Pollen aperture factor INP1 assembles gradually and asynchronously at specific membrane domains and is involved in keeping them adjacent to callose wall.
Abstract
Accurate placement of extracellular materials is a critical part of cellular development. To study how cells achieve this accuracy, we use formation of pollen apertures as a model. In Arabidopsis (Arabidopsis thaliana), three regions on the pollen surface lack deposition of pollen wall exine and develop into apertures. In developing pollen, Arabidopsis INAPERTURATE POLLEN1 (INP1) protein acts as a marker for the preaperture domains, assembling there into three punctate lines. To understand the mechanism of aperture formation, we studied the dynamics of INP1 expression and localization and its relationship with the membrane domains at which it assembles. We found that INP1 assembly occurs after meiotic cytokinesis at the interface between the plasma membrane and the overlying callose wall, and requires the normal callose wall formation. Sites of INP1 localization coincide with positions of protruding membrane ridges in proximity to the callose wall. Our data suggest that INP1 is a late-acting factor involved in keeping specific membrane domains next to the callose wall to prevent formation of exine at these sites.
In organisms across all kingdoms, cells rely on precise deposition of extracellular materials to make cell walls, cuticles, or extracellular matrices. The generation of these structures helps cells to control their morphology and growth, facilitates tissue formation, provides cues for cellular navigation, and allows cells to invade other organisms or protect themselves from environmental dangers (Cosgrove, 2005; Scheffers and Pinho, 2005; Bowman and Free, 2006; Moussian, 2010; Rozario and DeSimone, 2010; Underwood, 2012). Yet, despite the importance of extracellular structures, the questions of how cells decide where to place these structures and how they mark domains to be either covered with or protected from extracellular materials are still poorly understood.
Pollen presents an excellent model for studying mechanisms that control formation of extracellular structures at precise locations. Pollen grains are protected by a complex extracellular structure, pollen wall exine, which creates elaborate patterns on the pollen surface that are highly morphologically diverse across species, yet conserved within a species. In most plants, exine deposition on the pollen surface is not entirely uniform: in addition to the areas covered by exine, there are also areas without or with a reduced amount of exine (Furness and Rudall, 2004). These areas are called apertures, and they help adjust pollen volume to different levels of humidity, regulate the rate of water entry upon pollen hydration, and serve as sites of exit for pollen tubes during pollen germination (Wodehouse, 1935; Heslop-Harrison, 1976, 1979; Edlund et al., 2004; Prieu et al., 2016). The distribution of apertures on the pollen surface is usually not random, and there are indications that aperture positioning is under tight genetic control (Reeder et al., 2016). In many eudicot species, apertures tend to be equally spaced around the equator of pollen grains. The wild-type pollen of Arabidopsis, like pollen of many other eudicots, has three equidistant longitudinal apertures. The precise placement of apertures and the ease of aberrant pattern recognition make them a compelling model for cellular regulation of deposition of extracellular structures and for formation of distinct cellular and extracellular microdomains.
Previously, we showed that the product of the Arabidopsis (Arabidopsis thaliana) INAPERTURATE POLLEN1 (INP1) gene is absolutely required for aperture formation (Dobritsa and Coerper, 2012; Reeder et al., 2016). INP1 is a novel protein without recognizable domains of defined function and is currently the only factor known to be involved in aperture formation. Interestingly, the INP1 protein exhibits a dynamic and highly unusual localization: in microspores, the precursors of pollen grains, it assembles into puncta distributed along three equidistant, linear domains located near the plasma membrane (Dobritsa and Coerper, 2012). This unusual tripartite localization is highly reminiscent of the positions of the three Arabidopsis apertures. Indeed, our previous 3D reconstructions from confocal z-stacks of microspores demonstrated that the three lines of INP1 puncta underlie the positions of future apertures (Dobritsa and Coerper, 2012; Reeder et al., 2016). These results suggest that the proper localization and assembly of INP1 into three lines are crucial for aperture placement and formation. Understanding how INP1 gets localized to distinct membrane domains and what it does to protect these sites from exine deposition can reveal mechanisms that cells employ to form extracellular structures with precision. This ability is clearly essential for a variety of cellular functions in multiple organisms.
The mechanism that controls aperture formation and brings INP1 to its distinct positions at the microspore periphery is not understood. What is known is that when wild-type Arabidopsis pollen develops, male meiosis gives rise to four pollen progenitors, the microspores, that are initially held together as a tetrahedral tetrad, and it is during the tetrad stage that the first signs of apertures become visible (Dobritsa and Coerper, 2012; Reeder et al., 2016). At the end of meiotic cytokinesis, each of these microspores has three points of last contact with their three sisters. It was noticed long ago that in species like Arabidopsis, which have a tetrahedral tetrad arrangement and develop three equatorial apertures, positions in the vicinity of these last-contact points between sister microspores become the places where apertures will eventually develop (Wodehouse, 1935). Apertures forming near the points of last contact develop as characteristic pairs, with an aperture on one microspore facing an aperture on its sister microspore. The correlation between the positions of last-contact points and the positions of apertures had led to a hypothesis that the last-contact points may serve as landmarks that determine aperture placement (Wodehouse, 1935; Ressayre et al., 2002).
Two models could account for the formation of INP1 lines at three equidistant positions in each microspore and their alignment with the INP1 lines in sister microspores. The first model suggests that the process of male meiotic cytokinesis delivers INP1 to the three sites of last contact in each microspore. Later, during the tetrad stage, INP1 spreads out from these positions perpendicular to the division planes to form three longitudinal lines in each microspore, which will become aligned with lines on sister microspores (Dobritsa and Coerper, 2012). According to this model, INP1 would contribute to specification of distinct linear aperture domains. An alternative model is that the membrane domains where INP1 assembles into punctate lines are already specified by an as-yet-unknown mechanism prior to INP1 arrival. In this scenario, INP1 is not the master regulator but rather the executor of a preestablished aperture pattern.
To distinguish between these models, we have studied the dynamic process of INP1 assembly into the punctate lines premarking aperture positions. Here, we provide evidence that INP1 is produced by the microspore mother cells, and that later, in tetrad-stage microspores, it gradually, and not completely synchronously, assembles into puncta and lines along the three longitudinal domains of the microspore membrane surface. No evidence was found, however, for the initial delivery of INP1 to three spots per microspore corresponding to the points of last contact at the end of cytokinesis. Our results are consistent with the model suggesting that the prespecified membrane aperture domains in microspores exist before the gradual and asynchronous INP1 assembly at these domains.
To understand the mechanism through which INP1 punctate lines promote aperture formation, we asked where, relative to the plasma membrane, the INP1 puncta formed. We demonstrate that assembly of INP1 into punctate lines appears to take place at the interface between the plasma membrane and the callose wall (CW) that overlies it. We found that presence of the CW is necessary for the formation of INP1 lines at the correct positions. In turn, the presence of INP1 on the plasma membrane affects membrane morphology, causing formation of distinct protruding ridges in the membrane domains, visible with confocal microscopy and with transmission electron microscopy (TEM) as areas closely apposed to the CW. Our data suggest a mechanism in which INP1 is the late-acting aperture factor, not required for domain specification but involved in keeping the three longitudinal domains of the plasma membrane in proximity to the CW to prevent exine deposition at these three sites and facilitate formation of apertures.
RESULTS
Expression of INP1 in Microspore Mother Cells, but Not in Tapetum, Can Efficiently Restore Formation of Apertures in the inp1 Mutant
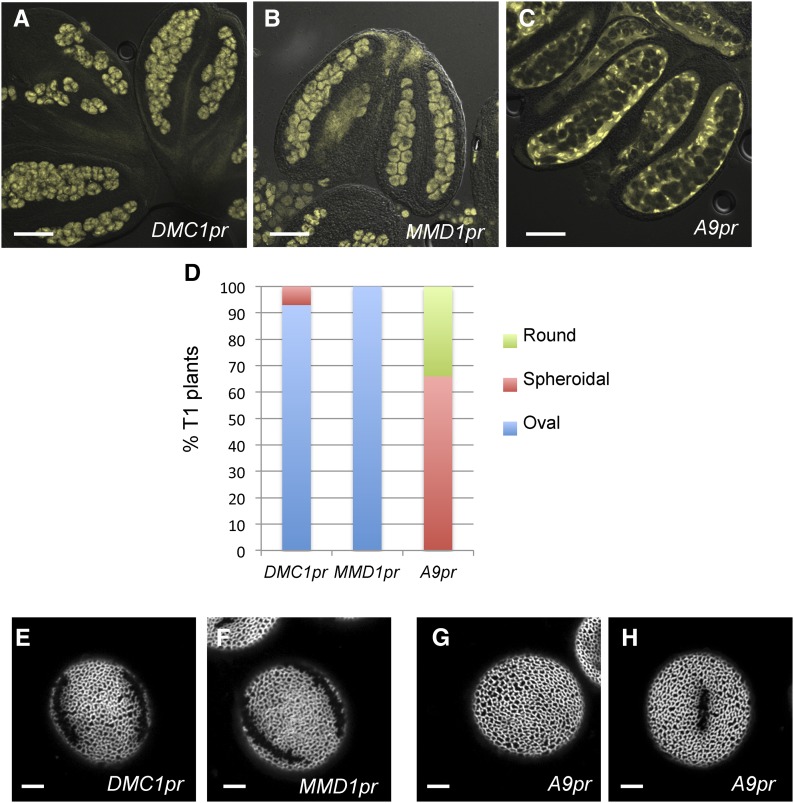
Our previous genetic analysis indicated that INP1 acts sporophytically and thus must be expressed from the diploid genome, yet the obvious INP1-YFP punctate signal was first visible in haploid tetrad-stage microspores (Dobritsa and Coerper, 2012). Therefore, INP1 protein or transcript is either inherited by microspores from the microspore mother cells (MMCs) during meiosis, or, less likely due to the presence of CWs that surround tetrads, is transported into microspores from the nearby diploid tapetal layer. To distinguish between these possibilities, we expressed INP1-YFP either in the inp1 MMC using the DMC1 and MMD1 promoters, both of which are strongly expressed in the MMC/microspores (Klimyuk and Jones, 1997; Yang et al., 2003; Fig. 1, A and B), or in the inp1 tapetum using the tapetum-specific A9 promoter (Paul et al., 1992; Feng and Dickinson, 2010; Fig. 1C). We then tested the abilities of these constructs to restore apertures.
