Abstract
Hyperlipidemia is considered to be a high lipid level in blood, can induce metabolic disorders and dysfunctions of the body, and results in some severe complications. Therefore, hunting for some metabolite markers and clarifying the metabolic pathways in vivo will be an important strategy in the treatment and prevention of hyperlipidemia. In this study, a rat model of hyperlipidemia was constructed according to histopathological data and biochemical parameters, and the metabolites of serum and urine were analyzed by UPLC-Q-TOF/MS. Combining pattern recognition and statistical analysis, 19 candidate biomarkers were screened and identified. These changed metabolites indicated that during the development and progression of hyperlipidemia, energy metabolism, lipid metabolism, amino acid metabolism and nucleotide metabolism were mainly disturbed, which are reported to be closely related to diabetes, cardiovascular diseases, etc. This study demonstrated that a UPLC-Q-TOF/MS based metabolomic approach is useful to profile the alternation of endogenous metabolites of hyperlipidemia.
Keywords: UPLC-Q-TOF/MS, Hyperlipidemia, Metabolomic, Pattern recognition
1. Introduction
Hyperlipidemia is characterized by an increase in plasma total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C) and/or a decrease in high-density lipoprotein cholesterol (HDL-C) [1]. It was found that hyperlipidemia is a major risk factor for atherosclerosis and cardiovascular diseases, such as hypercholesterolemia and hypertriglyceridemia, which are the leading causes of death in the United States and other western countries. Emerging data establish dyslipidemia as a significant contributor to the development of diabetic neuropathy, and accumulated data from several large-scale trials of patients with type 2 diabetes also point to early dyslipidemia as a major independent risk factor for the development of diabetic neuropathy [2]. In addition, one study suggests that hyperlipidemia and thereby atherosclerosis are the leading causes of cardiac illness and deaths, in which circumstance heart attack and stroke are responsible for more deaths than all other causes combined [3]. Therefore, it is imperative for us to diagnose these diseases at an early stage in order to improve the clinical outcome and reduce mortality. The progression of diseases is mainly due to genetic determinants, environmental factors, and their interactions [4]. Diet, as an important environmental factor, plays a pivotal role in the development of this metabolic syndrome [5]. In order to improve the clinical outcomes of hyperlipidemia and the related chronic diseases mentioned above, it is necessary for us to deeply investigate the pathophysiology of hyperlipidemia. As hyperlipidemia is a systemic progressive disorder, alterations of endogenous metabolites may provide a prognostic index at the initial stage of this disorder compared with those of the putative lipid levels at the symptomatic stage.
Serum- or plasma-based biochemical assays and histopathological evaluation are commonly used in the study of hyperlipidemia [6]. However, these standard approaches may be inadequate. Metabolomic, as a sensitive and unbiased analytical method that assesses all metabolites in biological samples, has been widely used in the biomedical sciences [7]. This approach shows a significant potential for the diagnosis of human diseases [8], specification of genetic modifications [9], physiological evaluations [10], biomarkers screening [11], and drug toxicity/safety assessments [12]. Metabolomics focuses on the comprehensive measurement of small molecular weight compounds and visualizes abnormal, disease-related physiological states in whole samples. Moreover, it not only determines the relationships between phenotype and metabolisms, but also identifies the key metabolites associated with a particular phenotype and investigates the biological function and metabolic changes in the organism [13]. Up to now, the major two analytical platforms used in the metabolomic analysis have been nuclear magnetic resonance (NMR) [14] and mass spectrometry (MS), which are usually coupled to gas chromatography (GC) [4], liquid chromatography (LC), and capillary electrophoresis (CE) [15].
In the present study, we constructed a Sprague-Dawley (SD) rat hyperlipidemia model and applied an ultra-performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF/MS) based metabolomic approach together with histopathological examination and a biochemical assay to investigate the biochemical abnormalities associated with hyperlipidemia in both serum and urine. The present work, as a compensation for the results reported previously, will be more beneficial to profile the alternation of endogenous metabolites of hyperlipidemia, to dissect the underlying mechanisms related to hyperlipidemia and the strong association with atherogenesis progression. The findings of the metabolic pathways and potential biomarkers should be further explored for clinical prevention and treatment of hyperlipidemia and the related chronic diseases in order to advance the progression of amelioration in subjects with hyperlipidemia-associated conditions and to improve the clinical outcome for these hyperlipidemia-related conditions.
2. Materials and methods
2.1. Ethics statement
All animal treatments were strictly in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Animal experiments were approved by the Administrative Committee of the Experimental Animal Care and Use of Second Military Medical University (SMMU, License no. 2011023).
2.2. Reagents and materials
HPLC-grade methanol and acetonitrile (ACN) were purchased from Merck (Darmstadt, Germany). Formic acid was obtained from Fluka (Buchs, Switzerland). Ultrapure water was purified using a Milli-Q system (Millipore, Bedford, MA, USA). The following compounds were obtained from Shanghai Jingchun Reagent Co.: linoleic acid, succinic acid and taurine. Hippuric acid was purchased from Larodan AB (Malmoö, Sweden). Sphinganine and C16 Sphinganine were purchased from Acros Organics (New Jersey, USA).
