Abstract
Background
Diffuse lower-grade gliomas (LGGs) are genetically classified into 3 distinct subtypes based on isocitrate dehydrogenase (IDH) mutation status and codeletion of chromosome 1p and 19q (1p/19q). However, the subtype-specific effects of additional genetic lesions on survival are largely unknown.
Methods
Using Cox proportional hazards regression modeling, we investigated the subtype-specific effects of genetic alterations and clinicopathological factors on survival in each LGG subtype, in a Japanese cohort of LGG cases fully genotyped for driver mutations and copy number variations associated with LGGs (n = 308). The results were validated using a dataset from 414 LGG cases available from The Cancer Genome Atlas (TCGA).
Results
In Oligodendroglioma, IDH-mutant and 1p/19q codeleted, NOTCH1 mutations (P = 0.0041) and incomplete resection (P = 0.0019) were significantly associated with shorter survival. In Astrocytoma, IDH-mutant, PIK3R1 mutations (P = 0.0014) and altered retinoblastoma pathway genes (RB1, CDKN2A, and CDK4) (P = 0.013) were independent predictors of poor survival. In IDH-wildtype LGGs, co-occurrence of 7p gain, 10q loss, mutation in the TERT promoter (P = 0.024), and grade III histology (P < 0.0001) independently predicted poor survival. IDH-wildtype LGGs without any of these factors were diagnosed at a younger age (P = 0.042), and were less likely to have genetic lesions characteristic of glioblastoma, in comparison with other IDH-wildtype LGGs, suggesting that they likely represented biologically different subtypes. These results were largely confirmed in the cohort of TCGA.
Conclusions
Subtype-specific genetic lesions can be used to stratify patients within each LGG subtype. enabling better prognostication and management.
Keywords: diffuse lower-grade glioma, genetic alteration, prognostic factor
Importance of the study
The clinical significance of genetic lesions within each LGG subtype has not been fully elucidated. In this study, we identified sets of subtype-specific genetic and clinicopathological markers for each World Health Organization subtype. The study subjects were a large cohort of patients who were genotyped for known or putative driver mutations and copy number variations associated with LGGs. Importantly, given that LGGs frequently have an indolent clinical history, the subjects were followed up for sufficiently long periods to accurately evaluate overall survival. In IDH-wildtype LGGs, the subsets of patients identified by these markers are likely to represent subtypes that differ in terms of overall survival, mean age, genetic profile, and patterns of DNA methylation. Our results could be used to establish a set of novel prognostic biomarkers, allowing patients within each LGG subtype to be further stratified for better clinical management.
Diffuse gliomas, the most prevalent primary malignant brain tumors, have been classified by the World Health Organization (WHO) into grades II–IV gliomas.1 Diffuse grade IV glioma, or glioblastoma (GBM), represents the most aggressive subtype, with a uniformly dismal prognosis: The 5-year overall survival (OS) rate is less than 5%.2 By contrast, diffuse grade II and III gliomas are generally less aggressive tumors with a median survival of more than 7 years.3 Although grades II and III are often collectively termed diffuse lower-grade gliomas (LGGs), there is substantial heterogeneity among these tumors in terms of pathological features and clinical outcome.1
In this regard, a significant advance in recent years has been the identification of a set of genetic lesions that are characteristic of LGGs and correlate well with histology and clinical outcome. These include highly recurrent mutations in the genes encoding isocitrate dehydrogenase (IDH) 1 and 2 and codeletion of 1p and 19q (1p/19q).4–8 In fact, LGGs can be more effectively classified into discrete subsets with unique profiles of histology and survival on the basis of these genetic lesions than based on histopathology alone. IDH-mutant LGGs are associated with a longer OS than IDH-wildtype LGGs.9,10 Among IDH-mutant LGGs, those with 1p/19q codeletion are predominantly oligodendroglial tumors (“Oligodendroglioma, IDH-mutant and 1p/19q codeleted,” hereafter called Oligodendroglioma IDH-mut/1p19q-codel) and are associated with significantly better survival than those without 1p/19q codeletion, which typically exhibit astrocytic histology (“Astrocytoma, IDH-mutant,” hereafter called Astrocytoma IDH-mut).11,12 Recently, 2 comprehensive molecular studies reported the landscape of genetic alterations in large cohorts of LGG patients.3,13 Both studies not only confirmed the aforementioned genetic subtypes and their impact on survival, but also demonstrated that each WHO subtype has a characteristic set of features, including additional genetic alterations, mean age, and DNA methylation and gene expression profiles. Thus, each subtype is considered to represent a discrete clinicopathological entity.
Given the high level of intertumor heterogeneity inferred from the presence of additional genetic lesions in each genetic subtype, it is possible that within each WHO subtype, we could find one or more subgroups that exhibit distinct biological behaviors and prognosis. In this regard, recent studies reported a number of genetic alterations that were implicated in poor clinical outcomes in particular subtypes, including CIC mutation in Oligodendroglioma IDH-mut/1p19q-codel14; loss of chromosome 9p, mutation of PIK3CA and PIK3R1, and deletion of CDKN2A in Astrocytoma IDH-mut15–17; and mutation of the TERT promoter in IDH-wildtype LGGs.18 However, the effects of these alterations on OS have not been systemically confirmed in a large cohort of patients who were fully genotyped for genetic alterations that are frequently found in LGGs and for whom long-term follow-up data were available; the latter point is essential for accurate evaluation of OS of a disease that frequently exhibits an indolent clinical history.
In this study, we investigated the effects of subtype-specific genetic alterations on OS, using datasets from 2 independent cohorts of LGG patients: one from Japan (JPN) for discovery, and one from The Cancer Genome Atlas (TCGA) for validation. All of the subjects had been fully genotyped for known or putative driver mutations and copy number variations (CNVs) associated with LGGs and annotated for relevant clinical characteristics and long-term survival. In the light of recent advances in our molecular understanding of diffuse gliomas, it remains to be determined how GBM and LGGs, especially anaplastic astrocytoma, differ from each other. In fact, Astrocytoma IDH-mut and IDH-wildtype LGGs share molecular and clinical features with GBM, IDH-mutant and GBM, IDH-wildtype, respectively.13,19 In this study, we used TCGA GBM data to compare the clinical, genetic, and epigenetic features of LGG subtypes with unfavorable prognostic factors with those of GBM.
