WRKY75 functions as a component of the regulatory network in Arabidopsis that modulates the onset and progression of floral initiation through GA signaling.
Abstract
Flowering time is tightly controlled by both endogenous and exogenous signals. Although several lines of evidence have suggested the involvement of WRKY transcription factors in floral initiation, the underlying mechanisms and signaling pathways involved remain elusive. Here, we newly identified Arabidopsis (Arabidopsis thaliana) WRKY DNA binding protein75 (WRKY75) as a positive regulator of flowering initiation. Mutation of WRKY75 resulted in a delay in flowering, whereas overexpression of WRKY75 significantly accelerated flowering in Arabidopsis. Gene expression analysis showed that the transcript abundance of the flowering time integrator gene FLOWERING LOCUS T (FT) was lower in wrky75 mutants than in the wild type, but greater in WRKY75-overexpressing plants. Chromatin immunoprecipitation assays revealed that WRKY75 directly binds to the promoter of FT. Both in vivo and in vitro biochemical analyses demonstrated that WRKY75 interacts with DELLA proteins. We found that both REPRESSOR OF ga1-3 (RGA) RGA-LIKE1 (RGL1) and GA INSENSITIVE (GAI) can repress the activation ability of WRKY75, thereby attenuating expression of its regulon. Genetic analyses indicated that WRKY75 positively regulates flowering in a FT-dependent manner and overexpression of RGL1 or gain-of-function of GAI could partially rescue the early flowering phenotype of WRKY75-overexpressing plants. Taken together, our results demonstrate that WRKY75 may function as a new component of the GA-mediated signaling pathway to positively regulate flowering in Arabidopsis.
In plants, floral initiation is tightly controlled by intricate networks of signaling pathways that integrate a variety of environmental conditions and endogenous developmental cues. In Arabidopsis (Arabidopsis thaliana), the molecular and genetic basis of the transition from vegetative to reproductive growth has been well documented (Andrés and Coupland, 2012; Dally et al., 2014; Blümel et al., 2015). Dedicated genetic and molecular biological studies have identified several genetic pathways that regulate the floral transition, including the photoperiod, autonomous, vernalization, gibberellic acid (GA), and the age pathways (Michaels, 2009; Amasino, 2010; Srikanth and Schmid, 2011; Wang, 2014). These pathways mediate both endogenous and environmental signals and then converge to modulate the expression of a set of floral integrators, including FLOWERING LOCUS T (FT), LEAFY (LFY), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1; Han et al., 2008; Kumar et al., 2012; Riboni et al., 2013; Hu et al., 2014; Wang, 2014). They in turn activate several floral meristem identity genes, including LFY, APETALA1, CAULIFLOWER, and FRUITFULL (FUL), thereby leading to the phase transition from vegetative to reproductive growth (Han et al., 2008; Davis, 2009; Wang, 2014).
In Arabidopsis, FT is a well-characterized floral integrator that moves from the leaves to the shoot apex to initiate floral evocation (Kardailsky et al., 1999). Numerous studies have demonstrated that the expression of FT can be activated by a wide range of transcription factors. For example, CONSTANS (CO), which integrates the photoperiod signal and the circadian clock, can directly induce the transcriptional activation of FT under long day (LD) conditions (Suárez-López et al., 2001; An et al., 2004; Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009). The CIB proteins were shown to function redundantly to promote CRY2-dependent flowering by activating the transcription of FT (Liu et al., 2008, 2013). Moreover, PHYTOCHROME INTERACTING FACTOR4 and WRKY71 also participate in flowering time regulation by positively modulating FT expression (Kumar et al., 2012; Yu et al., 2016). However, the transcription of FT can also be negatively regulated by transcriptional repressors, such as FLOWERING LOCUS C (Helliwell et al., 2006; Searle et al., 2006), SHORT VEGETATIVE PHASE (Lee et al., 2007), TEMPRANILLO1 (Castillejo and Pelaz, 2008), and SCHLAFMŰTZE (Mathieu et al., 2009). Thus, to ensure the appropriate timing of flowering, plants have evolved complex regulatory mechanisms to make sure the expression of FT is well controlled.
GAs are a class of tetracyclic diterpenoid hormones that modulate diverse developmental processes throughout the plant lifecycle (Achard and Genschik, 2009). In Arabidopsis, genetic screening has identified the major components involved in GA perception and signaling: three GIBBERELLIN INSENSITIVE DWARF1 (GID1) GA receptors (GID1a, b, and c), the F-box ubiquitin ligase SLEEPY1 (SLY1), and five DELLA repressors [GA INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3] (Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; McGinnis et al., 2003; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). Upon GA perception, GID1 and SLY1 (as an SCFSLY1 complex) recruit DELLA proteins for ubiquitination and subsequent degradation, leading to the release and activation of various transcription factors that subsequently regulate downstream signaling cascades and modulate responses to GAs (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Dill et al., 2004; Harberd et al., 2009; Claeys et al., 2014). Numerous studies have demonstrated that the GA pathway plays an important role in floral initiation under both short days (SDs) and LDs. Wilson et al. (1992) demonstrated the GA synthesis mutants ga1-3 and ga1-6 both fail to flower under SDs but show very weak late-flowering phenotypes under LDs, implying that GAs play a major role in flowering under SDs. However, recent studies have provided evidence that GAs also participate in flowering time control under LDs. For example, the GA-deficient mutant ga1, Columbia-0 (Col-0), showed greatly reduced expression of FT, but this reduction could be fully rescued by exogenous GA3 treatment (Wang et al., 2016; Hou et al., 2014). The gid1a, b, and c triple mutant also exhibited a remarkably late flowering phenotype under LDs (Griffiths et al., 2006; Willige et al., 2007), whereas the quintuple DELLA-deficient mutant (rga-t2 gai-t6 rgl1-1 rgl2-1 rgl3-1, loss-of function) della showed an early flowering phenotype (Wang et al., 2016).
WRKY transcription factors comprise a large family of regulatory proteins in plants. In Arabidopsis, the WRKY transcription superfamily comprises an estimated 74 members that can be divided into three major structural groups, based both on the number of WRKY domains and the features of their zinc fingerlike motifs (Eulgem et al., 2000, 2005; Rushton et al., 2010). Genetic and molecular biological studies have demonstrated that WRKY members play important roles in various biotic and abiotic stress responses (Rushton et al., 2010; Chen et al., 2012), and several developmental and physiological processes (Johnson et al., 2002; Lagacé and Matton, 2004; Song et al., 2010; Xu et al., 2004; Miao and Zentgraf, 2007; Zhang et al., 2004; Zou et al., 2004, 2011; Chen et al., 2017). Recent functional analyses have provided some evidence to demonstrate that WRKY proteins also participate in flowering time regulation. For example, WRKY71 is involved in flowering regulation by directly activating the expression of FT and LFY (Yu et al., 2016). WRKY12 and WRKY13 oppositely modulate flowering time under SD conditions via the direct regulation of FUL (Li et al., 2016).
Despite the evidence for WRKY transcription factor involvement in flowering time regulation described above, the molecular mechanisms underlying the effects of WRKY transcription factors during floral initiation are still largely unknown. In this study, we used a molecular genetic approach to investigate the role of WRKY75 in flowering time regulation. Our results demonstrated that altered expression of the WRKY75 gene affects floral initiation. Further investigation indicated that WRKY75 acts as a transcriptional activator to transmit GA-mediated flowering signals by directly binding to downstream target sequences such as FT. Moreover, investigation of the associated mechanisms revealed that DELLAs physically interact with WRKY75 and repress its activation ability. Our results thus provide evidence that WRKY75 functions as a positive regulator of flowering in Arabidopsis.
