Abstract
Diffuse gliomas are the most common human primary brain tumors and remain incurable. They are complex entities in which diverse genetic and nongenetic effects determine tumor biology and clinical course. Our current understanding of gliomas in patients is primarily based on genomic and transcriptomic methods that have profiled them as bulk, providing critical information yet masking the diversity of cells within each tumor. Recent advances in single-cell DNA and RNA profiling have paved the way to studying tumors at cellular resolution. Here, we review initial studies deploying single-cell analysis in clinical glioma samples, with a focus on RNA expression profiling. We highlight how these studies provide new insights into glioma biology, tumor heterogeneity, cancer cell lineages, cancer stem cell programs, the tumor microenvironment, and glioma classification.
Keywords: diffuse gliomas, heterogeneity, single-cell RNA sequencing
Introduction
Insufficient progress has been made in the management of gliomas over the last century, urging for a better understanding of their underlying biology. Gliomas are intricate ecosystems composed of diverse malignant cells and nonmalignant cells, whose behavior as a whole determines response to therapies and patient outcome.1–3 At least 3 determinants shape the biology of gliomas: (i) genetic alterations drive cellular transformation and the evolution of cancer cells; (ii) cellular lineages and their associated developmental pathways and epigenetic programs ascribe cancer cells with key phenotypic and functional features; programs such as those of neural stem/progenitor cells, oligodendrocyte progenitor cells, and mature glial cells (astrocytic, oligodendrocytic, ependymal) strongly influence cancer cell behavior; (iii) diverse nonmalignant cells, including microglia, macrophages, lymphocytes, endothelial, and other cells—collectively forming the tumor microenvironment (TME)—further influence glioma biology and response to therapy.
While it is critical to precisely measure all cellular elements in gliomas, standard genomic and transcriptomic methods profile these complex entities as bulk samples, measuring only the average signal and masking the inherent cellular diversity. It is thus key to develop a framework for the unbiased analysis of human samples at single-cell resolution. In this review, we describe initial studies applying single cell expression profiling and genomic techniques to dissect the composition and function of diffuse gliomas. We argue that single-cell techniques are particularly powerful at deciphering (i) genetic heterogeneity; (ii) cellular lineages and stem cell programs; (iii) the composition of the TME; (iv) tumor classification schemes. In the sections below we first briefly describe the single-cell genomics workflow and then discuss each of these aspects in the context of gliomas, describing the relevant questions in the field and the contribution of single-cell genomic approaches to solve them.
Single-Cell Tumor Profiling
In theory, all methods for genomic profiling may be performed at the single-cell level, thus providing information about cellular diversity, which is particularly important for heterogeneous samples such as clinical tumors. Indeed, single-cell genomic profiling is now possible for DNA,4 RNA,5,6 protein,7 epigenome,8 and even multi-omics.9 However, a fundamental limitation of all of those approaches is that the paucity of starting material within a single cell, coupled with the partial capture rate of the experimental protocols, invariably leads to noisy data with limited sensitivity. While this limitation affects all single-cell methods, the effect is less pronounced when profiling abundant molecules such as RNA or proteins, compared with DNA. Together with the relative ease of profiling RNA, compared with proteins, single-cell RNA sequencing (scRNA-seq) has recently become widely used across diverse biological disciplines, including cancer. Single-cell DNA sequencing, although more technically challenging, has also attracted significant attention due to the critical importance of cancer mutations and their heterogeneity in the context of tumor evolution. Application of additional single-cell genomic methods, such as assay for transposase-accessible chromatin,10 DNA methylation, and mass cytometry,11 has so far been more limited, yet as these provide complementary information to that of DNA and RNA profiling and are consistently improving, we anticipate increased use of these methods in the next 5 years.