Materials and Methods
Patients and Dataset
In total, 308 (JPN) and 414 (TCGA) patients aged ≥18 years with previously untreated supratentorial diffuse grade II and III gliomas were analyzed, along with 471 GBM patients from TCGA.20,21 Clinical and pathological characteristics of patients are summarized in Table 1 (also see Supplementary Table S1). Tumors were classified into 3 major subtypes according to the WHO classification, revised in 2016 (Oligodendroglioma IDH-mut/1p19q-codel, Astrocytoma IDH-mut, and IDH-wildtype LGGs),1 although for IDH-wildtype LGGs we did not distinguish between astrocytoma and oligodendroglioma. In the JPN cohort, the diagnosis of LGG was made by local pathologists in the participating centers. For 288 (93.5%) of the 308 JPN samples, histological specimens were centrally reviewed by 2 independent board-certified pathologists, as previously described.3 Data of preoperative MRI of Oligodendroglioma IDH-mut/1p19q-codel with contrast enhancement was available in 132 (93.6%) of JPN and 49 (35.2%) of TCGA patients (http://public.cancerimagingarchive.net/, accessed August 22, 2017). For the cohort from TCGA, we used DNA methylation-based subgroups data from Ceccarelli et al, who divided gliomas into 6 subgroups.20 The extent of tumor resection was unknown in 3 JPN and 10 TCGA cases. Informed consent was obtained from all JPN patients before tumor sampling by surgery, which was performed between 1990 and 2013. This study was approved by the ethics committees or institutional review boards of all participating institutes.
Table 1.
Clinical characteristics of patients
| Lower-Grade Gliomas | Glioblastoma | |||||||
|---|---|---|---|---|---|---|---|---|
| Cohort | JPN (n = 308) | TCGA (n = 414) | TCGA (n = 471) | |||||
| Subtype | Oligo, IDH-mut/ 1p19q-codel |
Astro, IDH-mut | IDH-WT LGGs | Oligo, IDH-mut/ 1p19q-codel |
Astro, IDH-mut |
IDH-WT LGGs | GBM, IDH-mut |
GBM, IDH-WT |
| Case, n (%) | 141 (46) | 109 (35) | 58 (19) | 139 (34) | 196 (47) | 79 (19) | 37 (8) | 434 (92) |
| Follow-up years—median (25th and 75th percentiles) | ||||||||
| 7.17 (3.56–10.75) |
5.03 (3.01–9.14) |
5.05 (3.24–8.85) |
1.71 (1.03–3.22) |
1.67 (1.10–3.33) |
1.35 (0.66–1.92) |
0.87 (0.48–1.78) |
0.70 (0.45–1.00) |
|
| No. of event | 32 | 43 | 40 | 21 | 43 | 43 | 22 | 318 |
| OS (year)—median (95% CI) | ||||||||
| 20.45 (16.4-NR) |
8.41 (7.10-NR) |
2.45 (2.10–4.11) |
11.19 (6.52-NR) |
7.29 (5.62–10.9) |
1.78 (1.54–2.23) |
3.23 (2.02–7.54) |
1.11 (1.02–1.24) |
|
| Age at diagnosis—median (25th and 75th percentile) | ||||||||
| 45 (36–54) | 37 (30–46) | 50 (41–65) | 45 (37–55) | 36 (30–43) | 55 (45–62) | 38 (28–45) | 60 (52–69) | |
| WHO grade, n (%) | ||||||||
| Grade II | 81 (57) | 77 (71) | 20 (34) | 77 (55) | 100 (56) | 16 (20) | ||
| Grade III | 60 (43) | 32 (29) | 38 (66) | 62 (45) | 96 (49) | 63 (80) | ||
| Grade IV | 37 (100) | 434 (100) | ||||||
| Tumor location (supratentorial), n (%) | ||||||||
| Frontal lobe | 113 (80) | 80 (73) | 34 (59) | 102 (73) | 117 (60) | 29 (37) | ||
| Occipital lobe | 2 (1) | 1 (1) | 3 (5) | 3 (2) | 1 (1) | 0 (0) | ||
| Parietal lobe | 13 (9) | 10 (9) | 3 (5) | 11 (8) | 20 (10) | 7 (9) | ||
| Temporal lobe | 12 (9) | 18 (17) | 17 (29) | 21 (15) | 57 (29) | 41 (52) | ||
| Unknown | 1 (1) | 0 (0) | 1 (2) | 2 (1) | 1 (1) | 2 (3) | 37 (100) | 434 (100) |
| Surgery, n (%) | ||||||||
| GTR | 99 (70) | 66 (61) | 27 (47) | 88 (63) | 115 (59) | 44 (56) | ||
| PR | 40 (28) | 42 (39) | 31 (53) | 49 (35) | 73 (37) | 35 (44) | ||
| Unknown | 2 (1) | 1 (1) | 0 (0) | 2 (1) | 8 (4) | 0 (0) | 37 (100) | 434 (100) |
Abbreviations: Oligo = oligodendroglioma; Astro = astrocytoma, WT = wildtype; NR = nor reached; GTR = gross total resection; PR = partial resection.
Mutations and Copy Number Variations
Detection of gene mutations and CNVs in JPN patients was performed as previously described.3 In brief, whole-exome sequencing (WES) and targeted sequencing data were obtained from 52 and 308 cases, respectively, of the JPN cohort. In targeted sequencing, we selected 185 genes, which included recurrently mutated genes in LGGs and related disorders as previously described.3 Somatic mutation calling was performed using the empirical Bayesian mutation calling method, in which we adopted variants with variant allele frequencies ≥0.05 in tumor samples.22 We analyzed single nucleotide polymorphism–array data to assess broad and focal CNVs based on a hidden Markov model using Copy Number Analyzer for GeneChip, as previously described.3,23 CNVs that involved over 70% of the affected chromosome arms were considered broad CNVs (Supplementary Figures S1 and S2). For the cases from TCGA, high-throughput sequencing/microarray data and follow-up clinical information as of July 15, 2016 were obtained from http://cancergenome.nih.gov/, accessed August 22, 2017.13 No data for CNVs were available for 39 JPN cases and 1 TCGA case. Mutation status of the TERT promoter was unknown for 24 TCGA cases. Used for subsequent analyses were gene mutations and focal or broad CNVs found in ≥10% of each WHO subtype and major signaling pathways (Notch; retinoblastoma [RB]; receptor tyrosine kinase/phosphoinositide 3-kinase/mammalian target of rapamycin; SWItch/sucrose non-fermentable; and histone methyltransferase). We selected the set of genes that constituted each signaling pathway as described in previous studies (Supplementary Table S2).3,24–26 Altered pathways were defined by mutations or focal CNVs of more than one corresponding gene. Other subtype-specific alterations previously implicated in clinical outcomes of patients were also included in the analysis: specifically, homozygous deletion of CDKN2A/B and mutations of PIK3R1 and PIK3CA in Astrocytoma IDH-mut (Supplementary Table S3).21 We excluded gain of chromosome 7q and loss of chromosome 10p in IDH-wildtype LGGs, because gain of chromosome 7p and 7q or loss of chromosome 10p and 10q almost always co-occurred (P = 4.16e-11 and 2.15e-8 in Fisher’s exact test, respectively) (Supplementary Figure S3).
