Improved technology and methodology for affinity purification of nuclei and analysis of nuclear transcriptomes, chromatin, and other nuclear components.
Abstract
Isolated nuclei provide access to early steps in gene regulation involving chromatin as well as transcript production and processing. Here, we describe transfer of the isolation of nuclei from tagged specific cell types (INTACT) to the monocot rice (Oryza sativa L.). The purification of biotinylated nuclei was redesigned by replacing the outer nuclear-envelope-targeting domain of the nuclear tagging fusion (NTF) protein with an outer nuclear-envelope-anchored domain. This modified NTF was combined with codon-optimized Escherichia coli BirA in a single T-DNA construct. We also developed inexpensive methods for INTACT, T-DNA insertion mapping, and profiling of the complete nuclear transcriptome, including a ribosomal RNA degradation procedure that minimizes pre-ribosomal RNA (pre-rRNA) transcripts. A high-resolution comparison of nuclear and steady-state poly(A)+ transcript populations of seedling root tips confirmed the capture of pre-messenger RNA (pre-mRNA) and exposed distinctions in diversity and abundance of the nuclear and total transcriptomes. This retooled INTACT can enable high-resolution monitoring of the nuclear transcriptome and chromatin in specific cell types of rice and other species.
Since the mid-1980s, gene expression studies in plants that sample steady-state levels of polyadenylated (poly(A)+) mRNAs obtained from organs have propelled the understanding of gene regulation and cellular processes. The sampling of poly(A)+ mRNAs from laser-captured tissue sections or cells sorted based on expression of a fluorescent marker has further refined knowledge of gene activity, particularly at the cell-type-specific level and even more recently at the single-cell level (reviewed by Bailey-Serres, 2013; Efroni et al., 2015). Poly(A)+ transcript levels provide a snapshot of the output of transcription and mRNA stability in the whole cell. RNA-sequencing (RNA-seq) on cell-type-specific transcriptomes additionally yields information about the sites where transcription initiates and polyadenylation occurs and provides insights into the alternative splicing and retention of introns. However, steady-state transcript abundance may not accurately predict nascent transcription or the transcripts undergoing translation, particularly as plants modulate these processes under environmental stress (Park et al., 2012). Early analyses of plant transcript dynamics that used hybridization kinetics gleaned that nuclear and polyribosomal (polysomal) mRNA populations are highly distinct in tobacco (Nicotiana tobaccum) leaves (Goldberg et al., 1978). Others have compared nuclear poly(A)+ (Zhang et al., 2008) and total nuclear RNA (Deal and Henikoff, 2010) to total poly(A)+ mRNA of Arabidopsis (Arabidopsis thaliana) using microarrays. More recently, two studies have used nuclear RNAs to evaluate their structure and RNA-protein interactions (Gosai et al., 2015; Foley et al., 2017). To date, there has been no comparison of nuclear RNA with total cellular poly(A)+ RNA using high-throughput RNA-seq for any plant. Technologies that facilitate these comparisons in model and crop plants could aid the study of regulatory processes that are obscured by exclusive sampling the total poly(A)+ mRNA. Genome-scale evaluation of nuclear RNA can be accomplished by isolation of nuclei using differential centrifugation, fluorescence activated nuclear sorting (FANS; Zhang et al., 2008) or isolation of nuclei tagged in specific cell types (INTACT; Deal and Henikoff, 2010, 2011; Bailey-Serres, 2013). The latter two techniques were established for Arabidopsis and take advantage of transgenes that encode GFP fused to the core nucleosome protein histone 2A or a biotinylated chimeric fusion protein with affinity to an outer nuclear membrane (ONM) protein, respectively. FANS requires preparation of nuclei from fresh tissue and the use of a fluorescent activated cell sorter. INTACT, on the other hand, is an inexpensive affinity-purification method that is routinely performed on frozen tissues, enabling studies of rapid changes in gene regulation. Because INTACT entails the use of a transgene that can be driven by spatially or temporally regulated promoters, it enables sampling of nuclei of subpopulations of cell types or tissues within intact organs (Deal and Henikoff, 2010).
INTACT provides access to nuclei, including DNA, histones, other nuclear proteins, preribosomal RNAs, pre-mRNA and other RNAs within the nucleoplasm. There are two required components for this technology. The first is a synthetic nuclear tagging fusion (NTF) protein comprised of an ONM-targeting domain (Trp-Pro-Pro [WPP]) from RAN GTPASE-ACTIVATING PROTEIN1 (RanGAP1; Rose and Meier, 2001), fused to GFP and a biotin ligase target peptide (BLRP). The second is an Escherichia coli biotin ligase (BirA) that is expressed in the same tissue (Deal and Henikoff, 2010, (2011). The WPP domain interacts with a coil-coiled domain of a WPP-interacting protein (WIP). WIP itself is anchored via a C-terminal transmembrane domain or KASH (Klarsicht/ANC-1/Syne-1 homology) to the ONM and interacts with other proteins of the inner nuclear membrane (reviewed by Zhou et al., 2015; Rose and Meier, 2001; Xu et al., 2007). The interaction between the coil-coiled domain of WIP and the WPP domain places the NTF on the exterior of the nucleus. The biotinylation of the NTF enables the use of streptavidin-coated magnetic beads to selectively purify tagged nuclei from crude extracts of plant tissues. As WIP is directly anchored to the ONM and WPP is not, we considered that replacing the WPP domain with a KASH domain would provide more stable anchoring of the NTF to nuclei.
We recently translated INTACT to tomato (Solanum lycopersicum; Ron et al., 2014). Here, we retooled both the NTF and BirA for efficient INTACT in monocots, specifically rice. We also refined inexpensive methods for preparation of RNA-seq libraries from the complete nuclear transcriptome by including subtraction of pre-rRNA To demonstrate one use of this technology, we used an INTACT construct driven by the near-constitutive CaMV 35S promoter (p35S) to isolate nuclei and performed a comparative RNA-seq analysis on nuclear and total poly(A)+ mRNA from root tips. The results demonstrate notable distinctions between the gene transcripts represented and their abundance in the nuclear and poly(A)+ transcriptomes. We discuss applications of INTACT to the study of nuclear processes of gene regulation, from chromatin to mRNA export, and at the organ to cell-specific level.
RESULTS AND DISCUSSION
Generation of Transgenic Rice Lines for Purification of Nuclei Using INTACT
The retooling of INTACT for rice included modifications to improve biotinylation and to establish a stable interaction with the ONM. First, due to the greater GC content of the third codon position of genes in rice as compared to Arabidopsis (Tatarinova et al., 2013), we synthesized a rice codon usage-optimized version of E. coli BirA. This was included in a binary T-DNA vector along with the NTF construct. In Arabidopsis and S. lycopersicum, to establish a tagged nucleus, the NTF used was the WPP of RanGAP1 (Deal and Henikoff, 2010; Ron et al., 2014). The rice ACTIN2 (ACT2; LOC_Os10g36650) promoter was selected to drive expression of BirA, as its transcript is detected across cell types based on available expression data (Jain et al., 2007; Jiao et al., 2009). Because WPP association with the ONM is developmentally regulated (Xu et al., 2007; Zhao et al., 2008), we tested whether the C-terminal region of OsWIP (c-OsWIP, including a coil-coiled and a C-terminal transmembrane domain; reviewed by Zhou et al., 2015) could be used for INTACT. We designed three NTF constructs using rice WPP and c-OsWIP domains for evaluation. NTF1 combined the OsWPP domain of the RanGAP1 ortholog (LOC_Os05g46560.1; Rose and Meier, 2001) with GFP and a C-terminal BLRP domain (Fig. 1A). For NTF2 and NTF3, we replaced the WPP domain with the c-OsWIP from two rice orthologs of Arabidopsis WIP1: LOC_Os09g30350.1 (OsWIP2) and LOC_Os08g38890.1 (OsWIP3) (Xu et al., 2007). To position this domain in the ONM so that the GFP and BLRP of the NTF would be on the cytoplasmic face of the nucleus, we placed the BLRP at the N terminus, fused to GFP and then followed by the c-OsWIP (Fig. 1A).
Figure 1.
Three NTF proteins and their subcellular localization. A, Three synthetic NTF proteins were tested. Each was composed of three functional regions: a BLRP, a GFP for visualization, and a WPP domain or a C-terminal region of WIP proteins for targeting to or anchoring on cytoplasmic side of the nuclear envelope, respectively. The nuclear targeting domains are OsWPP (1), c-OsWIP2 (2), and c-OsWIP3 (3). B, Diagrammatic representation of predicted NTF location in the nuclear-envelope-cytoplasm interface. OsNTF1 is associated with the nuclear envelope via interaction between its WPP domain and endogenous WIP proteins. The C-terminal region of WIP of OsNTF2 and OsNTF3 may be sufficient for integration into the outer nuclear envelope. C, Representative confocal images of GFP fluorescence in organs from 7-d-old plants grown in a growth chamber expressing a p35S:OsNTFs. a to f, root meristematic zone (Root MZ); g to i, root differentiated zone (Root DZ) 1 cm from the root tip, epidermal cells; j to l, blade region between the midvein and outer margin of a newly expanding leaf blade—guard cells and epidermal cells are visible. Leaves are maximum intensity Z-projections of a confocal Z series. INM, Inner nuclear membrane; ONM: outer nuclear membrane. Scale bars: a to c and g to l, 25 µm; d to f, 5 µm.
