Arabidopsis nonhost resistance gene PSS1 encoding an unknown glycine-rich plasma membrane protein has shown to enhance sudden death syndrome resistance in transgenic soybean plants.
Abstract
Nonhost resistance is defined as the immunity of a plant species to all nonadapted pathogen species. Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 is nonhost to the oomycete plant pathogen Phytophthora sojae and the fungal plant pathogen Fusarium virguliforme that are pathogenic to soybean (Glycine max). Previously, we reported generating the pss1 mutation in the pen1-1 genetic background as well as genetic mapping and characterization of the Arabidopsis nonhost resistance Phytophthora sojae-susceptible gene locus, PSS1. In this study, we identified six candidate PSS1 genes by comparing single-nucleotide polymorphisms of (1) the bulked DNA sample of seven F2:3 families homozygous for the pss1 allele and (2) the pen1-1 mutant with Columbia-0. Analyses of T-DNA insertion mutants for each of these candidate PSS1 genes identified the At3g59640 gene encoding a glycine-rich protein as the putative PSS1 gene. Later, complementation analysis confirmed the identity of At3g59640 as the PSS1 gene. PSS1 is induced following P. sojae infection as well as expressed in an organ-specific manner. Coexpression analysis of the available transcriptomic data followed by reverse transcriptase-polymerase chain reaction suggested that PSS1 is coregulated with ATG8a (At4g21980), a core gene in autophagy. PSS1 contains a predicted single membrane-spanning domain. Subcellular localization study indicated that it is an integral plasma membrane protein. Sequence analysis suggested that soybean is unlikely to contain a PSS1-like defense function. Following the introduction of PSS1 into the soybean cultivar Williams 82, the transgenic plants exhibited enhanced resistance to F. virguliforme, the pathogen that causes sudden death syndrome.
Nonhost resistance is defined as immunity of an entire plant species against all races or isolates of a nonadapted pathogen species. Examples include fungi and oomycete pathogens that fail to penetrate and propagate in the nonhost plants (Heath, 2000; Mysore and Ryu, 2004; Lipka et al., 2005; Senthil-Kumar and Mysore, 2013; Hadwiger, 2015; Lee et al., 2016). It is widely considered that nonhost resistance mechanisms are multilayered and are often elicited by pathogen-associated molecular patterns (PAMPs; Jones and Dangl, 2006). Upon failure of the pathogens to invade a nonhost due to the activation of basal host resistance triggered by PAMPs (PAMP-triggered immunity), effectors are secreted by the phytopathogens to derive nutrition and interfere with the host defense physiology, leading to the development of susceptibility known as effector-triggered susceptibility. Host plants then express cognate R genes encoding receptors that recognize one or more of these effectors and trigger immunity (effector-triggered immunity), which is manifested commonly as a hypersensitive reaction or programmed cell death (Jones and Dangl, 2006).
A mutant study identified PENETRATION DEFICIENT1 (PEN1), PEN2, and PEN3 genes that confer nonhost immunity of the Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) against the barley (Hordeum vulgare) powdery mildew pathogen Blumeria graminis f. sp. hordei (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). Study of the three PEN genes revealed two parallel nonhost resistance mechanisms that suppress the penetration of B. graminis f. sp. hordei. One mechanism, regulated by PEN1, entails vesicle-mediated secretion of free radicals such as hydrogen peroxide to invasion sites. In the other mechanism, PEN2 and PEN3 regulate the transport of antimicrobial glucosinolates and Trp-derived secondary metabolites to infection sites (Clay et al., 2009; Schulze-Lefert and Panstruga, 2011). Other genes, such as Enhanced Disease Susceptibility1, Phytoalexin-Deficient4, Senescence-Associated Gene101, Mildew Resistance Locus O2, the UDP-glucosyltransferase UGT84A2/BRT1, and the calcium sensor CaM7, were identified to be involved in Arabidopsis nonhost resistance (Lipka et al., 2005; Stein et al., 2006, Nakao et al., 2011; Langenbach et al., 2013; Campe et al., 2016). In addition to these nonhost resistance genes, spatial and temporal changes in the production of stress hormones play major roles in nonhost immunity. For example, the involvement of salicylic acid and jasmonic acid in the expression of nonhost defense in Arabidopsis against nonadapted fungal isolates has been reported (Mellersh and Heath, 2003).
Gly-rich proteins (GRPs) belong to a protein superfamily that is characterized by the presence of a Gly-rich domain arranged in (Gly)n-X repeats. The expression of genes encoding GRPs is tissue specific, and they are often developmentally regulated or modulated by biotic and abiotic factors (Mangeon et al., 2010). GRPs are involved in a variety of functions in plants, including cell wall structure, plant defense, pollen hydration, signal transduction, osmotic stress, cold stress, flowering time control, development, and cell elongation (Mousavi and Hotta, 2005; Mangeon et al., 2010). GRPs take part in plant defense responses by maintaining cell wall components and callose deposition (Ueki and Citovsky, 2002; Lin and Chen, 2014), modulating PR-1 expression (Park et al., 2001), and displaying antimicrobial activity to inhibit the growth of microbes (Park et al., 2000; Egorov et al., 2005; Tavares et al., 2012). Aside from the Gly-rich domain, some GRPs carry RNA-binding domains. Arabidopsis AtGRP7 is a Gly-rich RNA-binding protein that regulates callose deposition in the PAMP flg22-induced FLS2-mediated immunity (Fu et al., 2007). AtGRDP2 encodes a short Gly-rich domain protein containing a DUF1399 domain and a putative RNA recognition motif. Overexpression of AtGRDP2 resulted in higher tolerance of Arabidopsis to salinity stress (Ortega-Amaro et al., 2015).
Arabidopsis is a model plant with T-DNA mutants available for most of its genes, making it suitable for studying nonhost resistance mechanisms (Alonso et al., 2003; Rhee et al., 2003). We previously reported the identification of 30 Phytophthora sojae-susceptible mutants named pss1 through pss30 from screening of over 3,500 ethylmethane sulfonate (EMS)-induced M2 families. The pss1 mutant was shown to be susceptible also to Fusarium virguliforme (Sumit et al., 2012). PSS1 was genetically mapped to chromosome 3 by bulked segregant analysis (Sumit et al., 2012). In this study, we applied SHORE mapping to identify six candidate PSS1 genes (Schneeberger et al., 2009). Analyses of T-DNA insertion mutants of the candidate PSS1 genes led to the identification of the At3g59640 gene that complemented EMS-induced pss1 and two T-DNA insertion-induced pss1 mutants. We showed that PSS1 localizes to the plasma membrane. Furthermore, we identified that, upon stable transformation, PSS1 enhances resistance to the fungal pathogen F. virguliforme in transgenic soybean (Glycine max) plants.
RESULTS
The Nonhost Resistance PSS1 Gene Encodes a GRP
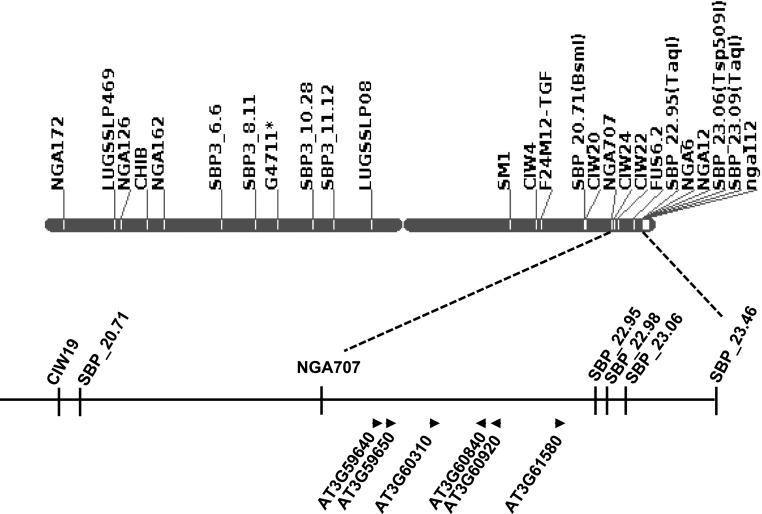
PSS1 was mapped to a 2.75-Mb genomic region between markers SBP_20.71 and SBP_23.46 on chromosome 3 (Sumit et al., 2012; Fig. 1). Comparison of the sequence of the PSS1 region in a bulked DNA sample generated from seven F2:3 homozygous families for the pss1 allele with that of the Col-0 genome sequence revealed 30 point mutations or single-nucleotide polymorphisms. Nine of these mutations were nonsynonymous. The pss1 mutant was generated in the Col-0 pen1-1 mutant background (Sumit et al., 2012). Three of the nine nonsynonymous mutations are common to both pen1-1 and pss1 mutants and were not considered for further study. The six candidate PSS1 genes, each carrying one pss1-specific nonsynonymous mutation, are presented in Figure 1 and Table I.
Figure 1.
Candidate nonhost resistance PSS1 genes. The six putative nonhost-resistant genes are shown in a 1.2-Mb genomic region flanked by NGA707 and SBP_22.95 markers mapped to chromosome 3. The arrowheads indicate the orientations of six candidate PSS1 genes on the Arabidopsis Col-0 genome sequence.
