Abstract
Background
Reproductive factors, particularly parity, have differential effects on breast cancer risk according to estrogen receptor (ER) status, especially among African American (AA) women. One mechanism could be through DNA methylation, leading to altered expression levels of genes important in cell fate decisions.
Methods
Using the Illumina 450K BeadChip, we compared DNA methylation levels in paraffin-archived tumor samples from 383 AA and 350 European American (EA) women in the Women’s Circle of Health Study (WCHS). We combined 450K profiles with RNA-seq data and prioritized genes based on differential methylation by race, correlation between methylation and gene expression, and biological function. We measured tumor protein expression and assessed its relationship to DNA methylation. We evaluated associations between reproductive characteristics and DNA methylation using linear regression.
Results
410 loci were differentially methylated by race, with the majority unique to ER− tumors. FOXA1 was hypermethylated in tumors from AA versus EA women with ER− cancer, and increased DNA methylation correlated with reduced RNA and protein expression. Importantly, parity was positively associated with FOXA1 methylation among AA women with ER− tumors (P = 0.022), as was number of births (P = 0.026), particularly among those who did not breastfeed (P = 0.008). These same relationships were not observed among EA women, although statistical power was more limited.
Conclusions
Methylation and expression of FOXA1 is likely impacted by parity and breastfeeding. Because FOXA1 regulates a luminal gene expression signature in progenitor cells and represses the basal phenotype, this could be a mechanism that links these reproductive exposures with ER− breast cancer.
Keywords: Breast cancer, African American, DNA methylation, FOXA1, ER−, breast cancer, Disparities
Introduction
Breast cancer rates in African American (AA) women continue to rise, and the gap in mortality between racial/ethnic groups is widening [1]. Part of the higher mortality for AA women with breast cancer may be due to the greater prevalence of tumors that are negative for receptors for estrogen (ER−), progesterone (PR−) and human epidermal factor 2 (HER2−), thus lacking therapeutic targets and resulting in poorer prognosis [2]. The reasons for this disparity have been, for the most part, unknown. In the African American Breast Cancer Epidemiology and Risk (AMBER) Consortium, with close to 4000 AA cases and more than 14,000 AA controls, we recently showed that having children, although inversely associated with risk of ER+ breast cancer, was positively associated with risk of ER− breast cancer, with risk greatest among women who did not breastfeed [3]. AA women are more likely than EAs to have children and to not breastfeed [4, 5], factors that could be related to their higher likelihood of ER− breast cancer.
These differential risk relationships indicate that there may be distinct etiologic pathways for ER− and ER+ breast cancer, and that these diverging pathways, perhaps influenced by epigenetics, are affected by events occurring at an early age. DNA methylation, a mitotically heritable epigenetic modification that occurs in temporally and spatially specific patterns throughout development, is affected by the milieu of hormonal changes that occur during breast development, pregnancy, and lactation in the human and mouse mammary gland [6–8]. DNA methylation changes could serve as markers to identify genes differentially regulated by reproductive factors which may contribute to the increased prevalence of ER− breast tumors in AA women.
One possible mechanism that could increase the frequency of ER− over ER+ breast cancer is the impairment of breast luminal cell differentiation. Reproductive events could influence divergence of luminal progenitor cells to ER+ and ER− populations through aberrant DNA methylation, altering expression of genes crucial to progenitor cell differentiation. Significantly, one recent study identified parity-associated hypermethylation of FOXA1 in normal breast tissue, suggesting that downregulation of this pioneer factor contributes to attenuation of ERα function, which may impact breast tumor development [9]. Thus, reproductive factors may affect breast cancer development through DNA methylation, and differential reproductive patterns between EA and AA women could influence the greater prevalence of ER− tumors in AAs through this mechanism. However, understanding the differences in tumor biology between EAs and AAs and by reproductive characteristics is still in early stages.
In a small pilot study using frozen tissue, we previously found that differences in DNA methylation patterns in breast tumor tissue between AAs and EAs were observed primarily in ER− breast cancer [10]. Here, in a large epidemiological study of breast cancer in EA and AA women, with risk factor information, we sought to identify differentially methylated loci (DML) by race in tumors stratified by ER status, with confirmation of the impact of methylation of top DML using RNA-seq and immunohistochemistry (IHC). Following up on one of our top prioritized DMLs, FOXA1, we assessed associations of parity and breastfeeding with DNA methylation to investigate the hypothesis that these reproductive exposures, which are linked to altered risk of ER− cancer, act, in part, through epigenetic modification of key transcriptional regulatory genes in the breast.
