Abstract
Base excision repair (BER) is a conserved and ubiquitous pathway that is initiated by DNA glycosylases, which recognize and remove damaged or mismatched nucleobases, setting the stage for restoration of the correct DNA sequence by follow-on BER enzymes. DNA glycosylases employ a nucleotide-flipping step prior to cleavage of the N-glycosyl bond, and most exhibit slow release of the abasic DNA product and/or strong product inhibition. As such, studying the catalytic mechanism of these enzymes requires care in the design, execution, and interpretation of single- and multiple-turnover kinetics experiments, which is the topic of this chapter.
Keywords: Enzyme kinetics, Single-turnover kinetics, Multiple-turnover kinetics, Catalytic turnover, DNA repair, DNA glycosylase, Base excision repair
1. INTRODUCTION
Damage to the nucleobases of DNA occurs continuously, due to endogenous and exogenous agents, and the resulting lesions fall largely into three categories, oxidation, deamination, and alkylation (Lindahl & Wood, 1999). To counter this threat, DNA glycosylases find and excise damaged bases, generating an abasic site, and the repair process is completed by other base excision repair (BER) proteins (Stivers & Jiang, 2003). There are two basic types of DNA glycosylases: monofunctional enzymes, which hydrolytically cleave the N-glycosyl bond to release the damaged base, and bifunctional enzymes, which remove the base and then nick the DNA backbone via a secondary lyase activity. While this chapter focuses on kinetic methods to study monofunctional glycosylases, the approaches can be adapted to study bifunctional glycosylases.
The reaction catalyzed by DNA glycosylases is more complex than might initially be imagined, which can complicate the collection and interpretation of enzymatic kinetics experiments. For example, the chemical step (base excision) is preceded by a reversible, base-flipping step (for most glycosylases), which transfers the target nucleotide out of the DNA helix and into the enzyme active site. Moreover, studies indicate that base excision is likely a stepwise process, with cleavage of the N-glycosyl bond preceding nucleophile addition (Drohat & Maiti, 2014). In addition, the catalytic turnover of DNA glycosylases is often limited by slow product release and/or strong product inhibition. This is consistent with the potentially toxic effect of releasing the abasic product, which can block DNA replication, stall transcription, and form interstrand crosslinks (Admiraal & O’Brien, 2015; Catalano et al., 2015; Price et al., 2014; Wilson & Bohr, 2007). As such, many glycosylases bind tightly to abasic DNA until they are displaced by a follow-on BER enzyme, typically an AP endonuclease (Waters, Gallinari, Jiricny, & Swann, 1999).
In this chapter, we discuss methods for the collection and analysis of single-turnover and multiple-turnover kinetic experiments, two approaches that are commonly used for studying the enzymatic activity of DNA glycosylases. Our laboratory studies thymine DNA glycosylase (TDG), which removes thymine and uracil bases that are mispaired with guanine (Neddermann & Jiricny, 1994; Wiebauer & Jiricny, 1990). TDG also removes 5-formyl- and 5-carboxylcytosine in a multienzyme pathway for active DNA demethylation (He et al., 2011; Maiti & Drohat, 2011b). Therefore, the methods described below are illustrated by data obtained for TDG.
2. KINETIC METHODS TO STUDY CATALYSIS BY DNA GLYCOSYLASES
2.1 Overview
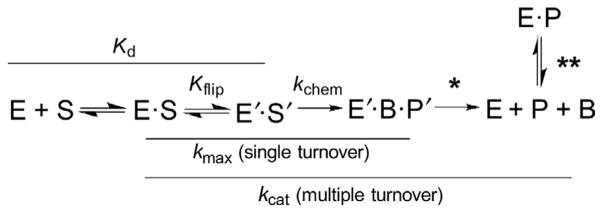
To set the stage, it is informative to consider the kinetic mechanism for a DNA glycosylase (monofunctional) and the reaction steps that influence the rate constants obtained from single- and multiple-turnover kinetics experiments (Fig. 1). Single-turnover experiments are performed with an excess of enzyme (E) relative to substrate (S) and yield an observed rate constant (kobs) that reports on steps from E–S association through the chemical step(s) to give enzyme-bound product. Under saturating enzyme conditions ([E] ≫ Kd), these experiments yield the maximal rate of base excision (kobs ≈ kmax), which is not impacted by E–S association or postchemical steps. Multiple-turnover experiments, performed with a large excess of S relative to E, yield a steady-state rate constant that reports on all steps, including product release and product inhibition; under saturating substrate conditions they give the maximal rate of catalytic turnover (kcat). For many glycosylases, the postchemical steps are strongly rate limiting (they dominate kcat), due to tight binding of the glycosylase to its reaction product, abasic DNA. Together, these two approaches provide important and complementary information about DNA glycosylase reactions, particularly when used to examine the effect of modifying catalytic groups of the enzyme, via site-directed mutagenesis, or modifying the substrate, through structure–activity relationships.
Fig. 1.
General kinetic mechanism for DNA glycosylases (monofunctional). The reversible nucleotide-flipping step, with an internal equilibrium of Kflip, involves a conformational change for both enzyme (E) and DNA substrate (S). The reaction steps that contribute to the two rate constants discussed here, kmax and kcat, are indicated (solid lines). The single asterisk denotes dissociation of the product complex, including liberation of the excised nucleobase (B); the double asterisk denotes product inhibition through binding of the glycosylase to abasic DNA (P). Each of these terms can contribute to kcat, depending upon the glycosylase.
