Abstract
Combined sewer overflows (CSOs) degrade water quality and end-of-pipe treatment is one potential solution for retrofitting this outdated infrastructure. The goal of this research was to evaluate peracetic acid (PAA) as a disinfectant for CSOs using viability based molecular methods for antibiotic resistance genes (ARGs), indicator organism marker gene BacHum, and 16S rRNA genes. Simulated CSO effluent was prepared using 23–40% wastewater, representing the higher end of the range of wastewater concentrations reported in CSO effluent. PAA residual following disinfection was greatest for samples with the lowest initial COD. Treatment of simulated CSO effluent (23% wastewater) with 100 mg∙min/L PAA (5 mg/L PAA, 20 min) was needed to reduce viable cell sul1, tet(G), and BacHum (1.0±0.63–3.2±0.25-log) while 25 to 50 mg•min/L PAA (5 mg/L PAA, 5–10 min) was needed to reduce viable cell loads (0.62±0.56–1.6±0.08-log) in 40% wastewater from a different municipal treatment plant. Increasing contact time after the initial decrease in viable cell gene copies did not significantly improve treatment. A much greater applied Ct of 1200 mg∙min/L PAA (20 mg/L PAA, 60 min) was required for significant log reduction of 16S rRNA genes (3.29±0.13-log). No significant losses of mexB were observed during the study. Data were fitted to a Chick-Watson model and resulting inactivation constants for sul1 and tet(G) > BacHum > 16S rRNA. Amplicon sequencing of the 16S rRNA gene indicated the initial viable and total microbial communities were distinct and that treatment with PAA resulted in marked increases of the relative abundance of select phyla, particularly Clostridia which increased by 1–1.5 orders of magnitude. Results confirm that membrane disruption is a mechanism for PAA disinfection and further treatment is needed to reduce total ARGs in CSO effluent.
Keywords: Disinfection, Antibiotic Resistance Genes (ARGs), Peracetic Acid (PAA), Chick-Watson
TOC image
Introduction
Combined sewer overflows (CSOs) degrade water quality and threaten human health by releasing viable fecal indicator bacteria,1 pathogen markers,2 antibiotic resistant bacteria (ARB),3 and antibiotic resistance genes (ARGs)4 into surface water bodies. Combined sewer systems are designed to collect both storm water runoff and wastewater (WW) and a portion of the untreated waste stream overflows into adjacent surface waters during heavy rainfall or snow melt when the flow exceeds plant capacity. The cost of upgrading combined sewer systems has been estimated at $40.8 billion for the US.5 Therefore, end-of-pipe treatment technologies for CSO may be attractive as lower cost alternatives to upgrading sewer infrastructure, with a variety of treatment trains under consideration.6, 7 Disinfection with peracetic acid (PAA) has been proposed for end-of-pipe treatment and is appealing because it has not been found to create toxic, mutagenic, or chlorinated by-products8 reducing the need for pre-treatment to remove the high load of organic matter present in CSO.1
During CSO events water quality varies across the hydrograph with WW accounting for 4–39% of the flow and runoff accounting for the remainder.1 Effective disinfection (3.4–5.6-log removal) of simulated CSO effluent (5% WW) was reported for E. coli with 5 mg/L of PAA treatment for 5 min.9 More researchers have reported disinfection of total coliforms as measured by cultivation based techniques in WW influent indicating treatment with 20 mg/L PAA 10 min may be optimal for reducing target organisms.10 Given the cost of PAA,8 PAA demand of organic matter in CSO effluent,11 and small footprint available for end-of-pipe treatment, understanding the impact of lower doses and shorter contact times for a range of CSO effluents is needed to provide economical recommendations for application.
CSOs are a source of ARGs4 and elevated levels of ARB were observed in CSO impacted surface waters during wet weather.3 In urban waters without significant agricultural impacts, WW is the dominant source of ARGs during baseflow conditions.12 The ARG concentrations observed in CSO effluent were 2.5–100 times lower than observed for WWTP effluent.4
However, untreated CSO effluent contributes viable ARB3 as opposed to the majority of the ARGs present in WWTP effluent that are extracellular DNA remaining after disinfection.13 ARGs in environmental matrices present a risk to human health.14 The effect of PAA on antibiotic resistant organisms and ARGs has been investigated for secondary WW but not CSO effluent. CSO effluent is unique in that it may receive no-pretreatment or treatment with different unit processes (e.g., hydrodynamic separation or rapid filtration) prior to disinfection as opposed to the settling and biological treatment that secondary WW effluent receives prior to disinfection. Two recent studies demonstrated that PAA is not effective at reducing the total concentrations of tet(A), blaTEM, qnrS, ermB, sul1, and sul2 genes encoding resistance to tetracycline, beta-lactams, quinolone, erythromycin and sulfonamides respectively, in secondary WW effluents.15, 16 Flow cytometry analysis with live-dead staining demonstrated damage to cell walls indicating that a portion of detected ARGs were extracellular or present in nonviable cells.15 Little data is available reporting disinfection kinetics for ARG carrying cells in WW,17 and none is available for CSO effluent. Understanding the partitioning of ARGs between viable and non-viable sources is critical for understanding the mechanisms and therefore the risk of ARG proliferation in the environment. That is, defining the partitioning of ARGs helps differentiate the risk from growth and horizontal gene transfer of ARGs in viable cells versus transformation (uptake of extracellular DNA) for extracellular ARGs and ARGs from non-viable cells. Viability based molecular techniques [i.e., propidium monoazide (PMA) qPCR also known as viability PCR (vPCR)] have been used for other disinfectants to demonstrate disinfection kinetics for indicator organism marker genes.18 Evaluating disinfection kinetics for PAA treatment of ARG carrying cells and shifts in microbial community following disinfection could further understanding of the risk associated with PAA-treated CSO effluent.
The goal of this study was to determine PAA disinfection kinetics for end-of-pipe treatment by monitoring a broader range of microbial contaminants (ARGs and indicator organism marker genes) and using simulated CSO effluent representing higher percentages of WW (and therefore, risk) than has been previously investigated. The ability of PAA disinfection to interfere with the membranes of ARG-carrying bacteria and human fecal indicator organisms of the Bacteroidales order and to destroy ARGs in CSO was investigated. Bacterial community analysis was performed to determine the impact of PAA on the “viable” microbial community structure. To differentiate between viable and total gene copies and better understand the mechanisms of PAA, viability-based qPCR targeting genes from cells only with sufficiently intact membranes was applied.19 Data were modeled using Chick-Watson kinetic model to determine inactivation coefficients to help inform end-of-pipe disinfection. Results can be used to better understand the risk posed by ARG in PAA disinfected CSO effluent.
