Auxin and ROP signaling affect nuclear migration in root epidermal cells.
Abstract
Polar nuclear migration is crucial during the development of diverse eukaryotes. In plants, root hair growth requires polar nuclear migration into the outgrowing hair. However, knowledge about the dynamics and the regulatory mechanisms underlying nuclear movements in root epidermal cells remains limited. Here, we show that both auxin and Rho-of-Plant (ROP) signaling modulate polar nuclear position at the inner epidermal plasma membrane domain oriented to the cortical cells during cell elongation as well as subsequent polar nuclear movement to the outer domain into the emerging hair bulge in Arabidopsis (Arabidopsis thaliana). Auxin signaling via the nuclear AUXIN RESPONSE FACTOR7 (ARF7)/ARF19 and INDOLE ACETIC ACID7 pathway ensures correct nuclear placement toward the inner membrane domain. Moreover, precise inner nuclear placement relies on SPIKE1 Rho-GEF, SUPERCENTIPEDE1 Rho-GDI, and ACTIN7 (ACT7) function and to a lesser extent on VTI11 vacuolar SNARE activity. Strikingly, the directionality and/or velocity of outer polar nuclear migration into the hair outgrowth along actin strands also are ACT7 dependent, auxin sensitive, and regulated by ROP signaling. Thus, our findings provide a founding framework revealing auxin and ROP signaling of inner polar nuclear position with some contribution by vacuolar morphology and of actin-dependent outer polar nuclear migration in root epidermal hair cells.
Diverse eukaryotic organisms display polar nuclear migration within various cell types, which often is crucial to the development of the organism, as abnormal nuclear placement can cause severe developmental aberrations (Burke and Roux, 2009; Gundersen and Worman, 2013). Components and regulators of the cytoskeleton as well as nuclear membrane proteins play important roles in polar nuclear migration (Gundersen and Worman, 2013). In the plant Arabidopsis (Arabidopsis thaliana), cells display polar nuclear migration in a multitude of developmental contexts (Griffis et al., 2014; Kawashima et al., 2014; Zhou and Meier, 2014). During lateral root development, the accumulation of the plant hormone auxin promotes the migration of the nuclei within two longitudinally adjacent xylem pole pericycle cells followed by an asymmetric cell division initiating the formation of lateral root primordia (De Rybel et al., 2010). This coordinated polar nuclear migration is mediated by a SOLITARY-ROOT (SLR)/INDOLE ACETIC ACID14 (IAA14)-AUXIN RESPONSE FACTOR7 (ARF7)-ARF19 nuclear auxin signaling module (De Rybel et al., 2010; Goh et al., 2012). Polar nuclear migration also occurs in root epidermal hair cells (trichoblasts), where the nucleus moves from the cell body into the elongating root hair (Chytilova et al., 2000; Ketelaar et al., 2002; Van Bruaene et al., 2003; Jones and Smirnoff, 2006).
During root hair elongation, the nucleus moves toward the growing tip and finally comes to rest at a fixed distance from the tip (Ketelaar et al., 2002; Zhang et al., 2015). Nuclear migration appears to be required for hair growth, because optical trapping of the nucleus induces growth arrest (Ketelaar et al., 2002). Nuclear migration in elongating root hairs of Arabidopsis relies on F-actin, as demonstrated by pharmacological studies (Chytilova et al., 2000; Ketelaar et al., 2002). Additionally, modulation of actin filaments regulated by plasma membrane-associated Ca2+-Binding Protein2 is required for the maintenance of nuclear positioning (Zhang et al., 2015). Microtubules do not appear to play a major role in the migration of the nucleus into the elongating root hair (Ketelaar et al., 2002; Zhang et al., 2015). Consistent with its association with actin filaments, the plant-specific Myosin XI-i localizes to the nuclear membrane. It may act as a nucleocytoplasmic linker that binds to both actin filaments and outer nuclear membrane proteins such as the WPP domain-interacting tail-anchored protein1 (WIT1) and WIT2 (Tamura et al., 2013; Zhou et al., 2015). Indeed, loss of function of these components results in the reduction of nuclear migration velocity in the elongating root hair (Tamura et al., 2013). Rho-of-Plant (ROP) GTPases accumulate early at the hair initiation site, where their position is instructed by auxin, and later remain at the tip of the growing root hair (Molendijk et al., 2001; Jones et al., 2002; Fischer et al., 2006; Ikeda et al., 2009; Nakamura et al., 2012). Overexpression and dominant-interference experiments indicate that ROP2 may regulate actin organization during root hair growth (Jones et al., 2002) and loss of SUPERCENTIPEDE1 (SCN1) Rho-GDI function induces the initiation of multiple hairs from cells (Parker et al., 2000; Carol et al., 2005).
During leaf epidermal cell differentiation, ROP2/4 are required for actin organization and leaf pavement cell morphogenesis (Fu et al., 2005). A potential function of AUXIN-BINDING PROTEIN1 (ABP1) during leaf pavement cell shape formation remains controversial (Xu et al., 2010; Gao et al., 2015), similar to a proposed developmental role of ABP1 in ROP activation as observed in protoplasts (Xu et al., 2010). The Arabidopsis SPIKE1 (SPK1) gene was originally identified based on the trichome and other leaf cell shape phenotypes of its mutant and was found to encode a DOCK family-like protein (Qiu et al., 2002). The SPK1 protein exerts Rho-Guanine Nucleotide Exchange Factor (Rho-GEF) activity toward plant ROP proteins and controls actin-dependent cell morphogenesis via the interaction with heteromeric WAVE and actin-related protein2/3 complexes (Basu et al., 2008). In seedling protoplasts, SPK1 also is required for ROP6 activation (Lin et al., 2012), and pharmacological stabilization of the actin cytoskeleton revealed a weak effect of the rop6-1 mutation on PIN2 auxin efflux carrier protein internalization in root meristematic cells, further indicating an interplay between ROP6 and actin function (Lin et al., 2012). However, it remains unknown whether and which components of auxin and ROP signaling contribute to actin-dependent nuclear migration in root trichoblasts. Here, we report an initial nuclear placement at the inner lateral membrane during cell elongation that precedes nuclear migration toward the outer lateral membrane into the elongating root hair. Since it remained unknown which factors influence nuclear placement at the inner membrane and which factors instruct the directionality of the nuclear movement into the root hair, we analyzed both events in more detail. Our study presents a framework for nuclear auxin signaling and ROP signaling-mediated, ACTIN7 (ACT7)-dependent nuclear movement during two distinct nuclear migration events.
RESULTS
Arabidopsis Root Trichoblasts Display Inner Polar Nuclear Position
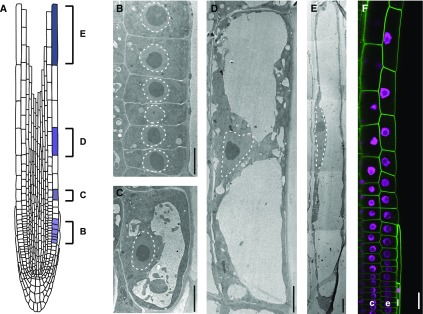
To examine the position of the nucleus in root trichoblasts throughout cell differentiation (Fig. 1A), we employed transmission electron microscopy (TEM). Nuclei resided in a central position in meristematic cells (Fig. 1B), whereas a gradual shift toward the inner lateral membrane took place throughout trichoblasts in the elongation zone (Fig. 1, C–E). To observe the nuclear position in vivo, we generated plants expressing the nuclear marker HISTONE2B (H2B)-mCherry and the plasma membrane marker EGFP-LTI6a. Consistent with the TEM data, we found that nuclei gradually changed their position from the center of the cell toward the inner lateral membrane during cell elongation (Fig. 1F), which we refer to as the inner polar nuclear position. To investigate whether inner polar nuclear localization can be observed in atrichoblasts, we analyzed nuclear position in both trichoblasts and atrichoblasts in aldehyde-fixed roots of the wild type. In both cell types, nuclei were found at the inner lateral membrane during cell elongation (Supplemental Fig. S1, A and B). Quantitative and statistical analyses revealed that the distribution of nuclei did not differ between trichoblasts and atrichoblasts at a cell length of 20 to 50 µm (Supplemental Fig. S1C).