2.2 Reagents and General Considerations
2.2.1 Substrates
While most DNA glycosylases act on duplex DNA, some act also on single-stranded DNA. Regardless, substrates are typically comprised of synthetic oligodeoxynucleotides (ODNs), which can be obtained from a commercial source or an academic facility; the latter source can be less expensive for ODNs that contain a noncanonical (modified) nucleobase. Synthetic ODNs can be purified by one of several methods, including reverse-phase HPLC (Malik, Coey, Varney, Pozharski, & Drohat, 2015), denaturing polyacrylamide gel electrophoresis (PAGE), or cartridge devices (e.g., Glen-Pak cartridges; Glen Research, Sterling, VA). The concentration of ODNs is determined by absorbance (260 nm), using a calculated extinction coefficient (ε260) that accounts for nearest-neighbor effects (Cavaluzzi & Borer, 2004; Tataurov, You, & Owczarzy, 2008), or, for greater accuracy, an ε260 that is determined experimentally via enzymatic hydrolysis of the ODN to the constituent NMPs (Kallansrud & Ward, 1996). ODNs should be stored in sterile, nuclease-free buffer (e.g., TE pH 7.5) at −20°C, in aliquots to avoid (or minimize) multiple freeze–thaw cycles.
If the glycosylase reaction will be monitored using electrophoretic methods (Section 3), the ODN containing the target (substrate) base must have a detection label, which can be a fluorophore, added during or after ODN synthesis, or a radiolabel, typically 32P, incorporated postsynthesis using standard methods. Labeling with a fluorophore avoids complications associated with handling and storing radioisotopes, but radiolabeling offers greater sensitivity, allowing experiments to be performed with far lower DNA substrate concentrations. Our laboratory has found that 0.1 pmol of fluorescein-labeled DNA (e.g., 10μL of 10 nM DNA) can be readily detected in a polyacrylamide gel using a Typhoon FLA 9500 imager (GE Healthcare).
Duplex DNA is produced by annealing two complementary ODNs, one containing a target (substrate) base, using standard procedures. To ensure that all molecules of the ODN-containing the substrate base are in duplex form, one can use 5%–10% molar excess of the complementary ODN.
2.2.2 Enzymes
Most DNA glycosylases can be recombinantly expressed in Escherichia coli and purified using standard chromatographic approaches. It is important that the enzymes are pure, ideally >95% as judged by SDS-PAGE, and accurately quantified, ideally by absorbance at 280 nm using a calculated extinction coefficient (Gill & von Hippel, 1989). Purified glycosylases should be stored at −80°C, in aliquots that can be used in a single day, to avoid multiple freeze–thaw cycles. TDG, the enzyme used as an example below, is purified as described (Bennett et al., 2006).
2.2.3 Reaction Conditions
The temperature used for kinetic assays should be considered carefully for several reasons. While it is preferable to collect experiments at physiological temperature, many DNA glycosylases are not stable in vitro at 37°C, at least not without a stabilizing agent, as shown for TDG (Maiti & Drohat, 2011a). Indeed, when a single-turnover reaction is found to reach a plateau prior to complete conversion of substrate product, loss of enzyme activity due to thermal instability is a potential explanation. Thus, it is important to establish that the enzyme is stable, or can be stabilized, at the desired experimental temperature. Some enzymes are stabilized by agents such as glycerol or BSA (~0.1 mg/mL) (Baldwin & O’Brien, 2009); the identity and required concentration of a stabilizing agent must be determined empirically. For some enzymes, other stabilizing agents are needed. For example, TDG is stabilized by the presence of excess (ideally saturating) amounts of DNA (can be nonspecific), which is already present when performing multiple-turnover experiments. Another consideration regarding temperature is the rate of a single-turnover reaction; some proceed so quickly that useful time points cannot be collected manually. This can potentially be addressed by using a lower temperature to slow the reaction, though some glycosylases (or substrates) will require use of a rapid chemical quenched-flow instrument (Bennett et al., 2006).
Other factors to consider include pH, which can substantially affect glycosylase activity, and the effects may be substrate dependent, particularly for reactions involving acid or base catalysis (Maiti, Michelson, Armwood, Lee, & Drohat, 2013). Ionic strength can also impact glycosylase activity, and this too can be substrate dependent. For example, TDG activity for a G·U substrate is relatively constant as ionic strength increases up to 0.20 M, while its G·T activity decreases substantially as ionic strength increases above 0.05 M (Maiti & Drohat, 2011a). In addition, a minimal ionic strength (~0.02 M) may be required to stabilize the enzyme and duplex DNA.