Independent transgenic rice lines were produced that express each NTF version under the control of the p35S (p35S:OsNTFs). There was no detectable phenotypic consequence of expression of any of these constructs on development or fertility under standard culture or greenhouse conditions. The sites of T-DNA insertion in 12 independent transgenics were determined using a high-throughput method that captures T-DNA border regions and flanking DNA by hybridization to biotinylated probes (Supplemental Table S1; Supplemental Results). Sites of insertion were validated by PCR. The p35S:OsNTF1, p35S:OsNTF2, and p35S:OsNTF3 lines used for subsequent analyses had moderate levels of GFP fluorescence and a single insertion in chromosomes 6, 1, and 1, respectively.
Detection of Nuclear Envelope-Associated and Anchored Biotinylated-GFP
Confocal microscopy was used to monitor the cellular distribution of the three OsNTF proteins in cells of roots and leaves of-7-d-old seedlings. OsNTF1 is predicted to associate with the ONM via protein-protein interaction with a WIP (Fig. 1B). Most of the GFP signal from OsNTF1 was localized at the periphery of nuclei in roots (meristematic and differentiated region) and expanding juvenile leaf blades (Fig. 1C; Supplemental Fig. S1). The GFP signal was also abundant in the cytoplasm of the root meristematic zone as well as the differentiated zone (epidermal cells), where it was displaced by the central vacuole. OsNTF2 and OsNTF3 were designed to be anchored to the ONM via the C-terminal transmembrane domain of WIP (Fig. 1B). The sharper halo of GFP signal around the nuclei in p35S:OsNTF2- and p35S:OsNTF3-expressing plants indicates a more integral association of WIP than WPP with the nuclear membrane (Fig. 1C). p35S:OsNTF2 and p35S:OsNTF3 plants had less GFP signal in the cytoplasm than p35S:OsNTF1 plants, even though the highly expressed p35S was used for all constructs. Lower cytoplasmic accumulation of NTF2 and NTF3 is quite evident in the guard cells of the leaf blade. The high cytoplasmic GFP signal in multiple p35S:OsNTF1 lines (Supplemental Fig. S1) suggest that the amount of protein produced exceeded available WIP or other nuclear anchoring proteins. Alternatively, OsNTF2 or OsNTF3 protein that does not integrate into the ONM may be more readily degraded than OsNTF1.
To confirm that OsNTFs were biotinylated in planta, root tissue was solubilized in a sodium dodecyl sulfate (SDS)-containing buffer and proteins were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), blotted to a membrane, and probed with streptavidin conjugated to horseradish peroxidase, which binds with high affinity to biotin and enables enzymatic detection, respectively. Biotinylated proteins with the expected electrophoretic mobility were detected in p35S:OsNTF1-, 2-, and 3-expressing plants (Supplemental Fig. S2). These results demonstrate that a chimeric GFP can associate with or be anchored to the ONM in rice, supporting the use of these OsNTF-expressing lines for affinity purification of nuclei.
OsNTFs with a WPP or KASH Domain Enable Rapid Purification of Nuclei
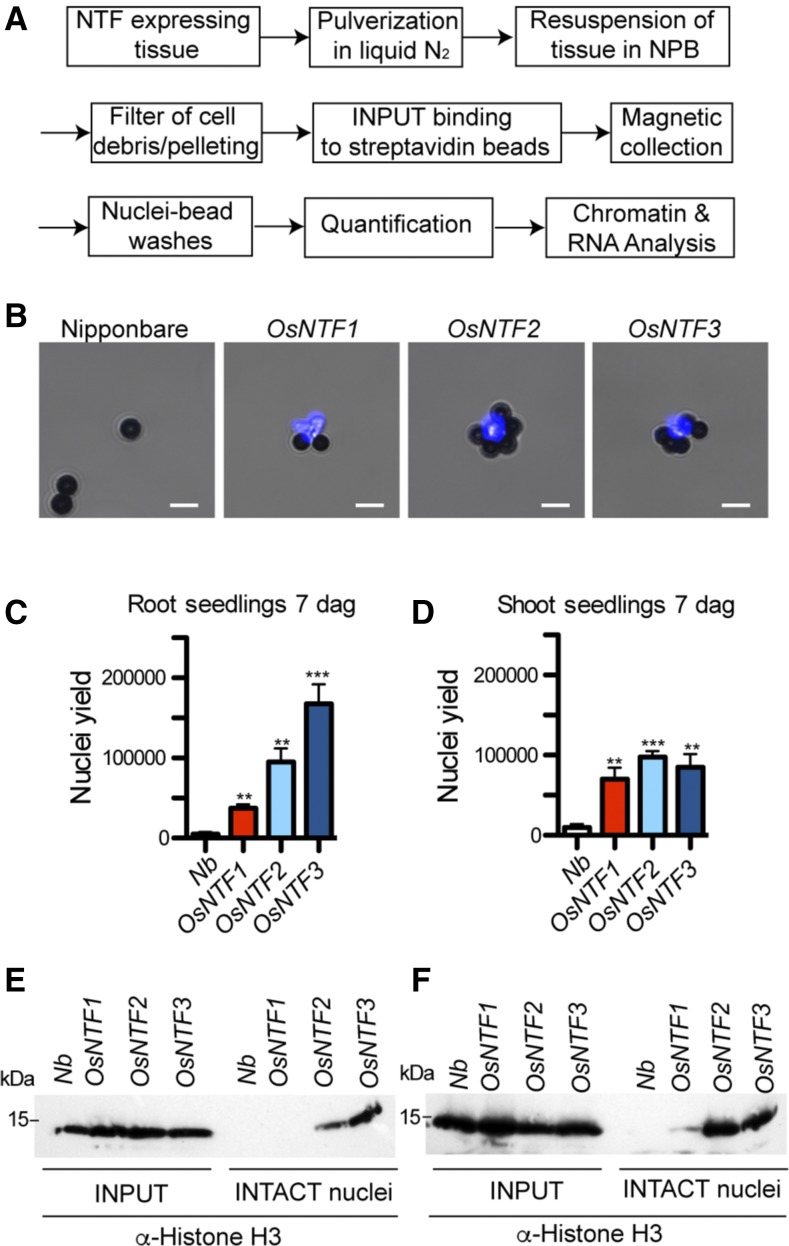
Nuclei from frozen tissue of 7-d-old seedlings expressing OsNTFs were purified by INTACT following the procedure described for Arabidopsis (Deal and Henikoff, 2010), with minor modifications (Fig. 2A; see “Materials and Methods”). After affinity capture by binding to an excess of streptavidin-coated magnetic beads and extensive washing, the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) to detect nucleic acids by fluorescent microscopy (Fig. 2B). Nuclei were purified with all three OsNTF versions in significant excess over nuclei captured by the same procedure from roots and shoots of nontransgenic seedlings (Fig. 2, C and D; Supplemental Fig. S3A). To further evaluate INTACT with these constructs, we examined the presence of chromatin by monitoring histone H3 abundance. Histone H3 levels were similar for all four genotypes in the input (Fig. 2, E and F), whereas H3 levels captured by INTACT were higher from p35S:OsNTF2 and p35S:OsNTF3 tissue in accordance with greater number of nuclei purified (Fig. 2, C and D). The same samples in Figure 2, C to F, indicate that histone H3 levels were efficiently purified from OsNTF2 and OsNTF3 seedlings. Thus, the INTACT constructs with a KASH domain display a more defined nuclear association and less cytoplasmic signal and were suitable for capture of nuclei and associated chromatin from seedling tissues. Nuclei yields presented some differences between NTF constructs expressed, transgenic lines evaluated, organs sampled and experiments performed. For example, nuclear yields were higher in roots than shoots for OsNTF3 but not OsNTF2 (Fig. 2, C and D), and high OsNTF1 accumulation as in the line presented in Supplemental Figure S1 provided similar levels of purified nuclei as OsNTF2 and OsNTF3 lines (Supplemental Fig. S3A). We conclude that all three NTF constructs prepared for and expressed in rice worked for the intended purpose. A p35S:OsNTF2 line that consistently gave good yields of nuclei from seedling and field-grown tissue was used for further evaluation of INTACT purified protein and RNA.
Figure 2.
