Abstract
Atomic force microscopy (AFM) is a scanning probe technique that allows visualization of single biomolecules and complexes deposited on a surface with nanometer resolution. AFM is a powerful tool for characterizing protein–protein and protein–DNA interactions. It can be used to capture snapshots of protein–DNA solution dynamics, which in turn, enables the characterization of the conformational properties of transient protein–protein and protein–DNA interactions. With AFM, it is possible to determine the stoichiometries and binding affinities of protein–protein and protein–DNA associations, the specificity of proteins binding to specific sites on DNA, and the conformations of the complexes. We describe methods to prepare and deposit samples, including surface treatments for optimal depositions, and how to quantitatively analyze images. We also discuss a new electrostatic force imaging technique called DREEM, which allows the visualization of the path of DNA within proteins in protein–DNA complexes. Collectively, these methods facilitate the development of comprehensive models of DNA repair and provide a broader understanding of all protein–protein and protein–nucleic acid interactions. The structural details gleaned from analysis of AFM images coupled with biochemistry provide vital information toward establishing the structure–function relationships that govern DNA repair processes.
1. INTRODUCTION
Atomic force microscopy
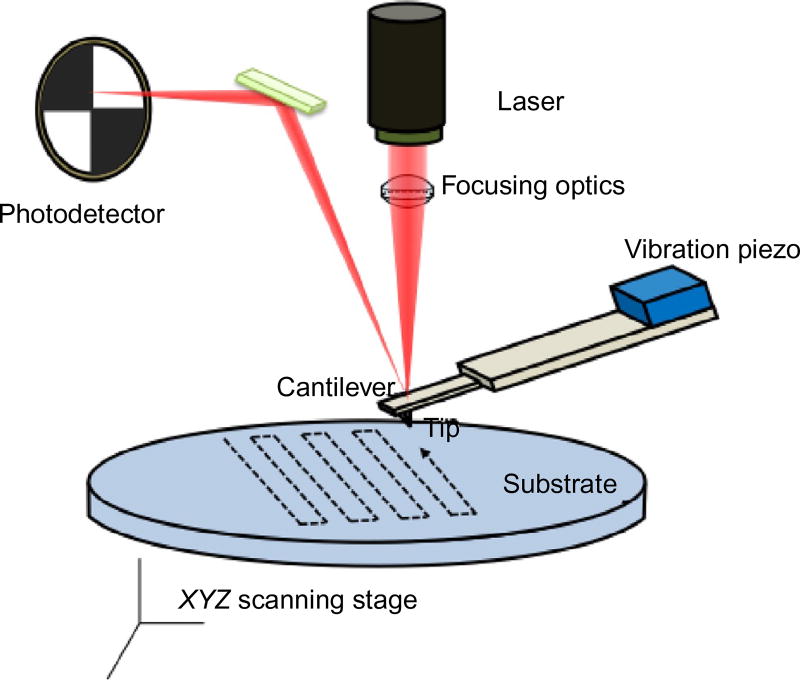
AFM is a three-dimensional imaging technique that allows the visualization of single molecules and nanoscale materials. As a variation of the scanning tunneling microscope, Binnig, Quate, and Gerber implemented the AFM in 1986 to obtain atomic resolution of nonconductive surfaces (Binnig, Quate, & Gerber, 1986). The AFM has since become a powerful tool to image soft biological materials both in air and under solution with nanometer resolution (Bustamante, Erie, & Keller, 1994; Bustamante & Rivetti, 1996; Czajkowsky & Shao, 1998; Hansma & Hoh, 1994; Muller, Fotiadis, Scheuring, Muller, & Engel, 1999). AFM utilizes a cantilever with a sharp tip, usually made of silicon, to scan over a sample deposited onto a flat surface (Fig. 1). When the tip and sample are in close proximity, interaction forces lead to a deflection in the cantilever. An incident laser reflects off the cantilever onto a photodiode that senses and measures the deflection. Topographic images are obtained by using feedback electronics that maintain constant either the deflection (contact mode) or the cantilever’s oscillation amplitude (or frequency) (intermittent contact). In intermittent contact mode, the phase of the cantilever oscillation provides additional information about the sample properties. Variants of AFM such as Kelvin probe force microscopy and electrostatic force microscopy (EFM) report electrostatic information about the sample by monitoring conductive tip-sample interactions under an applied bias. Our lab recently developed a dual-resonance-frequency enhanced EFM (DREEM) technique that produces topographic and electrostatic images simultaneously and allows visualization of the path of DNA within multiprotein complexes (Wu et al., 2016). A growing trend in structural biology combines high-resolution crystallography and NMR data with lower resolution data from techniques such as small-angle X-ray scattering, electron microscopy (EM), and AFM to determine the structure of individual multiprotein–DNA complexes. With comprehensive image analysis, AFM and DREEM may become quantitative tools to couple the structure of essential proteins and DNA to their functions in DNA repair.
Fig. 1.
Schematic diagram of the atomic force microscope.
DNA Repair
Many DNA repair pathways have evolved to safeguard the genome of all organisms from damage and mutations. In this paper, we focus on DNA mismatch repair (MMR); however, the methods discussed could be applied to most studies of DNA repair or other DNA metabolic processes, such as transcription. DNA MMR is a postreplicative process comprised of a system of molecular machines that intricately coordinate to repair base–base mismatches and insertion deletion loops (Modrich & Lahue, 1996). MMR is initiated by MutS and MutL homologs that have DNA binding and ATPase activity, and are conserved in all organisms (Kunkel & Erie, 2005). Germline mutations in MSH2 and MLH1 MMR genes underlie Lynch syndrome, which is characterized by a spectrum of cancers including hereditary nonpolyposis colorectal cancer predisposition (Fishel et al., 1993; Modrich & Lahue, 1996; Plotz, Zeuzem, & Raedle, 2006).
In vitro reconstitution of MMR by Escherichia coli and human proteins has provided many insights into the mechanism of MMR (Dzantiev et al., 2004; Kunkel & Erie, 2015; Lahue, Au, & Modrich, 1989); however, several questions still remain that can be answered with visualization and dynamic characterization of the multiple transient protein–protein and protein–DNA interactions in the pathway. Eukaryotic MMR involves the coordinated stepwise action of approximately 10 different proteins on a DNA molecule containing a mismatch. MutSα recognizes the mismatch by binding to DNA and then undergoes ATP-dependent conformational changes to interact with MutLα. This MutSα–MutLα complex then interacts with PCNA, which activates MutLα to nick the daughter strand. This nick (or possibly preexisting nicks/gaps in the strand) serves as an entry point for exonucleases and polymerase to complete repair. DNA ligase seals the nick. Structural, biochemical, and single-molecule experiments indicate that ATP binding and hydrolysis regulate conformational changes in MMR proteins to modulate transient interactions that coordinate repair. Many single-molecule studies to date have focused on MutS, MutL, and the initial recognition complex (Cho et al., 2012; Cristovao et al., 2012; DeRocco, Anderson, Piehler, Erie, & Weninger, 2010; DeRocco, Sass, Qiu, Weninger, & Erie, 2014; Gorman et al., 2012; Groothuizen et al., 2015; Jeong et al., 2011; Jiang & Marszalek, 2011; Josephs, Zheng, & Marszalek, 2015; Lee et al., 2014; Liu et al., 2016; Qiu et al., 2012; Sass, 2007; Sass, Lanyi, Weninger, & Erie, 2010; Tessmer et al., 2008; Wang et al., 2003; Yang, Sass, Du, Hsieh, & Erie, 2005), thus it is unclear how most of the MMR proteins interact on the molecular level throughout the pathway. These interactions are rare and dynamic events that are obscured in ensemble methods, and challenging to observe due to a requirement to synchronize with other steps of the reaction. Single-molecule methods such as AFM and its variations, single-molecule fluorescence resonance energy transfer (smFRET), and hybrid techniques are well equipped to complement biochemical experiments with molecular-level data (Erie & Weninger, 2014). A comprehensive molecular mechanism of MMR or any other type of DNA repair will include knowing the stoichiometries of each protein and the conformations of the proteins and the DNA during each step along the pathway. Knowledge of the conformations, stoichiometry, and dynamic properties of MMR proteins will facilitate the elucidation of the molecular mechanism of MMR.