RESULTS
Mutation of WRKY75 Delayed Flowering Time
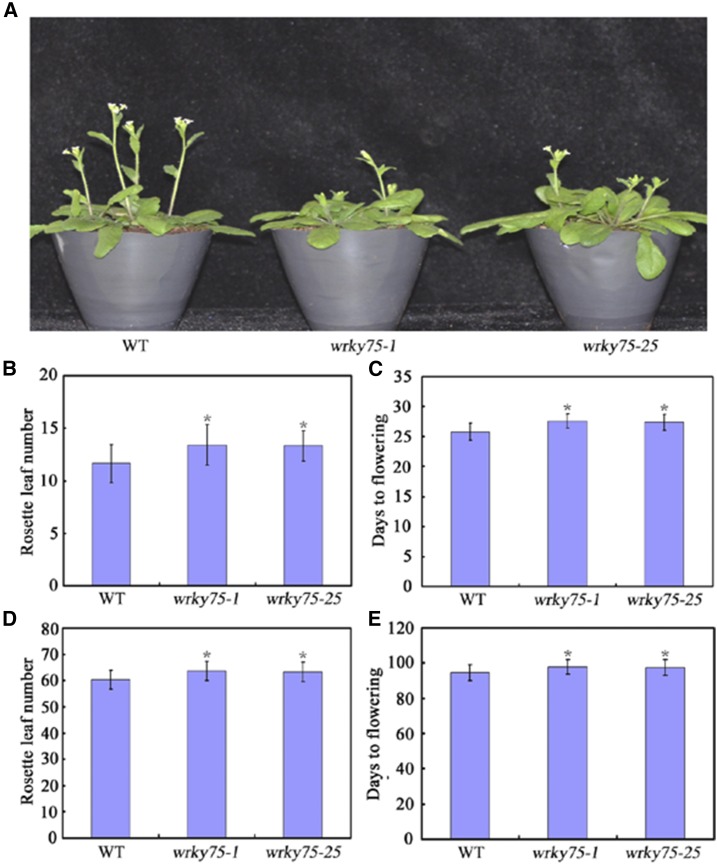
To investigate the possible regulatory roles of Arabidopsis WRKY transcription factors in plant developmental processes, we screened WRKY-associated T-DNA insertion mutants and RNAi lines to identify potential WRKY proteins that may be involved in flowering time regulation. From this screen, we identified two T-DNA insertion mutants (wrky75-1 and wrky75-25) with altered flowering time compared with the wild type. As shown in Supplemental Figure S1, quantitative real-time PCR (qRT-PCR) analysis demonstrated that WRKY75 was effectively knocked down by T-DNA insertion. Then, to determine the possible functions of WRKY75 in flowering-time regulation, we analyzed the flowering phenotypes of wrky75-1 and wrky75-25. Wild-type, wrky75-1, and wrky75-25 seeds were germinated simultaneously and grown in soil under the same growth conditions. As shown in Figure 1, A to C, under LD (16 h light/8 h dark) conditions, wrky75 mutant plants exhibited a relatively delayed flowering phenotype, as measured by the total rosette leaf number (RLN) and days from germination to flowering (DTF). Similarly, under SD conditions, wrky75 mutant plants also showed delayed flowering compared with their wild-type counterparts (Fig. 1, D and E). Thus, mutation of WRKY75 caused a delayed flowering time in Arabidopsis.
Figure 1.
Flowering phenotype of wrky75 mutant plants. A, Representative images of wrky75 mutant plants showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing more than 30 plants for each line. Representative plants were photographed. B and C, Flowering phenotype of wrky75 mutant plants assessed by RLN (B) and DTF (C) under LD conditions. D and E, Flowering phenotype of wrky75 mutant plants assessed by RLN (D) and DTF (E) under SD conditions. For (B) to (E), values are mean ± sd of approximately 30 plants (*P < 0.05). Asterisks indicate Student’s t-test significant differences. The experiments were repeated at least three times with similar results.
Ectopic Expression of WRKY75 Dramatically Accelerated Flowering Time
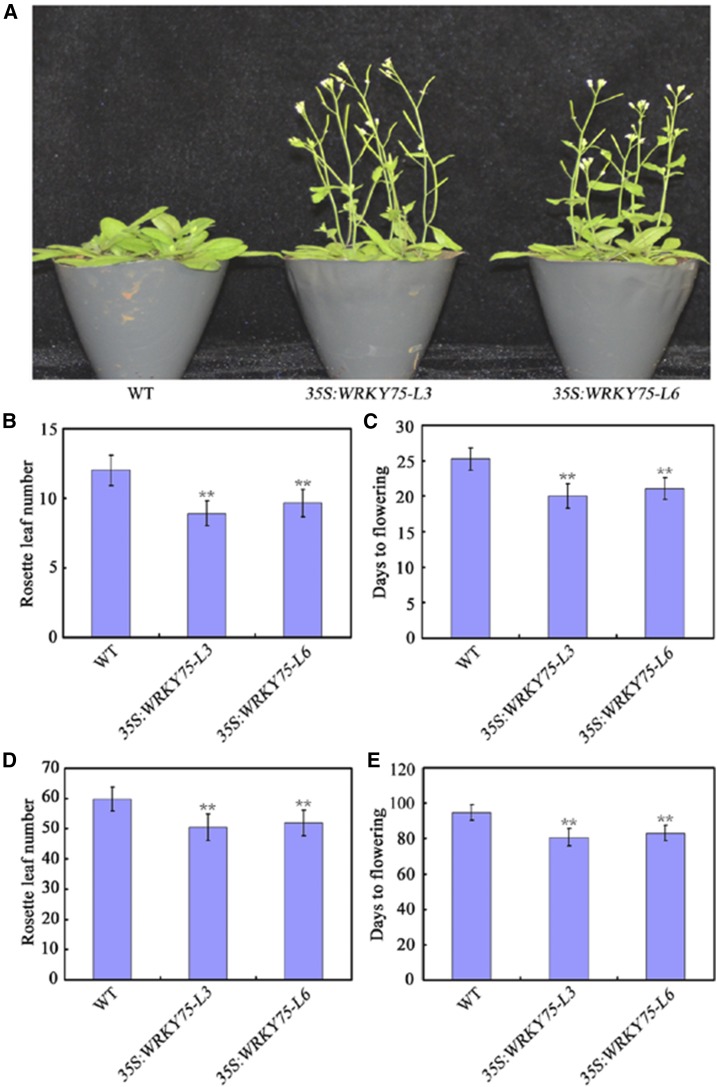
To further investigate the role of WRKY75 in flowering time regulation, we generated transgenic Arabidopsis plants constitutively expressing WRKY75 under the control of the Cauliflower mosaic virus 35S promoter. Two transgenic lines that showed obviously enhanced expression of WRKY75 were selected from 13 primary T1 WRKY75 overexpression lines for further analysis (Supplemental Fig. S2). In contrast to wrky75 mutant plants, flowering was clearly accelerated in 35S:WRKY75 plants compared with wild-type plants, as measured by total RLN and DTF, under both LD and SD conditions (Fig. 2). Constitutive overexpression of WRKY75 thus dramatically accelerated flowering time in Arabidopsis. These results confirmed that WRKY75 plays an important role in flowering time regulation.
Figure 2.
Flowering phenotype of 35S:WRKY75 transgenic plants. A, Representative images of 35S:WRKY75 transgenic plants showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing more than 30 plants for each line. Representative plants were photographed. B and C, Flowering phenotype of 35S:WRKY75 transgenic plants assessed by RLN (B) and DTF (C) under LD conditions. D and E, Flowering phenotype of 35S:WRKY75 transgenic plants assessed by RLN (D) and DTF (E) under SD conditions. For (B) to (E), Values are mean ± sd of approximately 30 plants (**P < 0.01). Asterisks indicate Student’s t-test significant differences. The experiments were repeated at least three times with similar results.