Nuclei isolation using INTACT. A, Scheme of INTACT purification. B, Capture of biotinylated nuclei from roots of transgenic lines carrying p35S:OsNTF1, 2, or 3 and pACT2:BirA transgenes by use of streptavidin-coated magnetic beads. Nuclei were stained with DAPI to visualize DNA. Viewed with mixed white light and DAPI-channel fluorescence illumination. Nuclei (bright blue) surrounded by 2.8 µm spherical beads (dark gray) are visible. Scale bar is 5 μm. C and D, Nuclei were purified from roots and shoots of 7-d-old seedlings expressing the different OsNTF versions, stained with propidium iodide for visualization and counted by use of a hemocytometer. Asterisks indicate significant differences in an unpaired two-tailed Student’s t test (**P < 0.01 and ***P < 0.001) in a comparison to nuclei purified with nontransgenic Nipponbare (Nb). E and F, Detection of Histone 3 (H3) protein in nuclei before (crude nuclei) or after purification by INTACT (INTACT nuclei) for roots (E) and shoots (F). For each sample, an equal amount of starting material (0.5 g) was used to obtain nuclei, and a proportional amount of each yield was solubilized in an SDS-containing buffer and fractionated by electrophoresis in a 12% (w/v) polyacrylamide-SDS gel, transferred to a membrane and processed with commercial monoclonal antibody against H3 (α-Histone H3). Molecular weight marker shown on the left. The expected size of Histone H3 is 15.27 kD.
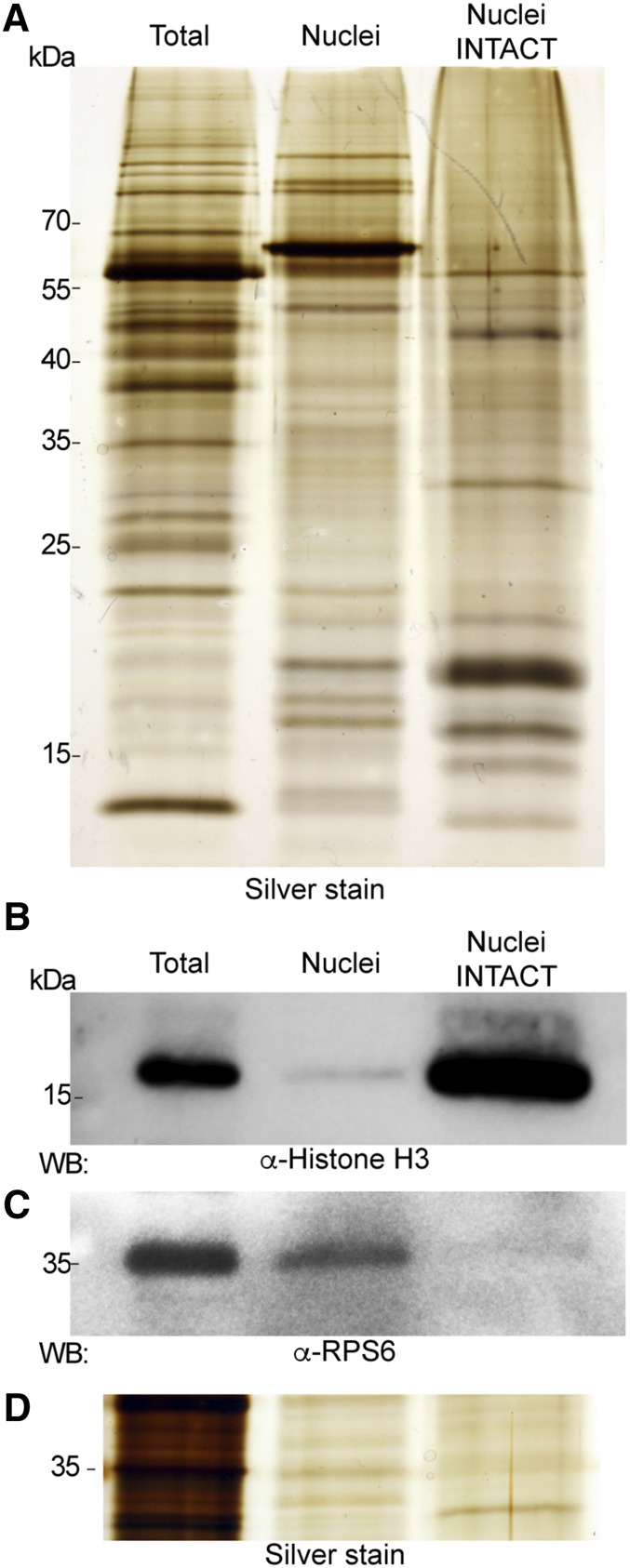
To compare and contrast the nuclei obtained by INTACT or conventional centrifugation methods, we monitored abundant proteins. First, histone H3 protein levels of INTACT and sedimented nuclei, of shoot tissue of 5-week-old plants grown under field conditions were compared. Total cellular and nuclear proteins were fractionated by SDS-PAGE and silver stained (Fig. 3A). As expected, a greater number of protein bands were detected in the total protein extract than in isolated nuclei. Both nuclear preparations were enriched in low-molecular mass proteins of 9.3 to 19.7 kD apparent mass, characteristic of the abundant core histones. INTACT nuclei showed an enrichment of histone H3 as compared to the conventionally isolated nuclei (Fig. 3B). The abundance of ribosomal protein S6 (RPS6) was higher in the conventionally purified nuclei than in the INTACT nuclei (Fig. 3C). We anticipated that RPS6 would also be detected in the nuclear fraction due to ribosome biogenesis in the nucleolus, but the level was far less than in the total extract, indicating that nuclear preribosomes are highly outnumbered by cytoplasmic ribosomes. The higher level of RPS6 relative to histone H3 in the conventionally purified nuclei could reflect chromatin loss, cytoplasmic contamination, or better maintenance of the nuclear membrane-rough endoplasmic reticulum connection. Altogether, these results confirm the effective purification of chromatin by INTACT with OsNTF2 in rice.
Figure 3.
Evaluation of protein composition from nuclei captured from the shoots of 5-week-old field grown plants using conventional methods and INTACT. A, Total proteins (Total); nuclei prepared by conventional sedimentation methods (Nuclei) and using INTACT (Nuclei INTACT) were separated by 12% (w/v) SDS-PAGE and visualized by silver staining. B to D, Total and nuclear proteins were analyzed by western blot using α-Histone H3 and ribosomal protein α-RPS6 antibodies or silver stained (bottom). Molecular weight markers are shown on the left. Protein loading is the same for B, C, and D, based on protein quantification (see “Materials and Methods”). Protein loading in A was adjusted to optimize the visual comparison between samples.
Preparation of Nuclear RNA Libraries with pre/rRNA Subtraction
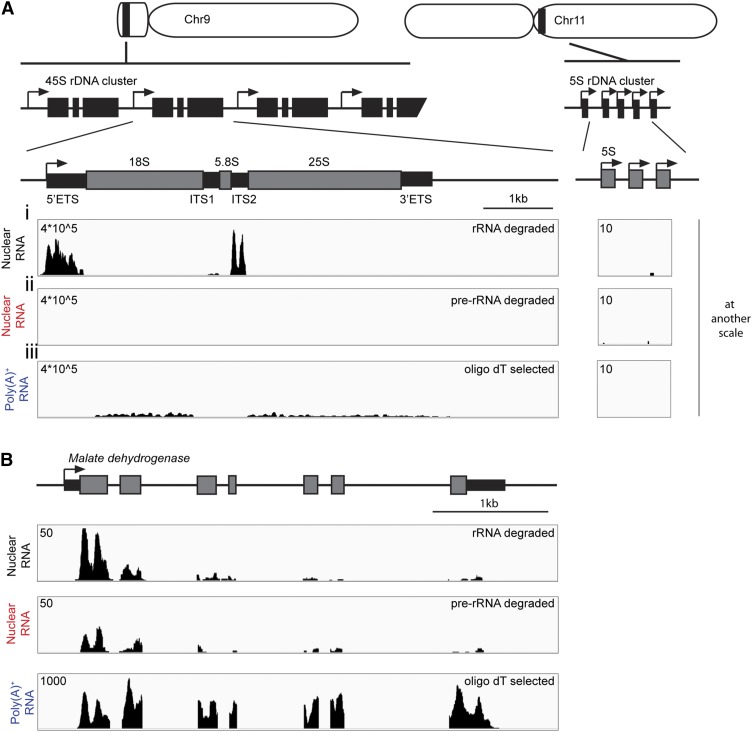
Next, we explored the use of INTACT to evaluate the complete nuclear transcriptome by RNA-seq using root tips of 7-d-old p35S:NTF2 seedlings. To selectively remove the abundant pre- and processed ribosomal RNAs (rRNAs) produced by the 45S rDNA and 5S rRNA clusters (Fig. 4A), we developed a method to remove rRNA that did not rely on a commercial kit. Nuclear RNA was hybridized to DNA oligos complementary to 25S, 18S, 5.8S, and 5S rRNAs, and the DNA:RNA hybrids were selectively degraded at high temperature with a thermostable RNase H. RNA-seq run confirmed this rRNA degradation method reduced reads corresponding to mature rRNAs but not the transcribed spacer sequences of pre-rRNA (Fig. 4Ai). As DNA oligos corresponding to the pre-rRNA external transcribed spacers (5′ ETS and 3′ ETS) and internal transcribed spacers (ITS1 and ITS2) were not included in the initial rRNA degradation assay, the presence of the 5′ ETS, ITS1, ITS2, and 3′ ETS regions confirmed that nuclear RNA had been isolated. We subsequently adjusted our method to include DNA oligos to subtract mature rRNA as well as pre-rRNA ETS, ITS1, and ITS2 regions (pre-rRNA degraded). This reduced the unprocessed pre-rRNA reads to below the level of rRNA reads in oligo(dT)-selected RNA (Fig. 4, Aii–Aiii). As a control, we show that relative coverage over MALATE DEHYDROGENASE gene is unaffected (Fig. 4B).