Experimental
Disinfection experiment
Grab samples of municipal WWTP influent from two different utilities were collected during baseflow conditions October 26, 2015 (10:30 AM, WWTPa) and November 16, 2016 (10:30 AM, WWTPb) and stored on ice during transport then at 4°C until the start of the experiment. Total suspended solids (TSS) in influent measured according to Environmental Sciences Section (ESS) Method 340.220 from WWTPa (228 ±109 mg/L) and WWTPb (63 ±31 mg/L) were within normally reported ranges for WW influent.21 Both WWTPa and WWTPb collect WW in separate sanitary sewer systems primarily from households with no major hospital or industrial inputs. Simulated CSO effluent was prepared by diluting municipal WWTP influent with sterile deionized water (23 or 40% WWTP influent, v/v). Dilution with sterile DI water was chosen given that the microbial community structure was similar in WW influent diluted with sterile DI water and water collected outside a CSO outfall during wet weather.4 These dilutions were selected to represent the higher range for percentages of WW observed during CSO events,1, 9 but would not address microbial loads in the highly urban runoff of many CSO cities that makes up the remaining portion of the CSO effluent. PAA (32% wt in acetic acid, Sigma Aldrich) was diluted to working stock solutions immediately prior to the disinfection experiments and concentrations were confirmed by colorimetric methods, described below. PAA is commercially available in the form of a quaternary equilibrium mixture containing acetic acid, hydrogen peroxide, PAA and water.8
In the first experiment, 23% WWTPa samples were treated with PAA (0, 5, and 20 mg/L) during rapid stirring on a stir plate in triplicate. Subsamples (170 mL) were collected at 0, 5, 10, 20, and 60 min. In the second experiment, 40% WWTPb samples were treated with 5 mg/L PAA while stirred, as in first experiment in triplicate and subsamples (250 mL) were collected at 0, 5 and 10 min, based on results from the first experiment indicating that incubations longer than 20 min did not result in significant changes for three of five genes tested. Reactor volumes of 400 mL were used. Larger subsamples were collected in the second experiment to improve qPCR detection limits. The disinfection reaction in the subsamples was quenched with sodium thiosulfate (100 mg/L) and catalase (50 mg/L). For the second experiment, chemical oxygen demand (COD) was analyzed according to Hach Method 8000 with Hach COD vials (20–1500 mg/L range) and a DR2700 spectrophotometer (Hach, Loveland, CO). Aliquots (80mL) were analyzed for TSS. Conductivity and pH were measured with a calibrated multimeter (Orion Star A329, Thermo Scientific) after quenching the reactions. Samples were analyzed using PAA test paper for 0–160 ppm (MicroEssential, Brooklyn, NY, reported precision +/- 10%) to evaluate PAA concentration before and after quenching.
PAA decay
To estimate PAA decay in simulated CSO and the impact of COD, a third experiment was performed on grab samples of influent from WWTPa and b (collected during baseflow 7/27/17 and 7/14/2017, respectively), in triplicate. Samples from WWTPa (11.5 and 23% dilutions) were treated with PAA (5 or 20 mg/L for 0, 10, 20, and 60 min) and WWTPb (20 and 40% dilutions) were treated with PAA (5 mg/L for 0, 5, and 10 min). TSS, pH, and COD were measured prior to treatment, as described above. Samples were analyzed for PAA using a commercial kit (Peracetic Acid Vacu-Vials, CHEMetrics, Midland, VA) at each sampling point and after quenching. PAA concentration was determined by measuring absorbance at 515 nm and using the calibration provided by the manufacturer to calculate PAA in mg/L.
Viability analysis
Cells were recovered from the batch disinfection experiments by centrifugation at 4000×g for 15 min and reserving the bottom ~10 mL. Aliquots of the centrifuge concentrated cells (500 μL) were either treated with 50 μM propidium monoazide (PMA) or preserved for DNA extraction. PMA is a dye that inhibits PCR of DNA originating from non-viable cells and extracellular DNA by intercalation with double stranded nucleic acids. It has been reported that the photo-induced cross-linkage renders the DNA insoluble and results in its loss during DNA extraction.22 Samples treated with PMA allowed for quantification of genes from cells with intact membranes (“viable cells”) only. Samples without PMA treatment allowed for quantification of total genes from cells with intact or compromised membranes along with extracellular DNA (viable and nonviable). PMA treated samples were incubated at room temperature in the dark for five minutes and then exposed in a PMA-Lite™ LED Photolysis Device (Biotium, Hayward, CA) for 15 min to facilitate cross linking of the dye prior to storage. Samples were stored at −20°C until DNA extraction. PMA methods were adapted from Nocker et al.19, 23
Cultivation
Heterotrophs were cultivated from subsamples from the second experiment to confirm the effectiveness of PAA treatments on reducing the concentration of viable cells. Aliquots of quenched triplicate subsamples were serially diluted (10−2–10−6, v:v), plated on LB Agar and incubated at 37°C until CFUs were visible (18 hrs). LB is a rich media that is used for cultivating E. coli and other heterotrophs. Results were reported as CFU/mL.
Molecular Methods
DNA was extracted from centrifuge concentrated cells with and without PMA treatment (500 μL) using a Fast DNA Spin Kit for Soil (MP Biomedicals, Solon, OH). DNA was diluted 1:50–1:100 to reduce inhibition and qPCR was performed to quantify the ARGs sul1,24 tet(G),25 and mexB,26 fecal indicator marker gene BacHum for Gram negative Bacteroidales,27 and 16S rRNA gene copies.28 sul1 encodes for a dihydropteroate synthase of Gram negative bacteria for resistance to sulfonamides. tet(G) is an efflux protein in Gram negative bacteria for tetracycline resistance found on plasmid and chromosome. mexB is a subunit of an efflux pump conferring antibiotic resistance typically associated with Gram negative Pseudomonas aeruginosa,29 which has been expressed in Gram positive organisms as well.30 These ARGs were selected because they are commonly observed in wastewater and represent qPCR targets of different amplicon lengths. qPCR reaction mixtures for the analysis of sul1, tet(G), mexB and 16S rRNA gene copies consisted of 5 μL SsoFastSuperMix (BioRad, Hercules, CA), 0.4μM forward and reverse primers, 2.4 μL molecular biology grade water, and 1μL diluted DNA extract. The qPCR reaction mixture for BacHum consisted of 5 μL SsoFastProbes SuperMix (BioRad, Hercules, CA), 0.07 μM probe, 0.22 μM forward and reverse primers, 1 μL molecular biology grade water, and 1 μL diluted DNA extract. Thermocycler (BioRad CFX96 Touch, Hercules, CA) conditions are summarized in Table S1. Samples were analyzed in triplicate with a seven-point standard curve (102–108 gene copies) and a no-template control with each qPCR plate. Melt curve analysis was performed for all ARG and 16S rRNA gene analyses and the length of select PCR products was checked via agarose gel electrophoresis for all gene targets. The quantification limits for targeted genes were below 104 copies/mL. qPCR was also performed for ARG associated with Gram positive microbes: vanA for vancomycin resistance31 and mecA for methilicillin resistant Staphylococcus aureus.32 Amplification was not observed for these genes in the simulated CSO samples, despite amplification in positive controls.
Amplicon sequencing (300-bp, paired end) was performed at a commercial laboratory (MrDNA, Shallowater, TX) targeting the V3–4 regions of the 16S rRNA gene. Sequences were analyzed using the mothur33 MiSeq Standard Operating Procedure. Rarefaction curves are included in Supplemental Information. Subsampling to obtain an equal number of sequences per sample resulted in 21,200 sequences/sample using a custom boot-strap.
Data Analysis
All statistical analyses were performed in R with an α<0.05 representing significant differences. A Kruskal-Wallis test was performed to test for differences (1) in total and viable log normalized ARG concentrations for a given PAA treatment, (2) in the viable cell log normalized ARG removal between different PAA treatments, (3) PAA concentrations across time for a given WW, and (4) PAA residual for the same treatment of different WW sources and dilutions. When significant differences were observed a post-hoc pair-wise t-test was performed with a Bonferroni correction for multiple comparisons. A Kruskal-Wallis test was performed on COD, pH, CFU, and inactivation constants to test for differences between treatments with a post-hoc test as above. A student’s t-test (for parametric data) or Wilcoxon rank sum test (for nonparametric data) was performed to test for differences in log removal of a given gene with equivalent treatment (Ct=50 mg·min/L) for the two wastewaters tested.