2.3 Single-Turnover Kinetics Experiments
2.3.1 Overview
Single-turnover experiments, performed with a large molar excess of enzyme relative to substrate, yield a rate constant (kobs) that reports on steps including E–S association, reversible nucleotide flipping, and the chemical step, and is not influenced by product release or product inhibition (Fig. 1). Briefly, the experiments are performed by adding concentrated enzyme to a buffered solution that contains substrate and all other components, incubating for specific time period, and then quenching to rapidly halt the reaction. Samples are taken at multiple time points (at least five), and the reaction progress (fraction product) for each sample is analyzed via electrophoresis (PAGE) or HPLC (Section 3). The rate constant for a given reaction is obtained by fitting the dependence of fraction product on time, using Eq. (1):
| (1) |
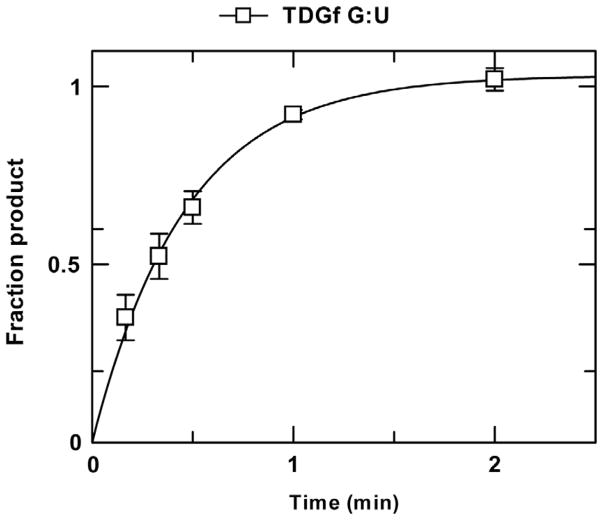
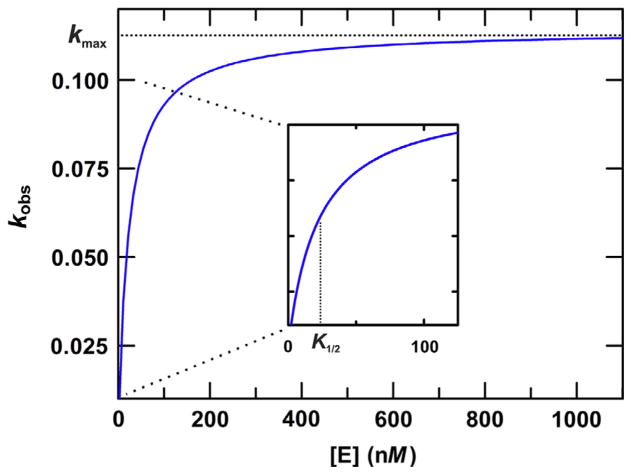
where A is the amplitude, k is the rate constant, and t is the reaction time. An example of fitted data obtained from a single-turnover experiment (for TDG) is shown (Fig. 2). When performed under saturating enzyme conditions ([E] ≫ Kd), the rate constant reflects the maximal rate of base excision (kobs ≈ kmax). While kmax is formally obtained by fitting the hyperbolic dependence of kobs on [E], from a series of single-turnover experiments performed with varying [E] (Fig. 3) (Johnson, 1992), kmax can be approximated with good accuracy by performing single-turnover experiments using an enzyme concentration that is known to be saturating ([E] ≫ Kd). This condition is confirmed by observation that kobs is unchanged for multiple enzyme concentrations that differ by twofold (or more) (Bennett et al., 2006).
Fig. 2.
Single-turnover kinetics experiment collected for TDG (1.5μM) and a G·U substrate (0.5μM) at 22°C. The progress curve (fraction product vs time) was fitted to Eq. (1), giving an amplitude of 1.03±0.03 and a rate constant of kobs = 2.17±0.14 min−1. Because the TDG concentration is 1000-fold greater than the Kd for TDG binding to a G·U substrate, the rate constant reflects the maximal rate of base excision (kmax), as confirmed by findings that single-turnover experiments performed with higher TDG concentrations (5 μM) yield similar kobs values.
Fig. 3.
Illustration of how kmax is determined by fitting the hyperbolic dependence of kobs on enzyme concentration. A series of single-turnover kinetics experiments are performed using a broad range of enzyme concentrations, including some below and some well above the expected Kd for the E–S complex, and a substrate concentration below that of the lowest enzyme concentration (to satisfy single-turnover conditions). The data are fitted to the following equation: kobs = kmax[E]/([E] + K1/2), where K1/2 is the enzyme concentration for which kobs = kmax/2, which can reflect the Kd for the productive E–S complex. The curve shown has a kmax of 0.113 min−1 and K1/2 of 22 nM.
2.3.2 Experimental Considerations
A few considerations are important regarding experimental design. First, the minimal substrate concentration that can be used depends on the detection method. For example, our lab uses an HPLC assay that requires at least 25 pmol of substrate (e.g., 50μL of 0.5μM DNA) for detection by absorbance (260 nm, unlabeled DNA) (Bennett et al., 2006). Electrophoretic assays require far less substrate, but the DNA must be labeled to enable detection (see Section 3). The enzyme concentration should exceed that of substrate by a least twofold, and should be saturating ([E]/Kd ≥ 10) to obtain the maximal rate of product formation (kobs ≈ kmax). At least five time points should be collected, with one at about the half-life (t1/2) and at least two each before and after the half-life (e.g., 0.25*t1/2, 0.5*t1/2, 1*t1/2, 2*t1/2, and 5*t1/2). If the time points are comfortably spaced (separated by ≥5 s), the individual samples can be taken from a single large reaction and quenched. For closely spaced time points, one can initiate and quench single reactions (for each time point). A rapid quenched-flow instrument is used to collect rapid time points, as short as 2 ms (Johnson, 1992). Because the final time points collected may differ from those initially planned, it is important to note exact time that each time point is quenched.
2.3.3 Solutions and Reagents
10× reaction buffer, filter sterilized (e.g., 0.2 M HEPES pH 7.5, 1 M NaCl, 2 mM EDTA)
DNA substrate (preferably ≥20× concentration)
DNA glycosylase enzyme (preferably ≥20× concentration)
BSA stock (10 mg/mL for use at 0.1 mg/mL; filter sterilized; can omit if [E] > 1μM)
Quench solution (e.g., 3× solution of 0.30 M NaOH, 0.03 M EDTA)
Sterile, deionized H2O (diH2O)
2.3.4 Procedure
Determine the total reaction volume, based on the number of time points and the sample size. For example, a reaction with six time points, 10μL each, requires a total volume of at least 60μL. For closely spaced time points, individual (single point) reactions may be collected.
