Abstract
Aim
The aim of our study is to focus on hTERT (human Telomerase Reverse Transcriptase) expression to identify tumoral tissue after a comparison to TP53 and KRAS. More than 85% of cancer cells contain genetic aberrations and also overexpression of hTERT, and, in fact, the promoter of hTERT characterizes all malignant cells.
Patients and methods
Our sample is composed of 18 patients, including 10 with CRC that underwent surgical procedure and 8 patients without CRC, which represent the control group. The hTERT gene expression, KRAS and p53 were evaluated by methodical Real Time - PCR on RNA extracted from tumor tissues, peritumoral tissue and control cases.
Results
Within the CRC group the evaluation of the tumor tissue showed an increase of hTERT expression with a statistical significance (> 0.1) in 5 of these, also associated with substantial increase of KRAS (> 0.2). The peritumoral tissue assessment showed important increase in KRAS in 4 patients (> 0.2), while hTERT is not found to be particularly increased. The value of p53 did not show any particular significance (<0.1).
Discussion
The analysis of our data leads us to consider that the increase of hTERT is evident in patients suffering from CRC and that some of them will become significant in relation to the increase of KRAS and independent of p53. In peritumoral tissues, however, KRAS increases considerably, instead hTERT maintains a low concentration and this is compatible with the cellular evolution of the neoplastic tissue adjacent to the tumor.
Conclusions
hTERT could be used for diagnosis and prognosis in the future, to be able to identify the risk of tumor progression and to set up an adequate therapy.
Keywords: Telomerase, Colorectal cancer, KRAS, P53
Introduction
Colorectal Cancer (CRC) is the third neoplasia and the fourth most common cause of oncological death worldwide (1, 2). Surgery is the first therapeutic choice to be performed from the diagnosis, associated with an oncological evaluation for pre or post-operative chemo or radio therapy.
CRC presents several mutations in oncogenes and oncosuppressors, such as APC, KRAS, BRAF, and in the latest studies also hTERT, which is modified or overexpressed in at least 85% of all CRC in the world. hTERT codifies for the catalytic subunit of telomerase, an enzyme whose role is crucial for the maintenance and the synthesis of chromosomal ends, the telomerase (3), that in the somatic cells face at each replication a progressive shortening of their sequences, reaching a length limit that let a further division impossible (4). It is essential for the maintenance of proliferative capacity, for preventing cellular senescence and death (5).
This activity is normally present in only a few cytotypes (stem cells, germ cells, activated lymphocytes, epithelial cells with a high turnover), while in the majority of somatic cells the gene is silenced, resulting in an inactivation of telomerase.
An overexpression of hTERT, already proved in several solid and hematologic tumors, acts in the prevention of cellular senescence and death causing the immortalization of the cell clone in which the gene is over activated, maintaining telomere length above the threshold that sets a limit to the replication (6).
The presence of hTERT could be an effective strategy in CRC treatment as gene-therapy target in association with surgery and chemotherapy protocols, considering its exclusive and specific presence in the neoplastic cells and its absence in the normal somatic ones.
Patients and methods
In this retrospective observational study, from January 2010 to December 2014, in our General Surgery Unit, we enrolled 10 patients with CRC and 8 control group patients, to evaluate the expression of hTERT in tumoral and in peritumoral tissues and also to study if hTERT could be a more specific tumor marker, compared to TP53 and KRAS.
CRC group (6 males and 4 females) consisted in 2 cancers on ascending colon, 3 on descending colon, 2 on sigmoid and 3 on rectum. 57 years (± SD) was the mean age (female range 55–75ys ; male range 50–70ys). 4 procedures were performed via traditional laparotomy and 6 laparoscopic procedures. Type of operation: 2 right hemicolectomies; 3 left hemicolectomies; 2 Hartmann’s procedure; 3 anterior resection of rectum. During surgical procedures, it was performed an intraoperative removal of tumor and peritumoral tissue (within 3cm from the cancer).
All patients were diagnosed with CT scan and colonoscopy with biopsy. According to TNM classification, staging has identified 4 pts stage I and 6 pts in stage II. Pathology examination revealed 3 poorly differentiated cancers, 3 moderately differentiated and 4 well differentiated. Exclusion criteria were: patients with TNM 3, colorectal cancer perforation and obstruction, metastatic colorectal cancers, patients with ASA IV–V score. All patients’ characteristics can be appreciated in Table 1.