In Vivo Interaction of WRKY75 with the FT Promoter
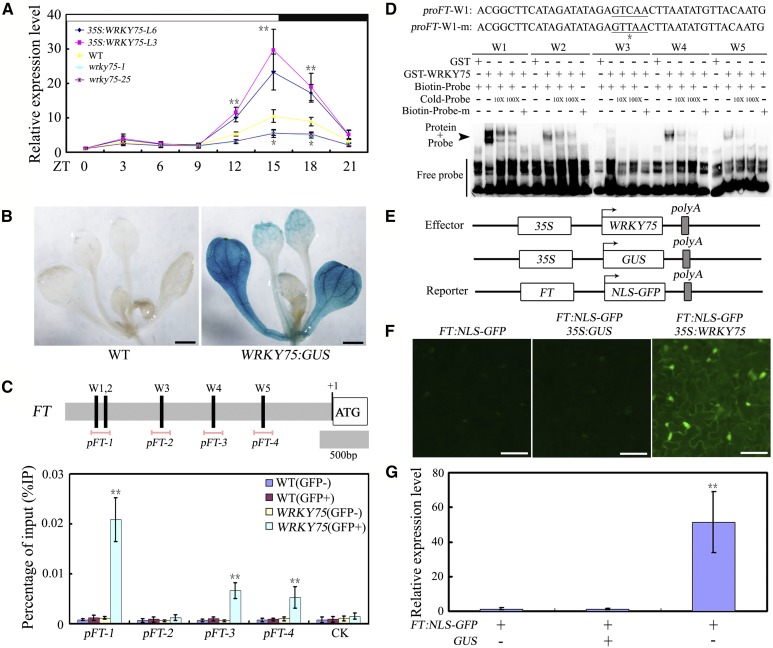
Considering that FT functions as a central node of floral integration and many external inputs are channeled into the transcriptional regulation of FT to regulate flowering (Corbesier et al., 2007; Tamaki et al., 2007; Andrés and Coupland, 2012; Pin and Nilsson, 2012), we then compared the FT expression levels among 35S:WRKY75 transgenic plants (L3 and L6), the wild type, and wrky75-1 and wrky75-25 mutant plants. As shown in Figure 3A, the FT transcript levels were substantially higher in 35S:WRKY75 transgenic plants but were relatively reduced in WRKY75 mutant plants compared with those in wild-type plants. Given that FT mainly expressed in the vascular tissues (Adrian et al., 2010), we then tested whether WRKY75 also has expression in the vascular tissues. Analyses of theβ-glucuronidase (GUS) reporter expression in transgenic plants expressing GUS under control of the WRKY75 promoter demonstrated that this promoter was active in the vascular bundle cells (Fig. 3B). These results suggested that WRKY75 may promote flowering through the activation of FT.
Figure 3.
WRKY75 promotes flowering by activating FT transcription. A, Expression of FT in the indicated genotypes. Ten-d-old plants grown under normal growth conditions (22°C, LD) were harvested at the indicated ZT for total RNA extraction and qRT-PCR assays. Transcript levels of FT in untreated Col-0 leaves at ZT0 were arbitrarily set to 1. Values are mean ± sd of three independent biological replicates. Two-way ANOVA was performed for statistical analysis; asterisks indicate significant differences as compared to controls, *P < 0.05, **P < 0.01. B, WRKY75:GUS expression in 12-d-old seedlings. The expression of WRKY75 was detected by GUS staining. Representative seedlings were photographed. Scale bar: 1 mm. C, The promoter structure of the FT gene and fragment used in the ChIP assay. The upper panel shows schematic representation of the FT promoter regions containing W-box clusters. The diagram indicates the number and relative position of the W-boxes in the respective promoters relative to the ATG start codon. In the promoter fragment names, the prefix “p” indicates promoter. Pink lines indicate the sequences detected by ChIP assays. ChIP assays were performed with chromatin prepared from WRKY75:YFP-WRKY75:3′-WRKY75 transgenic plants, using an anti-GFP antibody (immunoprecipitated). ChIP results are presented as a percentage of input DNA. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences as compared to controls, **P < 0.01. D, The EMSA analysis of the binding of recombinant WRKY75 protein to the promoter of FT. The oligonucleotides (proFT-W1/2/3/4/5 and proFT-W1/2/3/4/5-m) were used as the probes. Underlining signifies W-box sequence and asterisk represents the mutated base in the W-box element (as exampled with W1). GST, GST-WRKY75, biotin-probe, labeled mutated probe, and unlabeled probe at a 10× and 100× molar excess were present (+) or absent (−) in each reaction. E, Schematic of the FT:NLS-GFP reporter and WRKY75 and GUS effectors. F, Transient expression assays showed that WRKY75 activates the expression of FT. GFP fluorescence was detected 48 h after coinfiltration with the indicated constructs. The experiment was repeated three times with similar results and representative photos were displayed. Scale bar, 50 μm. G, qRT-PCR analysis of the accumulation of GFP transcripts. Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with combinations of various constructs in (E). The N. benthamiana ACTIN gene was used as an internal control. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences as compared to controls, **P < 0.01. Biotin-Probe-m, biotin-labeled probe with a single nucleic acid mutation from TGAC to TAAC; Cold-Probe, unlabeled probe.
WRKY transcription factors function by binding directly to a putative cis-element, the W-box (T/CTGACC/T), in the promoters of their target genes (Eulgem et al., 2000; Ulker and Somssich, 2004). Our data suggested that WRKY75 plays an important role in flowering time regulation by positively modulating the expression of FT. Interestingly, a search of the Arabidopsis genome sequence uncovered several putative W-box elements in the promoter of FT. The presence of these elements indicated that the modulation we observed may have been caused by the direct interaction of WRKY75 with the FT promoter. To examine whether FT is a direct target of WRKY75, we first conducted chromatin immunoprecipitation (ChIP) experiments using WRKY75:YFP-WRKY75:3′-WRKY75 transgenic plants (Rishmawi et al., 2014). The ChIP-qPCR results showed that WRKY75 could bind to the promoter of FT via the W-box sequence (pFT-1, pFT-3, and pFT-4, respectively; Fig. 3C). Additionally, we conducted an EMSA with the GST-WRKY75 recombinant protein to determine the in vitro binding of WRKY75 to these regions. As shown in Figure 3D, WRKY75 was capable of binding to the probes containing W1, W2, W4, or W5, but not W3. The binding signals decreased after the addition of unlabeled wild-type competitors. In contrast, the WRKY75 protein did not bind to the mutant probe carrying a mutated W box (Fig. 3D). The GST protein alone also did not bind to the W boxes (Fig. 3D). These data suggest that WRKY75 directly binds to the promoter of FT.
To further confirm the positive regulatory function of WRKY75, we generated and analyzed an FT promoter-driven nuclear localization signal (NLS)-GFP fusion protein (FT:NLS-GFP) in transient expression assays in tobacco (Nicotiana benthamiana) leaves. FT:NLS-GFP was used as a reporter plasmid. Effector plasmids were generated that contained either a WRKY75 or a GUS gene driven by the CaMV 35S promoter (35S:WRKY75 and 35S:GUS; Fig. 3E). Coexpression of the WRKY75 gene with the reporter plasmid resulted in significantly enhanced GFP signals compared with the control (Fig. 3, F and G). This result supported the hypothesis that WRKY75 acts as a positive regulator of flower time control.
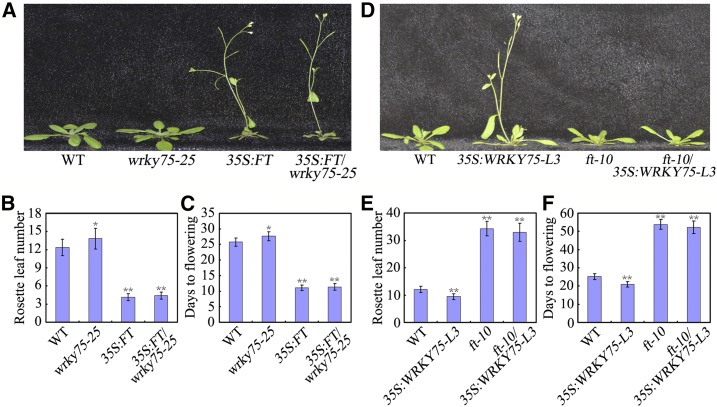
WRKY75 Promotes Flowering in an FT-dependent Manner
The phenotypic analysis, and biochemical and molecular evidence demonstrated that the transcription factor WRKY75 positively regulates flowering time through the direct activation of FT expression. To further confirm this conclusion, the genetic relationship between WRKY75 and FT was explored. The wrky75-25 mutant was crossed with 35S:FT transgenic plants, and the flowering phenotypes of wrky75-25 and 35S:FT/wrky75-25 were examined. Under our experimental conditions, we detected an early flowering phenotype in 35S:FT plants, and mutation of WRKY75 did not change this in terms of the RLN and DTF (Fig. 4, A to C), although the wrky75-25 mutant showed a delay in floral transition (Fig. 1). Additionally, we generated plants overexpressing WRKY75 in the ft-10 mutant background (ft-10/35S:WRKY75) by genetic crossing, and examined the floral phenotype of ft-10 and ft-10/35S:WRKY75. The mutation of FT changed the early flowering phenotype of 35S:WRKY75 (Fig. 4, D to F) so that it mimicked the clearly late flowering phenotype of ft-10. Thus, the genetic analysis indicated that WRKY75 acts upstream of FT and functions as a positive regulator of flowering time in an FT-dependent manner.