Gene log removal was calculated as
where [Gene]t=i is the gene concentration (copies/mL) at the time of interest i and [Gene]t=0 is the initial gene concentration (copies/mL) for a given replicate. Ct values, expressed as mg·min/L, were calculated as [PAA applied (mg/L)]×treatment time. Inactivation constants were calculated for data from both experiments combined for a given gene [except Ct=1200 mg·min/L PAA for BacHum, sul1, and tet(G)] using linear regression in Excel:
where α is the inactivation constant (L/mg·min), C is the concentration of PAA applied (mg/L), n is the constant of dilution (taken to be 1 based on goodness of fit), and t is time (min).
To compare microbial community structure for select total and viable samples, cluster analysis was performed in Primer7 (PrimerE, UK) on the Bray-Curtis dissimilarity matrix generated from log-normalized abundance data. To test for significant differences a SIMPROF test was performed.
Results
Disinfection of ARG carrying cells
Treating simulated CSO effluent consisting of 23% WW with a Ct [nominal PAA dose (mg/L) × time (min)] of at least 100 mg·min/L PAA and 40% WW with 25 mg·min/L PAA resulted in significant decreases in sul1 concentrations from viable cells compared to the initial viable cell sul1 concentrations (all p<1.8×10−4, Fig. 1a, b). Viable cell sul1 concentrations were lower than total sul1 concentrations observed at the same time for all tested Ct (all p<1.2×10−6) except for the 50 mg·min/L PAA treatment of either 23 and 40% WW (both p=1). Increased removal of sul1 concentrations from viable cells were not observed after 20 min with 23% WW and 5 mg/L PAA, after 10 min with 23% WW and 20 mg/L PAA, or after 5 min with 40% WW and 5 mg/L PAA (p=1.0). Average log removals of viable cell sul1 ranged from 0.74 to 2.8 (Table 1). Greater log removal was observed for sul1 with Ct of 50 mg·min/L PAA for 40% wastewater from WWTPb than for 23% wastewater with the same Ct from WWTPa (p=0.036).
Fig. 1.
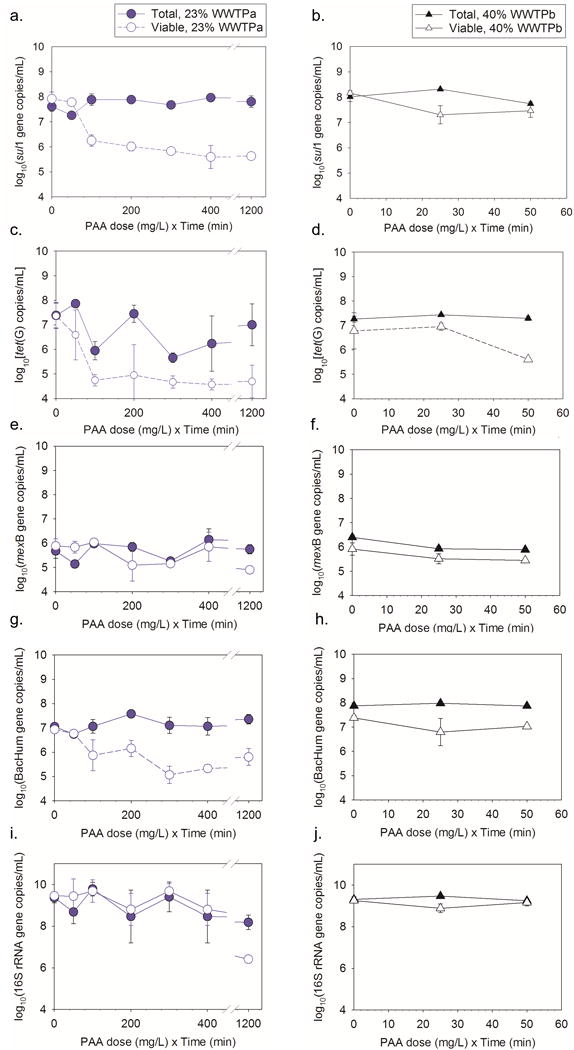
Comparison of total and viable cell gene copy numbers with different PAA treatment and exposure times for (a, b) sul1, (c,d) tet(G), (e,f) mexB, (g,h) BacHum, and (i,j) 16S rRNA genes. Experiments were performed with 23% wastewater from WWTPa or 40% wastewater from WWTPb to create the simulated CSO effluent. Experiments were performed for up to 60 min for WWTPa and 10 min for WWTPb. Error bars represent standard deviation of replicate (n=3, except as indicated in Table 1) samples.
Table 1.
Log removal of viable cell ARG, fecal indicator and 16S rRNA gene concentrations during treatment of simulated CSO with PAA. Results represent averages ± standard deviation (n=3) or ± maximum and minimum values (n=2). Bold results indicate a statistically significant decrease in genes originating from viable cells was observed from T=0 min (p<0.05).
| WW Fraction | [PAA] | Treatment Time | Ct | Log Removal | ||||
|---|---|---|---|---|---|---|---|---|
| (mg/L) | (min) | (mg∙min/L) | sul1 | tet(G) | mexB | BacHum | 16S rRNA | |
| 23% | 5 | 10 | 50* | 0.05±0.09 | 1.0±1.3 | −0.17±0.24 | 0.14±0.03 | 0.27±0.83 |
| 5 | 20 | 100* | 1.6±0.22 | 3.2±0.23 | −0.38±0.07 | 1.0±0.63 | 0.02±0.54 | |
| 5 | 60 | 300 | 2.0±0.12 | 3.2±0.25 | 0.50±0.06 | 1.8±0.36 | 0.03±0.36 | |
| 20 | 10 | 200 | 2.4±0.17 | 1.9 ±1.2 | 1.1 ±0.66 | 0.79±0.33 | 1.4±1.8 | |
| 20 | 20 | 400 | 2.8±0.46 | 2.3±0.22 | 0.30±0.60 | 1.5±0.11 | 0.91±0.77 | |
| 20 | 60 | 1200 | 2.8±0.03 | 2.2± 0.66 | 1.2± 0.01 | 1.1±0.35 | 3.29±0.13 | |
|
| ||||||||
| 40% | 5 | 5 | 25 | 0.90±0.36 | 0.32±0.15 | 0.49±0.20 | 0.62±0.56 | 0.34±0.21 |
| 5 | 10 | 50 | 0.74±0.26 | 1.6±0.08 | 0.55±0.05 | 0.38±0.09 | 0.06±0.17 | |
n=2
Significant losses of viable cell tet(G) compared to initial viable cell tet(G) concentrations were observed for 23% wastewater with select Ct (100 and 300 mg·min/L) (p<0.02, Fig. 1c, d). No significant losses in viable cell tet(G) were observed for Ct 200, 400, and 1200 mg·min/L (p>0.16). A larger standard deviation observed between replicates at 200 mg·min/L may explain the lack of significant differences observed for that treatment. Viable cell tet(G) concentrations were lower than the paired total tet(G) concentrations for 23% WW with 1200 mg·min/L PAA (p=0.04). Treatment of 23% WW resulted in 3.2 ± 0.30 log removal of viable tet(G) with 5 mg/L PAA (Table 1). For the 40% WW experiment after 10 min both the control and experimental samples (Ct =50 mg·min/L) had significantly less viable and total cell tet(G) than at the start of the experiment (all p<2.0×10−3). No difference was observed in log removal with Ct of 50 mg·min/L PAA for 40% wastewater from WWTPb than for 23% wastewater from WWTPa (p=0.7).