Figure 1.
Inner polar nuclear positioning during cell elongation of root trichoblasts. A, Schematic root structure. Cells highlighted in blue correspond to the cells shown in B to E. B to E, TEM images of root epidermal trichoblasts. B, Trichoblasts in the meristematic zone. C to E, Trichoblasts in the elongation zone. Dotted lines show outlines of the nuclei. Bars = 10 µm. F, Live image showing part of a lateral root cap layer (l), a root epidermal trichoblast cell file (e), and a cortical cell file (c). Nuclei, H2B-mCherry (magenta); PM, EGFP-LTI6a (green). Bar = 20 µm.
High Auxin Concentration Alters Inner Polar Nuclear Position
Auxin and ethylene promote polar root hair placement along root epidermal cells (Masucci and Schiefelbein, 1994; Fischer et al., 2006), and mutations in the CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) gene encoding a Raf-like kinase, a negative regulator of the ethylene signaling pathway, increase auxin concentration in the root tip (Ikeda et al., 2009). Thus, we addressed the potential contribution of CTR1 to polar nuclear positioning in root trichoblasts by analyzing the nuclear position in aldehyde-fixed roots of the well-characterized hypomorphic ctr1btk allele (Ikeda et al., 2009). While nuclei were positioned at the inner lateral membrane during cell elongation in the wild type (Fig. 2A), nuclei were significantly mispositioned in ctr1btk trichoblasts of the same cell length range (Fig. 2B; Supplemental Table S1), with a large population of nuclei shifted significantly toward both outer lateral and apical directions (Fig. 2I). To directly address the effect of auxin on this process, we analyzed the nuclear position in epidermal cells of seedlings grown on 300 nm 1-naphthaleneacetic acid (1-NAA)-containing medium. 1-NAA treatment caused a significant shift of nuclei toward both the outer lateral and the apical membrane (Fig. 2, E, F, and L), strongly resembling the nuclear position phenotype of ctr1btk (Fig. 2B). Together, our findings demonstrate that activated ethylene signaling and increased auxin concentration alter the inner polar nuclear position.
Figure 2.
Nuclear auxin signaling modulates inner polar nuclear positioning. A to H, Nuclear position in trichoblasts of aldehyde-fixed 5-d-old seedling roots. A, Wild-type Columbia-0 (Col-0). B, ctr1btk. C, ctr1btk;axr2-1. D, ctr1btk;arf7-1;arf19-1. E, A 1 μm NaOH solvent control (in MS-agar). F, Treatment with 300 nm 1-NAA in Col-0. G, Treatment with 300 nm 1-NAA in axr2-1. H, Treatment with 300 nm 1-NAA in arf7-1;arf19-1. Nuclei, 4′,6-Diamidino-2-phenylindole (DAPI; cyan) overlaid with a bright-field image. Dotted lines show inner lateral cell walls of trichoblasts. Asterisks indicate mispositioned nuclei. Arrowheads show basal and apical ends of cells. Bars = 20 µm. I to N, Quantitative analysis of nuclear position. I, Col-0 versus ctr1btk. J, ctr1btk versus ctr1btk;axr2-1. K, ctr1btk versus ctr1btk;arf7-1;arf19-1. L, A 1 μm NaOH solvent control in MS-agar versus 300 nm 1-NAA in Col-0. M, Treatment with 300 nm 1-NAA, Col-0 versus 300 nm 1-NAA, axr2-1. N, Treatment with 300 nm 1-NAA, Col-0 versus 300 nm 1-NAA, arf7-1;arf19-1. Distributions of relative nuclear position are plotted; n = 150 cells per genotype or treatment. Significances of differences between distributions were determined independently for distribution along the apical-basal axis as well as for distribution along the inner-outer axis employing a nonparametric, two-sample Kolmogorov-Smirnoff (KS) test with a significance threshold of P < 0.05. **, P = 0.000. Exact P values are shown in Supplemental Table S1.
Nuclear Auxin Signaling Contributes to Precise Inner Polar Nuclear Position
To investigate the involvement of AUX/IAA-ARF nuclear auxin signaling in inner nuclear positioning, we utilized the axr2-1 mutant, encoding a dominant-stabilized version of the IAA7/AUXIN RESISTANT2 (AXR2) protein, an AUX/IAA family transcriptional repressor and auxin coreceptor (Nagpal et al., 2000; Calderón Villalobos et al., 2012). In addition, we analyzed the arf7-1;arf19-1 double loss-of-function mutant (Okushima et al., 2005, 2007). Both the axr2-1 and arf7-1;arf19-1 mutants exhibited a subtle, but significant, basal shift of nuclear position (Supplemental Fig. S2). Strikingly, however, the strong nuclear position shift toward the outer and toward the apical membrane in the ctr1btk mutant was strongly suppressed by the axr2-1 mutation (Fig. 2, C and J), such that nuclear position in the ctr1btk;axr2-1 double mutant was restored to the wild-type position (Supplemental Fig. S4D). Consistent with these findings, the effect of exogenous auxin application on nuclear positioning was largely abolished by the axr2-1 mutation (Fig. 2, G and M). Similarly, the nuclear position shift of ctr1btk was suppressed significantly in the ctr1btk;arf7-1;arf19-1 triple mutant (Fig. 2, D and K), and the effect of exogenous auxin treatment was strongly suppressed in the arf7-1;arf19-1 double mutant (Fig. 2, H and N). Together, our results demonstrate that IAA7-ARF7-ARF19-dependent auxin signaling contributes to the inner polar nuclear position along the apical-basal axis at average endogenous and at elevated auxin concentrations or ethylene signaling.
ZIG/VTI11-Dependent Vacuole Morphology Mildly Contributes to the Inner Polar Nuclear Position
Auxin has been shown to affect vacuole morphology through the nuclear auxin signaling pathway in late meristematic cells (Löfke et al., 2015). This prompted us to address whether the mispositioning of the nucleus observed in the ctr1btk mutant is associated with an alteration of vacuole morphology. To this end, we analyzed vacuole size in trichoblasts during cell elongation, where nuclei are positioned at the inner lateral membrane at a cell length of 20 to 50 µm (Supplemental Fig. S3). We found that vacuolar size in ctr1btk was reduced significantly compared with that of the wild type (Supplemental Fig. S3, A, B, and L). The arf7-1;arf19-1 double mutant showed a similar vacuole size to the wild type (Supplemental Fig. S3, D and L). Strikingly, however, the reduction of vacuole size in the ctr1btk mutant was suppressed significantly by the arf7-1;arf19-1 mutations (Supplemental Fig. S3, C and L), such that vacuole size in the ctr1btk;arf7-1;arf19-1 triple mutant recovered to the wild-type size (Supplemental Fig. S3L). Together, our results suggest that there is a correlation between the reduction of vacuole size and the mispositioning of the nucleus observed at elevated auxin levels or ethylene signaling, both of which are mediated by ARF7-ARF19-dependent nuclear auxin signaling.