Calculate the required volume of substrate, enzyme, stabilizing agent (if needed), and diH2O. Add all components except enzyme to the reaction tube, mix thoroughly, and incubate at reaction temperature.
Prepare and label sample tubes containing the appropriate amount of quench solution, based on its concentration and the volume of samples to be taken for the time points.
Initiate the reaction by adding concentrated enzyme and mixing gently but thoroughly.
For each time point, transfer a sample from the reaction to a tube containing the quench solution and mix rapidly and thoroughly, noting the exact time of quench.
Incubate the samples at 85°C for 10 min to quantitatively hydrolyze the sugar–phosphate backbone at abasic sites generated by the glycosylase. Samples should be analyzed via denaturing PAGE or HPLC to determine the fraction product (Section 3), or stored at 4°C until analysis (−20°C if longer than a few hours).
2.3.5 Notes and Potential Problems
If smaller sample volumes are desired (after quenching), e.g., for analysis via PAGE, one can use a 10× quench solution (1 M NaOH, 0.1 M EDTA).
Some nucleobases are labile when heated under alkaline conditions (0.1 M NaOH), such as 5-hydroxyuracil (hoU), 5-hydroxymethyluracil (hmU), and 5-formylcytosine (fC). For hoU and hmU substrates the problem can be alleviated by quenching with 0.1 M piperidine with 0.03 M EDTA and heating at 85°C for 15 min to nick at abasic sites (Bennett et al., 2006). For fC substrates, our lab has found that heating for 3 min after the quench gives quantitative cleavage of DNA at abasic sites, while heating for 15 min chemically alters the fC nucleobase and leads to a small level of backbone nicking that is not associated with glycosylase activity.
Single-turnover reactions should proceed to completion (all substrates converted to product, A > 0.95); if not, one of several issues could be the explanation. One possibility is that the enzyme is not stable under the reaction conditions and its activity is diminishing with time (see Section 2.2.3). Another possibility is that the concentration of active enzyme is below that of substrate and that catalytic turnover is very slow relative to the formation of enzyme-bound product (chemical step). Another possibility is that some fraction of the ODN containing the target base is not paired with a complementary ODN, leading to incomplete substrate processing for glycosylase that requires duplex DNA. In this case, the duplex substrate should be prepared again. Alternatively, to avoid using individual ODNs to make duplex substrate, one can use a single, self-complementary ODN that anneals to give the desired duplex substrate (Hedglin, Zhang, & O’Brien, 2013; Schonhoft, Kosowicz, & Stivers, 2013).
2.4 Multiple-Turnover Kinetics Experiments
2.4.1 Overview
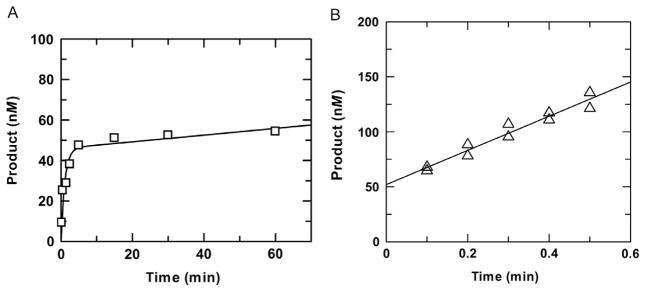
The general procedures for performing multiple-turnover kinetics experiments are similar to those used for single-turnover experiments, except that they are collected with a large molar excess of substrate relative to enzyme ([E] < [S]). Multiple-turnover experiments yield a steady-state velocity (v) that reports not only on steps up to and including the chemical step but also on postchemical steps, including product release and product inhibition (Fig. 1). When the experiments are performed with a saturating amount of substrate ([S] ≫ Kd), the steady-state velocity reaches maximal level (vmax), reflecting the maximal rate of catalytic turnover (kcat = vmax/[E]). For most DNA glycosylases, steps after base excision (chemistry) are strongly rate limiting, such that kcat ≪ kmax, and progress curves from multiple-turnover reactions exhibit “burst” kinetics, with an initial presteady-state (exponential) phase, reflecting the rapid formation of enzyme-bound product, followed by a steady-state (linear) phase, reflecting much slower enzymatic turnover (Fig. 4A). The kinetic parameters for the exponential and linear phases can be obtained by fitting a progress curve to Eq. (2):
| (2) |
where A and k are the amplitude and rate constant for the exponential phase, v is the initial steady-state velocity, and t is time. Note that Eq. (2) contains the same exponential term found in the equation used for fitting single-turnover data (Eq. 1), plus a linear (steady-state) term (vt).
Fig. 4.
Examples of multiple-turnover experiments for which the exponential (burst) phase is (A) or is not (B) observed. (A) Multiple-turnover experiment performed with 0.05μM TDG and a saturating concentration of G·T substrate (1.0μM) at 37°C. Fitting to Eq. (2) gives a rate constant of kobs =0.93±0.29 min−1 and amplitude of A=0.046±0.005 μM for the exponential phase and a rate constant of kcat = 0.0033 ± 0.0025 min−1 for the steady-state phase (using kcat = v/[E]). (B) Multiple-turnover experiment performed with 0.05μM TDG and a saturating concentration of G·5FU substrate (1.5μM). Due to the rapid chemical step for G·5FU substrates, the exponential phase is complete within 1 s and is not observed. A linear fitting of data from two independent experiments gives a steady-state rate constant of kcat = 3.1±0.3 min−1 and y-intercept of 52±4 nM. Close agreement between the amplitude (y-intercept) and the enzyme concentration used in the experiment indicates the enzyme is fully active.