There were no significant differences between the total and viable cell sul1 or tet(G) concentrations at the beginning of either experiment (all p=1). Changes in total ARG concentrations were not observed with treatment compared to initial total ARG during either experiment (all p>0.05). No significant change in ARG concentrations from total or viable cells was observed in untreated (0 mg/L PAA) controls (all p>0.30) except for tet(G) in 40% WW with no treatment and incubation of 10 min (p=2.3×10−5). For mexB differences were not observed between total and viable cells at any time points (all p>0.43, Fig. 1e, f) and changes in total or viable concentrations were not observed over time (all p>0.05).
Disinfection of indicator organism markers and cultivation
Treating simulated CSO effluent consisting of 23% WW for 20 min with 5 mg/L PAA (Ct≥100 mg·min/L) and 20 mg/L PAA (Ct≥400 mg·min/L) resulted in significant decreases in BacHum concentrations from viable cells compared to the initial viable cell BacHum concentrations (all p<0.02, Fig. 1g, h). Increasing contact time with 23% WW beyond 20 min with either PAA concentration tested did not further reduce BacHum concentrations from viable cells (p>0.31). For 40% WW, 25 mg·min/L of PAA treatment decreased BacHum in viable cells compared to the initial viable cell concentrations (p<0.02), but concentrations in viable cells treated with 50 mg·min/L were not significantly different from initial viable concentrations (p=1.0). There was no significant difference in the total and viable cell BacHum concentrations at the beginning of either experiment (p>0.17). Changes in total BacHum concentrations were not observed with PAA treatment compared to initial total BacHum during either experiment (p=1). No significant change in the total or viable cell BacHum concentrations was observed in untreated (0 mg/L PAA) controls across time during the experiment (p=1.0). Log removals for viable cell BacHum ranged from 0.62 ± 0.56 to 1.8 ± 0.36 (Table 1). No difference was observed in log removal with Ct of 50 mg·min/L PAA for 40% wastewater from WWTPb compared the same treatment for 23% wastewater from WWTPa (p=0.2).
Cultivation was performed to confirm the loss of viability in cells with PAA disinfection demonstrated using vPCR for the 40% WW samples. Average starting concentrations in the 40% WW experiment were 1.0×105 CFU/mL (Fig. S1). CFU concentrations in treated samples (average concentration of 5.2×104 CFU/mL) were significantly lower than control plates (average concentration 1.6×107 CFU/mL) (p<0.03). Similar to qPCR results for ARG and BacHum, significant losses of viable cell concentrations were observed with PAA treatments of 25 and 50 mg·min/L, resulting in 0.95±0.47 and 0.45±0.36 log removal of CFU, respectively.
PAA decay experiments
During Experiment 2, PAA test strips indicated the PAA concentration remained consistent over the course of treatment and disinfectant concentrations were 0 mg/L after quenching. PAA decay experiments were performed to better characterize the initial, final, and post quenching PAA doses and the impact of COD on PAA residual. For WWTPa, the experiment was performed on 23% (75±18 mg COD/L) and 11.5% (39±13 mg COD/L) WW (summarized in Table S2). Nominal concentrations of 5 and 20 mg/L PAA were comparable to the measured initial concentrations of 5.2±0.6 and 20±1.9 mg/L PAA, respectively. No change in PAA concentrations were observed in the 11.5% WWTPa reactor with treatment through 20 min (all p>0.18). At 60 min of treatment of 23% WWTPa, PAA concentration decreased compared to initial measured concentrations (both p<0.04) with 59±28% of the 5mg/L PAA and 82±0.7% of the 20 mg/L PPA applied remaining as residual (Fig 2). For WWTPb, the experiment was performed on 40% (158±34mg COD/L) and 20% (117±69mg COD/L) WW. The PAA dose applied in this experiment was 5 mg/L PAA for all samples and the measured initial concentrations were 4.7±0.1 mg/L PAA. Decreases in disinfectant concentrations were observed after 5 min or more of treatment for 40% WW (p<0.01) and 10 min for 20% WW (p=0.019, Fig. 2). After quenching, 0–3% of the initial PAA dose remained across Experiment 3. Comparing between the two WW sources for Ct 50 mg·min/L PAA, the percentage of PAA remaining was least for 40% WW from WWTPb (all p<0.02), which had the highest COD. The percent of PAA remaining at Ct of 50 mg·min/L PAA was slightly less for 23% WWTPa than 20% WWTPb (which had comparable COD) and for 11.5% WWTPa than 20% WWTPb (both p<0.037).
Fig. 2.
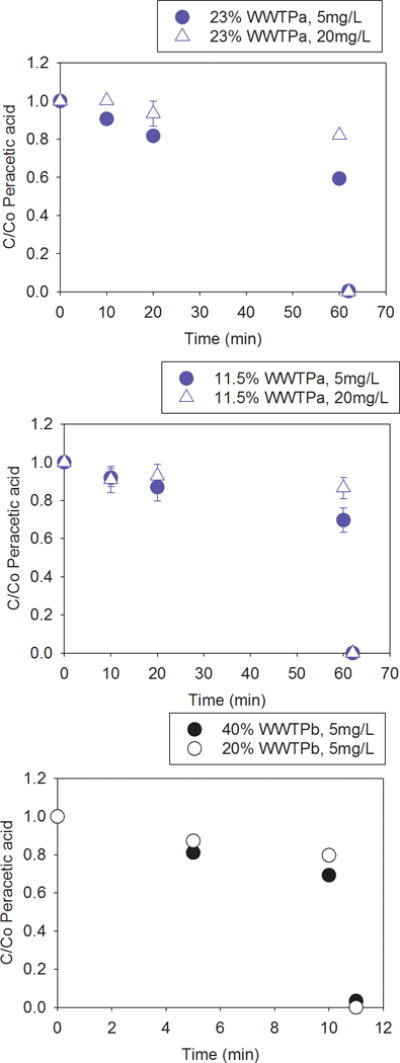
PAA concentration remaining (C) compared to initial PAA concentration (Co) for Experiment 3 during treatment of (a) 23% wastewater from WWTPa, (b) 11.5% wastewater from WWTPa, and (c) 20% or 40% wastewater from WWTPb and immediately after quenching. Treatment was performed with either 5 mg/L or 20 mg/L PAA. Error bars represent standard deviation of replicate (n=3) samples.
Water quality
Water quality was monitored throughout the 40% WW experiment (Experiment 2). Conductivity in samples treated with 25 mg·min/L PAA was greater than the no treatment controls (p=0.024) but not samples treated with 50 mg·min/L PAA (p=0.44, Fig. S2a). COD increased significantly after 5 to 10 min of treatment with 5 mg/L PAA (all p=8.5×10−6, Fig. S2b). COD in controls was unchanged across time (p=1.0). Differences were observed in pH for select samples across time and with treatment +/- 0.4 pH units (Fig. S2c). TSS measurements were similar in treated samples and controls (p=1.0, Fig. S2d).