Materials and methods
Patient samples
Breast tumor tissue samples were from participants in the Women’s Circle of Health Study (WCHS), a case–control study designed to investigate risk factors for aggressive breast cancer in AA and EA women [11]. Study participation rates have been described previously [11]. The protocol was approved by all relevant Institutional Review Boards. In-home interviews were conducted to obtain data on known and suspected risk factors for breast cancer and, as part of the informed consent, participants signed a release for pathology reports and specimens, which were obtained from participating hospitals. Of participants enrolled up until we identified cases for inclusion in this study, more than 95% signed a release for their tumor blocks. Not all hospitals provided tissue blocks when requested; this accounted for the greatest proportion of patients not included in the methylation analyses. Other reasons included insufficient tissue for DNA extraction, low yield during DNA extraction, and samples removed during QC of DNA methylation data.
Formalin-fixed paraffin embedded (FFPE) samples (punches, curls, or sections) [12] were deparaffinized, lysed, incubated until completely digested, and heated to inactivate the Proteinase K. Following bisulfite treatment, FFPE lysates were restored using the Infinium HD DNA Restoration Kit (Illumina). DNA was purified using the DNA Clean & Concentrator-5 kit (Zymo Research) for quantification by Quant-iT Picogreen (Invitrogen) and block randomized across plates with respect to self-reported race, sample type (slide, punch, or curl), patient age, and ER status using the OSAT program [13]. H&E sections of FFPE tumor specimens were viewed by the pathologist (T.K.) for selection of cores for construction of TMAs, with a minimum of three 0.6 mm cores taken and placed into TMA blocks for analysis. Completed TMAs were stored at room temperature under nitrogen to preserve antigenicity.
DNA methylation profiling and analysis
DNA methylation was surveyed at >485,000 CpG dinucleotides across each breast tumor genome using the Illumina Infinium HumanMethylation450 BeadChip (450K array), which has been extensively validated, providing reliable coverage of CpG dinucleotides across 99% of RefSeq genes and 96% of CpG islands in the human genome [14, 15]. Hybridized and processed arrays were scanned using Illumina BeadArray Reader with High-Density (HD) Technology and BeadScan software. The raw intensity was then extracted using GenomeStudio, and the data are summarized into BeadStudio IDAT files and processed by the minfi R package. The methylation level of each CpG site, expressed as a β-value, ranged from 0 (unmethylated) to 1 (methylated). The 450K array data were subjected to rigorous sample and locus specific quality control criteria, SWAN normalization, and correction for batch effects using the ComBat algorithm [16, 17]. In subsequent analysis, we removed probes and samples with poor detection P values using the IMA package [18]. We also removed probes known to map ambiguously, and those that contain SNPs, to reduce the likelihood that any observed methylation differences were due to genetic polymorphisms [19–21]. DNA methylation levels at 276,108 CpG loci in 733 tumor samples passed QC and were utilized in downstream analyses. Differential methylation analysis by race, stratified by ER status, at single CpG loci was performed using IMA [18], using the Wilcoxon rank-sum test and Benjamini and Hochberg multiple testing correction [22]. Differentially methylated loci (DML) were defined as CpG loci with a mean β-value difference (IΔβI) of at least 0.10 and FDR-adjusted P value <0.05. Sequenom EpiTYPER was used to confirm the methylation analysis of eight genes (assay primers in Table S1) in nine patient samples. Pearson’s correlations between 450K DNA methylation probes and DNA methylation of CpG site as determined by Sequenom were calculated.
RNA sequencing
To examine relationships between RNA-seq gene expression and DNA methylation, we used an independent collection of 50 fresh frozen breast tumor samples (Table S2) from Roswell Park Cancer Institute Pathology Network Shared Resource (PNSR). DNA samples from these cases were analyzed previously by 450K methylation profiling [10]. RNA from these same cases was extracted, and libraries were run on HiSeq 2500, generating 100-bp single-stranded paired-end reads. Reads were aligned to the human reference genome (build GRCh37/hg19) using TopHat software, allowing a maximum of 1 hit per read. Alignment quality control was done with RseQC software. Reads per kilobase million (RPKM) were calculated for each gene. For each identified raDML (450K probe) associated with an annotated gene in the Illumina 450K manifest, the Spearman correlation was computed between methylation levels (beta values) and mRNA expression (log counts per million, logCPM) of the nearest annotated gene. Differential expression between groups based on race and ER status was assessed using DESeq2.