Figure 4.
Nuclear transcriptome libraries generated including an rRNA degradation step. A, Location, organization of ribosomal DNA (rDNA), and coverage of nuclear transcriptome. Rice 45S rDNA cluster is located in the short arm of chromosome (Chr) 9 and consists of tandem repeats of 5′ external transcribed spacer (ETS), 18S rRNA, internal transcribed spacer1 (ITS1), 5.8S rRNA, ITS2, 25S rRNA, and 3′ ETS. 5S rDNA cluster is located in Chr 11 composed of repeats of 5S rRNA. i and ii show read coverage on a 45S and 5S region for nuclear RNA treated with mature rRNA complementary probes (i) or full pre-rRNA probes (ii). iii shows the coverage obtained from poly(A)+-selected libraries from total tissue lysates. B, Comparison of read coverage in the nuclear and poly(A)+ RNA transcriptomes for the gene MALATE DEHYDROGENASE (LOC_Os01g46070).
A systematic comparison was made of nuclear RNA and poly(A)+ RNA from root tips of seedlings. Nuclei were isolated using p35S:NTF2 from four biological replicate samples of 200 root tips and used to generate nuclear RNA-seq libraries by rRNA subtraction and random-primer-enabled cDNA synthesis (Townsley et al., 2015). Total RNA-seq libraries were constructed with oligo dT-selected RNA. After recognizing that the sequences included some reads in very high abundance that were of organellar origin or the ETS, ITS1, and ITS2 regions, in cases where oligos to these regions were not used, we established a pipeline for analysis that commenced with a filtration step. Sequence reads that mapped to mitochondria and chloroplast genomes were removed, and the remaining reads were mapped to exons with ≤2 mismatches to the Nipponbare genome (IRGSP-1.0.30). This yielded an average coverage of 26.7 (total [poly(A)+] RNA) and 1.52 million (nuclear RNA; Supplemental Table S2A). Despite the difference in read number, subsampling of the libraries confirmed that each library had reached a saturation of transcripts identified as present in each population (Supplemental Fig. S4). Based on reads that mapped to exons, both the poly(A)+ RNA libraries (R2 ≥ 0.978, Pearson correlation) and nuclear RNA (R2 ≥ 0.965) yielded highly consistent results, although there was somewhat greater variation in the nuclear RNA libraries (Supplemental Table S2A).
The Root-Tip Nuclear and poly(A)+ RNA Transcriptomes Differ in Complexity and Transcript Enrichment
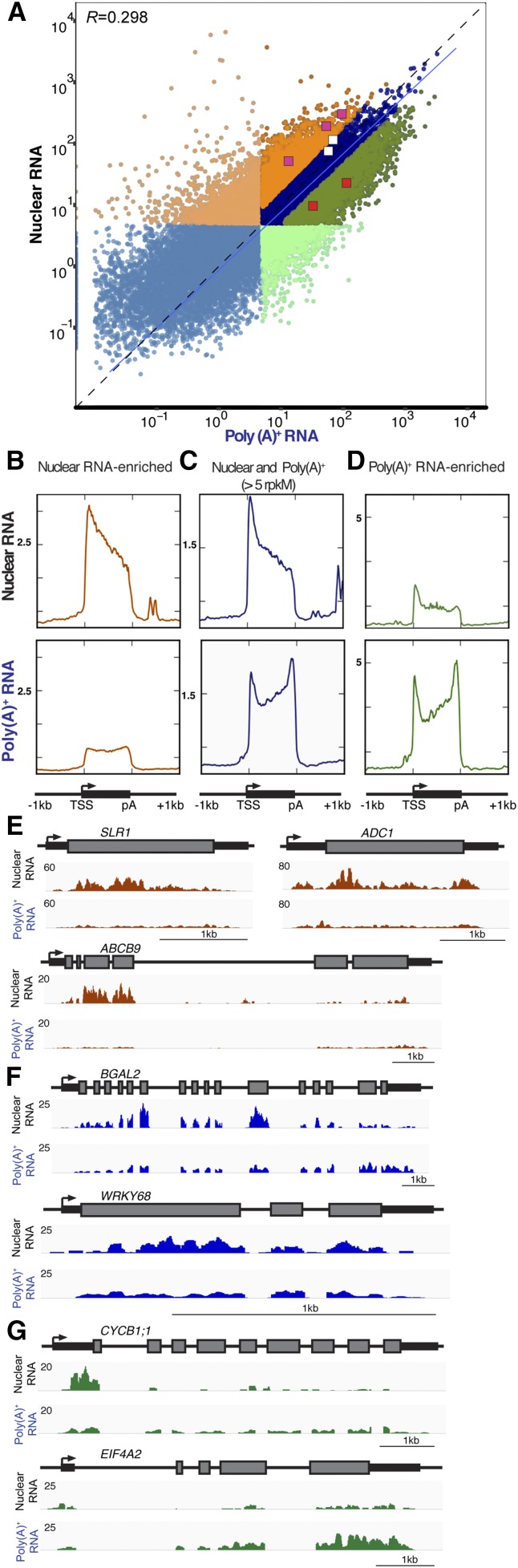
To compare the nuclear and poly(A)+ transcriptomes we determined the number of genes detected (complexity) and evaluated differences in transcript abundance using averaged data from the four biological replicate samples for each RNA population. First, we evaluated the portion of the transcript mapping to introns, coding regions and untranslated regions (UTRs; Supplemental Fig. S5). The proportion of intronic to exonic reads (coding regions, 5′ UTR, 3′ UTR) was 2-fold higher in the nuclear RNA in all of the biological replicates. To estimate complexity (i.e. the total number of gene transcripts detected), we applied a threshold of >5 reads per kilobase per million reads (rpkM) on exonic regions (Supplemental Table S2B). In the nuclear RNA about 18,100 and in the poly(A)+ RNA, about 16,500 protein-coding transcripts were detected, respectively, with 14,264 shared by both RNA populations (Fig. 5A). Gene Ontology enrichment analysis of the RNAs detected only above the 5 rpkM threshold in the nuclear RNA included meiotic division (Biological Process [BP]:adj. P value 0.00448), ribonucleoside binding (Molecular Function [MF]:0.0039), and peptide receptor activity (MF:0.0043), whereas those detected only in poly(A)+ RNA above the 5 rpkM threshold identified phospholipid biosynthetic process (BP:0.00606) among others (Supplemental Table S2C). This indicates that these mRNAs were highly enriched in the nucleus as compared to the poly(A)+ RNA pool, demonstrating that nuclear RNA and poly(A)+ measures of transcript abundance yield distinct results. The difference between these readouts was also evident from a weak positive correlation in abundance of RNAs in the two populations (R 0.298, Pearson correlation; Fig. 5A). We determined that this was due to both distinctions in transcriptome complexity and transcript abundance. Of the 14,264 transcripts above the 5 rpkM threshold in both populations, 6,234 were significantly enriched in one or the other population (nuclear, 3,152; poly(A)+, 3,082; |log2 fold difference| >1, FDR ≤ 0.01; Supplemental Table S2D). Higher nuclear abundance was found for genes associated with transferase activity (MF:2.87E-5) and cell wall organization or biogenesis (BP:0.0005), whereas higher poly(A)+ enrichment was noted for genes associated with translation (BP:1.25E-67) and intracellular transport (BP:1.18E-17; Supplemental Table S2E).
Figure 5.