Figure 4.
The genetic analysis of WRKY75 and FT. 35S:FT/wrky75-25 and ft-10/35S:WRKY75-L3 were generated by genetic crossing, and then the flowering time phenotype of these genotypes was examined. A and D, Representative images of the indicated genotypes showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing more than 30 plants for each line. Representative plants were photographed. B, C, E, and F, Flowering phenotype of the indicated genotypes assessed by RLN (B and E) and DTF (C and F) under LD conditions. For (B), (C), (E), and (F), Values are mean ± sd of approximately 30 plants (*P < 0.05,**P < 0.01). Asterisks indicate Student’s t-test significant differences. The experiments were repeated at least three times with similar results.
Physical Interaction of WRKY75 with DELLA Repressors
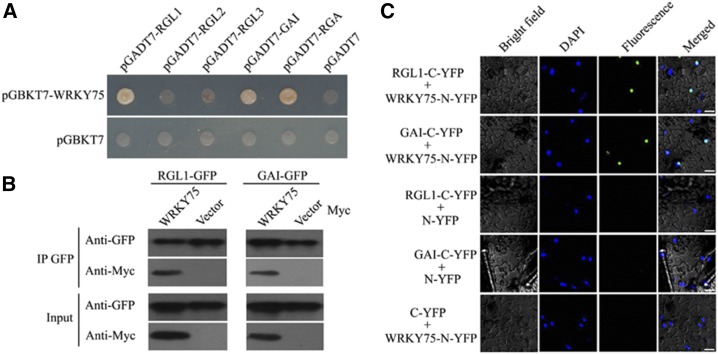
To understand how WRKY75 participates in flowering time regulation, we used the yeast two-hybrid system to identify its potential interaction partners. The full-length WRKY75 coding sequence was fused to the Gal4 DNA binding domain of a bait vector (BD-WRKY75). Yeast cells harboring the bait were transformed with a library of cDNAs containing inserts for prey proteins fused to GAL4-AD. After screening, three independent clones encoding RGL1 were identified by prototrophy for His and Ade. To confirm the interaction of these clones with WRKY75 in yeast, the open reading frame sequences of the clones were fused with the AD domain of the pGADT7 vector and used for further interaction experiments with WRKY75. The bait and prey vectors were cotransformed into yeast, and the protein-protein interactions were reconstructed (Fig. 5A). We further investigated the interactions of WRKY75 with all five Arabidopsis DELLA proteins in the yeast two-hybrid system. Besides RGL1, WRKY75 also interacted with GAI and RGA (Fig. 5A).
Figure 5.
Interaction between WRKY75 and DELLAs. A, Yeast two-hybrid assay analysis. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The GAL4 activation domain expressed by pGADT7 (shown as “AD”) was used as negative controls. The experiment was repeated three times with similar results and representative photos were displayed. B, CoIP analysis. GFP-fused RGL1 and GAI were immunoprecipitated using anti-GFP antibody, and coimmunoprecipitated Myc-WRKY75 was then detected using anti-Myc antibody. Protein input for GFP-RGL1 and GFP-GAI in immunoprecipitated complexes were also detected and are shown. The experiment was repeated three times with similar results and representative photos were displayed. C, BiFC analysis. Fluorescence was observed in nuclear compartments of N. benthamiana leaf epidermal cells; the fluorescence resulted from complementation of the C-terminal portion of YFP fused to RGL1 or GAI (RGL1-cYFP and GAI-cYFP) with the N-terminal portion of YFP fused to WRKY75 (WRKY75-nYFP). No signal was observed from negative controls. The experiment was repeated three times with similar results and representative photos were displayed. Scale bar: 25 μm. DAPI, 49,6-diamidino-2-phenylindole.
The interactions of WRKY75 with RGL1 and GAI were corroborated by coimmunoprecipitation (CoIP) and bimolecular fluorescence complementation (BiFC) assays. RGL1 and GAI were used as representatives in the CoIP and BiFC assays. For the CoIP analysis, Myc-WRKY75 and GFP-RGL1/GAI were coexpressed in Nicotiana benthamiana leaves. The protein complexes were incubated with anti-GFP and A/G-agarose beads, and then separated by SDS-PAGE for immunoblotting with an anti-MYC antibody. WRKY75 pulled down RGL1 and GAI (Fig. 5B). To determine whether these interactions also occur in plant cells, we then used BiFC. Full-length RGL1, GAI, and WRK75 proteins were fused to the N-terminal or C-terminal fragment of a yellow fluorescent protein (YFP), yielding RGL1-nYFP, GAI-nYFP, and WRKY75-cYFP, respectively. Agrobacterium cells harboring each interaction pair were infiltrated into N. benthamiana leaves. In parallel, empty vectors in combination with each fusion construct were coinfiltrated into N. benthamiana leaves. After 48-h incubation, the resultant YFP signals were observed by fluorescence microscopy. The samples coinfiltrated with an interaction pair showed YFP fluorescence in the cell nuclei, whereas none of the control samples yielded any signal (Fig. 5C). These results indicated that WRKY75 and its partners colocalize and interact in plant cell nuclei. Taken together, these results demonstrated that WRKY75 physically interacts with RGL1 and GAI, implying that WRKY75 functions as a direct target of DELLAs.
RGL1-mediated Repression of WRKY75 Activation Ability
Previous studies have revealed that DELLAs function as repressors by directly interacting with downstream transcription activators, such as MYC2, PHYTOCHROME INTERACTING FACTORS (PIFS), BRASSINAZOLE RESISTANT1 (BZR1), and ETHYLENE-INSENSITIVE3, to inhibit their DNA-binding and transcriptional activities (An et al., 2012; Hong et al., 2012; Xiang et al., 2012). Likewise, DELLAs can interfere with several components of hormonal and developmental signaling pathways through protein-protein interactions (Hou et al., 2010; Zhang et al., 2011; Qi et al., 2014). Several researches have also demonstrated that DELLAs can act as coactivators, for instance by interacting with INDETERMINATE DOMAINs (IDDs), including the GAF1 transcription factor, to mediate the GA responses (Fukazawa et al., 2014; Yoshida et al., 2014). Based on the fact that WRKY75 and RGL1/GAI play opposite roles in regulating floral transition, we proposed that interaction with DELLA repressors may repress the transcriptional activation ability of WRKY75 on target genes.
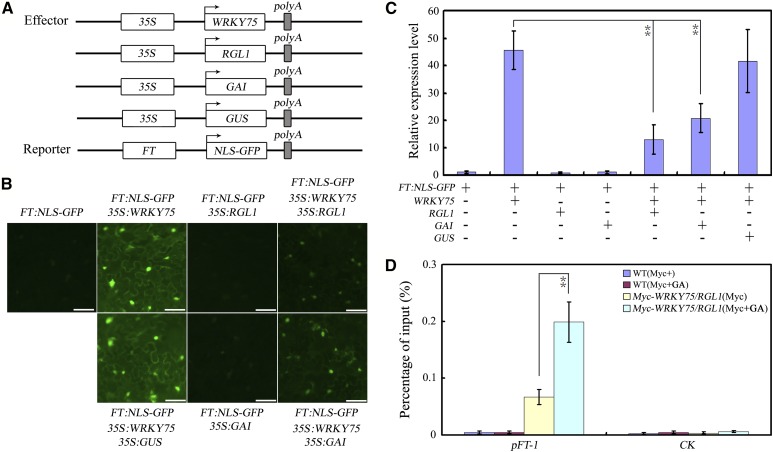
To test this possibility, FT:NLS-GFP was again used as a reporter plasmid. Effector plasmids were generated that contained either a WRKY75, RGL1, GAI, or GUS gene driven by the CaMV 35S promoter (35S:WRKY75, 35S:RGL1, 35S:GAI, and 35S:GUS; Fig. 6A). When the reporter construct was transformed into N. benthamiana leaves and incubated at 22°C, a very low fluorescence signal was observed (Fig. 6B). When FT:NLS-GFP and 35S:WRKY75 were coinfiltrated into N. benthamiana leaves, a much stronger fluorescence signal was observed (Fig. 6B). In contrast, coinfiltration of FT:NLS-GFP and 35S:RGL1 or 35S:GAI generated relatively lower fluorescence levels (Fig. 6B). Additionally, coinfiltration of FT:NLS-GFP with 35S:WRKY75 and 35S:RGL1 or 35S:GAI generated a dramatically weaker fluorescence signal than coinfiltration of FT:NLS-GFP with 35S:WRKY75 (Fig. 6B). As a control, coinfiltration of FT:NLS-GFP with 35S:GUS and 35S:WRKY75 was performed, but no obvious differences in fluorescence signal were observed compared with coinfiltration of FT:NLS-GFP and 35S:WRKY75 (Fig. 6B). Taken together, these results demonstrated that both GAI and RGL1 repressed the WRKY75 activity.