FOXA1 IHC
TMAs containing breast tumor samples from 190 of the 383 AA cases, and 156 of the 350 EA cases, were stained using the monoclonal primary antibody HNF-3α (FOXA1) from Santa Cruz Biotechnology (Catalog No. sc-101058). Stained slides were digitally imaged at ×20 magnification using the Aperio ScanScope XT (Aperio Technologies, Vista, CA); automated image analysis of IHC staining was performed using a Genie classifier. Tumor cores on TMAs were collapsed into case-level data using a cellularity-weighted approach, as previously described [23]. With the weighted average of percent positivity values, an H-score was calculated, which reflects the extent of nuclear immunoreactivity, ranging from 0 to 300 [24]. The Spearman correlation between DNA methylation at the indicated 450K probe and FOXA1 protein expression (H-score) was computed.
Linear regression analysis
Associations between reproductive risk factors (parity and breastfeeding) and DNA methylation were examined using linear regression. Parity was modeled dichotomously (yes/no), or as a grouped linear term among parous women (1, 2, or ≥3 births). Breastfeeding was modeled as a binary exposure (yes/no). Regression models used log-transformed beta values and adjusted for age at diagnosis. Statistical interactions were assessed, where indicated, by comparing nested regression models (with or without an interaction term) via the likelihood ratio test. Stata SE version 14.2 (College Station, TX) was used for these analyses.
Results
Demographic and reproductive characteristics of included study participants are shown in Table 1. Information on WCHS patients not included in the methylation analysis is shown in Table S3. No significant age differences were observed between included cases versus those not analyzed. AA individuals accounted for a slightly lower percentage of included (52%) versus not included (61%) cases, as tumor blocks were less readily available. Reproductive characteristics were generally similar for included versus non-included study participants.
Table 1.
Demographic and reproductive characteristics of study participants by ER status (±) and race (AA or EA)
| ER+ (n = 509) | ER− (n = 224) | |||
|---|---|---|---|---|
|
|
|
|||
| AA (n = 242, 48%) | EA (n = 267, 52%) | AA (n = 141, 63%) | EA (n = 83, 37%) | |
| Age at diagnosis | ||||
| <50 | 94 (39) | 110 (41) | 62 (45) | 41 (49) |
| ≥50 | 146 (61) | 156 (59) | 77 (55) | 42 (51) |
| Parity | ||||
| Parous | 200 (83) | 186 (70) | 117 (84) | 52 (63) |
| Nulliparous | 40 (17) | 80 (30) | 22 (16) | 31 (37) |
| Number of birthsa | ||||
| 1 | 51 (25) | 42 (23) | 33 (28) | 15 (29) |
| 2 | 75 (38) | 89 (48) | 39 (33) | 21 (40) |
| 3+ | 74 (37) | 55 (29) | 45 (39) | 61 (31) |
| Breastfeedinga | ||||
| Ever | 93 (47) | 110 (59) | 49 (42) | 28 (54) |
| Never | 107 (53) | 76 (41) | 68 (58) | 24 (46) |
Numbers do not add to total subjects due to missing data
Among parous women only
Differentially methylated loci by race (raDMLs) and ER status
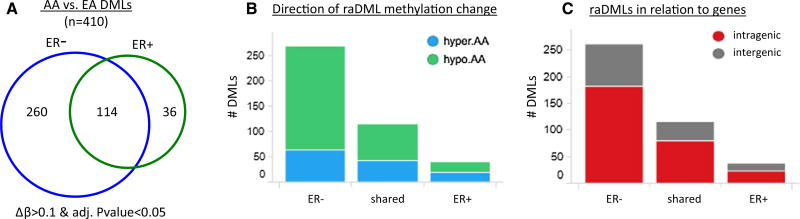
As shown in Fig. 1, we identified 410 raDMLs between breast tumors from AA and EA women (DML, |Δβ| ≥ 0.1 and FDR < 0.05), with the majority (n = 260) specific to ER− tumors, 36 specific to ER+ disease, and 114 raDMLs observed in both ER− and ER+ tumors (Fig. 1a). Most of the raDMLs specific to ER− tumors, and those independent of ER status, were hypomethylated in AA women, while there were roughly equal numbers of hypomethylated and hypermethylated raDMLs for those observed in ER + tumors (Fig. 1b). The majority of raDMLs for ER− tumors were contained within 167 unique genes, while gene-assigned raDMLs for ER+ or ER− independent tumors fell within 23 or 73 genes, respectively (Fig. 1c).
Fig. 1.
