Abstract
Aberrant neuronal DNA methylation patterns have been implicated in the promotion of obesity development; however, the role of neuronal DNA methyltransferases (Dnmts), enzymes that catalyze DNA methylation, in energy balance remains poorly understood. We investigated whether neuronal Dnmt1 regulates normal energy homeostasis and obesity development using a neuronal Dnmt1 knockout (ND1KO) mouse model, Dnmt1fl/fl Synapsin1Cre, which specifically deletes Dnmt1 in neurons. Neuronal Dnmt1 deficiency reduced adiposity in chow-fed mice and attenuated obesity in high-fat diet (HFD)–fed male mice. ND1KO male mice had reduced food intake and increased energy expenditure with the HFD. Furthermore, these mice had improved insulin sensitivity, as measured using an insulin tolerance test. The HFD-fed ND1KO mice had smaller fat pads and upregulation of thermogenic genes in brown adipose tissue. These data suggest that neuronal Dnmt1 plays an important role in regulating energy homeostasis. Notably, ND1KO male mice had elevated estrogen receptor-α (ERα) gene expression in the medial hypothalamus, which previously has been shown to control body weight. Immunohistochemistry experiments revealed that ERα protein expression was upregulated specifically in the dorsomedial region of the ventromedial hypothalamus, a region that might mediate the central effect of leptin. We conclude that neuronal Dnmt1 regulates energy homeostasis through pathways controlling food intake and energy expenditure. In addition, ERα expression in the dorsomedial region of the ventromedial hypothalamus might mediate these effects.
The present study demonstrates the importance of neuronal DNA methylation in regulating energy homeostasis.
Energy homeostasis is a complex process by which energy intake, expenditure, and storage are kept in balance to ensure that every organ has the energy needed to function. Although the liver, pancreas, and adipose organs (among others) all have distinct roles in regulating energy homeostasis, the central nervous system is required for the correct orchestration of these organs. The hypothalamus, in particular, controls eating behavior, energy storage, and expenditure; thus, hypothalamic dysfunction can lead to energy imbalance and obesity (1). DNA methylation, the addition of methyl groups to DNA, is associated with gene repression (2). Although DNA methylation is necessary to regulate temporal and tissue-specific expression of genes, aberrant methylation patterns are associated with disease (3, 4). Altered methylation patterns in the central nervous system have been implicated as a contributing factor in the development of obesity. For example, increasing methylation of the proopiomelanocortin (POMC) gene (a key hypothalamic peptide in energy regulation) via deletion of methyl-CpG-binding protein 2 in POMC neurons causes obesity (5). In contrast, overfeeding or a high-fat diet (HFD) can induce hypermethylation of the POMC gene (6, 7). An HFD increases histone deacetylases, which are associated with increased DNA methylation in the hypothalamus (8) and reduces DNA methyltransferase (Dnmt)3a expression in the paraventricular hypothalamus (9). Corroborative evidence in humans has shown an association between POMC hypermethylation and childhood obesity (10). Also, POMC methylation has been inversely correlated with the successful maintenance of weight loss in obese men (11). Thus, neuronal DNA methylation could play an important role in regulating energy homeostasis.
Dnmt1 has a critical function in maintaining DNA methylation patterns in dividing cells (12). It is, thus, surprising that Dnmt1 gene expression remains detectable in many regions of the adult brain (13–15), given that most of the brain is in a postmitotic state. Several lines of evidence have pointed to a role for Dnmt1 beyond just maintaining methylation (16). For example, neuronal deletion of both Dnmt1 and Dnmt3a (but not Dnmt3a alone) impairs synaptic plasticity and produces cognitive deficits in mice (17). Double-knockout Dnmt3a/Dnmt3b embryos still have some de novo methylation activity (18). It has been suggested that the combined actions of Dnmt3a and Dnmt1 synergistically produce complete de novo methylation on double-stranded DNA (19, 20). Deletion of Dnmt3a in the paraventricular nucleus of the hypothalamus causes metabolic dysfunction, hyperphagia, decreased energy expenditure, and obesity (2). It is less clear, however, whether neuronal Dnmt1 regulates energy homeostasis.
Our aim for the present study was to investigate the role of neuronal Dnmt1 in energy homeostasis and obesity. Using a knockout mouse model in which Dnmt1 was specifically deleted from neurons, we found that neuronal Dnmt1 deficiency altered metabolism and energy regulation in normal chow-fed mice and attenuated obesity and metabolic dysfunction in HFD-fed mice. We also found that neuronal Dnmt1 deficiency altered the expression of neuropeptides and hormones that have key roles in energy regulation. In particular, estrogen receptor-α (ERα) in the central nervous system is known to play a role in body weight in both male and female rodents, specifically through actions in the ventromedial hypothalamus (VMH) (21, 22). In line with this, we found that neuronal Dnmt1 deficiency altered the ERα expression pattern in the VMH in HFD-fed mice, indicating that ERα might be a part of the mechanism through which neuronal Dnmt1 mediates its effects on energy and metabolism.
Materials and Methods
Knockout mice
Neuronal Dnmt1-deficient mice were generated in-house by crossing Dnmt1fl/fl mice (23) with Dnmt1fl/fl mice that also expressed heterozygous Cre recombinase enzyme under the control of the Synapsin1 promoter (SynCre+/−) (24). Dnmt1fl/fl and SynCre+/− mouse lines were sourced from National Institutes of Health–supported Mutant Mouse Resources and Research Centers (Bethesda, MD) and Jackson Laboratories (Bar Harbor, ME), respectively. SynCre+/− mice express Cre recombinase enzyme in the brain, spinal cord, and dorsal root ganglion neurons beginning at embryonic day 12.5 (25). Male and female pups were either Dnmt1fl/flSynCre+/− [neuronal Dnmt1 knockout (ND1KO)] or Dnmt1fl/flSynCre−/− [no knockout; control (fl/fl)]. We also studied Dnmt1+/+SynCre+/− (normal Dnmt1 expression, but expressing Synapsin1-Cre) to confirm that the expression of SynCre alone does not produce any phenotype. At 6 weeks of age, male and female ND1KO and fl/fl littermates were fed ad libitum either a normal chow diet (catalog no. 5001; LabDiet; Purina, St. Louis, MO) or a HFD (60% fat; catalog no. 12492; Research Diets, New Brunswick, NJ). The Georgia State University institutional animal care and use committee approved all procedures, which were performed in accordance with the Public Health Service and US Department of Agriculture guidelines.
Physiological measurements
Body weight was measured weekly throughout the duration of the experiment. Glucose and insulin tolerance testing was performed after 16 to 18 weeks of the mice consuming the diets. For the glucose tolerance test (GTT), the mice were fasted for 16 hours overnight. Blood was obtained from a tail nick to measure the glucose concentration using a OneTouch Ultra Blood Glucose Meter and glucose test strips (LifeScan, Inc., Milpitas, CA). After a baseline blood glucose measurement, the mice were intraperitoneally injected with a 20% dextrose solution (1 g/kg), and subsequent blood glucose measurements were performed at 15, 30, 60, 90, and 120 minutes after the glucose injection. For the insulin tolerance test (ITT), the mice were fasted for 4 hours, and then the baseline blood glucose was measured. The mice were intraperitoneally injected with either a 1.0 U/kg (males) or a 0.8 U/kg (females) dose of 0.25 U/mL insulin (Humulin R; Eli Lilly, Indianapolis, IN), and blood glucose was measured at the same time points used for the GTT. Energy expenditure, oxygen consumption, carbon dioxide production, respiratory exchange ratio, and physical activity data were collected using a TSE PhenoMaster metabolic chamber system (TSE Systems, Chesterfield, MO) after 22 to 24 weeks of HFD feeding (males) or 26 to 28 weeks of HFD feeding (females). Data were collected after 3 days of acclimation to the metabolic chambers. The body composition of the relative lean and fat mass was obtained in vivo using time-domain nuclear magnetic resonance technology with a MiniSpec Body Composition Analyzer (Bruker, Spring, TX) after 24.5 weeks of the chow diet and 22 weeks of the HFD. The mice were euthanized using carbon dioxide inhalation and were decapitated (at 28.5 weeks of age for chow-fed, male and female; and 25 weeks for HFD-fed male or 29 weeks for HFD-fed female). Blood was collected for serum and stored at −80°C. Fat pads [brown, epididymal (perigonadal in females), retroperitoneal, and subcutaneous/inguinal] and liver were dissected and weighed and then frozen in liquid nitrogen. A sample of each fat pad and of the liver was stored in formalin for later histological analysis.
Cold exposure
Eight-week-old fl/fl (n = 8) and ND1KO (n = 15) male mice were housed singly with only corn cob bedding in the cage and were subjected to a 5°C environment for 7 days. A separate cohort of fl/fl (n = 6) and ND1KO (n = 9) male mice housed at room temperature were included in the gene expression assays. After 7 days, the mice were removed from the cold room individually and immediately euthanized by carbon dioxide inhalation until they were nonambulatory and then rapid cervical dislocation. We harvested and weighed the brown, epididymal, and subcutaneous fat tissues and formalin-fixed a sample of each fat pad. The remaining fat was frozen in liquid nitrogen for gene and protein analyses.
Gene and protein quantification
The fat tissues were removed from −80°C storage and placed in liquid nitrogen. Each fat sample was removed from the liquid nitrogen and immediately homogenized in TRI Reagent (Molecular Research Center, Cincinnati, OH) using a handheld homogenizer. Gene expression was quantified by quantitative reverse transcription polymerase chain reaction (PCR) using a Stratagene Mx3005P qPCR System (Agilent Technologies, Santa Clara, CA), ABI Universal PCR Master Mix (Applied Biosystems, Foster City, CA), and primer and probe sets purchased from Applied BioSystems. Each assay included the reference gene cyclophilin as an internal control. All data were analyzed using the ΔΔCt method (26).
Separate tissue samples were homogenized for protein quantification using radioimmunoprecipitation assay buffer with protease inhibitor cocktails (Sigma-Aldrich, St. Louis, MO) and a handheld homogenizer. The protein concentration was quantified using a DC Protein Assay (BioRad, Hercules, CA), and protein samples were denatured at 90° to 100° for 10 minutes. Western blots were performed by loading 20 to 30 µg of protein onto a 4% to 15% gradient polyacrylamide gel (Criterion TGX; BioRad), which was then transferred to a polyvinylidene difluoride membrane. We immunoblotted the membranes by first blocking with 5% nonfat milk, followed by antibodies against uncoupling protein 1 at a 1:500 dilution (catalog no. 23841; Abcam, Cambridge, MA), hormone-sensitive lipase (HSL), or phosphorylated HSL serine 660 at a 1:1000 dilution (catalog nos. 4107S and 4126S, respectively; Cell Signaling, Danvers, MA) overnight at 4°C, followed by Alexa Fluor goat anti-rabbit 680 secondary antibody (catalog no. A21109; Thermo Fisher Scientific, Waltham, MA) at 1:5000 concentration for 3 hours. Bands were visualized and quantified using an Odyssey Fc Imaging System (Li-Cor, Lincoln, NE) with background subtracted. All membranes were immunoblotted with α-tubulin antibody at 1:500 dilution (catalog no. 2144S; Cell Signaling), which was used as a normalization control.