Table 1.
PATIENT’S CHARACTERISTICS AND PROCEDURES.
| CRC Group | Control Group | |
|---|---|---|
| Gender | M 6 | M 4 |
| F 4 | F 4 | |
|
| ||
| Age | 57 | 55 |
|
| ||
| Surgical Procedure | 2 Right Colectomies | 8 Lap. Cholecistec. |
| 3 Left Colectomies | ||
| 2 Hartmann | ||
| 3 Rectum Ant. Res. | ||
|
| ||
| TNM | 4 Stage 1 | |
| 6 Stage 2 | ||
|
| ||
| Pathology | 3 Poorly Diff. | |
| 3 Moderately Diff. | ||
| 4 Well Diff. | ||
Furthermore, 8 pts (4 males and 4 females, mean age 55) were enrolled as a control group, which was free CRC and needed to be treated surgically because of cholelitiasis. It was necessary for our analysis a sample of tissue belonging to the gallbladder as healthy tissue.
The protocol was approved by the Ethical Committee and it was clearly explained to all participants. Written informed consent was obtained from all patients.
Samples of excised tissues were collected to evaluate gene expression of hTERT, KRAS and p53 and were cryopreserved in liquid nitrogen pending subsequent RNA extraction, to study their expression using Real Time PCR (qRT-PCR). We also calculated median values of three genes’ expression and we compared all groups (tumoral, peritumoral and normal tissue).
RNA extraction
Assumed the extreme instability of RNA and for the high risk of contamination with RNase, all materials have been pre-treated with H2O and DEPC (diethylpyrocarbonate). The technique used is a variant of the Chomczynski method (7). The cellular RNA from tissue samples was extracted by homogenisation in TRIzol® Reagent (Invitrogen). This methodology allows an extraction with less contamination by genomic DNA and proteins. The RNA quantity was determined by spectrophotometric examination at a wave length of 260 nm, while the total RNA obtained qualitative analysis is performed by electrophoresis on 1.7% agarose gel in which has been added the ‘ethidium bromide.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
For the reverse transcription was used the High-Capacity cDNA Reverse Transcription (Applied Biosystems Foster City, CA, USA) kit, following the protocol of the supplier company. A volume of 1 micrograms of total RNA was denatured at 75°C for 3 minutes then added to a reaction mix containing: 10X RT buffer, 25X dNTP mix, 10X RT random primers, of MultiScribe Reverse Transcriptase 50 U / ul, RNase inhibitor, (Applied Biosystems, Branchburg, NJ, USA). The samples were incubated (Gene Amp PCR 9700, Applied Biosystems) for 10 minutes at 25°C, for 120 minutes at 37°C, for 5 seconds at 85°C and for ∞ at 4°C.
qRT-PCR
The analysis of the expression of hTERT gene, TP53, and KRAS was determined using real-time PCR8. On the cDNA obtained after reverse transcription are the reactions of qRT-PCR were performed using IQTM SYBR Green Supermix (6mMMgCl2, dNTP, DNA polymerase iTaq, SYBR Green I, fluorescein and stabilizers) (Bio-Rad Laboratories, Hercules, CA, USA). The quantification of mRNA levels was performed on a MiniOpticon of Real-Time PCR Detection System (Bio-Rad Laboratories). The protocol used in the PCR reactions is the following: activation of the polymerase at 95°C for 3 min, followed by 45 cycles of 95°C for 10 s, 60°C for 30 s. The melting curves were generated through 60 additional cycles (65°C for 5 s with an increase of 0.5°C / cycle). The gene expression results were obtained as the average Ct (threshold cycle) of the samples in triplicate values. The expression was determined using the ΔΔCt method. The expression values were normalized to GAPDH.
Statistical analysis
Data on the clinical and histological characteristics of the study population were presented as absolute numbers, as the mean ± standard deviation or median. The comparison of the values of hTERT, TP53 and KRAS was performed using t-test Student and Mann Whitney test. A value of p-value <0.05 was considered statistically significant.