ER status-specificity, genomic distribution, and methylation of raDMLs. a Distribution of the 410 raDMLs (AA vs. EA) by tumor ER status. b Relative methylation levels of raDMLs in tumors from AA women compared to tumors from EA women, grouped by ER status. c The number of raDMLs mapping within known genes (red) and the number of raDMLs not located within or near an annotated gene (gray), segregated by tumor ER status
Confirmation of methylation at raDMLs
Ten raDMLs, chosen based on their large range of β-values across samples, were examined in 9 tumor samples from RPCI PNSR using Sequenom EpiTYPER assays as a means of technically validating 450K-derived methylation measurements; for all 10 CpG loci tested, there was high correlation between the two independent assays (average R2 = 0.886, Fig. S1).
Relationships between DNA methylation and mRNA expression
To evaluate the potential functional significance of raDMLs described above, we assessed the association between methylation of each probe and mRNA expression of its nearest annotated gene, where applicable (Table 2). RNA-seq analysis of an independent set of 50 fresh frozen tumors, for which 450K data were also available, provided expression levels for 245 of the 263 genes containing the 410 raDMLs. Of these, 78 (31.8%) had a Spearman’s test P value of ≤0.05, with 72/78 (92%) having a correlation coefficient |rho| ≥ 0.3. The large majority of these 72 genes (58/72, 80%) exhibited an inverse correlation between expression and methylation, while 14/72 (20%) showed a positive association. Two genes exhibited very strong correlations (|rho| > 0.80, P < 10−10) between expression and methylation at the corresponding DML: Forkhead Box A1 (FOXA1) and Thrombospondin Domain Containing 4 (THSD4). Because FOXA1 has a well-established important biological role in mammary cell fate specification, promoting luminal differentiation and repressing the basal phenotype [25–27], we focused our further analyses on this locus.
Table 2.
raDMLs ranked by P value for the correlation between DNA methylation and mRNA expression
| 450 K Probe | Gene | Region | Rho | P value | DML group | Direction | |
|---|---|---|---|---|---|---|---|
| 1 | cg04932551 | FOXA1 | Body | −0.82 | <2.2 × 10−16 | ER-neg | hyper.AA |
| 2 | cg05739476 | THSD4 | 3′UTR | 0.85 | <2.2 × 10−16 | ER-neg | hypo.AA |
| 3 | cg22301128 | ART3 | Body | 0.74 | 2.8 × 10−9 | ER-neg | hyper.AA |
| 4 | cg01673307 | TAP1 | Body | −0.72 | 1.5 × 10−8 | ER-neg | hypo.AA |
| 5 | cg14864167 | PDE7A | Body | −0.72 | 2.4 × 10−8 | ER-neg | hypo.AA |
| 6 | cg10296238 | SPATC1L | Promoter | −0.69 | 1.0 × 10−7 | ER-ind | hyper.AA |
| 7 | cg05342835 | SYNC | Body | −0.64 | 1.3 × 10−6 | ER-neg | hyper.AA |
| 8 | cg14014720 | DAPK1 | Body | −0.61 | 3.7 × 10−6 | ER-neg | hypo.AA |
| 9 | cg08742575 | SPATC1L | Promoter | −0.61 | 4.2 × 10−6 | ER-ind | hyper.AA |
| 10 | cg12123019 | IL12RB1 | Promoter | −0.61 | 4.5 × 10−6 | ER-neg | hypo.AA |