Total bacterial community disinfection
16S rRNA gene copies were quantified as a surrogate for total bacterial population. Treating 23% WW with 1200 mg·min/L PAA resulted in a significant decrease in viable cell 16S rRNA genes (p=8.9×10−5, Fig. 1i, j). Viable cell 16S rRNA gene concentrations were lower than total cell 16S rRNA concentrations in 40% WW with 25 mg·min/L PAA (p=4.3×10−5) but comparable to the starting viable cell 16S rRNA concentrations (p=0.05). Treating the 40% WW with 50 mg·min/L PAA resulted in no losses of viable or total 16S rRNA (p=1). No differences were observed between log removal with Ct of 50 mg·min/L PAA for 40% wastewater from WWTPb and 23% wastewater from WWTPa (p=0.91).
Treatment of samples with the DNA intercalating dye PMA resulted in a significant shift in microbial community structure between total and viable samples even without PAA treatment (p=0.001, Fig. 3a). The total community was dominated by Proteobacteria throughout the experiment (Fig. 3b). The viable community with 0 mg·min/L PAA was also dominated by Proteobacteria but had lower relative abundances of Fusobacteria, Actinobacteria, Clostridia, and Bacilli. The viable cell community observed with 1200 mg·min/L PAA treatment was least similar to the other samples (81% similar, p=0.001). This community was dominated by Clostridia and marked by increases in Bacilli, Actinobacteria, and Erysipelotrichia and characterized by losses of Flavobacteria and Bacteriodia. No significant differences were observed in the microbial community structure for any treatment among the samples representing the total community. Likewise, no significant differences were observed in the viable community across time with no PAA treatment. Replicate samples that were sequenced (total, Ct=1200 mg·min/L PAA) were 92.7% similar.
Fig. 3.
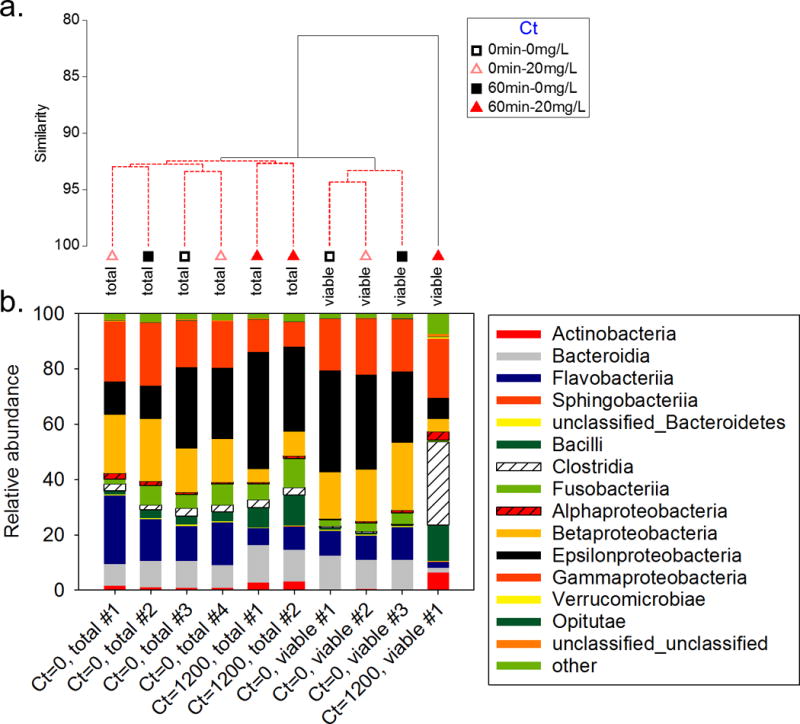
(a) Cluster analysis and (b) relative abundance of Bacterial phyla for different PAA treatments and exposure times (Ct values in mg·min/mL). Samples connected by red bars on the cluster tree do not have significantly different structures. #’s represent replicate samples that were sequenced.
Kinetics
Data were fitted to the Chick-Watson model and inactivation coefficients are listed in Table 2. R2 values for the Chick-Watson model fit ranged from moderate to substantial (0.45±0.17–0.76±0.21). Data were pooled across the experiments for the kinetic analysis given that reasonable model fits were observed with the combined data (note: comparable log removals were observed for the same Ct for four of the five genes tested with the WW from different sources and dilutions). Inactivation rates were greatest for sul1 and tet(G), which were not significantly different from one another (p=1). The inactivation coefficient for BacHum was significantly less than for these two ARGs (both p<0.001). The inactivation coefficient for the 16S rRNA gene was significantly less than all other genes tested (all p<0.024). Correlation was not observed between qPCR amplicon length and inactivation coefficient.
Table 2.
Average ± standard deviation for Chick-Watson disinfection coefficients (α) and R2 values of model fit combined for both 23% and 40% WW from different WWTP (n=3).
| BacHum | tet(G) | sul1 | 16S rRNA | |
|---|---|---|---|---|
| amplicon length (bp) | 89 | 134 | 163 | 202 |
| Chemistry | TaqMan | SybrGreen | SybrGreen | SybrGreen |
| α (L/mg min) | 1.1×10−2 ±1.7×10−3 | 1.9×10−2 ±1.4×10−3 | 1.8×10−2±1.3×10−3 | 6.1×10−3±1.2×10−3 |
| R2 | 0.76±0.21 | 0.45±0.17 | 0.67±0.19 | 0.67±0.14 |
Discussion
PAA for treating CSO
PAA treatment resulted in 0.62–3.3 log removal of the fraction of sul1, tet(G), and BacHum in viable cells in simulated CSO effluent with 25 mg·min/L PAA treatment or greater. The results of this study targeting viable cell genes can be compared to log removal of cultivable organisms in the literature given that PAA disinfection studies (and regulations) generally rely on cultivation or most probably number (MPN) evaluations. In a similar study with simulated CSO effluent, there was 3.4–5.6 log removal of E.coli compared to <2 log removal of Enterococcus in 5% WW with nominal PAA Ct of 25–300 mg∙min/L.9 The greater log removal observed with 5% WW may be due to there being less oxidizable organic matter than in the current study using 23–40% WW. Lower PAA doses were applied in a CSO treatment pilot study that achieved E.coli, fecal coliform, and Enterococcus log reductions of 1.7–2.3 with 2 mg/L PAA for 3 min (nominal Ct=6 mg∙min/L).7 However, for field studies where PAA was not reported to be quenched that have PAA, residual may result in potentially a longer incubation time than reported given the allowable hold time for these indicator organism techniques. Viable cell BacHum losses observed here (0.62–1.8-removal with 25–1200 mg∙min/L PAA) are lower than previously reported 3-log removal of cultivable total coliforms, fecal coliforms and fecal Streptococcus observed with a nominal Ct of 100 mg∙min/L PAA (PAA=10 mg/L) in disinfected wastewater effluent.34 Other reports of differences in chlorine disinfection efficiencies between vPCR and cultivation based techniques for the same organism have suggested viable but not cultivable organisms may play a role in this discrepancy.18
Having a low Ct is of interest when treating CSO effluent because (1) the footprint available for end-of-pipe treatment systems is often limited and (2) PAA was previously reported to be been more expensive than chlorination (estimated at five times the cost for treating wastewater in 2004),8 although more recent case studies (2012) suggest that the cost may be comparable.35, 36 Results of our study provide insight into a potential treatment for ARG carrying cells at CSO discharge locations. Further reductions in viable cell ARG concentrations were not observed for treatments longer than 10 min and tet(G) and 10 to 20 min for treatment of sul1. Thus, the rapid activity of PAA in reducing select viable ARG concentrations could minimize the treatment infrastructure requirements if PAA is supplied at sufficient dose (5–20 mg/L with residence time of 5 min).