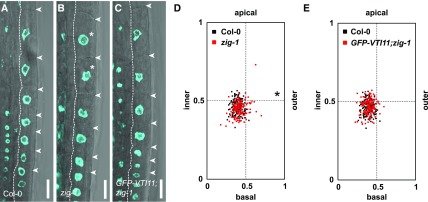
To directly investigate the impact of vacuole development on inner polar nuclear position, we analyzed the zigzag-1 (zig-1)/vti11 loss-of-function mutant defective in vacuole morphology, mutated in the VTI11 gene encoding a vacuolar Soluble NSF Attachment Protein Receptor (Kato et al., 2002). As reported previously (Löfke et al., 2015), we observed abnormal roundish vacuoles in zig-1 elongating trichoblasts (Supplemental Fig. S3, E and F). Moreover, vacuole size in zig-1 was reduced significantly compared with that of elongating cells in the wild type (Supplemental Fig. S3M). Interestingly, the distribution of nuclei was slightly but significantly shifted toward the outer lateral direction in the zig-1 mutant (Fig. 3, A, B, and D). In comparison, nuclear position in the proVTI11:GFP-VT11;zig-1 rescue line was restored to the wild-type position (Fig. 3, C and E). These results suggest that ZIG/VTI11 function required for vacuole development makes a subtle but significant contribution to the inner polar nuclear position in elongating root trichoblasts.
Figure 3.
ZIG/VTI11-dependent vacuole morphology contributes to inner polar nuclear position. A to C, Nuclear position in trichoblasts of aldehyde-fixed 5-d-old seedling roots. A, Wild-type Col-0. B, zig-1. C, proVTI11:GFP-VTI11/zig-1 (GFP-VTI11;zig-1) rescue line. Nuclei, DAPI (cyan) overlaid with a bright-field image. Dotted lines show inner lateral cell walls of trichoblasts. Asterisks indicate mispositioned nuclei. Arrowheads show basal and apical ends of cells. Bars = 20 µm. D and E, Quantitative analysis of nuclear position. D, Col-0 versus zig-1. E, Col-0 versus GFP-VTI11/zig-1. Distributions of nuclear position are plotted; n = 150 cells per genotype. *, P < 0.05. Exact P values from a nonparametric, two-sample KS test are shown in Supplemental Table S1.
SPK1-Dependent ROP Signaling Acts on Inner Polar Nuclear Position
To investigate whether ROP signaling contributes to inner polar nuclear position, we analyzed two loss-of-function alleles of SPK1 encoding a positive regulator of ROP signaling (Qiu et al., 2002; Lin et al., 2012). Nuclear position was shifted significantly from the inner membrane toward the outer lateral direction in spk1-1 and spk1-5 mutants (Fig. 4, B and I; Supplemental Fig. S4, B and C), suggesting that SPK1-dependent ROP signaling contributes to the correct inner polar nuclear position. However, although a mild reduction of vacuole size was observed in the spk1-5 mutant, this did not differ significantly from the wild type (Supplemental Fig. S3, G, H, and N), in contrast to the stronger and significant defects observed in the ctr1btk and zig-1 mutants (Supplemental Fig. S3, L and M). To investigate whether SPK1 action is linked to auxin and ethylene signaling, we generated and analyzed the ctr1btk;spk1-5 double mutant. The spk1-5 mutation only slightly suppressed the nuclear position shift of ctr1btk toward the inner lateral direction (Fig. 4, C, D, and J). This suggested that SPK1-dependent ROP signaling does not mainly mediate a response to ethylene signaling and enhanced auxin levels. Consistent with this view, the nuclear position shift of ctr1btk was hardly suppressed in the ctr1btk;spk1-5 double mutant when compared with the ctr1btk;axr2-1 double mutant (Fig. 2J; Supplemental Fig. S4, D and E) or the ctr1btk;arf7-1;arf19-1 triple mutant (Fig. 2K). Our results show that SPK1 is required for nuclear placement toward the inner membrane but render it unlikely that SPK1-dependent ROP activation acts as a main transducer of the auxin signal during this process.
Figure 4.
ROP activation modulates inner polar nuclear positioning synergistically with auxin/ethylene signaling. A to H, Nuclear position in trichoblasts of aldehyde-fixed 5-d-old seedling roots. A, Wild type Col-0. B, spk1-5. C, ctr1btk. D, ctr1btk;spk1-5. E, Col-0. F, scn1-1. G, ctr1btk. H, ctr1btk;scn1-1. Nuclei, DAPI (cyan) overlaid with a bright-field image. Dotted lines show inner lateral cell walls of trichoblasts. Asterisks indicate mispositioned nuclei. Arrowheads show basal and apical ends of cells. Bars = 20 µm. I to L, Quantitative analysis of nuclear position. I, Col-0 versus spk1-5. J, ctr1btk versus ctr1btk;spk1-5. K, Col-0 versus scn1-1. L, ctr1btk versus ctr1btk;scn1-1. Distributions of nuclear position are plotted; n = 150 cells per genotype. *, P < 0.05 and **, P = 0.000. Exact P values from a nonparametric, two-sample KS test are shown in Supplemental Table S1.
SCN1 and CTR1 Synergistically Act on Inner Polar Nuclear Position
We next investigated the effect of ROP overactivation utilizing the scn1-1 loss-of-function mutant defective in a negative regulator of ROP signaling (Parker et al., 2000; Carol et al., 2005). The nuclei of scn1-1 were located almost adjacent to the inner lateral membrane but displayed a significant basal shift when compared with the wild type (Fig. 4, F and K). In contrast, the vacuole size of the scn1-1 mutant did not differ significantly from that of the wild type (Supplemental Fig. S3, G, I, and N). Notably, however, the scn1-1 mutation significantly enhanced the nuclear position shift of ctr1btk toward the outer lateral direction (Fig. 4, G, H, and L), suggesting that overactivation of ROP signaling synergistically alters the inner positioning of the nucleus in combination with elevated ethylene signaling and auxin concentrations.
ACT7 Is Required for Precise Inner Polar Nuclear Position along the Apical-Basal Axis
Our results revealed that CTR1/auxin signaling, ZIG/VTI11-dependent vacuole morphology, and ROP signaling contribute to inner polar nuclear position. ROP signaling is thought to regulate actin organization in both the leaf and the root epidermis (Xu et al., 2010; Lin et al., 2012), although genetic evidence toward this is lacking. To investigate whether actin function is required for inner polar nuclear position, we analyzed nuclear position in the act7-6 loss-of-function mutant (Kiefer et al., 2015). Nuclear position in elongating act7-6 mutant cells was clearly misplaced along the apical-basal axis when compared with elongating wild-type cells, although the majority of the nuclei remained adjacent to the inner lateral membrane (Fig. 5, A–C). Hence, ACT7 function contributes to inner polar nuclear position along the apical-basal axis of the cell.
Figure 5.
ACT7 function is required for inner nuclear placement along the apical-basal axis and for outer polar nuclear migration. A and B, Nuclear position in trichoblasts of aldehyde-fixed 5-d-old seedling roots. A, Wild-type Col-0. B, act7-6. Nuclei, DAPI (cyan) overlaid with a bright-field image. Dotted lines show inner lateral cell walls of trichoblasts. Asterisks indicate mispositioned nuclei. Arrowheads show basal and apical ends of cells. Bars = 20 µm. C, Quantitative analysis of nuclear position in wild-type Col-0 versus act7-6. Distributions of relative nuclear position are plotted; n = 150 cells per genotype. Col-0 shown in Figure 2I is used as the control derived from the same experiments. **, P = 0.000. Exact P values from a nonparametric, two-sample KS test are shown in Supplemental Table S1. D, Bright-field image of trichoblasts of 5-d-old UAS:act7fiz1;UAS:H2B-mCherry reporter line seedling roots. Bars = 20 µm. E, Nuclear position marked by H2B-mCherry (magenta) fluorescence in trichoblasts of 5-d-old proAtMYC1:GAL4-VP16;UAS:mTFP-ER UAS:act7fiz1;UAS:H2B-mCherry (proAtMYC1>>act7fiz1) seedling roots. Trichoblast-specific proAtMYC1:GAL4-VP16 drives dominant-negative UAS:act7fiz1 concomitantly with UAS:mTFP-ER (cyan) and UAS:H2B-mCherry (magenta) fluorescence overlaid with a bright-field image. Asterisks indicate apically or basally mispositioned nuclei. Arrowheads show apical and basal ends of cells. Bar = 20 µm. F, Quantitative analysis of nuclear position in the UAS:act7fiz1;UAS:H2B-mCherry reporter line versus proAtMYC1>>act7fiz1. Distributions of relative nuclear position are plotted; n = 139 cells per line. *, P < 0.05. Exact P values from a nonparametric, two-sample KS test are shown in Supplemental Table S1. G to I, Nuclear position in aldehyde-fixed root trichoblasts in Col-0 (G) and act7-6 (H and I). Note the apical mispositioning in H and the basal mispositioning in I. Nuclei, DAPI (cyan) overlaid with a bright-field image. Arrowheads show apical and basal ends of cells. Bars = 50 µm.