Spearman correlation analysis was performed using beta values (450K) for the indicated probe and logCPM values (RNA-seq) for the indicated gene. The top ten raDMLs are shown based on smallest Spearman P value. DML group classifies the raDML as specific to ER− tumors (ER-neg), ER+ tumors (ER-pos), or independent of ER status (ER-ind). Direction indicates whether the DML was hypermethylated (hyper.AA) or hypomethylated (hypo.AA) in tumors from AA versus EA women
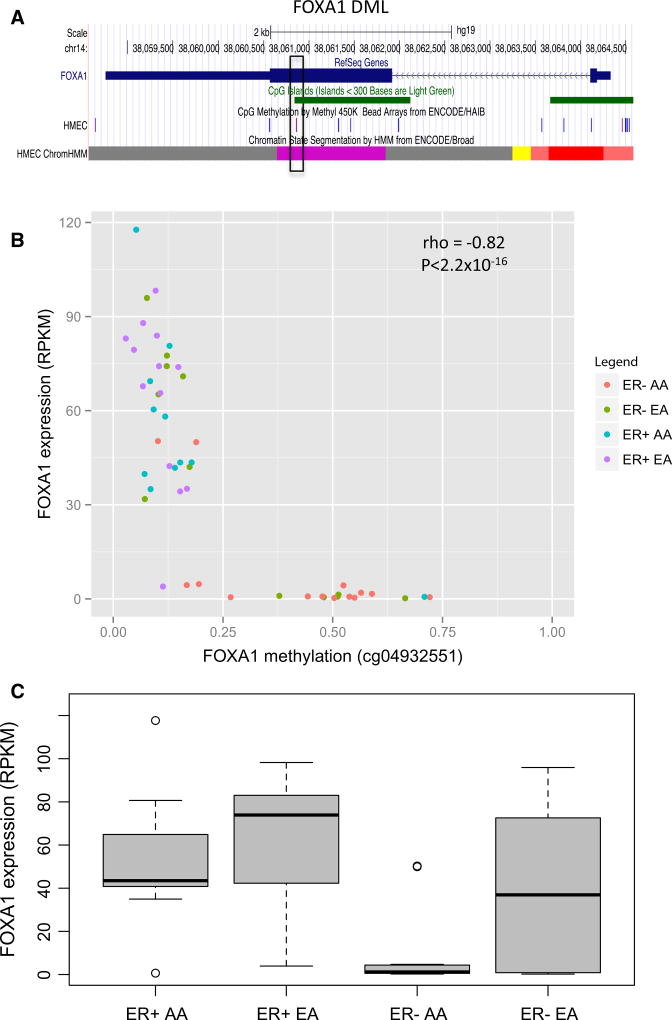
The FOXA1 raDML (cg04932551) maps to a CpG island located in the gene body which is annotated as a poised promoter in normal human mammary epithelial cells (HMEC), inferred from chromatin structure using a multivariate Hidden Markov Model (chromHMM) [28, 29] (purple block in chromHMM track, Fig. 2a). Methylation at this raDML was inversely associated with expression of FOXA1 (rho = −0.82, Table 2). In our primary 450K methylation analysis of 224 ER− tumors (Table 1), FOXA1 (cg04932551) was hypermethylated in tumors from AA versus EA women (Δβ = 0.13, adjusted P = 0.00151). Similarly, among the 26 fresh frozen ER− tumors with RNA-seq data available, mean methylation levels at cg04932551 trended higher in AA versus EA women (Fig. 2b; Δβ = 0.14, P = 0.057). This differential methylation by race tracked with lower expression of FOXA1 in AA versus EA women with ER− cancer (Fig. 2c; log2(Fold change) = −1.6, P = 0.003). Among women with ER+ cancer, FOXA1 expression differences by race were not statistically significant (P = 0.78).
Fig. 2.
Correlation of FOXA1 FOXA1 DML DNA methylation and mRNA expression. a Genome browser snapshot of FOXA1 RefSeq gene annotations, CpG islands (green), and HMEC 450 K probes. The HMEC chromHMM track indicates putative active (bright red), weak (light red), and poised (purple) promoters, as well as putative weak enhancers (yellow). b Scatter plot of FOXA1 DNA methylation (cg04932551) and mRNA expression. Red AA women with ER− tumors (n = 14); green EA women with ER− tumors (n = 12); blue AA women with ER+ tumors (n = 11); purple EA women with ER+ tumors (n = 13). c Boxplot of expression (RPKM) values for FOXA1 within each ER subtype by race
Relationship between FOXA1 DNA methylation and protein expression
To further establish the potential functional importance of DNA methylation in relation to FOXA1 expression, beyond our RNA-seq analysis, we performed IHC assays and digital pathology analysis for FOXA1 protein using TMAs containing breast tumor samples from a subset of the 141 AA women with ER− cancer. Consistent with our results comparing methylation and gene expression determined by RNA-seq, we observed a strong negative correlation between methylation levels at cg04932551 and FOXA1 H-score (rho = −0.64, P = 7.6 × 10−10) (Fig. 3). Representative imaging results are shown in Fig. 4, illustrating the inverse relationship between FOXA1 DNA methylation and FOXA1 protein expression. We obtained similar results in ER− tumors from EA women (Fig. S2C); weaker inverse correlations between methylation and expression were found in ER+ tumors, where FOXA1 DNA methylation levels were significantly skewed towards the lower end of the 0–100% range (Figs. S2B, D).
Fig. 3.