Statistical Analysis
To analyze the association of the numbers of broad CNVs and somatic mutations (6 and 34, respectively), the 75th percentile was chosen as a cutoff value. Older age was defined as ≥60 years, according to the classification and regression tree analysis (Supplementary Figure S4). Overall survival was calculated from the time of diagnosis until death or last follow-up and evaluated using the log-rank test and Cox proportional hazards regression modeling. Stratified log-rank tests were performed by introducing strata variables. The multivariate Cox regression analysis was performed using backward stepwise selection of variables based on the Akaike information criterion; candidate independent variables including clinicopathological factors (age, WHO grade, and extent of resection) and genetic alterations had P < 0.05 in Cox regression analyses adjusted for age and WHO grade. The proportional hazards assumption was checked before conducting multivariate analyses. In multivariate analyses, we performed multiple imputation of missing values using the bootstrap-based expectation-maximization method and created 5 imputed complete datasets in each WHO subtype. We performed separate survival analyses of all 5 datasets and combined the results using Rubin’s rule.27 Modeling was also performed using a Bayesian model averaging for 267 cases without missing data.28,29 Median follow-up time was assessed among individuals with censored data. Comparisons of frequencies were made using Fisher’s exact test. Differences in age and the number of CNVs and somatic mutations were analyzed using the Wilcoxon rank sum test. Fisher’s exact test with Benjamini–Hochberg correlation (Q-value) was used to investigate the co-occurrence among genetic alterations in IDH-wildtype LGGs. We used “survival” for Cox regression analysis and log-rank test, “MASS” for stepwise Cox regression analysis, “Amelia” for multiple imputation, “cat” for combined results from multiple imputation, “BMA” for Bayesian model averaging, and “rpart” and “rpart.plot” for the classification and regression tree analysis, all of which are included in the statistical software R version 3.1.3 (https://www.r-project.org/, accessed August 22, 2017). P-value and Q-value < 0.05 were taken to indicate statistical significance. Detailed statistical methods are provided in the Supplementary material.
Results
Clinical Features of Major LGG Subtypes in the JPN Cohort
In accordance with previous studies,3,13 patients with LGGs exhibited substantially different OS depending on the subtype: Oligodendroglioma IDH-mut/1p19q-codel had a significantly longer OS (median 20.45 y [95% CI, 16.40, not reached]) than Astrocytoma IDH-mut (8.41 y [7.10, not reached]) (P = 0.0012), which in turn had a significantly better clinical outcome than IDH-wildtype LGGs (2.45 y [2.10–4.11]) (P < 0.0001). In age- and WHO grade–stratified log-rank analysis, the effect of molecular subtype on OS was still significant: P = 0.00029 for Astrocytoma IDH-mut versus Oligodendroglioma IDH-mut/1p19q-codel and P = 0.00042 for IDH-wildtype LGGs versus Astrocytoma IDH-mut. The predominant tumor location was significantly different depending on WHO subtype (P = 0.0038) (Table 1), but no significant association was observed between predominant tumor location and OS in each subtype.
Association of Genetic Alterations with Clinicopathological Features
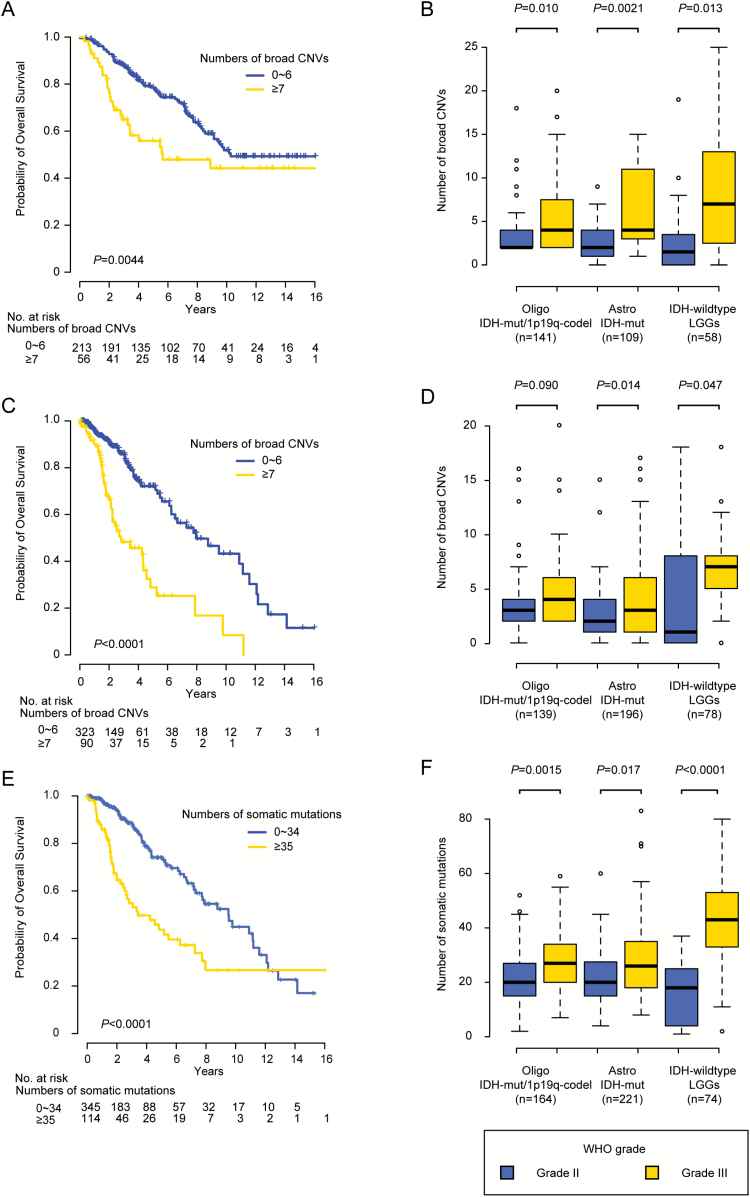