Table I. Six candidate PSS1 genes carrying nonsynonymous mutations between the NGA707 and SBP_22.95 markers mapped to Arabidopsis chromosome 3.
| Single-Nucleotide Polymorphism | Locus | Annotation | Base Change | Substitution |
|---|---|---|---|---|
| 22029832 | AT3G59640 | Gly-rich protein | G-A | Gly/Asp |
| 22033274 | AT3G59650 | Mitochondrial protein | G-A | Gly/Asp |
| 22290347 | AT3G60310 | Unknown protein | G-A | Ala/Thr |
| 22477739 | AT3G60840 | Microtubule-associated protein | G-A | Pro/Leu |
| 22504152 | AT3G60920 | BEACH domain proteins | G-A | Ala/Asp |
| 22786292 | AT3G61580 | Sphingoid LCB desaturase | G-A | Asp/Asn |
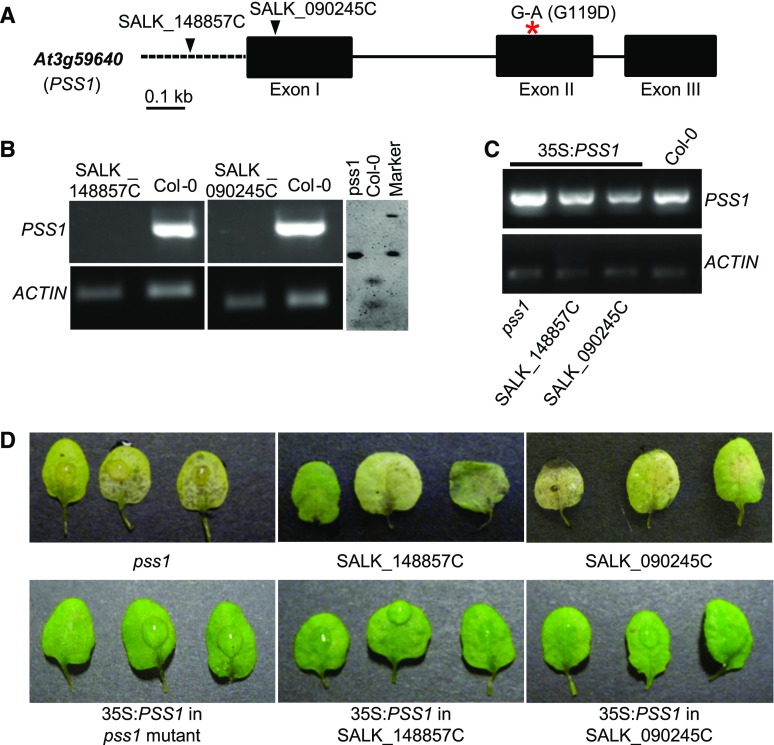
To identify the candidate PSS1 gene, 25 T-DNA knockout mutants for the six candidate PSS1 genes (Supplemental Table S1) were evaluated for response to P. sojae infection. Of the 25 lines tested, only two T-DNA lines, SALK_090245C and SALK_148857C, showed susceptibility to P. sojae. SALK_090245C and SALK_148857C contain T-DNA insertions in exon 1 and the promoter, respectively, of the At3g59640 gene (Fig. 2A). Reverse transcriptase (RT)-PCR failed to detect At3g59640 transcripts in either T-DNA insertion line (Fig. 2B). The pss1 and two T-DNA mutants were transformed with the At3g59640 cDNA fused to the cauliflower mosaic virus (CaMV) 35S promoter. PCR amplification confirmed the stable integration of the At3g59640 transgene in the pss1, SALK_148857C, and SALK_090245C mutants transformed with the 35S:At3g59640 cDNA fusion gene (Fig. 2C). The 35S:At3g59640-transformed pss1 and T-DNA insertion pss1 mutants expressed immunity to P. sojae, suggesting that At3g59640 complemented the lost immunity function of the pss1 mutants (Fig. 2D; Supplemental Fig. S1). Therefore, we concluded that At3g59640 is the PSS1 gene.
Figure 2.
Identification of PSS1 through mutant and complementation analyses. A, Analyses of T-DNA mutants in the At3g59640 gene. The locations of the T-DNA insertions in the At3g59640 gene between the two P. sojae-susceptible T-DNA mutants, SALK_090245C and 148857C, are shown by arrowheads. The red asterisk shows the nonsynonymous transition G-to-A mutation in exon II, which results in the substitution of Gly (G) to Asp (D) at position 119 in the pss1 mutant protein. Black boxes represent three exons, and the lines connecting exons represent introns. The promoter is shown with a dashed line. B, Molecular analyses of pss1 mutants. RT-PCR confirms the absence of PSS1 transcripts in two T-DNA mutants shown at left. The EMS-induced pss1 mutant is confirmed by AciI enzyme digestion of the PCR products of genomic DNA from pss1 and Col-0. Note that the transition mutation led to loss of the restriction site in the pss1 mutant. C, Molecular analyses of the PSS1 cDNA-transformed pss1 mutants. Electrophoresis is shown for PCR-amplified PSS1 gene sequences from the EMS-induced pss1 mutant and the SALK_148857C and SALK_090245C T-DNA mutants transformed with the 35S:PSS1 cDNA gene. D, PSS1 complemented the pss1 mutants. Phenotypes of the pss1 and two T-DNA insertion mutants in the At3g59640 gene and their respective complemented transgenic plants 3 d following P. sojae infection are shown.
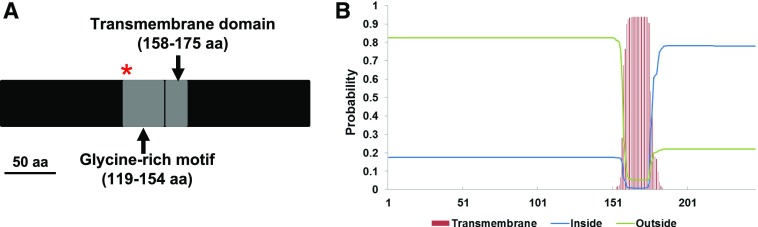
PSS1 encodes a GRP with unknown function. Apart from the Gly-rich motif (amino acids 119–154; Fig. 3A), PSS1 also contains a predicted transmembrane motif (amino acids 158–175; Fig. 3B). In the EMS-induced pss1 mutant allele, a Gly residue is substituted with an Asp residue at position 119. This mutation is located within the conserved Gly-rich domain (Fig. 3A). We hypothesized that the change in this conserved residue led to a change in the protein structure of PSS1 and a loss of the immunity function. To investigate this, we predicted structures of PSS1 and its mutant proteins using the I-TASSER program (Zhang, 2008). Pairwise structure alignment suggested that the pss1 mutant protein possesses low structural similarity to PSS1 (TM-align score = 0.26, less than the threshold of 0.5; Supplemental Fig. S2; Zhang and Skolnick, 2005).
Figure 3.
PSS1 encodes a GRP containing a putative Gly-rich motif and a transmembrane domain. A, Schematic diagram of the PSS1 protein. The red asterisk indicates the substitution of Gly-119 with the Asp residue in pss1 mutant, and two gray boxes represent a Gly-rich motif (amino acid residues [aa] 119–154) and a transmembrane domain (amino acid residues 158–175). B, Predicted transmembrane helix between amino acid residues 158 and 175 of PSS1 (greater than 90% certainty).
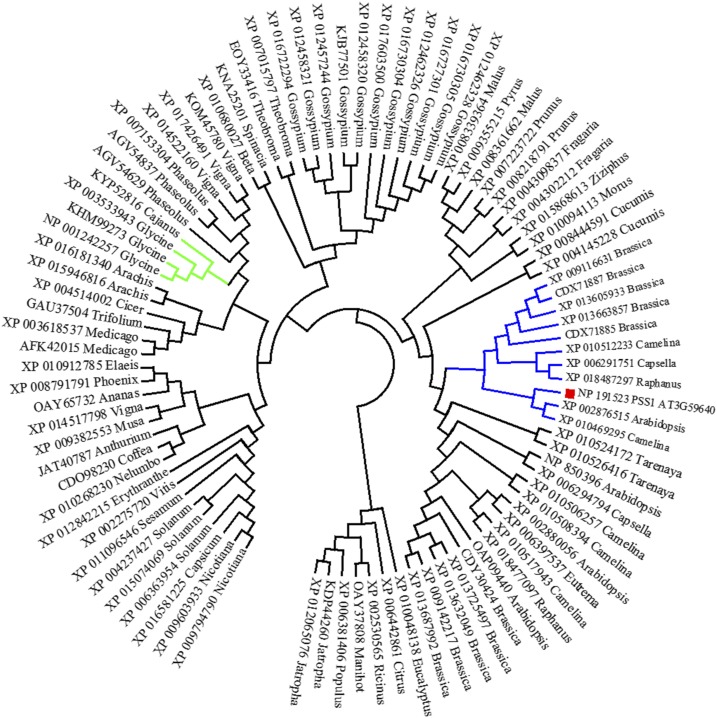
Based on the arrangement of Gly-rich units, PSS1 is classified as a member of the class VII GRPs that carry a mixed pattern of Gly-rich repeats (Mangeon et al., 2010). A sequence similarity search with BLASTP identified 93 plant proteins with amino acid identity greater than 33% to PSS1 with E < 1e-25. None of the 93 PSS1 homologs have been characterized. A few of them have been predicated to be Major Histocompatibility Complex Class II Regulatory Factor (XP_013615797; Brassica oleracea), Autophagy-Related Protein3 (JAT40787; Anthurium amnicola), and Nuclear Envelope Protein (NP_850396; Arabidopsis). The constructed neighbor-joining phylogenetic tree revealed that PSS1 clustered in a subclade with 10 homologs of the Brassicaceae family (Fig. 4). Alignment of these 10 PSS1 homologs and PSS1 revealed that the Gly-rich motifs and transmembrane domains were highly conserved among these GRPs (Supplemental Fig. S3). The soybean PSS1 homologs were placed in a distinct subclade (Fig. 4). Further study is warranted to determine if any of the genes is orthologous to PSS1 and governs any defense function.
Figure 4.
Phylogenetic tree of the PSS1 homologs. Ninety-three PSS1 homologs were used to construct the phylogenetic tree. PSS1 is denoted with the red rectangle. The subclade containing PSS1 is shown in blue, whereas the subclade with soybean PSS1 homologs is presented in green. The percentage identity between PSS1 and soybean homologs is 38% or less.