2.3. Animal study
A total of 12 adult male Sprague-Dawley rats (180±20 g) obtained from the SLAC Laboratory Animal Co., Ltd. (Shanghai, China) were maintained under standard laboratory conditions (temperature of 21–23 °C, relative humidity of 45–65%, and 12 h/12 h light/dark cycle) with aseptic food and tap water ad libitum. After one week of habituation, 6 rats, selected randomly, were fed a basic diet as the normal control group (NC), while the others were fed a high-fat diet (HFD) for 4 weeks during the experimental period. Before the end of the experiment, the rats were housed in metabolic cages and fasted overnight (allowed free access to water). Rat urine was collected in tubes. Then the rats were weighed and anesthetized with an intraperitoneal injection of 3% sodium pentobarbital (2 mL/kg). An abdominal incision was made to expose the liver and inferior vena cava. Blood (3–4 mL) was withdrawn from the abdominal aorta and collected in tubes. Serum was extracted by centrifugation at 3500 rpm for 15 min. The serum was divided into two parts, one for the lipid levels assay and the other for metabolomic analysis. The serum and urine were both stored at −80 °C before UPLC-Q-TOF/MS analysis.
2.4. Histopathological examination
After being weighed, the livers were washed with saline and put in a buffer solution of 10% formalin. Fixed tissues were processed routinely for paraffin embedding, and 4 μm sections were prepared and dyed with hematoxylin-eosin; the stained areas were viewed under an optical microscope at 200×. The hepatic index (HI) was estimated from the ratio of total liver weight to body weight.
2.5. Biochemical analysis
Serum was thawed and incubated at room temperature. Lipid levels, including total cholesterol (TC), triglycerides (TG), high-density lipoprotein-cholesterol (HDL-C) and low-density lipoprotein-cholesterol (LDL-C), were determined using an automatic biochemical analyzer. The arteriosclerosis index (AI) was calculated as follows:
| (1) |
2.6. Metabolomic study
2.6.1. Extraction of the metabolites in serum and urine
The serum samples were thawed at room temperature prior to analysis. Methanol (400 μL) was added to every 100 μL of serum. After vigorous shaking for 1 min and incubation on ice for 10 min, the mixture was centrifuged at 14,000 g for 15 min at 4 °C to precipitate the protein. The urine samples were thawed at room temperature prior to analysis. To reduce the effect of the solvent and get a good peak shape, a 100 μL sample of urine was diluted with 400 μL acetonitrile and centrifuged at 15,000 g for 10 min at 4 °C. All the clear supernatant was transferred to an autosampler vial and kept at 4 °C. An aliquot of 4 μL sample was made onto the column in each run. An in-house quality control (QC) was prepared by pooling and mixing the same volume of each sample. The QC sample was run six times prior to the start of the analytical run to “condition” the system and analyzed after every 3 samples to check for system stability.
2.6.2. UPLC-MS analysis
UPLC-MS analysis was performed using an Agilent 1290 Infinity LC system coupled to an Agilent 6530 Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) mass spectrometer (Agilent, USA). Chromatographic separations were performed using an ACQUITY UPLC HSS T3 column (2.1 mm×100 mm, 1.8 μm, Waters, Milford, Ireland) maintained at 45 °C. The mobile phase consisted of 0.1% formic acid (A) and ACN modified with 0.1% formic acid (B). The following gradient program was used: 0% B at 0–2 min, 0%–15% B at 2–10 min, 15%–30% B at 10–14 min, 30%–95% B at 14–17 min, 95% B at 17–19 min, 95%–0% B at 19–20 min and followed by re-equilibrated step of 5 min. The flow rate was 400 μL/min and the injection volume was 4 μL.
Mass spectrometry was performed using the Agilent 6530 Accurate Mass Quadrupole Time-of-Flight (Q-TOF) mass spectrometer (Agilent, USA), operating in both positive and negative ion modes. The capillary voltage was 3.5 kV, the drying gas flow was 11 L/min, and the gas temperature was 350 °C. The nebulizer pressure was set at 45 psig. The fragment voltage was set at 120 V and skimmer voltage was set at 60 V. All analyses were conducted using a mixture of 10 mM purine (m/z 121.0508) and 2 mM hexakis phosphazinen (m/z 922.0097) as internal standards to ensure mass accuracy and reproducibility. Data were collected in a centroid mode and the mass range was set at m/z 50–1000 using an extended dynamic range. MS/MS analysis was carried out to study the structure of the potential biomarkers. MS spectra were collected at 2 spectra/s and MS/MS spectra were collected at 0.5 spectra/s with a medium isolation window (4 m/z), and the collision energy was 20 V.
2.6.3. Data processing and statistical analysis
Data processing used the method previously published by our group with minor modifications [16]. The raw data in the instrument specific format (.d) were firstly converted to common data format (.mzdata) files by a conversion software program, in which the isotope interferences were excluded. The program XCMS (http://metlin.scripps.edu/download/) was used for nonlinear alignment of the data in the time domain and automatic integration and extraction of the peak intensities. The variables that did not present in at least 80% of the groups were filtered. The output data were imported into MATLAB 7.0 software (Math Works, Inc, USA) where data were normalized using the summation of response of all metabolites in one sample, in order to partially compensate for the concentration bias of metabolites between samples and to obtain the relative intensity of metabolites. The resulting three-dimensional matrix, including retention time and m/z pairs, sample names, and normalized ion intensities, was introduced to the multivariate data analysis.