We describe methods to image proteins and protein–DNA complexes involved in MMR with standard AFM and DREEM. We also describe our procedures for image analysis to characterize the conformations and stoichiometries in MMR complexes. Lastly, we discuss complementary and hybrid techniques that are essential to elucidating dynamic molecular mechanisms. The bulk of our work has been conducted with MMR proteins, but these methods can be extended to study other protein–nucleic acid interactions important in a variety of biological processes.
2. METHODS TO STUDY DNA REPAIR COMPLEXES WITH AFM
2.1 Preparation of Proteins
Elucidating the details of the interactions that govern the MMR pathway requires purified proteins. Because AFM is an imaging technique, no further modification of the protein, such as fluorescent labeling, is required. We will discuss experiments using proteins from Thermus aquaticus (Taq), yeast (Saccharomyces cerevisiae), and humans. Detailed protocols of the expression and purification of Taq MutS and MutL proteins can be found in our other recent Methods in Enzymology publication (Gauer et al., 2016). Yeast and human MutSα and MutLα proteins are purified as described previously (Antony & Hingorani, 2003; Geng et al., 2011; Hall & Kunkel, 2001; Kadyrov, Dzantiev, Constantin, & Modrich, 2006). Purified protein is dispensed into ~2 µL aliquots and snap-frozen for storage at −20°C or −80°C.
2.2 Preparation of DNA Substrates
Careful design of the DNA substrate for MMR investigations is key in getting the most information out of AFM studies. Generating a lesion in a designated position within the DNA allows us to determine mismatch specificity (Tessmer et al., 2008), the effect of protein binding on DNA geometry (Wang et al., 2003), and conformational changes that correspond to transient events that ultimately lead to DNA repair. One of the most useful recent advances for generating substrates containing lesions or mismatches is the commercial availability of nicking endonucleases, or “nickases” (Ando, Takagi, Kosawa, & Ikeda, 1969). These endonucleases are modified to cleave only one side of their restriction site. By engineering two or more nick sites in close proximity to each other, a gap can be formed, and an oligonucleotide that spans the nick sites containing a modified base, lesion, or extra bases can be introduced to anneal this gap, which can then be ligated to leave DNA that has a specific lesion at a known location. This method has been optimized by the Hsieh and Matson labs (Geng et al., 2011; Robertson & Matson, 2012), and the description below is adapted from their methods.
General protocol for preparing DNA substrates is described later (estimated time 2 days)
Plasmid/PCR preparation: A plasmid containing four Nt.BBvCI nickase sites across a 31 base span (pUC19-VSR) (Robertson & Matson, 2012) is purified from E. coli using a miniprep kit. Plasmid DNA obtained from mini/midi preps is suitable for longer substrates. For shorter substrates, PCR allows reliable generation of substrates with any desired length. PCR cleanup is necessary before proceeding to nicking.
-
Nicking DNA: Nicking is performed using a modified standard protocol for the enzyme Nt.BBvCI. Usually 10 or more reactions are run in parallel. To ensure consistency across all reactions and to account for pipetting errors, a master mix enough for 11 reactions is made, as shown below:
Component 1 × Reaction 11 × Reaction pUC19-VSR 1.5 µg 16.5 µg 10 × NEB CutSmart buffer 2 µL 22 µL Nanopure water Fill to 17.5 µL Fill to 192.5 µL Nt.BBvCI 2.5 µL 27.5 µL Total volume 20 µL 220 µL The reaction is incubated in thin-wall PCR tubes at 37°C for 2.5 h, followed by heating at 80°C for 20 min to promote the dissociation of the short nicked fragments and allow the complementary oligonucleotide containing the mismatch to anneal. No cleanup is necessary after heating. Nicking efficiency can be improved by increasing the incubation time and/or nickase concentration. This additional procedure increases the percent of molecules that are nicked at every nick site, and it is particularly helpful for DNAs with several nick sites. Additionally, increasing the amount of DNA in the reaction helps increase throughput. 20 µL reactions are optimal because keeping the reaction volumes small allows subsequent reactions to be performed directly.
-
Annealing: Assemble the annealing reaction as follows
Component Volume Nicking reaction 20 µL Complementary oligo (100 mM) 2.1 µL (50 × molar excess) 5 M NaCl 0.5 µL (50 mM total) Nanopure water Fill to 50 µL Total volume 50 µL In a thermocycler, run the following program:Step (°C) Duration (min) 80 2 70 20 65 40 55 20 45 20 4 20 Introducing additional nick sites beyond the two necessary to create a gap results in smaller fragments that are more easily displaced than a fragment of equal length to the oligonucleotide to be inserted. Additionally, extending the incubation time above the Tm for fragments, but below the Tm for the complementary oligo, greatly enhances the amount of complement that successfully outcompetes the native fragments. A “step-down” thermocycle profile, with incubations at increasingly lower temperatures is well suited for this annealing. Cleanup is not necessary after annealing but may increase efficiency of ligation in the following step. The trade-off between increasing efficiency and losing DNA from cleanup will undoubtedly vary based on substrate.
-
Ligation: At this point, 500 µL total volume of annealed sample is available for ligation. A master mix is recommended. A master mix for 20 reactions (plus 1 control) is shown below:
Component 1 × Reaction 21 × Reaction 10 × T4 ligase buffer 2 µL 42 µL Annealed sample Fill to 19.5 µL Fill to 409.5 µL T4 ligase 0.5 µL 10.5 µL Total volume 20 µL 420 µL Incubate the reaction at 16°C for 16 h. For increased ligation efficiency, add an additional 0.5 µL T4 Ligase at the halfway point of incubation. Both T4 and E. coli DNA ligase are suitable; however, E. coli DNA ligase has greater efficiency with sticky end ligations, which the substrate most closely resembles. Denature DNA ligase by heating at 65°C for 20 min and then clean up reaction using a PCR clean-up kit. Measure concentration by absorbance spectrometer. Store DNA in TE buffer.
2.3 Complex Formation
Typical concentrations for protein-only samples are 10–30 nM. For DNA-only imaging, ~2 µg/mL works well. We typically mix protein and DNA in high- or low-salt deposition buffer. The low salt buffer is 25 mM HEPES, 50 mM NaOAc, 10 mM Mg(OAc)2, and 5% glycerol at pH 7.5. The high salt buffer is the same except that it has 100 mM NaOAc. Complexes are allowed to incubate on ice or at room temperature for 1–10 min and then deposited on freshly cleaved or functionalized mica.