Serum assays
Serum free fatty acids were quantified using an HR Series NEFA [nonesterified (free) fatty acids] kit (Wako Diagnostics, Richmond, VA), and triglycerides were quantified using an L-Type Triglycerides M kit (Wako Diagnostics). Serum leptin and insulin were quantified using enzyme-linked immunosorbent assay kits (catalog nos. 90030 and 90080; CrystalChem, Downers Grove, IL).
Adipose tissue histology
After formalin fixation, fat tissues were dehydrated through a series of increasing isopropanol and were embedded in paraffin blocks. Brown, subcutaneous, and epididymal adipose tissues were cut manually on a rotating microtome into 6-µm sections and mounted on SuperFrost Plus slides (Thermo Fisher Scientific). Tissue morphology was visually analyzed using hematoxylin and eosin staining (Sigma-Aldrich). Fat histology images were captured using an Olympus DP73 photomicroscope and CellSens software (Olympus, Waltham, MA). Representative images were enhanced using ImageJ software (National Institutes of Health, Rockville, MD). All images compared with each other were collected and processed using the same parameters.
Adipocyte lipid droplet size and number analysis was conducted on 20× magnification of brown adipose tissue (BAT) and 10× magnification of subcutaneous white adipose tissue (WAT) using the freely available ImageJ Magnetic Resource Imaging Adipocytes macro tool (National Institutes of Health) (27). Lipid droplets were counted and sized using the default parameters suggested by the tool package. All images were identically treated by researchers unaware of the experimental conditions. The lipid droplet size and number correspond to the circular isolated droplets within each section of tissue (i.e., BAT lipid droplets are not equivalent to cell size owing to the multiple droplets residing within the cell membrane).
Hypothalamic gene expression
Four hypothalamic regions were dissected (arcuate, medial, paraventricular, and lateral) with the aid of a dissecting microscope according to the methods reported by Minokoshi et al. (28). First, the brain was placed in a block with 1-mm grooves, and a straight blade was placed in the exact midline of the brain to create a sagittal cut. Two additional blades were placed on each side at 1-mm intervals from the midline blade. The arcuate, medial, and paraventricular regions were dissected out from the right and left blades closest to the midline, and the lateral hypothalamus was dissected out from the farthest lateral right and left blades. The hypothalamic samples were frozen in liquid nitrogen and stored at −80°C for later gene analysis.
Immunofluorescence
ND1KO (n = 7) and fl/fl (n = 7) male mice were fed the chow diet, and a separate cohort of fl/fl (n = 8) and ND1KO (n = 8) male mice were fed the HFD for 12 weeks beginning at 6 weeks of age. Body weight was recorded weekly. After 11 to 12 weeks of the chow diet or HFD, the mice were euthanized, and the brains were processed for immunofluorescence. The mice were transcardially perfused with heparinized saline, followed by 4% paraformaldehyde. The brains were soaked in 4% paraformaldehyde in 4°C for 2 to 3 hours and then placed in 18% sucrose/phosphate-buffered saline until they sank. The brains were frozen and sectioned on a cryostat at 20 µm and mounted onto slides. Immunofluorescence was conducted using primary antibodies to target ERα (catalog no. 06-935; EMD Millipore, Merck KGaA, Darmstadt, Germany) at a 1:20,000 concentration and neuron-specific nuclear protein at a 1:1000 concentration (catalog no. MAB377; EMD Millipore) overnight at 4°C. Alexa Fluor secondary antibodies (catalog nos. A21207 and A21202; Thermo Fisher Scientific) were used at a 1:5000 concentration for 3 hours at room temperature. The slides were mounted with Prolong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Images were acquired the following day using an Olympus DP73 fluorescent photomicroscope and CellSens software (Olympus). ERα-positive cells in the ventrolateral portion of the VMH (VMHvl) and dorsomedial region of the VMH (VMHdm) were manually counted with the aid of the Photoshop CS3 count tool and using the neuron-specific nuclear protein image of the same section overlaid as a guide for the boundary of the VMH. Three sections per brain were analyzed. The cell counts per section were averaged for each brain, and the mean of these averages was calculated for each group. The Student t test was used to compare the genotypes.
Esr1 methylation
ND1KO and fl/fl male mice were fed either a chow diet (fl/fl, n = 9; ND1KO, n = 7) or HFD (fl/fl, n = 10; ND1KO, n = 10) for 12 weeks, beginning at 6 weeks of age. The mice were euthanized, and the hypothalamus was dissected according to the previously described methods. DNA was isolated from the ventromedial/dorsomedial hypothalamic nuclei (V/DMH) region using phenol/chloroform/isoamyl alcohol (25:24:1), and 2 µg of DNA from each sample was used for bisulfite conversion using an Epitect Bisulfite Kit (catalog no. 59104; Qiagen, Valencia, CA). The Esr1 promoter regions exon A and exon C were amplified using primers from EpigenDX (catalog nos. ADS910 and ADS911; Hopkinton, MA). Pyrosequencing and methylation analysis of these exons were performed by EpiGenDX. One-way analyses of variance (ANOVAs) were used to test for significance between the genotypes at each CpG site.
Statistical analysis
Data were analyzed using Excel 2013 and are presented as the mean ± the standard error of the mean. Areas under the curve (AUCs) were calculated for the GTT and ITT data using GraphPad Prism 5. Statistical significance between the genotypes was tested using Student t tests or one-way ANOVAs for each diet and sex, with SPSS Statistics 20 (IBM Corp., Armonk, NY). Repeated-measures ANOVAs were performed on data from the metabolic chamber experiments, GTTs and ITTs, and weekly body weight. Follow-up specific tests were performed on the GTT and ITT data using a Bonferroni correction if the main effect was relevant.
Results
Neuronal Dnmt1 deficiency reduces adiposity in chow-fed mice
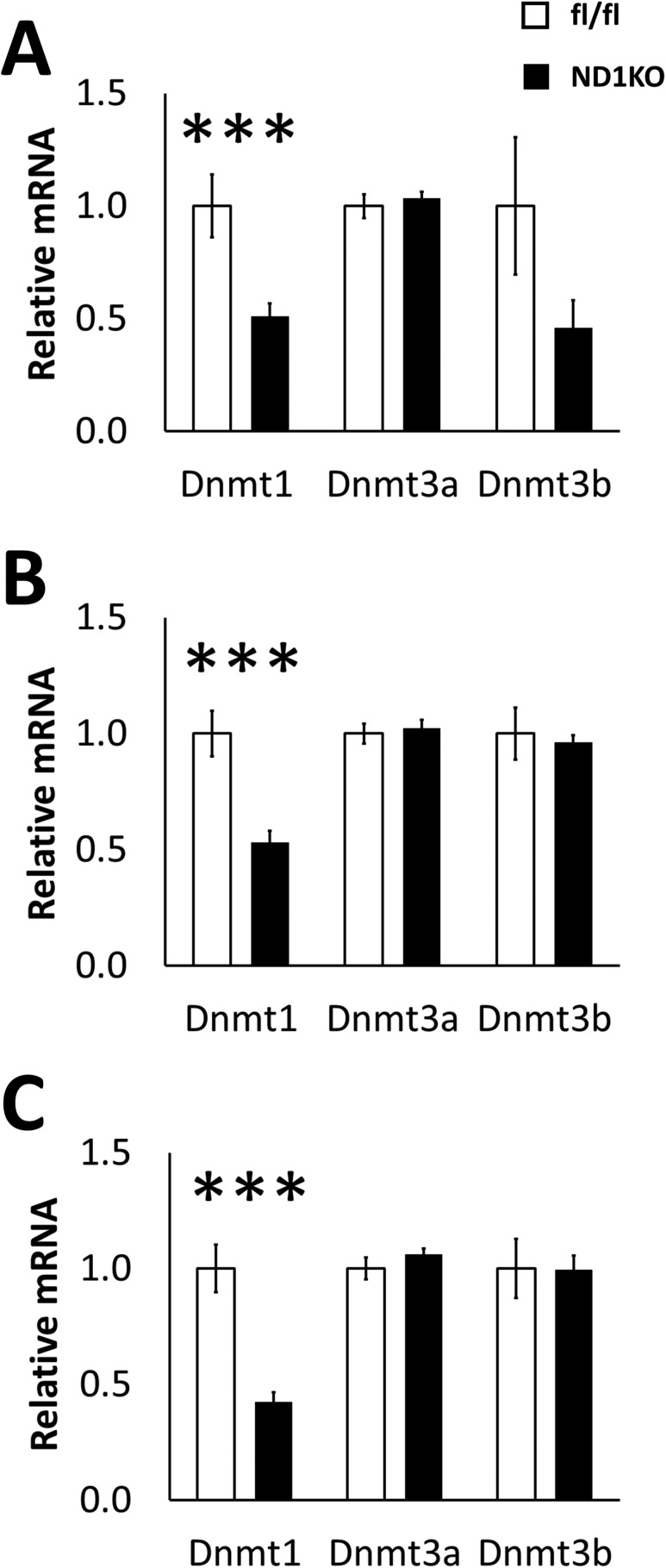
Dnmt1 was deleted in neurons by breeding Dnmt1fl/fl mice with SynCre+/− mice. We first quantified Dnmt1 gene expression in several different hypothalamic nuclei of HFD-fed male ND1KO mice to test the knockdown efficiency of our mouse model. Dnmt1 gene expression was significantly reduced in the arcuate, ventromedial/dorsomedial, and paraventricular regions (Fig. 1A–1C). To confirm that no compensatory increases had occurred in the other Dnmts as a result of Dnmt1 knockdown, we also quantified Dnmt3a and Dnmt3b gene expression and found no substantial changes (Fig. 1A–1C). Some Dnmt1 expression remained in these regions in the ND1KO mice, presumably owing to the presence of Dnmt1 expression in non-neuronal cells, such as glia.
Figure 1.
Hypothalamic gene expression of DNA methyltransferases. Dnmt1, Dnmt3a, and Dnmt3b messenger RNA (mRNA) expression in the (A) arcuate, (B) ventromedial/dorsomedial, and (C) paraventricular hypothalamic regions of male HFD-fed fl/fl and ND1KO mice. ***P < 0.001.