Statistically significant differences in mean values were tested using an independent sample t-test and p<0.05 was considered statistically significant. Prior to multivariate analysis, the resultant data matrices were mean-centered and scaled to Pareto variance. The data were analyzed by the orthogonal partial least squares discriminant (OPLS-DA) method using SIMCA-P (version 11, U metrics) which is extensively used to select the biomarkers by a variable importance plot (VIP) [17], [18]. Variables that had significant contributions to discrimination between groups were preferentially considered as the potential biomarkers.
3. Results
3.1. H&E staining and hepatic index
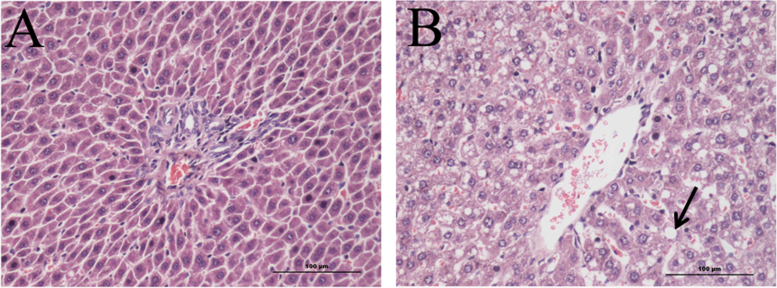
H&E staining was performed to access the fat content in the liver, and microscopic liver tissues are shown in Fig. 1. Liver histological examination of the NC group showed normal cell architecture (Fig. 1A), while significant morphological changes were observed in the HFD group. Liver sections in the HFD group showed less cells and lipid vacuolization, which are indicated with black arrow in Fig. 1B. These results were consistent with those of previous studies [19], and indicated that a high-fat diet accumulates fat in hepatic tissue cells, which can finally cause fatty liver. In addition, Table 1 shows significant differences in the liver index of the HFD group compared to the NC group.
Fig. 1.
Photomicrographs of hepatic tissue morphology of experimentally hyperlipidemic rats (×200). The features included (A) few fat droplets shown in the liver of normal control rats (NC) and (B) large fat droplets were indicated by the arrowhead in the liver of rats fed a high-fat diet (HFD).
Table 1.
Serum lipid levels of experimental hyperlipidemic rats induced by high-fat diet.
| Group | TC (mM) | TG (mM) | HDL-C (mM) | LDL-C (mM) | AI | HI |
|---|---|---|---|---|---|---|
| NC | 1.41±0.15 | 0.78±0.26 | 1.11±0.14 | 0.18±0.08 | 0.27±0.08 | 0.03±0.02 |
| HFD | 1.67±0.39 | 0.97±0.28 | 0.92±0.14⁎ | 0.52±0.31⁎ | 0.82±0.37⁎ | 0.04±0.02⁎⁎ |
Values are expressed as mean ±SD, n=6.
TC, total cholesterol; TG, triglycerides; HDL-C, high-density lipoprotein-cholesterol; and LDL-C, low-density lipoprotein-cholesterol.
p<0.05, compared with the control group (NC).
p<0.01, compared with the control group (NC).
3.2. Serum biochemical parameters
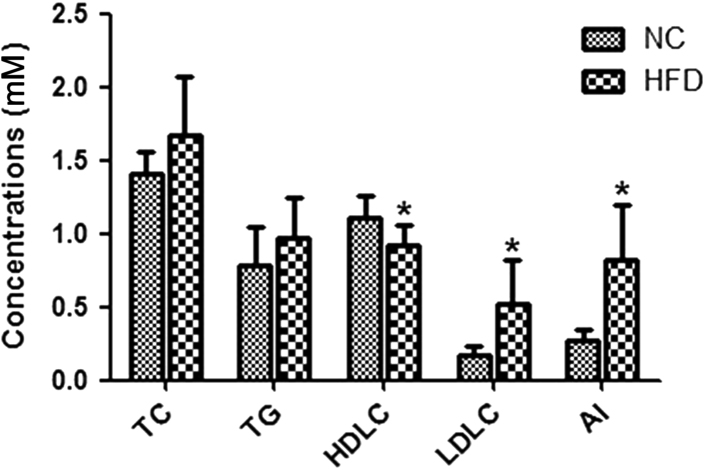
As shown in Table 1 and Fig. 2, the levels of HDL-C and LDL-C in the serum were significantly elevated in the HFD group compared with the NC group. However, no significant differences were found between the levels of TC and TG in the serum of the NC and HFD groups, but the contents of TC and TG were enhanced in the HFD group. The AI was also markedly elevated in the HFD group compared with the NC group. According to the definition of hyperlipidemia, the rat model was successfully constructed.
Fig. 2.
Serum lipids profiles of the control group (NC) and the high-fat diet group (HFD). TC, total cholesterol; TG, triglyceride; HDLC, high-density lipoprotein cholesterol; and LDLC, low-density lipoprotein cholesterol. *p<0.05 vs. NC.