Single-cell DNA or RNA profiling of tumors is conceptually similar to bulk profiling, with few important differences. First, tumor samples are acutely disaggregated into single-cell suspension using a combination of mechanical and enzymatic digestion protocols. Second, individual cells are separated either by flow cytometry into 96/384-well plates or by microfluidic devices into distinct chambers (eg, by Fluidigm C1) or droplets (eg, in Drop-Seq,12 In-Drop,13 or 10X platforms before they are profiled). Special attention should be given to the timing of dissociation and encapsulation or sorting to ensure that tumor cells do not alter their expression state or viability. Third, it should be noted that given the limited sensitivity of single-cell RNA or DNA profiling, the power of these methods primarily lies in interrogating signatures composed of multiple genes that are involved in coherent biological phenomena (see examples and discussions below). Further description of the single-cell methods is provided elsewhere,6,14,15 while here we focus on the lessons learned from applying these approaches to gliomas.
Genetic Heterogeneity
Genetic heterogeneity is an important determinant of treatment failure in cancer. In gliomas, it has been the most extensively highlighted in glioblastoma, isocitrate dehydrogenase (IDH)-wildtype. Glioblastoma is characterized by a complex genetic landscape with both inter- and intratumoral heterogeneity.16 Signature copy number variations include gain of chromosome 7 and loss of chromosome 10, events that are thought to represent the earliest genetic alteration in its pathogenesis.17 Glioblastoma is additionally characterized by amplifications and rearrangements of receptor tyrosine kinases (RTKs), including EGFR, PDGFRA, or more rarely MET. EGFR amplifications are frequently associated with deletions of exons that encode for the epidermal growth factor receptor (EGFR) extracellular domain.18–22 Gene rearrangements are also found in the extracellular domain or PDGFRA.18 While these genetic events have been studied for many years and shown to transduce aberrant signaling, their inhibition by tyrosine kinase inhibitors (TKIs) has yet to show significant clinical efficacy. There are many potential explanations for this lack of therapeutic effect: (i) Additional mutations (loss of phosphatase and tensin homolog in 15%–40% of glioblastomas and other mutations) activate the mitogen-activated protein kinase pathway, downstream of RTK, and thereby maintain the pathway active despite RTK inhibition.23 (ii) Adaptation to RTK inhibition occurs through upregulation of platelet derived growth factor receptor (PDGFR) β,24 elimination of double minute chromosomes (small fragments of extrachromosomal DNA) containing EGFR variant III during treatment,25 or other feedback mechanisms. (iii) RTK inhibition is insufficient.26 (iv) Through intratumoral heterogeneity in RTK alterations: resistance to TKIs can occur through distinct RTK alterations in different cells of the same tumor, as supported by the observation that EGFR-amplified glioblastomas also contain PDGFRA-amplified tumor cells.27 The latter explanation calls for further analysis of genetic intratumoral heterogeneity through the use of single-cell genomic approaches. In glioblastoma, single-cell DNA28 and RNA29,30 sequencing efforts have further extended our understanding of intratumoral heterogeneity and shown that (i) distinct cells in the same tumor express different EGFR or PDGFRA variants; (ii) the same cell can coexpress multiple EGFR oncogenic variants; (iii) specific transcriptional programs are associated with PDGFRA and EGFR alterations during tumor evolution—for example, PDGFRA alterations were associated with induction of an expression program reminiscent of oligodendrocyte progenitor cells, while EGFR alterations were linked to programs of invasion30; (iv) there is important cell-to-cell variability in the expression of receptors and ligands for critical signaling pathways.29 These observations are also supported by bulk and single-cell multifocal and longitudinal profiling16,31 and highlight the numerous challenges for TKIs in neuro-oncology.
Glioma Stem Cells
Cellular hierarchies are increasingly appreciated to play critical roles in different malignancies.1,32 Hematopoietic cancers as well as a number of solid tumors have been shown to contain subpopulations of cells endowed with tumor initiating potential and stem cell properties, including expression of embryonic or tissue stem cell genes. These cells, termed cancer stem cells (CSCs), are generally thought to underlie tumor growth, while the bulk of the tumor may be composed of more differentiated progeny. CSCs are also thought to be more resistant to existing anticancer therapies, thereby driving tumor regrowth following treatment.33,34 It is thought that the defining properties of CSCs are rooted in their epigenetic state, governed by the activity of transcription factors, chromatin regulators, and associated cellular networks.35 It follows that attempts to increase therapeutic effectiveness in tumors with a documented cellular hierarchy should aim at targeting or differentiating CSCs.