In the JPN cohort, a larger number of broad CNVs were significantly associated with poor prognosis in LGGs: 5-year OS of patients with 0–6 and ≥7 CNVs were 79% and 56%, respectively (P = 0.0044) (Fig. 1A). CNV number still had prognostic significance in LGGs, as determined by log-rank test stratified by molecular subtype (P = 0.017). The number of broad CNVs was also associated with histological grade in LGG: grade III tumors had significantly more broad CNVs than grade II tumors in all LGG subtypes (Fig. 1B). In the TCGA cohort, a larger number of broad CNVs was also significantly associated with reduced OS: 5-year OS of patients with 0–6 and ≥7 CNVs were 72% and 29%, respectively (P < 0.0001) (Fig. 1C), even in the analysis stratified by molecular subtype (P = 0.0065). Furthermore, in patients with Astrocytoma IDH-mut and IDH-wildtype LGGs, albeit not those with Oligodendroglioma IDH-mut/1p19q-codel, grade III tumors were more likely than grade II tumors to have larger numbers of broad CNVs (Fig. 1D). Next, we evaluated the association between the number of somatic mutations and clinicopathological features. For these analyses, we used WES data in the combined JPN and TCGA cohort (n = 459), because the number of JPN patients for whom WES data were available (n = 52) was too small to be assessed separately. A larger number of somatic mutations were significantly associated with clinical outcomes in LGGs: 5-year OS of patients having 0–34 and ≥35 somatic mutations were 74% and 44%, respectively (P < 0.0001) (Fig. 1E), even in the analysis stratified by molecular subtype (P = 0.0047). They were also associated with histological grade in all LGG subtypes (Fig. 1F). These results suggest that larger numbers of broad CNVs and somatic mutations could be associated with more aggressive LGG phenotypes.
Fig. 1.
Association of genetic alterations with clinicopathological features. (A) Kaplan–Meier curves of OS of LGG patients in the JPN cohort, classified based on the number of broad CNVs. (B) Mean numbers of broad CNVs in each molecular subtype in the JPN cohort plotted with 25% and 75% quartiles according to WHO grade. (C) Kaplan–Meier curves of LGG patients in the cohort from TCGA, classified based on the number of broad CNVs. (D) Mean numbers of broad CNVs of each molecular subtype in the cohort from TCGA, plotted with 25% and 75% quartiles according to WHO grade. (E) Kaplan–Meier curves of LGG patients in the combined JPN and TCGA cohort, classified based on the number of somatic mutations. (F) Mean numbers of somatic mutations of each molecular subtype in the combined JPN and TCGA cohort, plotted with 25% and 75% quartiles according to WHO grade. Oligo = oligodendroglioma; Astro = astrocytoma.
Association of Genetic Alterations with Overall Survival
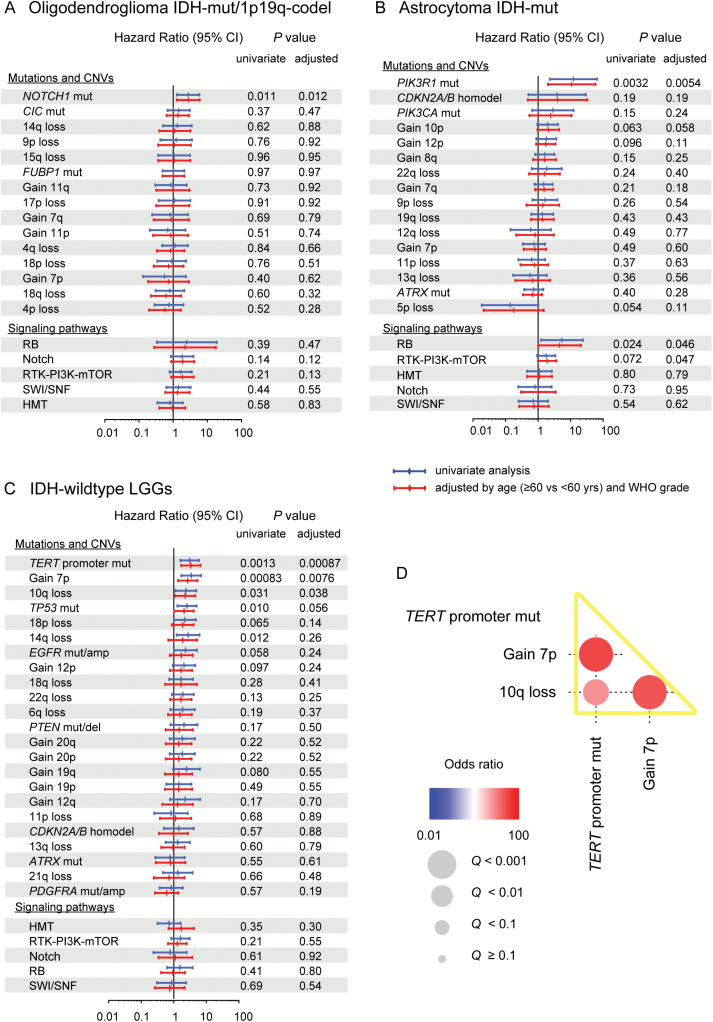
Next, we evaluated the effects of recurrent genetic alterations within each WHO subtype in the JPN cohort. In univariate analysis, NOTCH1 mutations were significantly associated with poor OS in Oligodendroglioma IDH-mut/1p19q-codel, and PIK3R1 mutations and altered RB pathway genes exhibited a similar pattern in Astrocytoma IDH-mut (Fig. 2A and B). In IDH-wildtype LGGs, 5 lesions, including mutation of the TERT promoter and TP53, gain of chromosome 7p, and loss of chromosome 10q and 14q, were shown to negatively affect OS (Fig. 2C). After adjustment for age and WHO grade, these genetic alterations had subtype-specific significant unfavorable prognostic values, except for TP53 mutation (P = 0.056) and loss of chromosome 14q (P = 0.26) in IDH-wildtype LGGs (Fig. 2A–C and Supplementary Figure S5). Mutations in CIC and FUBP1 and those in ATRX were commonly observed in Oligodendroglioma IDH-mut/1p19q-codel and Astrocytoma IDH-mut, respectively, but did not significantly affect OS in the JPN cohort.