3.3. Multivariate statistical analysis of metabolites
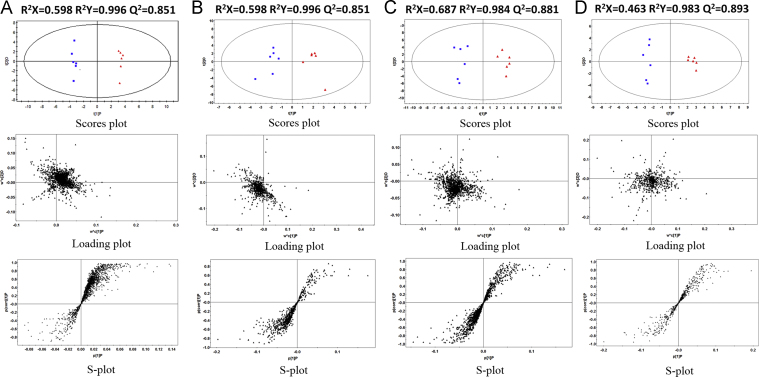
Before multivariate statistical analysis, peaks with a retention time of less than 0.5 min (near to the dead time) were excluded due to a high degree of ion suppression that they suffered. The datasets from positive and negative ionization modes contained 713 and 1746 variables with the NC and HFD groups in serum, and 589 and 1239 variables with the NC and HFD groups in urine, respectively. To determine whether the metabolite fingerprints in serum/urine differed between the NC and HFD groups in the metabolomic approach, we constructed orthogonal partial least squares discriminant analysis (OPLS-DA) models which have been widely used in metabonomic studies [17], [18]. In this study, four OPLS-DA models were established and employed to identify biomarkers which were related to hyperlipidemia in serum and urine under the ESI+/ESI− mode, which are shown in Fig. 3. Commonly, R2Y provides an estimate of how well the model fits the Y data, whereas Q2Y is an estimate of how well the model predicts the Y [20]. In order to gain high predictive ability, the values of R2Y and Q2Y should be close to 1. The related parameters of the four OPLS-DA models are given in Fig. 3. As Fig. 3 shows, there is a distinguished classification between the clustering of the NC and HFD groups. The R2Y and Q2Y were all above 0.98 and 0.85 in four models, respectively, which indicates that the models had good prediction characteristics.
Fig. 3.
OPLS-DA score plot (top panel), loading plot (middle panel) and S-plot (bottom panel) of the UHPLC/TOF-MS spectra from the NC group (red triangle) and HFD group (blue box). (A: serum in ESI− mode; B: serum in ESI+ mode; C: urine in ESI− mode; and D: urine in ESI+ mode).
3.4. Detection and identification of biomarker candidates
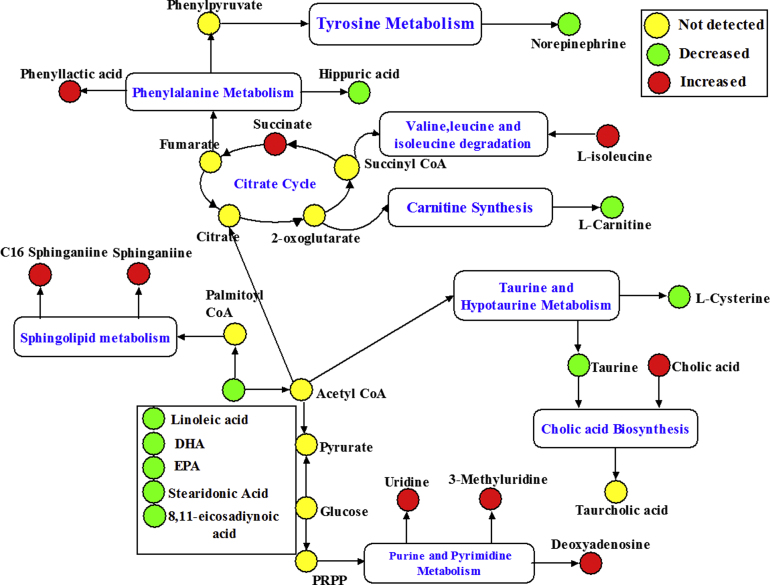
To select potential biomarkers related to hyperlipidemia, statistically significant differences for the variables between the NC and HFD groups were tested by an independent sample t-test. Metabolites that differed significantly between the NC and HFD groups (p<0.05) were identified as candidate biomarkers. Next, the variable importance plot (VIP) reflecting the importance of variables was applied to filter the important metabolites in the model. In this study, metabolite ions with a VIP value >1.5 were considered as potential biomarkers. Following the criteria above, a total of 19 metabolite ions were selected as potential biomarkers related to HFD induced hyperlipidemia. The detailed method for compound identification has been described in our previous work [16], [21]. A summary of biomarker candidates identified in hyperlipidemia rats is shown in Table 2, and the related pathway is shown in Fig. 4.
Table 2.