PSS1 Is Induced in Response to P. sojae Infection
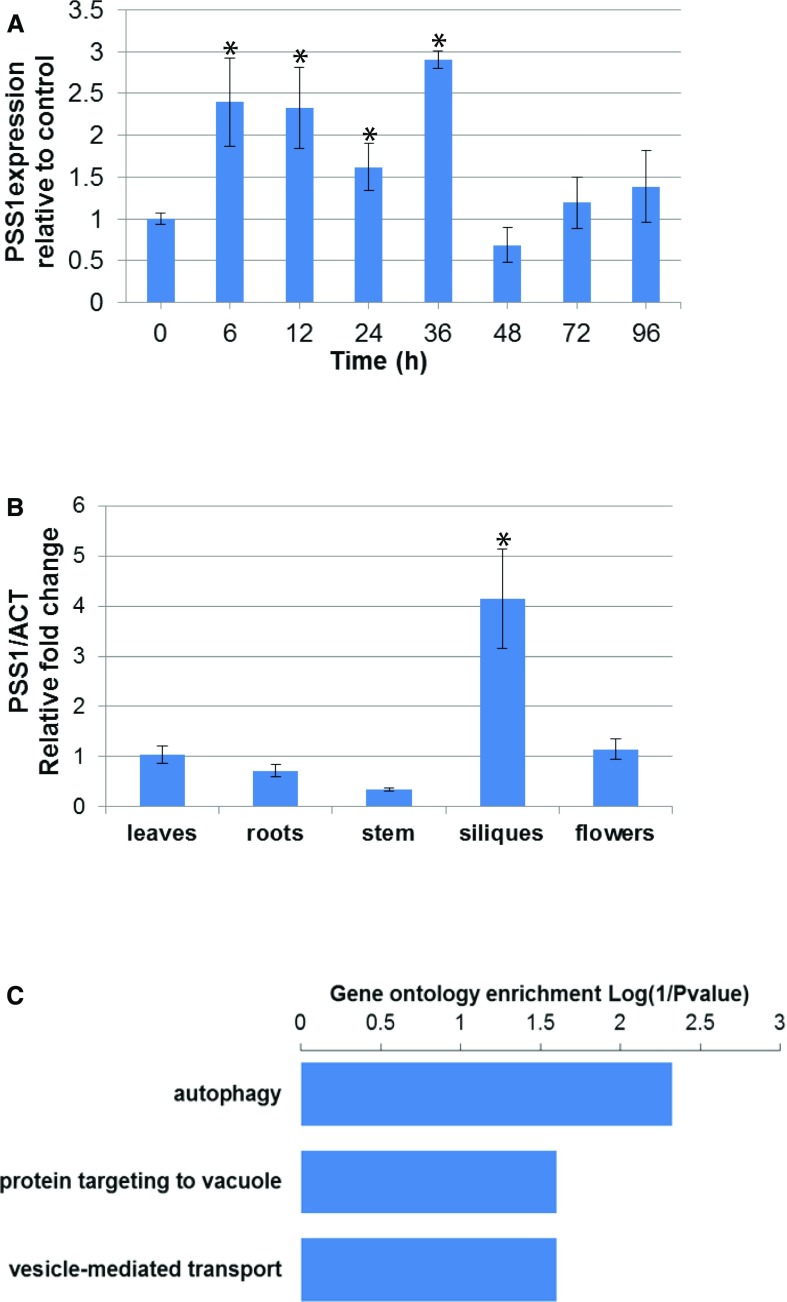
PSS1 has shown no homology to any protein with known function (searched on August 16, 2017). pss1 and knockout T-DNA insertion mutants did not show any discernible defects in general growth and root development (data not shown). To address the biological function of PSS1, quantitative reverse transcriptase (qRT)-PCR was performed. The PSS1 expression pattern also was examined by searching the Arabidopsis eFP Browser (http://bar.utoronto.ca/), which contains an extensive collection of gene expression microarray data (Winter et al., 2007). PSS1 is induced following infection not only with P. sojae (Fig. 5A) but also at least 1.5-fold following infection with several pathogens, including Golovinomyces orontii, Hyaloperonospora arabidopsidis, and Pseudomonas syringe, as well as in response to treatments with various elicitors, including Hrpz and flg22 (Supplemental Table S2). PSS1 expression is highest in siliques (Fig. 5B).
Figure 5.
Expression of PSS1 and genes that show expression patterns similar to PSS1. A, Expression of PSS1 following P. sojae infection. qRT-PCR of PSS1 was conducted following inoculation of Arabidopsis leaves with P. sojae in three independent experiments. The fold change values are relative to the mock control. PSS1 expression levels with asterisks were significantly induced (P < 0.05) when compared with the 0-h control. B, Expression patterns of PSS1 among various Arabidopsis tissues. qRT-PCR expression data of PSS1 were collected among Arabidopsis organs in three independent experiments. Expression comparison was against the levels in leaves (P < 0.05). Data in A and B are from three biological replications, and data were standardized against the transcript levels of the Actin gene. C, Gene Ontology enrichment (biological process) of PSS1 coexpression genes. The coexpression gene analysis was based on the mRNAseq data set using the software Genevestigator (Hruz et al., 2008).
Coexpressed genes with the same transcriptional regulatory pathway could be functionally related, or they could be members of the same biochemical or regulatory pathway or protein complexes. We conducted initial coexpression analysis using a data set from a microarray platform available at ATTED-II (http://atted.jp/; Obayashi et al., 2007). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (Kanehisa and Goto, 2000) enrichment analyses suggested that the coexpression network is related to three biological functions: autophagy, para-aminobenzoic acid metabolic process, and nuclear mRNA splicing via spliceosome (Supplemental Fig. S4; Kerrien et al., 2007). To avoid any biases resulting from the use of a single data set, coexpression analysis was conducted also for the mRNA sequencing (mRNAseq) data set available at Genevestigator (Hruz et al., 2008). Gene Ontology enrichment analysis of the top 25 coexpressed genes suggested that the genes were putatively involved in three biological processes: (1) autophagy, (2) protein targeting to the vacuole, and (3) vesicle-mediated transport (Fig. 5C). Utilization of different gene expression data sets is expected to yield reliable information (Ballouz et al., 2015). To validate the outcomes of the coexpression analyses (Fig. 5C; Supplemental Fig. S4), we conducted semiquantitative RT-PCR of eight genes (Supplemental Table S3) selected from both mRNAseq and microarray data sets and observed that the autophagy-related gene ATG8a (At4g21980) is induced upon P. sojae infection (Supplemental Table S3). ATG8a is the core gene in autophagy (Yoshimoto et al., 2004). These results indicate a possible connection of PSS1 to an autophagy-related defense mechanism (Liu et al., 2005).
PSS1 Was Localized to the Plasma Membrane
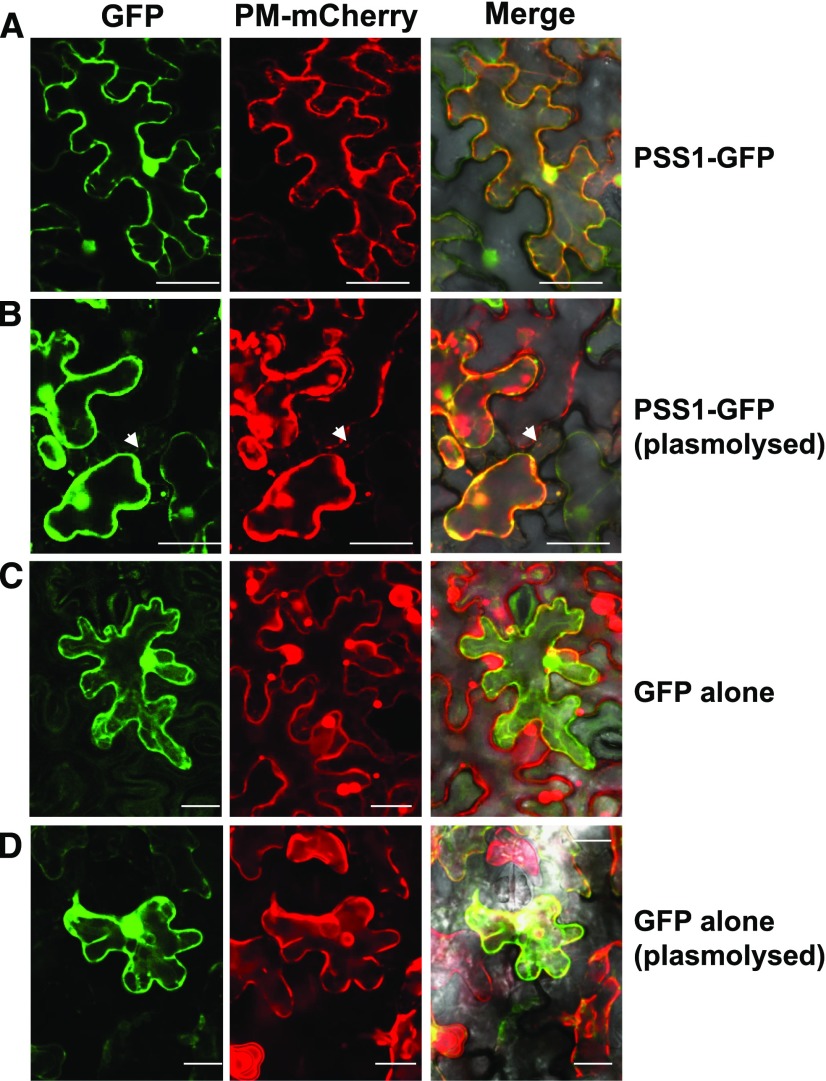
Investigation of PSS1 using a transmembrane prediction program (http://cbs.dtu.dk/services/TMHMM/) revealed that it contains a single transmembrane domain between residues 158 and 175 (Fig. 3B). The Cell eFP browser (http://bar.utoronto.ca/cell_efp/cgi-bin/cell_efp.cgi; Winter et al., 2007) predicated that PSS1 could reside in nuclei, mitochondria, chloroplast, and plasma membrane. SignalP 4.0, however, did not identify any secretory signal peptides in PSS1. We expressed enhanced GFP (eGFP)-fused PSS1 (GFP-PSS1 and PSS1-GFP) transiently in Nicotiana benthamiana (Fig. 6). The plasma membrane protein AtPIP2A fused to mCherry was used as a plasma membrane marker (Nelson et al., 2007) and eGFP alone as a control. Two days after coinfiltration, the GFP fluorescence signals of PSS1-GFP were detected as sharp, thin lines at the cell periphery and overlapped with the red fluorescence of the mCherry-fused plasma membrane marker (Fig. 6). After plasmolysis, colocalization of PSS1-GFP with the plasma membrane marker was retained and detached from cell wall, and obvious Hechtian strands were observed (Supplemental Fig. S5B), a characteristic of plasma membrane proteins. These results suggest that PSS1 is most likely an integral plasma membrane protein, not a cell wall protein. The GFP-PSS1 fusion protein with GFP at the N terminus exhibited loss of its plasma membrane localization; instead, it showed cytoplasmic localization, similar to eGFP (Supplemental Fig. S5A). This suggests that the signal for plasma membrane localization in PSS1 is most likely located at the N terminus.