Fig. 2.
Hazard ratios (HRs) for OS in univariate and adjusted Cox regression models, according to the presence or absence of mutations or CNVs on each LGG subtype. (A, B, C) Hazard ratios and their 95% CIs plotted for mutations, focal and broad CNVs, and signaling pathways in each LGG subtype, from univariate (blue) and age- and WHO grade–adjusted (red) Cox regression models. Results of HRs and 95% CIs are shown on a log10 scale. (D) Among genetic alterations significantly associated with poor prognosis in univariate Cox analysis of IDH-wildtype LGGs, positive (red) and negative (blue) correlations were detected. Size and color gradients of each circle indicate the level of significance as expressed by Q-values and odds ratios of correlations, respectively. TERT promoter mutation, gain of chromosome 7p, and loss of chromosome 10q were strongly correlated and are indicated by yellow bars. RTK-PI3K-mTOR = receptor tyrosine kinase-phosphoinositide 3-kinase-mammalian target of rapamycin; SWI/SNF = SWItch/sucrose non-fermentable; HMT = histone methyltransferase.
Multivariate Modeling of Overall Survival in LGG Subtypes
In line with previous reports,30,31 age, WHO grade, and extent of resection also significantly affected OS in patents with LGGs in the JPN cohort (Supplementary Table S4). Thus, to determine the contributions of genetic and clinicopathological factors to OS, we performed Cox proportional hazards regression modeling with backward stepwise selection of variables, incorporating the aforementioned clinicopathological factors, in addition to genetic abnormalities with age- and WHO grade–adjusted P-values < 0.05. In this analysis, we noted that gain of chromosome 7p, loss of chromosome 10q, and TERT promoter mutation were strongly mutually correlated in IDH-wildtype LGGs (Fig. 2D) and frequently co-occurred in GBM, IDH-wildtype.20,32 Hence, instead of using the individual lesions as separate variables, we adopted the co-occurrence of all 3 lesions as a single variable for the analysis of IDH-wildtype LGGs. A number of genetic alterations, together with clinicopathological features, were extracted as independent predictors of survival: NOTCH1 mutations (hazard ratio [HR] = 3.14 [95% CI, 1.44–6.84]; P = 0.0041) and extent of resection (partial resection vs gross total resection) (HR = 3.44 [1.59–7.47], P = 0.0019) in Oligodendroglioma IDH-mut/1p19q-codel; PIK3R1 mutations (HR = 16.2 [95% CI, 2.94–89.5]; P = 0.0014) and altered RB pathway genes (HR = 7.08 [95% CI, 1.51–33.2]; P = 0.013) in Astrocytoma IDH-mut; co-occurrence of gain of chromosome 7p, loss of chromosome 10q, and TERT promoter mutation in IDH-wildtype LGGs (HR = 2.53 [95% CI, 1.13–5.65], P = 0.024) and WHO grade (grade III vs grade II) (HR = 7.09 [2.94–17.1], P < 0.0001) (Table 2 and Fig. 3A, C, and E). To address the issue of uncertainty regarding the model ultimately selected through stepwise selection, we also performed an analysis based on Bayesian model averaging. For each WHO subtype, all variables significantly selected through the stepwise selection had larger posterior probabilities than the unselected variables in the Bayesian model averaging (Supplementary Figure S6), suggesting that the modeling was robust and not affected by the method of variable selection.
Table 2.
Hazard ratio for OS in stepwise multivariate survival model
| JPN Cohort | TCGA Cohort | ||||
|---|---|---|---|---|---|
| Subtype | Variables | Hazard Ratio (95% CI) |
P-value | Hazard Ratio (95% CI) |
P-value |
| Oligodendroglioma IDH-mut/1p19q-codel | |||||
| Clinical factors | |||||
| Extent of resection (PR vs GTR) | 3.44 (1.59–7.47) | 0.0019 | 1.40 (0.50–3.94) | 0.53 | |
| Age (≥60 y vs <60 y) | 2.25 (0.89–5.70) | 0.086 | 12.5 (3.84–40.4) | <0.0001 | |
| Genetic factors (present vs absent) | |||||
| NOTCH1 mut | 3.14 (1.44–6.84) | 0.0041 | 1.82(0.69–4.83) | 0.23 | |
| Astrocytoma IDH-mut | |||||
| Clinical factors | |||||
| Extent of resection (GTR vs PR) | 1.77 (0.95–3.29) | 0.072 | 1.16 (0.59–2.31) | 0.66 | |
| Genetic factors (present vs absent) | |||||
| PIK3R1 mut | 16.2 (2.94–89.5) | 0.0014 | 8.69 (1.90–39.7) | 0.0053 | |
| Altered RB pathway genes | 7.08 (1.51–33.2) | 0.013 | 20.5 (6.71–62.9) | <0.0001 | |
| IDH-wildtype LGGs | |||||
| Clinical factors | |||||
| WHO grade (grade III vs grade II) | 7.09 (2.94–17.1) | <0.0001 | 3.57 (1.06–12.0) | 0.040 | |
| Age (≥60 y vs <60 y) | 1.71 (0.86–3.38) | 0.12 | 2.18 (1.12–4.22) | 0.022 | |
| Genetic factors (present vs absent) | |||||
| Co-occurrence of 7p gain, 10q loss, and pTERT mut | 2.53 (1.13–5.65) | 0.024 | 2.11 (1.05–4.24) | 0.037 | |
Abbreviations: mut = mutant; pTERT = TERT promoter; GTR = gross total resection; PR = partial resection.