The CSC model has far-reaching implications, yet remains controversial primarily due to 3 unresolved questions: (i) CSCs from human tumors are mostly identified by functional assays such as transplantation into recipient hosts of a different species (eg, in vivo limiting dilution assay) or by assays performed in vitro (eg, colony or sphere forming assays); the relevance of these assays to tumor cells in situ is often debated and it thus remains unclear which tumor types contain CSCs and how to best define them. (ii) The relative contribution of genetic mutations to the putative hierarchies has remained difficult to probe, making it challenging to separate genetic evolution from nongenetic hierarchies. (iii) Potential dynamic state transitions complicate any static observation, as CSC-like features have been proposed to be acquirable by non-CSC populations.1,32,36 While the latter point is hard to address by single-cell profiling approaches, these novel methods pave the way to fully address the first 2 questions, by characterizing the diversity of cellular states within tumors, their genotype (at least partially), and their similarity to normal cell types.
Many groups have identified putative CSCs in glioblastoma, IDH-wildtype.32 Glioblastoma CSCs have been isolated using a variety of cell surface markers, suggesting that distinct CSC populations can be identified in different glioblastoma patients.34,37–40 Unbiased approaches, such as scRNA-seq, would be important to reveal the full expression programs underlying these subpopulations, understand the diversity of CSC programs, and identify potential targets. Single-cell RNA-seq in glioblastoma29 revealed a putative CSC program, yet the study was limited by the low number of single cells profiled per tumor (~100) and the size of the cohort (5 tumors). Additional studies with dramatically increased throughput (eg, with massively parallel single-cell profiling) and scale are warranted to refine our understanding of glioblastoma cellular architecture.
While multiple studies isolated and interrogated CSCs in IDH-wildtype glioblastoma, the existence of such cells is not well documented in IDH-mutant gliomas. This is mostly due to the difficulty of establishing the in vivo functional assays assessing tumor initiation, as these tumors tend not to grow in animal models when xenotransplanted. To overcome these limitations, analysis of the transcriptomes of individual cells by scRNA-seq represents a very compelling alternative strategy that can provide insight into the cellular architecture of IDH-mutant gliomas, in situ and directly in patients. A recent study leveraged scRNA-seq to explore the cellular programs of IDH-mutant oligodendroglioma, focusing on untreated grade II tumors—an early stage in IDH-mutant glioma development, in which cells might better resemble their normal counterparts.41 The study showed that most cancer cells are differentiated and reminiscent of one of 2 glial lineages (oligodendrocyte-like or astrocyte-like cells), while a smaller subset of cells appear undifferentiated and resemble neural stem/progenitor cells.41 These developmental programs were observed in all tumors analyzed and across multiple genetic subclones within the same tumor, suggesting that stemness and differentiation programs are at least in part independent of genetic evolution. Importantly, by examining gene expression programs which are activated during the cell cycle, the study found that proliferation is highly enriched in undifferentiated cells; these results together point to a model whereby a subpopulation of stemlike cells is responsible for fueling the growth of oligodendroglioma, while most cancer cells are differentiated and do not cycle. Notably, scRNA-seq analysis of IDH-mutant astrocytoma revealed a strikingly similar cellular composition to that observed in oligodendroglioma.42 This hints at shared developmental and lineage programs between these 2 types of IDH-mutant gliomas (see “Glioma Classification” section below) and suggests a common histogenesis, which is also supported by a similar age of onset for astrocytoma (IDH-A) and oligodendroglioma (IDH-O) in adults in their 30s and 40s.43 In sum, scRNA-seq analysis of IDH-mutant gliomas supports that they are stem cell–driven tumors and that targeting a specific cellular phenotype of stem cells or triggering stem cell differentiation could have a major impact on their management.