Comparison of the nuclear and total poly(A)+ transcriptome of root tips. Nuclear RNA was extracted from INTACT-purified root-tip nuclei, and total poly(A)+ RNA was captured using a biotinylated oligo dT. A, Scatterplot representing the mean rpkM values on rice annotated genes from four biological replicates for nuclear and poly(A)+ RNA. Light-orange and light-green dots indicate 3,831 and 2,215 transcripts with >5 rpkM only in nuclear or poly(A)+ RNA, respectively. More intense-colored dots indicate transcripts with >5 rpkM in both RNA populations. Dark-orange dots indicate 3,152 transcripts differentially enriched in nuclear RNA, whereas 3,082 dark-green dots represent transcripts enriched in poly(A)+ |log2 FC > 1; FDR < 0.01|. A linear model is indicated with a solid blue line presenting a 0.298 Pearson correlation value, and y = x is indicated by a dashed line. Pink, red, and white squares are genes evaluated in E to G. B, C, and D, Nuclear and poly(A)+ RNA coverage evaluation across transcription units. The x-axis represents a transcription unit indicating the transcription start site (TSS), the cleavage and polyadenylation site (pA), and 1-kb region upstream or downstream. The y-axis indicates the relative transcript coverage, using different scales for each panel. B, nuclear RNA-enriched transcripts from A (dark-orange dots). C, 14,262 transcripts detected in both nuclear and poly(A)+ populations at >5 rpkM. D, poly(A)+-enriched transcripts (dark-green dots). E, F, and G, Examples of coverage over individual transcripts. E, Nuclear-enriched transcripts showing the presence of 5′ bias in nuclear coverage include a DELLA PROTEIN SLR1 (SLR1, LOC_Os03g49990), ARG DECARBOXYLASE (ADC1, LOC_Os06g04070), ABC TRANSPORTER B FAMILY MEMBER9 (ABCB9, LOC_Os02g09720). These transcripts are indicated in A as pink squares. F, Examples of transcripts detected in both nuclear and poly(A)+ include BETA-GALACTOSIDASE2 (BGAL2, LOC_Os06g37560) and TRANSCRIPTION FACTOR WRKY68 (WRKY68, LOC_Os04g51560). These transcripts are indicated in A as white squares. G, Examples of poly(A)+-enriched transcripts showing a high 5′ bias in nuclear coverage include a CYCLIN-B1-1 (CYCB1;1, LOC_Os01g59120) and EUKARYOTIC INITIATION FACTOR4A-2 (EIF4A2, LOC_Os02g05330). These transcripts are indicated in A as red squares.
Technical and biological factors could contribute to the lack of congruence between the nuclear and poly(A)+ RNA transcriptomes measured by our approach. First, the NTF construct is designed to capture nuclei from cells expressing the 35S promoter, whereas poly(A)+ purification accesses all cells following disruption with a chaotropic agent. Therefore, some poly(A)+-enriched mRNAs could be those from cells that do not express the p35S. Although not exhaustively evaluated, the NTF GFP signal was detectable across cell types in the rice root (Fig. 1C). Second, the greater complexity of the nuclear transcriptome may reflect the absence of the requirement for polyadenylation, thereby including protein-coding RNAs at various points in transcription, processing, nuclear export, and splicing. Indeed, as mentioned, the nuclear RNA samples had greater proportions of introns and 5′ UTRs (flanking regions; Supplemental Figure S5).
To further explore if the nuclear RNA pool was enriched for nascent transcripts, we determined the average distribution of RNA-seq reads across all individual transcripts detected in nuclear and poly(A)+ RNA with >5 rpkM, from the annotated start site to the cleavage/poly(A) addition site. This illuminated a bias in RNA-seq reads near the 5′ end of nuclear transcripts in contrast to the more equally distributed reads across the entire transcription unit in poly(A)+ RNA (Fig. 5C). The read coverage averaged for all poly(A)+ RNAs uncovered a modest 5′-end bias and a more pronounced 3′-end bias. Genome browser views illustrate the average transcript read distribution for BETA-GALACTOSIDASE2 (BGAL2) and TRANSCRIPTION FACTOR WRKY68 (WRKY68; Fig. 5, A and F). Although these mRNAs have introns, few nuclear RNA reads map to intervening regions, suggesting that splicing occurs with fidelity.
To gain an understanding of the biological basis for nuclear-enriched transcripts, read coverage across the transcription unit was averaged for the RNAs that were significantly enriched in the nuclear or poly(A)+ populations. Intriguingly, the nuclear 5′-end bias was less prominent in transcripts with the highest nuclear abundance (Fig. 5B). By contrast, for these RNAs, the coverage for the polyadenylated transcript pool was more evenly distributed along the transcription unit. Three nucleus-enriched transcripts illustrate this read mapping pattern in genome browser views: DELLA protein SLENDER LEAF RICE1 (SLR1), ARG DECARBOXYLASE, and ABC TRANSPORTER B FAMILY MEMBER9 (Fig. 5, A and E). This indicates that export of some nuclear transcripts may be rate limited after completion of splicing and processing. Contrastingly, transcripts with significantly enriched poly(A)+ abundance had a 5′-end bias in the nuclear but not poly(A)+ read coverage, similar to the average transcript read coverage (Fig. 5D). These transcripts are present at higher levels in the poly(A)+ mRNA transcriptome. Genome browser views of CYCLIN B1-1 (CYCB1;1) and EUKARYOTIC INITIATION FACTOR 4A-2 (EIF4A-2) illustrate a pattern of nuclear depletion relative to poly(A)+ transcript accumulation (Fig. 5, A and G).
The distinct read coverage patterns between transcripts in the INTACT-isolated nuclear relative to the oligo(dT)-selected poly(A)+ RNA pool showed in Fig. 5B-D, could indicate differences in RNA processing and turnover in the nuclear and cytoplasmic compartments. For example, differences in read coverage could reflect distinctions in the timing and fidelity of transcription, cotranscriptional processing, nuclear degradation, mRNA export to the cytoplasm, translation, and cytoplasmic degradation. In this scenario, the protein-coding transcripts enriched in the nucleus and depleted in the poly(A)+ pool could undergo destabilization following export to the cytoplasm, whereas those enriched in the poly(A)+ pool, which are presumed to be predominantly cytoplasmic, could be stabilized by active translation. Alternatively, these could be translationally inactive but stabilized in mRNA ribonucleoprotein complexes. Those transcripts with similar abundance in the two populations assayed, such as BGAL2 and WRKY68 (Fig. 5, A and F), could be generally stable. The underlying reasons for the enrichment in nuclear pre-mRNA versus poly(A)+ transcript abundance could be explored by inhibiting transcription with the adenosine analog cordycepin or other compounds and a subsequent time-course evaluation of the RNA in the two populations. Alternatively, mutants defective in posttranscriptional processes such as alternative splicing, cleavage/polyadenylation, mRNA export, nonsense-mediated decay, decapping-dependent decay, or deadenylation-dependent decay could be combined with INTACT to explore specific posttranscriptional mechanisms.
CONCLUSION
This study introduces the use of INTACT for the first time in a monocot, tests a new generation of NTF constructs, and evaluates nuclear transcriptome RNA-seq in a plant. The original version of INTACT developed for Arabidopsis and applied to tomato utilizes a protein domain of a RanGAP to place a biotinylated protein at the nuclear envelope by binding to an ONM protein (Deal and Henikoff, 2010; Ron et al., 2014). We directly transferred INTACT to rice using the rice RanGAP WPP and confirmed that it could be used for capture of nuclei. We also redesigned the INTACT construct to anchor the biotinylated protein to the ONM (Fig. 1). These constructs proved useful for isolation of nuclei from root and shoot tissue of seedlings grown in sterile culture (Fig. 2) and shoot tissue of greenhouse and field-grown plants. INTACT-obtained nuclei may have less contamination with ribosomes than nuclei isolated by conventional differential centrifugation (Fig. 3), which could prove beneficial for proteomic and other analyses. However, we cannot rule out the possibility that the reduced association of ribosomes with INTACT purified nuclei reflects methodological distinctions in capture of ONM associated rough endoplasmic reticulum.
Here, we show that this method of nuclear isolation can be used to generate RNA-seq libraries to monitor nuclear protein-encoding RNAs, following selective removal of pre/rRNA. This subtraction of rRNA can be accomplished with commercial products, but also by the method demonstrated here that relies on hybridization of RNA to user-defined DNA oligos followed by a high-temperature DNA:RNA hybrid digestion. We also determined the insertion site of the INTACT transgenes into the rice genome using an efficient method for T-DNA insertion site detection, establishing OsNTF genetic stocks for crossing to mutant genotypes.
The application of the INTACT method to root tips of rice enabled a comparison of the nuclear transcriptome to that routinely analyzed, steady-state poly(A)+ mRNA. This was accomplished by production of RNA-seq libraries with random priming of cDNA synthesis, the latter RNA captured by oligo(dT) selection based on the attribute of a poly(A)+ tail. Deep sequencing of the libraries enabled a systematic comparison of the number, enrichment and RNA-seq read-distribution across individual transcripts. The results demonstrate that the two populations are not identical, as concluded from reassociation kinetic studies on tobacco nuclear and polysomal RNAs over 40 years ago (Goldberg et al., 1978). Of over 14,000 protein-coding gene transcripts detected in our study, >3,000 were enriched significantly in the nuclear versus total poly(A)+ transcriptome, respectively. We did not monitor nuclear polyadenylated transcripts, as their abundance is low and our goal was to assess the complete nuclear transcriptome. We hypothesize that nuclear-enriched mRNAs may be selectively nuclear-retained or unstable following their export to the cytoplasm, whereas poly(A)+-enriched mRNAs may be more rapidly synthesized, processed, and exported and maintained in the cytoplasm.