Figure 6.
RGL1 and GAI repress WRKY75 activation ability. A, Schematic of the FT:NLS-GFP reporter and WRKY75, RGL1, GAI, and GUS effectors. B, Transient expression assays showed that RGL1 and GAI repress transcriptional activation of WRKY75. GFP fluorescence was detected 48 h after coinfiltration with the indicated constructs. The experiment was repeated three times with similar results and representative photos were displayed. Scale bar, 50 μm. C, qRT-PCR analysis of the accumulation of GFP transcripts. Total RNAs were extracted from leaves of N. benthamiana coinfiltrated with combinations of various constructs in (A). The N. benthamiana ACTIN gene was used as an internal control. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences as compared to controls, **P < 0.01. D, RGL1 interferes with the binding of WRKY75 to its target genes (shown in Fig. 3A). 35S:Myc-WRKY75 was crossed with 35S:RGL1 to obtain Myc-WRKY75/RGL1 plants. ChIP assays were performed with chromatin prepared from Myc-WRKY75/RGL1 plants, using an antiMyc antibody (immunoprecipitated). ChIP results are presented as a percentage of input DNA. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences as compared to controls, **P < 0.01.
To further verify the effect of RGL1 or GAI on WRKY75’s transcriptional function, we analyzed relative GFP expression in N. benthamiana leaves. We detected high levels of GFP transcripts in FT:NLS-GFP- and 35S:WRKY75-coinfiltrated N. benthamiana leaves (Fig. 6C). In contrast, coexpression of the RGL1 or GAI protein with WRKY75 significantly suppressed GFP transcript accumulation (Fig. 6C). These results further supported the hypothesis that both RGL1 and GAI repress the transcriptional function of WRKY75.
To determine whether the binding of WRKY75 to its target genes is affected by RGL1 interaction, we crossed 35S:Myc-WRKY75 with 35S:RGL1, and named the progeny Myc-WRKY75/RGL1. Twelve-d-old Myc-WRKY75/RGL1 seedlings treated with GA or the corresponding solvent (mock) were used for ChIP-qPCR. The results showed that GA treatment enhanced the binding of WRKY75 to the FT promoter compared with mock treatment (Fig. 6D). Hence, this result suggested that RGL1 may decrease the binding ability of WRKY75 to its target genes in vivo.
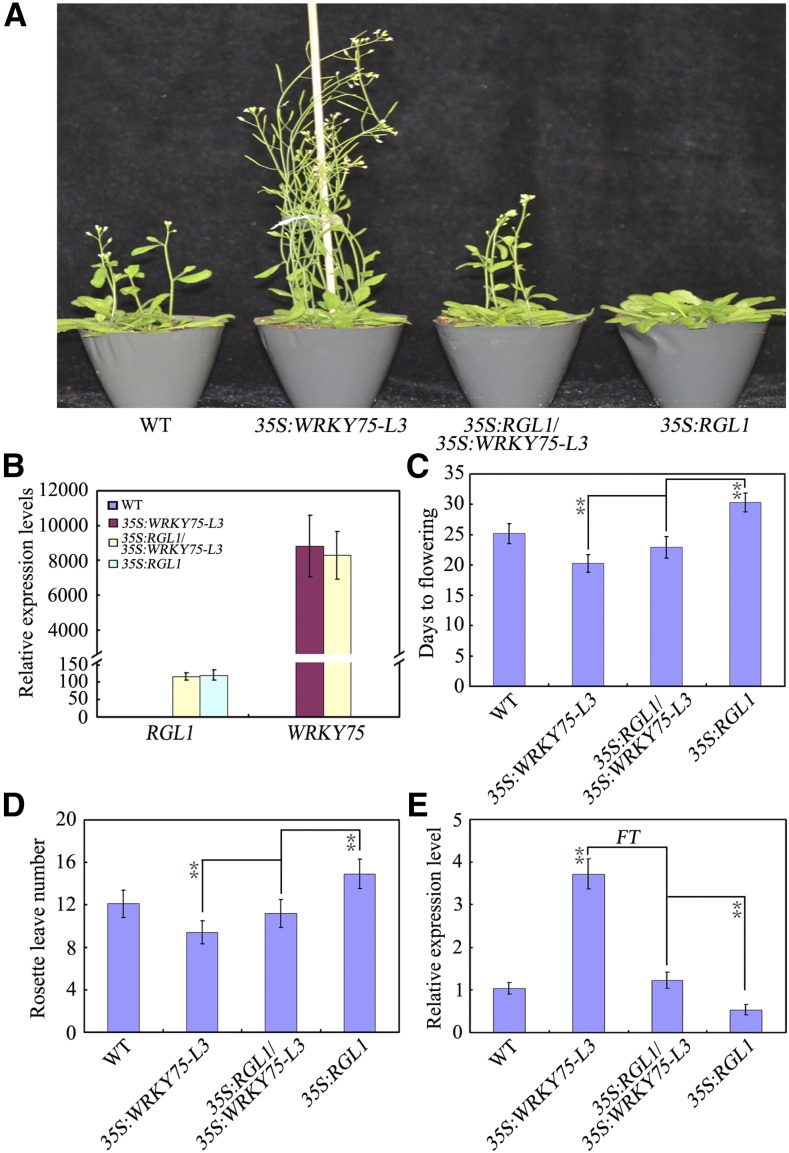
Overexpression of RGL1 or Gain of Function of GAI Partially Rescues the Early Flowering Phenotype of WRKY75-Overexpressing Plants
To further examine the regulatory effect of DELLA repressors on the transcriptional function of WRKY75 in Arabidopsis, we investigated whether overexpression of RGL1 repressed the early flowering phenotype induced by WRKY75 overexpression. We again used the Myc-WRKY75/RGL1 plants. The Myc-WRKY75/RGL1 plants showed a delayed flowering phenotype compared with 35S:RGL1 (Fig. 7, A to D), as measured by total RLN and DTF, under LD conditions. Furthermore, expression of FT was also higher in Myc-WRKY75/RGL1 plants than in 35S:RGL1 transgenic plants (Fig. 7E). We also saw similar results in Myc-WRKY75/gai-1 [a gain-of-function gai-1 (Col-0) mutant generated from the gai-1 (Ler) allele (a GA-insensitive mutant) by backcrossing gai-1 (Ler) with Col-0 three times] plants (Supplemental Fig. S3). These results suggested that overexpression of RGL1 or gain of function of GAI could partially suppress the early flowering phenotype in 35S:WRKY75 plants, and further supported the hypothesis that RGL1 and GAI repress the transcriptional function of WRKY75 in Arabidopsis.
Figure 7.