Figure 1.
Expression of INP1 in microspore mother cells, but not in tapetum, restores formation of apertures in the inp1 mutant. A to C, INP1-YFP is expressed in the sporogenic layer of anthers when driven with DMC1 (A) or MMD1 (B) promoters and is expressed in tapetum when driven with the A9 promoter (C). D, Percentage of plants producing round, spheroidal, or oval pollen grains among the T1 populations containing, respectively, DMC1pr:INP1-YFP, MMD1pr:INP1-YFP, or A9pr:INP1-YFP transgenes. E to H, Long apertures are restored in plants with the DMC1pr (E) and MMD1pr (F) constructs, while pollen with no apertures (G) or with short apertures (H) is produced by plants expressing the A9pr construct. Scale bars = 50 μm in A to C and 5 μm in E to H.
Shape of dry pollen grains can be used as a reliable proxy for the presence of apertures: wild-type Arabidopsis pollen with normal apertures has an oval shape, pollen without apertures looks round, and pollen with shorter apertures appears spheroidal or intermediate between round and oval (Dobritsa et al., 2011; Dobritsa and Coerper, 2012). Therefore, to evaluate presence of apertures in the DMC1pr-, MMD1pr-, and A9pr-driven lines, we first looked at the shape of dry pollen under a dissection microscope. We found that the DMC1pr:INP1-YFP and the MMD1pr:INP1-YFP constructs significantly outperformed the tapetum-specific A9pr-driven construct in restoring apertures (Fig. 1D): 100% of the T1 lines (n = 10) with the MMD1pr-driven construct had oval pollen, indicative of efficient aperture restoration. Similarly, 93% of the DMC1pr T1 lines (n = 28) had oval pollen and 7% had spheroidal pollen. However, none of the 18 A9pr T1 plants produced oval pollen, 34% had round pollen, and 66% had pollen classified as spheroidal (Fig. 1D).
Consistent with the observations of pollen shapes, using high-magnification confocal microscopy, we found that long apertures were indeed restored in the MMD1pr:INP1-YFP inp1 and DMC1pr:INP1-YFP inp1 plants expressing INP1 in MMC (Fig. 1, E and F). In contrast, only pollen grains without apertures or with short apertures were present in the A9pr:INP1-YFP inp1 lines (Fig. 1, G and H), indicating that expression of INP1 in tapetum does not effectively restore normal apertures. Presence of short apertures in some A9pr lines suggests that either the A9 promoter has some weak expression in MMC in those lines, or that some limited movement of INP1 from tapetum to microspores is possible. Independently of these possibilities, the data strongly suggest that MMC, and not the tapetal cells, are the sporophytic source of the INP1 product.
Formation of the Aperture-Marking Punctate Lines of INP1 Is a Gradual and Nonsynchronous Process That Occurs in Tetrad-Stage Microspores
To understand the dynamics of INP1 expression and localization, we used confocal microscopy to perform detailed monitoring of spatial and temporal patterns of INP1-YFP, initially using the previously described INP1pr:INP1-YFP line (Dobritsa and Coerper, 2012). We developed a protocol for imaging and 3D reconstruction of sporogenic cells that allowed us to simultaneously visualize the developmental stage of the precursors of pollen grains, status of CWs, plasma membranes, and INP1-YFP localization (Fig. 2, see below). Our objective was to answer the following questions: (1) Is there any INP1 signal visible in MMC or microspores before the three INP1 lines have formed? (2) Does the subcellular INP1 traffic get specifically directed toward the three equally spaced positions at each microspore periphery? (3) Does all INP1 initially congregate at three equidistant points at the microspore equator? (4) Does it later spread out longitudinally to define the length of apertures? Although the possibility of delivery of INP1 to three equidistant equatorial points and its spread along longitudinal domains from these positions to form the INP1 lines have been previously proposed based on the phenotypes of the inp1 mutants with short apertures (Dobritsa and Coerper, 2012), these phenomena have not yet been observed directly.
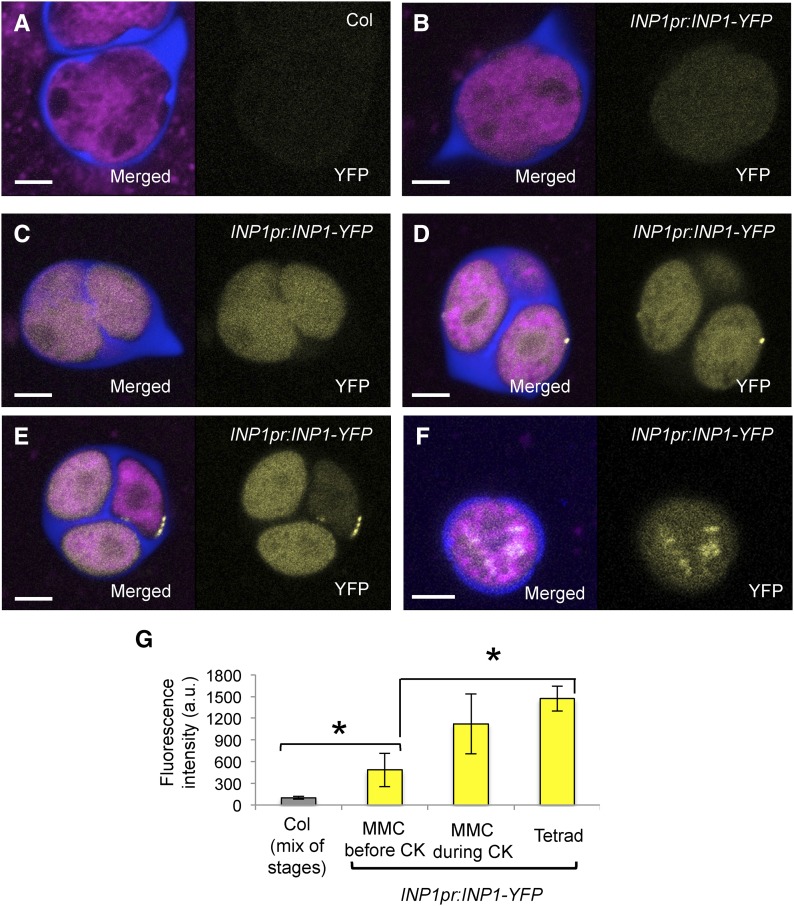
Figure 2.
Diffuse INP1-YFP fluorescence first appears in MMC, but puncta do not form until the tetrad stage. A to F, Confocal images of MMCs and tetrads. In each, image on the right shows YFP fluorescence and image on the left is the merged fluorescent signal from YFP (yellow), Calcofluor White (blue, CW), and CellMask Deep Red (magenta, membranous structures). A, Control Col-0 sporogenic cells exhibit essentially no background yellow fluorescence (MMC prior to cytokinesis is shown). B to F, Cytoplasmic and punctate YFP fluorescence in INP1pr:INP1-YFP sporogenic cells at different stages of development. B, MMC prior to cytokinesis. C, MMC during cytokinesis. D, Tetrad with some puncta. E, Tetrad with punctate lines forming. Note that the microspore with the INP1 line has the reduced amount of cytoplasmic YFP fluorescence compared to its sisters in which puncta have not yet formed. F, A young free microspore with cytoplasmic dots of YFP fluorescence. Scale bars = 5 μm. G, Quantification of mean diffused YFP fluorescence in control Col-0 sporogenic cells (Col, different developmental stages) and INP1pr:INP1-YFP in MMC before and during cytokinesis and at the tetrad stage (a.u., arbitrary units). Error bars indicate SD. Asterisks indicate statistical significance (P < 0.05).