Identification of significantly differential metabolites in serum/urine of hyperlipidemic rats and their related metabolic pathway.
| No | TR(min) | Ion(m/z) | Adduct | Formula | Identification | Trende | Related pathway |
|---|---|---|---|---|---|---|---|
| 1a | 16.65 | 279.2344 | [M−H]− | C18H32O2 | Linoleic acid | (⁎)↓ | Fatty acid metabolism |
| 1b | 16.63 | 281.2481 | [M+H]+ | C18H32O2 | Linoleic acid | (⁎)↓ | Fatty acid metabolism |
| 2a | 16.51 | 303.2346 | [M−H]− | C20H32O2 | 8,11-Eicosadiynoic acid | (⁎)↓ | Fatty acid metabolism |
| 3a | 15.74 | 301.2182 | [M−H]− | C20H30O2 | EPA | (⁎)↓ | Fatty acid metabolism |
| 3b | 15.72 | 303.2321 | [M+H]+ | C20H30O2 | EPA | (⁎)↓ | Fatty acid metabolism |
| 4a | 9.83 | 453.2871 | [M+COO]− | C24H40O5 | Cholic acid | (⁎)↑ | Cholic acid biosynthesis |
| 5a | 15.12 | 275.2018 | [M−H]− | C18H28O2 | Stearidonic acid | (⁎)↓ | Fatty acid metabolism |
| 6b | 0.71 | 119.0894 | [M+H]+ | C4H6O4 | Succinic acid | (⁎)↑ | Citrate cycle |
| 7b | 13.96 | 274.6561 | [M+H]+ | C16H35NO2 | C16 Sphinganine | (⁎)↑ | Sphingolipid metabolism |
| 8b | 16.29 | 329.2479 | [M+H]+ | C22H32O2 | Docosahexaenoic acid | (⁎)↓ | Fatty acid metabolism |
| 9b | 10.09 | 302.2149 | [M+H]+ | C18H39NO2 | Sphinganine | (⁎)↑ | Sphingolipid metabolism |
| 10c | 5.89 | 178.0507 | [M−H]− | C9H9NO3 | Hippuric acid | (⁎)↓ | Phenylalanine metabolism |
| 10d | 5.85 | 180.0656 | [M+H]+ | C9H9NO3 | Hippuric acid | (⁎)↓ | Phenylalanine metabolism |
| 11c | 8.87 | 165.0557 | [M−H]− | C9H10NO3 | Phenyllactic acid | (⁎)↑ | Phenylalanine metabolism |
| 12c | 0.69 | 124.0075 | [M−H]− | C2H7NO3S | Taurine | (⁎)↓ | Taurine and hypotaurine metabolism |
| 13c | 0.77 | 166.0182 | [M+COO]− | C3H7NO2S | l-Cysteine | (⁎)↓ | Taurine and hypotaurine metabolism |
| 14c | 8.88 | 166.0588 | [M+COO]− | C8H11NO3 | Norepinephrine | (⁎)↓ | Tyrosine metabolism |
| 15d | 0.72 | 132.0981 | [M+H]+ | C6H13NO2 | l-Isoleucine | (⁎)↑ | Valine, leucine and isoleucine degradation |
| 16d | 0.68 | 162.1121 | [M+H]+ | C7H15NO3 | l-Carnitine | (⁎)↓ | Carnitine synthesis |
| 17d | 14.16 | 269.1387 | [M+NH4]+ | C10H13N5O3 | DHA | (⁎)↑ | Purine metabolism |
| 18d | 0.73 | 259.0932 | [M+H]+ | C10H14N2O6 | 3-Methyluridine | (⁎)↑ | Pyrimidine metabolism |
| 19d | 0.85 | 245.1295 | [M+H]+ | C9H12N2O6 | Uridine | (⁎)↑ | Pyrimidine metabolism |
Identified in serum under ESI−.
Identified in serum under ESI+.
Identified in urine under ESI−.
Identified in urine under ESI+.
Change trend compared with the control group. (↑): up-regulated and (↓): down-regulated.
Represents a statistically significant difference (p<0.05).
Fig. 4.
The integrative plot of the metabolites and the relevant pathways׳ changing for high-fat diet induced hyperlipidemia according to the KEGG PATHWAY database.
4. Discussion
In this study, a parallel metabolite profiling of serum and urine based on the UPLC-Q-TOF/MS based metabolomic analysis was applied to provide comprehensive and complementary insights into hyperlipidemia based on the 19 selected biomarkers related to a high fat diet induced hyperlipidemia model. The results demonstrated that a four-week long high fat diet can markedly alter the endogenous metabolites in SD rats by partially regulating energy metabolism, lipid metabolism, amino acid metabolism and nucleotide metabolism. The major pathways related to these metabolites are presented in Fig. 4. In short, the combined analysis of serum and urine is meaningful and complementary, uncovering more comprehensive metabolomic data than either biofluid alone, providing a holistic method for determining the response of an intact system to physical perturbation, leading to a broader view of the metabolic network.
As we all know, succinic acid, which is localized mainly in the mitochondria, is a vital intermediate of the TCA cycle, which is the main pathway of glucose degradation and the primary energy supplier for universal organisms [22]. In this study, succinic acid was significantly increased in the HFD group compared to the NC group, suggesting that a high fat diet may disturb the TCA cycle markedly and ultimately alter energy metabolism. A wealth of evidence further supports the fact that an abnormal TCA cycle can significantly alter energy metabolism [23]. Our results displayed an increased amount of succinic acid in serum, which can be explained as a consequence of increased fatty catabolism, and it was possible that energy metabolism was disturbed in liver mitochondria in the process of hyperlipidemia [24].
