Abstract
Recent advances in proteomics and genomics have enabled discovery of thousands of previously nonannotated small open reading frames (smORFs) in genomes across evolutionary space. Furthermore, quantitative mass spectrometry has recently been applied to analysis of regulated smORF expression. However, bottom-up proteomics has remained relatively insensitive to membrane proteins, suggesting they may have been underdetected in previous studies. In this report, we add biochemical membrane protein enrichment to our previously developed label-free quantitative proteomics protocol, revealing a never-before-identified heat shock protein in Escherichia coli K12. This putative smORF-encoded heat shock protein, GndA, is likely to be ∼36–55 amino acids in length and contains a predicted transmembrane helix. We validate heat shock-regulated expression of the gndA smORF and demonstrate that a GndA-GFP fusion protein cofractionates with the cell membrane. Quantitative membrane proteomics therefore has the ability to reveal nonannotated small proteins that may play roles in bacterial stress responses.
Despite their varied and often essential functions, small proteins have been consistently underannotated in both prokaryotic and eukaryotic genomes.1 Small open reading frame (smORF)-encoded small proteins function in bacteria as regulators of sporulation, cell division, membrane transport, membrane-bound enzymes, protein kinases, and chaperones.1−7 In a study of 51 recently discovered small Escherichia coli proteins, 21 were upregulated under a specific stress or growth condition.8 Notably, 90% of the small proteins that exhibited regulated expression were predicted to contain single transmembrane helices.8 It is therefore reasonable to hypothesize that additional small, membrane-associated bacterial stress response proteins remain to be discovered. Of the three leading technologies for smORF discovery, computational genomics,9 ribosome footprinting,10,11 and liquid chromatography–tandem mass spectrometry proteomics (LC–MS/MS),12−15 LC–MS/MS has the advantage of direct detection of peptides derived from nonannotated proteins and has recently been extended to quantitative analysis.16−19 However, bottom-up LC–MS/MS proteomics affords relatively poor detection of membrane proteins due to their low abundance and hydrophobicity,20,21 suggesting that membrane-associated, nonannotated small proteins may have been missed by previous quantitative LC–MS/MS studies. To address this limitation, we present a workflow for quantitative membrane proteomics. We apply this methodology to the E. coli K12 heat shock response, enabling the discovery of a previously nonannotated, membrane-associated small heat shock protein, which we provisionally name GndA.
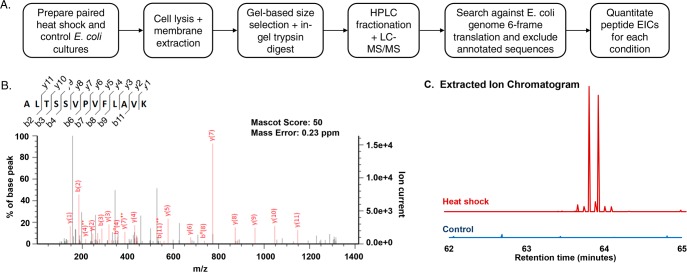
We and others recently reported a label-free quantitation protocol for comparative profiling of nonannotated peptides between two conditions.16,19Figure 1A provides an overview of a membrane-focused quantitative proteomic workflow. Briefly, E. coli K12 substr. MG1655 is grown under standard (control) conditions or subjected to heat shock, then lysed. Cell membranes are pelleted via ultracentrifugation, and the membrane proteome is resolubilized and separated on a peptide gel.15 Protein bands of low molecular weight are excised and subjected to trypsin digest. The digest is then fractionated by electrostatic repulsion hydrophilic interaction chromatography (ERLIC), and the fractions are analyzed by LC–MS/MS. Subsequently, the data are searched against a 6-frame translation of the E. coli K12 MG1655 genome using MASCOT, permitting identification of both known and nonannotated peptides. Annotated peptides are excluded with a string-matching algorithm14 via comparison to the E. coli K12 MG1655 proteome. For semiquantitative, comparative analysis of peptide abundance, sequences detected only in the heat shock sample and not the control are identified, then MS1 extracted ion chromatograph (EIC) peak intensities at the same retention time are compared.
Figure 1.