The CSO effluent source may impact the PAA dose required for significant removal of viable cell ARG and BacHum given that lower doses were required for significant removals for the 40% compared to 23% WW. For example, while the starting sul1 concentration was higher in the 40% WW compared to 23% WW (p=0.02), significant removal of viable cell sul1 was observed with a lower Ct for 40% WW (25 mg∙min/L) compared to the 23% WW (100 mg∙min/L). Greater log removal of sul1 was observed with the same Ct (50 mg∙min/L) for the 40% WW compared to 23% WW, but notably not the other genes analyzed. Measuring PAA residual and COD could help explain these observations, as was performed for Experiment 3 demonstrating that generally higher COD WW had a higher PAA demand. However, these results are not directly comparable given the sampling was performed at a different time. Others have observed that PAA concentrations up to 6 mg/L for 60 min (nominal Ct=360 mg∙min/L) could not consistently meet target fecal coliform levels in disinfected WW effluent due to day-to-day variability in effluent quality.37 Gehr et al.’s observation of variability in PAA performance may be due in part to the differences in PAA available for treating cells that would result from the demand of other organic constituents being oxidized by PAA. In contrast, WW concentration had little effect on disinfection efficiency of Enterococcus in 5, 15 and 40% WW treated with up to 30 mg/L for 10 min.9 Considering the variability in effluent quality across a storm,4 testing a range of CSO effluents is likely a pertinent consideration for end-of-pipe treatment.
The impact of PAA treatment on water quality was determined by monitoring pH, conductivity, COD, and TSS. Of these parameters, pH was slightly lower in treated samples, unlikely of practical significance, and COD was higher in treated samples. The increase in COD with PAA treatment is expected to result from reactions with transition metals, suspended and dissolved solids, and organic species in the wastewater.38 Others have reported increases in biological oxygen demand (BOD) and COD after disinfection with oxidants thought to be due to the oxidant causing modifications to more recalcitrant organic matter resulting in it being more readily oxidable by the COD test.39
PAA Degradation
PAA decay experiments demonstrated that 59–87% of the initial PAA dose was present after 10–60 min of treatment. Extended exposure times of 60 min (nominal Ct=1200 mg min/L) were needed before significant decreases in PAA were observed for 11.5 and 23% WWTPa. However, significant PAA decay was observed after only 5 min (nominal Ct=25mg min/L) with 40% WW and only 10 min (nominal Ct=50mg min/L) with 20% WW from WWTPb. The generally greater PAA residual remaining with lower dilutions of WW and lower COD are consistent with greater PAA consumption by higher WW concentrations and COD observed by others.9, 37 PAA concentrations measured after quenching indicated that the concentrations of sodium thiosulfate and catalyze were sufficient to reduce the remaining amount of disinfectant. For 23% WWTPa and 5 or 20 mg/L PAA, compared to nominal doses of PAA multiplied by time of 50, 100, 200, 300, 400 and 1200, observed values were 51±9.0, 91±14, 189±7.7, 199±36, 351±15 and 927±32 mg∙min/L, respectively. For 40% WWTPb treated with 5 mg/L PAA the nominal dose of PAA multiplied by time of 25 and 50 mg∙min/L resulted in observed Ct of 19±0.49 and 33±0.47 mg∙min/L, respectively. These differences in nominal Ct and observed should be considered when comparing to other WWs.
Mode of action
PAA was effective at reducing the fraction of sul1 and tet(G) from viable cells present in simulated CSO effluent. This result is consistent with reports that PAA did not enhance overall ARG removal (i.e., reduce total ARG concentrations measured by qPCR) in post-secondary treated WW effluent after disinfection.15,16 The application of vPCR in this study incorporates the specificity of qPCR and overcomes the limitation of qPCR to differentiate between DNA originating from viable and nonviable sources. PAA’s mode of action is presumed to be oxidization of sulfhydryl and sulfur bonds in proteins, enzymes and other metabolites, including those in microbial membranes.8 Because treatment of cells with PMA reduces the qPCR signal from extracellular DNA and DNA from cells with compromised membranes, it can be reasoned that PAA was effective at disrupting the membranes of cells carrying sul1 and tet(G) genes resulting in significant decreases in viable cell ARG concentration. Thus, following treatment, the majority of the sul1, tet(G), and BacHum gene concentrations present in simulated CSO samples originated from nonviable cells. Live-dead staining with flow cytometry was previously used to demonstrate the membrane destroying mechanism of PAA for WW disinfection.15 At the beginning of treatment in the current study, there were no differences between total and viable ARG concentrations indicating ARGs were predominantly in viable cells, consistent with reports that bacterial cells with intact membranes comprised approximately 70% of the bacterial community in municipal wastewater influent.15 A previous study differentiated intra- and extracellular ARGs in environmental matrices using a washing technique, observing extracellular DNA generally made up only less than 0.5% of total DNA in animal sludge samples.40 In contrast, extracellular ARGs were found in higher concentration than intracellular ARGs in river sediments including those receiving treated wastewater.41
The viability based method applied here may overestimate the dose required for inactivation of ARG carrying cells given that it is possible for cells with intact membranes to not be viable (i.e., if DNA is sufficiently damaged). Although the use of PMA is a more conservative method to measure loss of viability because it relies only on the permeability of the membrane, linear correlations were observed between reduction in cultivability and of PMA-qPCR signals in samples treated with various disinfectants.42 While UV damage to cells can be repaired, there are not known reports of repairs of membrane damage from oxidants. Nonetheless, the method does allow for better estimation of the risk of extracellular DNA versus intracellular DNA, which would be exchanged between cells using different mechanisms, compared to qPCR.
PAA was not effective at reducing total ARG concentrations in treated simulated CSO effluent consistent with a recent investigation for its use for disinfection of WW.15 ARGs from non-viable sources are of interest given that the rate of ARG propagation may be expected to be different for ARGs in viable cells (which can spread via growth and horizontal gene transfer) compared to extracellular ARGs/ARGs in cells with compromised membranes (which can spread via transformation).14 The ARGs investigated here may be found on chromosomal DNA or plasmids.43–45 Transformation of extracellular ARGs (plasmids carrying genes for kanamycin-resistance) and expression was previously demonstrated with sediment cells in a controlled experiment.13 Given that the primer sets used in our study targeted PCR inserts (134–244 bp) that were not the full length of the ARG, it is possible that disinfection with PAA resulted in ARG damage to the given gene outside of the target amplicon and therefore not detectable with our protocols. PAA was reported to reduce the concentration of nucleic acids as measured by optical density in a study looking for the mechanisms of fungicidal effects.46 However, the observation that PAA disinfection did not reduce total ARGs in treated simulated CSO is consistent with other disinfection studies with oxidants.47 For example, Fahrenfeld et al.48 observed that chlorination did not reduce total sul1, tet(G), or tet(O) concentrations in secondary WW effluent (losses of sul2 were observed post-chlorination). In contrast, UV disinfection reduces total ARG concentrations at higher doses of UV than is required for loss of viability of antibiotic resistant bacteria.32 Therefore, treatment combining PAA to damage cell membranes inactivating microbes and providing better exposure for UV to disrupt DNA may be a strategy for mitigating the risk of ARG propagation in CSO impacted environments.