A number of previous studies suggest that lipid-based metabolism is essential to many biochemistry reactions and related to many biological functions [21]. Our results found that the levels of five unsaturated fatty acids (linoleic acid, 8,11-eicosadiynoic acid, docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA) and stearidonic acid) were significantly down-regulated in the HFD group, which is indicative of enhanced peroxidation and oxidative stress [6]. Such peroxidation and oxidative stress can further lead to apolipoprotein B proteolysis, thus impairing the secretion of very low-density lipoproteins (VLDL) so that exports of TG from the liver will decrease contribution to TG accumulation in the liver [20], which was further confirmed by our liver pathological results (Fig. 1). This lipid β-oxidation can generate large amounts of electrons entering the mitochondrial respiratory chain to produce excess reactive oxygen species (ROS). For example, linoleic acid, which cannot be synthesized in vivo and has to be obtained from the diet [25], is one of the polyunsaturated fatty acids that were reported to be associated with atherosclerotic and inflammatory diseases because they are major components of the cytoplasmic membrane and precursor fatty acids for prostaglandins and leukotrienes [26]. On the whole, the downgraded unsaturated fatty acid in our study suggested that a long term high fat diet induced hyperlipidemia altered fatty acid metabolism and induced oxidative stress; however, the detailed mechanism in the process of this lipid disorder still needs to be further investigated.
Cholic acid, a major primary bile acid, which plays an important role in lipolysis and cholesterol catabolism, showed an increased trend in our study. A previous study reported that bile acids served many important physiological functions, including cholesterol homeostasis, lipid absorption, and generation of bile flow [8]. Meanwhile, another report suggested that the rise of bile acid caused the dysfunction of hepatic and intestinal [11]. Therefore, the elevated cholic acids may be explained in part to that high fat diet causing the injury of liver and intestine, which was partly supported by H&E staining (Fig. 1). Moreover, cholic acid is synthesized in the liver and secreted in the gallbladder or intestine. The bile acids show an important physiological function for both absorption of nutrients and regulation of whole-body lipid metabolism. The increased cholic acid level in the plasma of model rats indicated that intestinal absorption of liposoluble nutrients was disturbed in diet induced hyperlipidemic rats [27]. Of note, Guo et al. investigated the activity of hydroxymethyl glutarate coenzyme A (HMG-CoA) reductase and microsomal cytochrome P-450 cholesterol 7α-hydroxylase (CYP7A1) in hyperlipidemic rats and found that the rats fed HFD showed a slightly higher level of liver total bile acid compared with those fed a normal diet, mainly due to up-regulation of the gene expression of CYP7A1 in the liver of the HFD rats as compared with those of the NC group rats [28].
Among these metabolites, sphingasine and C16 sphinganine, two ceramide-related metabolites detected in this study, were significantly increased, indicating that sphingolipid metabolism turned to be abnormal in response to hyperlipidemic. One study reported that an obviously increased level of sphinganine in the urine of rats may be due to the abnormal accumulation of esterification of cholesterol [29]. Intensive studies pointed out the important role of sphingolipids in cell growth, cell differentiation, cell apoptosis and vital signal transduction pathways [30], [31]. To better understand these metabolic disorders, we originated from some key enzymes involved in sphingolipid metabolism. Sphingomyelinases (SMases) are the key enzyme involved in the generation of ceramide and a cascade of bioactive lipids [32]; we implicated that the up-regulating of SMases should be one factor associated with the accumulation of sphingasine and C16 sphinganine. However, in many cases, further study still needs to be conducted to explore the clear mechanism of sphingolipid-mediated etiology accompanying hyperlipidemia.
Another prominent finding in the present study is the marked elevation levels of free nucleosides, including deoxyadenosine, uridine, and 3-methyluridine, in the aqueous urine samples from rats fed a high fat diet. It is indicated that lipid oversupply may cause the degradation of nucleic acids or inhibit the synthesis of DNA and RNA as these metabolites are the basic structural units for nucleic acids [33]. Furthermore, a previous study supported that the up-regulated level of uridine may be due to insufficient protein synthesis and increased compensatory RNA synthesis [34].
The study also found that amino acid metabolism was affected in the HFD group, leading to changes in the levels of many amino acids, including l-leucine, l-cysteine, l-carnitine, taurine, and hippuric acid. Taurine has been reported to be the highest abundance β-amino acid present in many tissues of humans and animals, and processes many physiological functions such as bile acid conjugation, antioxidation and detoxication [22]. In the present study, the level of taurine was decreased after high fat exposure, which could be explained in two ways: taurine acted as an antioxidant to prevent ROS produced by oxidative stress; moreover, taurine could bind cholic acid to improve its secretion. Meanwhile, taurine is an end product of cysteine catabolism; large consumption of taurine in the HFD group indirectly reflects the decreased trend of cysteine in the model group [35], which could be supported by the decline of l-cysteine acid in our results. Hippuric acid is the glycine conjugate of benzoic acid which is derived from phenylalanine [26]. In addition, the alteration of urinary hippurate is frequently reported to be related to modulation of the activity or population of gut microbiota. Therefore, high fat exposure may have also caused changes in gut microbiota [8], [33], which needs further investigation to overcome the challenges to the microbiome. Carnitine is a quaternary ammonium compound that occurs both in the kidney and the liver and is transported in the blood for use by the muscles [36]. The reason for the alternation of the level of carnitine might be the accumulation in serum TG levels in the model group. In our study, l-Isoleucine was also found to be up-regulated in the HFD group and further investigation is needed to explain this outcome.