Results
From January 2010 to December 2014 in our General Surgery Unit, 10 patients (6 males and 4 females, mean age of 57) with a preoperative CRC diagnosis and 8 pts (4 males and 4 females; mean age of 55) without CRC were treated surgically and enrolled for our study to evaluate and compare hTERT, TP53 and KRAS’ expression from tumoral, peritumoral and healthy tissues.
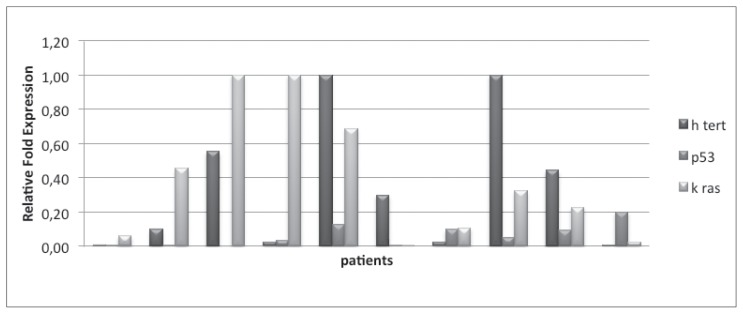
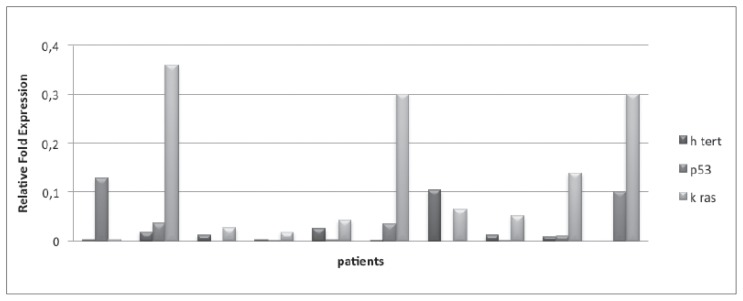
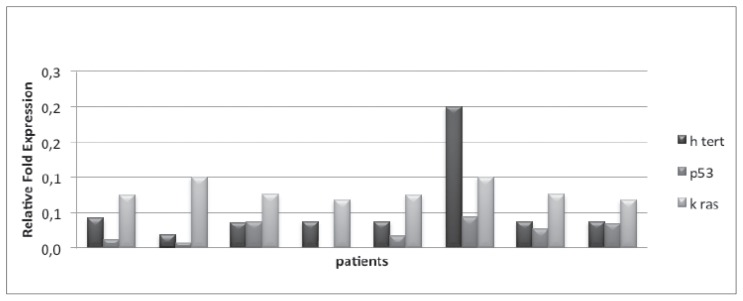
In CRC group, the median values of hTERT, TP53, KRAS were respectively of 0.2 (0–1), 0.044 (0–0.2) and 0.275 (from 0.004 to 1). In peritumoral tissue, median values of 3 genes were respectively of 0.009 (0 to 0.104), 0.006 (0 to 0.127) and 0.057 (0.003 to 0.359). For what concerns the control group, the median values were respectively 0.037 (0.019 to 0.2), 0.022 (0 to 0.044) and 0.075 (0.068 to 0.1) (Figures 1, 2, 3).
Figure 1.
Gene expression of hTERT, p53, Kras in tumor tissue.
Figure 2.
Gene expression of hTERT, p53, Kras in peritumoral tissue.
Figure 3.
Gene expression of hTERT, p53, Kras in healthy patients.
In CRC group, the evaluation of tumor tissue showed an increase of hTERT in all samples, with values higher than 0.1 and higher than 0.2 (respectively 5 and 5 pts). Peritumoral tissue assessment showed a significant increase (> 0.2) of KRAS levels in four patients, while hTERT is not found to be particularly increased. Finally, the value of TP53 did not show any statistical significance (<0.1). Within the control group it was detected an increase of hTERT in one pt, with uncertainty.
The comparative study of hTERT values, TP53 and KRAS in tumor, peritumoral and healthy tissues showed that the comparison between hTERT tumor and peritumoral values (p = 0.018) has statistical significance (p <0.05); while between tumor/healthy samples (p = 0.059) and peritumoral/healthy ones (p = 0.095) there is no statistical significance. KRAS value in healthy/tumor tissue samples has statistical significance (p = 0.04); while tumor/peritumoral samples (p = 0.061) and peritumoral/healthy (p = 0.334) did not demonstrate any significance. Finally, TP53 values were not statistically significant in any comparison.