Correlation of FOXA1 DNA methylation and protein expression. Scatter plot of FOXA1 protein expression in ER− tumors from AA women (n = 74), measured by immunohistochemistry (IHC), versus FOXA1 DNA methylation at cg04932551. The H-score is an aggregate weighted measure of weak, moderate, and strong staining, as described in the Methods
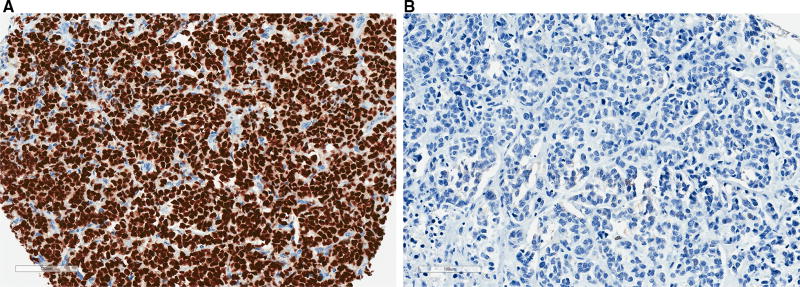
Fig. 4.
Representative images of FOXA1 IHC in TMA cores derived from ER− breast cancers in AA women. a High FOXA1 protein expression (H-score = 288.6) in a tumor with low FOXA1 DNA methylation (beta = 0.08). b Low FOXA1 protein expression (H-score = 0.2) in a tumor with high FOXA1 DNA methylation (beta = 0.78)
FOXA1 methylation in relation to reproductive risk factors
To investigate the basis for increased FOXA1 DNA methylation and reduced FOXA1 mRNA/protein expression in AA versus EA ER− tumors, we examined the relationship between reproductive history and FOXA1 DNA methylation in both AA and EA women with ER− cancer. Among AA women with ER− tumors, parous women had higher mean methylation levels at FOXA1 cg04932551 relative to nulliparous women (P = 0.022) (Table 3(A)). By contrast, no significant relationship with parity was observed among EA women with ER− tumors (P = 0.403); an interaction was observed between parity and race in relation to FOXA1 DNA methylation (P = 0.03). Among parous AA women with ER− tumors, we also found evidence of a statistically significant positive linear trend between number of births and methylation levels at cg04932551 (coefficient = 0.082, P = 0.026); a trend of smaller magnitude was suggested among parous EA women but was not statistically significant (coefficient = 0.039, P = 0.481) (Table 3(B)). Of particular interest, the positive association between number of births and FOXA1 methylation among parous AA women with ER− tumors remained significant, and appeared stronger (coefficient = 0.115, P = 0.008), in those who did not breastfeed; in those who did breastfeed, a trend of smaller magnitude was suggested but non-significant (coefficient = 0.058, P = 0.412) (Table 3(C)). Similar results were also observed for three additional CpGs located within the CpG island containing cg04932551 (Table S4). In contrast to our findings for AA women with ER− tumors, we did not find evidence for associations between reproductive factors and FOXA1 DNA methylation levels in ER+ tumors from AA women (Table S5).
Table 3.
FOXA1 DNA methylation in relation to reproductive factors among women with ER− cancer
| N | Coefficient | 95% CI | P | |
|---|---|---|---|---|
| (A) Parous vs. Nulliparous | ||||
| AA | 139 | 0.172 | (0.025, 0.319) | 0.022 |
| EA | 83 | −0.062 | (−0.208, 0.085) | 0.403 |
| (B) Number of Births (1, 2, 3+) | ||||
| AA | 117 | 0.082 | (0.010, 0.155) | 0.026 |
| EA | 52 | 0.039 | (−0.072, 0.151) | 0.481 |
| (C) Number of Births (1, 2, 3+) stratified by history of breastfeeding (BF) | ||||
| AA, no BF | 68 | 0.115 | (0.031, 0.199) | 0.008 |
| AA, yes BF | 49 | 0.058 | (−0.083, 0.198) | 0.412 |
| EA, no BF | 24 | 0.031 | (−0.120, 0.181) | 0.679 |
| EA, yes BF | 28 | 0.052 | (−0.130, 0.234) | 0.563 |
Linear regression was performed to evaluate associations between DNA methylation levels at cg04932551 (log-transformed beta values) and (A) parity (yes/no); (B) number of births among parous women (1, 2, or 3+); and (C) birth count (1, 2, or 3+) stratified by history of breastfeeding (yes/no). AA and EA women were assessed separately. Regression models were adjusted for age at diagnosis. Regression coefficients are listed for the indicated comparisons in A, B, and C
Discussion
This study sought to examine molecular pathological factors that could account for the more frequent occurrence of ER− breast cancer in AA women, particularly in relation to parity and not breastfeeding, known to increase risk of ER− disease. Genome-wide methylation analysis identified seven times as many DML by race in ER− versus ER+ tumors. While our previous pilot work [10] was conducted using a much smaller sample set and analyzed DNA from fresh frozen, as opposed to FFPE, tissue specimens, we found that a substantial number of the 410 raDMLs identified in the current study were also detected in this previous analysis (N = 197 of 410, 48%). Of the top raDMLs highly correlated with gene expression, FOXA1 was found to be hypermethylated in tumors from AA versus EA women with ER− cancer, and methylation levels showed strong inverse relationships with both mRNA and protein levels. Of particular interest, we observed a significant positive association between parity and FOXA1 methylation in tumors from AA women who did not breastfeed, which was attenuated and non-significant among those who did breastfeed.