Future works applying similar strategies will interrogate the developmental lineages and putative CSC programs in a range of malignancies. In addition, when feasible, identification of putative CSCs by scRNA-seq should be followed by functional studies to test their tumor initiation capacity and drug resistance (due to a lack of functional models, this could not be tested in IDH-mutant gliomas). While conceptually similar to previous studies in the CSC field, these renewed efforts will leverage single-cell approaches to better define CSC markers in an comprehensive manner.
Glioma Classification
Patients with similar tumors often present dissimilar responses to treatments, motivating their classification into meaningful subtypes that can guide precision medicine. Indeed, impactful tumor classification has been one of the main goals of cancer research and of large-scale tumor profiling studies such as The Cancer Genome Atlas (TCGA). In diffuse gliomas, genetic characterization has revolutionized our understanding of tumor types and their pathogenesis and is a cornerstone of the 2016 World Health Organization (WHO) classification of brain tumors.44 While genomics have been critical, the role of transcriptional classification remains debated. In glioblastoma, TCGA analysis suggested the existence of 4 transcriptional subtypes (proneural, classical, neural, and mesenchymal), with partially distinct genetic aberrations, clinical behavior, and response to therapy.45 For example, patients with the classical or mesenchymal subtype were shown to benefit the most from concurrent chemotherapy/radiation, whereas those with other subtypes did not.18,45 However, subsequent studies found that distinct subtypes may be represented in different areas of the same glioblastoma31 and that subtypes can change at recurrence, suggesting influences from the microenvironment and from tumor evolution. Single-cell analysis further demonstrated that individual tumors consistently contain cancer cells that resemble distinct subtypes.29 Thus, while the bulk-level classification is driven by the most common of those programs, single-cell analysis shows that distinct subtypes are observed in individual patient tumors, even within the same spatial region. Additional single-cell studies with larger numbers of samples and cells, and improved technologies (eg, profiling DNA + RNA from the same single cell46; retaining spatial information of individual cells47,48) will further interrogate these programs and their relationship to genetic and regional influences. Furthermore, profiling of epigenetic states may help to identify the underlying regulatory circuits that maintain these coexisting states.
Beyond subtypes of the “same” type of glioma, single-cell approaches can shed light on the classification to distinct classes of gliomas by elucidating the differences between them in terms of genetics, developmental lineages, and TME. For example, a recent study focused on IDH-mutant gliomas, interrogating genetically defined IDH-O and IDH-A. IDH-O and IDH-A differ by morphology and signature genetic events—such as 1p/19q codeletion, hTERT promoter mutation in IDH-O and TP53, and ATRX mutations in IDH-A. To comprehensively understand the differences between IDH-O and IDH-A, the study examined the expression differences between them using a large cohort of bulk TCGA samples and a smaller cohort profiled by scRNA-seq.42 Strikingly, most gene expression differences that were found between IDH-O and IDH-A bulk samples were not recapitulated in the single-cell analysis of cancer cells and were instead reflecting the TME. IDH-O bulk samples are associated with much higher expression of neuronal genes (possibly reflecting a preferential cortical location for IDH-O), while IDH-A bulk samples are associated with much higher expression of genes expressed by microglia/macrophages (see the “Tumor Microenvironment” section below), and neither of these sets of genes were observed in scRNA-seq of cancer cells.42 Additionally, differences between IDH-O and IDH-A that were positively reproduced in single-cell analysis were primarily accounted by genetics: Most are located on chromosome arm 1p or 19q or are known targets of the protein capicua homolog (CIC) or P53, such that their differential expression is expected based on the genetic differences between IDH-O and IDH-A. Therefore, the widespread expression differences between bulk IDH-O and IDH-A reflect TME and genetics. In contrast, no differences were observed in oligodendrocytic and astrocytic lineage-differentiation genes, arguing against the common notion that IDH-O and IDH-A are of distinct glial lineages. Indeed, as noted above, the study found a similar hierarchy in IDH-O and IDH-A, with proliferating neural stem/progenitor cells and 2 arms of differentiations toward both the oligodendrocytic and astrocytic lineages within each tumor type. Taken together, this work redefined the distinction between IDH-O and IDH-A, highlighting previously unappreciated differences in TME and questioning the notion of distinct glial lineages. Future analysis of additional glioma types and brain tumors will extend this approach and provide a systematic framework to understand differences between subclasses of gliomas.