Discussion
Many studies demonstrated that hTERT expression is highly specific for neoplastic cells and associated with increased enzyme activity. In fact, telomerase activity more than the actual length of telomeres is the key element of providing the cells the ability to replicate indefinitely without incurring senescence and apoptosis (9).
Despite all the attempts to understand the mechanism of telomerase in neoplastic cells, the factors that allow neoplastic cells to gain the reactivation of telomerase remain obscure and the changes that regulate its expression are still unknown.
hTERT presence has been already determined in almost all neoplastic tissue and absent in normal tissues and in peri-tumoral tissues. This feature let telomerase be a very attractive and therapeutic target.
In particular, in literature it has been established the role of hTERT in the initiation, promotion and progression of cancer, as well as the acquisition of an invasive and metastatic phenotype.
Tang et al. (10) investigated the role of telomerase activation in carcinogenesis, examining a group of patients with adenomatous polyps and a second group of patients with a colonic neoplasia. They observed high levels of the enzyme expression in 20% of intermediate polyps, in 73% of polyps larger than 2cm and in 96% of adenocarcinoma samples, while the levels found in normal mucosa and in the smaller polyps (<1cm) were respectively of 18% and 16%.
Roger et al. (11) found that, among all the other mechanisms of carcinogenesis, telomere dysfunction was the biggest driving force in chromosomal rearrangements’ generation in tumor progression. Although the mechanisms are not well defined yet, it was cleared that telomere shortening that occurs in normal mucosal cells into senescence, also occurs in colorectal cancer, but telomeric erosion has been detected only in adenomatous polyps and not in high degrees of dysplasia.
Tanaka et al. (12) have instead demonstrated that alterations in telomere maintenance, related to hTERT activity, do not seem to be so significant in initiation of colonic neoplasia as in its progression and acquisition of an invasive phenotype. A p53 defect and the shortening of telomere length were not enough in determining telomere fusion. In some cases, telomeric fusion was observed even in normal tissues and in peritumoral ones, suggesting that chromosomal instability is present in a non-neoplastic region. The erosion of telomeric regions is still extended and increases with tumoral invasiveness and lymph nodal infiltration.
Boldrini et al. (13) also investigated the role of hTERT overexpression in local and metastatic invasiveness. They evaluated the levels of transcripts of the hTERT gene in different samples from adenomatous polyps, dysplastic polyps, tumoral and two metastatic tissues from sporadic CRC. The results obtained by RT-PCR showed the highest levels of transcription in correspondence of all tissues examined. HTERT could then simplify the progression to metastatic disease.
Therefore, in literature the relationship between hTERT activity and the lifestyle of the population under study has been evaluated, particularly with regard to alimentary habits and smoking. Pellet et al. demonstrated that BMI and smoking were significantly and inversely associated with telomere length in comparison to control group, that instead, with a greater telomere length, had a lower risk of CRC onset (14). The study group was subsequently associated to higher BMI and presence of three SNPs of hTERT that predispose to CRC (15).
In our study we wanted to evaluate the expression of hTERT in tumoral and in peritumoral tissues, as well as in the control group of patients without CRC and our purpose was to understand if hTERT could be a more specific indicator of disease, compared to well-studied markers such as TP53 and KRAS.
We calculated median values of the expression of the three genes and we compared the different groups (tumoral, peritumoral and normal tissue). We noticed that medians were higher in cancer group than in two ones. From the analysis of the individual values of hTERT, TP53 and KRAS, hTERT in tumor tissues presented in all patients values greater than 0.1 and, in five of these, greater than 0.2; while in the peritumoral tissue, hTERT did not appear to have increased particularly. KRAS showed a significant increase (> 0.2) of its levels in four patients in the peritumoral sample. Finally, the TP53 value was not particularly relevant in any of the samples analysed. These data may indicate that hTERT should be considered a more specific marker of cancer than KRAS and TP53. Within the control group, an increase of hTERT value in a single subject was also detected.