The Cts reported to result in significant losses of antibiotic resistant cultivable organisms in treated WW were comparable or higher than minimum found to reduce vPCR signals from ARG carrying cells in simulated CSO effluent in this study. PAA disinfection significantly decreased detection of cultivable ARG carrying uropathogenic E. coli isolates (detected via microarray following cultivation) in treated WW effluent at a dose of 0.9–2 mg/L and a contact time of 30 min to meet a goal of 200 CFU/100 mL.17 Exposure to PAA was associated with 1.6–3.7 log reductions in uropathogenic E. coli concentrations with 30, 50 and 60 mg∙min/L PAA (applied dose × time).17
Gene to gene comparisons
There was gene-to-gene variation in the required disinfectant dose for removal of viable cell gene copies. For example, 50 mg∙min/L PAA resulted in significant loss of viable cell tet(G) for 40% WW compared to 25 mg∙min/L PAA for significant loss of viable cell sul1. However, the inactivation coefficients for these two ARGs were comparable. In contrast, mexB concentrations did not change with PAA treatment, although at the highest PAA treatment viable cell concentrations were almost different compared to the initial viable cell concentration (p=0.05), thus behaving most similarly to the 16S rRNA gene. There were significant differences observed between the inactivation coefficients with sul1 and tet(G) > BacHum >16S rRNA (inactivation coefficients were not evaluated for mexB given that significant losses in viable cell concentrations were not observed across the experiment). Notably, 16S rRNA genes originating from viable cells exhibited a significant decrease only after 60 min of treatment at the highest Ct tested (1200 mg∙min/L PAA). qPCR product lengths and temperature have were previously reported to influence PMA-qPCR49 and could contribute to these differences. Short DNA fragments (<200 bp) were not found to have completely reduced signals for certain heat-killed strains.50 However, the 16S rRNA amplicon length was greater than those for sul1 and tet(G) where viable cell gene copy losses were observed with lower PAA Ct (and all genes quantified using SybrGreen chemistry). mexB had the longest targeted amplicon length and its concentrations were unchanged following PAA disinfection. Further, BacHum had the shortest amplicon (quantified with TaqMan probe chemistry) and behaved similarly to sul1 and tet(G). Correlation between inactivation coefficient and amplicon length were not observed. Therefore, factors other than amplicon length (e.g., temperature, target cell, reaction chemistry) are relevant for understanding PAA disinfection efficiency.
The impact of membrane structure (Gram positive versus Gram negative) is of interest for PAA disinfection. The ARGs sul1 and tet(G) measured in this study are associated with Gram negative organisms and the action of PAA on outer membrane lipoproteins may facilitate its effectiveness against Gram negative cells.8 Bacteriodales, the host order for BacHum genes, are also Gram negative organisms. The lower Ct values required for removal of viable sul1 and tet(G) and fecal indicator compared to universal 16S rRNA gene may be partially explained by the membrane composition of cells associated with these primer targets given that both Gram negative and Gram positive cells contain 16S rRNA. There is evidence that much longer PAA treatment times (240 and 360 min) were required for 3 and 4-log removal of cultivable Enterococcus, a Gram positive organism, compared with E.coli, a Gram negative organism requiring only 10 min treatment time for similar removals.9 This would suggest that large increases in treatment time regardless of PAA concentration may be required for specific targets, as observed here for 16S rRNA and mexB. Other researchers have reported that PMA may need to be optimized for different cell types and enhancing kits are available for targeting Gram positive bacteria. In fact, in the PMA and PAA treated sample for amplicon sequencing, marked increases in Clostridia, Actinobacteria, and Bacilli were observed. All three of these phyla are Gram positive, thus supporting that either or both PAA and PMA were less effective at reducing vPCR signals from Gram positive bacteria using our protocol. Given that these phyla had higher relative abundances in the total than the viable populations with Ct of 0 mg∙min/L PAA, differences in PAA’s ability to disrupt membranes between Gram positive and negative bacteria may contribute. The initial bacterial community was >90% Gram negative at the start of the experiment therefore differences in membrane structure are unlikely the only contributing factor. ARG associated with Gram positive microbes (vanA and mecA) were tested for but not observed in the samples. However, mexB, which has mostly been associated with both Gram negative bacteria, was not impacted by PAA, highlighting the potential for specific disinfection efficiencies across targets.
In the current study, differences in microbial community structures for samples treated with PMA were observed compared to untreated samples from the initial Ct of 0 mg∙min/L. Community shifts have been observed in response to PAA treatment of drinking water biofilms,51 UV disinfection of biologically treated wastewater52 and in the viable community of ozonated municipal wastewater sludge investigated by PMA-modified Miseq sequencing.53 Similar to this experiment, Proteobacteria was a dominant phylum in the ozonation study although there was a decrease in the relative abundance in the viable population.53
Conclusions
This work provides insight into the partitioning of ARGs between cells with intact membranes versus cells with compromised membranes or extracellular DNA from PAA treated CSO effluent. The viability method applied allowed for demonstration that PAA was effective at reducing viable cell sul1, tet(G), and fecal indicator with at least 25 or 100 mg∙min/L PAA and 16S rRNA genes with 1200 mg∙min/L PAA. The PAA disinfection efficiency for sul1 carrying cells varied by wastewater source with a higher nominal Ct required for more dilute simulated CSO effluent indicating that treatment of select targets may be a greater function of source water quality than dilution factor. Inactivation coefficients for sul1 and tet(G) were greater than for BacHum and inactivation coefficients for these ARG and BacHum were greater than for 16S rRNA. These inactivation coefficients may be used to estimate removal of viable concentrations of these gene targets in CSO effluent. Overall, results indicate that further steps are necessary to remove total ARGs in PAA treated CSO effluent (e.g., hydrodynamic separation,4 UV disinfection32).
Supplementary Material
Acknowledgments
Laboratory assistance was provided by Hannah Delos Reyes, Sophia Blanc, Reba Oduro and Raquele Strickland. Thanks to our utility partners for providing access to influent samples. Funding for this project was provided by grants from the New Jersey Water Resources Research Institute, the National Science Foundation (#1510461), NIH Bridges Grant: R25GM058389, and a Mark B. Bain Fellowship from the Hudson River Foundation to AE.
References
- 1.Passerat J, Ouattara NK, Mouchel JM, Rocher V, Servais P. Water Res. 2011;45:893–903. doi: 10.1016/j.watres.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 2.Donovan E, Unice K, Roberts JD, Harris M, Finley B. Appl Environ Microbiol. 2008;74:994–1003. doi: 10.1128/AEM.00601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young S, Juhl A, O’Mullan GD. J Water Health. 2013;11:297–310. doi: 10.2166/wh.2013.131. [DOI] [PubMed] [Google Scholar]
- 4.A. Eramo, H. Delos Reyes and N. Fahrenfeld, (in review).