Since actin organization impacts vacuole morphology in late meristematic cells (Scheuring et al., 2016), we addressed whether vacuole morphology would be strongly altered in act7-6, as observed for nuclear mispositioning in elongated trichoblasts. Strikingly, however, the mean vacuole size between the wild type and act7-6 did not differ significantly in elongating trichoblasts, although vacuole size in the act7-6 mutant was somewhat more variable than in the wild type (Supplemental Fig. S3, J, K, and O). Thus, the contribution of ACT7 function to inner nuclear positioning is unlikely to be mostly dependent on action via actin-dependent vacuole morphology. Consistent with this view, the act7-6 mutant displayed a much stronger deviation of nuclear position along the apical-basal axis, which differed from the ctr1btk and zig-1 mutant phenotypes (Figs. 2I and 3D), displaying clearly reduced vacuole size.
Since ACT7 is expressed in most root cell types (McDowell et al., 1996), it is possible that the loss of ACT7 function in other cell types than the epidermis could be responsible for the nuclear mispositioning observed in root hair cells. To investigate whether local interference with actin function was sufficient to induce nuclear displacement in epidermal cells, we designed a GAL4/UAS system (Brand and Perrimon, 1993) for tissue-specific expression of a dominant-negative ACT7 version carrying the frizzy1 (fiz1) mutation altering Glu-272 to Lys-272 (Kato et al., 2010) under the control of the lateral root cap- and trichoblast-specific proAtMYC1 promoter (Zhao et al., 2012). Indeed, root trichoblasts expressing act7fiz1 displayed significantly perturbed nuclear position mostly along the apical-basal axis of the cell (Fig. 5, D–F; Supplemental Fig. S5), suggesting a local requirement for actin function during nuclear placement in root epidermal cells.
Root Trichoblasts Display ACT7-Dependent Outer Polar Nuclear Migration
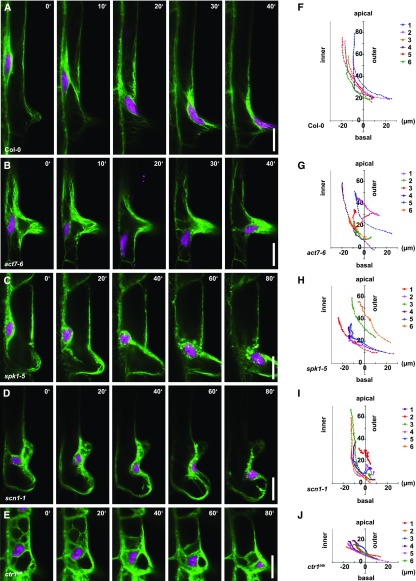
Since ACT7 function is required for polar placement of the root hair along the apical-basal axis of the cell (Kiefer et al., 2015), we were interested in whether ACT7 also contributes to outer nuclear migration during root hair outgrowth. In wild-type cells, nuclei were commonly observed at the base of the outgrowing hair, indicating that they changed location from an inner position toward the outgrowing root hair (Fig. 5G). In the act7-6 mutant, nuclei were sometimes observed at the extreme apical or basal end of the cells (Fig. 5, H and I), where they were never observed in the wild type, indicating that nuclear placement was impaired. To analyze the dynamics of outer polar nuclear migration, we performed simultaneous time-lapse imaging of the nucleus and actin filaments in plants expressing both H2B-mCherry and the GFP-ACTIN-BINDING DOMAIN2 (ABD2)-GFP reporter for F-actin (Wang et al., 2008). In wild-type trichoblasts, the nucleus surrounded by actin bundles was initially located in proximity to the inner lateral membrane (Fig. 6A; Supplemental Movie S1). During root hair outgrowth, the nucleus started to migrate toward the outer lateral membrane and moved into the hair (Fig. 6, A and F; Supplemental Movie S1). Actin bundles surrounded the nucleus during this movement (Fig. 6A; Supplemental Movie S1), indicating an association of the nucleus with F-actin throughout this process. Consistent with these findings, the velocity of nuclear movement was reduced significantly in the act7-6 mutant compared with the wild type (Table I; Supplemental Fig. S7A). Moreover, loss of ACT7 function clearly altered the trajectories of nuclear migration (Fig. 6, B and G) when compared with the wild type (Fig. 6, A and F). In extreme cases, for example in a cell displaying an apical shift of root hair positioning, the nucleus first moved toward the basal end of the cell and then moved back apically toward the hair (Fig. 6, B and G; Supplemental Movie S2). These findings show that the directionality and velocity of outer polar nuclear migration require ACT7 function.
Figure 6.
Dynamics of outer polar nuclear migration in ACT7, ROP pathway, and ethylene signaling mutants. A to E, Time-lapse images of polar nuclear migration in wild-type Col-0 (A), act7-6 (B), spk1-5 (C), scn1-1 (D), and ctr1btk (E). Nuclei, H2B-mCherry (magenta); actin filaments, GFP-ABD2-GFP (green). Bars = 20 µm. F to J, Trajectories of polar nuclear migration in Col-0 (F), act7-6 (G), spk1-5 (H), scn1-1 (I), and ctr1btk (J). n = 6 migration events per genotype. Coordinates represent the tracks of nuclei in 1-min intervals over 40 min. The zero reference position represents the outer basal-most corner of the cell.
Table I. Average velocity of nuclear movement in root hair cells.
The average velocity was calculated from the same data set used in the trajectory shown in Figure 6 and the histogram displayed in Supplemental Figure S7. In total, six movements from independent roots and experiments per genotype were subjected to calculation of average velocity. Tracks indicate the movement of the nucleus at 1-min intervals. Average velocities ± sd are displayed.
| Genotype | Total No. of Tracks | Average Velocity |
|---|---|---|
| µm min−1 | ||
| Col-0 | 252 | 1.51 ± 0.82 |
| act7-6 | 455 | 0.77 ± 0.69 |
| spk1-5 | 410 | 0.80 ± 0.75 |
| scn1-1 | 555 | 0.67 ± 0.43 |
| ctr1btk | 632 | 0.36 ± 0.38 |
ROP Signaling Acts on Outer Polar Nuclear Migration
Our results showing that the nucleus moves directionally toward the root hair in an ACT7-dependent manner imply the existence of a signaling source directing nuclear migration toward the hair outgrowth. Since ROP proteins accumulate at the hair initiation site and at the tip of the growing hair (Molendijk et al., 2001; Jones et al., 2002), we examined the role of ROP signaling in outer polar nuclear migration. To this end, we performed time-lapse imaging analysis of nuclear migration in spk1-5. Intriguingly, the velocity of nuclear movement was reduced significantly and movement trajectories displayed irregularities in the spk1-5 mutant (Fig. 6, C and H; Table I) when compared with the wild type (Fig. 6, A and F; Table I; Supplemental Fig. S7B). For example, the nucleus in spk1-5 paused at the inner lateral membrane for longer time periods than in the wild type, consistent with the overall reduction of nuclear migration velocity observed in the spk1-5 mutant (Fig. 6, C and H; Supplemental Movie S3).
