. Similarly, in adult brains histone deacetylation was shown to occur during the first steps of myelin repair and to be necessary for differentiation of progenitors into myelinating oligodendrocytes [8]. Using the cuprizone model of demyelination consequent to oligodendrogliopathy, remyelination was shown to be characterized by the HDAC1 mediated repression of genes serving as “transcriptional brakes” (e.g. SOX2 and HES5) of myelin genes. Indeed, HDAC inhibitors impaired remyelination efficiency and this could be attributed to a failure to remove acetyl histone marks at the promoters of Sox2 and Hes5, which resulted in increased binding of RNA Pol II to these regions and increased transcription [8]. To define whether these findings could inform the discovery of therapeutic targets, immunohistochemical studies were also conducted on post-mortem MS brains. In normal appearing white matter (NAWM) of MS brains, acetyl H3 marks were found to have an opposite pattern to that described for normal brain development, and correlated with increased HAT levels (e.g. CBP and P300) and unchanged HDAC levels. In the same brains, expression of TCF7L2, a downstream Wnt effector and negative regulator of oligodendrocyte differentiation [11], was also increased and this correlated with histone acetylation at the promoter region [12]. It is important to note that impaired histone acetylation was detected in NAWM rather than at lesions and peri-lesions and suggested dysregulated homeostatic balance resulting from increased HAT activity. Presumably strategies decreasing HAT activity or interfering with its downstream effectors, might be considered as potential therapeutic targets [13],[14].
Histone acetylation in T cell activation
While HDAC-mediated oligodendrocyte differentiation may play a beneficial role in MS, HDAC-mediated activation of T cells may initiate pathogenesis and exacerbate disease severity. HDAC transcripts increase with immune cell activation [15] suggesting a regulatory role for these enzymes. Specifically, HDACs were shown to promote Th1 differentiation, as their inhibition suppressed production of IFN gamma [16] and promoted differentiation into regulatory T cells (Treg) by upregulation of FOXP3 [17]. Since imbalanced ratio of effector T cells (Teff) to Treg is characteristic of MS pathogenesis [18], HDAC inhibitors have been proposed as therapeutic strategies to regain immune homeostasis. Several studies using Experimental Autoimmune Encephalitis (EAE), highlighted the therapeutic potential of HDAC inhibitors [19]–[21]. When administered prophylactically (pre-clinical) or semi-therapeutically (at onset), these inhibitors improved clinical disability and decreased clinical severity. The treatment also reduced inflammation and immune cell infiltration in the spinal cord, thereby reducing the extent of demyelination and axonal damage. While distribution of inflammatory infiltrates was not assessed, treated mice had lower Th1 and Th17 cell counts and higher Th2 and Treg cell numbers, suggesting a predominant effect on T cell mediated demyelination [19]–[21].
Therefore HDAC inhibitors alleviated EAE disease severity by reducing pathogenic T cell activation and/or differentiation.
Links between histone acetylation and DNA methylation
Epigenetic modifications work in synchrony to organize chromatin structure and alter gene expression. Specifically, histone modifications and DNA methylation bi-modally influence each other to implement a unidirectional change in gene expression. Histone acetylation and unmethylated DNA are often found at promoters of actively transcribed genes. In contrast, deacetylation or repressive methylation of histones and DNA are found at the promoters of repressed genes [22]. Here, we present evidence related to the role of the DNA methyltransferase enzymes (DNMTs) in modulating myelin formation and T cell function.
“Writers” and “erasers” of DNA methylation
DNA methylation refers to the addition of methyl groups to cytosines. If occurring near transcriptional start sites, DNA methylation prevents transcription factor binding, resulting in stable transcriptional repression. Gene-body methylation, in contrast, is less well understood and could potentially affect transcription through regulation of splicing events and recruitment of chromatin modifiers [23]. “Writers” of DNA methylation include the DNA methyltransferases (DNMTs): DNMT1, DNMT3a and DNMT3b. Canonically, DNMT1 is involved in maintenance methylation, which guarantees faithful transmission of methylation marks from mother to daughter cells. By contrast, DNMT3a and DNMT3b are involved in de novo methylation or the establishment of new methylation marks on previously unmethylated regions [23]. DNA methylation can be “erased” passively or actively. Passive DNA demethylation occurs in the absence of methylation by DNMT1 onto newly synthesized DNA. On the other hand, active DNA demethylation is carried out by the ten-eleven-translocation enzymes (Tets), Tet1-3. Demethylation is initiated by the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which can be further converted into 5-formylcytosine (5fC) and then 5-carboxylcytosine (5caC) for complete removal from DNA by thymine-DNA glycosylase (TDG). While 5hmc is considered a transient intermediary step towards complete DNA demethylation, high 5hmc levels in certain tissues such as the brain indicate it may be a more stable and integral part of the epigenetic code [24]. We review the role of DNMTs in oligodendrocyte and immune cell function and discuss the implications for MS.