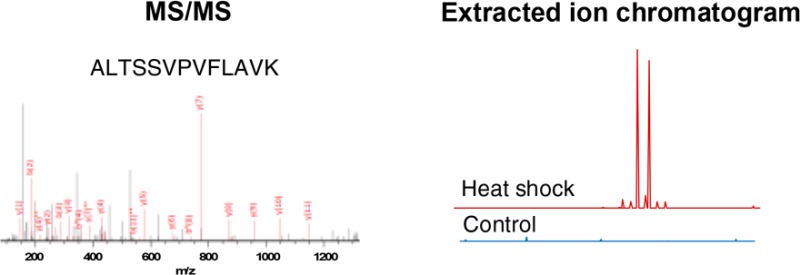
Discovery of small open reading frame (smORF)-encoded membrane proteins through quantitative proteomics. (A) Overview of membrane-targeted quantitative proteomic discovery protocol. (B) MS/MS spectrum corresponding to an unannotated tryptic peptide fragment detected only in the heat shock sample is shown. Identified y-ions and b-ions are shown in red on the spectrum and indicated on the peptide sequence to which the spectrum was matched. (C) Extracted ion chromatograms (EICs) comparing peaks (shown in stick mode) corresponding to the peptide ion m/z value detected in (B) in heat shock and control conditions at the same retention time. The same y-axis scale is used in both conditions. A viewing window of 1 Da around the parent ion mass is used.
Prior to analysis of nonannotated sequences, we first validated our workflow’s ability to quantify differential expression of peptides from an annotated heat shock protein. Comparing the EICs for selected tryptic fragments of the heat shock protein DnaJ (Hsp40) and a known nonheat shock protein, 50S ribosomal subunit protein L6 (RplF), verified that the DnaJ peptide was detected only during heat shock, while the RplF peptide was detected equally under both normal growth and heat shock (Supporting Information (SI), Figure S1), as expected. Therefore, we reliably distinguished heat shock responsive vs constitutive expression using label-free quantitation.
Second, we analyzed our workflow’s size selectivity. To do so, we first plotted the sizes of all annotated proteins identified in both our heat shock and control samples that were subjected to membrane enrichment. This analysis revealed a clear enrichment of small proteins, with the most commonly detected protein sizes ranging from 10 to 20 kDa (SI, Figure S2A), similar to size distributions obtained for soluble proteins in past LC–MS/MS proteomics studies of smORFs.14−16
Finally, we confirmed that we obtained an enrichment in peptides derived from membrane proteins by comparing our membrane-enriched control sample (not subjected to heat shock) to a previously reported sample grown under similar conditions that was not subjected to membrane preparation.19 We compared all of the annotated proteins identified using our membrane-enriched sample and the sample without membrane enrichment against a list of all E. coli K12 substr. MG1655 membrane proteins obtained from EcoCyc. These searches showed that 412/1208, or 34%, of annotated proteins detected from the membrane enrichment workflow had a membrane localization annotation, as opposed to 488/1849, or 26%, of annotated proteins detected from the regular workflow without enriching for membrane proteins (SI, Figure S2B). Of these proteins with membrane annotation, we detected peptides from 135 of them only in the workflow with membrane enrichment. These results suggest that our workflow provides an enhancement in the detection of peptides derived from membrane proteins while retaining small size selectivity.
The results of our proteomic analysis of heat shock and control samples are presented in SI, Proteomic results, and protein-level identifications are ranked according to sequence coverage. Because we focused on molecular genetic validation rather than statistical analysis of replicates to identify GndA as a heat shock protein (vide infra), we note that only a single experimental replicate is presented, so any other candidate heat shock-specific peptides must be considered putative. Nevertheless, our data set may aid hypothesis generation about regulated expression of predicted proteins. For example, peptides mapping to four known or predicted small proteins without currently annotated heat shock functions were detected in the heat shock sample but not in the control sample (SI, Figure S3 and S4). Two of these proteins are known or predicted to localize to the membrane (YfgG and YghG), and three currently lack functional characterization. Further experiments will be required to test heat shock responsive expression of these proteins.
In our heat shock data set, we identified precisely one nonannotated tryptic peptide exhibiting excellent sequence coverage (Figure 1B). Comparative analysis of the extracted ion chromatogram for this nonannotated tryptic peptide revealed MS1 ion intensity in our heat shock sample and not in the control (Figure 1C). This nonannotated peptide maps uniquely to an open reading frame (ORF) that is contained entirely within the gene gnd in the +2 reading frame (Figure 2). The putative protein that would be produced by translation of this ORF would therefore be completely different from Gnd at the amino acid level. Because of its coencoding with gnd, we refer to the smORF as gndA. There are two in-frame ATG codons upstream of the sequence putatively encoding the peptide detected by LC–MS/MS, either of which could plausibly initiate translation of GndA (Figure 2B). The length of GndA would thus most likely be 36–54 amino acids. Because bottom-up proteomics does not provide full sequence coverage for this putative protein, we have not yet confirmed the start codon or complete primary sequence for GndA, and it remains possible that neither in-frame ATG codon is the correct start site for this protein.