Overexpression of RGL1 partially rescues the early flowering phenotype of WRKY75 overexpressing plants. A, The flowering phenotypes of the indicated genotypes. Plants were grown under long days at 22°C. Three independent experiments were performed with each replica containing more than 30 plants for each line. Representative plants were photographed. B, Expression of WRKY75 and RGL1 in the indicated genotypes. RNA was isolated form 3-week-old leaves of the indicated genotype. ACTIN2 gene was used as an internal control. Values are mean ± sd of three independent biological replicates. C and D, Flowering phenotype of the indicated genotypes assessed by RLN (D) and DTF (E) under LD conditions. Values are mean ± sd of approximately 30 plants (**P < 0.01). Asterisks indicate Student’s t-test significant differences. The experiments were repeated at least three times with similar results. E, Expression of FT in the indicated genotypes. Ten-d-old plants grown under normal growth conditions (22°C, LD) were harvested at ZT15 for total RNA extraction and qRT-PCR assays. Transcript levels of FT in untreated Col-0 leaves were arbitrarily set to 1. Values are mean ± sd of three independent biological replicates. Asterisks indicate Student’s t-test significant differences (**P < 0.01).
Involvement of WRKY75 in GA-mediated Flowering Time Regulation
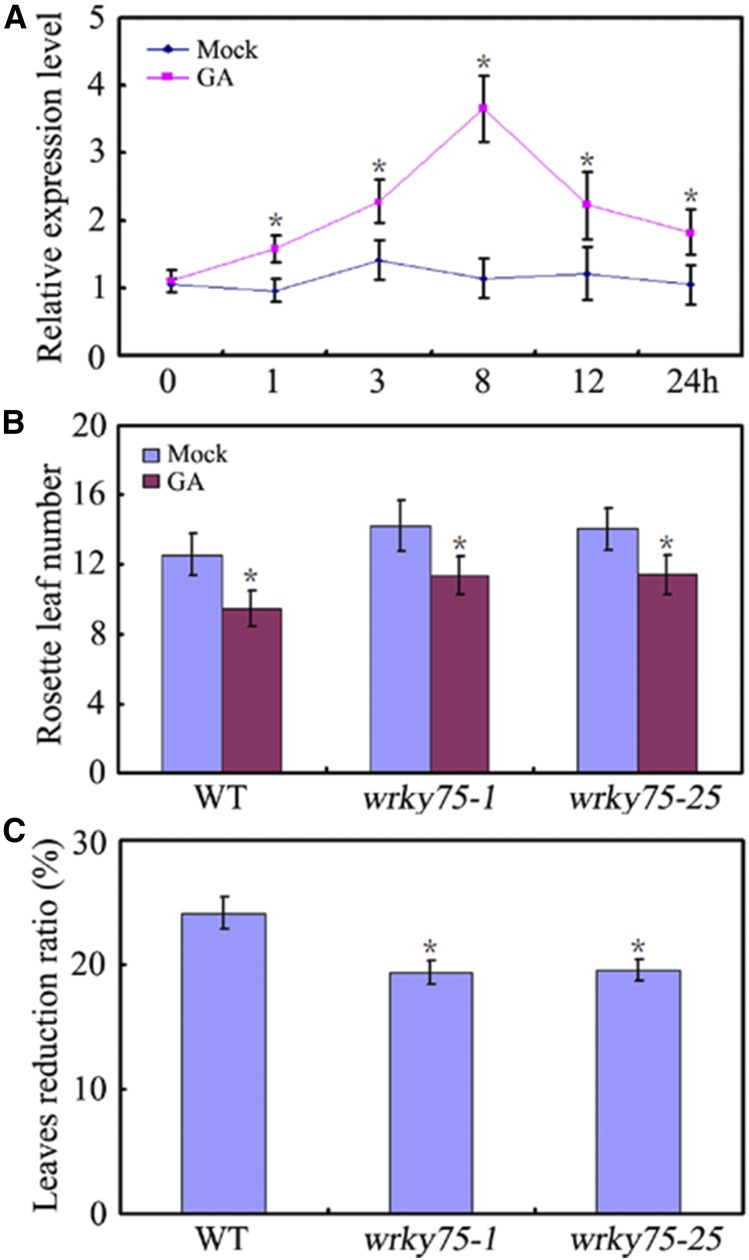
Based on the above results, we wondered whether knock-down of WRKY75 altered the response to GA compared with the wild type. If GA promotes flowering independent of WRKY75, flowering should be similarly accelerated in the wild type and wrky75 mutants. We first investigated the expression level of WRKY75 upon GA treatment. GA3 treatment induced the expression of WRKY75 (Fig. 8A), implying the possible involvement of WRKY75 in GA-mediated flowering time regulation. Consistent with this, knock-down of WRKY75 led to delayed GA-mediated flowering time compared with the wild type, as indicated by a relative decrease in leaf number (Fig. 8, B and C). Thus, our results demonstrated that WRKY75 is at least partially involved in the regulation of GA-mediated flowering time control.
Figure 8.
GA effects on WRKY75 mutants. A, Expression of WRKY75 upon GA treatment. RNA was isolated form the leaves treated with GA3 or Mock. Transcript levels of WRKY75 in untreated Col-0 leaves were arbitrarily set to 1. ACTIN2 gene was used as an internal control. Values are mean ± sd of three independent biological replicates (*P < 0.05). B, Flowering phenotype of the indicated genotypes grown under normal or GA3-treated conditions assessed by RLN under LD conditions. C, The reduction ratio in response to GA. The reduction ratio was calculated as [number of leaves (Mock) − number of leaves (GA3)]/number of leaves (Mock). Values are mean ± sd of approximately 20 plants (*P < 0.05). Asterisks indicate Student’s t-test significant differences. The experiments were repeated at least three times with similar results.
DISCUSSION
The proper timing of the floral transition is critical for reproductive success, and plants have evolved sophisticated and elaborate regulatory mechanisms to coordinately control flowering so that it occurs at the optimal time. Flowering time is controlled by both endogenous and exogenous signals. GA serves as an important endogenous signal to regulate flowering under both LD and SD conditions. Our results demonstrated that WRKY75 may function as a positive regulator in flowering time control via the GA pathway.
WRKY75 Positively Regulates Flowering in Arabidopsis
Although the WRKY factors comprise one of the largest groups of transcription factors in plants, and substantial progress in functional research on WRKY transcription factors has been achieved over the past 20 years (Rushton et al., 2010; Chen et al., 2012), little is known about their possible involvement in flowering time regulation. In Arabidopsis, several WRKY transcription factors, such as WRKY12, WRKY13 (Li et al., 2016), and WRKY71 (Yu et al., 2016), have been reported to participate in the regulation of flowering. Our investigation demonstrated that wrky75 mutant plants show delayed flowering compared with wild-type plants, whereas WRKY75-overexpressing plants exhibit early flowering (Figs. 1 and 2), implying that WRKY75 is also involved in flowering time regulation. Our findings demonstrate that WRKY75 may function as a new WRKY member to positively regulate flowering in Arabidopsis.
As a central node of floral integration, the transcription of FT is regulated by a large number of factors, some of which act as repressors and others as activators. In our study, FT expression was decreased in wrky75-1 and wrky75-25 plants and increased in 35S:WRKY75 transgenic plants (Fig. 3A), as was also the case for the TWIN SISTER OF FT (TSF; Supplemental Fig. S4A). This expression model is perfectly consistent with the flowering time behavior of these lines (Figs. 1 and 2). This observation suggests that WRKY75 is able to positively regulate the expression of FT. Furthermore, genetic analysis demonstrated that ft-10/35S:WRKY75 could not alter the ft-10 mutant phenotype whereas 35S:FT/wrky75-25 showed a markedly earlier flowering phenotype (Fig. 4), which mimicked that of 35S:FT plants, implying that WRKY75 acts to accelerate flowering in a FT-dependent manner. Previous studies have demonstrated that GA also controls flowering initiation by regulating the expression of LFY and SOC1 (Blazquez et al., 1998; Moon et al., 2003). Analysis of LFY and SOC1 expression indicated that SOC1, but not LFY, was positively regulated by WRKY75 (Supplemental Fig. S4, B and C). Furthermore, the expression of FLOWERING LOCUS C (FLC), a core transcriptional repressor in vernalization pathway (Sheldon et al., 2000), seems also not obviously affected by WRKY75 (Supplemental Fig. S4D).