DNA methylation in myelin repair: evidence of a beneficial role in MS
Similar to histone deacetylation, DNA methylation is critical for late stage oligodendrocyte differentiation in neonatal and adult mice. During brain development, oligodendrocyte progenitor differentiation into myelinating oligodendrocytes is characterized by global DNA hypermethylation and upregulation of Dnmt1. These changes are necessary to coordinate the initiation of a transcriptional program with exit from the cell cycle. Lineage specific ablation of Dnmt1 in progenitor cells arrested proliferation and impaired differentiation [25], resulting in dramatic hypomyelination, which was manifested by impaired motor coordination and decreased survival by the third postnatal week. Surprisingly, despite defective proliferation and differentiation, progenitors lacking Dnmt1 did not undergo cell death. Rather, they activated an ER stress response possibly due to the accumulation of aberrant splicing events, demonstrating a multifaceted role for Dnmt1 in the regulation of transcription [25]. DNA methylation was shown to be equally important for oligodendrocyte progenitor differentiation following a demyelinating event in the adult brain. Dnmt1 was primarily detected in OPCs whereas Dnmt3a was mainly detected in differentiated oligodendrocytes. Loss of Dnmt1 and Dnmt3a impaired differentiation into mature oligodendrocytes following lysolecithin-induced focal demyelination and resulted in thinner myelin and inefficient remyelination [26]. Interestingly, loss of a single Dnmt was less dramatic than loss of both Dnmt1 and Dnmt3a, suggesting compensatory or redundant roles. Deletion of Dnmt1 reduced proliferation only in the developing but not in the adult brain. Since the neonatal and adult studies employed different genetic strategies and used distinct lines of Cre recombinase targeted to early progenitors (i.e. Olig1+ cells) or newly formed oligodendrocyte (i.e. Plp+ cells), there is a possibility that the effects on cell cycle may simply reflect the distinct proliferative potential of these two targeted cell populations. Future studies comparing the contribution of DNA methylation to neonatal and adult progenitor function are needed to fully address these issues.
Genome-wide studies in MS patients suggest that aberrant DNA methylation may also contribute to some neuropathological features. Analysis of genome-wide DNA methylation changes in NAWM of MS brains revealed potential DNA hypermethylation in oligodendrocytes. Specifically, several genes specific to mature myelinating oligodendrocytes including MBP, NDRG1, and BCL2L2 showed DNA hypermethylation that was associated with decreased mRNA and protein expression. Additionally, some of the hypermethylated genes, such as NDRG1 and BCL2L2, are implicated in oligodendrocyte survival, suggesting a role for DNA methylation in resilience to myelinotoxic factors [27]. The current literature suggests that enhancement of DNMT activity in oligodendrocyte progenitors but not myelinating oligodendrocytes may serve as an effective therapeutic strategy for myelin repair in MS.
DNA methylation in immune cell activation: evidence of a detrimental role in MS
DNMT inhibitors ameliorate disease course in EAE models, suggesting their therapeutic potential for treatment of MS. Low doses of the DNMT inhibitor 5-aza-2′-deoxycytidine (5-aza) administered prophylactically completely prevented EAE onset when mice were immunized with a mild dose of myelin peptide and significantly reduced disease severity in mice immunized with a higher dose of myelin peptide [28],[29]. More interestingly, administration of 5-aza therapeutically (at onset) also reduced disease severity. In both models, the distribution of T cell subsets was explored and revealed reduced Th1 and Th17 cell numbers, reduced immune cell infiltration and demyelination in the spinal cord of 5-aza treated mice. In these mice, the promoter of Foxp3 was hypomethylated, thereby suggesting upregulated transcript levels and a potential shift towards regulatory T cells (Tregs). 5-aza treatment also affected effector (Teff) cell function. A comparison of T cells isolated from EAE mice either untreated or treated with 5-aza revealed differential effects in a suppression assay of T cell proliferation, evaluating Treg induced suppression of Teff function. While the function of Teff isolated from untreated EAE control mice was not suppressed by Tregs, those Teff cells isolated from the 5-aza treated EAE mice were suppressed by both Tregs isolated from untreated controls or from 5-aza treated EAE mice. Therefore 5-aza directly affected Teff cell function but had no effect on Treg function [28],[29].