Cross-linking is a useful tool for imaging complexes that dissociate rapidly and/or involve multiple proteins because it allows the capture of transient complexes that would otherwise be difficult to observe in sufficient amounts for quantitative analysis. Cross-linking effectively increases the number of complexes observed in a sample deposition by trapping complexes in solution. For AFM studies involving DNA and proteins, the cross-linker conjugates free amines of proteins to the amino groups of nucleic bases. The most common cross-linkers are aldehydes. Formaldehyde is the simplest molecule in this class, but others with long spacer arms between bifunctional aldehyde groups are also available and useful for conjugating across larger distances. We have successfully used glutaraldehyde to cross-link and image multiprotein complexes on DNA. To prevent artifacts, optimization is necessary to determine the shortest cross-linking time and least amount of cross-linking agent needed. We typically add glutaraldehyde to a final concentration of 0.85% for 30 s–2 min and quench by either diluting with deposition buffer or adding Tris buffer to the reaction. If possible, it is best to also perform control experiments without cross-linker, which can often be done at lower protein concentrations, where the cross-linked and uncross-linked data can be compared directly.
2.4 Sample and Surface Preparation and Sample Deposition
Proper sample preparation is a key step in achieving reproducible deposition and quality AFM images. Currently, varying deposition quality is one of the major bottlenecks in obtaining high-quality quantitative data from AFM images, though our lab is working to alleviate this problem. Although all single-molecule techniques require careful handling to avoid potential contaminations, AFM is particularly sensitive to impurities. Nanopure water that is suitable for standard biochemical procedures can lead to indeterminate deposits when imaged. As such, it is recommended that all solutions used for sample preparation be passed through a 0.02 filter. It is also useful to deposit and image all solutions to confirm purity. AFM readily detects protein aggregation, and these aggregates can complicate analysis; therefore, single-use aliquots of protein that are snap-frozen at time of purification are strongly suggested. DNA contamination is less frequent, but the only way to confirm contamination is via AFM imaging. Samples that may suggest good purity by bulk measurements can lead to unpleasant surprises at the single-molecule level in AFM images. Another useful discussion of sample preparation of DNA repair complexes can be found in Ristic, Sanchez, and Wyman (2011).
Mica is the most common surface used for deposition of proteins and DNA because of its near-perfect basal cleavage, which leaves a surface that is atomically flat. Mica can be ordered precut to specified dimensions, or it can be cut or punched, but the latter methods are not recommended for smaller pieces of mica. For mounting on the AFM, the mica is usually affixed to disks/slides with double-sided tape or glue from a hot glue gun. Be careful not to get the glue on the edge of the mica, which will make the mica difficult or impossible to peel. Mica disks with a 0.25-in. (6.35 mm) or 0.5-in. (12.7 mm) diameter are sufficient for most AFM applications. Mica that has been attached can be peeled multiple times, allowing many depositions using one mica disc.
Prior to deposition, the mica is freshly peeled mica using Scotch tape. Because the top layer of mica that peeled away on the tape is a mirror of the surface that has been exposed, any cracks, scratches, or other surface abnormalities on the tape can be seen with the naked eye. If defects are present, the mica should be peeled again until a “perfect” surface is obtained. The sample can be deposited directly onto the freshly peeled mica, or the surface can be chemically modified before deposition.
Deposition of proteins on unmodified mica can be done in most any buffer, as long as it does not contain BSA. For DNA, the solution must contain divalent metal ions because mica is negatively charged. Cation-assisted methods use multivalent cations to create a salt-bridge between negatively charged DNA molecules and the mica surface. While several different cations are viable options, Mg2+ is the most common and best choice, unless it would allow unwanted catalytic activity, in which case, Ca2+ can be used instead. The cation-assisted method works well for deposition of DNA samples in which the monovalent ion concentration is less than ~70 mM, but at higher concentrations, the monovalent cations in the solution weaken binding to the surface by DNA. For higher salt depositions of DNA-containing samples, it is necessary to modify the surface to render it positively charged prior to deposition.
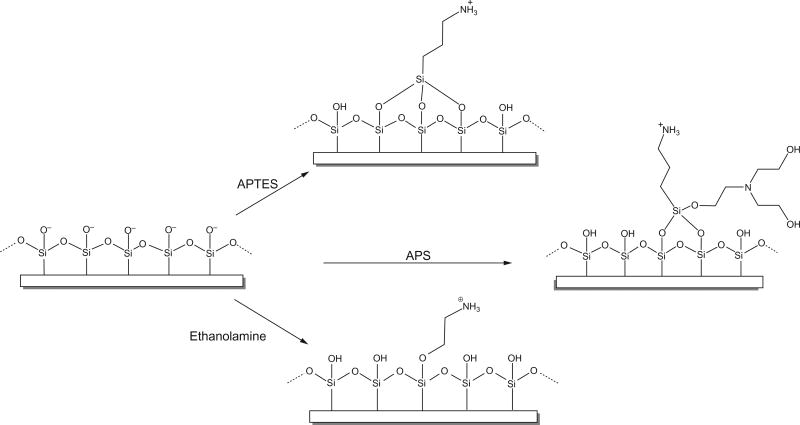
Chemical functionalization is accomplished by modifying exposed hydroxyl groups on the mica surface with a chemical that changes its charge from negative to positive. Modification can be achieved using either 3-aminopropyltriethoxysilane (APTES) or 1-(3-aminopropyl)silatrane (APS) to covalently attach aminopropyl groups directly onto the surface of the mica (Liu et al., 2005; Shlyakhtenko et al., 2003; Shlyakhtenko, Gall, & Lyubchenko, 2013). These functional groups are stable up to pH 10, and they allow for a broad range of pH and salt concentrations to be used in the deposition buffer, without the need for divalent cations. It is important to note that both APTES and APS render the mica surface hydrophobic, and therefore, the sample may not spread evenly, which can be a concern if the complexes are not cross-linked because deposition artifacts are more likely. The choice between APTES and APS is irrelevant for imaging in air, but imaging in solution requires APS because APTES results in large aggregates appearing on the surface over time (Shlyakhtenko et al., 2003). Ethanolamine can also be used, and it works similarly to APTES, exposing primary amines, but it is less hydrophobic. Fig. 2 shows the different surface functionalizations.
Fig. 2.
Surface functionalization of mica.
Another approach that has shown promise is the use of surface coatings that do not covalently modify the mica surface but still leave a positively charged functional group on the surface to interact with DNA. Chains of either poly-l-lysine or poly-l-ornithine incubated on the surface will result in the primary amine side chains of these polymers free to interact and successfully hold the DNA to the surface (Podesta et al., 2004).
2.4.1 General Procedures for the Surface Treatments
APTES or ethanolamine
In a clean desiccator, place 30 µL of APTES or ethanolamine on a strip of parafilm alongside the freshly peeled mica and incubate for 15 min. After 15 min, remove the parafilm; the mica can be left in the desiccator until ready to deposit samples.