For reactions performed with a very low concentration of enzyme relative to substrate ([E]/[S] < 0.01), the amplitude of the exponential phase might be too small to observe. Similarly, for some reactions, the exponential phase will not be observed because the chemical step is too rapid (kmax > 1 s −1). In this case, the initial steady-state phase of the progress curve ([product] vs time) is fitted to a linear equation, where the slope gives the initial velocity (v0).
2.4.2 Considerations for Experimental Design
A few items should be considered regarding experimental design, including the concentration of enzyme and substrate. The enzyme concentration should be relatively low (ideally [E]/[S] < 0.05) to minimize the consumption of substrate and the formation of product (a potentially strong inhibitor) during the presteady-state (burst) phase, which can help to extend the duration of the steady-state (linear) phase. However, some glycosylases exhibit extremely slow turnover such that a relatively high enzyme concentration may be needed to give a measurable steady-state velocity (v) in a reasonable timeframe, as exemplified by kcat (vmax/[E]) of 0.0003 min−1 for TDG acting on a G/T DNA substrate (Fitzgerald & Drohat, 2008). For most enzymes, the maximal rate of catalytic turnover, kcat = vmax/[E]), is obtained by fitting the dependence of v0 on [S] to the Michaelis–Menten equation. However, this is not practical for many DNA glycosylases due to extremely slow catalytic turnover. For such glycosylases, the maximal turnover rate kcat can be obtained from the initial velocity obtained under saturating substrate conditions, where v0 ≈ vmax, as confirmed by observation that v0 is unchanged for several substrate concentrations that are considered to be above the saturating level. For conditions in which the presteady-state phase is observed, time points should be chosen such that at least four fall in each of the exponential and the steady-state phases, and they may need to be optimized after based on initial experiments. Otherwise, the procedure for performing a multiple-turnover experiment is quite similar to that for a single-turnover experiment (Section 2.3).
2.4.3 Application for Determining Active Enzyme Concentration
For glycosylase reactions in which kmax ≫ kcat (e.g., kmax/kcat > 100), the amplitude (A) obtained from fitting multiple-turnover data to Eq. (2) can provide a measure of the concentration of active enzyme under the reaction conditions. If the exponential phase is too rapid to be observed (kmax > 1 s−1), the active enzyme concentration can be taken as the y-intercept observed for a linear fitting of the initial steady-state phase (Fig. 4B) (Morgan, Maiti, Fitzgerald, & Drohat, 2011; Porello, Leyes, & David, 1998). If the result of such experiments indicates that the concentration of active enzyme is lower than expected, it may indicate that the concentration of the enzyme stock is not accurate or that some fraction is inactive, at least not under the reaction conditions. Of course, this approach requires an accurate determination of the substrate concentration.
2.5 Characterizing the Enhancement of DNA Glycosylase Activity by AP Endonucleases
2.5.1 Overview
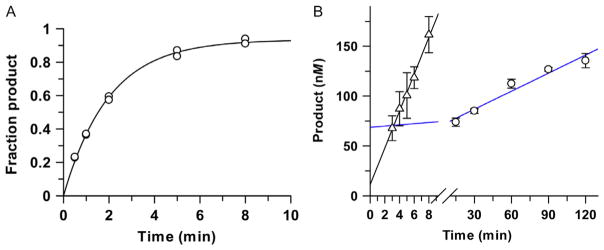
Given the very tight binding of many glycosylases to their abasic DNA product, a cytotoxic intermediate of BER, there has been much interest in characterizing and quantifying the effect of AP endonucleases in stimulating the catalytic turnover (kcat) of glycosylases. The studies typically involve multiple-turnover experiments using both the glycosylase and an AP endonuclease. Studies of TDG provide an excellent example, as AP endonuclease 1 (APE1) dramatically enhances its catalytic turnover (kcat) (Fitzgerald & Drohat, 2008; McLaughlin, Coey, Yang, Drohat, & Matunis, 2016; Sassa, Caglayan, Dyrkheeva, Beard, & Wilson, 2014; Waters et al., 1999) (Fig. 5). Similarly, APE1 enhances the catalytic turnover of alkyladenine-DNA glycosylase (Baldwin & O’Brien, 2009) and many other DNA glycosylases.
Fig. 5.
Enhancement of TDG steady-state turnover by APE1. (A) Single-turnover experiments, performed with 1.0 μM TDG and 0.3μM G·caC substrate, yield a rate constant of kobs = 0.50 ±0.02 min−1, reflecting the maximal rate of product formation (kmax). (B) Multiple-turnover experiments performed with 0.05 μM TDG and 1.0 μM G·caC substrate give a slow steady-state rate constant of kcat = 0.011 ±0.002 min−1 in the absence of APE1 (blue line, circles). Steady-state turnover is much (32-fold) higher for TDG in the presence of 1.2 μM APE1, (triangles), and approaches the theoretical maximum ( ).