Access of nuclear contents enables monitoring of nuclear RNA, posttranslational histone modifications associated with chromatin, DNA methylation, and protein abundance, which can aid exploration of epigenetic and transcriptional regulatory mechanisms. INTACT can enable profiling all of these levels of regulation from the same preparation of nuclei, as shown for mouse neuronal cells (Mo et al., 2015). Access to nuclei can also improve chromatin immunopurification (Wang and Deal, 2015) and chromosome conformation studies (Rodriguez-Granados et al., 2016), enable RNA-protein interaction analyses (Foley et al., 2017), as well as refine proteomic analyses by allowing access to nuclei of discrete cell- or tissue-types (Amin et al., 2014). INTACT and other methods for isolation of nuclei vary in their advantages and disadvantages. INTACT has similar applications as FANS, used to assay nuclear poly(A)+ RNA, endoreduplication (Zhang et al., 2008), and regions of open chromatin by assay for transposase accessible chromatin-sequencing (Lu et al., 2017; Bajic et al., 2018). INTACT constructs can be driven by near-constitutive, cell-type- and region-specific promoters, enabling assay of subpopulation of nuclei of multicellular organs (Deal and Henikoff, 2010). Users can take advantage of the GFP signal from the NTF to define the expression patterns and confirm nuclear localization in cells of interest. As distinctions in nuclear and poly(A)+ mRNA populations (Fig. 5) could reflect differences in regulation of individual mRNAs, INTACT could be a good companion method for TRAP (translating ribosome affinity purification) that allows evaluation of RNAs engaged in translation (Zanetti et al., 2005; Mustroph et al., 2009). Such studies could advance understanding of the multiple levels of regulation that fine tune the expression of individual genes. Moreover, use of the same promoters to drive INTACT and TRAP constructs might be used to target the same regions and cell types to obtain a comparative readout of nuclear pre-mRNA and translated mRNAs. The laser capture microdissection of tissue performed on rice (Jiao et al., 2009; Takehisa et al., 2012) and other species is not limited by genotype, whereas INTACT, fluorescent-activated cell sorting, and TRAP require transgenic plants (reviewed by Bailey-Serres, 2013). This disadvantage may be outweighed by the advantage of INTACT and TRAP over other methods of isolation of nuclei and transcripts from defined cells, as tissues may be cryopreserved prior to biochemical purifications. These technologies will likely complement single-cell transcriptomic and epigenomic analyses developed for animals but not yet reported in plants (Tanay and Regev, 2017). We propose that INTACT and TRAP are appropriate for evaluation of gene dynamics in individual cell types at multiple scales of regulation in response to environmental stimuli, which can help define genetic mechanisms critical to plant acclimation and adaptation to extremes in environment.
MATERIALS AND METHODS
Plant Material, Growth Conditions and Transformation
The rice genotype Oryza sativa japonica cv Nipponbare was used for gene cloning and plant transformation. Agrobacterium-mediated transformation of embryogenic calli derived from rice mature seed embryos and plant regeneration were performed as described by Sallaud et al., 2003. After transfer to soil containing pots, plants were grown in a greenhouse (28°C day/20°C night under natural light conditions) at the University of CA, Riverside to obtain leaf tissue used for DNA analyses and the production of seeds. For isolation of nuclei and RNA, seeds from transgenic lines were dehulled and surface sterilized in 50% (v/v) bleach solution for 30 min and then rinsed with sterile distilled water. For nuclei isolation, seedlings were grown on plates (10 cm × 10 cm) containing Murashige and Skoog standard medium agar (1% w/v) and 1% w/v Suc, during 7 d in a growth chamber (16 h day/8 h night; at 28°C/25°C day/night; 110 μEm−2s−1), in a greenhouse (28°C day/20°C night under natural light conditions) or in the field during the autumn at the University of CA, Riverside, Agricultural Experiment Station. The apical zone of the seminal and crown roots (1 cm root tips) or the shoots were harvested directly into liquid nitrogen, pulverized and stored at −80°C until use.
NTF Plasmids
A binary vector for INTACT in rice (O. sativa L.) was constructed as described in Ron et al. (2014) using the Multisite Gateway vector pH7WG instead of pK7WG (https://gateway.psb.ugent.be/search). All primers used for construction are listed in Supplemental Table S3A. The NTF version 1 (OsNTF1) construct was made by replacing the Arabidopsis (Arabidopsis thaliana) WPP domain for the one of rice (OsWPP; LOC_Os05g46560.1, AK242655; Rose and Meier, 2001). This was comprised of a 345-nucleotide region of the main open reading frame (amino acids 1–115). These were introduced via partial digestion (StuI/MfeI) and ligation. The epitope-tagged Escherichia coli biotin ligase (mBirA + 3xmyc) was replaced by a synthesized (GenScript) codon use-optimized version (KpnI/SnaBI), using the most frequently used codons in rice (http://www.kazusa.or.jp/codon/). For versions 2 and 3 (OsNTF2 and OsNTF3), the WPP domain was replaced with a C-terminal region of WIP to anchor the synthetic protein to the nuclear envelope. Enhanced GFP (eGFP) was amplified with primers to introduce StuI/MfeI sites at the 5′- and SnaBI/PacI/AvrII at the 3′-end, respectively, and incorporated to pTOPO-TA (Life Technologies). The BLRP was amplified from the OsNTF1 binary vector to place StuI/MfeI sites at the 5′- and 3′-ends, respectively. This product was ligated upstream of eGFP. Inverted T-DNA nos and 35S CaMV terminators were amplified with SnaBI/PacI sites and ligated downstream of eGFP. This cassette was digested and ligated to replace OsWPP-GFP-BLRP in the OsNTF1 binary vector using the sites SnaBI/StuI. The correct orientation was checked by PCR. C-terminal domains from KASH protein coding regions OsWIP2 (LOC_Os09g30350.1, AK241295.1) and OsWIP3 (LOC_Os08g38890.1, AK071174) were amplified from root O. sativa L. cv Nipponbare cDNA with primers to generate PacI/AvrII sites (Xu et al., 2007). The PCR products were ligated to generate BLRP-GFP-OsWIP2 (OsNTF2) and BLRP-GFP-OsWIP3 (OsNTF3) nuclear envelope-tagging proteins. For all binary constructs, Os-codon BirA was made by coupling the promoter of OsACT2 (LOC_Os10g36650; from −2550 bp to the start codon, O. sativa L. cv IR64) using a unique KpnI site for cloning 3′ to the NTF and in the opposite orientation in the binary vector pH7WG backbone to generate the sequence verified constructs p35S:OsNTF1/OsWPP, p35S:OsNTF2/OsWIP2, and p35SOsNTF3/OsWIP3. Constructs were electroporated into Agrobacterium tumefaciens EHA105A prior to use in plant transformation.
Generation of T-DNA-Enriched Libraries for Mapping of the Insertion Site
Young leaf tissue from transgenic lines was frozen in liquid nitrogen and stored at -80°C. For DNA extraction, 25 mg of sample was placed in a 2 mL microcentrifuge tube along with three small stainless-steel ball bearings. The samples were kept in liquid nitrogen before and after tissue disruption using a Tissue Lyser II (Qiagen) for three 1 min cycles at 30 Hz, stopping every minute to chill the samples in liquid nitrogen. Tissue Lyser II blocks were prechilled in –20°C before use. Genomic DNA (gDNA) was extracted as described in (Edwards et al., 1991), including two chloroform extractions. Extracted gDNA quality was assessed by electrophoresis in 1.2% (w/v) agarose gels and quantified using a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technology). When different samples contain regions near one T-DNA border that can be identifiable after sequencing, it is possible to pool DNA from different samples before library preparation. Pools of 1 µg gDNA were made and diluted to 100 µL with DNAse free water, then sheared by sonication (Bioruptor, Diagenode) in a 4°C water/ice bath at low intensity for 25 min with intervals set to 30 s on/30 s off. The ice bath was replaced every 5 min. DNA fragments between 300 and 500 bp were selected using Agencourt AMPure XP beads according to the manufacturer’s instructions (Beckman Coulter). DNA binding to these beads depends upon the concentration of PEG-8000 in 1.25 m NaCl. Captured gDNAs were evaluated for the desired fragment size by electrophoresis in 2% (w/v) agarose gels.
Illumina-compatible libraries of 420 to 620 bp fragments were prepared by performing end repair, dA tailing, adapter ligation, and amplification as described in Wang et al. (2011) with the following modifications. gDNA fragments were ligated with an universal adapter and amplified for 13 cycles of PCR using unique Tru-seq (Illumina) barcode adapters. To avoid the presence of adapter dimers, three rounds of AMPure XP bead purifications using 17.5% (w/v) PEG-8000 were performed, two before amplification and one after. Library quality was assessed using a Qubit fluorometer (Life Technologies), and the DNA 1000 Assay on an 2100 BioAnalyzer (Agilent Technologies). Library concentration ranged from 0.5 to 45 ng/μL and averaged 2.8 ng/μL, with and average size ∼500 bp. Based on concentrations obtained by DNA 1000 Assay, libraries were multiplexed to obtain a total of 500 ng and concentrated using a Speedvac concentrator (Savant).