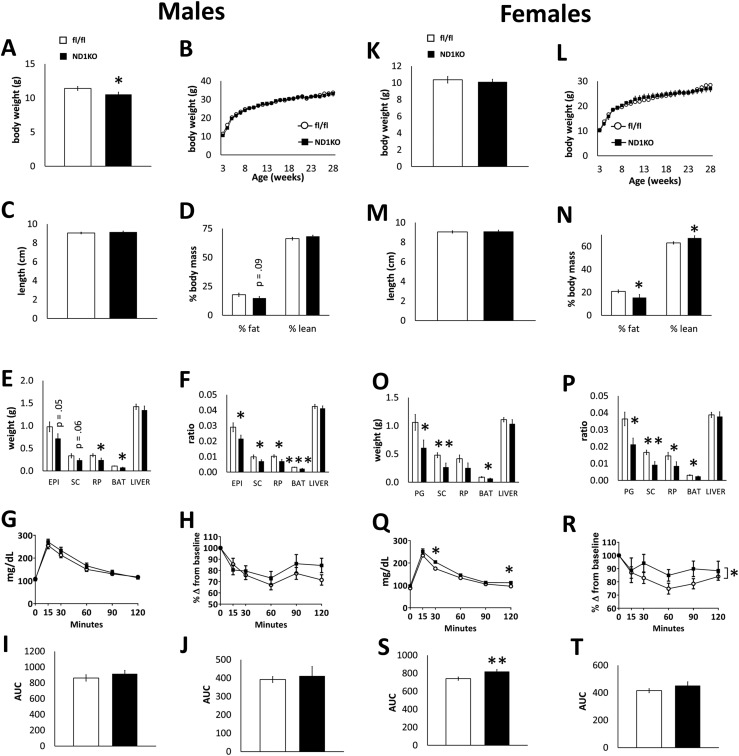
We first tested the role of neuronal Dnmt1 in energy regulation under normal metabolic demands. Male and female ND1KO and fl/fl control littermates were fed a standard chow diet from weaning onward. We found ND1KO mice were born with expected frequency (data not shown). ND1KO male mice weighed less at weaning (3 weeks old; Fig. 2A); however, this difference had disappeared by the time they were 5 weeks old (Fig. 2B). Female ND1KO mice showed no difference in body weight (Fig. 2K and 2L). Neuronal Dnmt1 deficiency did not affect body length in either male or female mice (Fig. 2C and 2M). We measured in vivo body composition and found that male ND1KO mice had a tendency for a reduced fat percentage (Fig. 2D), and female ND1KO mice had a significantly lower fat percentage and a higher percentage of lean mass (Fig. 2N). In male ND1KO mice, retroperitoneal tissue and BAT pads weighed significantly less (Fig. 2E), and the epididymal and subcutaneous (inguinal) adipose tissue tended to weigh less. When normalized to body weight, all fat pads weighed less in the male ND1KO mice (Fig. 2F). In the female ND1KO mice, perigonadal, subcutaneous, and brown adipose tissue pads weighed significantly less (Fig. 2O). When normalized to body weight, all fat pads weighed less in the female ND1KO mice (Fig. 2P). Neuronal Dnmt1 deletion did not affect the liver mass in male or female mice (Fig. 2E, 2F, 2O, and 2P).
Figure 2.
Phenotype of neuronal Dnmt1 deficiency in chow-fed mice. Chow-fed fl/fl and ND1KO (left) male and (right) female mice. (A, K) Body weight at 3 weeks old and (B, L) weekly body weight. (C, M) Body length and (D, N) body composition. (E, O) WAT (epididymal/perigonadal, subcutaneous, retroperitoneal), BAT, and liver weights and (F, P) the same normalized to body weight. (G, Q) Blood glucose measurements during an intraperitoneal GTT and (I, S) AUC of blood glucose during the test. (H, R) Blood glucose measurements during an intraperitoneal ITT and (J, T) AUC of blood glucose during the test. *P < 0.05; **P < 0.01; ***P < 0.001.
We next tested whether ND1KO mice had altered glucose or insulin dynamics. Glucose and insulin tolerance did not differ between the male genotypes (Fig. 2G–2J). Female ND1KO mice had a slightly greater blood glucose concentration during the GTT and a substantially greater blood glucose AUC (Fig. 2Q and 2S). Despite a substantial main effect of genotype throughout the ITT, female ND1KO mice showed no difference in the blood glucose AUC (Fig. 2R and 2T).
Neuronal Dnmt1 deficiency attenuates obesity in HFD-fed mice
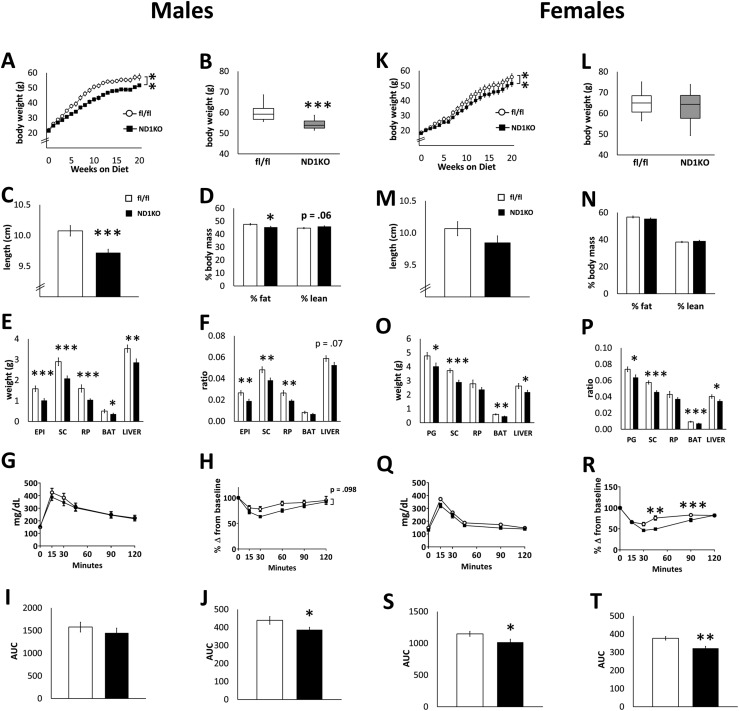
We next tested whether neuronal Dnmt1 deficiency alters obesity development by feeding male and female ND1KO and fl/fl mice the an HFD beginning at 6 weeks of age. Male ND1KO mice weighed significantly less after 2 weeks of the HFD (Fig. 3A) and continued to weigh less throughout the duration of the experiment (the maximal weight difference occurred at ∼12 weeks of the HFD). HFD-fed female ND1KO mice had significantly reduced body weight throughout the duration of the HFD, as indicated by repeated-measures two-way ANOVA (Fig. 3K). However, the final body weight at the end of the study did not differ substantially between the genotypes (Fig. 3L). The distribution and variability of body weight of the HFD-fed males and females at 31 to 34 weeks of age are illustrated in Fig. 3B and 3L, respectively. Unlike the chow-fed ND1KO mice, the HFD-fed ND1KO male mice had a reduced body length (Fig. 3C). Male ND1KO mice also had a substantially reduced percentage of body fat, with a tendency seen for an increase in lean mass (Fig. 3D). Female ND1KO mice showed no relevant differences in body length or body composition compared with female fl/fl mice (Fig. 3M and 3N). In addition, we confirmed that the body weight difference in male and female ND1KO mice fed an HFD resulted from the lack of Dnmt1 specifically, in that the presence of Synapsin1-Cre recombinase by itself did not affect body weight substantially in male or female mice fed an HFD (Supplement Fig. 1 (2.7MB, docx) ).
Figure 3.
Phenotype of neuronal Dnmt1 deficiency in HFD-fed mice. HFD-fed fl/fl and ND1KO (left) male and (right) female mice. (A, K) weekly body weight and (B, L) final body weight at end of study. (C, M) Body length and (D, N) body composition. (E, O) WAT (epididymal/perigonadal, subcutaneous, retroperitoneal), BAT, and liver weights and (F, P) the same normalized to body weight. (G, Q) Blood glucose measurements during an intraperitoneal GTT and (I, S) AUC of blood glucose during the test. (H, R) Blood glucose measurements during an intraperitoneal ITT and (J, T) AUC of blood glucose during the test. *P < 0.05; **P < 0.01; ***P < 0.001.
HFD-fed male ND1KO mice had significantly smaller epididymal, subcutaneous, and retroperitoneal fat pads, even when normalized to body weight (Fig. 3E and 3F). BAT was substantially smaller but not when normalized to body weight (Fig. 3E and 3F). Female ND1KO mice had substantially smaller perigonadal, subcutaneous, and brown adipose tissue fat pads, and these differences remained relevant even after body weight normalization (Fig. 3O and 3P). Both male and female ND1KO mice had a smaller liver weight (Fig. 3E and 3O), although after adjusting for body weight, male ND1KO mice only showed a tendency for reduced liver weight (Fig. 3F and 3P). In the male mice, no change was seen in the ability to clear a glucose infusion from the blood between the two genotypes (Fig. 3G and 3I). In response to an insulin injection, however, male ND1KO mice had a lower AUC of blood glucose (Fig. 3H–3J), indicating improved insulin sensitivity. HFD-fed female ND1KO mice had a significantly reduced AUC of blood glucose during the GTT (Fig. 3Q and 3S). Like the males, female ND1KO mice also showed improved insulin tolerance (Fig. 3R and 3T).
Given that male HFD-fed ND1KO mice exhibited the greatest reduction in body weight, we continued our analysis of other metabolic features in the male mice only. The serum concentrations of insulin, leptin, triglycerides, and nonesterified (free) fatty acids of ND1KO and fl/fl male mice are displayed in Table 1. Neuronal Dnmt1 deficiency did not alter serum leptin or insulin concentrations in chow-fed mice, although a trend was seen for ND1KO mice to have lower concentrations of both hormones. HFD-fed ND1KO mice had significantly lower serum leptin concentrations compared with HFD-fed fl/fl mice, as might be expected from the reduced adiposity, because leptin is secreted in direct proportion to the amount of fat stores in the body (29). Serum insulin was not different in the HFD-fed ND1KO or fl/fl mice, however. Neuronal Dnmt1 deficiency did not affect serum triglycerides or nonesterified fatty acids in either chow-fed or HFD-fed mice.
Table 1.
Serum Hormones and Lipids of Male ND1KO and fl/fl Mice Fed Ad Libitum
| Diet | Genotype | Insulin, ng/mL | Leptin, ng/mL | Triglycerides, mg/dL | NEFA, mmol/L |
|---|---|---|---|---|---|
| Chow | fl/fl | 1.86 ± 0.4 | 9.69 ± 1.3 | 123.9 ± 10.4 | 0.65 ± 0.06 |
| ND1KO | 1.22 ± 0.2 | 7.42 ± 1.6 | 109.4 ± 14.4 | 0.58 ± 0.08 | |
| HFD | fl/fl | 24.6 ± 7.9 | 71.1 ± 4.4 | 96.8 ± 16.0 | 0.912 ± 0.07 |
| ND1KO | 31 ± 5.4 | 57.6 ± 2.4a | 112.3 ± 7.7 | 0.857 ± 0.04 |
Abbreviation: NEFA, nonesterified (free) fatty acids.
P < 0.01.