Before the meiotic cytokinesis, low levels of diffuse YFP fluorescence were visible inside MMC (Fig. 2, B and G). These levels, however, were significantly higher than the background level in untransformed plants (Fig. 2, A and G), supporting the idea that INP1 is expressed in MMC. At this stage, the YFP fluorescence was spread uniformly throughout MMC and most of their organelles, including nuclei, without significant enrichment at any cellular region. During cytokinesis, the signal increased but remained diffuse without any visible puncta formation (Fig. 2, C and G). Very rarely, some puncta were present at the end of cytokinesis, but in the majority of cases we have not observed any peripheral puncta until after the cytokinesis was completed. Later, at the tetrad stage, three types of INP1-YFP-expressing tetrads were observed: (1) tetrads with diffuse uniform fluorescence throughout the microspores, which lacked any visible INP1 puncta (Supplemental Movie S1); (2) tetrads with a small number of INP1 puncta located at one to three positions at the microspore periphery (Fig. 2D; Supplemental Movie S2); and (3) tetrads with multiple INP1 puncta that were becoming organized into three aperture-marking lines in each microspore (Fig. 2E; Supplemental Movie S3).
In comparison with the relatively weak signal produced by INP1-YFP driven by the INP1 native promoter, INP1-YFP expressed from the DMC1 promoter exhibited much stronger fluorescence and had a very similar pattern of protein localization at the tetrad stage (see below). Therefore, to facilitate imaging, most of the subsequent observations were performed using the DMC1pr:INP1-YFP line.
DMC1pr:INP1-YFP-expressing MMC that had completed meiosis but not yet cytokinesis already exhibited strong YFP fluorescence, yet no peripheral puncta were observed and the signal remained diffuse and was located throughout the cells (Supplemental Fig. S1A). In many of these cells, in addition to the diffuse INP1-YFP signal throughout the MMC, there was a noticeable nuclear enrichment of the INP1-YFP signal (Supplemental Fig. S1A), which may be due to the overexpression of the protein.
Although the DMC1pr line exhibited significantly higher levels of INP1 expression compared to the INP1pr line (approximately 3- to 5-fold increase in the transcript levels and YFP signal intensity; Reeder et al., 2016), it also did not exhibit distinct INP1 puncta at the cell periphery until the tetrad stage. This suggests that either the signals necessary for the formation of INP1 puncta and lines are not present at the periphery until after the completion of meiotic cytokinesis or that INP1 does not get delivered to the positions where its assembly into puncta can begin until the tetrad stage.
At the tetrad stage, three types of tetrads, similar in phenotypes to the ones described above for the INP1pr:INP1-YFP plants, were observed in the DMC1pr:INP1-YFP plants (Fig. 3; Supplemental Movie S4–6). Careful observation of hundreds of tetrads provided evidence that INP1 is not initially delivered to very specific positions at the microspore periphery. Instead, it appears to initially cover the entire microspore and then, gradually, assemble first into spots and later into three equally spaced lines. The process of the assembly into puncta and lines does not occur completely synchronously either within a single microspore or between the sister microspores in a tetrad. In multiple tetrads, puncta were present at some, but not all, of the three areas in a microspore or in some, but not all, microspores in a tetrad, indicating that formation of puncta does not occur simultaneously at every possible site in a tetrad but instead happens independently at each site. In many cases, we observed microspores with only one or two INP1 spots. In other cases, INP1 lines had nearly finished forming in some microspores in a tetrad, but not in their sisters (Fig. 3D; Supplemental Movie S7). In a number of cases, especially for INP1pr:INP1-YFP, we observed tetrads in which well-formed INP1 lines in a microspore correlated with low levels of general diffuse fluorescence in this microspore, whereas its sisters contained high levels of diffuse fluorescence and lacked INP1 lines (Fig. 2E). These observations support the notion that the INP1 aggregating into punctate lines at the microspore periphery is likely derived from the pool of cytoplasmic INP1.
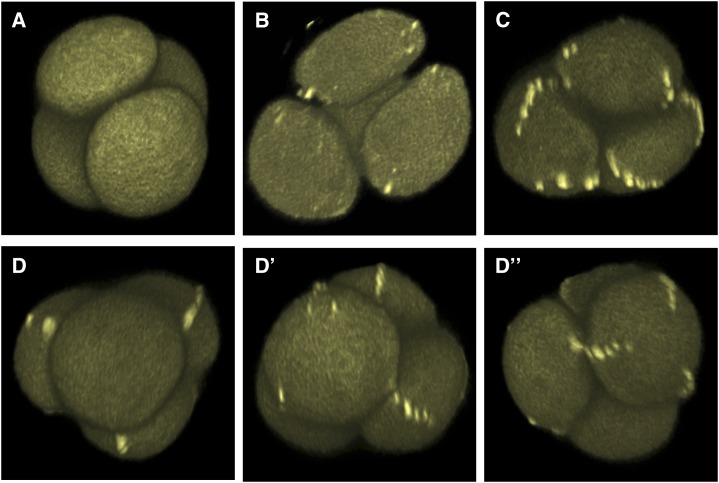
Figure 3.
Assembly of punctate INP1 lines in tetrad-stage microspores is gradual and nonsynchronous. 3D reconstructions of tetrad-stage microspores from the DMC1pr:INP1-YFP plants showing INP1-YFP lines at different stages of assembly. YFP fluorescence is shown. A, A tetrad with no puncta. See also Supplemental Movie S4. B, A tetrad with several puncta forming. See also Supplemental Movie S5. C, A tetrad with assembled INP1 lines. See also Supplemental Movie S6. D to D”, Microspores in the same tetrad can have INP1 lines at different stages of assembly. Three views of the same tetrad with a different microspore facing forward on each panel. D, The front microspore has no INP1 puncta or lines. D’, The front microspore has some INP1 puncta. D”, The front microspore has three INP1 punctate lines. See also Supplemental Movie S7.
When only a single INP1 punctum was found at one of the three future aperture sites, it was not always closely associated with the positions adjacent to the last-contact points (i.e. the membrane areas in contact with the division plane). Instead, these single INP1 spots were often observed at more distal positions relative to the last-contact points (Supplemental Movie S3). However, the INP1 puncta were almost exclusively observed along the three longitudinal lines of plasma membrane that would correspond to the regions of future apertures, suggesting that these domains of plasma membrane possess specific features that make them distinct from other membrane areas and cause INP1 to aggregate at these regions. Notably, the higher levels of INP1-YFP expression from the DMC1 promoter compared to the native promoter did not change the morphology or number of INP1 lines, strongly suggesting that the aperture domains are prespecified before the arrival of INP1.
At the early free microspore stage, in both INP1pr and DMC1pr:INP1-YFP plants only a small fraction of microspores exhibited INP1-YFP lines at the cell periphery (Supplemental Fig. S1B), indicating that the lines formed by INP1 are short lived and disappear soon after tetrad dissolution. The majority of the early free microspores had either a uniform YFP fluorescence distributed throughout the cell or showed large YFP-positive intracellular aggregates (Fig. 2F). The YFP signal disappeared at the later stages. Together, these observations suggest that after microspore release from tetrads, INP1 undergoes rapid endocytosis and degradation.
INP1 Puncta and Lines in Tetrad-Stage Microspores Appear to Form between the Plasma Membrane and the CW
INP1 is a novel protein without obvious signal peptides, sites for lipid-anchoring, or clear transmembrane domains (Dobritsa and Coerper, 2012). To determine where in relation to the plasma membrane INP1 puncta are localized, we labeled INP1-YFP-expressing tetrads with Calcofluor White, which stains CWs (Hughes and McCully, 1975; Krishnamurthy, 1999), and with the lipophilic dye CellMask Deep Red, which stains membranous structures, including plasma membrane (Wilson et al., 2014). We have not observed direct colocalization of the YFP punctate signal with either the plasma membrane or with the CW. In contrast, YFP puncta appeared to be present on the surface of the plasma membrane, underneath the CW (e.g. Fig. 2E): this was especially apparent in 3D volume reconstructions from z-stacks of tetrads (Figs. 4C and 5D) or in signal intensity profiles of the three fluorophores along the line that passed through two INP1-YFP puncta (Fig. 4, A and B).