In an analysis with more than 4000 AA women with breast cancer and 14,000 controls, we previously showed differential associations between reproductive factors and risk of ER+ or ER− breast cancer. Parity was associated with reduced risk of ER+ cancer, but increased risk of ER− disease particularly among women who did not breastfeed [3]. This may be especially relevant for AA women, who are more likely to be parous [4] and not to breastfeed [5]. Previous studies have shown that reproductive factors are associated with DNA methylation differences in breast tissue, both normal and tumor [6, 7, 9]. Our data now indicate that DNA methylation of FOXA1 positively correlates with parity in AA women with ER− tumors, especially among those who did not breastfeed, providing some insight into potential etiologic mechanisms. We did not find evidence of such associations among AA women with ER+ tumors, the majority of which exhibited low levels of FOXA1 DNA methylation. Weaker or absent associations between parity and FOXA1 methylation were observed among EA women with ER− tumors. Larger sample sizes in future studies will be required to validate and further characterize these relationships, particularly specificity based on race, given that past work has reported similar parity and breastfeeding associations with ER− cancer risk across multiple ethnic groups [30].
FOXA1 is a pioneer transcription factor and an essential regulator of breast development, specifically in luminal cells. As an established regulator of both ESR1 and its target genes, nearly half of FOXA1 binding sites co-localize with estrogen response elements across the genome (reviewed in Bernardo et al. [26]); binding of FOXA1 protein to heterochromatic DNA facilitates the binding of ER and other transcription factors [31]. Notably, single nucleotide polymorphisms (SNPs) found to be associated with breast cancer risk are enriched in FOXA1 binding sites and have been shown to modulate protein binding [32]. Most breast tumors, both luminal (ER+) and basal-like (ER−), are thought to arise from luminal progenitor cells (reviewed in Gross et al. [33]). FOXA1 has been shown to be a pivotal transcription factor in regulating the transition from progenitor cells, by inducing luminal cell-specific genes and repressing the basal cell phenotype [25]. Therefore, we hypothesize that parity-associated methylation and repression of FOXA1 blocks differentiation, thereby generating a pool of abnormal luminal progenitor cells with increased plasticity, and that it is these aberrant progenitors that can give rise to ER− breast cancer following transforming genetic and epigenetic alterations.
Taken together, these data suggest that early reproductive events contribute to the more frequent occurrence of aggressive ER− breast cancer among AA women, at least in part, through methylation and consequent reduced expression of FOXA1. The higher prevalence of reproductive exposures associated with ER− cancer among AA as compared to EA women may explain, in part, the increased rate of ER− disease, but these same risk factors (e.g., parity without breastfeeding) may also exert more pronounced biological effects in AA women. Consistent with this notion, we observed a significant linear trend of higher FOXA1 DNA methylation associated with multiple births among AA women, but only the suggestion of a weaker, non-significant trend among EA women (albeit with a smaller sample). Whether this suggestive difference between AA and EA women can be confirmed remains to be determined. Larger studies with epidemiological data, as well as experimental studies with mouse models, are needed to assess if, indeed, methylation of FOXA1 is influenced by reproductive factors and integrally involved in the development of aggressive ER− breast cancer, particularly in AA women.
Supplementary Material
Acknowledgments
The authors would like to thank William McCann, Gregory Ciupak, Warren Davis and Priya Nair for administrative and technical assistance. This work was supported by the Breast Cancer Research Foundation (C.B.A.) and grants from the National Institutes of Health/National Cancer Institute (R01 CA1332641, P01 CA151135, R01 CA100598), the US Army Medical Research and Material Command (DAMD-17-01-1-0334), and a gift from the Philip L. Hubbell family. The RPCI DataBank and BioRepository and the Genomics Core Facility are RPCI CCSG Shared Resources (NIH P30 CA016056-27)
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-017-4418-y) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest The authors declare no conflicts of interest.