Figure 6.
PSS1 is localized to the plasma membrane. A, PSS1-GFP fusion and mCherry-tagged plasma membrane (PM) marker Arabidopsis PIP2A colocalized to plasma membrane of the epidermal cells of N. benthamiana. B, The colocalized PSS1-GFP and PIP2A-mCherry fluorescent proteins remain as a complex following plasmolysis with 1 m NaCl. C, Control GFP fluorescent protein was localized to cytoplasm. D, Plasmolysis of the cell coexpressing the GFP and PIP2A-mCherry proteins. White arrowheads indicate Hechtian strands (for details, see Supplemental Fig. S5). Bars = 50 μm for PSS1-GFP and 25 μm for GFP alone.
PSS1 Transgenic Soybean Lines Exhibited Enhanced Sudden Death Syndrome Resistance
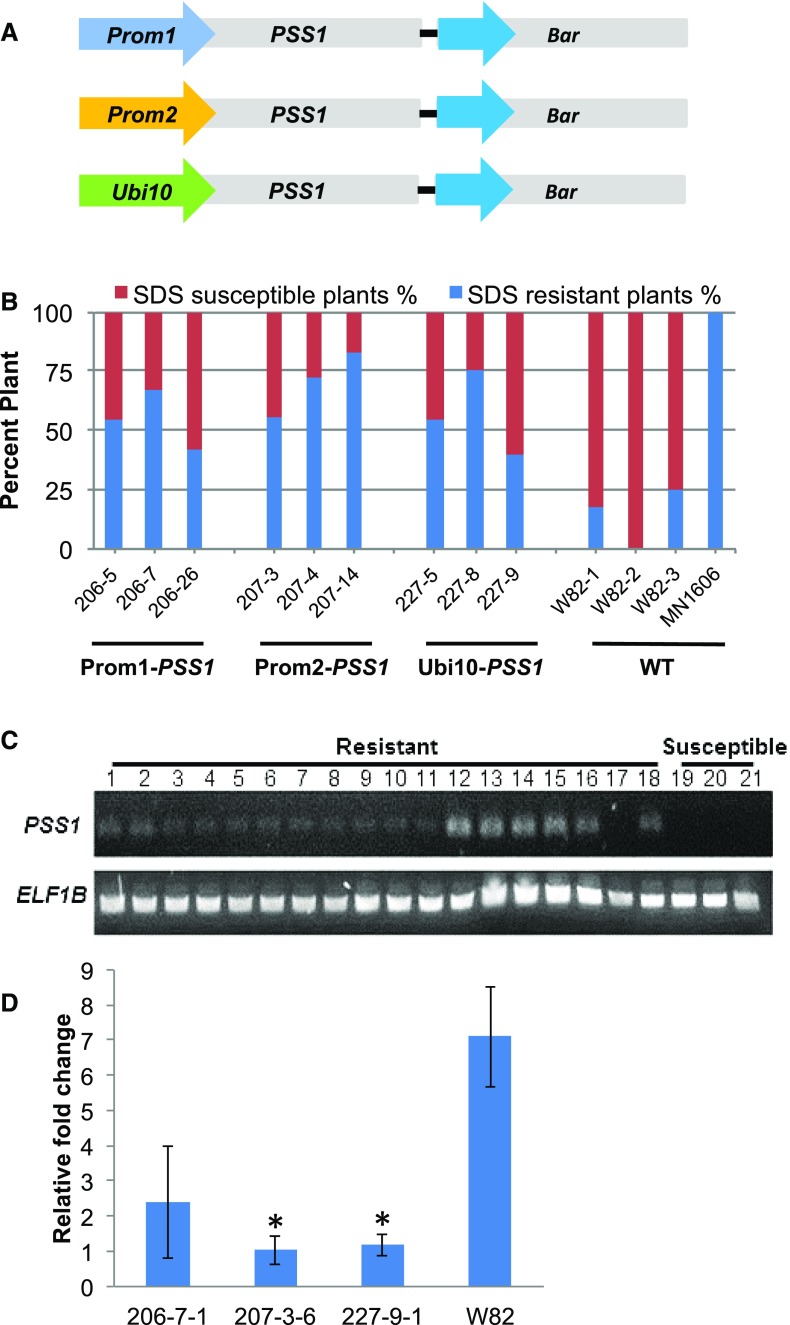
The soybean cv Williams 82 was transformed with the PSS1 cDNA fused individually to three promoters: (1) Prom1, a soybean F. virguliforme Mont-1 infection-inducible promoter (Glyma18g47390; B.B. Sahu and M.K. Bhattacharyya, unpublished data); (2) Prom2, a soybean root-specific promoter (Glyma10g31210; http://www.oardc.ohio-state.edu/SURE/GmROOT/GmRoot.htm; Ngaki et al., 2016); and (3) Ubi10, an Arabidopsis constitutive promoter (At4g05320; Norris et al., 1993; Fig. 7A). R1 seeds were collected from R0 soybean plants grown in the greenhouse. Transgenic soybean plants carrying PSS1 did not show any obvious changes in morphology compared with the nontransgenic recipient cv Williams 82. To examine the responses of transgenic plants to F. virguliforme, seeds were planted in a soil premixed with F. virguliforme Mont-1 isolate and the seedlings were grown in growth chambers. The cv Williams 82 is a moderately susceptible line, while MN1606 is a sudden death syndrome (SDS)-resistant line. Foliar SDS symptoms were recorded 4 weeks after planting. Approximately one-third to two-thirds of the selected R1 plants showed enhanced SDS resistance, with disease severity ratings of less than 2.0 (Fig. 7B). The nontransgenic cv Williams 82 line exhibited SDS severity ratings of over 4.0 among 90% of the plants, and 90% of MN1606 plants showed disease ratings of less than 2.0. RT-PCR analysis indicated that PSS1 transcripts were present in all SDS-resistant R1 progeny but not in the SDS-susceptible R1 progeny (Fig. 7C).
Figure 7.
Expression of PSS1 enhances SDS resistance in transgenic soybean plants under growth chamber conditions. A, Schematic depiction of promoter-PSS1 fusion genes along with the CaMV 35S promoter-fused bar gene in three binary plasmids used to generate transgenic soybean plants. B, Responses of the transgenic lines to root infection with F. virguliforme Mont-1 in growth chambers. Plants with foliar SDS scores of 2 or less were considered resistant, and those with scores greater than 2 were considered susceptible. Percentage resistant and susceptible R1 progeny are presented for each of the PSS1 transgenes generated by fusing the PSS1 gene to Prom1, Prom2, and Ubi10 promoters. For each transgenic event, 15 R1 plants were studied. The experiment was repeated two more times and showed similar results. Foliar SDS symptoms for individual plants were scored 4 weeks following planting. MN1606, SDS-resistant control; WT, transgene recipient nontransgenic cv Williams 82 (W82) as the SDS-susceptible control. C, RT-PCR analysis of the transgenic R1 plants for PSS1 transcripts. Lanes 1 to 6, RT-PCR products from F. virguliforme-infected roots of three independent lines carrying promoter Prom1 fused to PSS1; lanes 7 to 12, RT-PCR products from F. virguliforme-infected roots of three independent lines carrying promoter Prom2 fused to PSS1; and lanes 13 to18, RT-PCR products from F. virguliforme-infected roots of three independent lines carrying promoter Ubi10 fused to PSS1. For each independent transgenic line, two R1 SDS-resistant plants (lanes 1–18) were analyzed. Lanes 19 to 21, RT-PCR products from F. virguliforme-infected roots of three independent R1 progeny plants that were SDS susceptible. D, Relative biomasses of F. virguliforme measured by genomic DNA qPCR of the FvTox1 gene among three independent transgenic lines. Root samples were collected 2 weeks following infection with the F. virguliforme Mont-1 isolate in a growth chamber. 206-7-1, Transgenic line carrying Prom1-PSS1; 207-3-6, transgenic line carrying Prom21-PSS1; 227-9-1, transgenic line carrying the Ubi10-1-PSS1 fusion gene; W82, cv Williams 82. Asterisks indicate statistical significance at P < 0.05 when compared with the biomass of F. virguliforme in cv Williams 82.