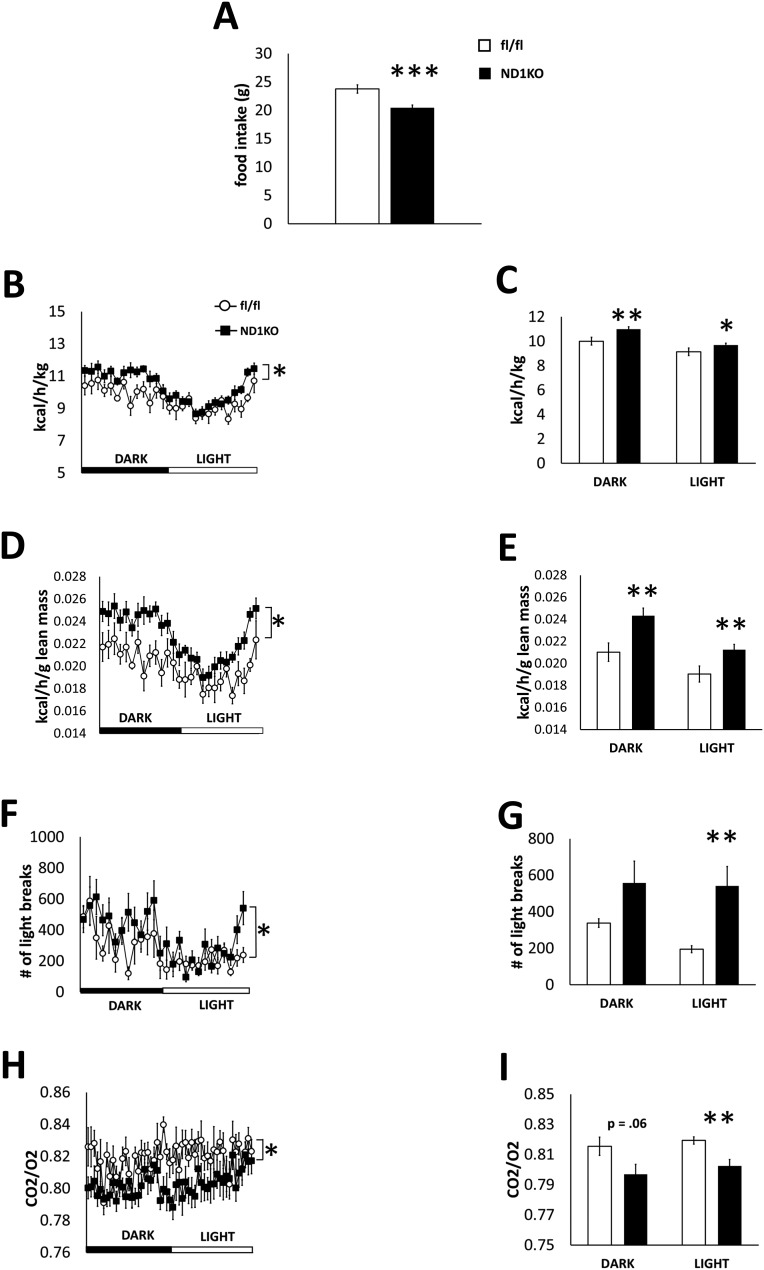
Consistent with their reduced obesity, HFD-fed male ND1KO mice consumed less food during a 7-day period (Fig. 4A). We placed the male mice in metabolic cages and found that the energy expenditure was substantially increased in ND1KO mice compared with fl/fl mice when normalized to either body weight (Fig. 4B and 4C) or lean mass (Fig. 4D and 4E). ND1KO mice had a decreased respiratory exchange ratio (Fig. 4F and 4G) during one light-dark cycle of the 7-day metabolic cage experiment, indicating a preferential usage of fatty acids over carbohydrates as an energy source. In addition, ND1KO mice had a higher physical activity level (Fig. 4H and 4I) during a 24-hour period, as analyzed using repeated-measures ANOVA. However, only the activity level during the light period differed substantially between the genotypes when the overall mean values for the light and dark periods were analyzed by t test.
Figure 4.
Food intake and metabolic parameters in HFD-fed fl/fl and ND1KO mice. (A) Cumulative food intake for 7 days, (B, C) energy expenditure normalized to body mass, (D, E) energy expenditure normalized to lean mass, (F, G) respiratory exchange ratio, and (H, I) physical activity for a 24-hour period in male HFD-fed fl/fl and ND1KO mice. *P < 0.05; **P < 0.01; ***P < 0.001.
Neuronal Dnmt1 deficiency alters the gene profile of BAT
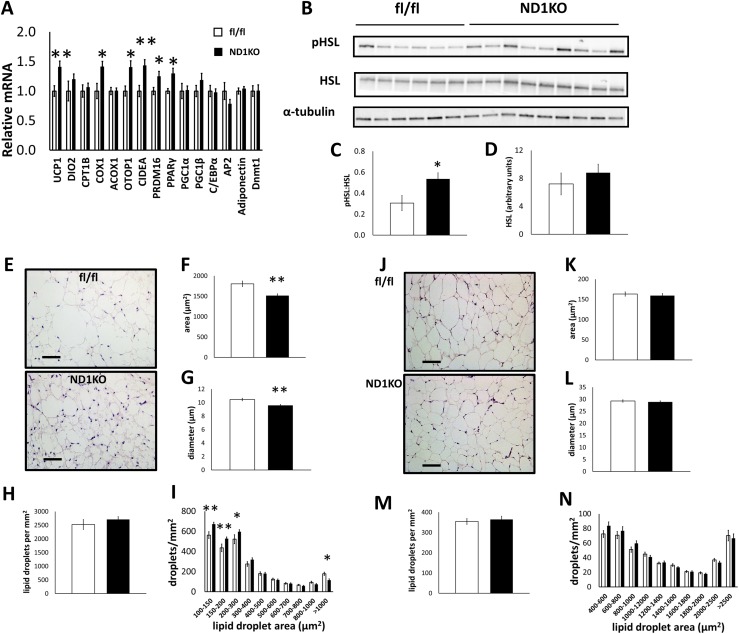
We characterized the ND1KO phenotype in further detail only in the HFD-fed males because neuronal Dnmt1 deficiency produced the most robust body weight differences in these mice. With an HFD, ND1KO mice showed upregulated Ucp1, Cox1 (cytochrome c oxidase-1), Otop1 (otopetrin-1), Cidea (cell death-inducing DFFA-like effector A), Pparγ (peroxisome proliferator-activated receptor gamma), and Prdm16 (PR domain-containing 16) gene expression in BAT (Fig. 5A). Collectively, these gene alterations suggest that neuronal Dnmt1 deficiency increased mitochondrial oxidative and uncoupling capacity in BAT, which can occur from elevated sympathetic nervous system (SNS) drive to BAT (30, 31). Phosphorylation of HSL, a key enzyme in SNS-mediated lipolysis, was substantially increased in the BAT of HFD-fed ND1KO mice compared with that in fl/fl mice (Fig. 5B–5D), indicating increased lipolytic activity in ND1KO mice. Consistent with the changes in gene and protein expression, visual observation of hematoxylin and eosin (H&E) staining of BAT from all mice in the experiment revealed smaller BAT lipid droplets in the HFD-fed ND1KO mice than in the fl/fl mice, indicating less lipid accumulation, or greater lipid usage, in BAT (Fig. 5E). We quantified the BAT lipid droplet size and observed that the lipid droplet area and diameter were substantially reduced in the ND1KO mice (Fig. 5F–5G), with no change in lipid droplet number (Fig. 5H). Analysis of the distribution of lipid droplet size demonstrated that ND1KO mice had significantly more small lipid droplets and fewer large lipid droplets (Fig. 5I). No major differences were seen in the gene profile of subcutaneous or epididymal WAT of HFD-fed male ND1KO and fl/fl mice (data not shown). Also, although the WAT pads weighed less, H&E staining demonstrated no visible differences in cell size between the genotypes (Fig. 5J), indicating that fewer adipocytes might be present in the fat pads. We confirmed this observation by quantifying the subcutaneous WAT lipid droplets, which showed no substantial differences in area, diameter, or density between the genotypes (Fig. 5K–5N).
Figure 5.
Adipose tissue analyses of male HFD-fed fl/fl and ND1KO mice. (A) BAT gene expression. (B) Western blot images of phosphorylated HSL (pHSL), total HSL (α-tubulin as a control). (C) Densitometry quantification of pHSL normalized to total HSL and (D) total HSL protein. (E) BAT H&E staining at 20× magnification (scale bar = 50 µm). BAT lipid droplet (F) area, (G) diameter, (H) density, and (I) size distribution. WAT (subcutaneous) H&E staining at 10× magnification (scale bar = 100 µm). WAT lipid droplet (F) area, (G) diameter, (H) density, and (I) size distribution. *P < 0.05; **P < 0.01.
To more thoroughly test the role of neuronal Dnmt1 in regulating BAT function, we placed a separate cohort of male ND1KO and fl/fl mice in a cold environment for 7 days and analyzed the gene expression, adipose tissue morphology, and immunohistochemistry. However, neuronal Dnmt1 deficiency failed to produce any major effect on body weight, fat pad mass, or gene expression when the mice were exposed to cold. Although 8-week-old male ND1KO mice housed at room temperature showed a tendency for reduced body weight and had a substantially reduced epididymal fat mass compared with fl/fl mice (Supplement Fig. 2A–2D (2.7MB, docx) ), cold-exposed ND1KO mice did not show any differences in fat pad size compared with cold-exposed fl/fl mice (Supplement Fig. 2E, and 2F (2.7MB, docx) ), although we did observe decreased body weight in ND1KO mice after cold exposure (Supplement Fig. 2B (2.7MB, docx) ). Furthermore, no major morphologic changes were seen in the BAT or WAT of ND1KO mice, as demonstrated by H&E staining (Supplement Fig. 2G–2J (2.7MB, docx) ). Cold-exposed ND1KO mice exhibited only a tendency for increased Ucp1 and Dio2 messenger RNA (mRNA) in subcutaneous fat, and BAT showed no such cold-induced changes (Supplement Fig. 3A–3D (2.7MB, docx) ). It is interesting that we observed genotype-dependent differences in BAT (at the gene, protein, and cell level) in the context of an HFD but not in the context of cold exposure. This raises the possibility that separate upstream pathways in the central nervous system might affect BAT function, whether in a state of energy excess (HFD) or increased energy usage (cold exposure conditions) (32).
Hypothalamic gene expression of ND1KO mice
The hypothalamus controls homeostatic processes, including energy balance, and does so via the expression of numerous peptides and neurotransmitters. Changes in methylation and expression of hypothalamic genes, such as POMC and NPY, are associated with weight gain and obesity (6, 7, 10, 11). Therefore, we investigated whether neuronal Dnmt1 deletion altered the expression of hypothalamic genes involved in energy regulation. However, in the chow-fed ND1KO mice, no substantial changes were found in gene expression in the arcuate (ARC), paraventricular (PVH), V/DMH, or lateral (LH) hypothalamic nuclei (Supplement Fig. 4A–4D (2.7MB, docx) ). Similarly, in HFD-fed mice, no change was seen in ARC gene expression (Supplement Fig. 4E (2.7MB, docx) ). In the PVH of HFD-fed mice, however, neuronal Dnmt1 deficiency caused upregulation of the vGat (vesicular GABA transporter gene) (Supplement Fig. 4F (2.7MB, docx) ). In the V/DMH, Crh (corticotropin-releasing hormone) and the vGlut2 (vesicular glutamate transporter 2) genes were upregulated (Supplement Fig. 4G (2.7MB, docx) ). Finally, in the LH, HFD-fed ND1KO mice had increased Crh gene expression (Supplement Fig. 4H (2.7MB, docx) ).