References
- 1.DeSantis CE, et al. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 2.Palmer JR, Ambrosone CB, Olshan AF. A collaborative study of the etiology of breast cancer subtypes in African American women: the AMBER consortium. Cancer Causes Control. 2014;25:309–319. doi: 10.1007/s10552-013-0332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer JR, et al. Parity, lactation, and breast cancer subtypes in African American women: results from the AMBER Consortium. J Natl Cancer Inst. 2014;106:dju237. doi: 10.1093/jnci/dju237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin JA, Hamilton BE, Osterman MJ, Curtin SC, Matthews TJ. Births: final data for 2013. Natl Vital Stat Rep. 2015;64:1–65. [PubMed] [Google Scholar]
- 5.McDowell MM, Wang C-Y, Kennedy-Stephenson J. Breastfeeding in the United States: findings from the national health and nutrition examination surveys, 1999–2006. NCHS Data Brief. 2008;5:1–8. [PubMed] [Google Scholar]
- 6.Huh SJ, et al. Age- and pregnancy-associated DNA methylation changes in mammary epithelial cells. Stem Cell Rep. 2015;4:297–311. doi: 10.1016/j.stemcr.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izadi P, Noruzinia M, Fereidooni F, Mostakhdemine Hosseini Z, Kamali F. Epigenetic marks in estrogen receptor alpha CpG island correlate with some reproductive risk factors in breast cancer. Mol Biol Rep. 2014;41:7607–7612. doi: 10.1007/s11033-014-3650-3. [DOI] [PubMed] [Google Scholar]
- 8.Dos Santos CO, Dolzhenko E, Hodges E, Smith AD, Hannon GJ. An epigenetic memory of pregnancy in the mouse mammary gland. Cell Rep. 2015;11:1102–1109. doi: 10.1016/j.celrep.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, et al. Genome-wide DNA methylation profiling reveals parity-associated hypermethylation of FOXA1. Breast Cancer Res Treat. 2014;147:653–659. doi: 10.1007/s10549-014-3132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrosone CB, et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget. 2014;5:237–248. doi: 10.18632/oncotarget.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambrosone CB, et al. Conducting molecular epidemiological research in the age of HIPAA: a multi-institutional case-control study of breast cancer in African-American and European-American women. J Oncol. 2009;2009:871250. doi: 10.1155/2009/871250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinal AC, et al. A methodological study of genome-wide DNA methylation analyses using matched archival formalin-fixed paraffin embedded and fresh frozen breast tumors. Oncotarget. 2017;8:14821–14829. doi: 10.18632/oncotarget.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan L, et al. OSAT: a tool for sample-to-batch allocations in genomics experiments. BMC Genomics. 2012;13:689. doi: 10.1186/1471-2164-13-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova M, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 16.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 17.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, et al. IMA: an R package for high-throughput analysis of Illumina’s 450K Infinium methylation data. Bioinformatics. 2012;28:729–730. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair JD, Price EM. Illuminating potential technical artifacts of DNA-methylation array probes. Am J Hum Genet. 2012;91:760–762. doi: 10.1016/j.ajhg.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Mu W, Zhang W. On the analysis of the illumina 450k array data: probes ambiguously mapped to the human genome. Front Genet. 2012;3:73. doi: 10.3389/fgene.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 23.Allott EH, et al. Performance of three-biomarker immunohistochemistry for intrinsic breast cancer subtyping in the AMBER consortium. Cancer Epidemiol Biomark Prev. 2016;25:470–478. doi: 10.1158/1055-9965.EPI-15-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirsch FR, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 25.Bernardo GM, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32:554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32:113–130. doi: 10.1042/BSR20110046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernardo GM, et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Work ME, et al. Reproductive risk factors and oestrogen/progesterone receptor-negative breast cancer in the Breast Cancer Family Registry. Br J Cancer. 2014;110:1367–1377. doi: 10.1038/bjc.2013.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeckhoute J, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19:372–380. doi: 10.1101/gr.084582.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowper-Sal lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gross K, Wronski A, Skibinski A, Phillips S, Kuperwasser C. Cell fate decisions during breast cancer development. J Dev Biol. 2016;4:4. doi: 10.3390/jdb4010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.