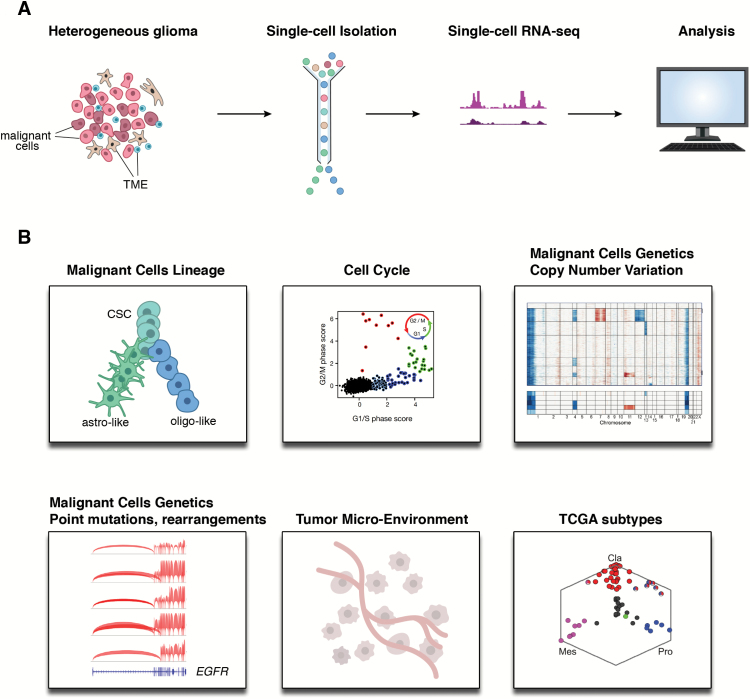
Fig. 1.
Single-cell RNA-seq in clinical gliomas. (A) Pipeline for single-cell expression profiling and analysis from tumor samples. (B) Single-cell RNA-seq can provide information on malignant cell lineages,41,42 cancer stem programs,41,42 cell cycle, malignant cell genetics (copy number variation, point mutations, genetic rearrangements),29,41,42 the tumor microenvironment,42 and tumor classification (eg, TCGA subtypes).29 Abbreviations: Cla: classical; Mes: mesenchymal; Pro: proneural. Figure adapted from references.29,41,42
Tumor Microenvironment
Gliomas contain a high proportion of non-malignant cell types that constitute the TME. The TME not only confounds the interpretation of bulk glioma profiles, but also directly influences the biology of gliomas. Cells in the TME secrete various ligands which are sensed by cancer cells and influence various pathways. Furthermore, cells in the TME physically interact with malignant cells, shape tissue organization, and determine nutrient availability. Notably, cells in the TME are in turn affected by cancer cells in ways that remain poorly understood. Thus, the entire tumor ecosystem must be considered in order to enable a complete understanding of tumor biology.
In scRNA-seq experiments, malignant cells can be distinguished from non-malignant cells based on their genetics and gene expression patterns. Mutations in transcribed regions may be identified from scRNA-seq, although this approach has limited sensitivity41; due to partial capture of transcribed regions, low or absent expression of many mutated genes and allele-specific expression of mutated genes, scRNA-seq was estimated to have, on average, only ~1% sensitivity for detection of a specific mutation. As an alternative, recent studies developed an approach to infer chromosomal aberrations based on the overall expression of genes in each chromosomal region.29,41,42,49 As most types of glioma harbor signature chromosomal events, this approach has proven very robust and informative. In the absence of chromosomal aberrations, the distinct gene expression clustering of malignant and non-malignant cells, along with at least partial genetic information on point mutations, is sufficient to enable a robust classification of cells into malignant and non-malignant. Once non-malignant cells are identified, these can be clustered to identify distinct cell types. This approach has been successful in identifying all major cell types in the TME of melanoma.49 Application to gliomas revealed primarily 2 cell types in the TME: microglia/macrophages (which are discussed further below) and oligodendrocytes. This highlights a potential difficulty in recovering other cell types in the TME of gliomas—such as neurons, astrocytes, endothelial cells, and T cells. While T cells are found at a low frequency in gliomas, they can be successfully recovered by enrichment for CD3 (unpublished data); the other cell types appear to reflect a distinct challenge, as they are not recovered despite their expected high frequency. One hypothesis is that these cells are especially sensitive to tumor dissociation (possibly due to their complex cellular structure, eg, neurons) or not optimally dissociated by protocols tailored for cancer cell separation (eg, blood vessels). Future studies will explore additional approaches, such as single nuclei sequencing,50 that avoid tumor digestion and dissociation and can potentially recover all cellular constituents.