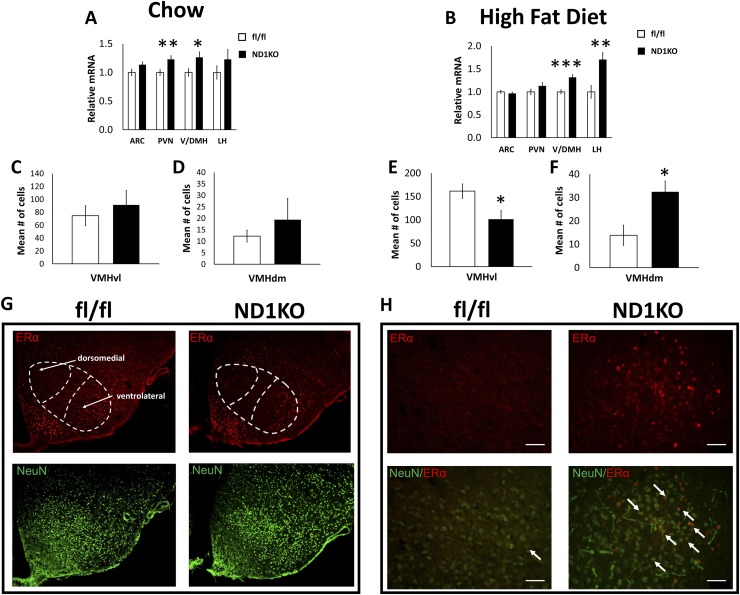
In addition to the standard players in hypothalamic energy regulation, another candidate protein that might be regulated by Dnmt1 is ERα. ERα expression (encoded by the Esr1 gene) in the VMH is involved in body weight control in both male and female mice (21, 22), and Esr1 expression is inversely associated with Esr1 gene promoter DNA methylation in the cerebral cortex (14). Furthermore, Esr1 negatively correlates with Dnmt1 expression (33, 34). As a first step in testing whether ERα might mediate the lean phenotype in our ND1KO male mice, we quantified Esr1 expression in the hypothalamic nuclei of male ND1KO and fl/fl mice. Chow-fed ND1KO mice had substantially increased Esr1 mRNA expression in the V/DMH and PVH (Fig. 6A). Similarly, HFD-fed ND1KO mice had increased ESR1 expression in the V/DMH and LH but not the PVH (Fig. 6B).
Figure 6.
Hypothalamic ERα analysis. Esr1 mRNA expression in the ARC, V/DMH, PVH, and LH of (A) chow-fed and (B) HFD-fed fl/fl and ND1KO male mice. ERα-positive cell counts in the VMHvl of (C) chow-fed and (E) HFD-fed fl/fl and ND1KO mice. ERα-positive cell counts in the VMHdm of (D) chow-fed and (F) HFD-fed fl/fl and ND1KO mice. (G) Representative images of ERα (red) and neuron-specific nuclear (NeuN) (green) staining at 10× magnification in the VMHdm and VMHvl of HFD-fed male fl/fl and ND1KO mice. (H) ERα-positive cells overlaid with NeuN at 40× magnification in the dorsomedial VMH of HFD-fed fl/fl and ND1KO mice. *P < 0.05; **P < 0.01; ***P < 0.001.
Esr1 promoter methylation in ND1KO mice
ERα expression in the cerebral cortex is regulated via DNA methylation of the Esr1 promoter region (14). Because ND1KO mice are neuronally deficient in Dnmt1, an enzyme that methylates DNA, we tested whether ND1KO mice had reduced methylation of the Esr1 promoter region, which might explain the increase in Esr1 mRNA exhibited by ND1KO mice. The mouse Esr1 gene promoter has five alternative splice sites upstream of the protein translation start site (exons A, B, C, F, and H) (35). The transcript variant spliced at exon C is the major transcript variant expressed in the brain (35, 36) (Supplement Fig. 5A (2.7MB, docx) ). However, increases in methylation on exons A and C have been associated with decreased Esr1 expression in the cerebral cortex throughout development in mice (33). We microdissected the V/DMH region, the same region in which we observed increases in Esr1 gene expression, and processed this tissue for bisulfite conversion and pyrosequencing. Chow-fed ND1KO mice had modest but substantial decreases in methylation at CpG sites 6 and 7 of exon A (Supplement Fig. 5B (2.7MB, docx) ). No difference was found in the overall methylation status on exon A between the genotypes (data not shown). In addition, no relevant differences were found in methylation at the CpG sites on exon A between ND1KO and fl/fl mice fed the HFD (Supplement Fig. 5B (2.7MB, docx) ). We also quantified methylation of five CpG sites on exon C; however, ND1KO mice did not have any decreases in methylation, just as we predicted (Supplement Fig. 5C (2.7MB, docx) ). Chow-fed ND1KO mice showed increases in methylation at CpG sites 4 and 5, and HFD-fed ND1KO mice showed an increase at CpG site 3.
ERα expression undergoes a “remodeling” in the VMH of ND1KO male mice
We next sought to precisely locate where the ERα protein was upregulated in the V/DMH of ND1KO mice. Previous studies have demonstrated that VMH ERα protein expression is greater in the VMHvl than in the central or dorsal portions of the VMH (37). In addition, VMH ERα has been implicated in the regulation of body weight (21), but we could not find any published evidence of ERα expression in the DMH regulating body weight. Thus, we predicted that ERα protein expression would be upregulated specifically in the VMH. Two separate cohorts of male ND1KO and fl/fl mice were fed either a normal chow diet or an HFD for 12 weeks (the time point that coincides with the largest body weight reduction; Fig. 3A), beginning at 6 weeks of age. The brains were processed for immunohistochemistry using an antibody targeting ERα protein. Consistent with previous reports, we confirmed strong ERα expression in the medial preoptic area (38, 39) and almost no ERα expression in the thalamus (39) (data not shown). We investigated whether ERα protein expression was increased in VMHvl (where ERα is expressed most highly) or the VMHdm, an area in the VMH mediating leptin’s effects on energy regulation (40, 41). ERα-positive cells were manually counted in the VMHvl, VMHdm, and arcuate nucleus (as a negative control, because ND1KO mice show no change in arcuate Esr1 expression). We chose sections within bregma −1.58 to −1.82 (42) because this area has been reported to have the greatest number of immunohistochemically labeled ERα-positive cells (21). The pattern of staining was consistent with those from previous reports of ERα immunoreactivity in the VMH, with the greatest expression in the VMHvl (Fig. 6C–6G) (43, 44). Although chow-fed ND1KO mice showed no change in the number of ERα-positive cells in the VMHdm (Fig. 6D), HFD-fed ND1KO mice had significantly more ERα-positive cells in VMHdm than did the HFD-fed fl/fl mice (Fig. 6F). HFD-fed ND1KO mice also showed a substantial reduction in the number of ERα-positive cells in the VMHvl (Fig. 6E), although chow-fed ND1KO mice showed no change (Fig. 6C). As a negative control, we found no difference in the number of ERα-positive cells in the arcuate nucleus of HFD-fed ND1KO and fl/fl mice (data not shown), consistent with our Esr1 expression data in the arcuate nucleus. Figure 6G and 6H illustrate ERα-stained cells colocalized with neurons in the VMHdm of HFD-fed ND1KO and fl/fl mice.
Discussion
Dysregulated neuronal DNA methylation patterns have been associated with obesity in both rodent and human studies (7, 11). In addition, high-calorie diets are associated with increased methylation and reduced expression, of the anorexigenic peptide POMC, which inhibits energy intake and promotes energy expenditure (6, 7). In the present study, we have demonstrated that neuronal Dnmt1 regulates energy homeostasis under normal and obese conditions. Specifically, deleting neuronal Dnmt1 in mice reduced adiposity under normal metabolic demands and attenuated body weight, adiposity, and glucose and insulin intolerance caused by an HFD. The phenotype seen in the HFD-fed mice was driven by both reduced food consumption and increased energy expenditure.
ND1KO mice ate less of the HFD compared with fl/fl mice. The degree to which reduced food intake alone attenuated obesity in ND1KO mice, separate from elevations in energy expenditure, was not specifically tested in the present study. An important follow-up study would be to pair-feed ND1KO and fl/fl mice during the first three months of the HFD, when the reductions in obesity are starting to become evident in ND1KO mice. Pair-feeding would distinguish between the relative contributions of reduced energy intake vs elevated energy expenditure in the attenuation of obesity. Likely, alterations in food intake and energy expenditure contribute to similar degrees to the leaner phenotype of ND1KO mice, because neither had particularly large effects compared with the other. Furthermore, we do not know whether the changes in food intake and energy expenditure occurred consistently throughout the entire HFD experiment or whether these effects were dynamic throughout the 25-week HFD. Various time point studies of ND1KO mice consuming an HFD should be conducted to gain greater detail of the ND1KO mouse phenotype.
The SNS innervates both BAT and WAT and regulates nonshivering thermogenesis and lipolysis in BAT and WAT (45–47). Activating the SNS drive to WAT also inhibits proliferation of energy-storing WAT cells (48, 49). Consistent with this, BAT from HFD-fed ND1KO male mice showed an upregulation of BAT-specific genes, including Ucp1 and other genes involved in mitochondrial oxidative function, suggesting an upregulated BAT thermogenic program. BAT in HFD-fed ND1KO mice also had elevated phosphorylation of HSL (phosphorylated HSL) and smaller lipid droplets, both of which suggest enhanced SNS-stimulated lipolysis. In contrast, WAT pads from ND1KO mice were smaller, although the lipid droplets did not differ in size, indicating that the WAT pads from ND1KO mice might have had less adipocytes overall. This could have resulted from reduced WAT proliferation, which can occur as a result of SNS stimulation to WAT, because it has been reported that norepinephrine inhibits WAT precursor cell proliferation in vitro (48). Collectively, these observations indicate that neuronal Dnmt1 deficiency might have elevated SNS drive to adipose tissues in HFD-fed mice, which would contribute to greater BAT thermogenesis and increased energy expenditure. Future experiments are needed to directly test whether ND1KO mice do have an elevated SNS drive to BAT and WAT.
Despite the evidence for enhanced BAT metabolism in HFD-fed ND1KO mice, little evidence was found of increased BAT function in cold-exposed ND1KO mice. In the context of increased energy needs (cold environment), the purpose of SNS drive to WAT or BAT is to induce changes in the tissue that allow it to use energy to produce heat and maintain the temperature stability of the animal. This is in contrast to a positive energy balance in diet-induced obesity, in which increased heat is not necessary, but energy expenditure increases owing to diet-induced thermogenesis, a phenomenon in which excess caloric consumption increases the metabolic rate and stimulates BAT thermogenesis (30). Thermogenesis might be stimulated upstream via different mechanisms, depending on whether it is triggered through cold or other factors (32). For example, cold is sensed through the peripheral sensory nerves that project to the preoptic area, which then releases the DMH from inhibition, leading to increased sympathetic drive to BAT (50). Other factors, such as ingesting a meal (postprandial thermogenesis) or consuming a HFD (diet-induced thermogenesis) also can increase sympathetic activation of BAT (30). Cold exposure and HFD feeding also can lead to differential gene expression in both BAT and WAT (51, 52). Thus, neuronal Dnmt1 deficiency could alter the neural pathways that regulate energy expenditure and BAT activity in the context of excess energy consumption but not in the context of a cold environment. A final thought is that cold effects changes in BAT depending on how long the animal has been acclimated to the cold (53). It is possible that a difference in BAT function between the genotypes would be evident if we were to analyze the mice at a different time point of cold exposure.