2.5.2 Experimental Considerations
The following items should be considering during experimental design for a multiple-turnover reaction with a DNA glycosylase and an AP endonuclease. First, many AP endonucleases, including APE1, require Mg2+ or another divalent cation as a cofactor, which must be present in the reaction buffer (~2 mM Mg2+ typically used for APE1). Conveniently, APE1, like many AP endonucleases, is stable at 37°C. Notably, DNA glycosylases can potentially be inhibited by the product of the AP endonuclease reaction, nicked abasic DNA. The use of an excess concentration of AP endonuclease relative to DNA can alleviate the inhibition, enabling the glycosylase turn-over rate (kcat) to approach the theoretical limit of kmax, the maximal rate of base excision. For example, under such conditions, APE1 enhances the catalytic turnover of TDG by 32-fold, such that kcat/kmax = 0.7 (Fig. 5).
2.5.3 Procedure
Multiple-turnover reactions are performed essentially as described above (Section 2.4), with the AP endonuclease added after other components, but prior to (or with) the glycosylase.
Although AP endonucleases nick the DNA backbone at AP sites, the quench and heating steps (Section 2.3.4) are still necessary, to halt the activity of both enzymes and nick any AP sites that have not been acted on by the AP endonuclease.
3. Analytical Methods for Monitoring DNA Glycosylase Activity
3.1 Overview
Here, we discuss the strengths and weaknesses of two methods to detect activity of DNA glycosylases: PAGE and anion-exchange HPLC, both performed under denaturing conditions. More widely used, PAGE can be highly sensitive, particularly for 32P-labeled DNA, requires a small sample size (~10μL), is performed using relatively inexpensive equipment, and is similar to other electrophoretic assays. However, the DNA must be labeled (substrate strand), with a fluorophore or 32P, and imaging equipment is needed to detect the labeled DNA. The gels must be prepared, equilibrated (prerun), handled carefully after electrophoresis to avoid damage, and analyzed to quantify the signal for substrate and product in each lane. The process also generates some hazardous waste that must be properly handled (polyacrylamide, formamide). PAGE can also be sensitive to contaminants, such as residual amounts of SDS that can remain in a gel box that has been used for protein analysis by SDS-PAGE. On the other hand, anion-exchange HPLC can be automated (using an autosampler), does not generate hazardous waste, and does not require labeling since the DNA can be detected by absorbance (260 nm). However, HPLC systems are expensive, and detection by absorbance requires high amounts of DNA substrate (e.g., 50μL of 0.50μM substrate) relative to PAGE (e.g., 10μL of ≤0.01μM). Each sample requires ~30 min to be analyzed by HPLC, so, while it can be automated, it is time-consuming, and the HPLC system and analytical column must be properly maintained.
3.2 Electrophoretic (Gel-Based) Analysis
3.2.1 Reagents
2× Sample loading buffer (formamide with 5% (v/v) 0.1 M EDTA, pH 8.0)
1 L of 5× TBE running buffer (54 g Tris–base, 27.5 g boric acid, 20 mL 0.5 M EDTA pH 8.0; adjust final volume to 1 L with deionized H2O)
15% TBE-urea denaturing PAGE mini-gels (Fisher Scientific)
3.2.2 Procedure
Place a mini-gel in a gel box with an internal heating/cooling core and fill the upper and lower chambers with 1× TBE.
Connect a circulating water bath (set to 55°C) and preheat the mini-gel and buffer.
Remove the comb from the mini-gel, taking care to not disturb the lanes.
Using TBE, gently but completely flush all urea from the wells.
Prerun the mini-gel for 30 min according to the manufacturer’s specifications. For a 15% TBE-urea gel, this corresponds to 180 V (constant) with a maximum current of 20 mA.
Add loading buffer to the quenched samples (from kinetics experiments), mix, incubate at 90°C for 5 min, then spin briefly.
Repeat step 4 to ensure that all excess urea is flushed out of each well.
Load a small volume (≤10μL) of each sample into the appropriate well.
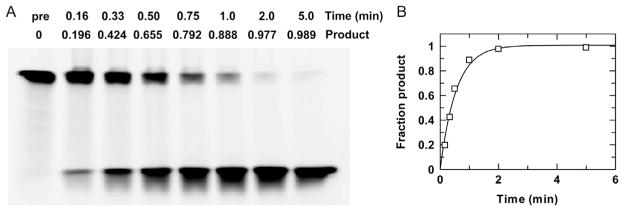
Run the gel according to manufacturer’s specifications. The time needed to achieve a quality separation depends on DNA fragment size and the percentage of acrylamide in the gel. For the 15% gel shown in Fig. 6, the intact and cleaved ODNs are a 40-mer and a 20-mer, respectively, and the running time was 40 min.
For 32P-labeled DNA, expose gels overnight to a phosphor screen before imaging.
Analyze gels using an imager optimized for the given probe. For the gel in Fig. 6, 3′-6-FAM-labeled DNA was detected using an LPB filter (GE Healthcare) with excitation at 473 nm, a photomultiplier value of 500, and preprogrammed correction for FAM.
The relative amounts of substrate and product DNA in each lane are quantified using the software available with the imaging system (e.g., ImageQuant for imagers from GE Healthcare).
Fig. 6.
Electrophoretic (gel-based) analysis of glycosylase activity. (A) Gel-based analysis of a single-turnover reaction, collected with 0.25 μM TDG and 0.025 μM G·U substrate (3′-6-FAM labeled) at 22°C. Samples quenched at indicated time points were analyzed to determine the fraction product, based on the intensity of imaged bands corresponding to substrate (upper) and product (lower). (B) Fitting the resulting progress curve to Eq. (1) yields a rate constant of kobs = 1.9 ±0.2 min−1.