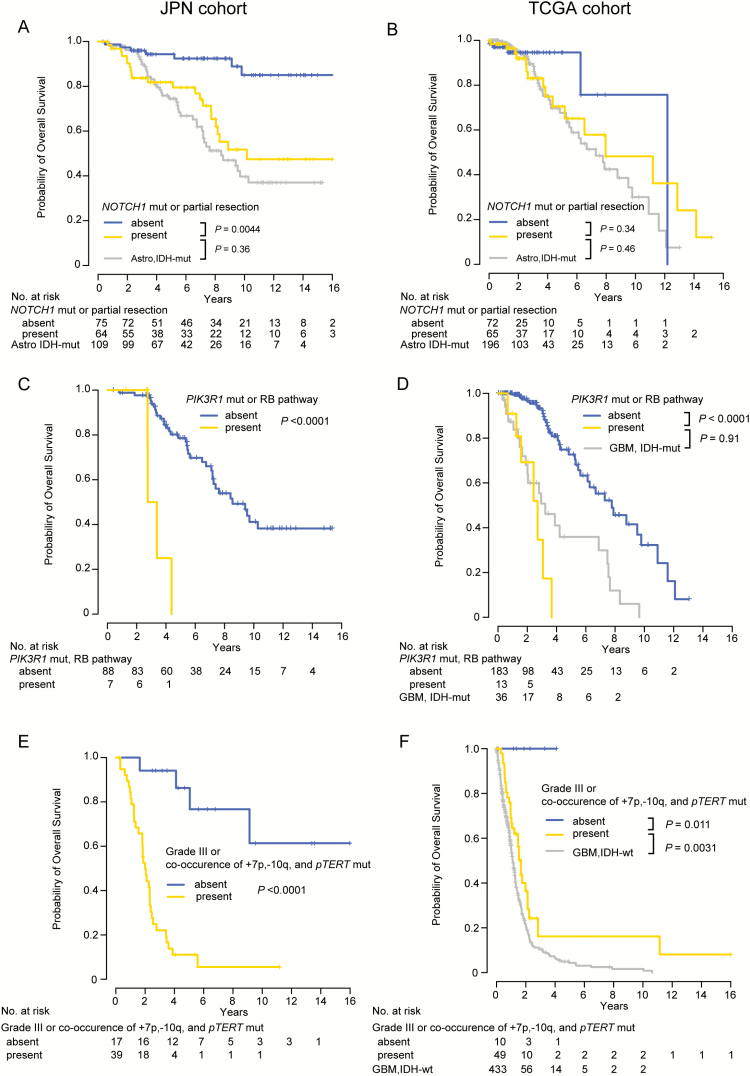
Fig. 3.
OS in each LGG subtype, according to prognostic factors. (A, B) OS of patients with Oligodendroglioma IDH-mut/1p19q-codel according to the presence or absence of prognostic factors, including NOTCH1 mutations and extent of resection (partial resection), in the (A) JPN and (B) TCGA cohorts. For comparison, survival curves for patients with Astrocytoma IDH-mut are also presented. (C, D) OS of patients with Astrocytoma IDH-mut according to the presence and absence of prognostic factors, including PIK3R1 mutations and altered RB pathway genes, in the (C) JPN and (D) TCGA cohorts. For comparison, survival curves for patients with GBM, IDH-mutant from the cohort from TCGA are also presented. (E, F) OS of patients with IDH-wildtype LGGs according to the presence and absence of prognostic factors, including WHO grade (grade III) and co-occurrence of gain of chromosome 7p, loss of chromosome 10q, and TERT promoter mutation (pTERT) in the (E) JPN and (F) TCGA cohorts. For comparison, survival curves for patients with GBM, IDH-wildtype from the cohort from TCGA are also presented. P-values were calculated using the log-rank test.
The results in the JPN cohort were validated in an independent cohort, using the publicly available dataset from TCGA. The JPN and TCGA cohorts were largely similar with regard to most of the clinically relevant demographic features, except for follow-up time, which was substantially longer in the JPN cohort (median 6.17 y) than in TCGA (median 1.59 y) (P < 0.0001). The subtype-specific effects of these genetic alterations and clinicopathological factors on survival that were identified in the JPN cohort were largely confirmed in the cohort from TCGA on the basis of univariate and multivariate analysis, except for the negative effect of NOTCH1 mutations and extent of resection (partial resection) in Oligodendroglioma IDH-mut/1p19q-codel, which were not statistically significant in the cohort from TCGA (Tables 2 and 3).
Table 3.
Hazard ratio for OS in univariate survival model
| JPN Cohort | TCGA Cohort | ||||
|---|---|---|---|---|---|
| Subtype | Variables | Hazard Ratio (95% CI) |
P-value | Hazard Ratio (95% CI) |
P-value |
| Oligodendroglioma IDH-mut/1p19q-codel | |||||
| Clinical factors | |||||
| Extent of resection (PR vs GTR) | 3.05 (1.49–6.26) | 0.0024 | 2.07 (0.77–5.54) | 0.15 | |
| Genetic factors (present vs absent) | |||||
| NOTCH1 mut | 2.64 (1.25–5.59) | 0.011 | 2.16 (0.83–5.64) | 0.11 | |
| Astrocytoma IDH-mut | |||||
| Genetic factors (present vs absent) | |||||
| PIK3R1 mut | 12.3 (2.32–64.8) | 0.0032 | 6.08 (1.42–26.1) | 0.015 | |
| Altered RB pathway genes | 5.47 (1.25–23.9) | 0.024 | 18.8 (6.24–56.5) | <0.0001 | |
| IDH-wildtype LGGs | |||||
| Clinical factors | |||||
| WHO grade (grade III vs grade II) | 7.05 (2.99–16.6) | <0.0001 | 3,83 (1.18–12.5) | 0.026 | |
| Genetic factors (present vs absent) | |||||
| Co-occurrence of 7p gain, 10q loss, and pTERT mut | 2.59 (1.22–5.53) | 0.014 | 2.26 (1.06–4.82) | 0.035 | |
Abbreviations: GTR = gross total resection; PR = partial resection.