ND1KO mice had a greater physical activity level, which likely contributed to their elevated energy expenditure. Previous studies have highlighted certain hypothalamic genes as being important in controlling spontaneous physical activity and energy intake and expenditure. TrkB (the brain-derived neurotrophic factor receptor) (54) or FoxO1 (55) expression in the hypothalamus, Rock1 expression in AgRP neurons (56), and Esr1 expression in the VMHvl (57, 58) all have been reported to drive physical activity levels, in addition to energy homeostatic processes. Thus, increased glucose uptake and usage in muscle from elevated physical activity most likely contributed to the elevated energy expenditure in ND1KO mice, although this was not directly measured. The finding of elevated physical activity in ND1KO mice raises the question of whether these mice have increased anxiety or other conditions that might be associated with increased movement. It was not within the scope of our study, however, to investigate the behavior in detail.
Changes in the adipose organs, whether brown or white, can strongly affect glucose and insulin dynamics. With a chow diet, ND1KO mice showed no major change in the ability to clear a glucose injection from the blood or to correct blood glucose levels after an insulin injection. Both male and female HFD-fed ND1KO mice, however, showed improvements in insulin tolerance (females also had improved glucose tolerance). Whether neuronal Dnmt1 affects insulin signaling directly, perhaps through altering innervation and drive to the liver or pancreas, or whether these improvements simply resulted from reductions in adiposity is not clear.
Energy expenditure and food intake behavior is regulated by the central nervous system and, in particular, the melanocortin system (59). Given the previous studies linking Pomc hypermethylation with obesity, we were surprised to find no changes in the expression of Pomc or any other major ARC genes as a result of neuronal Dnmt1 deficiency. An important consideration is that our analysis of the hypothalamic gene profile was conducted after a long duration of HFD feeding. Thus, our “snapshot” of the hypothalamic gene profile we have reported is one that includes any compensatory changes occurring over time with consumption of an HFD or with older age. It would be interesting to analyze the hypothalamic gene expression profile in both young adult ND1KO mice (8 weeks old) and ND1KO mice with a shorter exposure to the HFD. In contrast, converging evidence has pointed to ERα as an energy-regulating factor in the VMH (21, 22). In the present study, neuronal Dnmt1 deficiency upregulated the ERα gene, Esr1, in the V/DMH region of both chow- and HFD-fed mice. Furthermore, we discovered that neuronal Dnmt1 deficiency increased the number of ERα-positive cells in the VMHdm region. Although leptin receptors are expressed throughout the VMH (60), leptin-stimulated pSTAT3, a downstream leptin signaling protein, is more robustly induced in the VMHdm (40). Additionally, cFos (a marker of neuronal activation) is upregulated in VMHdm neurons within 2 hours after an intravenous leptin injection (41). Thus, ERα upregulation in the VMHdm also might suggest enhanced central leptin signaling. The precise VMH subregion and neuronal types through which ERα regulates body weight in male mice are not known. Deletion of ERα in SF1 neurons of the VMH produces obesity in female mice but not in male mice (22). However, ERα knockdown using adeno-associated virus delivery of a short hairpin RNA into the VMH does produce obesity in male mice (21). Our data suggest that ERα in the VMHdm could be involved in body weight regulation in male mice, although we could not rule out the possibility that the changes of ERα expression in VMHdm might be compensatory to Dnmt1 deletion or indirectly (mediated by additional factors) by Dnmt1 deletion owing to the use of the pan-neuronal Dnmt1 deletion approach. Thus, further studies are necessary to test whether ERα has a causative role in mediating the protective effects of neuronal Dnmt1 deletion during the development of obesity. For example, ERα could be immunohistochemically labeled along with other proteins of interest (e.g., leptin receptor, brain-derived neurotrophic factor) to determine the neuronal subtype of VMHdm ERα cells. Importantly, specific gain- and loss-of-function experiments should be performed to test whether VMHdm ERα expression is involved in the mechanism of the phenotype seen in ND1KO mice. In addition, approaches are available to specifically target and alter the methylation levels at promoters of interest (61–63). This has been achieved using a deactivated/catalytically inactive CRISPR-associated protein 9 endonuclease (dCas9) fused with the catalytic domain of the DNA methyltransferase Dnmt3a or the DNA demethylase ten-eleven translocation methylcytosine dioxygenase 1 (Tet1) (64). Targeting dCas9-Dnmt3a or dCas9-Tet1 to gene promoters using guide RNAs specific to these promoters can achieve gene promoter-specific alteration of DNA methylation (65). Thus, in future studies, we could deliver adeno-associated virus-encoded dCas9-Dnmt1 or dCas9-Tet1 combined with Esr1-specific guide RNAs to VMHdm regions to confirm whether DNA methylation at the Esr1 promoter in this region is responsible for regulating energy homeostasis.
Neuronal Dnmt1 deficiency also reduced the number of ERα-positive cells in the VMHvl in HFD-fed ND1KO males. ERα expression in the VMHvl positively regulates sexual behavior and aggression in males (21, 66). These results indicate that neuronal Dnmt1 deficiency might have altered these behaviors, although they were not measured in the present study. In chow-fed mice, we did not observe any relevant effects from neuronal Dnmt1 deficiency on ERα expression in the VMHvl or VMHdm. Although a trend was seen for the ND1KO mice to have greater ERα expression in the VMHdm, the variability was too great. A robust (twofold) increase was seen in ERα protein expression in the VMHvl in the HFD-fed fl/fl control mice compared with the chow-fed fl/fl control mice (Fig. 6E vs 6C), although the chow and HFD experiments were not conducted at the same time. A review of the published data yielded only one report in which a 4-month 42% HFD reduced ERα protein and mRNA in the whole hypothalamus of male mice (67). Although we did not conduct a count of ERα-positive cells nor did we quantify Esr1 expression in the gross hypothalamus, our quantitative PCR analyses of separate hypothalamic nuclei did not demonstrate decreases in gene expression in any of the nuclei (data not shown). Similarly, a time-course study of mice fed a purified control diet or HFD also showed only some increases, but no decreases, in Esr1 expression (unpublished data from Dr. Bingzhong Xue’s laboratory at Georgia State University). It is possible that an HFD decreases ERα expression in the entire hypothalamus but upregulates ERα expression in the smaller region of the VMHvl. Nevertheless, it is more meaningful to study ERα in the discrete nuclei of the hypothalamus rather than the hypothalamus as a whole, owing to the heterogeneous functions of these nuclei.
The neuronal expression of Esr1 is regulated via methylation of its promoter region in the cerebral cortex of mice throughout postnatal development (14). We tested whether Esr1 upregulation in the V/DMH of ND1KO mice was associated with hypomethylation of these same promoter regions. In exon C, the major Esr1 transcript expressed in the brain (35), chow-fed ND1KO mice surprisingly had increases in methylation at several CpG sites. On exon A, these mice had decreased methylation at CpG sites 6 and 7 compared with the fl/fl mice. It has been shown that even a single CpG site, if it is in a region critical to the binding of a transcription factor or other coactivator, might control expression of that gene. For example, in piglets, the tissue-specific expression of Esr1 is controlled by a single CpG site located at a transcription factor binding site (14, 68). Thus, it is possible that these two CpG sites on exon A might be important in regulating Esr1 expression in the hypothalamus. Although HFD-fed fl/fl mice also had slightly reduced methylation at two exon A CpG sites compared with chow-fed fl/fl mice, no further reduction was found in DNA methylation as a result of Dnmt1 deletion. One explanation might be that the reduced DNA methylation at the two CpG sites on exon A might regulate the increased Esr1 expression in ND1KO mice fed a chow diet. However, other factors might explain the increased Esr1 expression seen in ND1KO mice fed an HFD. Reduced methylation of a gene promoter usually correlates with an increased likelihood of that gene being expressed. Increased gene expression, however, might not necessarily correlate with reduced methylation. AN HFD could affect other aspects of the transcriptional machinery at the Esr1 promoter and, thereby, upregulate Esr1 expression without further reducing methylation. In addition, because we observed differential regulation of ERα protein expression in different regions within the VMH, it is possible that robust differences in Esr1 promoter methylation exist between the genotypes, although only within the discrete VMHvl or the VMHdm regions. Consequently, differential methylation patterns within these two regions could be masked in the methylation analysis of the entire V/DMH hypothalamic region. Therefore, although we did not have the techniques to do so, it would be important to perform more spatially precise analyses of Esr1 methylation to determine whether ERα protein upregulation in the VMHdm is associated with reduced methylation of the Esr1 promoter in the VMHdm.
Diet-induced obesity is a well-known model of hyperleptinemia and central leptin resistance (69, 70). In the present study, neuronal Dnmt1 deficiency reduced serum leptin concentrations but upregulated ERα expression in the VMHdm, a region that mediates the effects of leptin on energy regulation (40, 41). Although leptin is secreted in proportion to fat stores (29) and ND1KO mice showed reductions in both leptin and adiposity, it is possible that ND1KO mice also maintained leptin sensitivity better than fl/fl mice throughout the course of the HFD. The present finding that neuronal Dnmt1 deficiency reduced food intake and body weight in HFD-fed mice is consistent with enhanced leptin sensitivity (71). Our future studies will test this specifically by quantifying food intake and body weight in response to leptin injections. If ND1KO mice do have enhanced leptin sensitivity with an HFD, we will conduct further investigations of the neuronal leptin signaling cascade (e.g., phosphorylated STAT3) (72), with a particular focus on the VMHdm, in the ND1KO mouse.
In female mice fed a normal chow diet, neuronal Dnmt1 deficiency significantly reduced body fat and slightly elevated blood glucose concentrations during the GTTs and ITTs. Unlike the males, ND1KO female mice fed a HFD did not have significantly attenuated body weight at the end of the study, although they did show differences in overall body fat composition and reduced fat pad weights. HFD-fed ND1KO females showed significantly lower blood glucose levels at several points during an ITT, and the ND1KO males did not show a statistically significant effect of genotype throughout the test (despite a lower AUC). In the interpretation of these findings, it is important to consider that a number of differences are present in lipid metabolism between the sexes. Women have more body fat than men (73), which also is distributed differently, in that women store more fat in subcutaneous regions, and men store more fat in the visceral/abdominal region (74). Although the rate of lipid oxidation is similar between the sexes, females have greater lipolysis rates (75), are more sensitive than males to norepinephrine-induced lipolysis (75), have greater triglyceride secretion and clearance, and greater fasting free fatty acid levels than males (76). In the HFD-fed females, we might speculate that neuronal Dnmt1 deficiency could enhance the protective effects of estrogen (e.g., anti-inflammatory, anorectic) in females fed a HFD. However, female ND1KO mice showed no substantial differences in body weight at the end of the HFD. A possibility is that perhaps estrogen had already provided the maximum protective effect in the HFD-fed females and that this could not be further enhanced by neuronal Dnmt1 deficiency. The contribution of estrogen to the neuronal Dnmt1 phenotype seen in HFD-fed female mice could be tested by comparing the phenotype of ovariectomized vs intact ND1KO female mice. Additionally, ERα expression in the VMH of ND1KO female mice was not quantified in the present study; however, this would be a critical experiment to conduct to further understand the effects of neuronal Dnmt1 deficiency in females.