We next analyzed the scn1-1 mutant that displays two or multiple hairs emerging from a single cell (Parker et al., 2000; Carol et al., 2005). Strikingly, the trajectories of nuclear movement in scn1-1 clearly differed from those in the wild type and the velocity of nuclear movement was reduced significantly (Fig. 6, D and I; Table I; Supplemental Fig. S7C). The nucleus often moved along the inner lateral membrane toward the inner basal-most corner of the cell and then moved over into the basal-most hair bulge (Fig. 6I). Intriguingly, the nucleus sometimes moved randomly between the ectopically formed apical and basal hair bulges in the scn1-1 mutant (Fig. 6, D and I; Supplemental Movie S4), indicating competition between two sites directing nuclear migration in these cells. Consistent with these observations, we also found nuclear placement at the ectopically formed apical or at the basal-most bulge in scn1-1 roots fixed with aldehyde prior to nuclear staining (Supplemental Fig. S6). Together, our findings reveal that both overactivation and reduced ROP activation affect the velocity and the directionality of outer polar nuclear migration, strongly suggesting a regulatory role for ROP signaling during this polar nuclear movement.
High Auxin Concentration Alters Outer Polar Nuclear Migration
Since our findings revealed that enhanced ethylene signaling and high auxin concentration modulate inner polar nuclear migration, we investigated the effect on outer polar nuclear migration in ctr1btk employing time-lapse imaging analysis. The trajectories of nuclear movement in ctr1btk differed from those in the wild type: after outgrowth of the root hair bulge, the nucleus remained in the center of the cell, then moved basally and into the hair (Fig. 6, E and J; Supplemental Movie S5), suggesting that high auxin concentration alters the trajectory of movement toward the basal-most direction of the hair outgrowth. Strikingly, the velocity of nuclear movement in ctr1btk was reduced significantly compared with that in the wild type (Table I; Supplemental Fig. S7D; Supplemental Movie S5). These results suggest that increased auxin concentration in the ctrbtk mutant mainly affects the velocity of nuclear movement and additionally diverts nuclear migration toward the basal-most ends of trichoblasts.
DISCUSSION
Our study unravels previously unnoticed contributions to inner polar nuclear position and to outer polar nuclear migration events in root trichoblasts (Supplemental Fig. S8). While nuclear movement inside the hair had been described to some extent (Ketelaar et al., 2002; Tamura et al., 2013; Zhang et al., 2015), early inner polar nuclear position remained uncharacterized, and little was known about the factors regulating directional movement toward the hair during outer polar nuclear migration.
Our findings reveal that endogenously elevated ethylene signaling as well as exogenous auxin application alter inner nuclear placement. Since CTR1-dependent ethylene signaling represses local auxin biosynthesis in the root tip (Ikeda et al., 2009), auxin homeostasis balanced by ethylene signaling likely modulates inner nuclear placement. This hypothesis is extended by our findings that mutations in IAA7/AXR2 and ARF7/ARF19 cause subtle deviations of nuclear position in the basal direction by themselves, demonstrating a requirement of nuclear auxin signaling for correct nuclear placement. More strikingly, mutations in ARF7 and ARF19 or IAA7/AXR2 strongly or fully repress the deviation of nuclear position observed in the ctr1btk mutant and upon auxin application. Hence, partial derepression of endogenous auxin biosynthesis and exogenous auxin act on nuclear placement via the nuclear auxin signaling pathway. Our findings reveal that reduced CTR1 function decreases vacuolar size during cell elongation via the ARF7-ARF19-dependent nuclear auxin signaling pathway, indicating a correlation between mispositioning of the nucleus from the inner epidermal membrane and a reduction of vacuole size in the ctr1btk mutant. Interestingly, loss of ZIG/VTI11 function caused slight nuclear mispositioning as well as a clear reduction of vacuole size during cell elongation, suggesting that vacuole morphology influences inner nuclear positioning to some extent. Together, our findings suggest that vacuole development additionally mildly contributes to positioning of the nucleus and that part of this may be mediated by the nuclear auxin signaling pathway, at least at elevated endogenous auxin levels.
In leaf pavement cells, an auxin signal was thought to activate ROP signaling via a presumptive nonnuclear auxin signaling pathway (Xu et al., 2010, 2014), but this view has recently been challenged (Gao et al., 2015). We tested the effect of elevated ethylene signaling and auxin production via ROP signaling by analyzing the ctr1btk;spk1-5 double mutant, revealing only a very minor suppression of ctr1btk-induced nuclear displacement by the spk1-5 mutation. By contrast, mutations in the nuclear auxin signaling module strongly or fully corrected the nuclear displacement caused by the ctr1btk mutation. Hence, an AUX/IAA-ARF nuclear auxin signaling module rather than SPK1-dependent ROP signaling transduces the auxin signal modulating early inner nuclear placement. Furthermore, interference with SCN1-dependent ROP signaling enhances nuclear displacement in the ctr1btk background toward a more outer lateral direction. Our in vivo evidence strongly suggests that auxin and ROP signaling synergistically act on inner polar nuclear placement rather than in a linear pathway, as proposed, for example, for actin-dependent endocytic trafficking (Lin et al., 2012). SPK1-dependent ROP signaling influences actin organization during trichome morphogenesis (Qiu et al., 2002; Uhrig et al., 2007; Basu et al., 2008) and has been implicated in actin organization in the root epidermis (Lin et al., 2012). Hence, the nuclear displacement observed in the spk1 mutants opened the possibility for a requirement of actin organization in inner nuclear placement. Indeed, analyses of the act7-6 loss-of-function mutant and of act7fiz1 dominant-negative interference in root hair cells revealed the contribution of ACT7 to inner nuclear placement along the apical-basal axis.
An apical and basal deviation of nuclear placement also was found in cells at the later stage of root hair differentiation, where time-lapse imaging of the act7-6 mutant revealed alterations of the directionality and the velocity of outer polar nuclear migration. Notably, similar deviations of nuclear movement were observed in the spk1-5 and scn1-1 mutants defective in positive and negative regulators of ROP, respectively. Hence, ROP signaling likely regulates the directionality and velocity of ACT7-dependent outer polar nuclear migration. This is consistent with the findings that YFP-ROP2 is ectopically misplaced at hair initiation sites in the scn1-1 mutant (Carol et al., 2005) and that ROP recruitment to the hair initiation site is shifted toward the basal-most ends of cells in ctr1btk and other auxin-overproducing mutants (Fischer et al., 2006; Ikeda et al., 2009). Strikingly, our results reveal that ectopically formed apical hair bulges in the scn1-1 mutant perturb nuclear migration in competition with basal hair bulges, suggesting competition between several sites of ROP activation. Together, our findings suggest that a ROP signaling source confers a directional cue for the nucleus to move from the inner membrane toward the hair in an ACT7-dependent manner. Dominant interference with F-actin dynamics revealed them to be essential for directional migration of the sperm nucleus within the female gametophyte of Arabidopsis, where dominant-negative interference with and localization of ROP8 suggested its requirement for actin dynamics (Kawashima et al., 2014). High redundancy within the ROP protein family rendered it difficult to genetically determine the contribution of respective individual ROP-GTPases in gametophytes and in root epidermal cells by loss-of-function studies (Lin et al., 2012; Kawashima et al., 2014).