Figure 2.
Location of the nonannotated gene, gndA, within the E. coli MG1655 genome. (A) A gene locus diagram shows the coordinate of the stop codon downstream of a frame-shifted sequence within the annotated gnd gene. Sizes are proportional to gene lengths and directionality of coding sequences is indicated with arrows. (B) The coding sequence of gnd is shown with the sequence corresponding to the tryptic peptide fragment detected by MS/MS bolded and underlined. Highlighted in red are two upstream, in-frame candidate ATG start codons.
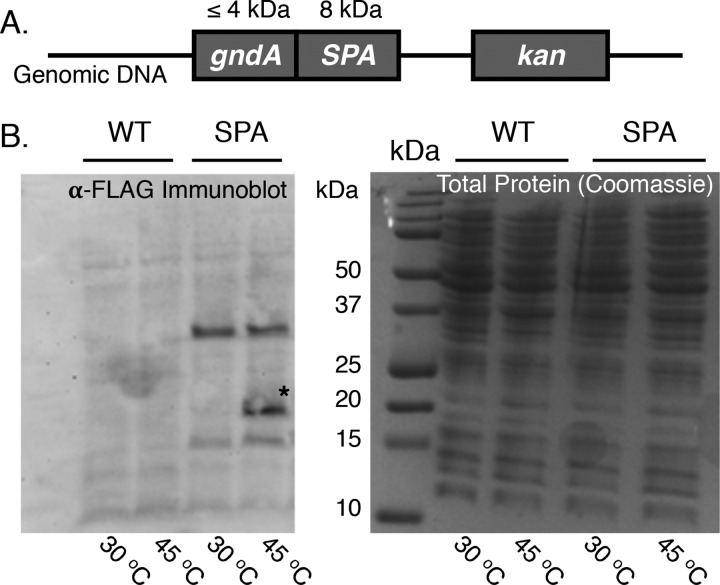
Because we identified only a single tryptic peptide that mapped to GndA, rigorous molecular genetic confirmation of its expression was required. We verified that gndA was expressed and upregulated during heat shock by generating a chromosomally tagged strain with the coding sequence for the tandem epitope tag SPA8 integrated at the 3′ end of the predicted gndA smORF. We confirmed the site of SPA tag insertion via integration check PCR and sequencing (SI, Figure S5). We grew the SPA-tagged strain under control and heat shock conditions and specifically detected expression of an immunoreactive band during heat shock (Figure 3). (Many membrane proteins exhibit anomalous mobility in SDS-PAGE,22 so the apparent migration of GndA-SPA may not exactly correlate with its molecular weight.) This result is consistent with expression of a small protein in the gndA reading frame during heat shock.
Figure 3.
gndA is expressed and upregulated during heat shock. (A) An E. coli MG1655 strain was generated with the SPA epitope tag (followed by a kanamycin selection marker, kan) introduced at the C-terminus of GndA. (B) Cell lysates of SPA-tagged and wild-type E. coli MG1655 strains grown at 30 °C (control) and 45 °C (heat shock) were separated on a 16% tricine gel and stained with Coomassie blue (right). Western blotting was performed on the same samples using anti-FLAG antibody to detect a portion of the SPA tag (left).
In the absence of a complete assignment of the gndA coding sequence, it remained possible that the observed peptide was generated via an alternative mechanism, such as ribosomal frameshifting during translation of 6-phosphogluconate dehydrogenase (Gnd), the protein product of gnd. We therefore confirmed that GndA can be translated independently. We generated pET21a plasmids containing the genomic sequence comprising the annotated ATG start codon of gnd to the stop codon of gndA. GFP was fused to the C-terminus of GndA to enhance stability and enable immunoblotting. We also deleted the start codon of gnd from this construct. We observed that expression of both of these constructs in BL21 cells produces the same product, which migrates at a slightly higher apparent molecular weight than GFP alone (SI, Figure S6). This result is consistent with independent translation of GndA, although it does not exclude all alternative interpretations.