APS
APS is not commercially available but can be synthesized with standard organic chemistry lab equipment (Shlyakhtenko et al., 2003). A 50-mM APS stock solution in water is stable at 4°C for at least 6 months. Prepare a 1:300 dilution working solution, which is stable at room temperature for several days. To functionalize the mica, place enough of the working solution to fully cover the surface and let it sit in a covered container for 30 min. Afterward, rinse with deionized water and dry under inert gas stream. APS mica remains useable for several days.
Poly-l-ornithine/lysine (PO) (Podesta et al., 2004)
Prepare a working solution of 0.012 µg/mL PO. This solution has a shelf life of at least 1 year stored at 4°C. Deposit the working solution onto freshly peeled mica; 10 µL is sufficient for mica that is about 15 mm in diameter. Tilting and gentle agitation may be necessary to make the solution spread evenly across the entire surface. Take care not to touch the mica surface while handling. After 2 min of incubation, rinse with 400 µL deionized water, wick away the majority of the water by touching a filter paper to the edge of the mica, then dry under inert gas stream.
2.4.2 Deposition
To prepare a deposition, dilute the protein to a final concentration of 10–70 nM in deposition buffer, with a DNA concentration of 1–5 µg/mL. Deposit 5–20 µL of the diluted protein (or protein–DNA) onto freshly cleaved ruby mica (Spruce Pine Mica Company, Spruce Pine, NC). Rinse the sample immediately with nanopure water by gently dropping water onto the surface, blot excess water by touching filter paper to the edge of the mica and then dry the sample using a stream of nitrogen. Take care not to touch the mica surface while handling.
An example cross-linking experiment
Prepare the surface of the mica as desired (see above). Prepare a working solution of glutaraldehyde by diluting 50% glutaraldehyde stock solution with deposition buffer to a final concentration of 8.5%.
Combine the following:
| Component | Amount |
|---|---|
| Linearized DNA | 400 ng |
| Taq MutS | 125 nM |
| ATP | 1 mM |
| Total volume | 18 µL |
Incubate the reaction for a given time at room temperature, then add 2 µL 8.5% glutaraldehyde and incubate for an additional 30 s–1 min. Dilute the reaction 10-fold (and/or quench with Tris buffer) and make 15 µL depositions onto the mica prepared earlier. Gently wash with ~1 mL water, wick away excess water with Whatman filter paper, dry under inert gas, and store in desiccator.
2.5 Data Acquisition
Begin by turning the AFM on and allowing it to warm up. Warming up helps to minimize piezo drift. Insert a cantilever into the holder for the AFM, and align the laser and tune the cantilever to a drive frequency so that it is slightly to the left or right of peak maximum, along the steep portion of the resonance peak. Place a prepared sample into the sample holder and engage the tip. Imaging parameters will vary based the instrument; however, it is most important to image with the minimal force at which quality images can be obtained. In addition, the proper image size and surface coverage are necessary for specific analyses. For protein volume analysis (Ratcliff & Erie, 2001; Yang, Wang, & Erie, 2003), a 1 µm × 1 µm image at 512 × 512 pixels (=2 nm × 2 nm/pixel) is sufficient. For DNA length analysis, 4 nm/pixel works well.
3. IMAGE ANALYSIS
3.1 Background
The power of AFM to dissect biological processes at the molecular level is harnessed by analyzing four main characteristics of protein–DNA complexes extracted from images. These include specificity, binding affinity, stoichiometry, and conformation. Taken together, these quantitative data inform us about the molecular mechanisms of DNA repair processes. Achieving statistical confidence in our analysis requires evaluating many complexes over multiple images. Extracting these quantitative data on several complexes across multiple images can be time consuming. A typical workflow is as follows: (1) process (correct) raw image data, (2) identify protein–protein and/or protein–DNA complexes, and (3) analyze individual complexes for the characteristics described earlier. An overview of the software available for AFM analysis is outlined in Table 1. Some of the software packages are general-purpose basic image analysis programs, while others have strengths in tackling specific analysis tasks. Often, supplemental applications to many specialized analyses have to be written in-house via various programming languages (i.e., MATLAB, Java, and Python). We are developing a user-friendly MATLAB software that significantly streamlines the analysis, and it will be available when it is complete.
Table 1.
Overview of Software for Single-Molecule AFM Study
| Software | Image Formats | Platforms | Image Processing |
Particle Analysis |
Single- Molecule Analysis |
Batch Analysis |
Extensions | Open Source |
Free |
|---|---|---|---|---|---|---|---|---|---|
| WSxM (http://www.wsxmsolutions.com) | Various | Windows | Yes | No | No | Yes | No | No | Yes |
| Gwyddion (http://gwyddion.net) | Various | Multiple | Yes | Yes | No | Yes | Via C or Python | Yes | Yes |
| Nanoscope Analysis (http://nanoscaleworld.bruker-axs.com/) | Nanoscope format | Windows | Yes | Yes | No | Yes | No | No | Yes |
| Asylum Research (http://support.asylumresearch.com) | Asylum Research format | Windows, Mac OS | Yes | Yes | Yes | Yes | Via Igor Pro | Yes | Yes |
| ImageSXM (https://www.liverpool.ac.uk/~sdb/ImageSXM) | Various | Mac OS | Yes | Yes | No | No | No | No | Yes |
| ImageJ (http://imagej.net) | Nanoscope format | Multiple | Yes | Yes | No | Yes | Via Java | Yes | Yes |
| SPIP (http://imagemet.com) | Various | Windows | Yes | Yes | Yes | Yes | Via C++ | No | No |
3.2 Image Correction
AFM images usually require correction before they can be analyzed. Before further analysis, images often need to be flattened, as described previously (Ratcliff & Erie, 2001). A polynomial curve fitting approach is typically used to normalize the surface height. High surface features, such as large proteins, can be masked from the flattening procedure using a height-based threshold. Over-flattening with a high degree polynomial can sometimes introduce artifacts and should be avoided; first-order flattening is optimal.
3.3 Deposition and Imaging Artifacts
In single-molecule studies, statistical analyses are only as convincing as the quality and amount of the data that are analyzed. For a single deposition, many images on different areas of the sample should be collected. It is important that a sample looks the same irrespective of the image area. A variation in protein and/or DNA concentration in different regions of the same sample indicates a nonuniform deposition and should not be used for quantitative data analysis. For example, determining binding affinity depends greatly on accurate counting of free and bound biomolecules (Ratcliff & Erie, 2001; Yang et al., 2005).
Other AFM image artifacts derive from factors that impact the tip-surface interaction. Anything that influences that interaction will trigger the AFM feedback system to adjust the tracking of the surface. Some possibilities include tip degradation and contamination, or imaging forces that are too large or too small, which both result in the tip improperly tracking the surface features. For example, chemical treatment on the surface and residual salt or water from buffer can lead to the appearance of a patchy surface due to the additional attractive and/or capillary forces added on the tip when scanning over those areas. In addition, if excessive imaging force is used, features can be flattened and removed of their fine details. Biomolecules are particularly vulnerable to excessive imaging force.