The localization of ROP2, ROP4, and ROP6 to the root hair initiation site (Molendijk et al., 2001; Jones et al., 2002; Stanislas et al., 2015) suggests the involvement of at least three ROPs in nuclear migration in trichoblasts compared with at least ROP8 in the female gametophyte (Kawashima et al., 2014). Therefore, we resorted to loss-of-function mutants in positive and negative regulators of ROP activation to genetically elucidate the role of endogenous ROP signaling in polar nuclear migration. A role for auxin or ethylene signaling during nuclear migration has not been addressed in the female gametophyte, while the SLR/IAA14 and ARF7/ARF19 auxin signaling module contributes to coordinated nuclear migration during the initiation of lateral root primordia (De Rybel et al., 2010; Goh et al., 2012). However, a role for ROP signaling remains unaddressed during nuclear migration in this developmental process. Our study reveals a role for ROP and auxin/ethylene signaling in nuclear positioning events in both inner and outer actin-dependent nuclear placement. In addition, the signaling mechanism of the inner polar nuclear position mediated by both ROP and the auxin/ethylene signaling pathway appears to sequentially override actin-dependent outer polar nuclear migration during early to late stages of trichoblast differentiation. While precise nuclear placement within the root hair contributes to its tip growth, as revealed by optical trapping experiments (Ketelaar et al., 2002), the physiological role of nuclear placement to the inner membrane during root trichoblast elongation remains to be established in future studies. Thus, our work provides an initial framework for auxin and ROP signaling during actin-dependent nuclear migration in epidermal hair cells that can serve as a stepping stone for further analyses of mechanisms underlying polar nuclear migration in diverse contexts of plant cell differentiation and development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Col-0 accession of Arabidopsis (Arabidopsis thaliana) was used as the wild type as described (Kiefer et al., 2015). The Wassilewskija accession of Arabidopsis was used as the wild type only for spk1-1. spk1-5 (N822069; Lin et al., 2012) was obtained from the European Nottingham Arabidopsis Stock Centre. Additional mutants or marker lines were provided by the corresponding authors of the following articles: act7-6 (Kiefer et al., 2015), scn1-1 (Parker et al., 2000), spk1-1 (Qiu et al., 2002), ctrbtk (Ikeda et al., 2009), axr2-1 (Timpte et al., 1994), arf7-1;arf19-1 (Okushima et al., 2005), zig-1 (Kato et al., 2002), proVTI11:GFP-VTI11;zig-1 (Niihama et al., 2005), pro35S:EGFP-LTI6a (Grebe et al., 2003), and pro35S:GFP-ABD2-GFP (Wang et al., 2008).
Growth conditions were as described (Kiefer et al., 2015). Seeds were surface sterilized and stratified as described (Fischer et al., 2006) and/or surface sterilized with minor modifications. In brief, seeds were surface sterilized with 70% (v/v) ethanol for 1 min, incubated in 3% (w/v) sodium hypochlorite for 12 min, and washed twice with sterilized distilled water. Seedlings were vertically grown for 5 d on Murashige and Skoog (MS)-agar medium (1× MS salt [Duchefa], 10 g L−1 Suc, 8 g L−1 plant agar [Duchefa], and 0.5 g L−1 MES [Sigma-Aldrich], pH 5.7). For the experiments of scn1-1 and ctr1btk;scn1-1 shown in Figure 4, E to H, K, and L and Supplemental Figure S6, all genotypes were vertically grown on MS-agar medium without MES. MS-agar medium containing 300 nm 1-NAA (Sigma-Aldrich) was prepared as described (Grebe et al., 2002).
TEM
Roots from 5-d-old Col-0 seedlings were immersed in freezing medium (200 mm Suc, 10 mm trehalose, and 10 mm Tris buffer, pH 6.6), transferred into planchettes (Leica Microsystems; 3 × 0.5 mm, Al, types A and B), and frozen in a high-pressure freezer (HPM100; Leica Microsystems). Freeze substitution was conducted in a Leica EM AFS2 freeze substitution unit (Leica Microsystems) in dry acetone supplemented with 0.4% (w/v) uranyl acetate at −85°C for 16 h, followed by a 5-h warmup to −50°C. After washing with 100% (v/v) ethanol for 60 min, the roots were infiltrated and embedded in Lowicryl HM20 resin at −50°C (intermediate steps with 30% [v/v], 50% [v/v], and 70% [v/v] HM20 in ethanol, 60 min each) and polymerized for 3 d with UV light in the freeze substitution apparatus. Ultrathin sections were cut on a Leica Ultracut UCT ultramicrotome, poststained with aqueous uranyl acetate and lead citrate, and examined with a CM100 (Philips) transmission electron microscope operating at 100 kV. A 9000F Mark II system (Canon) was used for scanning of negatives (MACO EM films TYP S, 6.5 cm × 9 cm, ES 206).
DAPI Staining
Microtubule-stabilizing buffer (MTSB) was prepared as described (Grebe et al., 2003). Whole 5-d-old seedlings were fixed in 4% (w/v) paraformaldehyde (Sigma-Aldrich) in MTSB for 1 h on ice and washed three times with MTSB buffer. DNA was stained with 1 µg mL−1 DAPI (Sigma-Aldrich) in MTSB buffer for 30 min and washed four times with MTSB buffer and three times with distilled water. Samples were mounted in distilled water or Citifluor AF1 (Citifluor).
Quantitative and Statistical Analyses of Nuclear Positioning
Trichoblasts were identified in comparison with atrichoblasts based on trichoblast-specific properties, including a shorter cell length, a denser cytoplasm (Dolan et al., 1994), and a higher cell division rate (Berger et al., 1998). In all cases, trichoblasts in the elongation zone displaying a length of 20 to 50 µm derived from three independent experiments were analyzed. DAPI fluorescence images overlaid with a bright-field image were subjected to quantitative analysis. A minimum rectangle was placed on a nucleus surrounding the entire DAPI fluorescence signal using ImageJ (https://imagej.nih.gov/ij/), and the centroid of the rectangle was obtained. The centroid position of the nucleus, the distance between the basal membrane and the centroid, and the distance between the inner lateral membrane and the centroid were determined as well as the cell length and width employing ImageJ. The relative distance from the basal membrane to the centroid position along the apical-basal axis (y axis) and the relative distance from the inner membrane to the centroid position along the inner-outer axis (x axis) were calculated. Relative values of each nuclear position were plotted in a graph with a ratio of the y axis to the x axis of 2:1. A total of 150 elongating cells were analyzed, except for the proAtMYC1:GAL4-VP16;UAS:mTFP-ER driver line, the UAS:act7fiz1;UAS:H2B-mCherry reporter line, and proAtMYC1:GAL4-VP16;UAS:mTFP-ER UAS:act7fiz1;UAS:H2B-mCherry with n = 139 cells. For comparison of the inner-outer and basal-apical distributions, differences in distributions of relative nuclear positioning between genotypes or treatments were tested on the pooled sample (n = 139 or 150 cells) using the nonparametric, two-sample KS test (http://www.physics.csbsju.edu/stats/KS-test.n.plot_form.html) with a significance threshold of P < 0.05. For calculation of significances based on two-dimensional distributions, differences in the distribution of relative nuclear position between genotypes or treatments were tested by the two-sample, two-dimensional KS test using Matlab (The MathWorks) with a significance threshold of P < 0.05.