The phenotype in ND1KO mice fed a HFD is relatively moderate compared with other models of metabolic disorders or obesity attenuation. For example, the leptin-deficient ob/ob mouse displays massive, early-onset obesity that is accompanied by the progressive development of insulin resistance and adipocyte hypertrophy and hyperplasia, which occurs even with a chow diet (1, 77). The effects of leptin deficiency in the ob/ob mouse on energy expenditure are hugely robust compared with the effects of neuronal Dnmt1 deficiency (78). This is not surprising, given that ND1KO mice have pan-neuronal deletion of a ubiquitously expressed enzyme that likely regulates multiple aspects of energy homeostasis in different brain regions. Thus, widespread deletion of neuronal Dnmt1 might produces less robust effects owing to compensation of different brain regions. Site-specific PVH deletion of Dnmt3a produces obesity with a chow diet (9), and this effect is much more robust than that of the ND1KO mouse. Thus, site-specific deletion of Dnmt1 might yield different, and perhaps more robust, phenotypes than that of the ND1KO mouse.
In the present study, although the use of pan-neuronal Dnmt1 deletion has provided some insight into how neuronal DNA methylation regulates energy balance and further helped us to narrow down the specific brain regions that might be the target of neuronal DNA methylation in regulating energy balance, we acknowledge that this approach might lack some specificity, because Dnmt1 is ubiquitously expressed and likely regulates multiple pathways in various neurons. Using site-specific and neuron-specific deletions of Dnmt1 or manipulating DNA methylation levels in a site-specific and neuron-specific manner in future studies will be necessary to identify more in-depth mechanisms. Furthermore, the present study used Synapsin-Cre, which would delete Dnmt1 in neurons during early development. This makes it difficult to examine whether the effects we observed in ND1KO mice resulted from developmental defects or physiological effects in adulthood, or both. Thus, future studies using an inducible Cre system are necessary to elucidate more clearly the role of neuronal Dnmt1 in adult animals in the regulation of energy homeostasis.
In conclusion, neuronal Dnmt1 deficiency attenuated obesity and improved insulin tolerance in HFD-fed male mice by reducing food intake and increasing energy expenditure. Overall, our findings imply that neuronal Dnmt1 might function to enhance the deleterious effects of an HFD. These metabolic changes might be driven in part by mechanisms involving enhanced SNS drive to adipose tissues, increased ERα expression in the VMHdm, and enhanced leptin sensitivity, which we will test in future studies. To the best of our knowledge, the present study has, for the first time, demonstrated the importance of neuronal Dnmt1 expression in energy regulation and has highlighted ERα as a promising target for future obesity therapeutics. These data add to the growing body of evidence implicating aberrant DNA methylation in obesity development. It will be necessary moving forward to pinpoint the factors that cause such altered methylation to help prevent obesity and the associated metabolic disorders.
Acknowledgments
We thank Drs. Nancy Forger at Georgia State University and Yong Xu at Baylor College of Medicine for their ERα immunohistochemistry protocols and antibody samples.
Financial Support: This work was supported by American Heart Association Predoctoral Grant 16PRE27260157 (to E.C.B.), American Diabetes Association Grant 7-13-BS-159 (to H.S.), American Heart Association Grant 11GRNT7370080 (to H.S.), National Institutes of Health Grant R01DK084172 (to H.S.), American Heart Association Grants 10SDG3900046 and 17GRNT33670492 (to B.X.), National Institutes of Health Grants R01DK107544 and R01HL107500 (to B.X.), and Georgia State University Brains & Behavior Seed Grant and Center for Obesity Reversal Seed Grant (to B.X.).
Author Contributions: B.X. and E.C.B. designed the experiments. E.C.B. performed the experiments and analyses. B.X. supervised experiment performance, data analysis, and manuscript writing and performed brain dissections for the animal models. H.S. provided expertise on the study design and performed part of the organ dissections for the animal studies. J.T.G. had an integral function in the design and analysis of the ERα immunofluorescence experiments and performed all adipose tissue lipid droplet quantification and analyses. R.W. provided critical expertise for the Esr1 methylation study.
Current Affiliations: E.C. Bruggeman’s current affiliation is the Department of Human Genetics, Emory University School of Medicine, Atlanta, Georgia 30322. J.T. Garretson’s current affiliation is Albert Einstein College of Medicine, Bronx, New York 10461.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Appendix.
Antibody Table
| Peptide/Protein Target | Antigen Sequence (if Known) | Name of Antibody | Manufacturer, Catalog No. | Species Raised in; Monoclonal or Polyclonal | Dilution Used | RRID |
|---|---|---|---|---|---|---|
| ERα | Anti-ERα antibody | Millipore, 06-935 | Rabbit; polyclonal | 1:20,000 | AB_310305 | |
| NeuN | Anti-NeuN, clone A60 antibody | Millipore, MAB377 | Mouse; monoclonal | 1:1,000 | AB_2298772 |
Abbreviations: NeuN, neuron-specific nuclear protein; RRID, Research Resource Identifier.
Footnotes
- ANOVA
- analysis of variance
- ARC
- arcuate nucleus of the hypothalamus
- AUC
- area under the curve
- BAT
- brown adipose tissue
- dCas9
- deactivated/catalytically inactive CRISPR-associated protein 9 endonuclease
- Dnmt
- DNA methyltransferase
- ERα
- estrogen receptor-α
- GTT
- glucose tolerance test
- HFD
- high-fat diet
- H&E
- hematoxylin and eosin
- HSL
- hormone-sensitive lipase
- ITT
- insulin tolerance test
- LH
- lateral hypothalamic nucleus
- mRNA
- messenger RNA
- ND1KO
- neuronal Dnmt1 knockout
- PCR
- polymerase chain reaction
- POMC
- proopiomelanocortin
- PVH
- paraventricular hypothalamic nucleus
- SNS
- sympathetic nervous system
- Tet1
- ten-eleven translocation methylcytosine dioxygenase 1
- V/DMH
- ventromedial/dorsomedial hypothalamic nucleus
- VMH
- ventromedial hypothalamus
- VMHdm
- dorsomedial region of the ventromedial hypothalamus
- VMHvl
- ventrolateral portion of the ventromedial hypothalamus
- WAT
- white adipose tissue.
References
- 1.Bray GA, York DA. Hypothalamic and genetic obesity in experimental animals: an autonomic and endocrine hypothesis. Physiol Rev. 1979;59(3):719–809. [DOI] [PubMed] [Google Scholar]
- 2.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44(4):535–543. [DOI] [PubMed] [Google Scholar]
- 3.de Mello VD, Pulkkinen L, Lalli M, Kolehmainen M, Pihlajamäki J, Uusitupa M. DNA methylation in obesity and type 2 diabetes. Ann Med. 2014;46(3):103–113. [DOI] [PubMed] [Google Scholar]
- 4.Koukoura O, Spandidos DA, Daponte A, Sifakis S. DNA methylation profiles in ovarian cancer: implication in diagnosis and therapy (review). Mol Med Rep. 2014;10(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Lacza Z, Sun YE, Han W. Leptin resistance and obesity in mice with deletion of methyl-CpG-binding protein 2 (MeCP2) in hypothalamic pro-opiomelanocortin (POMC) neurons. Diabetologia. 2014;57(1):236–245. [DOI] [PubMed] [Google Scholar]
- 6.Marco A, Kisliouk T, Weller A, Meiri N. High fat diet induces hypermethylation of the hypothalamic Pomc promoter and obesity in post-weaning rats. Psychoneuroendocrinology. 2013;38(12):2844–2853. [DOI] [PubMed] [Google Scholar]
- 7.Plagemann A, Harder T, Brunn M, Harder A, Roepke K, Wittrock-Staar M, Ziska T, Schellong K, Rodekamp E, Melchior K, Dudenhausen JW. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol. 2009;587(Pt 20):4963–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funato H, Oda S, Yokofujita J, Igarashi H, Kuroda M. Fasting and high-fat diet alter histone deacetylase expression in the medial hypothalamus. PLoS One. 2011;6(4):e18950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohno D, Lee S, Harper MJ, Kim KW, Sone H, Sasaki T, Kitamura T, Fan G, Elmquist JK. Dnmt3a in Sim1 neurons is necessary for normal energy homeostasis. J Neurosci. 2014;34(46):15288–15296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuehnen P, Mischke M, Wiegand S, Sers C, Horsthemke B, Lau S, Keil T, Lee YA, Grueters A, Krude H. An Alu element-associated hypermethylation variant of the POMC gene is associated with childhood obesity. PLoS Genet. 2012;8(3):e1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crujeiras AB, Campion J, Díaz-Lagares A, Milagro FI, Goyenechea E, Abete I, Casanueva FF, Martínez JA. Association of weight regain with specific methylation levels in the NPY and POMC promoters in leukocytes of obese men: a translational study. Regul Pept. 2013;186:1–6. [DOI] [PubMed] [Google Scholar]
- 12.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011;12(2):206–222. [DOI] [PubMed] [Google Scholar]
- 13.Simmons RK, Stringfellow SA, Glover ME, Wagle AA, Clinton SM. DNA methylation markers in the postnatal developing rat brain. Brain Res. 2013;1533:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westberry JM, Trout AL, Wilson ME. Epigenetic regulation of estrogen receptor alpha gene expression in the mouse cortex during early postnatal development. Endocrinology. 2010;151(2):731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56(1-2):39–44. [DOI] [PubMed] [Google Scholar]
- 16.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–318. [DOI] [PubMed] [Google Scholar]
- 17.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13(4):423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. [DOI] [PubMed] [Google Scholar]
- 19.Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem. 2002;269(20):4981–4984. [DOI] [PubMed] [Google Scholar]
- 20.Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21(15):4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sano K, Tsuda MC, Musatov S, Sakamoto T, Ogawa S. Differential effects of site-specific knockdown of estrogen receptor α in the medial amygdala, medial pre-optic area, and ventromedial nucleus of the hypothalamus on sexual and aggressive behavior of male mice. Eur J Neurosci. 2013;37(8):1308–1319. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Nedungadi TP, Zhu L, Sobhani N, Irani BG, Davis KE, Zhang X, Zou F, Gent LM, Hahner LD, Khan SA, Elias CF, Elmquist JK, Clegg DJ. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 2011;14(4):453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27(1):31–39. [DOI] [PubMed] [Google Scholar]
- 24.Hoesche C, Sauerwald A, Veh RW, Krippl B, Kilimann MW. The 5′-flanking region of the rat synapsin I gene directs neuron-specific and developmentally regulated reporter gene expression in transgenic mice. J Biol Chem. 1993;268(35):26494–26502. [PubMed] [Google Scholar]