Although current scRNA-seq studies of glioma were limited in their coverage of the TME, they revealed surprising patterns of diversity among microglia/macrophages. Microglia are the predominant resident immune cells in the brain and are therefore present within glioma. Macrophages may also enter glioma through the circulation.51 Since microglia and macrophages share a similar expression profile, the composition of immune cells in gliomas has been the subject of much debate. Through profiling of >1000 CD45+ cells from 13 IDH-A and IDH-O samples,42 the expression diversity among immune cells in these tumors was shown to be consistent with the differences between microglia and macrophages, as defined by other studies.52 However, instead of a model whereby 2 expression states can be discerned (microglia and macrophages), this study highlighted a continuum in which many cells reflect an intermediate state between a microglia and a macrophage expression program. Thus, even though there might only be 2 tissues of origin (brain resident vs circulating), other factors such as the microenvironment might generate a continuum of states, as recently suggested.52 Furthermore, each tumor was associated with only a limited range of states within this continuum, with some tumors enriched for microglia-like and others enriched for macrophage-like cells. Both within the single-cell data and within the TCGA bulk datasets, this pattern was associated with tumor grade: Tumors of low grade tend to have microglia-like cells, while tumors of higher grade tend to be enriched in macrophage-like cells. These results suggest that early in their development gliomas primarily contain brain-resident microglia cells and are later infiltrated by macrophages as they progress, concomitant with the increase in vascularity. Indeed, an endothelial expression signature correlates with the macrophage-like, but not microglia-like, expression program in the TCGA cohorts.42 Taken together, these results hint at the dynamics of the glioma immune composition and at the reprogramming of immune cells within the TME, which blurs the differences between microglia and macrophages and creates a continuum of intermediate cellular states. Recent work comparing macrophages between glioma and healthy brains also suggested that, in contrast to the traditional classification of tumor macrophages to M1 and M2 polarization, the macrophages in glioblastoma may reflect a nonpolarized M0 state.53 Future studies will be required to explore the significance of these cellular states to glioma progression and the interactions of microglia/macrophages with cancer cells on one hand and with T cells on the other hand.
Concluding Remarks
Research in human gliomas has traditionally focused on cellular models or on bulk tissue sample characterization. While these approaches have revolutionized our understanding of these tumors, they have masked aspects of the complexity of glioma ecosystems. Single-cell methods are now enabling a new kind of studies, combining the authenticity of patient samples with the power of single-cell resolution to address unresolved fundamental questions. While the studies described here reflect the first iteration of such an approach, we anticipate that continued improvements in single-cell technologies, decreased costs, and their deployment to a large number of glioma samples and in a variety of clinical contexts will provide a basis for deeper understanding of tumor biology. In particular, such efforts will redefine genetic heterogeneity, cellular lineages, CSC programs, and the TME across different types of glioma and uncover potential targets for future therapies.