Foliar SDS is induced by toxins produced by F. virguliforme in infected roots (Ji et al., 2006; Brar et al., 2011). We hypothesized that overexpressed PSS1 in roots conferred enhanced root resistance against the pathogen. To test our hypothesis, seeds of three SDS-resistant R2 plants, each representing one of the three transgenes (Prom1-PSS1, Prom2-PSS1, and Ubi10-PSS1), were planted in soil mixed with F. virguliforme Mont-1 inoculum in a growth chamber. Genomic DNA quantitative PCR (qPCR) was conducted for the single-copy F. virguliforme FvTox1 toxin gene (Brar et al., 2011). The qPCR revealed that the levels of F. virguliforme growth in the roots of SDS-resistant transgenic soybean plants expressing the PSS1 gene under the control of Prom1, Prom2, or Ubi10 was decreased up to 85% as compared with that in cv Williams 82 (Fig. 7D). These results suggest that PSS1 suppressed the extent of F. virguliforme’s spread in the infected roots.
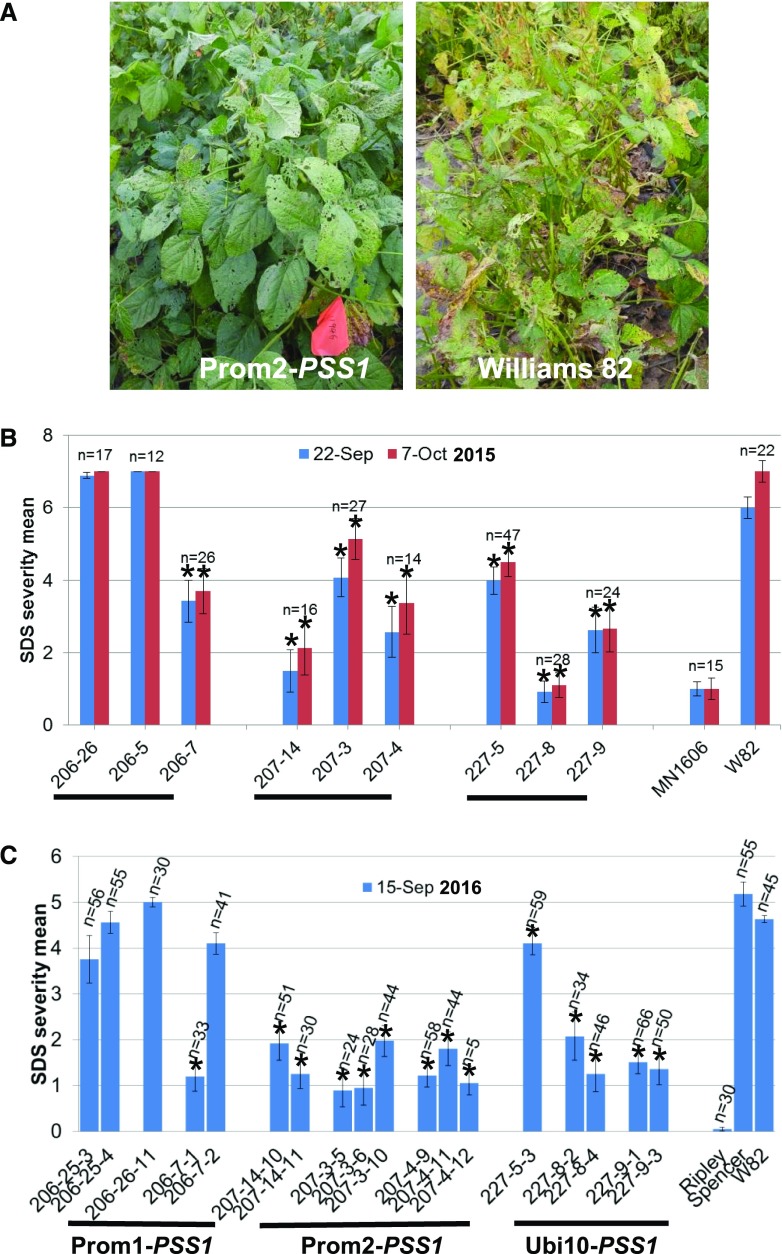
To determine if transgenic SDS-resistant plants showed enhanced SDS resistance under field conditions, field trials were conducted at Hinds Farms, Iowa State University, in the 2015 and 2016 seasons. The transgenic soybean lines carrying PSS1 transgenes showed enhanced SDS resistance under field conditions (Fig. 8A). In each season, transgenic seeds were planted along with the control lines cv Williams 82, MN1606, or Ripley. The F. virguliforme NE305S isolate grown on sorghum (Sorghum bicolor) grains was mixed with seeds prior to sowing. In 2015, R1 seeds were sown. Three weeks after seed germination, Basta was sprayed on transgenic lines to eliminate any azygous progeny. We sprayed Basta at early vegetative growth stages. Foliar SDS symptoms appear after flowering, and foliar symptom severity was recorded at the reproductive stage (R6 stage, the stage at which the weight of developing pods peaks). There was a gap of 4 weeks between Basta spray and SDS symptom development. Once symptoms started to appear, the SDS severity of each plant was scored. Transgenic lines, 207-4 and 207-14 expressing Prom2-driven PSS1, 227-8 and 227-9 carrying Ubi10 promoter-fused PSS1, and 206-7 carrying promoter Prom1-fused PSS1, showed significantly enhanced SDS resistance under field conditions (P < 0.05; Fig. 8B). Leaves of SDS-resistant plants were collected just after the first scoring of foliar SDS severity to determine the transgene copy number (Supplemental Table S4). The harvested R2 seeds of putative homozygous R1 plants were grown in the 2016 field trial. In 2016, Basta herbicide was applied to eliminate any possible azygous segregants from heterozygous R1 plants. The SDS severity index indicated that transgenic lines with promoter Prom2- and Ubi10-driven PSS1 showed enhanced SDS resistance (Fig. 8C).
Figure 8.
Expression of PSS1 enhances SDS resistance in transgenic soybean plants under field conditions. A, Representative field plot showing SDS-resistant transgenic and cv Williams 82 control plants. B and C, Mean foliar SDS severity for individual transgenic lines in the 2015 and 2016 field trials. Each line comprised 12 to 66 Basta-resistant R1 or R2 seedlings. The experiment was conducted in a randomized block design. Asterisks indicate significant reductions in foliar SDS scores between transgenic lines and the nontransgenic recipient cv Williams 82 (W82) control at P < 0.05.
DISCUSSION
In this investigation, we applied a map-based cloning approach to isolate the Arabidopsis nonhost resistance PSS1 gene. The gene was mapped to a 2.75-Mb genomic region. A bulked DNA sample of seven F2:3 lines homozygous for the pss1 allele was sequenced to identify the candidate PSS1 gene through SHORE mapping (Schneeberger et al., 2009). Analyses of T-DNA insertion lines and complementation analyses confirmed that PSS1 encodes a GRP. Mutations in this gene led to a loss of immunity of Arabidopsis to two soybean pathogens, P. sojae and F. virguliforme, but not to the bacterial pathogen, Pseudomonas syringae pv glycinea, which causes bacterial blight in soybean (Sumit et al., 2012).
The pss1 mutant was created in the pen1-1 genetic background because P. sojae can penetrate single cells of the pen1-1 mutant (Sumit et al., 2012). Ecotype Col-0, on the other hand, is not penetrated by P. sojae. Therefore, we expected to observe an epistatic effect of PEN1 on PSS1 if PSS1 was to encode a second layer of plant defense. Surprisingly, a 3:1 segregating ratio was observed for the pss1 mutation (PSS1:pss1::3:1), suggesting a single gene action with no epistasis effect of PEN1 on PSS1 (Sumit et al., 2012). This observation was further supported by responses of two T-DNA insertion lines, SALK_090245C and SALK_148857C. Both mutants are susceptible to P. sojae, although both carry the PEN1 allele (Fig. 2). Together with the previous study (Sumit et al., 2012), our study suggests that PSS1 may act at both prehaustorial and posthaustorial levels, while PEN1 acts at the prehaustorial level against this soybean pathogen.
PSS1 encodes GRP1. GRPs are classified into seven classes based on the pattern of their Gly-rich domain. PSS1 belongs to group VII, which carry a mixed arrangement of Gly repeats with no other conserved domains (Mangeon et al., 2010).
PSS1 confers broad-spectrum nonhost immunity of Arabidopsis to two soybean pathogens (Sumit et al., 2012). Transgenic studies in soybean have suggested the utility of this gene in enhancing disease resistance in crop plants. We have localized the protein to the plasma membrane through its transient expression in N. benthamiana (Fig. 6). The broad-spectrum disease resistance function and its putative plasma membrane location suggest a possible recognition/signaling role for the protein in the activation of host defense responses. However, we cannot rule out the possibility of other mechanisms, including possible structural and/or chemical barriers mediated by PSS1.
GRPs are involved in multiple functions in the plant defense response, such as blocking virus movement, interacting with kinases, and modulating the transcription of defense genes (Park et al., 2001, 2008; Ueki and Citovsky, 2002, 2005; Tao et al., 2006; Kim et al., 2007, 2015; Nicaise et al., 2013). PSS1 is induced by many pathogens. PAMPs such as flagellin (flg22), harpin (HrpZ), necrosis-inducing proteins, and lipopolysaccharide also can induce its expression. Surprisingly, the bacterial PAMP HrpZ is Gly rich and triggers the hypersensitive response at the infection site (Choi et al., 2013).
The subcellular localization and predicted protein structure suggest that PSS1 is an integral plasma membrane protein carrying one membrane-spanning domain. Plant immunity is regulated at both transcriptional and posttranscriptional levels. Pre-RNAs of target regulatory genes must be processed correctly to regulate defense responses. GRPs with an RNA-binding domain have been suggested to play a role in RNA processing (Woloshen et al., 2011). Alternate splicing has been documented as essential for the expression of effector-triggered immunity in tobacco (Dinesh-Kumar and Baker, 2000). Whether PSS1 has any role in RNA splicing has yet to be investigated.