Bioinformatic and biochemical analyses suggest that the predicted primary sequence of GndA may correspond to a small transmembrane protein. A portion of the putative GndA sequence (Figure 4A), highlighted in red, was predicted by three programs (TMPred, Phobius,23,24 and PredictProtein25) to form a transmembrane helix. Using the GFP fusion construct employed in SI, Figure S6, we performed a membrane fractionation. We verified by Western blotting that GndA-GFP is highly enriched in the membrane pellet after ultracentrifugation as compared to total clarified lysate and the soluble fraction, consistent with membrane localization (Figure 4C). A BLAST search against the NCBI nonredundant protein database did not reveal significant homology between GndA and known proteins (data not shown), and the predicted primary sequence of GndA lacks a signal sequence. Therefore, determination of the full sequence, function, mechanism of membrane insertion, inner vs outer membrane localization, and orientation of GndA in the membrane will require further study.
Figure 4.
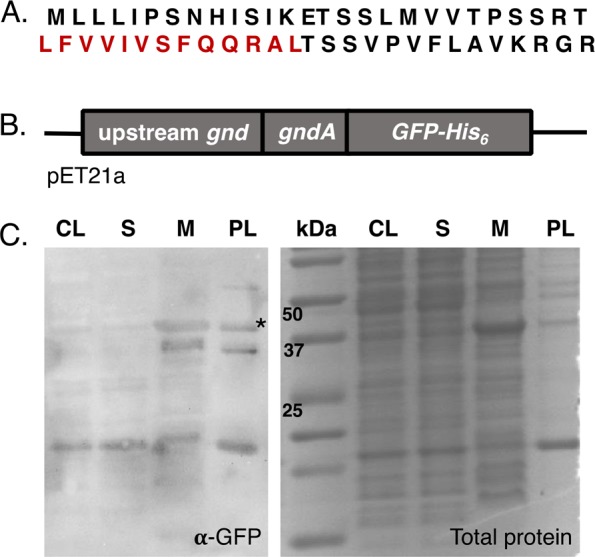
GndA is enriched in the membrane fraction. (A) The hypothetical primary sequence of GndA contains a predicted transmembrane helix (red). (B) BL21 cells were transformed with a pET21a plasmid encoding a GndA-GFP fusion protein. (C) Cell lysates were fractionated, separated on a 16% tricine gel, and stained with Coomassie blue as a loading control (right). Western blotting was performed on the same samples, probing using anti-GFP antibody (left). kDa, molecular weight ladder; CL, clarified lysate; S, soluble fraction; M, membrane pellet; PL, preclarified lysate.
In summary, we have developed an LC–MS/MS method to detect a peptide derived from a nonannotated small membrane protein regulated by heat shock, GndA. Notably, gndA would have been difficult to identify through alternative approaches to smORF discovery, including bioinformatics and ribosome footprinting, because the frameshifted gndA coding sequence is completely contained within the larger gnd sequence. Thus, our method presents a complementary approach to new gene discovery. In the future, we anticipate that this method can be extended to profiling of nonannotated membrane proteins expressed under different stress conditions and in other organisms.
Acknowledgments
We thank Jason Crawford for E. coli strain MG1655, plasmid pKD46, and advice on bacterial genetics.
Glossary
Abbreviations
- EIC
extracted ion chromatogram
- ERLIC
electrostatic repulsion hydrophilic interaction chromatography
- GFP
green fluorescent protein
- kDa
kilodalton
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- ORF
open reading frame
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- smORF
small open reading frame.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00864.
Author Contributions
P.Y. designed and performed experiments and wrote the manuscript. N.G.D. performed mass spectrometry. S.A.S. designed experiments and wrote the manuscript. All authors have given approval to the final version of the manuscript.
This work was supported in part by an American Cancer Society Institutional Research Grant Individual Award for New Investigators (IRG-58-012-57), the Leukemia Research Foundation, the Searle Scholars Program, the NIH (R01GM122984), and Yale University West Campus start-up funds (to S.A.S.). P.Y. was in part supported by an NIH Predoctoral Training Grant (5T32GM067543-12). N.G.D. was supported in part by a Rudolph J. Anderson postdoctoral fellowship from Yale University.
The authors declare no competing financial interest.