To further confirm our hypothesis, we performed a comparison between the median values of the three genes in the three groups (tumoral, peritumoral and normal tissue). This study showed statistically significance only between tumoral and peritumoral group for what concerns hTERT gene and between tumoral and normal tissue for KRAS. Then we investigated the possible correlation between gene’s expression and cancer grading (G1, G2 and G3) and an increased expression of hTERT emerged and it is parallel to the increase of the degree; on the contrary, there was no detectable correlation for KRAS.
Our results agree with results reported by Tang and Roger: increased activity of telomerase has a crucial role in acquisition and progression of colonic neoplasia, in agreement with the significantly higher values of hTERT.
Furthermore, the comparison between hTERT overexpression and the cancer grading showed an increase in the most undifferentiated lesions (G3), and consequently in more invasive CRC.
Conclusions
The aim of our study is to focus on hTERT’s expression to identify its expression in tumoral tissue after a comparison to TP53 and KRAS. Because of its specificity as tumoral tissue’s marker hTERT could be an important therapeutic target to silence and regulate and modulate tumor progression.
Our study considers a scant number of patients. For this reason we prefer to improve our data and to obtain more accurate results. hTERT could be used for diagnosis and prognosis in the future and also to identify the risk of tumor progression and to set up an adequate therapy.
Contributor Information
Collaborators: C. DEGIORGI, G. D’ELIA, F. FERRARESE, and N. PALASCIANO
References
- 1.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Yu-Zhou Q, Xue-Cheng X, Hai-Zhou L, et al. Screening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancer. Oncology reports. 2015;33:2733. doi: 10.3892/or.2015.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zecchin D, Boscaro V, Medico E, Barault L, et al. BRAF V600E Is a Determinant of Sensitivity to Proteasome Inhibitors. Mol Cancer Ther. 2013;12(12):2950–2961. doi: 10.1158/1535-7163.MCT-13-0243. [DOI] [PubMed] [Google Scholar]
- 5.Kelland L. Targeting the limitless replicative potential of cancer: the telomerase/telomere pathway. Clin Cancer Res. 2007;13:4960–4963. doi: 10.1158/1078-0432.CCR-07-0422. [DOI] [PubMed] [Google Scholar]
- 6.Yang ZR, Wang HF, Zhao J, Peng YY, Wang J, et al. Recent developments in the use of adenoviruses and immunotoxins in cancer gene therapy. Cancer Gene Ther. 2007;14:599–615. doi: 10.1038/sj.cgt.7701054. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Cawthon RM. Telomere lenght measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Rse. 2009:37. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He S, Sun XJ, Zheng JB, et al. Recombinant lentivirus with enhanced expression of caudal related homeobox protein 2 inhibits human colorectal cancer cell proliferation in vitro. Mol Med Rep. 2015;12(2):1838–1844. doi: 10.3892/mmr.2015.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang, Cheng, et al. Close correlation between Telomerase expression and adenomatous polyp progression in multistep colorectal carcinogenesis. Cancer Res. 1998;58:4052. [PubMed] [Google Scholar]
- 11.Roger L, Jones RE, Heppel NH. Extensive telomere erosion in the iniation of colorectal adenomas and its association with chromosomal instability. J Natl Cancer Inst. 2013;105(16) doi: 10.1093/jnci/djt191. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka H, Matthew J, et al. The precence of telomere fusion in sporadic colon cancer indipendently of disease stage, TP53/KRAS mutation status, mean telomere lenght, and telomerase activity. Neoplasia. 2014;16:814–23. doi: 10.1016/j.neo.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldrini L, Faviana S, Gisfredi Y, Zucconi D. Evaluation of telomerase mRNA (hTERT) in colon cancer. Int J Oncol. 2002 Sep;21(3):493–7. doi: 10.3892/ijo.21.3.493. [DOI] [PubMed] [Google Scholar]
- 14.Pellatt AJ, Wolff RK, Lundgreen A. Genetic and lifestyle influence on telomere length anda subsequent risk of colon cancer in a case control study. Int J Mol Epidemiol Genet. 2012;3(3):184–94. [PMC free article] [PubMed] [Google Scholar]
- 15.Pellatt AJ, Wolff RK, Herrick J. TERT’s role in colorectal carcinogenesis. Mol Carcinog. 2013;52(7):507–13. doi: 10.1002/mc.21885. [DOI] [PMC free article] [PubMed] [Google Scholar]