3.2.3 Notes
Sample loading buffer can take several forms; many include formamide to denature the DNA and carry the sample to the bottom of the well. Bromophenol blue (0.025%) and xylene cyanol (0.025%) can be included to visually monitor progress, for 32P-labeled DNA but not fluorescently labeled DNA (due to potential interference with emission).
The optimal percentage of polyacrylamide in the gel depends on length of the ODNs for substrate and product.
For short reactions, it can be convenient to perform the kinetics reactions while prerunning the gel.
Formamide is a liquid at −20°C; aliquots that appear frozen should be discarded.
Dispose of formamide and polyacrylamide in appropriate hazardous waste containers.
If loading dyes are omitted from the samples, it is helpful to run dye in the first lane to monitor progress, skipping a lane between this and any fluorescently labeled sample. It is also useful to label the lanes (permanent marker) to indicate those containing samples.
Load any unused lanes with formamide (same amount used in samples) to obtain tighter bands and avoid bleeding at the edges of the gel.
3.3 Chromatographic Analysis via HPLC
3.3.1 Equipment
While there is no requirement for a particular HPLC system, a specific HPLC column gives excellent results. Our laboratory uses a DNAPac PA200 analytical anion exchange and guard column (4 × 250 and 4 × 50 mm, respectively; ThermoFisher Scientific). Our HPLC system is a Varian Prostar 500 equipped with a UV/Vis detector and a Prostar 410 autosampler, which greatly facilitates analysis of multiple samples. Samples are loaded into 12 × 32 mm, 600μL vials with screw caps (Sun SRI) for sampling.
3.3.2 Solutions
Mobile phase A: 0.02 M dibasic sodium phosphate, 0.03 M sodium perchlorate, pH 12
Mobile phase B: 0.02 M dibasic sodium phosphate, 0.50 M sodium perchlorate, pH 12
3.3.3 Procedure
Power on the HPLC system and UV lamp, and set the detection wavelength to 260 nm.
-
Enter the programs and needed for automated loading and analysis of multiple samples (gradients, etc.). We typically use the following gradient program (flow rate of 1.2 mL/min):
Inject the sample and continue with 7% B for 1 min
Linear gradient from 7% to 30% B over 29 min
Increase to 100% B and hold for 2 min (1 column volume)
Return to 7% B for 6 min (3 column volumes)
Flush the system lines with the two mobile phases (B first, followed by A).
Connect the analytical column (DNAPac PA200), avoiding the introduction of air, and equilibrate with 10 column volumes of mobile phase (7% B, 1.2 mL/min).
Transfer quenched samples from kinetics reactions (Section 2) into clean HPLC sample vials and seal with a cap.
Load samples into the autosampler and start the analysis.
After analysis of all samples, rinse the column with water to remove the mobile phase.
3.3.4 Notes
While it is preferable to use new HPLC sample vials and caps, they can be reused if carefully cleaned between use; vial caps showing extensive wear should be discarded.
Running a blank sample before samples of interest is recommended.
Mobile phase solutions should be degassed and filtered (0.2μm).
The two DNA strands of a duplex are sometimes not resolved by the DNAPac PA200, which complicates the quantification (integration) of the peak corresponding to the substrate strand. The problem can be solved by increasing the content of dG and/or dT in the complementary ODN (decreasing dG and/or dT in the target ODN), which increases the elution time of the complement and decreases that of the target (due to ionization of imino sites of dG and dT). On a similar note, the target ODN should be designed such that the two product fragments differ sufficiently in length to be resolved as two distinct peaks.
4. DETERMINING RATE CONSTANTS FROM KINETICS EXPERIMENTS
4.1 Determining the Fraction Product for Samples in Kinetics Reactions
The first step in determining the rate constants for a particular progress curve is to calculate the amount of product (fraction product) for each time point (quenched sample) in a progress curve. The approach depends on whether the samples were analyzed by HPLC or electrophoretic methods. For the latter (Section 3.2), the first step is to quantify the bands in each lane that correspond to substrate and product, typically using software that comes with the imaging system (e.g., ImageQuant, GE Healthcare). This requires background subtraction, which can be performed in several different ways. Our lab uses the “Image Rectangle” option by highlighting a section of the gel outside of the sample lanes, where no discernible signal is present. Bands for substrate and product are then selected within a given lane. Even if a band for substrate or product is not readily seen, the corresponding region should be quantified, based on its position in other lanes, and included in the analysis. ImageQuant generates a report for each lane, giving the absolute intensity and the fractional intensity of bands corresponding to substrate or product (Fig. 6). Thus, the ImageQuant analysis yields directly the information required for generating a progress curve (fraction product vs time).
For HPLC analysis, peaks in the chromatogram (A260 vs time) arising from ODNs corresponding to substrate (S) and products (P1, P2) are identified, based on standards of the expected length, and integrated to obtain the peak area for each. Because retention time depends on the charge of an ODN, shorter ODNs (e.g., products) will elute before longer ones (substrate). The fraction product is determined using Eq. (3):
| (3) |
4.2 Obtaining Rate Constants From the Progress Curves
Once the fraction product has been determined for all samples of a given experiment, the rate constants are determined using nonlinear regression and a computer program such as GraFit (Erithacus Software). When fitting data from single-turnover experiments (Eq. 1), we treat the amplitude (A) as a variable (rather than fixed) parameter, because the fitted amplitude gives an indication of whether the reaction has gone to completion (Section 2.3). The same is done when fitting data from multiple-turnover experiments to Eq. (2), because the fitted amplitude can be informative, perhaps giving an indication of the concentration of active enzyme in the reaction.