The functions of coexpressed genes showing the same transcriptional regulatory pathway could be used in predicting the functions of genes. RT-PCR of seven genes that were coexpressed with PSS1 in mRNA sequencing or transcript hybridization to microarrays studies indicated that PSS1 coexpresses with the core autophagy gene ATG8a (At4g21980; Supplemental Table S3; Yoshimoto et al., 2004). Autophagy is a conserved intracellular trafficking and degradation process and has been shown to be linked to the induction of programmed cell death or the hypersensitive response as part of basal plant immunity (Liu et al., 2005; Teh and Hofius, 2014). It will be important to determine if PSS1 is involved in autophagy-mediated plant immunity.
In recent years, SDS has emerged as the second most serious soybean disease after soybean cyst nematodes in the United States; in certain years, it can cause yield suppression valued up to $0.7 billion (Bradley and Allen, 2014). Although first reported in 1971, the fungal pathogen F. virguliforme causing SDS has spread to all soybean-growing states in the United States and Canada (Ngaki et al., 2016). Currently, the use of SDS-resistant cultivars is the only option available to manage this disease. However, breeding SDS-resistant cultivars is not trivial, since the SDS resistance is partial and governed by more than 40 quantitative trait loci (Swaminathan et al., 2016). The pathogen is soil borne and remains in infected roots, where it produces toxins that cause the foliar SDS (Brar et al., 2011; Brar and Bhattacharyya, 2012; Pudake et al., 2013; Chang et al., 2016). The development of transgenic SDS-resistant lines is a possible alternative to combat this disease. The transgenic expression of plant antibodies or interacting peptides that bind to foliar SDS-inducing toxins has shown some promise in enhancing SDS resistance in soybean (Brar and Bhattacharyya, 2012; Wang et al., 2015; B. Wang and M.K. Bhattacharyya, unpublished data).
Our study suggests that nonhost disease resistance governed by PSS1 can enhance SDS resistance in transgenic soybean plants by restricting the spread of fungal growth in the infected roots (Fig. 7D). It is very unlikely that the enhanced SDS resistance in the transgenic lines was induced by Basta spray, as was observed in an earlier study conducted in transgenic rice (Oryza sativa; Ahn, 2008), because of the following reasons. In the growth chamber assays, we never sprayed Basta (Fig. 7). Second, although we sprayed Basta in both the 2015 and 2016 growing seasons on the field-grown soybean plants, not all transgenic soybean lines were SDS resistant; some were as susceptible as the nontransgenic cv Williams 82 plants (Fig. 8).
The PSS1-encoded resistance mechanism could complement the natural SDS resistance mechanisms and be suitable in breeding SDS-resistant soybean lines. Considering the widespread cultivation of transgenic soybean worldwide (e.g. about 94% of the soybean crop grown in the United States and 81% worldwide are transgenic; Perry et al., 2016), the development of SDS-resistant transgenic plants could be a good alternative to facilitate soybean breeding programs for SDS resistance.
In summary, PSS1 encodes a novel unknown mechanism to confer nonhost resistance of Arabidopsis against two important soybean pathogens, P. sojae and F. virguliforme. Its plasma membrane localization and induction in response to infection by multiple pathogens and treatment with PAMPs suggest its possible regulatory role in plant defenses. It is possible that PSS1 may confer its plant immunity function through autophagy. The transgenic study of PSS1 revealed that the transfer of nonhost resistance genes could be an important strategy in engineering disease resistance in crop plants.
MATERIALS AND METHODS
Plants and Pathogens
Arabidopsis (Arabidopsis thaliana) plants including the wild-type ecotypes Col-0 and Niederzenz as well as mutants were grown on LC1 soil (Sun Gro Horticulture) in growth chambers at 21°C and 60% humidity with a dark/light cycle of 8/16 h and a light intensity of 100 μmol m−2 s−1. Soybean (Glycine max) ‘Williams 82’ and transgenic lines were grown on Metro Mix 910 (Sun Gro Horticulture) at 23°C and 60% humidity with a dark/light cycle of 8/16 h and a light intensity of 300 μmol m−2 s−1 in growth chambers. The Phytophthora sojae NW strain was maintained on V8 agar plates, and Fusarium virguliforme isolates were maintained on PDA plates.
Nonhost-Resistant Gene Cloning
The PSS1 gene was mapped previously to the lower arm of chromosome 3 between markers SBP_20.71 and SBP_23.46 by conducting bulked segregation analysis in a segregating population developed from a cross between the pss1 mutant and Niederzenz (Sumit et al., 2012). Subsequently, genomic DNA of seven homozygous susceptible pss1/pss1 F2:3 families was extracted using the CTAB method (Murray and Thompson, 1980) and bulked for sequencing on the Illumina HiSeq 2500 platform at the Iowa State University DNA Facility. The short sequencing reads were assembled into contigs, which were aligned to the reference Col-0 sequence to identify mutations in the 2.75-Mb pss1 region using the SHORE program (Schneeberger et al., 2009). Because the pss mutants were developed in the pen1-1 mutant, any mutations originating from pen1-1 were not considered for further analysis.
Homozygous T-DNA knockout lines for the candidate PSS1 genes carrying nonsynonymous mutations were obtained from the Arabidopsis Biological Resource Center located at Ohio State University, and individual T-DNA mutant lines were verified by PCR amplification (Supplemental Table S1). Leaves of 3-week-old T-DNA insertion mutant lines, pss1, pen1-1, and Col-0 plants were inoculated with 20 µL of P. sojae NW zoospores (5 × 105 mL−1) as described previously (Sumit et al., 2012). Symptoms were scored 3 and 4 d after inoculation.
To complement pss1 and T-DNA insertion mutant lines, the nonhost-resistant cDNA was amplified by RT-PCR from the Col-0 transcripts and inserted into the binary vector pTF101.1 under the control of the CaMV 35S promoter. Sequencing was performed to confirm the identity of the PSS1 gene. The resulting construct was transformed into Agrobacterium tumefaciens strain EHA101 by following the freeze-thaw method. pss1 and T-DNA insertion mutant lines were transformed by conducting floral dip of the mutants with the A. tumefaciens EHA101 isolate carrying the candidate PSS1 gene (Weigel and Glazebrook, 2006). T1 and T2 progeny were screened for Basta resistance by spraying with Liberty (80 µg mL−1) herbicide. T3 plants along with the controls Col-0, pss1 mutant, and T-DNA insertion lines were inoculated with P. sojae spores to examine their disease phenotypes.
qRT-PCR
For qRT-PCR analyses of the PSS1 gene, three leaves of 3-week-old pss1 mutant plants were inoculated with 20 µL of P. sojae NW zoospores (5 × 105 mL−1) or water. Leaf samples were collected 6, 12, 24, 36, 48, 72, and 96 h after inoculation in three independent experiments. Total RNA was extracted using the SV Total RNA isolation kit (Promega). The isolated RNAs were reverse transcribed into cDNA using the SuperScript first-strand synthesis system (Thermo Fisher Scientific). Transcript amounts of the PSS1 and Actin genes were examined by conducting qRT-PCR with PSS1- and Actin-specific primers (Supplemental Table S5). qRT-PCR was conducted using SYBR Green master mixes (Thermo Fisher Scientific) by following the manufacturer’s instruction manual. For the study of tissue-specific expression of PSS1 in various tissues, including stem, roots, flowers, leaves, and siliques, RNA extraction was conducted as described earlier for leaves. The induced fold changes in PSS1 expression were calculated against the mock control.
To quantify the pathogen biomass in infected soybean roots, a genomic DNA-PCR was conducted for the DNA isolated from the transgenic and nontransgenic cv Williams 82 soybean plants infected with F. virguliforme Mont-1. DNA was diluted to 20 ng μL−1 for qPCR to quantify the single-copy F. virguliforme FvTox1 gene (Brar et al., 2011) as a measure of fungal biomass. The single-copy soybean gene, Glyma.05G014200, was used as an internal control. qPCR was run in an iQ5 Bio-Rad instrument using the SYBR Green protocol. The primers used for qPCR of Glyma.05G014200 were evaluated earlier (Ngaki et al., 2016) and are presented in Supplemental Table S5. For qPCR of FvTox1, primers developed previously for the quantification of FvTox1 and F. virguliforme biomass were used (Mbofung et al., 2011; Supplemental Table S5).
RT-PCR
To investigate the expression of PSS1 and identified genes that are coexpressed with PSS1, leaves of Col-0 and pss1 were inoculated with 20 µL of P. sojae NW isolate zoospore suspension (105 zoospores mL−1). Inoculated leaves were harvested in a time course (0, 6, 12, and 24 h postinoculation), and the RNAs were isolated and subjected to RT-PCR using primers specific for each gene (Supplemental Table S5). The Arabidopsis Actin gene was used as an internal control. The intensity of PCR bands of individual samples was quantified by using ImageJ (http://rsb.info.nih.gov/ij/index.html). We followed the procedure outlined in the ImageJ document to collect the pixelated data (https://imagej.nih.gov/ij/docs/user-guide-A4booklet.pdf, p. 129). The data from three independent experiments were analyzed for statistically significant differences for P. sojae-infected and uninfected water control leaf tissues for eight genes, including PSS1, using the open-source R program (Supplemental Table S3).