Supplementary Material
References
- Storz G.; Wolf Y. I.; Ramamurthi K. S. (2014) Annu. Rev. Biochem. 83, 753–777. 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin P. A.; Fan N.; Ricca E.; Driks A.; Losick R.; Cutting S. (1993) Mol. Microbiol. 9, 761–771. 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Wong R. S. Y.; McMurry L. M.; Levy S. B. (2000) Mol. Microbiol. 37, 364–370. 10.1046/j.1365-2958.2000.01998.x. [DOI] [PubMed] [Google Scholar]
- Wadler C. S.; Vanderpool C. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 20454–20459. 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOrsdel C. E.; Bhatt S.; Allen R. J.; Brenner E. P.; Hobson J. J.; Jamil A.; Haynes B. M.; Genson A. M.; Hemm M. R. (2013) J. Bacteriol. 195, 3640–3650. 10.1128/JB.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippa A. M.; Goulian M. (2009) PLoS Genet. 5, e1000788. 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S.; Sandoval M.; Storz G. (2011) J. Bacteriol. 193, 5887–5897. 10.1128/JB.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm M. R.; Paul B. J.; Miranda-Rios J.; Zhang A. X.; Soltanzad N.; Storz G. (2010) J. Bacteriol. 192, 46–58. 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm M. R.; Paul B. J.; Schneider T. D.; Storz G.; Rudd K. E. (2008) Mol. Microbiol. 70, 1487–1501. 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T.; Ghaemmaghami S.; Newman J. R. S.; Weissman J. S. (2009) Science 324, 218–223. 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T.; Lareau L. F.; Weissman J. S. (2011) Cell 147, 789–802. 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G.; Van Criekinge W.; Notelaers T.; Koch A.; Crappe J.; Gevaert K.; Van Damme P. (2013) Mol. Cell. Proteomics 12, 1780–1790. 10.1074/mcp.M113.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderperre B.; Lucier J. F.; Bissonnette C.; Motard J.; Tremblay G.; Vanderperre S.; Wisztorski M.; Salzet M.; Boisvert F. M.; Roucou X. (2013) PLoS One 8, e70698. 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S. A.; Mitchell A. J.; Schwaid A. G.; Cabili M. N.; Ma J.; Levin J. Z.; Karger A. D.; Budnik B. A.; Rinn J. L.; Saghatelian A. (2013) Nat. Chem. Biol. 9, 59–64. 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Ward C. C.; Jungreis I.; Slavoff S. A.; Schwaid A. G.; Neveu J.; Budnik B. A.; Kellis M.; Saghatelian A. (2014) J. Prot. Res. 13, 1757–1765. 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Diedrich J. K.; Jungreis I.; Donaldson C.; Vaughan J.; Kellis M.; Yates J. R. 3rd; Saghatelian A. (2016) Anal. Chem. 88, 3967–3975. 10.1021/acs.analchem.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza J. A.; Pina-Villalonga J. M.; Bosch R.; Nogales B.; Armengaud J. (2012) Mol. Cell. Proteomics 11, M111.013110. 10.1074/mcp.M111.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H.; Hahne H.; Ulbrich S. E.; Schnieke A.; Rottmann O.; Frishman D.; Kuster B. (2017) J. Prot. Res. 16, 2887–2898. 10.1021/acs.jproteome.7b00184. [DOI] [PubMed] [Google Scholar]
- D’Lima N. G.; Khitun A.; Rosenbloom A. D.; Yuan P.; Gassaway B. M.; Barber K. W.; Rinehart J.; Slavoff S. A. (2017) J. Prot. Res. 16, 3722. 10.1021/acs.jproteome.7b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers A. E.; Wu C. C. (2007) Chem. Rev. 107, 3687–3714. 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- Lai X. (2013) Electrophoresis 34, 809–817. 10.1002/elps.201200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath A.; Glibowicka M.; Nadeau V. G.; Chen G.; Deber C. M. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 1760–1765. 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L.; Krogh A.; Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method,. J. Mol. Biol. 338, 1027–1036. 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kall L.; Krogh A.; Sonnhammer E. L. (2007) Nucleic Acids Res. 35, W429–432. 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachdav G.; Kloppmann E.; Kajan L.; Hecht M.; Goldberg T.; Hamp T.; Honigschmid P.; Schafferhans A.; Roos M.; Bernhofer M.; Richter L.; Ashkenazy H.; Punta M.; Schlessinger A.; Bromberg Y.; Schneider R.; Vriend G.; Sander C.; Ben-Tal N.; Rost B. (2014) Nucleic Acids Res. 42, W337–343. 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.