Hybridization and sequence capture was achieved using a modification of the xGen Target Capture Protocol (IDT Technologies). This method provides a means for enriching and sequencing specific regions of interest in a kit-less option to the procedures provided by Inagaki et al. (2015) and Jupe et al. (2014). Four 5′-biotinylated oligos targeting the right and left border sequences were used for the hybridization. Supplemental Table S3B lists primers used for target capture. Blocking oligos were designed to avoid cross hybridization of universal adapters. Salmon sperm DNA was substituted for Cot-human DNA. The hybridization mix was incubated at 62°C for 48 h. DNA hybrids were captured for 30 min at room temperature using Streptavidin MyOne C1 Dynabeads (Life Technologies). Beads were washed for 5 min at 62°C with 1× SSC (150 mm NaCl, 15 mm Na-Citrate, pH 7.0), 0.1% (w/v) SDS, and then with 0.1× SSC, 0.1% (w/v) SDS twice at 62°C and once at room temperature. The final wash was performed with 0.2× SSC. After bead capture, DNA was eluted by incubation for 10 min in 50 µL 125 nm NaOH, diluted with 50 µL Tris-HCl (pH 8.8) and purified using AMPure XP beads. T-DNA enriched DNA and was amplified by 14 cycles of PCR using primers P5 and P7. The quality of the final multiplexed libraries was assessed by use of the Qubit fluorometer and an Agilent Bioanalyzer DNA 1000 assay. The efficiency of hybridization and capture was quantified by quantitative real-time PCR. Primers corresponding to the left and right T-DNA border sequences were used to quantify the abundance of insertion site fragments before and after hybridization and after final amplification. For a full bench protocol, see Supplemental Appendix I.
Sequencing and Data Analysis for T-DNA Insertion Mapping
The Illumina-compatible library was sequenced on MiSeq (Illumina) using 500 cycles of MiSeq reagent kit v2 at the Institute for Integrative Genome Biology at the University of California Riverside. The resulting 2 × 250 bp sequence data were analyzed using R statistical software. Paired-end reads containing overlapping sequences were merged to obtain longer reads using the FLASH (fast length adjustment of short reads; http://ccb.jhu.edu/software/FLASH/) module. The GREP (globally search a regular expression and print; http://www.gnu.org/software/grep/) module was used to filter out reads containing the left and right T-DNA border sequences, and the BOWTIE2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) module was used to map the border sequences against the rice genome (MSU Rice Genome Annotation Project Release 7). Regions of candidate insertions were identified using the chipseq module (http://www.bioconductor.org/packages/release/bioc/html/chipseq.html). See Supplemental Appendix I.
Laser-Scanning Confocal Microscopy
Laser-scanning confocal microscopy imaging was performed using a confocal Leica SP5. For images in Figure 1 and S1, settings were 20× objective, 488 nm excitation laser at 50% power, a 56.7 µm pinhole and 900 smart gain (SG), except in Supplemental Figure S1 for A1, G1: 800 SG, A2, G2: 650 SG. For images in Figure 2, settings were 20× objective (zoom 10×), 405 nm excitation laser at 15% power, a 56.7 µm pinhole and 1050 SG.
Nuclei Purification by INTACT
Nuclei were purified from frozen and pulverized tissue as described previously for Arabidopsis (Wang and Deal, 2015) with only minor modifications, including use of a 30 µm filter to exclude 30 to 70 µm cellular debris from the crude extract and extend centrifugation times. Tissue was resuspended in an ice-cold mortar containing 10 mL of freshly prepared Nuclei Purification Buffer (NPB: 20 mm 3-(N-morpholino) propanesulfonic acid (MOPS), 40 mm NaCl, 90 mm KCl, 2 mm EDTA, 0.5 mm EGTA, 0.5 mm spermidine, 0.2 mm spermine, pH, 7.0) containing 200 µL Protease Inhibitor Cocktail (0.4×, Sigma, P9599) per 50 mL of buffer. The homogenized extracts were filtered through a 30 µm nylon mesh (Celltrics) to remove cell debris and centrifuged at 1000g for 15 min at 4°C to pellet nuclei. Nuclei were resuspended in 1 mL of NPB (this suspension is considered input of the purification), 25 µL were kept for microscopic visualization, and 200 µL for protein electrophoresis and western blot detection. Twenty-five microliters of M-280 streptavidin-coated Dynabeads (∼1.5 × 107 beads; Life Technologies, catalog # 11205D) were added to the nuclei. This mixture was slowly rotated to allow capture of the nuclei in a cold room at 4°C for 30 min. The nuclei/beads suspension was diluted to 14 mL with NPB supplemented with 0.1% (v/v) Triton X-100 (NPB-T), in a 15 mL Falcon tube, mixed thoroughly and placed in a 15 mL magnet (adapted 50 mL tube magnet New England Biolabs (NEB, S1507S)) to capture bead-bound nuclei for 7 min at 4°C. The supernatant was carefully removed using a plastic Pasteur pipette, taking care to remove bubbles to avoid disturbing the beads. Beads were resuspended in 14 mL of cold NPB-T, placed on a rotating mixer for 30 s, and then placed back in the 15 mL magnet to capture the beads-nuclei at 4°C for 7 min. This wash step was repeated and bead-bound nuclei were resuspended in 1 mL of NPB-T and transferred to a new tube. Estimation of nuclei yield and analysis of purity via protein detection and western blot was performed as described previously (Deal and Henikoff, 2011). For RNA extraction, the bead-bound nuclei mixture was placed on a 2 mL tube magnet for 4 min at 4°C to capture the nuclei, which were resuspended in 20 µL NPB before proceeding to the extraction. For comparison, nuclei were isolated without the aid of INTACT according to the protocol described by Choudhary et al. (2009).
Protein Detection by Western Blotting
Crude cellular protein, input of INTACT, and purified nuclei were separated by 12% (w/v) SDS-PAGE and silver stained or used in immunoblot analyses. Proteins loaded in Figure 3, B to D, were quantified using Qubit Protein Assay Kit. This dye-based method overestimated the concentration of nuclear samples because of their higher concentration of proteins with elevated pI such as histones. Biotinylated proteins were detected using horseradish peroxidase (HRP)-conjugated streptavidin (1:2000; Thermo Fisher S911). Histone H3 was immunodetected using rabbit polyclonal antiserum (1:1,000; Abcam 1791). The small ribosomal subunit protein RPS6 was immunodetected using a rabbit polyclonal antiserum against Zea mays RPS6 (1:5,000; Williams et al., 2003) as primary antibody and an HRP-conjugated goat anti-rabbit IgG as secondary antibody (1:10,000; Bio-Rad). Visualization was performed using a chemiluminescence reagent according to the manufacturer’s instructions (Luminata Crescendo western HRP Substrate, Millipore, WBLUR0500) followed by exposure to x-ray film (Bioland, A03-02). Molecular mass markers (Pageruler 10-180 kD, Thermo Scientific) were used for each run.
Total and Nuclear RNA Extraction, rRNA Degradation, and Short-Read Library Preparation
Total poly(A)+ RNA was extracted from frozen tissue using polysome extraction buffer (Mustroph et al., 2009), followed by direct capture of poly(A)+ RNA using a biotinylated oligo dT according to Townsley et al. (2015). Nuclear RNA was extracted from 66,000 to 117,000 (Supplemental Table S2A) INTACT-purified nuclei using the RNeasy Micro kit (Qiagen), DNase-treated with Turbo DNase I (Thermo Fisher Scientific) and processed with Agencourt RNAClean XP beads (Beckman Coulter), per the manufacturer's’ instructions. rRNA was removed from the RNA using degradation method described by Morlan et al. (2012), which employs an RNase that specifically degrades RNA species in RNA:DNA duplexes, targeted by use of DNA oligomers that correspond to precursor and mature rRNA encoded in the nucleus.
To accomplish removal of specific abundant rRNA, 60-bp DNA probes were designed and synthesized (Integrated DNA Technologies) to complement the rice (O. sativa) nuclear rRNA sequences and their transcribed spacers. A maximum of six mismatches per probe was allowed for targeting O. sativa, Medicago truncatula, and Solanum lycopersicum, using additional species-specific probes when common probes were not possible due to sequence variation. Rice probes are listed in Supplemental Table S3C and were based on GenBank accessions KM036282 (18S rRNA - 5.8S rRNA - 26S rRNA, including the complete ITS and ETS regions) and DQ152232.1 (5S rRNA).
For degradation, probes were hybridized with up to 1 µg RNA in a 6-μL reaction containing 0.1 μm of each probe and 1 μL of 5× hybridization buffer (0.5 m Tris-HCl, pH 7.0, 1 m NaCl). The probe concentration was scaled down proportionally for lower amounts of RNA. For denaturation and hybridization, the sample was heated to 95°C for 2 min, cooled to 45°C at 0.1°C/s, and incubated at 45°C for 5 min. For RNase treatment, the hybridization reaction at 45°C was increased to 10 μL by adding 5 U of thermostable RNase H Hybridase (Epicentre) and 1 μL of Hybridase buffer (500 mm Tris-HCl, pH 7.4, 1 m NaCl, 200 mm MgCl2), and incubated at 45°C for 30 min. To remove the DNA probes and degraded RNA from the samples, the sample was retreated with Turbo DNase I (Thermo Fisher Scientific) and cleaned up with Agencourt RNAClean XP beads (Beckman Coulter). For a full bench protocol, see Supplemental Appendix II. RNA-seq libraries of pre/rRNA subtracted nuclear and poly(A)+ selected total mRNA were prepared as conventional non-strand specific libraries described by Townsley et al. (2015).