Additional factors that impact the performance of the AFM scanner and feedback system include environmental noise, piezo drift, and overly aggressive gain settings. The tip can significantly affect image quality because the AFM image is a dilation of sample by the tip, making the image resolution highly dependent on the tip radius. It is also important to examine the images for potential tip artifacts. Readers can refer to Atomic Force Microscopy for Biologists (Morris, 2010), chapter 3.5, by V.J. Morris et al. for additional reading on AFM image artifacts.
3.4 Characterizing Protein–DNA Complexes
For an unbiased analysis of AFM images, it is essential to examine all species present, unless there is a clear and valid reason for exclusion. Quantitative analysis of AFM images not only allows one to characterize the conformations of proteins and protein–DNA complexes, but it also permits the determination of the specificity and binding affinity of proteins for DNA and the stoichiometries and association constants for protein complexes (Ratcliff & Erie, 2001; Yang et al., 2005, 2003).
Specificity
The specificity of proteins binding to DNA can be determined by measuring the position distribution of proteins along the DNA, without the need to determine the binding affinities for the specific and nonspecific sites (Tessmer et al., 2008; Yang et al., 2005). In addition, the binding affinity is estimated by counting the number of protein–DNA complexes and free DNA molecules (Yang et al., 2005). Care must be taken to validate the binding affinity, however, because an overestimate of the binding affinity can occur if the protein surface coverage is too high because it increases the probability that proteins randomly land on the DNA. Detailed procedures for determining the specificity and binding affinities for proteins binding to DNA are described in Yang et al. (2003).
Stoichiometry
There are several ways to define stoichiometry, including (1) how many proteins per complex, (2) how many protein complexes per DNA, and (3) how many proteins per DNA molecule. To extract the number of proteins per complex, we use volume analysis (Ratcliff & Erie, 2001; Yang et al., 2003). For protein–protein complexes, the volume is directly proportional to the protein molecular weight (Ratcliff & Erie, 2001; Yang et al., 2003), and this relationship also appears to hold reasonably well for most protein–DNA complexes. Volume analysis has been used to determine the stoichiometry for many repair proteins (e.g., see Chelico, Sacho, Erie, & Goodman, 2008; Wang et al., 2006; Wang, Tessmer, Croteau, Erie, & Van Houten, 2008; Xue et al., 2002). For protein–DNA complexes, the first peak in the volume distribution usually represents the volume of a single protein on the DNA. This volume can be used to estimate the number of proteins per complex by dividing higher volume peaks by the peak volume corresponding to a single bound protein; however, there may not be an exactly 1:1 correlation. The total number of proteins bound to the DNA fragment is determined by summing all the proteins whether individual or in multimeric complexes on the DNA. It should be noted that the volume of a protein–DNA complex could vary due to conformation in the complex. Because volume analysis depends directly on the height and area measurements, any factors that influence those measurements also affect the volume measurement. For example, the height may be inaccurate if the surface height is not measured properly at the local level, or if tip-surface interactions change (discussed in Section 3.3).
Conformation
Perhaps the most outstanding strength of AFM in single-molecule protein–DNA studies is that we can directly visualize the conformation of a protein–DNA (or protein) complex at physiological conditions with relative ease. The conformation of a complex can be qualitatively described using parameters such as whether a complex loops the DNA or how a complex binds to the DNA. Physical parameters of the conformation, such as size (volume, height, area, etc.), geometries (orientation, eccentricity, fiber length, etc.), and the DNA bend angles in the complex are also qualitative metrics. Some examples demonstrating the characterization of protein-induced DNA conformational changes (Tessmer et al., 2005; Wang et al., 2003; Xue et al., 2002; Yang et al., 2005, 2003) and ligand-induced protein conformational changes (Harrison et al., 2016; Lemaire, Tessmer, Craig, Erie, & Cole, 2006; Sacho, Kadyrov, Modrich, Kunkel, & Erie, 2008) have been published. Combining AFM data with biochemical function studies allows the correlation between structure and function. Van Houten and coworkers describe methods for characterizing the conformations of nucleotide excision repair (NER) complexes in an accompanying chapter in this issue.
4. DREEM IMAGING
4.1 Principles
Visualization of multiprotein–DNA complexes is readily achievable with AFM as described in the previous section; however, in the normal AFM imaging mode, one cannot localize DNA within the multiprotein–DNA complex. The recently developed DREEM imaging technique, which allows the visualization of DNA in protein–DNA complexes (Wu et al., 2016), overcomes this problem. This method simultaneously provides a classic topographic image and an electrostatic force gradient image, which allows visualization of the protein–DNA interaction. DREEM has been used to reveal the path of DNA wrapping around histones, and as it passes through single MMR proteins and multiprotein complexes (Benarroch-Popivker et al., 2016; Kaur et al., 2016; Wu et al., 2016). The DNA appears to be visible in the DREEM images of these complexes due to charge neutralization where the protein and DNA interact. Similar to EFM, DREEM images are produced by monitoring the amplitude and/or phase of the induced vibration. A full theoretical treatment of DREEM concepts and the details of the procedure can be found in Wu et al. (2016) supplemental information. This sensitive, high-resolution technique is capable of simultaneously collecting both topography and electrostatic images at the nanometer scale.
4.2 Methods
The instrumental setup uses an AFM instrument (Asylum Research MFP3D) with an external lock-in amplifier (Sanford Research System, Sunnyvale, CA, model SR844 RF) and a function generator (Sanford Research System, model DS335). The mica is attached to glass slides with colloidal liquid silver (Ted Pella, Inc.) Highly doped silicon cantilevers (Nanosensors, PPP-FMR, force constant ~2.8 N/m) are used for imaging, and the cantilevers are made conductive by first removing the nonconductive oxide layer using plasma cleaning, scraping the cantilever chip, and lastly coating it with colloidal Ag solution. Protein–DNA complexes are formed in solution as described earlier and deposited on a freshly cleaved mica surface. The sample is grounded by continuing the Ag streak to the back of the glass slide connected to the metal in the AFM base for grounding.
The topographic and DREEM images are collected by simultaneously mechanically vibrating the cantilever near its fundamental resonance requency (ω1), and applying a bias voltage (VDC and VAC) to the tip at the first overtone (ω2). An AC bias (10–20 V) is used to generate a vibration at ω2, and a DC bias (−2.5 to +2.5 V) is applied to optimize the amplitude at ω2. The vibration amplitude (Aω2) and phase (φω2) are monitored as a function of sample positions using an external lock-in amplifier.