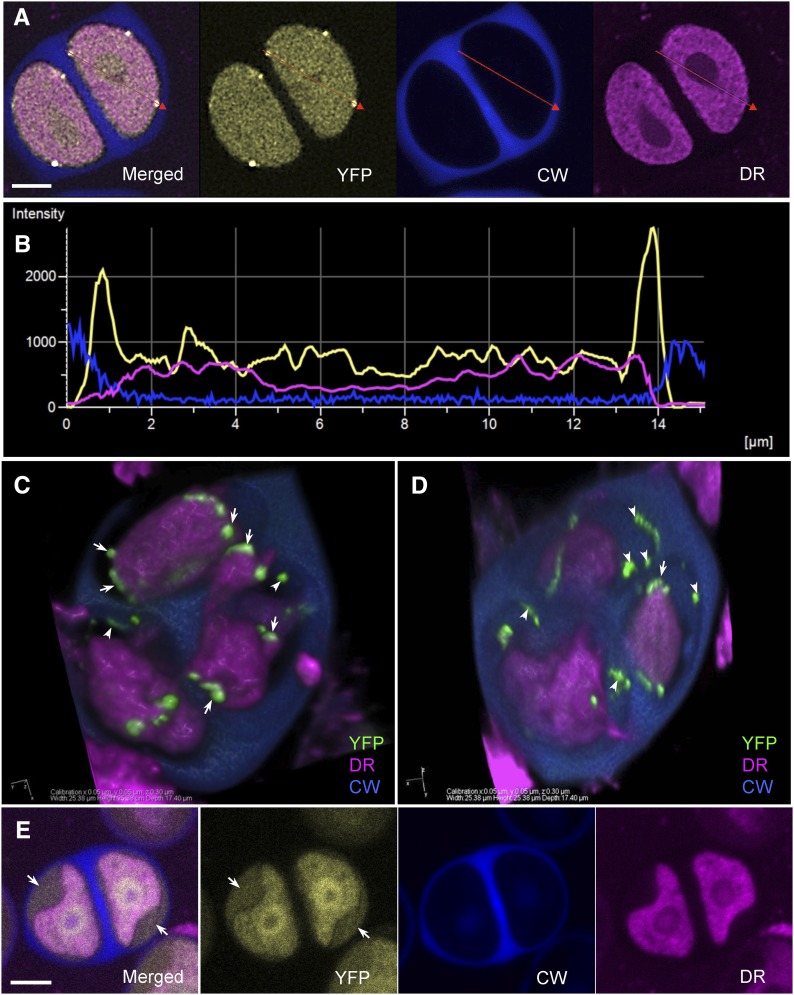
Figure 4.
INP1-YFP puncta and lines form between plasma membrane and CW. A, Confocal images of tetrad-stage microspores showing YFP (yellow), Calcofluor White (CW, blue), and CellMask Deep Red (DR, magenta) fluorescence. A red line is drawn through two INP1 puncta. B, Signal intensity profile of the three fluorophores along the red line shown in A. Yellow peaks coincide with the drop in magenta fluorescence and are followed by the increase in blue fluorescence. C to D, A 3D reconstruction of a plasmolyzed DMC1pr: INP1-YFP tetrad. Two different views of the same tetrad are shown. INP1-YFP puncta (green) are visible in association with both plasma membrane (arrows) and CWs (arrowheads). CW is partially removed to reveal the microspores. E, In microspores in which membranes got separated from the CW, the diffused INP1-YFP signal appears to occupy the space between the surface of plasma membranes and the CWs (arrows). Fluorophore colors as the same as in A. Scale bars in A and E = 5 μm.
Figure 5.
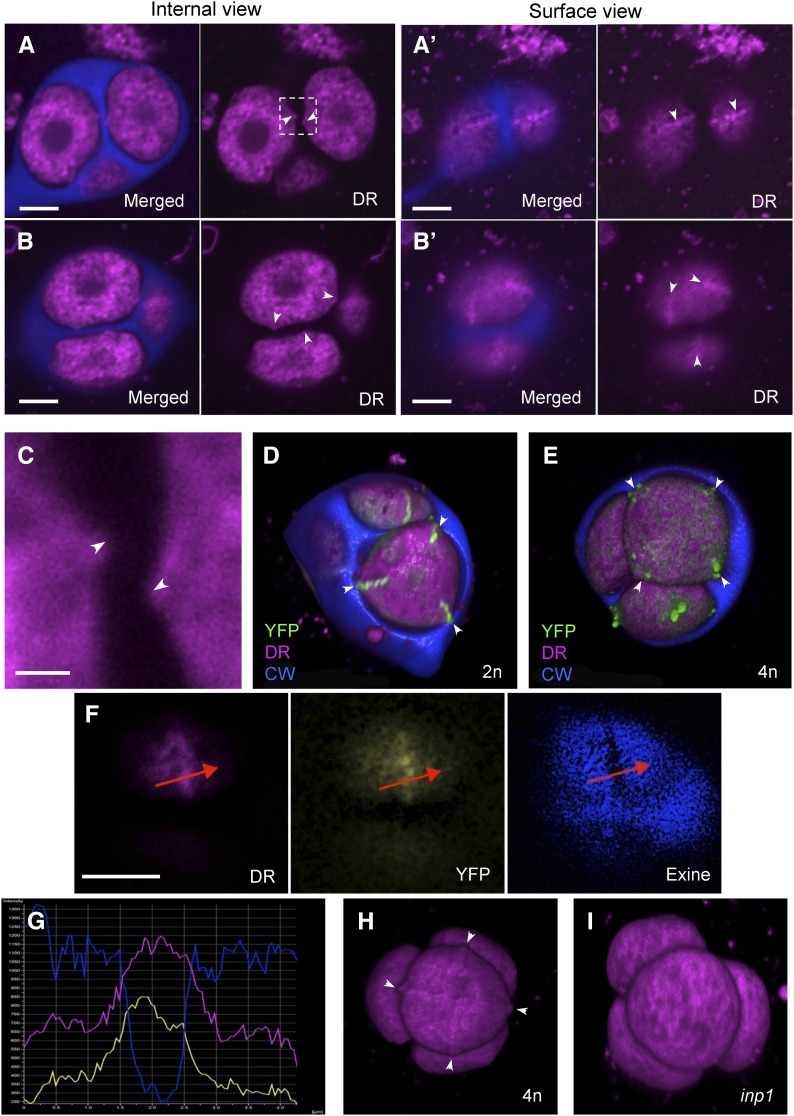
Microspore plasma membrane forms protrusions and ridges at the aperture sites. A to B’, Confocal images of two tetrads showing membranous structures (magenta) and CW (blue). Membrane protrusions visible on internal optical sections (A and B, arrowheads) correspond to ridges visible on surface views of the same tetrads (A’ and B’, arrowheads). C, A magnified view of the boxed region from A of membrane protrusions (arrowheads) facing each other in a way typical of aperture placement. D, A 3D reconstruction of a tetrad from a diploid plant. Microspores have triangular outline, with three corners coinciding with three INP1 lines (green). CW is partially removed to reveal microspores. E, A 3D reconstruction of a tetrad from a tetraploid plant. Microspores have rectangular outlines, with four corners coinciding with four INP1 lines (green). CW is partially removed to reveal microspores. F, A surface view of a late tetrad-stage microspores shows that the position of the membrane ridge visible with membrane dye DeepRed (DR) coincides with positions of the INP1-YFP line (YFP) and the aperture (visible as a gap in the developing exine). G, Signal intensity profile of the three fluorophores shown in H along the red line drawn through the aperture in H. Peaks of the magenta and yellow signal correspond to the dip in the blue signal. H, A 3D reconstruction of a tetrad from a tetraploid plant. Four membrane ridges (arrowheads) are visible in the front-facing microspore. See also Supplemental Movie S8. I, In inp1 tetrads, no ridges were apparent. Scale bars = 5 μm in (A to B’ and F) and 1 μm in C.
To establish if INP1 puncta were attached to the plasma membrane, we performed plasmolysis experiments by incubating DMC1pr:INP1-YFP tetrads in a solution of 25% Suc. In the plasmolyzed tetrads, some spots and lines of INP1-YFP were found in association with plasma membrane, while others appeared attached to CWs (Fig. 4, C and D; Supplemental Fig. S2). Taken together, our observations are consistent with the interpretation that in normal tetrads INP1-containing aggregates are in close contact with both structures, possibly serving as a bridge that connects the microspore membrane to the overlying CW. In support of the idea that INP1 is delivered to the plasma membrane surface, in many microspores in which membranes got separated from the CW during sample preparation, a diffuse INP1 signal also appeared to occupy the space between the surface of plasma membranes and the CWs (Fig. 4E; Supplemental Fig. S2).
INP1-Decorated Membrane Regions Form Ridges at the Aperture Positions
While performing 3D reconstructions of the INP1-YFP-expressing tetrads, we noticed that in many tetrads microspores had triangular outlines, with plasma membrane anchored at three positions corresponding to the positions of INP1 lines (Fig. 5D). We have recently demonstrated that in plants producing diploid rather than normal haploid pollen, INP1 often localizes to four, and not to three, areas per microspore. The pollen of such plants usually develops four apertures (Reeder et al., 2016). We therefore checked whether in those plants, instead of having triangular outlines, tetrad-stage microspores would look rectangular. Indeed, the plasma membranes in these microspores were “anchored” at four positions, which also coincided with the INP1-YFP lines (Fig. 5E) and along which membrane ridges could be observed.
The ridges formed by the membrane of microspores could often be recognized even in the absence of the INP1-YFP signal by the increased CellMask Deep Red signal when compared to the adjacent sites and by their characteristic positions, perpendicular to the centers of intersporal CWs and facing each other in the sister microspores (Fig. 5, A–C), and were especially apparent in 3D reconstructions of large microspores produced by tetraploid plants (Fig. 5H; Supplemental Movie S8). In the cases when tetrads have already developed early visible exine and apertures, we were able to confirm that the plasma membrane ridges, INP1 lines, and apertures all colocalize (Fig. 5, F and G).
Presence of these distinct membranous ridges prompted us to ask whether they were the cause or the effect of the INP1 line formation. To answer this question, we checked whether these ridges could still form in the inp1 mutant. In the absence of the INP1 function, the distinct membrane ridges were no longer recognizable (Fig. 5I), suggesting that their formation requires the presence of INP1.