Quantitative and Statistical Analyses of Vacuole Size
MDY-64 (Life Technology, Molecular Probes) was used for vacuolar membrane (tonoplast) staining. A 0.25 mm stock solution was prepared in dimethyl sulfoxide. Whole 5-d-old seedlings were incubated in 1× MS liquid medium (1× MS salt, 10 g L−1 Suc, and 0.5 g L−1 MES, pH 5.7) containing 0.25 µm MDY-64 for 5 min in the dark. After staining, seedlings were mounted on coverslips in fresh 1× MS liquid medium for observation by confocal laser scanning microscopy (CLSM). In all cases, vacuoles in trichoblasts displaying a cell length of 20 to 50 µm, the same length range observed for nuclear positioning analysis, were examined. MDY-64 fluorescence images overlaid with an H2B-mCherry fluorescence image or a bright-field image were subjected to quantitative analysis of vacuole size. The largest vacuole in a single trichoblast cell was subjected to quantitative analysis as described (Löfke et al., 2015), the area size of the largest vacuole was measured using ImageJ, and mean values and se were calculated. A total of 40 cells from 18 roots derived from three independent experiments (n = 3) were analyzed per genotype. Student’s two-tailed t test with equal variance was employed to assess the significance of differences.
Generation of H2B-mCherry;EGFP-LTI6a and H2B-mCherry;GFP-ABD2-GFP Lines
The multiple cloning site was removed from pGreenII0179 (John Innes Centre; Hellens et al., 2000) employing KpnI and SacI, and overhangs were blunted. Subsequently, the 35S-cauliflower mosaic virus cassette excised from p35S-2 (John Innes Centre; Hellens et al., 2000) using EcoRV was inserted by blunt-end ligation. An Arabidopsis HISTONE2B cDNA fragment followed by a poly-Ala linker and the Venus coding sequence was synthesized (Genscript) and cloned between the 35S and the cauliflower mosaic virus cassettes employing XbaI and EcoRI. The Venus coding region was replaced by mCherry cDNA amplified from pmCherry (Clontech) employing primers H2B_Cherry_F and H2B_Cherry_R with NotI and BsrGI restriction sites. All primer sequences employed in this study are displayed in Supplemental Table S2. The resulting construct was transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) and introduced into Col-0 plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected based on resistance to 20 µg mL−1 hygromycin (Duchefa). A T3 homozygous H2B-mCherry line was crossed with an EGFP-LTI6a line (Grebe et al., 2003) or a GFP-ABD2-GFP line (Wang et al., 2008). Double homozygous F3 lines were obtained and analyzed.
Generation of proAtMYC1:GAL4-VP16;UAS:mTFP-ER Driver and UAS:act7fiz1;UAS:H2B-mCherry Reporter Lines
For the construction of proAtMYC1:GAL4-VP16;UAS:mTFP-ER, a 2,581-bp AtMYC1 promoter fragment was amplified from Col-0 genomic DNA by PCR employing primers AtMYC1_Sac_F and AtMYC1_Sac_R and cloned into pGreenII0029 (John Innes Centre; Hellens et al., 2000) with SacI, giving rise to pGreenII0029_proAtMYC1. The GAL4VP16/UAS:GFP-ER fragment was amplified from pCB2 (Heidstra et al., 2004) by PCR employing primers GAL4 VP16_Not_F and GAL4 VP16_Not_R and subcloned into pBluescriptII SK(−) (Stratagene), resulting in plasmid pBS_GAL4VP16/UAS:GFP-ER. The GFP-coding region was deleted by PCR employing an antisense primer annealing prior to the 5′ end of the GFP cDNA and a sense primer binding just after the 3′ end of the GFP-coding region (primers chitinase_F and chitinase_R), resulting in pBS_GAL4VP16/UAS:ER. An mTFP cDNA fragment amplified by PCR employing primers mTFP_Blunt_F and mTFP_Blunt_R was ligated into the linearized pBS_GAL4VP16/UAS:ER plasmid. The resulting plasmid pBS_GAL4VP16/UAS:mTFP-ER was digested with NotI, and the excised GAL4VP16/UAS:mTFP-ER cassette was cloned into pGreenII0029_proAtMYC1 via the NotI site. For the construction of UAS:act7fiz1;UAS:H2B-mCherry, the ACT7 cDNA fragment was subcloned into pBluescriptII SK(−). The act7fiz1 cDNA was produced by PCR with mutagenic primers ACT7_fiz1_F and ACT7_fiz1_R. The act7fiz1 fragment amplified by PCR (primers ACT7_Blunt_F and ACT7_Blunt_R) was subcloned into pBnUAS-Ptn (Heidstra et al., 2004) using SmaI. The UAS:act7fiz1 fragment was digested and cloned into pGreenII0029 with NotI, giving rise to pGreenII0029_UAS:act7fiz1. The H2BmCherry fragment amplified by PCR (primers H2B_Blunt_F and H2B_Blunt_R) was subcloned into pBnUAS-Ptn using SmaI. The UAS:H2BmCherry fragment amplified by PCR (primers UASH2B_Kpn_F and UASH2B_Kpn_R) was cloned into pGreenII0029_UAS:act7fiz1 with KpnI. The resulting constructs were transformed into Col-0 plants as described above. Transgenic plants were selected based on resistance to 30 µg mL−1 kanamycin (Duchefa). T3 homozygous proAtMYC1:GAL4-VP16;UAS:mTFP-ER lines were crossed with T2 UAS:act7fiz1;UAS:H2B-mCherry lines. F2 lines were obtained and subjected to analysis.
CLSM
All analyses using CLSM were conducted with a Zeiss LSM780 laser scanning microscope (Zeiss) and a Zeiss LSM710 laser scanning microscope (Zeiss). Samples were imaged using a water-corrected C-Apochromat 40× objective with a numerical aperture of 1.2 (Zeiss) or an oil-corrected Plan-Apochromat 40× objective with a numerical aperture of 1.4 (Zeiss).
Image Processing
Images were processed and analyzed using ZEN2010 (Zeiss), ZEN2011 (Zeiss), ImageJ, and Adobe Photoshop CS4 (Adobe Systems) software and assembled in Adobe Illustrator CS3 (Adobe Systems).
Time-Lapse Imaging of Outer Polar Nuclear Migration
Roots of 5-d-old H2B-mCherry;GFP-ABD2-GFP seedlings were analyzed. For sample preparation, 3 mL of sterile, hot, liquid MS-agar medium was pipetted into a Lab-Tek II coverglass chamber system (no. 155360; Nalge Nunc International). After solidification, an MS-agar segment with an area of approximately 10 × 20 mm was removed from one edge of the chamber employing a sterile razor blade. Sterilized seeds were placed at the edge between the agar and the coverglass from where the MS-agar segment had been removed. Seedlings were left for 5 d at an approximately 70° angle, allowing for vertical root growth into the space between the MS-agar and the coverglass. The chamber was placed horizontally during time-lapse imaging analysis only. Samples were imaged using an oil-corrected Plan-Apochromat 40× objective with a numerical aperture of 1.4 (Zeiss). Time-lapse imaging analysis was carried out at 1-min intervals employing ZEN2011 (Zeiss) for imaging and the LSM780 microscope described above.
Quantitative and Statistical Analyses of Outer Polar Nuclear Migration
Data from six independent time-lapse experiments, each monitoring nuclear migration over 40 min per genotype, were subjected to quantitative analysis. To define the centroid of the nucleus in the image frame, images were processed and analyzed using ImageJ. The centroid of the nucleus was defined as the centroid of the H2B-mCherry fluorescence signal, and the coordinates of the centroid position of the nucleus in the image frames were recorded. For trajectory analyses, each nuclear centroid coordinate was normalized to the coordinate of the outer basal-most corner of its respective cell as the zero reference. Normalized centroid coordinates of the nucleus at 1-min intervals were plotted in a graph and connected with a single line. For velocity analyses, the distance of each movement was calculated within 1-min intervals. The frequency of a given velocity (µm min−1) was presented in a histogram. Differences in distributions of velocity between wild-type and mutant nuclei were tested using a nonparametric, two-sample KS test with a significance threshold of P < 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CTR1 (At5g03730), IAA7/AXR2 (At3g23050), ARF7 (At5g20730), ARF19 (At1g19220), ZIG/VTI11 (At5g39510), SPK1 (At4g16340), SCN1 (At3g07880), and ACT7 (At5g09810).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nuclear position in wild-type root trichoblasts and atrichoblasts.