These studies suggest DNA methylation as a key element skewing the Teff to Treg ratio in MS patients [30]–[32]. However, to date, no large DNA methylation changes in CD4 T cells have been identified. A genome-wide study on CD4 T cells from monozygotic twins, did not detect prominent and consistent differences in DNA methylation between affected and unaffected siblings [32]. This finding was striking since it highlighted an important element of epigenetic variability, suggesting heterogeneous patterns of silencing in distinct twin pairs. Given the relatively small sample size of the study, additional twin studies would be necessary to evaluate the existence of common findings or shared pathways.
Case-control studies on DNA methylation performed in CD4 T cells from MS patients using whole genome CpGs arrays revealed only minor differences in DNA methylation and reached distinct conclusions based on the type of analyses conducted [30],[33]. Graves et al, for instance, included the analysis of CpG within SNPs and highlighted the enrichment of differential DNA methylation in the MHC region. The identification of hypomethylation in regions containing MS risk alleles, such as HLA-DRB5, HLA-DRB6, and HLA-DQB1 [31], was interpreted as suggestive of a role of DNA methylation in favoring expression of genetic susceptibility traits. Bos et al, in contrast, removed these regions from the analysis, in order to reduce genetic variation [30]. To further define the relationship between the haplotype DRB1*1501 and the possibility of bearing an “MS-like” DNA methylation signature, Graves et al evaluated the association between differentially methylated CpGs and expression of the MS risk haplotype. Regardless of disease status the majority of “MS-associated” CpGs displayed DNA hypomethylation in risk haplotype carriers compared to non-carriers. However, among risk haplotype carriers, the same “MS-associated” CpGs still showed significantly different methylation in MS patients compared to controls, suggesting the occurrence of additional epigenetic events [31]. Of note, this preference for differential DNA methylation in the HLA-DRB region was not observed in CD8 T cells even when the analysis included regions containing SNPs [30],[33]. Taken together, these results highlight a unique interaction between genetic susceptibility and epigenetic modifications that is specific to CD4 T cells. These findings underline the importance of the interplay of multiple risk factors (i.e. genetic and epigenetic) in MS pathogenesis and support a pathogenic role for CD4 T cells in MS.
Interestingly, the overall DNA methylation state in the periphery changes with MS disease subtype. In both peripheral blood mononuclear cells (PBMCs) and cell-free plasma, MS patients show increased DNA methylation relative to healthy controls [34],[35]. In a study conducted by Kulakova et al, MS patients were further stratified by disease type: relapsing-remitting or primary progressive. Peripheral blood mononuclear cells (PBMCs) from primary progressive (PP) patients were characterized by higher levels of genome-wide DNA methylation than those from relapsing-remitting (RR) patients [26]. Since PBMCs constitute a mixed group of cell populations, future studies are needed to delineate the contribution of each cell type to the DNA methylation profile of PP-MS patients and evaluate their function in this hypermethylated state. All together, DNMT inhibitors have been shown to skew the immune response towards more regulatory T cells. As these inhibitors are administered systemically, future studies should evaluate its effects on other immune cell types such as monocytes, dendritic cells, or B cells.
Conclusion
Histone deacetylation and DNA methylation work together to repress transcription. However, these marks lead to distinctive functional outcomes depending on cell type. In oligodendrocytes, the removal of acetyl marks on histones and addition of repressive methylation marks on DNA are crucial to repress transcription of myelin “brakes” and initiate the differentiation of progenitors into mature myelin-forming oligodendrocytes. Enhancing this process could be beneficial for MS by promoting myelin repair. On the other hand, the same epigenetic events occurring in T cells may have a detrimental role. Inhibition of HDAC or DNMT activity is protective in an immune-mediated preclinical model of MS, EAE, suggesting that histone deacetylation and DNA methylation in immune cells are important for their pro-inflammatory functions. In conclusion, therapeutic strategies that target epigenetic modifications are highly attractive for treatment of MS but cell specificity must be addressed.