Plasma Membrane at the Developing Aperture Sites Is in Close Contact with the CW and Is Protected from Undulations and Primexine Deposition
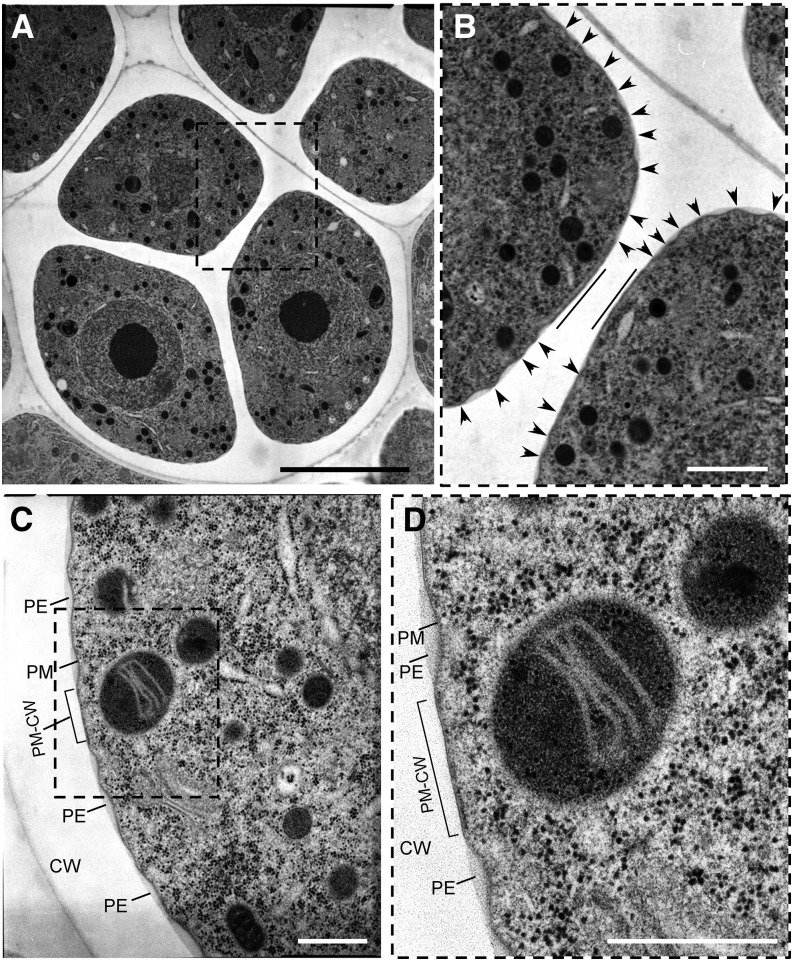
To look in more detail at the plasma membrane-CW interface in tetrad-stage microspores, we used TEM. We observed that microspores exhibited three equidistantly distributed flattened areas at which plasma membrane was apposed to the CW (Fig. 6; Supplemental Fig. S3A). In many tetrads, a flattened membrane area in one microspore was positioned directly opposite from a similar area on a sister microspore (Fig. 6B), consistent with apertures facing each other in sister microspores (Dobritsa and Coerper, 2012; Reeder et al., 2016). In contrast to the proximity between the membrane and CW at the putative aperture sites, the plasma membrane in other areas was separated from the CW and exhibited characteristic undulation and primexine deposition (Fig. 6, B–D; Supplemental Fig. S3, B and C), considered to be the first steps in the formation of future exine (Fitzgerald and Knox, 1995; Ariizumi and Toriyama, 2011; Quilichini et al., 2014).
Figure 6.
Plasma membrane at the developing aperture sites is in close contact with CW and is protected from undulations and primexine deposition. A to D, TEM sections of tetrad-stage microspores at different magnifications. A, A tetrad of microspores with a boxed area containing two flattened membrane regions on sister microspores facing each other. B, A magnified view of the region boxed in A with straight lines demarcating the flattened membrane regions that face each other and arrowheads pointing at the peaks of the undulating plasma membranes. C, A portion of the microspore from another tetrad with a boxed area showing a flattened region of plasma membrane in juxtaposition to the CW (PM-CW). D, A magnified view of the boxed area from C. PE, primexine; PM, plasma membrane. The sites of PM-CW close contacts are denoted by brackets in C and D. Scale bars: 5 μm (A); 1 μm (B); 500 nm (C and D).
It was proposed a long time ago (Heslop-Harrison, 1963, 1968) that the sites of prospective apertures might be protected from exine deposition by the presence of a layer of endoplasmic reticulum (ER) underneath the microspore plasma membrane, the so-called colpal shield. We therefore paid close attention to the possibility of a colpal shield presence at the developing aperture sites. Although we have not found clear indications of the presence of ER underneath the flattened membrane areas, on several occasions we have observed straight lines of ribosomes located directly below those membrane sites, with all the ribosomes equally positioned relative to the flattened membrane (Supplemental Fig. S3, B and C). A possible interpretation of these observations is that the line-forming ribosomes are present on the surface of the electron-lucent rough ER that indeed underlies plasma membrane at the positions of developing apertures. However, whether presence of ER next to the aperture membrane sites is functionally significant for aperture formation will require further investigation.
Callose Deposition Is Necessary for the Formation of INP1-Decorated Domains on the Membrane Surface
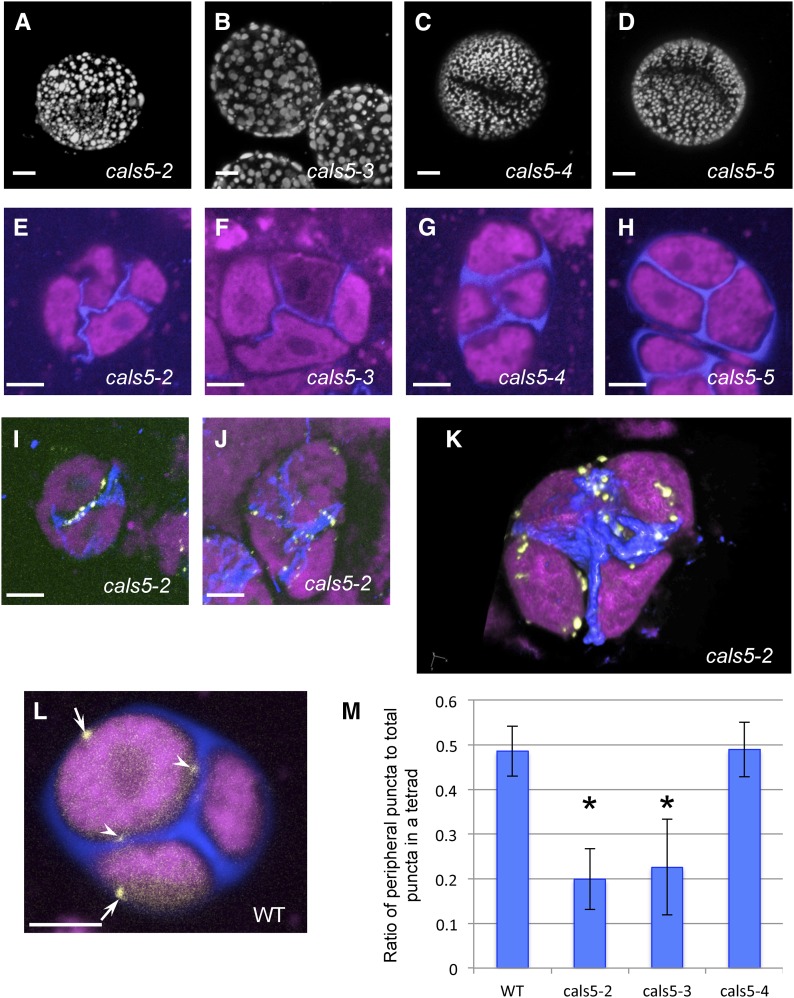
The finding that the INP1 lines develop in close contact with the CW prompted us to examine whether presence of the wall was necessary for the formation of the INP1-decorated domains at the microspore membrane. We started by looking at aperture presence in the mature pollen of the mutants defective in the callose synthase CALS5/GSL2 that is involved in the formation of the CW around MMC (Dong et al., 2005; Nishikawa et al., 2005). The allelic series for the CALS5 gene contains several alleles of various strengths, including strong alleles cals5-2 and cals5-3, an intermediate-strength allele cals5-4, and a mild allele cals5-5. These alleles result in different amounts of callose deposition around MMC (Dong et al., 2005; Nishikawa et al., 2005). While the reticulate pattern of exine was greatly disrupted in all these mutants (Fig. 7, A–D), we found that recognizable apertures were still present in the weaker alleles of CALS5 (cals5-4 and cals5-5; Fig. 7, C and D), but not in the stronger ones (cals5-2 and cals5-3; Fig. 7, A and B), indicating that presence of normal CW is necessary for aperture formation.
Figure 7.
