Figure 1.
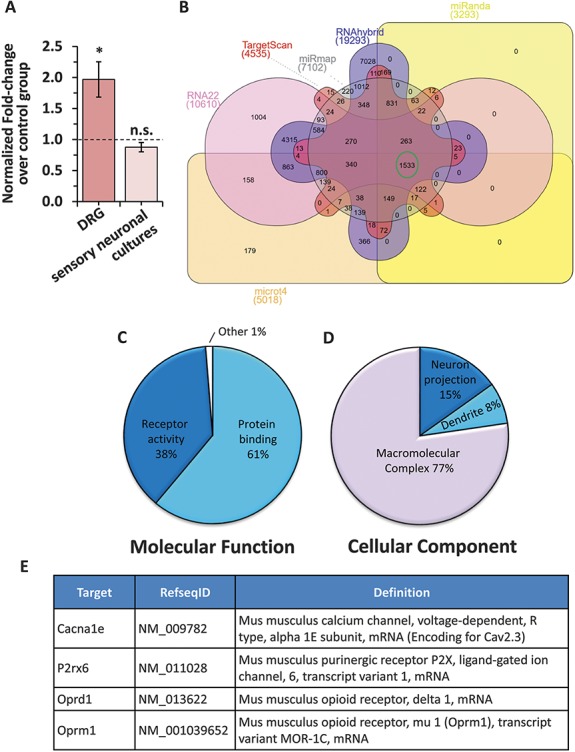
Analyses of previously validated and predicted targets for miR-34c-5p. (A) Change in the expression of Prex2, one of the previously validated targets for miR-34c-5p in DRGs isolated from tumor-bearing mice as compared to sham controls (dark colored bar) or in sensory neurons transfected with a miR-3c-5p specific inhibitor as compared to scramble-inhibitor transfected controls (light colored bar). (B) Venn diagram representation of predicted targets for miR-34c-5p by 6 different target prediction algorithms. The number of genes consistently predicted as targets for miR-34c-5p by all programs is highlighted in a green circle. (C, D) Pie-chart representation of significantly enriched (Adj. P ≤ 0.05 calculated by the multiple test adjustment method as compared to number of reference genes in the category genome-wide) molecular function (C) and cellular component (D) Gene ontology terms by unique genes from the list of 1533 genes commonly predicted as targets for miR-34c-5p by 6 independent algorithms. (E) Annotation details for 4 genes prioritized as putative targets for miR-34c-5p in sensory neurons.