As we all know, norepinephrine is known to promote lipid metabolism; norepinephrine consumption would be increased when a high lipid diet is ingested, and is supported by the present study [25]. Epinephrine is derived from phenylalanine and tyrosine, and it could indicate the alternation of phenylalanine and tyrosine metabolism in the HFD group.
5. Conclusion
In this study, a metabolomic approach based on UPLC-Q-TOF/MS has been developed to profile endogenous metabolic changes in the serum and urine of HFD rats. Totally 19 sensitive biomarkers were identified, primarily involved in energy metabolism, lipid metabolism, amino acid metabolism and nucleotide metabolism. By integrated histological assessment and biomedical analysis, it was found that lipid oversupply caused significant alternation of endogenous metabolites, which might result in some other complications. This present study demonstrated that a UPLC-Q-TOF/MS based metabolomic approach is useful to profile the alternation of endogenous metabolites of hyperlipidemia, to achieve a “snapshot” for the mechanism of the pathogenesis of hyperlipidemia and strong association with atherogenesis progression.
Acknowledgments
The authors appreciate the financial support from the National Natural Science Foundation of China (No. 81273472) and the Science and Technology Commission of Shanghai Municipality (No. 12401900802).
Footnotes
Peer review under responsibility of Xi׳an Jiaotong University.
References
- 1.Song X., Wang J., Wang P. 1H NMR-based metabolomics approach to evaluate the effect of Xue-Fu-Zhu-Yu decoction on hyperlipidemia rats induced by high-fat diet. J. Pharm. Biomed. Anal. 2013;78–79:202–210. doi: 10.1016/j.jpba.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A.M., Hinder L.M., Pop-Busui R. Hyperlipidemia: a new therapeutic target for diabetic neuropathy. J. Peripher. Nerv. Syst. 2009;14:257–267. doi: 10.1111/j.1529-8027.2009.00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain K.S., Kathiravan M.K., Somani R.S. The biology and chemistry of hyperlipidemia. Bioorg. Med. Chem. 2007;15:4674–4699. doi: 10.1016/j.bmc.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 4.Zha W., J. A, Wang G. Metabonomic characterization of early atherosclerosis in hamsters with induced cholesterol. Biomarkers. 2009;14:372–380. doi: 10.1080/13547500903026401. [DOI] [PubMed] [Google Scholar]
- 5.Kelly R.B. Diet and exercise in the management of hyperlipidemia. Am. Fam. Physician. 2010;81:1097–1102. [PubMed] [Google Scholar]
- 6.Jiang C., Yang K., Yang L. A 1H NMR-based metabonomic investigation of time related metabolic trajectories of the plasma, urine and liver extracts of hyperlipidemic hamsters. PloS One. 2013;8:e66786. doi: 10.1371/journal.pone.0066786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dettmer K., Aronov P.A. Mass spectrometry-based metabolomics. Mass. Spectrom. Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X., Fritsche J., Wang J. Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics. 2010;6:362–374. doi: 10.1007/s11306-010-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams R.E., Lenz E.M., Evans J.A. A combined 1H NMR and HPLC-MS-based metabonomic study of urine from obese (fa/fa) Zucker and normal Wistar-derived rats. J. Pharm. Biomed. Anal. 2005;38:465–471. doi: 10.1016/j.jpba.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Bernini P., Bertini I., Luchinat C. The cardiovascular risk of healthy individuals studied by NMR metabonomics of plasma samples. J. Proteome Res. 2011;10:4983–4992. doi: 10.1021/pr200452j. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Lin Z., Tan G. Metabonomic studies on potential plasma biomarkers in rats exposed to ionizing radiation and the protective effects of Hong Shan Capsule. Metabolomics. 2013;9:1082–1095. [Google Scholar]
- 12.Coen M. A metabonomic approach for mechanistic exploration of pre-clinical toxicology. Toxicology. 2010;278:326–340. doi: 10.1016/j.tox.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Wang C., Feng R., Sun D. Metabolic profiling of urine in young obese men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC/Q-TOF MS) J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011;879:2871–2876. doi: 10.1016/j.jchromb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Lu C., Wang Y., Sheng Z. NMR-based metabonomic analysis of the hepatotoxicity induced by combined exposure to PCBs and TCDD in rats. Toxicol. Appl. Pharmacol. 2010;248:178–184. doi: 10.1016/j.taap.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 15.Balderas C., Villasenor A., Garcia A. Metabolomic approach to the nutraceutical effect of rosemary extract plus Omega-3 PUFAs in diabetic children with capillary electrophoresis. J. Pharm. Biomed. Anal. 2010;53:1298–1304. doi: 10.1016/j.jpba.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 16.Wu S., Gao Y., Dong X. Serum metabonomics coupled with Ingenuity Pathway Analysis characterizes metabolic perturbations in response to hypothyroidism induced by propylthiouracil in rats. J. Pharm. Biomed. Anal. 2013;72:109–114. doi: 10.1016/j.jpba.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 17.Huang M., Joseph J.W. Metabolomic analysis of pancreatic beta-cell insulin release in response to glucose. Islets. 2012;4:210–222. doi: 10.4161/isl.20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L., Huang J., Wang Y. Metabonomic analysis reveals the CCl4-induced systems alterations for multiple rat organs. J. Proteome Res. 2012;11:3848–3859. doi: 10.1021/pr3003529. [DOI] [PubMed] [Google Scholar]