Defects in CW formation lead to defective aperture formation and abnormal localization of INP1 puncta. A to D, Although the reticulate pattern of pollen exine is disrupted in all mutant alleles of CALS5, aperture formation is specifically disrupted in strong, but not weak alleles of cals5. A and B, Pollen grains of cals5-2 and cals5-3 mutants lack apertures. C and D, In cals5-4 and cals5-5 mutants, apertures are still present. E to H, Tetrads in cals5 mutants stained with Calcofluor White (blue, CW) and CellMask Deep Red (magenta, membranous structures). Callose deposition is absent around the tetrad periphery in cals5-2 (E) and cals5-3 (F), yet weak intersporal walls still form. In cals5-4 and cals5-5, although the deposition of callose is also reduced compared to wild-type (shown in L), peripheral walls are formed and the intersporal walls are thicker and straighter than in cals5-2 and cals5-3. I to K, Strong cals5 mutants show abnormal localization of INP1 puncta. I and J, Maximum intensity projection of z-stacks of cals5-2 DMC1pr:INP1-YFP tetrads. INP1-YFP puncta exhibits strong colocalization with remaining CWs. K, A 3D reconstruction of a cals5-2 tetrad showing most INP1-YFP puncta in the proximity of intersporal CWs. (L) An example of a confocal section of a wild-type DMC1pr:INP1-YFP tetrad showing puncta localized both at the periphery of a tetrad (arrows) and at the central positions (arrowheads). Scale bars in A to J and L = 5 μm. M, Quantification of INP1 puncta localization in tetrads using serial optical sections. The ratio of peripheral puncta to the total number of puncta in a tetrad was calculated. Error bars indicate SD. Asterisks indicate statistically significant difference from wild type (P < 0.05).
Consistent with the previous observations on cals5 tetrads (Dong et al., 2005; Nishikawa et al., 2005), the intersporal CWs that develop during meiotic cytokinesis were present in the center of tetrads in all alleles (Fig. 7, E–H), although in the strong cals5 alleles these internal walls were thinner than normal and positioned irregularly (Fig. 7, E–F). The most significant difference was in the outer wall, which normally develops around MMC prior to meiosis: it was completely absent in the two strong alleles but was still recognizable in the two weaker alleles (Fig. 7, G–H).
To test whether the formation of punctate INP1-YFP lines at the three membrane domains was affected in the CW-defective mutants, we crossed the DMC1pr:INP1-YFP line with each of the four cals5 alleles. In the weaker cals5-4 and cals5-5 alleles, the punctate lines of INP1-YFP were formed at fairly normal positions around the microspore periphery (Supplemental Fig. S4). However, in the strong cals5-2 and cals5-3 alleles, INP1-YFP assembled into puncta at the abnormal sites. In these two mutants, the INP1-YFP puncta were found in conspicuous association with the remaining internal walls that separate microspores and were mostly absent from the outer tetrad periphery where the CW was missing (Fig. 7, I–K). This placement was different from the placement observed in the wild type and the weaker cals5 mutants, where INP1 puncta could be found both at the tetrad periphery and at the center. To quantify this effect, we used confocal sections and calculated the ratio of the INP1 puncta observed at the tetrad periphery (i.e. adjacent to the positions where the outer CW is normally found) to the total number of puncta in a tetrad (Fig. 7, L and M). While in the wild-type tetrads around 50% of the INP1 puncta were present at the periphery of tetrads, in the strong cals5-2 and cals5-3 mutants only about 20% of the puncta were found at the periphery and the majority of spots were found in close proximity to the internal CWs (Fig. 7M; Supplemental Fig. S, 5 and 6).
The internal walls in cals5-2 have highly irregular morphology and placement, and, strikingly, positions of the INP1 puncta often closely trace these wall irregularities (Fig. 7, I–J). 3D reconstructions of wild-type tetrads show that INP1 lines are positioned perpendicular to the nearest internal CW (Fig. 5D). In contrast, in the reconstructions of cals5-2 tetrads, rather than assembling into lines perpendicular to the walls, the INP1 puncta often align with the direction of internal walls (Fig. 7K; Supplemental Fig. S6). Taken together, these observations indicate that the normal formation of the CW is a necessary prerequisite for the correct formation of INP1 lines at specific membrane domains.
DISCUSSION
INP1 is a novel protein of unknown function that exhibits an unusual localization pattern, assembling into three equidistant punctate lines that premark positions of future apertures at the surface of tetrad-stage microspores. Even more intriguingly, the assembly of the INP1 lines takes place within a complex 3D arrangement of tetrahedral tetrads with the result that the INP1 lines and, eventually, apertures on one microspore usually exhibit a close alignment with lines and apertures on sister microspores. Such seemingly nonindependent patterns of INP1 positioning observed in sister microspores at late stages of tetrad development had previously led us to propose a working model for initial delivery of INP1 at the end of cytokinesis to three last-contact points between each microspore and its three sisters, followed by INP1 spreading from these focal points along three equidistant longitudinal domains and assembling into lines located perpendicular to the division planes (Dobritsa and Coerper, 2012).
The results presented here, however, are consistent with an alternative model in which INP1 gets assembled at the domains that are already specified. First, we have not found evidence for the initial delivery of INP1 to the points of last contact during cytokinesis. Rather, the assembly of INP1 into puncta and lines takes place after the completion of cytokinesis, at the tetrad stage. This is also consistent with our recent data that did not support the Wodehouse model and the importance of last-contact points for aperture formation in Arabidopsis (Reeder et al., 2016). Second, no evidence was found for the initial tripartite trafficking of INP1 in microspores. Third, the asynchronous formation of INP1 lines within a microspore and between the sister microspores in many tetrads argues against the interdependence of these lines and the perfect coordination of timing of INP1 distribution along the lines that were suggested by the first model.
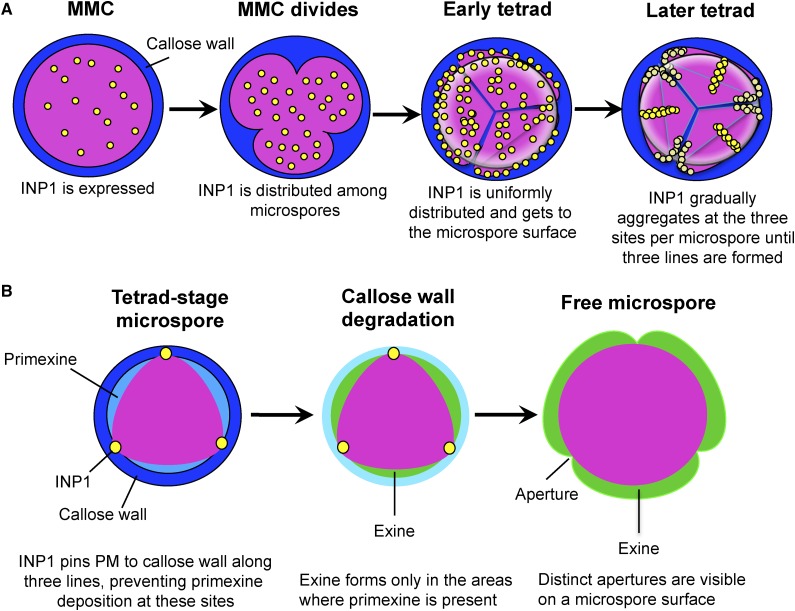
Based on the observations presented here, we propose the following mechanism (Fig. 8A): (1) INP1 protein is produced by MMC before and during cytokinesis and is initially distributed uniformly inside the cell; (2) during the tetrad stage, INP1 gets delivered to the cell surface, where it is also initially uniformly distributed; (3) later, INP1 gradually assembles first into individual puncta and then into punctate lines at the three membrane domains. The process of line formation is not absolutely synchronous and seems to happen somewhat independently at the three sites within a microspore, as well as between sister microspores. The first spots of INP1 can be found at positions that are distal to the areas where the last-contact points have been present, yet they are still located in the vicinity of what will become the pollen equator. This provides a possible explanation for the frequent positioning of short apertures in plants with reduced INP1 levels close to the pollen equator (Dobritsa and Coerper, 2012).
Figure 8.
A model of INP1 assembly into lines and formation of apertures on microspore surface. A, INP1 is produced by an MMC and initially has diffused internal localization. At the tetrad stage, INP1 gets to the microspore surface and gradually assembles first into puncta and then into punctate lines at the three preformed membrane domains. B, INP1 lines pin three domains of plasma membrane to the overlying CW, preventing primexine deposition at these regions. In the absence of primexine, exine fails to form in these regions but is deposited everywhere else, thus leading to the formation of three distinct apertures on a microspore surface.
The fact that higher levels of INP1 production do not lead to puncta assembling at more sites or to formation of wider apertures also indicates that the size and the positions of domains are already prespecified before INP1 arrives. Taken together, our results strongly suggest that INP1 acts as an executor of aperture formation, but not as the mastermind that determines where apertures should be placed and (with the possible exception of aperture length) how they should look. Therefore, there must be additional factors that act upstream of INP1 and specify aperture domains.