Subcellular Localization of PSS1
The PSS1 gene was fused at the N and C termini of GFP and cloned in pISUAgron5 vector (S. Li, N.N. Narayanan, and M.K. Bhattacharyya, unpublished data), in which eGFP is already fused to the CaMV 35S promoter. pISUAgron5 vector was used as the GFP control. A plasma membrane marker, Arabidopsis PIP2A fused to the mCherry tag, was obtained from the Arabidopsis Biological Resource Center (Nelson et al., 2007). For A. tumefaciens-mediated transient transformation, individual A. tumefaciens isolates containing each of the two GFP fusion constructs or control GFP construct were coinfiltrated with plasma membrane marker into leaves of 4-week-old Nicotiana benthamiana plants (Shamloul et al., 2014). Two days following infiltration, small leaf pieces were mounted in either water or 1 m NaCl. Samples were observed with a 20× oil-immersion lens mounted to a Leica SP5 X MP confocal/multiphoton inverted microscope. To monitor GFP fluorescence, a 488-nm argon laser and PMT detector with emission bandwidth set to 495 to 550 nm were used. To monitor the mCherry signal, a HeNe 561 laser (561 nm) and a third PMT detector (587–610 nm) were used (Schweiger and Schwenkert, 2014).
Generation of Transgenic Soybean Lines
The gene PSS1 was first cloned into vector pGEM-T (Promega) and sequenced to confirm its identity. The gene was then released from the pGEM-T vector and cloned in the modified binary pTF102 vectors carrying one of three promoters: Prom1, Prom2, and Ubi10. Prom1 is a soybean infection-inducible promoter (Glyma18g47390; B.B. Sahu and M.K. Bhattacharyya, unpublished data). Prom2 is a soybean root-specific promoter (Glyma10g31210; http://www.oardc.ohio-state.edu/SURE/GmROOT/GmRoot.htm; Ngaki et al., 2016). The Ubi10 promoter was isolated from the Arabidopsis At4g05320 gene (Norris et al., 1993). The resulting three constructs were transformed into A. tumefaciens strain EHA101 to generate stable transformants in the soybean cv Williams 82 at the Plant Transformation Facility, Iowa State University (Paz et al., 2004). Basta (glufosinate)-resistant R0 plants were tested for incorporation of the bar gene by PCR. For each PSS1 construct, at least three transgenic events were generated. Basta-resistant R0 plants were grown in a greenhouse to maturity for harvesting R1 seeds.
Evaluation of PSS1 Transgenic Lines in a Growth Chamber and under Field Conditions for SDS Resistance
R1 progeny derived from self-pollinated R0 plants were investigated for possible enhanced SDS resistance under growth chamber conditions. The F. virguliforme inocula were prepared, and colony-forming units of the inocula were determined as described previously (Li et al., 2009). To prepare the inoculum, 500 g of sorghum (Sorghum bicolor) grains was first soaked in distilled water overnight and then washed five times to remove sorghum seeds and debris that were floated. The excess water was drained, and grains were autoclaved for 40 min at 121°C. Each of the flasks containing sterilized sorghum grains was inoculated with F. virguliforme isolate Mont-1 by transferring 10 20-mm-diameter plugs from one-third-strength PDA plates containing 2-week-old F. virguliforme Mont-1 culture. Flasks were then incubated at room light and temperature and shaken gently by hand every other day for 2 weeks to ensure uniform fungal growth. After 1 month, the sorghum grains, infested with the fungus, were dried for 24 h under a fume hood. Infested kernels were then stored at 4°C until further use, typically no longer than 3 months.
For growth chamber assays, a 2:1 mixture of sand and soil was mixed with the inocula at a ratio of 19:1::soil mix:inocula and placed in 237-mL Styrofoam cups for sowing three seeds of each genotype. Fifteen seeds of each soybean line were evaluated in five Styrofoam cups. Plants were grown in a growth chamber at 22°C to 23°C and with light intensity of 300 μmol photons m−2 s−1 (Luckew et al., 2013). Foliar symptoms were scored 3 and 4 weeks following planting (Hartman et al., 1997).
R1 and R2 seeds were evaluated in field trials in two consecutive seasons, from June 11 to October 30, 2015, and from June 1 to October 15, 2016, at Hinds Research Farm, Iowa State University, located 4 miles north of Ames, Iowa. Field trials were carried out using a completely randomized block design with two replications in the 2015 trial and three replications in the 2016 trial. Twenty-four seeds of each genotype were mixed with approximately 5 mL of inocula of the F. virguliforme NE305S isolate and sown using a hand-push planter. At the unifoliate stage, transgenic lines were sprayed with glufosinate herbicide (250 mg L−1) mixed with 0.1% Tween 20. The spray was repeated 3 d later. The field was heavily irrigated to generate favorable conditions for SDS symptom development. In the 2016 trial, only homozygous transgenic lines showing 100% herbicide resistance were scored for foliar SDS. Individual plants were scored for foliar SDS symptoms using a scale of 1 to 7 (modified from Hartman et al., 1997) as follows: 1, no symptoms; 2, slight symptom development, with mottling and mosaic on leaves (1%–20% foliage affected); 3, moderate symptom development, with interveinal chlorosis and necrosis on foliage (21%–50% foliage affected); 4, heavy symptom development, with interveinal chlorosis and necrosis (51%–80% foliage affected); 5, severe interveinal chlorosis and necrosis (81%–100% foliage affected); 6, whole leaf necrosis; and 7, death of plants.
Molecular Characterization of Transgenic Plants
To verify gene expression in transgenic lines or Arabidopsis lines, total RNA was extracted using the SV Total RNA isolation system (Promega), following the manufacturer’s instructions, and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The isolated RNAs were reverse transcribed into cDNAs using the SuperScript first-strand synthesis system for RT-PCR (Thermo Fisher Scientific). Semiquantitative RT-PCR was conducted for the PSS1 gene or coexpressed genes along with ELF1B as the internal control.
Transgene copy numbers were determined by qPCR. Young leaves of transgenic plants were collected in the field, and genomic DNA was extracted at the Iowa State University DNA Facility using the Autogenprep 740 DNA extraction robot (AutoGen). The DNA amount of each sample was measured with a NanoDrop spectrophotometer and then was diluted to 20 ng μL−1 for qPCR. qPCR was conducted on a Biomark HD system using the 192.24 Taqman CNV protocol (Fluidigm). Two Taqman assays were designed (Supplemental Table S5): the bar gene (target) and the reference gene (an endogenous single-copy gene, Glyma.05G014200). Reporter/quencher dyes used were FAM/MGB-NFQ for bar and VIC/TAMRA for the reference gene. Data were analyzed using a Biomark HD data collection software, and from the analyzed data, the copy number for the bar gene was calculated.
Statistical Analysis
All data are presented as means ± se from at least three biological replications. The statistical significance of the difference was determined by conducting Student’s t test. Differences between treatments were considered significant at P < 0.05 in a two-tailed test. The statistical analysis for significant differences also was conducted using the open-source R program.
Bioinformatics Analyses
The conserved motifs in the PSS1 protein were predicated using the MyHits program (http://myhits.isb-sib.ch/cgi-bin/motif_scan). The prediction of signal peptide was conducted using SignalP 4.0 (Petersen et al., 2011). The transmembrane domain was predicted at the TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Genes coexpressed with PSS1 were identified from the ATTED-II database (http://atted.jp/) and Genevestigator (Hruz et al., 2008) using microarray and mRNAseq data sets, respectively. The phylogenetic tree of PSS1 homologs was generated by the neighbor-joining method with 1,000 bootstrap replications using the MEGA7 program (Kumar et al., 2016). Protein structure prediction of PSS1 and its mutant and their pairwise structure alignment were accomplished by using the I-TASSER and TM-align programs, respectively (http://zhanglab.ccmb.med.umich.edu/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Complementation analysis of the pss1 mutant with the CaMV 35S promoter-fused PSS1 cDNA.
Supplemental Figure S2. Structure comparison of PSS1 and its mutant pss1 protein.
Supplemental Figure S3. Alignment of PSS1 homologs.
Supplemental Figure S4. Coexpression gene analysis based on the microarray data set.
Supplemental Figure S5. Subcellular localization of N- or C-terminal GFP-tagged PSS1.
Supplemental Table S1. T-DNA insertional lines used for the identification of the candidate PSS1 gene.
Supplemental Table S2. Top 15 biotic stresses that induce the expression of PSS1.
Supplemental Table S3. Induction of the gene (At4g21980) encoding an autophagy-related protein 8A coexpressed with PSS1 in pss1.
Supplemental Table S4. pss1 transgene copy number among the R1 plants.
Supplemental Table S5. Primers used in this study.
Acknowledgments
We thank David Grant for reviewing the article and Jordan Baumbach for contributions in managing soybean transgenic plants; we also thank the Iowa State University Plant Transformation Facility for the generation of transgenic soybean plants and the Iowa State University DNA Facility for sequencing and preparation of soybean DNA samples.
Footnotes
This work was supported by USDA NIFA (grant no. 2013-68004-20374), the Iowa Soybean Association, and the Consortium of Plant Biotechnology Research.
Articles can be viewed without a subscription.