Stratification of Patients in LGG Subtypes
Although not statistically significant in the cohort from TCGA, NOTCH1 mutations and the extent of resection (partial resection) were independently associated with poor clinical outcomes in JPN patients with Oligodendroglioma IDH-mut/1p19q-codel. Survival of patients with either or both of these risk factors was significantly shorter than survival of those with neither of these factors in the JPN cohort (P = 0.00044), which did not differ significantly from the survival of patients with Astrocytoma IDH-mut in both the JPN (P = 0.36) and TCGA cohorts (P = 0.46) (Fig. 3A and B). NOTCH1 mutations were strongly associated with positive gadolinium enhancement in preoperative MRI (P = 0.011) and grade III (vs grade II) histology (P = 0.017) in the combined JPN and TCGA cohort, suggesting that NOTCH1 mutations could be associated with more aggressive phenotypes in Oligodendroglioma IDH-mut/1p19q-codel. PIK3R1 mutations and altered RB pathway genes were significantly associated with poor prognosis in Astrocytoma IDH-mut in both the JPN and TCGA cohorts (Tables 2 and 3). In the cohort from TCGA, patients with one or more of these genetic alterations had a prognosis similar to that of GBM, IDH-mutant in the cohort from TCGA (P = 0.91) (Fig. 3D). In IDH-wildtype LGGs, not only the co-occurrence of gain of chromosome 7p, loss of chromosome 10q, and TERT promoter mutation (HR = 2.26 [95% CI, 1.06–4.82], P = 0.035) but also histological grade (WHO grade III) (HR = 3.83 [95% CI, 1.18–12.5], P = 0.026) were extracted as significant risk factors predicting poor prognosis in the cohort from TCGA (Table 3). Patients with IDH-wildtype LGGs with grade III histology or co-occurrence of the high-risk genetic lesions (high-risk group) had a poor prognosis: median OS was 1.83 years in the JPN cohort and 1.66 years in the cohort from TCGA, which was only about 6 months longer than that of GBM, IDH-wildtype in the cohort from TCGA (1.11 y) (Fig. 3F). The high-risk tumors frequently had genetic alterations characteristic of glioblastoma and were diagnosed at significantly older age than those with none of these risk factors (low-risk group) (P = 0.042 and 0.0021 in the JPN and TCGA cohorts, respectively) (Supplementary Figure S7A and S7B).1,3,13 By contrast, patients with low-risk tumors exhibited excellent survival, similar to that of IDH-mutant LGGs (Fig. 3E and F). These low-risk patients with IDH-wildtype LGGs might be closely related to an entity described as pilocytic astrocytoma-like, a subtype of IDH-wildtype diffuse gliomas with a unique DNA methylation profile and a favorable outcome.20 Actually, among TCGA patients with IDH-wildtype LGGs, those with a pilocytic astrocytoma-like DNA methylation pattern were significantly enriched in the low-risk patients described above (8/8 vs 9/49; P < 0.0001) (Supplementary Figure S7C).20
Discussion
The major strength of this study is its analysis of a large number of patients who were clinically well annotated and fully genotyped for driver mutations and CNVs associated with LGG. This allowed for successful detection of subtype-specific genetic markers that reliably predict poor clinical outcome in each WHO subtype: NOTCH1 mutations in Oligodendroglioma IDH-mut/1p19q-codel; PIK3R1 mutations and altered RB pathway genes in Astrocytoma IDH-mut; and co-occurrence of gain of chromosome 7p, loss of chromosome 10q, and TERT promoter mutation in IDH-wildtype LGGs.
NOTCH1 mutations have been reported in a wide variety of human cancers, including gliomas. Depending on cancer type, these mutations result in different functional consequences, corresponding to discrete mutational hotspots.3,13,33–36 In LGGs most frequently observed in Oligodendroglioma IDH-mut/1p19q-codel, NOTCH1 mutations almost invariably affect the epidermal growth factor–like domain, leading to loss of protein function; consistent with this, inactivation of Notch or its mediators can induce accelerated glioma growth.3,13,37,38 These mutations were more frequently found in relapsed tumors than diagnostic samples38 and were significantly associated with positive gadolinium enhancement in preoperative MRI and grade III (vs grade II) histology, suggesting their roles in aggressive phenotypes. In accordance with this, NOTCH1 mutations were found to be independent predictors of poor clinical outcomes in the JPN cohort. The negative effect was statistically significant only in Oligodendroglioma IDH-mut/1p19q-codel cases from the JPN cohort, but not in the cohort from TCGA or in other LGG subtypes. However, the low frequencies of NOTCH1 mutations in Astrocytoma IDH-mut (n = 5 [5%]) and IDH-wildtype LGGs (n = 3 [5%]) precluded a precise evaluation of their effects in these LGG subtypes. Also, the significantly shorter median follow-up time in the cohort from TCGA (median 1.71 y) might not be sufficient to detect the effect on long-term survival of NOTCH1 mutations, which made a substantial contribution to the statistical difference in the JPN cohort (Table 1). Future studies are warranted to address these points, using a larger cohort of patients with long-term follow-up periods.
In Astrocytoma IDH-mut, PIK3R1 mutations and altered RB pathway genes were significantly and independently associated with a poor clinical outcome, which was similar to that in GBM, IDH-mutant. This is in agreement with previous studies reporting negative effects of PIK3R1 mutations and CDKN2A deletions on survival in this LGG subtype.15,17 These lesions accelerate cellular proliferation and lead to genomic/chromosomal instability, likely by cell-cycle dysregulation or aberrant receptor tyrosine kinase signaling.39,40 WHO grade has long been correlated with OS among patients with LGGs. However, some studies did not confirm this in IDH-mutant molecular subtypes.41,42 In fact, in our JPN cohort, WHO grade was not associated with patient prognosis in either univariate or multivariate Cox analyses in Oligodendroglioma IDH-mut/1p19q-codel or Astrocytoma IDH-mut, suggesting that it may not be appropriate to use WHO grade as a prognostic factor for these LGG subtypes.
As for IDH-wildtype LGGs, we previously demonstrated that this subtype could be further classified into 2 subcategories, tumors of WHO grade II and grade III histology, based on the fact that grades II and III tumors have substantially different clinical outcomes.3 In this study, we not only confirmed our previous result, but also identified high-risk genetic markers significantly associated with a poor prognosis, independently of WHO grade. In combination with WHO grade, the co-occurrence of these high-risk genetic lesions (ie, gain of chromosome 7p, loss of chromosome 10q, and mutation of the TERT promoter) can be used to stratify patients with IDH-wildtype LGGs into 2 discrete subsets with substantially different clinical outcome, genetic profile, median age at diagnosis, and pattern of DNA methylation.
In conclusion, we delineated a set of subtype-specific markers that predict poor clinical outcomes in LGGs. The subsets of each LGG subtype with these markers represent high-risk tumors, accounting for 46%–47% of Oligodendroglioma IDH-mut/1p19q-codel, 7% of Astrocytoma IDH-mut, and 53%–75% of IDH-wildtype LGGs in the JPN and TCGA cohorts. In particular, the prognosis of high-risk tumors in Astrocytoma IDH-mut and IDH-wildtype LGGs was extremely poor. These tumors may actually represent bona fide GBM, even though they lack some of its hallmark features, including necrosis and vascular proliferation.1,43 Patients with these high-risk LGGs could benefit from intensive therapy conventionally used for GBM, and this possibility should be tested in clinical trials.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by grants-in-aid for Scientific Research (22134006 and 15H05909 to S.O.; 23107010 to A.N.), a grant-in-aid for Young Scientists (Start-up) (15H06339 to K.A.), a grant-in-aid from the Japan Brain Foundation (to H.S.), the Funding Program for World-Leading Innovative Research and Development on Science and Technology (to S.O.), the Project for Development of Innovative Research on Cancer Therapeutics from the Japan Agency for Medical Research and Development, AMED (15cm0106056h0005 and 16cm0106501h0001 to S.O.), and the High Performance Computing Infrastructure System Research Project (hp150232 to S.O.).