The three plasma membrane domains that become decorated with INP1 aggregates likely possess specific features, resulting in entrapment or stabilization of INP1 molecules and, therefore, leading to INP1 aggregation specifically along these sites. Finding out what exactly makes these domains special and how they differ from the nearby regions is an exciting challenge. In recent years, more research has focused on formation of distinct areas of the plasma membrane that acquire specific repertoires of proteins and/or lipids (Furt et al., 2010; Gao et al., 2011; Spira et al., 2012; van den Bogaart et al., 2013; Bailey and Prehoda, 2015). Among the better-studied examples of distinct membrane domains in plants are the Casparian strips in root endodermis, domains associated with secondary wall formation in xylem vessel cells, apical and basal domains in root epidermis, tips of the growing pollen tubes, and the interdigitated lobes and indentations in the puzzle-shaped epidermal pavement cells in leaves (Zheng and Yang, 2000; Yang, 2008; Alassimone et al., 2010; Kleine-Vehn et al., 2011; Roppolo et al., 2011; Oda and Fukuda, 2012a; Roppolo and Geldner, 2012; Yang and Lavagi, 2012). The view that emerges from studying these systems suggests that signaling through small GTP-binding proteins, such as Rho-of-plants (ROPs), as well as presence of membrane-associated kinases and protein phosphorylation, are the frequent features important for asymmetry formation in membrane domains (Fu et al., 2005; Hwang et al., 2008; Kleine-Vehn et al., 2008; Li et al., 2008; Zourelidou et al., 2009; Oda and Fukuda, 2012b; Chang et al., 2013; Alassimone et al., 2016). Additionally, unequal distribution of membrane lipids could be associated with formation of specific domains (Fischer et al., 2004; Ischebeck et al., 2013; Tejos et al., 2014; Stanislas et al., 2015; Barbosa et al., 2016; Simon et al., 2016). Whether similar mechanisms are involved in the formation of pollen apertures remains to be discovered. Nevertheless, it is clear that INP1 cannot be the sole player in this process.
Based on the evidence presented here, INP1 is a later-acting aperture factor involved in the protection of membrane domains from exine depositions. Presence of INP1 punctate lines strongly correlates with the formation of distinct ridges visible on the plasma membrane. These ridges seem to disappear in the absence of INP1, suggesting that INP1 is necessary for their formation. The linear membrane domains along these ridges remain in close contact with the CW when the rest of membrane separates from the CW around the time of primexine deposition. We propose that INP1 may act as a bridge that tethers three plasma membrane regions in each wild-type microspore to the overlying wall and that these contacts between the wall and the membrane serve to prevent or limit exine deposition at these sites, resulting in aperture formation (Fig. 8B). The close apposition of the aperture plasma membrane domains to the CW is reminiscent of the situation with the Casparian strip membrane domain that also firmly adheres to the Casparian strip cell wall in root endodermis (Roppolo et al., 2011). Interestingly, the presence of normal CWs (or, potentially, presence of functional CALS5 or callose/CALS5-interacting proteins) is required not only for formation of apertures but also for proper assembly of INP1 lines: in the strong cals5 mutants that lack outer CWs but still have remnants of intersporal walls, INP1 puncta are found in conspicuous association with the remaining CWs. It has been previously noticed that, in multiple species, positions of apertures correlate with the last sites of callose deposition at the end of meiotic cytokinesis, so-called additional callose deposits (Albert et al., 2010, 2011; Prieu et al., 2017), providing further support to our conclusions on the importance of callose for formation of apertures and INP1-decorated domains. However, it will require further investigation to find out whether INP1 can directly interact with callose or whether this interaction depends on other molecular players.
MATERIALS AND METHODS
Plant Material and Growth Conditions
In addition to the Arabidopsis (Arabidopsis thaliana) lines created for this study, plants of the following genotypes were used: Col-0, Wassilewskija, inp1-1, INP1pr:INP1-YFP inp1 (Dobritsa and Coerper, 2012), tetraploid INP1pr:INP1-YFP (Reeder et al., 2016), cals5-2 (Dong et al., 2005), cals5-3, cals5-4, and cals5-5 (Nishikawa et al., 2005). Plants were grown at 20°C to 22°C with the 16-h-light:8-h-dark cycle in growth chambers or in a greenhouse.
Transgenic Constructs
To create DMC1pr:INP1-YFP, MMD1pr:INP1–YFP, and A9pr:INP1-YFP constructs, the INP1 open reading frame without the stop codon was first amplified with primers AgeI-INP1-EF and NcoI-INP1-FR (Supplemental Table S1), digested with AgeI and NcoI, and cloned into the previously described modified pGreenII02229 binary vector (pGR111; Dobritsa et al., 2010; Dobritsa and Coerper, 2012) upstream of the YFP gene. The promoters were amplified from Col-0 genomic DNA (for MMD1pr and A9pr) or Landsberg genomic DNA (for DMC1pr) using primers indicated in Supplemental Table S1, digested with SacI-AgeI, and cloned upstream of INP1-YFP. All constructs were verified by sequencing prior to transformation into the Agrobacterium strain GV3101. inp1 plants were transformed by the floral dip method (Clough and Bent, 1998); transgenic plants were selected with BASTA, and the presence of transgenes was confirmed with specific primers. At least 10 T1 plants per construct were examined for phenotypes.
Confocal Microscopy
Confocal microscopy of mature pollen grains was performed as described previously (Reeder et al., 2016). For imaging MMC and tetrads, anthers were dissected out of stage-9 flower buds (Smyth et al., 1990) and placed into Vectashield antifade solution (Vector Labs) supplemented with 0.02% Calcofluor White and 5 μg/mL membrane stain CellMask Deep Red (Molecular Probes). To release sporogenic cells, a coverslip was placed over anthers and a gentle pressure was applied on it. MMC and tetrads were imaged on a Nikon A1+ confocal microscope with a 100× oil-immersion objective (NA = 1.4) and 5× confocal zoom. YFP was excited with a 514-nm laser line and emission was collected at 522 to 555 nm, Calcofluor White was excited with a 405-nm laser line and emission was collected at 424 to 475 nm, and CellMask Deep Red dye was excited with a 640-nm laser line and emission was collected at 663 to 738 nm. Z-stacks of tetrads were obtained with a step size of 300 or 500 nm and volume-reconstructed using NIS Elements v.4.20 (Nikon). To create signal intensity profiles of fluorophores, the multichannel images were first processed using the Smooth filter in NIS Elements; then lines were drawn and profiles were obtained using the Signal Intensity Profile tool in NIS Elements.
TEM
TEMs for this study were captured from material prepared for a previous study, according to the methods presented in it (Paxson-Sowders et al., 2001). Briefly, wild-type Arabidopsis flower buds (Wassilewskija ecotype) were teased open and cryofixed by high-pressure freezing with a Balzers HPM 010, freeze substituted in 2% OsO4, and embedded in Spurr's resin. Silver sections were cut, placed onto copper grids, stained with methanolic uranyl acetate followed by Reynold's lead citrate, and examined with a Hitachi H-600 TEM operating at 75 kV. Images were captured on Kodak 4489 film, and developed negatives were scanned using an Epson Perfection V750 Pro flatbed scanner.
Accession Numbers
The Arabidopsis Genome Initiative accession numbers for the genes used in this study are the following: At4g22600 (INP1) and At2g13680 (CALS5).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used in this study.
Supplemental Figure S1. INP1-YFP localization in microspore mother cells and young free microspores in DMC1pr:INP1-YFP plants.
Supplemental Figure S2. INP1-YFP puncta and lines form between plasma membrane and CW.
Supplemental Figure S3. Plasma membrane at the developing aperture sites is in close contact with CW and is protected from primexine and exine deposition.
Supplemental Figure S4. Localization of INP1-YFP puncta is fairly normal in the weaker cals5 alleles.
Supplemental Figure S5. Localization of INP1-YFP puncta in the wild-type tetrads.
Supplemental Figure S6. Localization of INP1-YFP puncta in the cals5-2 tetrads.
Supplemental Movie S1. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing INP1pr:INP1-YFP (yellow). No INP1-YFP puncta are visible in this tetrad.
Supplemental Movie S2. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing INP1pr:INP1-YFP (yellow). Some INP1-YFP puncta and lines are visible in this tetrad. Note that the top microspore lacks any puncta.
Supplemental Movie S3. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing INP1pr:INP1-YFP (yellow). More advanced INP1-YFP lines are present in this tetrad.
Supplemental Movie S4. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing DMCpr:INP1-YFP (yellow) corresponds to the tetrad in Figure 3A. No INP1-YFP puncta are visible in this tetrad.
Supplemental Movie S5. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing DMCpr:INP1-YFP (yellow) corresponds to the tetrad in Figure 3B. Some INP1-YFP puncta are visible in this tetrad.
Supplemental Movie S6. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing DMCpr:INP1-YFP (yellow) corresponds to the tetrad in Figure 3C. More advanced INP1-YFP lines are present in this tetrad.
Supplemental Movie S7. Volume reconstruction of a z-stack of confocal sections through a wild-type tetrad of microspores expressing DMCpr:INP1-YFP (yellow) corresponds to the tetrad in Figure 3D to D”. Note the asynchronous development of INP1-YFP lines between the sister microspores.
Supplemental Movie S8. Volume reconstruction of a z-stack of confocal sections through a tetrad of microspores produced by a tetraploid lsq6 plant. Microspores were labeled with the membrane-staining CellMask Deep Red dye (magenta). Note the presence of four membrane protrusions in each of the microspores.