- 25.Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15(7):859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 27.Parlee SD, Lentz SI, Mori H, MacDougald OA. Quantifying size and number of adipocytes in adipose tissue. Methods Enzymol. 2014;537:93–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, Mu J, Foufelle F, Ferré P, Birnbaum MJ, Stuck BJ, Kahn BB. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature. 2004;428(6982):569–574. [DOI] [PubMed] [Google Scholar]
- 29.Jéquier E. Leptin signaling, adiposity, and energy balance. Ann N Y Acad Sci. 2002;967(1):379–388. [DOI] [PubMed] [Google Scholar]
- 30.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. [DOI] [PubMed] [Google Scholar]
- 31.Néchad M, Nedergaard J, Cannon B. Noradrenergic stimulation of mitochondriogenesis in brown adipocytes differentiating in culture. Am J Physiol. 1987;253(6 Pt 1):C889–C894. [DOI] [PubMed] [Google Scholar]
- 32.Peterson CM, Lecoultre V, Frost EA, Simmons J, Redman LM, Ravussin E. The thermogenic responses to overfeeding and cold are differentially regulated. Obesity (Silver Spring). 2016;24(1):96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westberry JM, Wilson ME. Regulation of estrogen receptor alpha gene expression in the mouse prefrontal cortex during early postnatal development. Neurogenetics. 2012;13(2):159–167. [DOI] [PubMed] [Google Scholar]
- 34.Wang YS, Chou WW, Chen KC, Cheng HY, Lin RT, Juo SH. MicroRNA-152 mediates DNMT1-regulated DNA methylation in the estrogen receptor α gene. PLoS One. 2012;7(1):e30635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kos M, O’Brien S, Flouriot G, Gannon F. Tissue-specific expression of multiple mRNA variants of the mouse estrogen receptor alpha gene. FEBS Lett. 2000;477(1-2):15–20. [DOI] [PubMed] [Google Scholar]
- 36.Prewitt AK, Wilson ME. Changes in estrogen receptor-alpha mRNA in the mouse cortex during development. Brain Res. 2007;1134(1):62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra SW, Hoskin E, Yudkovitz J, Pear L, Wilkinson HA, Hayashi S, Pfaff DW, Ogawa S, Rohrer SP, Schaeffer JM, McEwen BS, Alves SE. Immunolocalization of estrogen receptor beta in the mouse brain: comparison with estrogen receptor alpha. Endocrinology. 2003;144(5):2055–2067. [DOI] [PubMed] [Google Scholar]
- 38.Murphy AZ, Hoffman GE. Distribution of gonadal steroid receptor-containing neurons in the preoptic-periaqueductal gray-brainstem pathway: a potential circuit for the initiation of male sexual behavior. J Comp Neurol. 2001;438(2):191–212. [DOI] [PubMed] [Google Scholar]
- 39.Simerly RB, Chang C, Muramatsu M, Swanson LW. Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. J Comp Neurol. 1990;294(1):76–95. [DOI] [PubMed] [Google Scholar]
- 40.Elmquist JK, Ahima RS, Elias CF, Flier JS, Saper CB. Leptin activates distinct projections from the dorsomedial and ventromedial hypothalamic nuclei. Proc Natl Acad Sci USA. 1998;95(2):741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elmquist JK, Ahima RS, Maratos-Flier E, Flier JS, Saper CB. Leptin activates neurons in ventrobasal hypothalamus and brainstem. Endocrinology. 1997;138(2):839–842. [DOI] [PubMed] [Google Scholar]
- 42.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2nd ed. San Diego, CA: Academic Press; 2001. [Google Scholar]
- 43.Brock O, De Mees C, Bakker J. Hypothalamic expression of oestrogen receptor α and androgen receptor is sex-, age- and region-dependent in mice. J Neuroendocrinol. 2015;27(4):264–276. [DOI] [PubMed] [Google Scholar]
- 44.Yu CJ, Fang QQ, Tai FD. Pubertal BPA exposure changes central ERα levels in female mice. Environ Toxicol Pharmacol. 2015;40(2):606–614. [DOI] [PubMed] [Google Scholar]
- 45.Bartness TJ, Liu Y, Shrestha YB, Ryu V. Neural innervation of white adipose tissue and the control of lipolysis. Front Neuroendocrinol. 2014;35(4):473–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartness TJ, Shrestha YB, Vaughan CH, Schwartz GJ, Song CK. Sensory and sympathetic nervous system control of white adipose tissue lipolysis. Mol Cell Endocrinol. 2010;318(1-2):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartness TJ, Vaughan CH, Song CK. Sympathetic and sensory innervation of brown adipose tissue. Int J Obes. 2010;34(Suppl 1):S36–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones DD, Ramsay TG, Hausman GJ, Martin RJ. Norepinephrine inhibits rat pre-adipocyte proliferation. Int J Obes Relat Metab Disord. 1992;16(5):349–354. [PubMed] [Google Scholar]
- 49.Foster MT, Bartness TJ. Sympathetic but not sensory denervation stimulates white adipocyte proliferation. Am J Physiol Regul Integr Comp Physiol. 2006;291(6):R1630–R1637. [DOI] [PubMed] [Google Scholar]
- 50.Contreras C, Gonzalez F, Fernø J, Diéguez C, Rahmouni K, Nogueiras R, López M. The brain and brown fat. Ann Med. 2015;47(2):150–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bozaoglu K, Bolton K, McMillan J, Zimmet P, Jowett J, Collier G, Walder K, Segal D. Chemerin is a novel adipokine associated with obesity and metabolic syndrome. Endocrinology. 2007;148(10):4687–4694. [DOI] [PubMed] [Google Scholar]
- 52.Hansen IR, Jansson KM, Cannon B, Nedergaard J. Contrasting effects of cold acclimation versus obesogenic diets on chemerin gene expression in brown and brite adipose tissues. Biochim Biophys Acta. 2014;1841(12):1691–1699. [DOI] [PubMed] [Google Scholar]
- 53.Yu XX, Lewin DA, Forrest W, Adams SH. Cold elicits the simultaneous induction of fatty acid synthesis and beta-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J. 2002;16(2):155–168. [DOI] [PubMed] [Google Scholar]
- 54.Ozek C, Zimmer DJ, De Jonghe BC, Kalb RG, Bence KK. Ablation of intact hypothalamic and/or hindbrain TrkB signaling leads to perturbations in energy balance. Mol Metab. 2015;4(11):867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren H, Plum-Morschel L, Gutierrez-Juarez R, Lu TY, Kim-Muller JY, Heinrich G, Wardlaw SL, Silver R, Accili D. Blunted refeeding response and increased locomotor activity in mice lacking FoxO1 in synapsin-Cre-expressing neurons. Diabetes. 2013;62(10):3373–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang H, Lee SH, Ye C, Lima IS, Oh BC, Lowell BB, Zabolotny JM, Kim YB. ROCK1 in AgRP neurons regulates energy expenditure and locomotor activity in male mice. Endocrinology. 2013;154(10):3660–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musatov S, Chen W, Pfaff DW, Mobbs CV, Yang XJ, Clegg DJ, Kaplitt MG, Ogawa S. Silencing of estrogen receptor alpha in the ventromedial nucleus of hypothalamus leads to metabolic syndrome. Proc Natl Acad Sci USA. 2007;104(7):2501–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Correa SM, Newstrom DW, Warne JP, Flandin P, Cheung CC, Lin-Moore AT, Pierce AA, Xu AW, Rubenstein JL, Ingraham HA. An estrogen-responsive module in the ventromedial hypothalamus selectively drives sex-specific activity in females. Cell Reports. 2015;10(1):62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Y, Elmquist JK, Fukuda M. Central nervous control of energy and glucose balance: focus on the central melanocortin system. Ann N Y Acad Sci. 2011;1243(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caron E, Sachot C, Prevot V, Bouret SG. Distribution of leptin-sensitive cells in the postnatal and adult mouse brain. J Comp Neurol. 2010;518(4):459–476. [DOI] [PubMed] [Google Scholar]
- 61.Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, Lombardo A. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell. 2016;167(1):219–232.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, Sakai A, Nakashima H, Hata K, Nakashima K, Hatada I. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol. 2016;34(10):1060–1065. [DOI] [PubMed] [Google Scholar]
- 63.Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu GL, Hu R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44(12):5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee H, Kim DW, Remedios R, Anthony TE, Chang A, Madisen L, Zeng H, Anderson DJ. Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature. 2014;509(7502):627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morselli E, Fuente-Martin E, Finan B, Kim M, Frank A, Garcia-Caceres C, Navas CR, Gordillo R, Neinast M, Kalainayakan SP, Li DL, Gao Y, Yi CX, Hahner L, Palmer BF, Tschöp MH, Clegg DJ. Hypothalamic PGC-1α protects against high-fat diet exposure by regulating ERα. Cell Reports. 2014;9(2):633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fürst RW, Kliem H, Meyer HH, Ulbrich SE. A differentially methylated single CpG-site is correlated with estrogen receptor alpha transcription. J Steroid Biochem Mol Biol. 2012;130(1-2):96–104. [DOI] [PubMed] [Google Scholar]
- 69.Maffei M, Halaas J, Ravussin E, Pratley RE, Lee GH, Zhang Y, Fei H, Kim S, Lallone R, Ranganathan S, Kern PA, Friedman JM. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat Med. 1995;1(11):1155–1161. [DOI] [PubMed] [Google Scholar]
- 70.Münzberg H, Flier JS, Bjørbaek C. Region-specific leptin resistance within the hypothalamus of diet-induced obese mice. Endocrinology. 2004;145(11):4880–4889. [DOI] [PubMed] [Google Scholar]
- 71.Campfield LA, Smith FJ, Guisez Y, Devos R, Burn P. Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science. 1995;269(5223):546–549. [DOI] [PubMed] [Google Scholar]
- 72.Bjørbaek C, Kahn BB. Leptin signaling in the central nervous system and the periphery. Recent Prog Horm Res. 2004;59(1):305–331. [DOI] [PubMed] [Google Scholar]
- 73.Gallagher D, Heymsfield SB, Heo M, Jebb SA, Murgatroyd PR, Sakamoto Y. Healthy percentage body fat ranges: an approach for developing guidelines based on body mass index. Am J Clin Nutr. 2000;72(3):694–701. [DOI] [PubMed] [Google Scholar]
- 74.Palmer BF, Clegg DJ. The sexual dimorphism of obesity. Mol Cell Endocrinol. 2015;402:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmidt SL, Bessesen DH, Stotz S, Peelor FF III, Miller BF, Horton TJ. Adrenergic control of lipolysis in women compared with men. J Appl Physiol (1985). 2014;117(9):1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ter Horst KW, Gilijamse PW, de Weijer BA, Kilicarslan M, Ackermans MT, Nederveen AJ, Nieuwdorp M, Romijn JA, Serlie MJ. Sexual dimorphism in hepatic, adipose tissue, and peripheral tissue insulin sensitivity in obese humans. Front Endocrinol (Lausanne). 2015;6:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coleman DL, Hummel KP. The influence of genetic background on the expression of the obese (Ob) gene in the mouse. Diabetologia. 1973;9(4):287–293. [DOI] [PubMed] [Google Scholar]
- 78.Hwa JJ, Fawzi AB, Graziano MP, Ghibaudi L, Williams P, Van Heek M, Davis H, Rudinski M, Sybertz E, Strader CD. Leptin increases energy expenditure and selectively promotes fat metabolism in ob/ob mice. Am J Physiol. 1997;272(4 Pt 2):R1204–R1209. [DOI] [PubMed] [Google Scholar]