Numerous studies have demonstrated that the WRKY proteins perform their biological functions by directly binding to the W box (TTGACC/T) present in their target promoters. FT was found to contain several WRKY-specific W box cis-elements in its promoter. Our ChIP experiments revealed that WRKY75 can directly bind to the FT promoter (Fig. 3, C and D), suggesting that FT is a direct target of WRKY75. The opposite expression patterns of WRKY75 and FT in wrky75 mutants and overexpression lines, and the up-regulation of the GFP signal in transient expression assays (Fig. 3), further suggest that WRKY75 is a positive regulator of FT. Further investigation showed that WRKY75 do not directly associate with the SOC1 or TSF promoter (Supplemental Fig. S5). Our study thus provides evidence that WRKY75 functions as a promoter of flowering and triggers flowering via the direct activation of FT. Considering that WRKY12 and WRKY13 function oppositely in controlling flowering time under SD conditions by the direct regulation of FUL (Li et al., 2016) and WRKY71 accelerates flowering via the direct activation of FT and LFY (Yu et al., 2016), different WRKY proteins may participate in their distinct flowering pathways by modulating the expression of different floral integrators or floral meristem identity genes. Several previous studies have demonstrated that WRKY proteins can act as both positive and negative regulators. For example, our previous study revealed that WRKY8 regulates ABI4 positively and ACS6 negatively upon crucifer-infecting tobacco mosaic virus infection (Chen et al., 2013), and that WRKY57 directly represses the transcription of SENESCENCE4 and SENESCENCE-ASSOCIATED GENE12 in jasmonic acid-induced leaf senescence (Jiang et al., 2014). WRKY40 directly inhibits the transcription of diverse ABA-responsive genes, such as ABI5, during seed germination and postgermination growth (Shang et al., 2010). One recent study demonstrated that WRKY45 directly activates the expression of PHOSPHATE TRANSPORTER1;1 upon P starvation (Wang et al., 2014). Thus, we can deduce that WRKY proteins extensively participate in the fine-tuning and tight control of the complex signaling and transcriptional networks that mediate plant growth and stress responses by functioning as both repressors and activators.
WRKY75 Partially Mediates GA Signaling in Flowering Time Regulation
GA is an essential hormone that regulates diverse aspects of plant growth and development. Genetic screening has identified several key molecular components in the upstream signaling pathway of GA; however, the downstream signaling pathway of GA remains unclear. Several key transcription factors have recently been identified to function downstream of DELLA proteins. For example, PIFS (de Lucas et al., 2008; Feng et al., 2008), BZR1 (Bai et al., 2012; Gallego-Bartolomé et al., 2012), MYC2 (Hong et al., 2012; Wild et al., 2012), the WD-repeat/bHLH/MYB complex (Qi et al., 2014), and class 1 TCP transcription factors (Davière et al., 2014) function as direct targets of DELLA proteins to regulate photomorphogenesis, sesquiterpene synthase, JA signaling, trichome development, and plant height, respectively. DELLAs can also competitively interact with JAZ proteins and fine-tune JA signaling (Hou et al., 2010). Here, WRKY75 was identified as a new downstream component of the GA signaling pathway.
Although GA accelerates the transition from vegetative growth to flowering through degradation of transcriptional repressors (DELLAs), the underlying mechanisms involved remain elusive. A recent study revealed that the DELLA proteins interact with the microRNA156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors to repress the transcriptional activation of MADS box genes and miR172 (Yu et al., 2012). We also provided evidence that GA regulates plant flowering via interactions between DELLAs and CO under LD conditions (Wang et al., 2016) and via interactions between DELLAs and WRKY12/13 under SD conditions (Li et al., 2016). Thus, DELLAs transduce the GA signal via interactions with various transcription factors. In this study, we demonstrated that WRKY75 participates in flowering time regulation through physical interactions with DELLA proteins (Fig. 5). It is likely that DELLAs interfere with the transcriptional function of WRKY75. RT-qPCR analysis showed that WRKY75 responds to exogenous GA treatment, implying that WRKY75 can be regulated by GA at the transcriptional level (Fig. 8A). Transient expression assays also confirmed that both RGL1 and GAI can suppress the transcriptional activation of FT by WRKY75. ChIP-qPCR revealed that RGL1 decreases the binding ability of WRKY75 to its target genes in vivo (Fig. 6), and genetic analysis demonstrated that RGL1 alleviates the early flowering phenotype observed in WRKY75-overexpressing plants (Fig. 7). These data suggest that DELLA proteins can repress the transcriptional regulatory activity of WRKY75.
This study has revealed the molecular mechanisms underlying the regulation of flowering by WRKY75. Our findings indicate that WRKY75 functions as a new component of the flowering regulatory network and participates in the GA-mediated flowering time regulation via interactions with DELLAs. It will be interesting to further investigate how WRKY75’s transcription and translation is regulated during the transition from vegetative growth to reproductive growth, and whether this mechanism is conserved for other flowering-associated WRKY transcription factors. Overall, we have shown that WRKY75 functions as a new component of the GA-mediated signaling pathway to modulate the onset and progression of floral initiation. The molecular mechanisms revealed in this study add to our understanding of the involvement of WRKY transcription factors in flowering time regulation.
MATERIALS AND METHODS
Materials and Arabidopsis Growth Conditions
Taq DNA polymers were purchased from Takara Biotechnology. Other chemicals were obtained from Shanghai Sangon Biotechnology. The wrky75-1 (SALK_101367) and wrky75-25 were kindly provided by Dong-Tao Ren and Martin Hülskamp, respectively. To generate WRKY75, RGL1, and GAI overexpression transgenic plants, full-length cDNAs of WRKY75 and RGL1 were cloned into a pOCA30 or pOCA30-GFP vector in the sense orientation behind a CaMV 35S promoter. Plants used in this study were derived from Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0). Arabidopsis plants were grown in an artificial growth chamber at 22°C under LDs (16-h light/8-h dark cycle) or SDs (8-h light/16-h dark cycle). Primers used for identification of mutants or clones are listed in Supplemental Table S1.
qRT-PCR Analysis
For qRT-PCR analysis, total RNA was extracted using TRIzol reagent (Invitrogen) and was treated with RNase-free DNase (Fermentas), according to the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed in a 20 μL reaction mixture using the Superscript II (Invitrogen). After the reaction, 1 μL aliquots were used as templates for qRT-PCR. Half-reactions (10 μL each) were performed with the LightCycler FastStart DNA Master SYBR Green I Kit (Roche) on a LightCycler 480 real-time PCR machine (Roche), according to the manufacturer’s instructions. ACT2 (AT3G18780) was used as a control in quantitative RT-PCR. The gene-specific primers are listed in Supplemental Table S1.
Construction of Transgenic Overexpression Lines
To generate the 35S:WRKY75 and 35S:RGL1 construct, the cDNA fragment containing the full coding sequence was excised from a cloning plasmid and subcloned into the same restriction sites of the Agrobacterium transformation vector pOCA30 in the sense orientation behind the CaMV 35S promoter. Arabidopsis transformation was performed by the floral dip procedure. The seeds were collected from the infiltrated plants and selected on 1/2 Murashige & Skoog medium containing 50 μg/mL kanamycin. Kanamycin-resistant plants were transferred to soil 8 d after germination and were grown in a growth chamber.
GUS Staining
Histochemical detection of GUS activity was performed with 5-bromo-4-chloro-3-indolyl b-d-GlcA (X-gluc) as the substrate. Plant tissues were first prefixed in ice-cold 90% (v/v) acetone for 20 min, then washed three times with GUS staining buffer (without X-gluc) before incubation in X-gluc solution [1 mm X-gluc, 50 mm NaPO4, pH 7, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, and 0.05% (w/v) Triton X-100] under a vacuum for 10 min at room temperature, then incubated overnight at 37°C. Chlorophyll was removed using several changes of 70% (v/v) ethanol and the tissues were photographed.
Yeast Two-Hybrid Screening and Confirmation
The full-length WRKY75 CDS was cloned into the bait vector pGBKT7 and then transformed into the yeast strain Y2HGold (Clontech). Two-hybrid screening was performed via the mating protocol described in Clontech’s Matchmaker Gold Yeast Two-Hybrid user manual. To confirm protein-protein interactions, the full-length RGL1 coding sequences (CDS) were cloned into the prey vector pGADT7.