- 5.U.S.E.P Agency. Journal. 2016 [Google Scholar]
- 6.Chhetri RK, Bonnerup A, Andersen HR. J Indust Eng Chem. 2016;37:372–379. [Google Scholar]
- 7.Patoczka J, Dening J, Rolak J. presented in part at the WEFTEC; New Orleans, LA. September 24–28, 2016. [Google Scholar]
- 8.Kitis M. Environ Int. 2004;30:47–55. doi: 10.1016/S0160-4120(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 9.Chhetri RK, Thornberg D, Berner J, Gramstad R, Ojstedt U, Sharma AK, Andersen HR. Sci Total Environ. 2014;490:1065–1072. doi: 10.1016/j.scitotenv.2014.05.079. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Ruiz C, Martinez-Royano S, Tejero-Monzon I. Water Sci Technol. 1995;32:8. [Google Scholar]
- 11.González A, Gehr R, Vaca M, López R. Water Environ Res. 2012;84:247–253. doi: 10.2175/106143012x13347678384765. [DOI] [PubMed] [Google Scholar]
- 12.Storteboom H, Arabi M, Davis JG, Crimi B, Pruden A. Environ Sci Technol. 2010;44:7397–7404. doi: 10.1021/es101657s. [DOI] [PubMed] [Google Scholar]
- 13.Mao D, Luo Y, Mathieu J, Wang Q, Feng L, Mu Q, Feng C, Alvarez PJ. Environ Sci Technol. 2014;48:71–78. doi: 10.1021/es404280v. [DOI] [PubMed] [Google Scholar]
- 14.Ashbolt NJ, Heberer T, van den Eede C, Schönfeld J, Krone S, Backhaus T, Li XY, Borriello P, Amezquita A, Larsson DGJ, Brandt K, Smalla K, Silley P, Lawrence J, Ryan J. Environ Health Perspect. 2013;121:993–1001. doi: 10.1289/ehp.1206316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Cesare A, Fontaneto D, Doppelbauer J, Corno G. Environ Sci Technol. 2016;50:10153–10161. doi: 10.1021/acs.est.6b02268. [DOI] [PubMed] [Google Scholar]
- 16.Luprano ML, De Sanctis M, Del Moro G, Di Iaconi C, Lopez A, Levantesi C. Sci Total Environ. 2016;571:809–818. doi: 10.1016/j.scitotenv.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Biswal BK, Khairallah R, Bibi K, Mazza A, Gehr R, Masson L, Frigon D. Appl Environ Microbiol. 2014;80:3656–3666. doi: 10.1128/AEM.00418-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varma M, Field R, Stinson M, Rukovets B, Wymer L, Haugland R. Water Res. 2009;43:4790–4801. doi: 10.1016/j.watres.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. Appl Environ Microbiol. 2007;73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wisconsin State Lab of Hygiene. Journal. 1993 [Google Scholar]
- 21.Karvelas M, Katsoyiannis A, Samara C. Chemosphere. 2003;53:1201–1210. doi: 10.1016/S0045-6535(03)00591-5. [DOI] [PubMed] [Google Scholar]
- 22.Nocker A, Cheung CY, Camper AK. J Microbiol Methods. 2006;67:310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Nocker A, Richter-Heitmann T, Montijn R, Schuren F, Kort R. Int Microbiol. 2010;13:59–65. doi: 10.2436/20.1501.01.111. [DOI] [PubMed] [Google Scholar]
- 24.Pei R, Kim SC, Carlson KH, Pruden A. Water Res. 2006;40:2427–2435. doi: 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Aminov RI, Chee-Sanford JC, Garrigues N, Teferedegne B, Krapac IJ, White BA, Mackie RI. Appl Environ Microbiol. 2002;68:1786–1793. doi: 10.1128/AEM.68.4.1786-1793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoneda K, Chikumi H, Murata T, Gotoh N, Yamamoto H, Fujiwara H, Nishino T, Shimizu E. FEMS Microbiol Lett. 2005;243:125–131. doi: 10.1016/j.femsle.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 27.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. Water Res. 2007;41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 28.Muyzer G, De Waal EC, Uitterlinden AG. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Deng Z, Yan A. Biochem Biophys Res Commun. 2014;453:254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 30.Welch A, Awah CU, Jing S, van Veen HW, Venter H. Biochem J. 2010;430:355–364. doi: 10.1042/BJ20091860. [DOI] [PubMed] [Google Scholar]
- 31.Clark N, Cooksey R, Hill B, Swenson J, Tenover F. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinney CW, Pruden A. Environ Sci Technol. 2012;46:13393–13400. doi: 10.1021/es303652q. [DOI] [PubMed] [Google Scholar]
- 33.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazarova V, Janex ML, Fiksdal L, Oberg C, Barcina I, Pommepuy M. Water Sci Technol. 1998;38:9. [Google Scholar]
- 35.U S Environmental Protection Agency. 2012 [PubMed] [Google Scholar]
- 36.M. S. Graham, G. Lomax and G. Skipp, New Orleans, LA, 2012.
- 37.Gehr R, Wagner M, Veerasubramanian P, Payment P. Water Res. 2003;37:4573–4586. doi: 10.1016/S0043-1354(03)00394-4. [DOI] [PubMed] [Google Scholar]
- 38.P. Block, presented in part at the WEFTEC, New Orleans, LA, 2016, 2016.
- 39.El-Rehaili AM. Water Res. 1995;29:1571–1577. [Google Scholar]
- 40.Zhang Y, Snow DD, Parker D, Zhou Z, Li X. Environ Sci Technol. 2013;47:10206–10213. doi: 10.1021/es401964s. [DOI] [PubMed] [Google Scholar]
- 41.Mao D, Luo Y, Mathieu J, Wang Q, Feng L, Mu Q, Feng C, Alvarez PJJ. Environ Sci Technol. 2013;48:71–78. doi: 10.1021/es404280v. [DOI] [PubMed] [Google Scholar]
- 42.Nocker A, Sossa KE, Camper AK. J Microbiol Methods. 2007;70:252–260. doi: 10.1016/j.mimet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Skold O. Drug Resist Updat. 2000;3:155–160. doi: 10.1054/drup.2000.0146. [DOI] [PubMed] [Google Scholar]
- 44.Chopra I, Roberts M. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. second page, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sondergaard A, San Millan A, Santos-Lopez A, Nielsen SM, Gonzalez-Zorn B, Norskov-Lauritsen N. Antimicrob Agents Chemother. 2012;56:4958–4960. doi: 10.1128/AAC.00408-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tutumi M, Imamura K, Hatano S, Watanade T. Food Hygiene Saf Sci (Shokuhin Eiseigaku Zasshi) 1973;14:443–447. [Google Scholar]
- 47.Dodd MC. J Environ Monitor. 2012;14:1754–1771. doi: 10.1039/c2em00006g. [DOI] [PubMed] [Google Scholar]
- 48.Fahrenfeld NL, Ma Y, O’Brien M, Pruden A. Frontiers Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Contreras PJ, Urrutia H, Sossa K, Nocker A. J Microbiol Methods. 2011;87:89–95. doi: 10.1016/j.mimet.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 50.Banihashemi A, Dyke M, Huck P. J Appl Microbiol. 2012;113:863–873. doi: 10.1111/j.1365-2672.2012.05382.x. [DOI] [PubMed] [Google Scholar]
- 51.Roeder RS, Lenz J, Tarne P, Gebel J, Exner M, Szewzyk U. Int J Hyg Environ Health. 2010;213:183–189. doi: 10.1016/j.ijheh.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Hu Q, Zhang XX, Jia S, Huang K, Tang J, Shi P, Ye L, Ren H. Water Res. 2016;101:309–317. doi: 10.1016/j.watres.2016.05.092. [DOI] [PubMed] [Google Scholar]
- 53.Tian S, Tian Z, Yang H, Yang M, Zhang Y. Water. 2017;9:166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