RNA-seq and Data Analysis
Total poly(A)+ and nuclear RNA libraries were sequenced on HiSeq 3000 to obtain 50-nucleotide single-end reads. All data analysis steps were performed on the University of California Riverside Institute for Integrative Genome Biology high-performance bioinformatics cluster (http://www.bioinformatics.ucr.edu/), supported by NSF MRI DBI 1429826 and NIH S10-OD016290. R packages from Bioconductor, including systemPipeR (Girke, 2014) were used. Quality reports of raw reads were generated with the FastQC package (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Raw reads were mapped with the splice junction aware short read alignment suite Bowtie2/Tophat2 to the mitochondrial and chloroplast genome as a first filter before mapping to IGRSP1.0-30 rice sequence (http://plants.ensembl.org/Oryza_sativa/Info/Index), allowing unique alignments with ≤2 nucleotide mismatches. Expression analyses were performed by generating read count data for features of exons-by-genes using the summarizeOverlaps function from the GenomicRanges package (Juntawong et al., 2014). Statistical analysis of differentially expressed genes and other feature types was performed with the Generalized Linear model approach from the EdgeR package (http://bioconductor.org/packages/release/bioc/html/edgeR.html) to obtain fold change (fold difference) and false discovery rate (FDR) confidence filters, applied as indicated. To increase robustness, only genes identified as differentially expressed with both DEseq2 and EdgeR were used for downstream analysis. Enrichment analysis of Gene Ontology terms was performed with systemPipeR using the BioMart database. Gene feature read counts were determined over ranges produced with the function genFeatures on systemPipeR. Coverage plot over transcripts were calculated and produced with the functions computeMatrix and plotHeatmap from deepTools (http://deeptools.readthedocs.io/en/latest/index.html). Genes containing a high intron/exon coverage ratio were visually inspected using IGV and filtered from the analysis of feature counting and coverage plots, as these were high abundance short sequences that spuriously mapped to introns (Supplemental Table S4).
Data Visualization and Clustering
BAM files were generated for each sample. These were processed into bedgraphs using to bedtools genomecov command (Quinlan and Hall, 2010) and scaled by number of aligned reads to exonic regions of genes in each bam using the −scale argument. Bedgraph files were compressed to bigwigs using bedGraphToBigWig (http://genome.cse.ucsc.edu/index.html) and visualized using the Integrative Genome Browser (http://software.broadinstitute.org/software/igv/). Scatter plots and density plots were made using the ggplot2 (Wickham, 2006) library for R.
Accession Numbers
RNA-seq data were deposited in the Gene Expression Omnibus database from the National Center for Biotechnology Information under accession number GSE99122.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NTFs subcellular localization for a second independent transformation event.
Supplemental Figure S2. Detection of biotinylated OsNTF proteins in rice.
Supplemental Figure S3. Nuclei isolation from roots using INTACT from a second independent event.
Supplemental Figure S4. Coverage of gene features for nuclear and poly(A)+ RNA libraries.
Supplemental Figure S5. Complexity evaluation of nuclear and poly(A)+ RNA libraries.
Supplemental Table S1. Transgenic lines insertion sites.
Supplemental Table S2. Nuclear RNA and total poly(A)+ transcriptome dataset.
Supplemental Table S3. Primer and probe lists.
Supplemental Table S4. List of transcripts with high intron/exon coverage ratio
Supplemental Appendix I. Mapping of T-DNA insertion sites using sequence capture and Illumina-Miseq.
Supplemental Appendix II. Method for rRNA degradation.
Acknowledgments
We thank Maureen Hummel, Michael Covington, Marko Bajic, Donnelly West, and Kristina Zumstein for active discussions throughout this project.
Footnotes
This work was supported by the United States National Science Foundation (NSF) Plant Genome grant no. IOS-1238243 to R.D., N.R.S., S.M.B., and J.B.-S. and by a Finnish Cultural Foundation postdoctoral fellowship to K.K. A NSF Research Experiences for Undergraduates supplement (IOS-1238243) and site program (DBI-1461297) supported S.C. and J.V., respectively.
Articles can be viewed without a subscription.
References
- Amin NM, Greco TM, Kuchenbrod LM, Rigney MM, Chung M-I, Wallingford JB, Cristea IM, Conlon FL (2014) Proteomic profiling of cardiac tissue by isolation of nuclei tagged in specific cell types (INTACT). Development 141: 962–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J. (2013) Microgenomics: Genome-scale, cell-specific monitoring of multiple gene regulation tiers. Annu Rev Plant Biol 64: 293–325 [DOI] [PubMed] [Google Scholar]
- Bajic M, Maher KA, Deal RB (2018) Identification of open chromatin regions in plant genomes using ATAC-Seq. Methods Mol Biol 1675: 183–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary MK, Basu D, Datta A, Chakraborty N, Chakraborty S (2009) Dehydration-responsive nuclear proteome of rice (Oryza sativa L.) illustrates protein network, novel regulators of cellular adaptation, and evolutionary perspective. Mol Cell Proteomics 8: 1579–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S (2010) A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell 18: 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S (2011) Histone variants and modifications in plant gene regulation. Curr Opin Plant Biol 14: 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Ip P-L, Nawy T, Mello A, Birnbaum KD (2015) Quantification of cell identity from single-cell gene expression profiles. Genome Biol 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley SW, Gosai SJ, Wang D, Selamoglu N, Sollitti AC, Köster T, Steffen A, Lyons E, Daldal F, Garcia BA, et al. (2017) A global view of RNA-protein interactions identifies post-transcriptional regulators of root hair cell fate. Dev Cell 41: 204–220.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T. (2014) SystemPipeR: NGS workflow and report generation environment. UC Riverside. https://github. com/tgirke/systemPipeR [DOI] [PMC free article] [PubMed]
- Goldberg RB, Hoschek G, Kamalay JC, Timberlake WE (1978) Sequence complexity of nuclear and polysomal RNA in leaves of the tobacco plant. Cell 14: 123–131 [DOI] [PubMed] [Google Scholar]
- Gosai SJ, Foley SW, Wang D, Silverman IM, Selamoglu N, Nelson ADL, Beilstein MA, Daldal F, Deal RB, Gregory BD (2015) Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol Cell 57: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Henry IM, Lieberman MC, Comai L (2015) High-throughput analysis of T-DNA location and structure using sequence capture. PLoS One 10: e0139672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J (2014) Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 111: E203–E212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe F, Chen X, Verweij W, Witek K, Jones JDG, Hein I (2014) Genomic DNA library preparation for resistance gene enrichment and sequencing (RenSeq) in plants. Methods Mol Biol 1127: 291–303 [DOI] [PubMed] [Google Scholar]
- Lu Z, Hofmeister BT, Vollmers C, DuBois RM, Schmitz RJ (2017) Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015) Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86: 1369–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlan JD, Qu K, Sinicropi DV (2012) Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS One 7: e42882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-H, Chung PJ, Juntawong P, Bailey-Serres J, Kim YS, Jung H, Bang SW, Kim Y-K, Do Choi Y, Kim J-K (2012) Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol 159: 1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Granados NY, Ramirez-Prado JS, Veluchamy A, Latrasse D, Raynaud C, Crespi M, Ariel F, Benhamed M (2016) Put your 3D glasses on: Plant chromatin is on show. J Exp Bot 67: 3205–3221 [DOI] [PubMed] [Google Scholar]
- Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, et al. (2014) Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol 166: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I (2001) A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA 98: 15377–15382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al. (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, Minami H, Namiki N, Inukai Y, Nakazono M, et al. (2012) Genome-wide transcriptome dissection of the rice root system: Implications for developmental and physiological functions. Plant J 69: 126–140 [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A (2017) Scaling single-cell genomics from phenomenology to mechanism. Nature 541: 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarinova T, Elhaik E, Pellegrini M (2013) Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol Evol 5: 1443–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR (2015) BrAD-seq: Breath Adapter Directional sequencing: A streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front Plant Sci 6: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Deal RB (2015) Epigenome profiling of specific plant cell types using a streamlined INTACT protocol and ChIP-seq. Methods Mol Biol 1284: 3–25 [DOI] [PubMed] [Google Scholar]
- Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP (2011) A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS One 6: e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2006) ggplot: An implementation of the grammar of graphics in R. R package version 0.4.0
- Williams AJ, Werner-Fraczek J, Chang I-F, Bailey-Serres J (2003) Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol 132: 2086–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Meulia T, Meier I (2007) Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol 17: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang I-F, Gong F, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Barthelson RA, Lambert GM, Galbraith DW (2008) Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol 147: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Brkljacic J, Meier I (2008) Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Meier I (2015) The plant nuclear envelope as a multifunctional platform LINCed by SUN and KASH. J Exp Bot 66: 1649–1659 [DOI] [PubMed] [Google Scholar]