Acknowledgments
This work was supported by grant GM072711 from the National Institutes of Health (to A.C.D.).
References
- Admiraal SJ, O’Brien PJ. Base excision repair enzymes protect abasic sites in duplex DNA from interstrand cross-links. Biochemistry. 2015;54(9):1849–1857. doi: 10.1021/bi501491z. http://dx.doi.org/10.1021/bi501491z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin MR, O’Brien PJ. Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48(25):6022–6033. doi: 10.1021/bi900517y. http://dx.doi.org/10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. Journal of the American Chemical Society. 2006;128(38):12510–12519. doi: 10.1021/ja0634829. http://dx.doi.org/10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano MJ, Liu S, Andersen N, Yang Z, Johnson KM, Price NE, et al. Chemical structure and properties of interstrand cross-links formed by reaction of guanine residues with abasic sites in duplex DNA. Journal of the American Chemical Society. 2015;137(11):3933–3945. doi: 10.1021/jacs.5b00669. http://dx.doi.org/10.1021/jacs.5b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Research. 2004;32(1):e13. doi: 10.1093/nar/gnh015. http://dx.doi.org/10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drohat AC, Maiti A. Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA. Organic & Biomolecular Chemistry. 2014;12(42):8367–8378. doi: 10.1039/c4ob01063a. http://dx.doi.org/10.1039/c4ob01063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald ME, Drohat AC. Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. The Journal of Biological Chemistry. 2008;283(47):32680–32690. doi: 10.1074/jbc.M805504200. http://dx.doi.org/10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. http://dx.doi.org/10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M, Zhang Y, O’Brien PJ. Isolating contributions from inter-segmental transfer to DNA searching by alkyladenine DNA glycosylase. The Journal of Biological Chemistry. 2013;288(34):24550–24559. doi: 10.1074/jbc.M113.477018. http://dx.doi.org/10.1074/jbc.M113.477018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. 1 Transient-state kinetic analysis of enzyme reaction pathways. In: Sigman DS, editor. The enzymes. Vol. 20. Academic Press; 1992. pp. 1–61. [Google Scholar]
- Kallansrud G, Ward B. A comparison of measured and calculated single- and double-stranded oligodeoxynucleotide extinction coefficients. Analytical Biochemistry. 1996;236(1):134–138. doi: 10.1006/abio.1996.0141. http://dx.doi.org/10.1006/abio.1996.0141. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286(5446):1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Dependence of substrate binding and catalysis on pH, ionic strength, and temperature for thymine DNA glycosylase: Insights into recognition and processing of G.T mispairs. DNA Repair (Amst) 2011a;10(5):545–553. doi: 10.1016/j.dnarep.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. The Journal of Biological Chemistry. 2011b;286(41):35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Michelson AZ, Armwood CJ, Lee JK, Drohat AC. Divergent mechanisms for enzymatic excision of 5-formylcytosine and 5-carboxylcytosine from DNA. Journal of the American Chemical Society. 2013;135(42):15813–15822. doi: 10.1021/ja406444x. http://dx.doi.org/10.1021/ja406444x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik SS, Coey CT, Varney KM, Pozharski E, Drohat AC. Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA. Nucleic Acids Research. 2015;43(19):9541–9552. doi: 10.1093/nar/gkv890. http://dx.doi.org/10.1093/nar/gkv890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin D, Coey CT, Yang WC, Drohat AC, Matunis MJ. Characterizing requirements for SUMO modification and binding on base excision repair activity of thymine DNA glycosylase in vivo. The Journal of Biological Chemistry. 2016;291(17):9014–9024. doi: 10.1074/jbc.M115.706325. http://dx.doi.org/10.1074/jbc.M115.706325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT, Maiti A, Fitzgerald ME, Drohat AC. Stoichiometry and affinity for thymine DNA glycosylase binding to specific and nonspecific DNA. Nucleic Acids Research. 2011;39(6):2319–2329. doi: 10.1093/nar/gkq1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(5):1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37(42):14756–14764. doi: 10.1021/bi981594+. http://dx.doi.org/10.1021/bi981594+ [DOI] [PubMed] [Google Scholar]
- Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS. Inter-strand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. Journal of the American Chemical Society. 2014;136(9):3483–3490. doi: 10.1021/ja410969x. http://dx.doi.org/10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa A, Caglayan M, Dyrkheeva NS, Beard WA, Wilson SH. Base excision repair of tandem modifications in a methylated CpG dinucleotide. The Journal of Biological Chemistry. 2014;289(20):13996–14008. doi: 10.1074/jbc.M114.557769. http://dx.doi.org/10.1074/jbc.M114.557769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoft JD, Kosowicz JG, Stivers JT. DNA translocation by human uracil DNA glycosylase: Role of DNA phosphate charge. Biochemistry. 2013;52(15):2526–2535. doi: 10.1021/bi301561d. http://dx.doi.org/10.1021/bi301561d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chemical Reviews. 2003;103(7):2729–2759. doi: 10.1021/cr010219b. http://dx.doi.org/10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- Tataurov AV, You Y, Owczarzy R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophysical Chemistry. 2008;133(1–3):66–70. doi: 10.1016/j.bpc.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. The Journal of Biological Chemistry. 1999;274(1):67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- Wiebauer K, Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6(4):544–559. doi: 10.1016/j.dnarep.2006.10.017. http://dx.doi.org/10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]