Funding
This work was supported by grants-in-aid for Scientific Research (22134006 and 15H05909 to S.O.; 23107010 to A.N.), a grant-in-aid for Young Scientists (Start-up) (15H06339 to K.A.), a grant-in-aid from the Japan Brain Foundation (to H.S.), the Funding Program for World-Leading Innovative Research and Development on Science and Technology (to S.O.), the Project for Development of Innovative Research on Cancer Therapeutics from the Japan Agency for Medical Research and Development, AMED (15cm0106056h0005 and 16cm0106501h0001 to S.O.), and the High Performance Computing Infrastructure System Research Project (hp150232 to S.O.).
Conflict of interest statement. We declare no competing interests.
Supplementary Material
Acknowledgments
We are grateful to all patients who generously agreed to participate in this study. We gratefully acknowledge the consortium of TCGA and all of its members for making these invaluable data publicly available.
References
- 1. Louis DN, Perry A, Reifenberger G et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Ostrom QT, Gittleman H, Fulop J et al. . CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Aoki K, Chiba K et al. . Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–468. [DOI] [PubMed] [Google Scholar]
- 4. Ohgaki H, Kleihues P. Genetic profile of astrocytic and oligodendroglial gliomas. Brain Tumor Pathol. 2011;28(3):177–183. [DOI] [PubMed] [Google Scholar]
- 5. Smith JS, Perry A, Borell TJ et al. . Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18(3):636–645. [DOI] [PubMed] [Google Scholar]
- 6. Yan H, Parsons DW, Jin G et al. . IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. [DOI] [PubMed] [Google Scholar]
- 8. Hartmann C, Meyer J, Balss J et al. . Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. [DOI] [PubMed] [Google Scholar]
- 9. Sanson M, Marie Y, Paris S et al. . Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 10. Gorovets D, Kannan K, Shen R et al. . IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18(9):2490–2501. [DOI] [PubMed] [Google Scholar]
- 11. van den Bent MJ, Brandes AA, Taphoorn MJ et al. . Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344–350. [DOI] [PubMed] [Google Scholar]
- 12. Cairncross G, Wang M, Shaw E et al. . Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31(3):337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brat DJ, Verhaak RG, Aldape KD et al. ; Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gleize V, Alentorn A, Connen de Kérillis L et al. ; POLA network. CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann Neurol. 2015;78(3):355–374. [DOI] [PubMed] [Google Scholar]
- 15. Draaisma K, Wijnenga MM, Weenink B et al. . PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients. Acta Neuropathol Commun. 2015;3:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reis GF, Pekmezci M, Hansen HM et al. . CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization grades II-III) astrocytomas. J Neuropathol Exp Neurol. 2015;74(5):442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roy DM, Walsh LA, Desrichard A et al. . Integrated genomics for pinpointing survival loci within arm-level somatic copy number alterations. Cancer Cell. 2016;29(5):737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Killela PJ, Pirozzi CJ, Healy P et al. . Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. [DOI] [PubMed] [Google Scholar]
- 20. Ceccarelli M, Barthel FP, Malta TM et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brennan CW, Verhaak RG, McKenna A et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shiraishi Y, Sato Y, Chiba K et al. . An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41(7):e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nannya Y, Sanada M, Nakazaki K et al. . A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65(14):6071–6079. [DOI] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer. 2005;41(16):2381–2402. [DOI] [PubMed] [Google Scholar]
- 26. Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8(1):e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubin DB. Multiple imputation for nonresponse in surveys. Hoboken, NJ: Wiley-Interscience; 2004. [Google Scholar]
- 28. Hoeting JA, Madigan D, Raftery AE, Volinsky CT. Bayesian model averaging: a tutorial. Stat Sci. 1999;14(4):382–401. [Google Scholar]
- 29. Raftery AE, Madigan D, Hoeting JA. Bayesian model averaging for linear regression models. J Am Stat Assoc. 1997;92(437):179–191. [Google Scholar]
- 30. Pignatti F, van den Bent M, Curran D et al. ; European Organization for Research and Treatment of Cancer Brain Tumor Cooperative Group; European Organization for Research and Treatment of Cancer Radiotherapy Cooperative Group. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20(8):2076–2084. [DOI] [PubMed] [Google Scholar]
- 31. Smith JS, Chang EF, Lamborn KR et al. . Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J Clin Oncol. 2008;26(8):1338–1345. [DOI] [PubMed] [Google Scholar]
- 32. Ozawa T, Riester M, Cheng YK et al. . Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agrawal N, Frederick MJ, Pickering CR et al. . Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weng AP, Ferrando AA, Lee W et al. . Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. [DOI] [PubMed] [Google Scholar]
- 35. Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J Exp Med. 2011;208(10):1931–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rampias T, Vgenopoulou P, Avgeris M et al. . A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20(10):1199–1205. [DOI] [PubMed] [Google Scholar]
- 37. Giachino C, Boulay JL, Ivanek R et al. . A tumor suppressor function for notch signaling in forebrain tumor subtypes. Cancer Cell. 2015;28(6):730–742. [DOI] [PubMed] [Google Scholar]
- 38. Bai H, Harmancı AS, Erson-Omay EZ et al. . Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hernando E, Nahlé Z, Juan G et al. . Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430(7001):797–802. [DOI] [PubMed] [Google Scholar]
- 40. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. [DOI] [PubMed] [Google Scholar]
- 41. Olar A, Wani KM, Alfaro-Munoz KD et al. . IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129(4):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reuss DE, Mamatjan Y, Schrimpf D et al. . IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol. 2015;129(6):867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kros JM, Huizer K, Hernández-Laín A et al. . Evidence-based diagnostic algorithm for glioma: analysis of the results of pathology panel review and molecular parameters of EORTC 26951 and 26882 trials. J Clin Oncol. 2015;33(17):1943–1950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.