- 19.Feng L., Yu C., Ying K. Hypolipidemic and antioxidant effects of total flavonoids of Perilla Frutescens leaves in hyperlipidemia rats induced by high-fat diet. Food. Res. Int. 2011;44:404–409. [Google Scholar]
- 20.An Y., Xu W., Li H. High-fat diet induces dynamic metabolic alterations in multiple biological matrices of rats. J. Proteome Res. 2013;12:3755–3768. doi: 10.1021/pr400398b. [DOI] [PubMed] [Google Scholar]
- 21.Cai Y., Gao Y., Tan G. Myocardial lipidomics profiling delineate the toxicity of traditional Chinese medicine Aconiti Lateralis radix praeparata. J. Ethnopharmacol. 2013;147:349–356. doi: 10.1016/j.jep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L.C., Zhang X.D., Liao S.X. A metabonomic comparison of urinary changes in Zucker and GK rats. BMC Biotechnol. 2010;2010:431894. doi: 10.1155/2010/431894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun B., Li L., Wu S. Metabolomic analysis of biofluids from rats treated with Aconitum alkaloids using nuclear magnetic resonance and gas chromatography/time-of-flight mass spectrometry. Anal. Biochem. 2009;395:125–133. doi: 10.1016/j.ab.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Wu H., Zhang X., Li X. Comparison of metabolic profiles from serum from hepatotoxin-treated rats by nuclear-magnetic-resonance-spectroscopy-based metabonomic analysis. Anal. Biochem. 2005;340:99–105. doi: 10.1016/j.ab.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q., Wang G.J., A J.Y. Application of GC/MS-based metabonomic profiling in studying the lipid-regulating effects of Ginkgo biloba extract on diet-induced hyperlipidemia in rats. Acta Pharmacol. Sin. 2009;30:1674–1687. doi: 10.1038/aps.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y.Y., Lei P., Chen D.Q. Renal metabolic profiling of early renal injury and renoprotective effects of Poria cocos epidermis using UPLC Q-TOF/HSMS/MSE. J. Pharm. Biomed. Anal. 2013;81–82:202–209. doi: 10.1016/j.jpba.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 27.Huang D., Yang J., Lu X. An integrated plasma and urinary metabonomic study using UHPLC-MS: intervention effects of Epimedium koreanum on ‘Kidney-Yang Deficiency syndrome’ rats. J. Pharm. Biomed. Anal. 2013;76:200–206. doi: 10.1016/j.jpba.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Guo J., Bei W., Hu Y. A new TCM formula FTZ lowers serum cholesterol by regulating HMG-CoA reductase and CYP7A1 in hyperlipidemic rats. J. Ethnopharmacol. 2011;135:299–307. doi: 10.1016/j.jep.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Lv H., Zhang A. Metabolite profiling and pathway analysis of acute hepatitis rats by UPLC-ESI MS combined with pattern recognition methods. Liver Int. 2014;34:759–770. doi: 10.1111/liv.12301. [DOI] [PubMed] [Google Scholar]
- 30.Rao R.P., Yuan C., Allegood J.C. Ceramide transfer protein function is essential for normal oxidative stress response and lifespan. Proc. Natl. Acad. Sci. USA. 2007;104:11364–11369. doi: 10.1073/pnas.0705049104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotstein N.P., Miranda G.E., Abrahan C.E. Regulating survival and development in the retina: key roles for simple sphingolipids. J. Lipid Res. 2010;51:1247–1262. doi: 10.1194/jlr.R003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavoine C., Pecker F. Sphingomyelinases: their regulation and roles in cardiovascular pathophysiology. Cardiovasc. Res. 2009;82:175–183. doi: 10.1093/cvr/cvp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Ye Y., An Y. Systems responses of rats to Aflatoxin B1 exposure revealed with metabonomic changes in multiple biological matrices. J. Proteome Res. 2010;10:614–623. doi: 10.1021/pr100792q. [DOI] [PubMed] [Google Scholar]
- 34.Tan G., Liao W., Dong X. Metabonomic profiles delineate the effect of traditional Chinese medicine sini decoction on myocardial infarction in rats. PloS One. 2012;7:e34157. doi: 10.1371/journal.pone.0034157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., Sun B., Zhang Q. Metabonomic study on the toxicity of Hei-Shun-Pian, the processed lateral root of Aconitum carmichaelii Debx. (Ranunculaceae) J. Ethnopharmacol. 2008;116:561–568. doi: 10.1016/j.jep.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Kim M.J., Yang H.J., Kim J.H. Obesity-related metabolomic analysis of human subjects in black soybean peptide intervention study by ultraperformance liquid chromatography and quadrupole-time-of-flight mass spectrometry. J. Obes. 2013;2013:874981. doi: 10.1155/2013/874981. [DOI] [PMC free article] [PubMed] [Google Scholar]