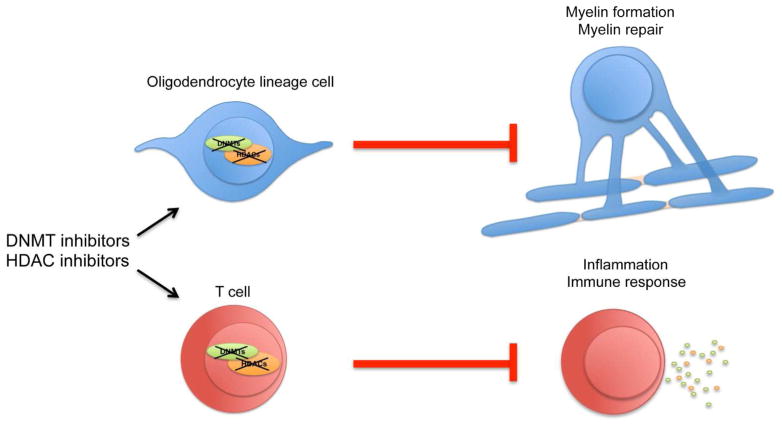
Figure 1. Cell-specific effects of DNMT and HDAC inhibitors.
Pharmacological inhibitors of DNA methyltransferases (DNMTs) or histone deacetylases (HDACs) have cell-specific effects. In oligodendrocyte lineage (OL) cells, they impair myelin formation and myelin repair, while in T cells they dampen inflammation and activation of immune responses
References
- 1.Ebers GC. Environmental factors and multiple sclerosis. Lancet Neurol. 2008;7(3):268–277. doi: 10.1016/S1474-4422(08)70042-5. [DOI] [PubMed] [Google Scholar]
- 2.Huynh JL, Casaccia P. Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment. Lancet Neurol. 12(2):195–206. doi: 10.1016/S1474-4422(12)70309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frohman EM, Racke MK, Raine CS. Multiple Sclerosis — The Plaque and Its Pathogenesis. N Engl J Med. 2006;354(9):942–955. doi: 10.1056/NEJMra052130. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the Histone Code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Popko B. Epigenetic control of myelin repair. Nat Neurosci. 2008;11(9):987–988. doi: 10.1038/nn0908-987. [DOI] [PubMed] [Google Scholar]
- 7.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169(4):577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen S, Sandoval J, Swiss VA, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11(9):1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Dupree J, Wang J, et al. The Transcription Factor Yin Yang 1 Is Essential for Oligodendrocyte Progenitor Differentiation. Neuron. 2007;55(2):217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye F, Chen Y, Hoang T, et al. HDAC1 and HDAC2 Regulate Oligodendrocyte Differentiation By Disrupting β-Catenin-TCF Interaction. Nat Neurosci. 2009;12(7):829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Neuronal Glial Cell Biol New Technol. 2009;19(5):479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedre X, Mastronardi F, Bruck W, et al. Changed Histone Acetylation Patterns in Normal-Appearing White Matter and Early Multiple Sclerosis Lesions. J Neurosci. 2011;31(9):3435. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gacias M, Gerona-Navarro G, Plotnikov AN, et al. Selective Chemical Modulation of Gene Transcription Favors Oligodendrocyte Lineage Progression. Chem Biol. 2014;21(7):841–854. doi: 10.1016/j.chembiol.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ntranos A, Casaccia P. Bromodomains: Translating the words of lysine acetylation into myelin injury and repair. Neurosci Lett. 2016;625:4–10. doi: 10.1016/j.neulet.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dangond F, Hafler DA, Tong JK, et al. Differential Display Cloning of a Novel Human Histone Deacetylase (HDAC3) cDNA from PHA-Activated Immune Cells. Biochem Biophys Res Commun. 1998;242(3):648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 16.Dangond F, Gullans SR. Differential Expression of Human Histone Deacetylase mRNAs in Response to Immune Cell Apoptosis Induction by Trichostatin A and Butyrate. Biochem Biophys Res Commun. 1998;247(3):833–837. doi: 10.1006/bbrc.1998.8891. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Tao R, Hancock WW. Using histone deacetylase inhibitors to enhance Foxp3+ regulatory T-cell function and induce allograft tolerance. Immunol Cell Biol. 2009;87(3):195–202. doi: 10.1038/icb.2008.106. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher JM, Lalor SJ, Sweeney CM, Tubridy N, Mills KHG. T cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Clin Exp Immunol. 