BiFC Assays
The cDNA sequences of enhanced YFP fragments, 173 amino acids located in the N terminus (nYFP), and 64 amino acids located in the C terminus (cYFP), were PCR-amplified and cloned into the XbaI-XhoI and BamHI-XhoI sites of pFGC5941 to generate pFGC-nYFP and pFGC-cYFP, respectively. The full-length RGL1 and GAI CDS were inserted into pFGC-cYFP to generate a C-terminal in-frame fusion with cYFP, whereas WRKY75 CDS were introduced into pFGC-nYFP to form N-terminal in-frame fusions with nYFP. The resulting plasmids were introduced into Agrobacterium tumefaciens (strain EHA105), and then infiltration of Nicotiana benthamiana leaves was performed. Infected tissues were analyzed 48 h after infiltration. YFP and 49,6-diamidino-2-phenylindole fluorescence were observed under a confocal laser scanning microscope (Olympus). The primers used for BiFC are listed in Supplemental Table S1.
CoIP Assays
For CoIP assays, WRKY75, RGL1, and GAI were individually cloned into tagging plasmids behind the Myc or GFP tag sequence. Myc-fused WRKY75 and GFP-fused RGL1/GAI were then transiently coexpressed in N. benthamiana leaves. All infected leaves were treated with 10 μM MG132 and 20 μM paclobutrazol (a GA biosynthesis inhibitor) 40 h after infiltration. After 8 h, those leaves were homogenized in an extraction buffer containing 50 mm 400 Tris-HCl pH7.5, 150 mm NaCl, 1 mm EDTA, 0.1% (w/v) Trition-X-100, and 1× Complete Protease Inhibitor Cocktail (Roche). Then, GFP-fused GAI/RGL1 and GFP was immunoprecipitated using an anti-GFP antibody, and coimmunoprecipitated proteins were detected using an anti-Myc antibody.
ChIP Assays
For the ChIP assay, 12-d-old seedlings of Myc-WRKY75 or GFP-WRKY75 and Col-0 seedlings were used as materials. The ChIP experiment was performed as described previously (Saleh et al., 2008). The Myc and GFP antibody was used to immunoprecipitate the protein-DNA complex, and the precipitated DNA was purified using a PCR purification kit for qRT-PCR analysis. The ChIP experiments were performed three times. Chromatin precipitated without antibody was used as the negative control, whereas the isolated chromatin before precipitation was used as the input control. ChIP results are presented as a percentage of input DNA. The primers used for qRT-PCR amplification of different promoters are listed in Supplemental Table S1.
EMSA Assays
The EMSA assay was conducted using a Chemiluminescent EMSA Kit (Beyotime) following the manufacturer’s protocol. The recombinant GST-WRKY75 protein and the GST protein were purified from Escherichia coli. The DNA fragments of the FT promoter were synthesized and biotin was labeled to the 5′ terminal of DNA. Biotin-unlabeled fragments of the same sequences or mutated sequences were used as competitors, and the GST protein alone was used as the negative control.
Transient Expression Assays
The transient expression assays were performed in N. benthamiana leaves. The NLS was fused with GFP reporter gene behind the native promoter of FT. The full-length CDS of RGL1, GAI, GUS, and WRKY75 were driven by the CaMV 35S promoter. These constructs were then introduced into the A. tumefaciens (strain EHA105). Infected tissues were analyzed 48 h after infiltration. The GFP signal was observed under a confocal laser scanning microscope (Olympus). All experiments were repeated with five independent biological replicates with similar results.
GA Treatments
GA3 (Sigma-Aldrich) was stored in 100% ethanol. Two solutions were then prepared: 1) GA3 100 μM, Tween 0.1% (v/v); and 2) pure ethanol 1% (v/v), Tween 0.1% (v/v). The soil grown plants were sprayed with solution 1 (GA3) or solution 2 (MOCK) every other day until the formation of the first silique. Twelve-d-old Myc-WRKY75/RGL1 seedlings were incubated in liquid 1/2 Murashige & Skoog supplemented with 50 μm GA or mock for 8 h and then were harvested.
Accession Numbers
The Arabidopsis Genome Initiative identifiers for the genes described in this article are as follows: WRKY75 (At5G13080), RGL1 (At1G66350), RGL2 (At3G03450), RGL3 (At5G17490), RGA (At2G01570), GAI (At1G14920), FT (At1G65480), SOC1 (At2g45660), TSF (At4g20370), LFY (At5g61850), FLC (At5g10140), and ACTIN2 (At3g18780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. T-DNA insertion of Arabidopsis wrky75 mutants.
Supplemental Figure S2. Real-time PCR analysis of 35S:WRKY75 transgenic plants.
Supplemental Figure S3. Gain of function of GAI partially rescues the early flowering phenotype of WRKY75 overexpressing plants.
Supplemental Figure S4. Expression of multiple flowering-related genes was regulated by WRKY75.
Supplemental Figure S5. WRKY75 does not associate with SOC1 and TSF promoters.
Supplemental Table S1. Primers used in this article.
Acknowledgments
We thank Xiaoya Chen (Institute of Plant Physiology and Ecology, SIBS, CAS), Dong-Tao Ren (China Agricultural University), and Martin Hülskamp (Cologne University) for sharing research materials.
Footnotes
This work was supported by the Natural Science Foundation of China (31401042, 31671275, 31200915, and 91417307), the National Key R & D Plan (2016YFD0101006), the West Light Foundation of CAS, Yong Innovation Promotion Association of CAS, Candidates of the Young and Middle Aged Academic Leaders of Yunnan Province (2015HB094), the Natural Science Foundation of Yunnan Province of China (2017FB047), and Innovation Team of Yunnan Province (2014HC017).
References
- Achard P, Genschik P (2009) Releasing the brakes of plant growth: how GAs shut down DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D (1998) Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10: 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümel M, Dally N, Jung C (2015) Flowering time regulation in crops—what did we learn from Arabidopsis? Curr Opin Biotechnol 32: 121–129 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D (2017) Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10: 1174–1189 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D (2013) WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 110: E1963–E1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, De Bodt S, Inzé D (2014) Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci 19: 231–239 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dally N, Xiao K, Holtgräwe D, Jung C (2014) The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc Natl Acad Sci USA 111: 10365–10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Davis SJ. (2009) Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ 32: 1201–1210 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schäfer E, Fu X, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y (2014) DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26: 2920–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H (2008) AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J 55: 253–265 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 5: 4601. [DOI] [PubMed] [Google Scholar]
- Hu JY, Zhou Y, He F, Dong X, Liu LY, Coupland G, Turck F, de Meaux J (2014) miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 26: 2024–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D (2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagacé M, Matton DP (2004) Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219: 185–189 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang H, Yu D (2016) The Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol Plant 9: 1492–1503 [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C (2013) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. (2009) Flowering time regulation produces much fruit. Curr Opin Plant Biol 12: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, Maeda T, Matsuoka M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Nilsson O (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ 35: 1742–1755 [DOI] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D (2014) Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26: 1118–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L, Pesch M, Juengst C, Schauss AC, Schrader A, Hülskamp M (2014) Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol 165: 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova A (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, Zhang XF, Zhao R, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Ai CR, Jing SJ, Yu DQ (2010) Research progress on function analysis of rice WRKY gene. Rice Sci 17: 60–72 [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:033–1036 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- Wang H, Pan J, Li Y, Lou D, Hu Y, Yu D (2016) The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol 172: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF (2014) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol 164: 2020–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW. (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Li X, Qin D, Guo F, Wu C, Miao L, Sun C (2012) Functional analysis of FLOWERING LOCUS T orthologs from spring orchid (Cymbidium goeringii Rchb. f.) that regulates the vegetative to reproductive transition. Plant Physiol Biochem 58: 98–105 [DOI] [PubMed] [Google Scholar]
- Xu YH, Wang JW, Wang S, Wang JY, Chen XY (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene+-Δ-cadinene synthase-A. Plant Physiol 135: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, Tada Y, Ohme-Takagi M, et al. (2014) DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci USA 111: 7861–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Liu Z, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park CM, Xiang F (2016) WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J 85: 96–106 [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, Lim J, Kamiya Y, Yamaguchi S, Sun TP (2011) Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134: 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Seemann JR, Neuman D, Shen QJ (2004) A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J Biol Chem 279: 55770–55779 [DOI] [PubMed] [Google Scholar]