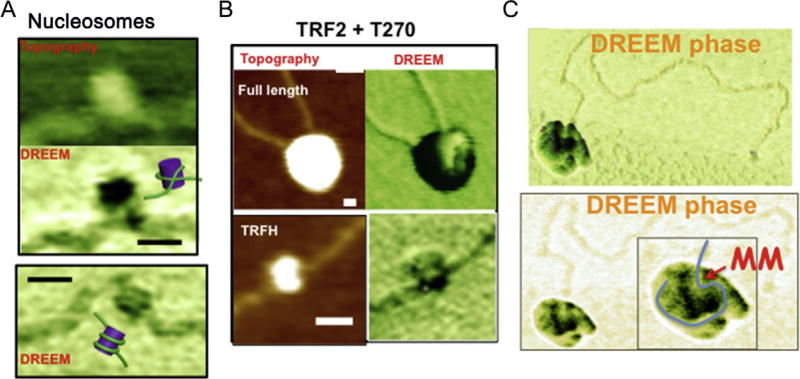
This technique has been applied to several different protein–DNA samples. DREEM images of both protein and DNA show a decrease in phase. However, proteins show a higher contrast as compared to DNA, making it possible to distinguish DNA from protein in a protein–DNA complex. Fig. 3 shows AFM and DREEM images of a nucleosome, and Taq MutS and human MutSα–MutLα proteins in complex with a 2-kbp DNA substrate containing a GT mismatch. Topographic images show smooth peaks typical of MutS on DNA (Tessmer et al., 2008; Wang et al., 2003). DREEM images reveal the path of DNA in the complexes (Wu et al., 2016). DREEM data combined with structural data on MutS (Obmolova, Ban, Hsieh, & Yang, 2000) allowed the modeling of the general orientation of MutS dimers in the complexes. In large MutSα–MutLα–mismatch DNA complexes (~8 proteins), the path of DNA can be resolved (Fig. 3), illustrating the potential power of the DREEM technique. DREEM also has been used to study the role of the shelterin proteins in the mechanism of t-loop formation at the end of the telomeres (Benarroch-Popivker et al., 2016; Kaur et al., 2016) (Fig. 3). Looped conformations of DNA inside large TRF2–DNA complexes observed with DREEM imaging gave new insights into the mechanism of DNA compaction and t-loop formation. The images show that the DNA is compacted inside, not wrapped around the outside, the large TRF2 protein–DNA complexes, with a DNA loop protruding out from the complex (Benarroch-Popivker et al., 2016). In addition, DREEM shows that the dimerization domain of the TRF2 protein, TRFH, wraps ~90 bps of telomeric DNA around it.
Fig. 3.
DREEM imaging of protein–DNA complexes. (A) DREEM phase image reveals DNA wrapping around histone proteins in nucleosomes reconstituted in vitro on DNA containing the 601 nucleosomal positioning sequences. (B) DREEM phase image of TRF2 on DNA containing 270 TTAGGG repeats (T270) reveals portions of folded DNA appearing at the edge (lighter regions) of large multimeric full-length TRF2 complexes (top) and wrapping around a subdomain of TRF2 (TRFH). Scale bars: 20 nm. (C) DREEM phase (top: surface plot; bottom: top view) images of a large MutSα–MutLα–DNA complex containing ~10 proteins on a linear DNA substrate containing a mismatch (MM).
4.3 Limitations of DREEM
Using sharp (tip radius 5–8 nm) highly doped silicon cantilevers and operating in repulsive intermittent contact mode maximizes the resolution in both topographic and electrostatic images. Doped silicon cantilevers can become oxidized, reducing their conductivity. Variability in the oxidation layers on the silicon cantilevers limits quantitative comparison of DREEM data collected with different cantilevers, or the same cantilever after several collected images. Typically, 10–12 high-quality DREEM images can be acquired from a single AFM tip before oxidation degrades the electrostatic signal. In addition, ~30% of doped silicon cantilevers do not generate sufficient contrast between the protein and DNA in DREEM images. Argon plasma cleaning appears to improve the quality of cantilevers for DREEM imaging. This technique is also subject to tip artifacts, as with conventional AFM, due to the asymmetries in the electric field between the tip and sample surface. Finally, DREEM is currently limited to imaging in air.
5. COMPLEMENTARY TECHNIQUES
Scanning probe microscopies such as AFM and DREEM are exceptional at imaging surface-bound protein–DNA complexes with nanoscale resolution. This direct visualization provides structural and stoichiometric details that are vital to understanding the molecular mechanisms of DNA repair pathways. Typical AFM studies offer a snapshot of solution dynamics, but biochemical studies indicate that DNA repair processes involve multiple transient interactions. Pushing the limits, emerging high-speed AFM is capable of subsecond temporal resolution (Ando et al., 2001; Endo & Sugiyama, 2014; Miyagi, Ando, & Lyubchenko, 2011; Sanchez, Suzuki, Yokokawa, Takeyasu, & Wyman, 2011). Using high-speed AFM, chromatin dynamics have been characterized (Miyagi et al., 2011). Although impressive, some complementary single-molecule fluorescence techniques are able to capture submillisecond solution dynamics. DNA complexes with the NER protein human XPA have been studied with scanning confocal fluorescence microscopy, (Segers-Nolten et al., 2002) capable of characterizing interaction dynamics on a time scale of 10 µs. We have employed TIRF-based single-molecule FRET as a complement to AFM studies to achieve simultaneous structural and dynamic characterization of DNA repair complexes (DeRocco et al., 2014; Gauer et al., 2016; Qiu et al., 2012, 2015; Sass et al., 2010). By fluorescently labeling protein domains or DNA, dynamic conformations of MMR proteins and DNA can be investigated. Our methods for conducting and analyzing smFRET experiments are outlined in another recent Methods in Enzymology review (Gauer et al., 2016). Combining these techniques with AFM allows for the dynamic smFRET data to be interpreted in terms of the conformations of the complexes.
Hybrid methods combine AFM with fluorescence microscopy (Fronczek et al., 2011; Sanchez, Kanaar, & Wyman, 2010; Sanchez, Kertokalio, van Rossum-Fikkert, Kanaar, & Wyman, 2013). These techniques overcome a serious limitation of AFM (and electron microscopy) to distinguish different proteins. One can label proteins of interest with different fluorescent dyes and collect fluorescence and scanning force data simultaneously. Fluorescence and AFM images of UvrA–UvrB–quantum dot complexes bound to UV-damaged DNA have been aligned with ~8 nm accuracy using FIONA-AFM (Fronczek et al., 2011).
6. CONCLUSIONS
We have outlined the methods to study DNA repair complexes with AFM. Such studies have revealed important details about the MMR mechanism. The combination of AFM and DREEM with single-molecule fluorescence to characterize the conformations and dynamics of DNA repair complexes has already yielded mechanistic insights of DNA repair processes, and this combination has the potential to be a powerful tool to characterize a wide array of DNA repair pathways. Development of higher-throughput methods of deposition and analysis, along with the continued improvement of the AFM instrumentation would greatly increase the experimental space that could be explored with AFM. AFM is well suited for the study of DNA repair because these processes involve several proteins that act together to orchestrate repair and there are often long-range interactions. AFM allows the determination not only of conformations but also stoichiometries, specificities, and affinities, making it a powerful tool for the quantitative characterization of protein–DNA complexes.
Acknowledgments
This work was supported by Grants from the National Institutes of General Medical Sciences (NIGMS) GM080294 and GM109832 to D.A.E.