Supplemental Figure S2. Nuclear position in root trichoblasts of axr2-1 and arf7-1;arf19-1 mutants.
Supplemental Figure S3. Vacuole morphology in root trichoblasts of ethylene signaling, auxin signaling, zig/vti11, ROP signaling, and act7-6 mutants.
Supplemental Figure S4. Nuclear position in spk1-1 and comparison between ctr1btk;axr2-1 and ctr1btk;spk1-5.
Supplemental Figure S5. Nuclear position upon root hair cell-specific expression of dominant-negative act7fiz1.
Supplemental Figure S6. Apical and basal positioning of the nucleus in scn1-1.
Supplemental Figure S7. Velocity of polar nuclear migration in act7-6, ROP pathway, and ethylene signaling mutants.
Supplemental Figure S8. Hypothetical model of inner and outer polar nuclear migration in Arabidopsis root epidermal cells.
Supplemental Table S1. Significance levels of differences between the distributions of relative nuclear position.
Supplemental Table S2. List of primers used for plasmid construction.
Supplemental Movie S1. Time-lapse movie of polar nuclear migration in the wild type (Col-0).
Supplemental Movie S2. Time-lapse movie of polar nuclear migration in act7-6.
Supplemental Movie S3. Time-lapse movie of polar nuclear migration in spk1-5.
Supplemental Movie S4. Time-lapse movie of polar nuclear migration in scn1-1.
Supplemental Movie S5. Time-lapse movie of polar nuclear migration in ctr1btk.
Acknowledgments
We thank Elison Blancaflor, Mark Estelle, Claire S. Grierson, Renze Heidstra, Miyo T. Morita, Daniel B. Szymanski, and Athanasios Theologis for sharing published materials; the European Nottingham Arabidopsis Stock Centre for distributing seed stocks; Hailiang Mao for reading the article; Anna Gustavsson for support concerning CLSM; and Maike Stange for support concerning statistical analysis in Matlab. We thank Jürgen Hartmann at the Max Planck Institute of Colloids and Interfaces for providing access to high-pressure freezing equipment.
Footnotes
This work was supported by ERC-2010-STG Green-Lat-Pol, grant number 260699 to M.G., and EMBO long-term fellowship ALTF 278-2010 to R.H.
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB (2008) A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc Natl Acad Sci USA 105: 4044–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Hung CY, Dolan L, Schiefelbein J (1998) Control of cell division in the root epidermis of Arabidopsis thaliana. Dev Biol 194: 235–245 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Burke B, Roux KJ (2009) Nuclei take a position: managing nuclear location. Dev Cell 17: 587–597 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Chytilova E, Macas J, Sliwinska E, Rafelski SM, Lambert GM, Galbraith DW (2000) Nuclear dynamics in Arabidopsis thaliana. Mol Biol Cell 11: 2733–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010) A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Fischer U, Ikeda Y, Ljung K, Serralbo O, Singh M, Heidstra R, Palme K, Scheres B, Grebe M (2006) Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr Biol 16: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA 112: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893 [DOI] [PubMed] [Google Scholar]
- Grebe M, Friml J, Swarup R, Ljung K, Sandberg G, Terlou M, Palme K, Bennett MJ, Scheres B (2002) Cell polarity signaling in Arabidopsis involves a BFA-sensitive auxin influx pathway. Curr Biol 12: 329–334 [DOI] [PubMed] [Google Scholar]
- Grebe M, Xu J, Möbius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Griffis AH, Groves NR, Zhou X, Meier I (2014) Nuclei in motion: movement and positioning of plant nuclei in development, signaling, symbiosis, and disease. Front Plant Sci 5: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ (2013) Nuclear positioning. Cell 152: 1376–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M (2009) Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol 11: 731–738 [DOI] [PubMed] [Google Scholar]
- Jones M, Smirnoff N (2006) Nuclear dynamics during the simultaneous and sustained tip growth of multiple root hairs arising from a single root epidermal cell. J Exp Bot 57: 4269–4275 [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14: 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M (2002) SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Morita MT, Tasaka M (2010) Defects in dynamics and functions of actin filament in Arabidopsis caused by the dominant-negative actin fiz1-induced fragmentation of actin filament. Plant Cell Physiol 51: 333–338 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Maruyama D, Shagirov M, Li J, Hamamura Y, Yelagandula R, Toyama Y, Berger F (2014) Dynamic F-actin movement is essential for fertilization in Arabidopsis thaliana. eLife 3: e04501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, de Ruijter NC, Grierson CS, Dogterom M, Emons AM (2002) Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. Plant Cell 14: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer CS, Claes AR, Nzayisenga JC, Pietra S, Stanislas T, Hüser A, Ikeda Y, Grebe M (2015) Arabidopsis AIP1-2 restricted by WER-mediated patterning modulates planar polarity. Development 142: 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, et al. (2012) A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol 22: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Dünser K, Scheuring D, Kleine-Vehn J (2015) Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. eLife 4: e05868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB (1996) The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol 111: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Kiefer CS, Grebe M (2012) Planar polarity, tissue polarity and planar morphogenesis in plants. Curr Opin Plant Biol 15: 593–600 [DOI] [PubMed] [Google Scholar]
- Niihama M, Uemura T, Saito C, Nakano A, Sato MH, Tasaka M, Morita MT (2005) Conversion of functional specificity in Qb-SNARE VTI1 homologues of Arabidopsis. Curr Biol 15: 555–560 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Löfke C, Krüger F, Kittelmann M, Eisa A, Hughes L, Smith RS, Hawes C, Schumacher K, Kleine-Vehn J (2016) Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc Natl Acad Sci USA 113: 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T, Hüser A, Barbosa IC, Kiefer CS, Brackmann K, Pietra S, Gustavsson A, Zourelidou M, Schwechheimer C, Grebe M (2015) Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat Plants 1: 15162. [DOI] [PubMed] [Google Scholar]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 23: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M (1994) The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig JF, Mutondo M, Zimmermann I, Deeks MJ, Machesky LM, Thomas P, Uhrig S, Rambke C, Hussey PJ, Hülskamp M (2007) The role of Arabidopsis SCAR genes in ARP2-ARP3-dependent cell morphogenesis. Development 134: 967–977 [DOI] [PubMed] [Google Scholar]
- Van Bruaene N, Joss G, Thas O, Van Oostveldt P (2003) Four-dimensional imaging and computer-assisted track analysis of nuclear migration in root hairs of Arabidopsis thaliana. J Microsc 211: 167–178 [DOI] [PubMed] [Google Scholar]
- Wang YS, Yoo CM, Blancaflor EB (2008) Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, et al. (2014) Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kang E, Yuan M, Fu Y, Zhu L (2015) PCaP2 regulates nuclear positioning in growing Arabidopsis thaliana root hairs by modulating filamentous actin organization. Plant Cell Rep 34: 1317–1330 [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang X, Zhu D, Cui S, Li X, Cao Y, Ma L (2012) A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J Biol Chem 287: 14109–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Meier I (2015) The plant nuclear envelope as a multifunctional platform LINCed by SUN and KASH. J Exp Bot 66: 1649–1659 [DOI] [PubMed] [Google Scholar]
- Zhou X, Meier I (2014) Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc Natl Acad Sci USA 111: 11900–11905 [DOI] [PMC free article] [PubMed] [Google Scholar]