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center at the Ohio State University for seed stocks, to Edwin de Feijter and Orin Hemminger (Nikon Instruments Inc.) for advice on confocal microscopy, and to Erich Grotewold and Mark Johnson for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB-1517511 to A.A.D. and H.A.O.), by the Department of Molecular Genetics and Center for Applied Plant Sciences at OSU (to A.A.D.) and by a scholarship from the China Scholarship Council (to P.L.).
Articles can be viewed without a subscription.
References
- Alassimone J, Fujita S, Doblas VG, van Dop M, Barberon M, Kalmbach L, Vermeer JEM, Rojas-Murcia N, Santuari L, Hardtke CS, et al. (2016) Polarly localized kinase SGN1 is required for Casparian strip integrity and positioning. Nat Plants 2: 16113. [DOI] [PubMed] [Google Scholar]
- Alassimone J, Naseer S, Geldner N (2010) A developmental framework for endodermal differentiation and polarity. Proc Natl Acad Sci USA 107: 5214–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B, Nadot S, Dreyer L, Ressayre A (2010) The influence of tetrad shape and intersporal callose wall formation on pollen aperture pattern ontogeny in two eudicot species. Ann Bot (Lond) 106: 557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B, Ressayre A, Nadot S (2011) Correlation between pollen aperture pattern and callose deposition in late tetrad stage in three species producing atypical pollen grains. Am J Bot 98: 189–196 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Prehoda KE (2015) Establishment of Par-polarized cortical domains via phosphoregulated membrane motifs. Dev Cell 35: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa ICR, Shikata H, Zourelidou M, Heilmann M, Heilmann I, Schwechheimer C (2016) Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143: 4687–4700 [DOI] [PubMed] [Google Scholar]
- Bowman SM, Free SJ (2006) The structure and synthesis of the fungal cell wall. BioEssays 28: 799–808 [DOI] [PubMed] [Google Scholar]
- Chang F, Gu Y, Ma H, Yang Z (2013) AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol Plant 6: 1187–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Coerper D (2012) The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine. Plant Cell 24: 4452–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Geanconteri A, Shrestha J, Carlson A, Kooyers N, Coerper D, Urbanczyk-Wochniak E, Bench BJ, Sumner LW, Swanson R, et al. (2011) A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol 157: 947–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16 (Suppl): S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Dickinson HG (2010) Tapetal cell fate, lineage and proliferation in the Arabidopsis anther. Development 137: 2409–2416 [DOI] [PubMed] [Google Scholar]
- Fischer U, Men S, Grebe M (2004) Lipid function in plant cell polarity. Curr Opin Plant Biol 7: 670–676 [DOI] [PubMed] [Google Scholar]
- Fitzgerald MA, Knox RB (1995) Initiation of primexine in freeze-substituted microspores of Brassica campestris. Plant Reprod 8: 99–104 [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Furness CA, Rudall PJ (2004) Pollen aperture evolution--a crucial factor for eudicot success? Trends Plant Sci 9: 154–158 [DOI] [PubMed] [Google Scholar]
- Furt F, König S, Bessoule J-J, Sargueil F, Zallot R, Stanislas T, Noirot E, Lherminier J, Simon-Plas F, Heilmann I, et al. (2010) Polyphosphoinositides are enriched in plant membrane rafts and form microdomains in the plasma membrane. Plant Physiol 152: 2173–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lowry PR, Zhou X, Depry C, Wei Z, Wong GW, Zhang J (2011) PI3K/Akt signaling requires spatial compartmentalization in plasma membrane microdomains. Proc Natl Acad Sci USA 108: 14509–14514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1963) An ultrastructural study of pollen wall ontogeny in Silene pendula. Grana Palynol 4: 7–24 [Google Scholar]
- Heslop-Harrison J. (1968) Pollen wall development. The succession of events in the growth of intricately patterned pollen walls is described and discussed. Science 161: 230–237 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1976) The adaptive significance of the exine. In Ferguson IK, Muller J, eds., The evolutionary significance of the exine (Linn Soc London Symp Series No 1), Academic Press, London, pp. 27–37 [Google Scholar]
- Heslop-Harrison J. (1979) An interpretation of the hydrodynamics of pollen. Am J Bot 66: 737–743 [Google Scholar]
- Hughes J, McCully ME (1975) The use of an optical brightener in the study of plant structure. Stain Technol 50: 319–329 [DOI] [PubMed] [Google Scholar]
- Hwang J-U, Vernoud V, Szumlanski A, Nielsen E, Yang Z (2008) A tip-localized RhoGAP controls cell polarity by globally inhibiting Rho GTPase at the cell apex. Curr Biol 18: 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Werner S, Krishnamoorthy P, Lerche J, Meijón M, Stenzel I, Löfke C, Wiessner T, Im YJ, Perera IY, et al. (2013) Phosphatidylinositol 4,5-bisphosphate influences PIN polarization by controlling clathrin-mediated membrane trafficking in Arabidopsis. Plant Cell 25: 4894–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Langowski L, Wisniewska J, Dhonukshe P, Brewer PB, Friml J (2008) Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. (2011) Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI, Jones JDG (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J 11: 1–14 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy KV. (1999) Methods in Cell Wall Cytochemistry. CRC Press, Boca Raton, Florida [Google Scholar]
- Li S, Gu Y, Yan A, Lord E, Yang Z-B (2008) RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol Plant 1: 1021–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B. (2010) Recent advances in understanding mechanisms of insect cuticle differentiation. Insect Biochem Mol Biol 40: 363–375 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D (2005) Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2012a) Secondary cell wall patterning during xylem differentiation. Curr Opin Plant Biol 15: 38–44 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2012b) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Paul W, Hodge R, Smartt S, Draper J, Scott R (1992) The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant Mol Biol 19: 611–622 [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Prieu C, Matamoro-Vidal A, Raquin C, Dobritsa A, Mercier R, Gouyon P-H, Albert B (2016) Aperture number influences pollen survival in Arabidopsis mutants. Am J Bot 103: 452–459 [DOI] [PubMed] [Google Scholar]
- Prieu C, Toghranegar Z, Matamoro-Vidal A, Nadot S, Albert B (2017) Additional callose deposits are located at the future apertural regions in sulcate, ulcerate, porate, colporate, colpate and syncolpate pollen grains. Bot J Linn Soc 183: 1–9 [Google Scholar]
- Quilichini TD, Douglas CJ, Samuels AL (2014) New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana. Ann Bot (Lond) 114: 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder SH, Lee BH, Fox R, Dobritsa AA (2016) A ploidy-sensitive mechanism regulates aperture formation on the Arabidopsis pollen surface and guides localization of the aperture factor INP1. PLoS Genet 12: e1006060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressayre A, Godelle B, Raquin C, Gouyon PH (2002) Aperture pattern ontogeny in angiosperms. J Exp Zool 294: 122–135 [DOI] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Dénervaud Tendon V, Pfister A, Alassimone J, Vermeer JEM, Yamazaki M, Stierhof Y-D, Beeckman T, Geldner N (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383 [DOI] [PubMed] [Google Scholar]
- Roppolo D, Geldner N (2012) Membrane and walls: who is master, who is servant? Curr Opin Plant Biol 15: 608–617 [DOI] [PubMed] [Google Scholar]
- Rozario T, DeSimone DW (2010) The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol 341: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers D-J, Pinho MG (2005) Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69: 585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MLA, Platre MP, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud M-C, Jaillais Y (2016) A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Söldner R (2012) Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol 14: 640–648 [DOI] [PubMed] [Google Scholar]
- Stanislas T, Hüser A, Barbosa ICR, Kiefer CS, Brackmann K, Pietra S, Gustavsson A, Zourelidou M, Schwechheimer C, Grebe M (2015) Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat Plants 1: 15162. [DOI] [PubMed] [Google Scholar]
- Tejos R, Sauer M, Vanneste S, Palacios-Gomez M, Li H, Heilmann M, van Wijk R, Vermeer JEM, Heilmann I, Munnik T, et al. (2014) Bipolar plasma membrane distribution of phosphoinositides and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26: 2114–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W. (2012) The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogaart G, Lang T, Jahn R (2013) Microdomains of SNARE proteins in the plasma membrane. Curr Top Membr 72: 193–230 [DOI] [PubMed] [Google Scholar]
- Wilson JN, Ladefoged LK, Babinchak WM, Schiøtt B (2014) Binding-induced fluorescence of serotonin transporter ligands: a spectroscopic and structural study of 4-(4-(dimethylamino)phenyl)-1-methylpyridinium (APP(+)) and APP(+) analogues. ACS Chem Neurosci 5: 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodehouse RP. (1935) Pollen Grains. Their Structure, Identification and Significance in Science and Medicine. McGraw-Hill, New York and London [Google Scholar]
- Yang X, Makaroff CA, Ma H (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2008) Cell polarity signaling in Arabidopsis. Annu Rev Cell Dev Biol 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lavagi I (2012) Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Curr Opin Plant Biol 15: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z (2000) The Rrop GTPase switch turns on polar growth in pollen. Trends Plant Sci 5: 298–303 [DOI] [PubMed] [Google Scholar]
- Zourelidou M, Müller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C (2009) The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636 [DOI] [PubMed] [Google Scholar]