References
- Ando T, Kodera N, Takai E, Maruyama D, Saito K, Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(22):12468–12472. doi: 10.1073/pnas.211400898. http://dx.doi.org/10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Takagi J, Kosawa T, Ikeda Y. Isolation and characterization of enzymes with nicking action from phage T4-infected Escherichia coli. Journal of Biochemistry. 1969;66(1):1–10. doi: 10.1093/oxfordjournals.jbchem.a129107. [DOI] [PubMed] [Google Scholar]
- Antony E, Hingorani MM. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42(25):7682–7693. doi: 10.1021/bi034602h. http://dx.doi.org/10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, et al. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Molecular Cell. 2016;61(2):274–286. doi: 10.1016/j.molcel.2015.12.009. http://dx.doi.org/10.1016/j.molcel.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Physical Review Letters. 1986;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. http://dx.doi.org/10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Erie DA, Keller D. Biochemical and structural applications of scanning force microscopy. Current Opinion in Structural Biology. 1994;4(5):750–760. doi: 10.1016/s0959-440x(97)80082-6. http://dx.doi.org/10.1016/s0959-440x(94)90175-9. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Rivetti C. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annual Review of Biophysics and Biomolecular Structure. 1996;25:395–429. doi: 10.1146/annurev.bb.25.060196.002143. [DOI] [PubMed] [Google Scholar]
- Chelico L, Sacho EJ, Erie DA, Goodman MF. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. The Journal of Biological Chemistry. 2008;283(20):13780–13791. doi: 10.1074/jbc.M801004200. http://dx.doi.org/10.1074/jbc.M801004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Jeong C, Kim D, Chang M, Song KM, Hanne J, et al. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20(7):1264–1274. doi: 10.1016/j.str.2012.04.017. http://dx.doi.org/10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristovao M, Sisamakis E, Hingorani MM, Marx AD, Jung CP, Rothwell PJ, et al. Single-molecule multiparameter fluorescence spectroscopy reveals directional MutS binding to mismatched bases in DNA. Nucleic Acids Research. 2012;40(12):5448–5464. doi: 10.1093/nar/gks138. http://dx.doi.org/10.1093/nar/gks138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Shao ZF. Submolecular resolution of single macromolecules with atomic force microscopy. FEBS Letters. 1998;430(1–2):51–54. doi: 10.1016/s0014-5793(98)00461-x. http://dx.doi.org/10.1016/s0014-5793(98)00461-x. [DOI] [PubMed] [Google Scholar]
- DeRocco V, Anderson T, Piehler J, Erie DA, Weninger K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. BioTechniques. 2010;49(5):807–816. doi: 10.2144/000113551. http://dx.doi.org/10.2144/000113551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRocco V, Sass LE, Qiu R, Weninger KR, Erie DA. Dynamics of MutS-mismatched DNA complexes are predictive of their repair phenotypes. Biochemistry. 2014;53(12):2043–2052. doi: 10.1021/bi401429b. http://dx.doi.org/10.1021/bi401429b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. Defined human system that supports bidirectional mismatch-provoked excision. Molecular Cell. 2004;15(1):31–41. doi: 10.1016/j.molcel.2004.06.016. http://dx.doi.org/10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Endo M, Sugiyama H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Accounts of Chemical Research. 2014;47(6):1645–1653. doi: 10.1021/ar400299m. http://dx.doi.org/10.1021/ar400299m. [DOI] [PubMed] [Google Scholar]
- Erie DA, Weninger KR. Single molecule studies of DNA mismatch repair. DNA Repair (Amst) 2014;20:71–81. doi: 10.1016/j.dnarep.2014.03.007. http://dx.doi.org/10.1016/j.dnarep.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. http://dx.doi.org/10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Fronczek DN, Quammen C, Wang H, Kisker C, Superfine R, Taylor R, et al. High accuracy FIONA-AFM hybrid imaging. Ultramicroscopy. 2011;111(5):350–355. doi: 10.1016/j.ultramic.2011.01.020. http://dx.doi.org/10.1016/j.ultramic.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauer JW, LeBlanc S, Hao P, Qiu R, Case BC, Sakato M, et al. Single-molecule FRET to measure conformational dynamics of DNA mismatch repair proteins. Methods in Enzymology. 2016;581:285–315. doi: 10.1016/bs.mie.2016.08.012. http://dx.doi.org/10.1016/bs.mie.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Du CW, Chen SY, Salerno V, Manfredi C, Hsieh P. In vitro studies of DNA mismatch repair proteins. Analytical Biochemistry. 2011;413(2):179–184. doi: 10.1016/j.ab.2011.02.017. http://dx.doi.org/10.1016/j.ab.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, et al. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proceedings of the National Academy of Sciences. 2012;109(45):E3074–E3083. doi: 10.1073/pnas.1211364109. http://dx.doi.org/10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groothuizen FS, Winkler I, Cristovao M, Fish A, Winterwerp HH, Reumer A, Sixma TK. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife. 2015;4:e06744. doi: 10.7554/eLife.06744. http://dx.doi.org/10.7554/eLife.06744.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Kunkel TA. Purification of eukaryotic MutL homologs from Saccharomyces cerevisiae using self-cleaving affinity technology. Protein Expression and Purification. 2001;21(2):333–342. doi: 10.1006/prep.2000.1379. http://dx.doi.org/10.1006/prep.2000.1379. [DOI] [PubMed] [Google Scholar]
- Hansma HG, Hoh JH. Biomolecular imaging with the atomic force microscope. Annual Review of Biophysics and Biomolecular Structure. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. http://dx.doi.org/10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- Harrison JS, Cornett EM, Goldfarb D, DaRosa PA, Li ZMM, Yen F, et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife. 2016;5 doi: 10.7554/eLife.17101. http://dx.doi.org/10.7554/eLife.17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, et al. MutS switches between two fundamentally distinct clamps during mismatch repair. Nature Structural & Molecular Biology. 2011;18(3):379–385. doi: 10.1038/nsmb.2009. http://dx.doi.org/10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Marszalek PE. Atomic force microscopy captures MutS tetramers initiating DNA mismatch repair. The EMBO Journal. 2011;30(14):2881–2893. doi: 10.1038/emboj.2011.180. http://dx.doi.org/10.1038/emboj.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs EA, Zheng T, Marszalek PE. Atomic force microscopy captures the initiation of methyl-directed DNA mismatch repair. DNA Repair (Amst) 2015;35:71–84. doi: 10.1016/j.dnarep.2015.08.006. http://dx.doi.org/10.1016/j.dnarep.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126(2):297–308. doi: 10.1016/j.cell.2006.05.039. http://dx.doi.org/10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kaur P, Wu D, Lin J, Countryman P, Bradford KC, Erie DA, et al. Enhanced electrostatic force microscopy reveals higher-order DNA looping mediated by the telomeric protein TRF2. Scientific Reports. 2016;6:20513. doi: 10.1038/srep20513. http://dx.doi.org/10.1038/srep20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annual Review of Biochemistry. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. http://dx.doi.org/10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. In: Bassler BL, editor. Annual Review of Genetics. Vol. 49. 2015. pp. 291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245(4914):160–164. doi: 10.1126/science.2665076. http://dx.doi.org/10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lee JB, Cho WK, Park J, Jeon Y, Kim D, Lee SH, et al. Single-molecule views of MutS on mismatched DNA. DNA Repair (Amst) 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. http://dx.doi.org/10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45(30):9074–9084. doi: 10.1021/bi060567d. http://dx.doi.org/10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hanne J, Britton BM, Bennett J, Kim D, Lee J-B, et al. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature. 2016;539:583–587. doi: 10.1038/nature20562. http://dx.doi.org/10.1038/nature20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li Z, Zhou HL, Wei G, Song YH, Wang L. Imaging DNA molecules on mica surface by atomic force microscopy in air and in liquid. Microscopy Research and Technique. 2005;66(4):179–185. doi: 10.1002/jemt.20156. http://dx.doi.org/10.1002/jemt.20156. [DOI] [PubMed] [Google Scholar]