References
- 1. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. [DOI] [PubMed] [Google Scholar]
- 2. Filbin MG, Suvà ML. Gliomas genomics and epigenomics: arriving at the start and knowing it for the first time. Annu Rev Pathol. 2016;11:497–521. [DOI] [PubMed] [Google Scholar]
- 3. Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. [DOI] [PubMed] [Google Scholar]
- 4. Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17(3):175–188. [DOI] [PubMed] [Google Scholar]
- 5. Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541(7637):331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziegenhain C, Vieth B, Parekh S et al. . Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65(4):631–643.e4. [DOI] [PubMed] [Google Scholar]
- 7. Wu M, Singh AK. Microfluidic flow cytometry for single-cell protein analysis. Methods Mol Biol. 2015;1346:69–83. [DOI] [PubMed] [Google Scholar]
- 8. Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16(12):716–726. [DOI] [PubMed] [Google Scholar]
- 9. Macaulay IC, Ponting CP, Voet T. Single-cell multiomics: multiple measurements from single cells. Trends Genet. 2017;33(2):155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buenrostro JD, Wu B, Litzenburger UM et al. . Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165(4):780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macosko EZ, Basu A, Satija R et al. . Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klein AM, Mazutis L, Akartuna I et al. . Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610–620. [DOI] [PubMed] [Google Scholar]
- 15. Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34(11):1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee JK, Wang J, Sa JK et al. . Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng YK, Beroukhim R, Levine RL et al. . A mathematical methodology for determining the temporal order of pathway alterations arising during gliomagenesis. PLoS Comput Biol. 2012;8(1):e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brennan CW, Verhaak RG, McKenna A et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87(21):8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwechheimer K, Huang S, Cavenee WK. EGFR gene amplification–rearrangement in human glioblastomas. Int J Cancer. 1995;62(2):145–148. [DOI] [PubMed] [Google Scholar]
- 21. Biernat W, Huang H, Yokoo H, Kleihues P, Ohgaki H. Predominant expression of mutant EGFR (EGFRvIII) is rare in primary glioblastomas. Brain Pathol. 2004;14(2):131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wikstrand CJ, Reist CJ, Archer GE, Zalutsky MR, Bigner DD. The class III variant of the epidermal growth factor receptor (EGFRvIII): characterization and utilization as an immunotherapeutic target. J Neurovirol. 1998;4(2):148–158. [DOI] [PubMed] [Google Scholar]
- 23. Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15(5):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akhavan D, Pourzia AL, Nourian AA et al. . De-repression of PDGFRbeta transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3(5):534–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nathanson DA, Gini B, Mottahedeh J et al. . Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343(6166):72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vivanco I, Robins HI, Rohle D et al. . Differential sensitivity of glioma versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snuderl M, Fazlollahi L, Le LP et al. . Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 28. Francis JM, Zhang CZ, Maire CL et al. . EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4(8):956–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel AP, Tirosh I, Trombetta JJ et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Müller S, Liu SJ, Di Lullo E et al. . Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol. 2016;12(11):889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sottoriva A, Spiteri I, Piccirillo SG et al. . Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bao S, Wu Q, McLendon RE et al. . Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 34. Chen J, Li Y, Yu TS et al. . A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339(6127):1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gupta PB, Fillmore CM, Jiang G et al. . Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. [DOI] [PubMed] [Google Scholar]
- 37. Anido J, Sáez-Borderías A, Gonzàlez-Juncà A et al. . TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18(6):655–668. [DOI] [PubMed] [Google Scholar]
- 38. Lathia JD, Gallagher J, Heddleston JM et al. . Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6(5):421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh SK, Hawkins C, Clarke ID et al. . Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 40. Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tirosh I, Venteicher AS, Hebert C et al. . Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Venteicher AS, Tirosh I, Hebert C et al. . Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355(6332):pii: eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cancer Genome Atlas Research, N., Brat DJ, Verhaak RG et al. . Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK.. WHO Classification of Tumors of the Central Nervous System (revised 4th ed.). Lyon: IARC; 2016. [DOI] [PubMed] [Google Scholar]
- 45. Verhaak RG, Hoadley KA, Purdom E et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Macaulay IC, Haerty W, Kumar P et al. . G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12(6):519–522. [DOI] [PubMed] [Google Scholar]
- 47. Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11(4):360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92(2):342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tirosh I, Izar B, Prakadan SM et al. . Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Habib N, Li Y, Heidenreich M et al. . Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353(6302):925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lavin Y, Winter D, Blecher-Gonen R et al. . Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gabrusiewicz K, Rodriguez B, Wei J et al. . Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight. 2016;1(2):pii: e85841. [DOI] [PMC free article] [PubMed] [Google Scholar]