References
- Ahn IP. (2008) Glufosinate ammonium-induced pathogen inhibition and defense responses culminate in disease protection in bar-transgenic rice. Plant Physiol 146: 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ballouz S, Verleyen W, Gillis J (2015) Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics 31: 2123–2130 [DOI] [PubMed] [Google Scholar]
- Bradley C, Allen T (2014) Estimates of soybean yield reductions caused by diseases in the United States. http://extension.cropsciences.illinois.edu/fieldcrops/diseases/ yield_reductions.php (November 1, 2017)
- Brar HK, Bhattacharyya MK (2012) Expression of a single-chain variable-fragment antibody against a Fusarium virguliforme toxin peptide enhances tolerance to sudden death syndrome in transgenic soybean plants. Mol Plant Microbe Interact 25: 817–824 [DOI] [PubMed] [Google Scholar]
- Brar HK, Swaminathan S, Bhattacharyya MK (2011) The Fusarium virguliforme toxin FvTox1 causes foliar sudden death syndrome-like symptoms in soybean. Mol Plant Microbe Interact 24: 1179–1188 [DOI] [PubMed] [Google Scholar]
- Campe R, Langenbach C, Leissing F, Popescu GV, Popescu SC, Goellner K, Beckers GJM, Conrath U (2016) ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytol 209: 294–306 [DOI] [PubMed] [Google Scholar]
- Chang HX, Domier LL, Radwan O, Yendrek CR, Hudson ME, Hartman GL (2016) Identification of multiple phytotoxins produced by Fusarium virguliforme including a phytotoxic effector (FvNIS1) associated with sudden death syndrome foliar symptoms. Mol Plant Microbe Interact 29: 96–108 [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim W, Lee C, Oh CS (2013) Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol Plant Microbe Interact 26: 1115–1122 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Baker BJ (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA 97: 1908–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorov TA, Odintsova TI, Pukhalsky VA, Grishin EV (2005) Diversity of wheat anti-microbial peptides. Peptides 26: 2064–2073 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR (2007) A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288 [DOI] [PubMed] [Google Scholar]
- Hadwiger LA. (2015) Anatomy of a nonhost disease resistance response of pea to Fusarium solani: PR gene elicitation via DNase, chitosan and chromatin alterations. Front Plant Sci 6: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman GL, Huang YH, Nelson RL, Noel GR (1997) Germplasm evaluation of Glycine max for resistance to Fusarium solani, the causal organism of sudden death syndrome. Plant Dis 81: 515–518 [DOI] [PubMed] [Google Scholar]
- Heath MC. (2000) Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol 3: 315–319 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Scott MP, Bhattacharyya MK (2006) Light is essential for degradation of ribulose-1,5-bisphosphate carboxylase-oxygenase large subunit during sudden death syndrome development in soybean. Plant Biol (Stuttg) 8: 597–605 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. (2007) IntAct: open source resource for molecular interaction data. Nucleic Acids Res 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Kim NH, Hwang BK (2015) GLYCINE-RICH RNA-BINDING PROTEIN1 interacts with RECEPTOR-LIKE CYTOPLASMIC PROTEIN KINASE1 and suppresses cell death and defense responses in pepper (Capsicum annuum). New Phytol 205: 786–800 [DOI] [PubMed] [Google Scholar]
- Kim JY, Park SJ, Jang B, Jung CH, Ahn SJ, Goh CH, Cho K, Han O, Kang H (2007) Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J 50: 439–451 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbach C, Campe R, Schaffrath U, Goellner K, Conrath U (2013) UDP-glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian soybean rust pathogen Phakopsora pachyrhizi. New Phytol 198: 536–545 [DOI] [PubMed] [Google Scholar]
- Lee S, Whitaker VM, Hutton SF (2016) Potential applications of non-host resistance for crop improvement. Front Plant Sci 7: 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hartman GL, Chen Y (2009) Evaluation of aggressiveness of Fusarium virguliforme isolates that cause soybean sudden death syndrome. J Plant Pathol 91: 77–86 [Google Scholar]
- Lin CH, Chen CY (2014) Characterization of the dual subcellular localization of Lilium LsGRP1, a plant class II glycine-rich protein. Phytopathology 104: 1012–1020 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Luckew AS, Leandro LF, Bhattacharyya MK, Nordman DJ, Lightfoot DA, Cianzio SR (2013) Usefulness of 10 genomic regions in soybean associated with sudden death syndrome resistance. Theor Appl Genet 126: 2391–2403 [DOI] [PubMed] [Google Scholar]
- Mangeon A, Junqueira RM, Sachetto-Martins G (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbofung GCY, Fessehaie A, Bhattacharyya MK, Leandro LFS (2011) A new TaqMan real-time polymerase chain reaction assay for quantification of Fusarium virguliforme in soil. Plant Dis 95: 1420–1426 [DOI] [PubMed] [Google Scholar]
- Mellersh DG, Heath MC (2003) An investigation into the involvement of defense signaling pathways in components of the nonhost resistance of Arabidopsis thaliana to rust fungi also reveals a model system for studying rust fungal compatibility. Mol Plant Microbe Interact 16: 398–404 [DOI] [PubMed] [Google Scholar]
- Mousavi A, Hotta Y (2005) Glycine-rich proteins: a class of novel proteins. Appl Biochem Biotechnol 120: 169–174 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Ryu CM (2004) Nonhost resistance: how much do we know? Trends Plant Sci 9: 97–104 [DOI] [PubMed] [Google Scholar]
- Nakao M, Nakamura R, Kita K, Inukai R, Ishikawa A (2011) Non-host resistance to penetration and hyphal growth of Magnaporthe oryzae in Arabidopsis. Sci Rep 1: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ngaki MN, Wang B, Sahu BB, Srivastava SK, Farooqi MS, Kambakam S, Swaminathan S, Bhattacharyya MK (2016) Tanscriptomic study of the soybean-Fusarium virguliforme interaction revealed a novel ankyrin-repeat containing defense gene, expression of whose during infection led to enhanced resistance to the fungal pathogen in transgenic soybean plants. PLoS ONE 11: e0163106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, Joe A, Jeong BR, Korneli C, Boutrot F, Westedt I, Staiger D, Alfano JR, Zipfel C (2013) Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J 32: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J (1993) The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol 21: 895–906 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Amaro MA, Rodríguez-Hernández AA, Rodríguez-Kessler M, Hernández-Lucero E, Rosales-Mendoza S, Ibáñez-Salazar A, Delgado-Sánchez P, Jiménez-Bremont JF (2015) Overexpression of AtGRDP2, a novel glycine-rich domain protein, accelerates plant growth and improves stress tolerance. Front Plant Sci 5: 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK (2001) Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem 276: 26688–26693 [DOI] [PubMed] [Google Scholar]
- Park CJCB, Park CB, Hong SS, Lee HS, Lee SY, Kim SC (2000) Characterization and cDNA cloning of two glycine- and histidine-rich antimicrobial peptides from the roots of shepherd’s purse, Capsella bursa-pastoris. Plant Mol Biol 44: 187–197 [DOI] [PubMed] [Google Scholar]
- Park JH, Suh MC, Kim TH, Kim MC, Cho SH (2008) Expression of glycine-rich protein genes, AtGRP5 and AtGRP23, induced by the cutin monomer 16-hydroxypalmitic acid in Arabidopsis thaliana. Plant Physiol Biochem 46: 1015–1018 [DOI] [PubMed] [Google Scholar]
- Paz MM, Shou H, Guo Z, Zhang Z, Banerjee AK, Wang K (2004) Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136: 167–179 [Google Scholar]
- Perry ED, Ciliberto F, Hennessy DA, Moschini G (2016) Genetically engineered crops and pesticide use in U.S. maize and soybeans. Sci Adv 2: e1600850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pudake RN, Swaminathan S, Sahu BB, Leandro LF, Bhattacharyya MK (2013) Investigation of the Fusarium virguliforme fvtox1 mutants revealed that the FvTox1 toxin is involved in foliar sudden death syndrome development in soybean. Curr Genet 59: 107–117 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Panstruga R (2011) A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci 16: 117–125 [DOI] [PubMed] [Google Scholar]
- Schweiger R, Schwenkert S (2014) Protein-protein interactions visualized by bimolecular fluorescence complementation in tobacco protoplasts and leaves. J Vis Exp 85: 51327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu Rev Phytopathol 51: 407–427 [DOI] [PubMed] [Google Scholar]
- Shamloul M, Trusa J, Mett V, Yusibov V (2014) Optimization and utilization of Agrobacterium-mediated transient protein production in Nicotiana. J Vis Exp 86: 51204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumit R, Sahu BB, Xu M, Sandhu D, Bhattacharyya MK (2012) Arabidopsis nonhost resistance gene PSS1 confers immunity against an oomycete and a fungal pathogen but not a bacterial pathogen that cause diseases in soybean. BMC Plant Biol 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Abeysekara NS, Liu M, Cianzio SR, Bhattacharyya MK (2016) Quantitative trait loci underlying host responses of soybean to Fusarium virguliforme toxins that cause foliar sudden death syndrome. Theor Appl Genet 129: 495–506 [DOI] [PubMed] [Google Scholar]
- Tao TY, Ouellet T, Dadej K, Miller SS, Johnson DA, Singh J (2006) Characterization of a novel glycine-rich protein from the cell wall of maize silk tissues. Plant Cell Rep 25: 848–858 [DOI] [PubMed] [Google Scholar]
- Tavares LS, Rettore JV, Freitas RM, Porto WF, Duque AP, Singulani JdeL, Silva ON, Detoni MdeL, Vasconcelos EG, Dias SC, et al. (2012) Antimicrobial activity of recombinant Pg-AMP1, a glycine-rich peptide from guava seeds. Peptides 37: 294–300 [DOI] [PubMed] [Google Scholar]
- Teh OK, Hofius D (2014) Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J Exp Bot 65: 1297–1312 [DOI] [PubMed] [Google Scholar]
- Ueki S, Citovsky V (2002) The systemic movement of a tobamovirus is inhibited by a cadmium-ion-induced glycine-rich protein. Nat Cell Biol 4: 478–486 [DOI] [PubMed] [Google Scholar]
- Ueki S, Citovsky V (2005) Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature. Proc Natl Acad Sci USA 102: 12089–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Swaminathan S, Bhattacharyya MK (2015) Identification of Fusarium virguliforme FvTox1-interacting synthetic peptides for enhancing foliar sudden death syndrome resistance in soybean. PLoS ONE 10: e0145156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2006) Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc 2006: 1031–1036 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshen V, Huang S, Li X (2011) RNA-binding proteins in plant immunity. J Pathogens 2011: 278697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33: 2302–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]