2010;162(1):1–11. doi: 10.1111/j.1365-2249.2010.04143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164(1–2):10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Zhang Z-Y, Wu Y, Schluesener HJ. Valproic acid ameliorates inflammation in experimental autoimmune encephalomyelitis rats. Neuroscience. 2012;221:140–150. doi: 10.1016/j.neuroscience.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Ge Z, Da Y, Xue Z, et al. Vorinostat, a histone deacetylase inhibitor, suppresses dendritic cell function and ameliorates experimental autoimmune encephalomyelitis. Exp Neurol. 2013;241:56–66. doi: 10.1016/j.expneurol.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 23.Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213(2):384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 24.Hahn MA, Szabó PE, Pfeifer GP. 5-Hydroxymethylcytosine: A stable or transient DNA modification? Genomics. 2014;104(5):314–323. doi: 10.1016/j.ygeno.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyon S, Huynh JL, Dutta D, et al. Functional Characterization of DNA Methylation in the Oligodendrocyte Lineage. Cell Rep. 2016;15(4):748–760. doi: 10.1016/j.celrep.2016.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyon S, Ma D, Huynh JL, et al. Efficient Remyelination Requires DNA Methylation. eNeuro. 2017;4(2) doi: 10.1523/ENEURO.0336-16.2017. ENEURO.0336-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynh JL, Garg P, Thin TH, et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci. 2014;17(1):121–130. doi: 10.1038/nn.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangano K, Fagone P, Bendtzen K, et al. Hypomethylating Agent 5-Aza-2′-deoxycytidine (DAC) Ameliorates Multiple Sclerosis in Mouse Models. J Cell Physiol. 2014;229(12):1918–1925. doi: 10.1002/jcp.24641. [DOI] [PubMed] [Google Scholar]
- 29.Chan MWY, Chang C-B, Tung C-H, et al. Low-Dose 5-Aza-2′-deoxycytidine Pretreatment Inhibits Experimental Autoimmune Encephalomyelitis by Induction of Regulatory T Cells. Mol Med. 2014;20(1):248–256. doi: 10.2119/molmed.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bos SD, Page CM, Andreassen BK, et al. Genome-Wide DNA Methylation Profiles Indicate CD8+ T Cell Hypermethylation in Multiple Sclerosis. PLOS ONE. 2015;10(3):e0117403. doi: 10.1371/journal.pone.0117403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graves M, Benton M, Lea R, et al. Methylation differences at the HLA-DRB1 locus in CD4+ T-Cells are associated with multiple sclerosis. Mult Scler J. 2014;20(8):1033–1041. doi: 10.1177/1352458513516529. [DOI] [PubMed] [Google Scholar]
- 32.Baranzini SE, Mudge J, van Velkinburgh JC, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464(7293):1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maltby VE, Graves MC, Lea RA, et al. Genome-wide DNA methylation profiling of CD8+ T cells shows a distinct epigenetic signature to CD4+ T cells in multiple sclerosis patients. [Accessed May 17, 2017];Clin Epigenetics. 2015 :7. doi: 10.1186/s13148-015-0152-7. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4635618/ [DOI] [PMC free article] [PubMed]
- 34.Kulakova OG, Kabilov MR, Danilova LV, et al. Whole-Genome DNA Methylation Analysis of Peripheral Blood Mononuclear Cells in Multiple Sclerosis Patients with Different Disease Courses. Acta Naturae. 2016;8(3):103–110. [PMC free article] [PubMed] [Google Scholar]
- 35.Liggett T, Melnikov A, Tilwalli S, et al. Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J Neurol Sci. 2010;290(1–2):16. doi: 10.1016/j.jns.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]