- Miyagi A, Ando T, Lyubchenko YL. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50(37):7901–7908. doi: 10.1021/bi200946z. http://dx.doi.org/10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annual Review of Biochemistry. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. http://dx.doi.org/10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Morris VJ. Atomic force microscopy for biologists [electronic resource] London: Imperial College Press; 2010. [Google Scholar]
- Muller DJ, Fotiadis D, Scheuring S, Muller SA, Engel A. Electrostatically balanced subnanometer imaging of biological specimens by atomic force microscope. Biophysical Journal. 1999;76(2):1101–1111. doi: 10.1016/S0006-3495(99)77275-9. http://dx.doi.org/10.1016/s0006-3495(99)77275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407(6805):703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Plotz G, Zeuzem S, Raedle J. DNA mismatch repair and Lynch syndrome. Journal of Molecular Histology. 2006;37(5–7):271–283. doi: 10.1007/s10735-006-9038-5. http://dx.doi.org/10.1007/s10735-006-9038-5. [DOI] [PubMed] [Google Scholar]
- Podesta A, Imperadori L, Colnaghi W, Finzi L, Milani P, Dunlap D. Atomic force microscopy study of DNA deposited on poly L-ornithine-coated mica. Journal of Microscopy. 2004;215:236–240. doi: 10.1111/j.0022-2720.2004.01372.x. http://dx.doi.org/10.1111/j.0022-2720.2004.01372.x. [DOI] [PubMed] [Google Scholar]
- Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, Erie DA, et al. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. The EMBO Journal. 2012;31(11):2528–2540. doi: 10.1038/emboj.2012.95. http://dx.doi.org/10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, Sakato M, Sacho EJ, Wilkins H, Zhang X, Modrich P, et al. MutL traps MutS at a DNA mismatch. Proceedings of the National Academy of Sciences. 2015;112(35):10914–10919. doi: 10.1073/pnas.1505655112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff GC, Erie DA. A novel single-molecule study to determine protein-protein association constants. Journal of the American Chemical Society. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- Ristic D, Sanchez H, Wyman C. Sample preparation for SFM imaging of DNA, proteins, and DNA-protein complexes. In: Peterman EJG, Wuite GJL, editors. Single molecule analysis: Methods and protocols. Vol. 783. Totowa: Humana Press Inc; 2011. pp. 213–231. [DOI] [PubMed] [Google Scholar]
- Robertson AB, Matson SW. Reconstitution of the very short patch repair pathway from Escherichia Coli. The Journal of Biological Chemistry. 2012;287(39):32953–32966. doi: 10.1074/jbc.M112.384321. http://dx.doi.org/10.1074/jbc.M112.384321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Molecular Cell. 2008;29(1):112–121. doi: 10.1016/j.molcel.2007.10.030. http://dx.doi.org/10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Kanaar R, Wyman C. Molecular recognition of DNA-protein complexes: A straightforward method combining scanning force and fluorescence microscopy. Ultramicroscopy. 2010;110(7):844–851. doi: 10.1016/j.ultramic.2010.03.002. http://dx.doi.org/10.1016/j.ultramic.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Kertokalio A, van Rossum-Fikkert S, Kanaar R, Wyman C. Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(28):11385–11390. doi: 10.1073/pnas.1306467110. http://dx.doi.org/10.1073/pnas.1306467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez H, Suzuki Y, Yokokawa M, Takeyasu K, Wyman C. Protein-DNA interactions in high speed AFM: Single molecule diffusion analysis of human RAD54. Integrative Biology. 2011;3(11):1127–1134. doi: 10.1039/c1ib00039j. http://dx.doi.org/10.1039/c1ib00039j. [DOI] [PubMed] [Google Scholar]
- Sass LE. Ensemble and single-molecule fluorescence studies of DNA mismatch repair initiation by MutS. [Doctor of Philosophy] Chapel Hill, NC: UNC Chapel Hill; 2007. [Google Scholar]
- Sass LE, Lanyi C, Weninger K, Erie DA. Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry. 2010;49(14):3174–3190. doi: 10.1021/bi901871u. http://dx.doi.org/10.1021/bi901871u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers-Nolten GMJ, Wyman C, Wijgers N, Vermeulen W, Lenferink ATM, Hoeijmakers JHJ, et al. Scanning confocal fluorescence microscopy for single molecule analysis of nucleotide excision repair complexes. Nucleic Acids Research. 2002;30(21):4720–4727. doi: 10.1093/nar/gkf599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhtenko L, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97(1–4):279–287. doi: 10.1016/S0304-3991(03)00053-6. http://dx.doi.org/10.1016/s0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko L, Gall Alexander A, Lyubchenko Yuri L. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy. Methods in Molecular Biology (Clifton, N.J.) 2013;931:295–312. doi: 10.1007/978-1-62703-056-4_14. http://dx.doi.org/10.1007/978-1-62703-056-4_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmer I, Moore T, Lloyd RG, Wilson A, Erie DA, Allen S, et al. AFM studies on the role of the protein RdgC in bacterial DNA recombination. Journal of Molecular Biology. 2005;350(2):254–262. doi: 10.1016/j.jmb.2005.04.043. http://dx.doi.org/10.1016/j.jmb.2005.04.043. [DOI] [PubMed] [Google Scholar]
- Tessmer I, Yang Y, Zhai J, Du C, Hsieh P, Hingorani MM, et al. Mechanism of MutS searching for DNA mismatches and signaling repair. The Journal of Biological Chemistry. 2008;283(52):36646–36654. doi: 10.1074/jbc.M805712200. http://dx.doi.org/10.1074/jbc.M805712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. The Journal of Biological Chemistry. 2006;281(22):15227–15237. doi: 10.1074/jbc.M601476200. http://dx.doi.org/10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
- Wang H, Tessmer I, Croteau DL, Erie DA, Van Houten B. Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Letters. 2008;8(6):1631–1637. doi: 10.1021/nl080316l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang Y, Schofield MJ, Du CW, Fridman Y, Lee SD, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):14822–14827. doi: 10.1073/pnas.2433654100. http://dx.doi.org/10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Kaur P, Li ZM, Bradford KC, Wang H, Erie DA. Visualizing the path of DNA through proteins using DREEM imaging. Molecular Cell. 2016;61(2):315–323. doi: 10.1016/j.molcel.2015.12.012. http://dx.doi.org/10.1016/j.molcel.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ratcliff GC, Wang H, Davis-Searles PR, Gray MD, Erie DA, et al. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3’–5’ exonuclease and 5’-protruding strand endonuclease activities. Biochemistry. 2002;41(9):2901–2912. doi: 10.1021/bi0157161. http://dx.doi.org/10.1021/bi0157161. [DOI] [PubMed] [Google Scholar]
- Yang Y, Sass LE, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Research. 2005;33(13):4322–4334. doi: 10.1093/nar/gki708. http://dx.doi.org/10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang H, Erie DA. Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods. 2003;29(2):175–187. doi: 10.1016/s1046-2023(02)00308-0. http://dx.doi.org/10.1016/s1046-2023(02)00308-0. [DOI] [PubMed] [Google Scholar]