A secreted mucin-like protein in the rice brown planthopper (Nilaparvata lugens) enables insect feeding and induces plant immune responses.
Abstract
The brown planthopper, Nilaparvata lugens, is a pest that threatens rice (Oryza sativa) production worldwide. While feeding on rice plants, planthoppers secrete saliva, which plays crucial roles in nutrient ingestion and modulating plant defense responses, although the specific functions of salivary proteins remain largely unknown. We identified an N. lugens-secreted mucin-like protein (NlMLP) by transcriptome and proteome analyses and characterized its function, both in brown planthopper and in plants. NlMLP is highly expressed in salivary glands and is secreted into rice during feeding. Inhibition of NlMLP expression in planthoppers disturbs the formation of salivary sheaths, thereby reducing their performance. In plants, NlMLP induces cell death, the expression of defense-related genes, and callose deposition. These defense responses are related to Ca2+ mobilization and the MEK2 MAP kinase and jasmonic acid signaling pathways. The active region of NlMLP that elicits plant responses is located in its carboxyl terminus. Our work provides a detailed characterization of a salivary protein from a piercing-sucking insect other than aphids. Our finding that the protein functions in plant immune responses offers new insights into the mechanism underlying interactions between plants and herbivorous insects.
Plants are subjected to attack by diverse herbivorous insects, which are generally classified based on their feeding strategies as chewing or piercing-sucking insects. Chewing insects, such as caterpillars and beetles, can cause serious mechanical damage to plant tissues, whereas piercing-sucking insects feed on plants through specially adapted mouthparts known as stylets and cause only limited physical damage to plant tissues (Walling, 2000). Insects also can injure plants indirectly by transmitting viral, bacterial, and fungal pathogens. Plants use sophisticated perception systems to detect insect feeding through cues derived not only from damage caused by feeding (Reymond et al., 2000) but also from insect saliva, oral secretions, eggs, volatiles, and microbes associated with the insects (Reymond, 2013; Felton et al., 2014).
When phloem-feeding insects feed on plants, their stylets transiently puncture the epidermis and penetrate plant cell walls. The insects then ingest the phloem sap. During this process, insects secrete both gelling and watery saliva from their salivary glands into plant cells. The secreted gelling saliva quickly solidifies and forms a continuous salivary sheath in the plant, encasing the full length of the stylet. The salivary sheath provides mechanical stability and protection for the insect against plant chemical defenses. For example, inhibiting the expression of structural sheath protein (SHP), a salivary protein secreted by Acyrthosiphon pisum aphids, reduces their reproduction by disrupting salivary sheath formation and, hence, their feeding from host sieve tubes (Will and Vilcinskas, 2015). Watery saliva contains digestive and cell wall-degrading enzymes. Plant immune responses to insect attack may be elicited or suppressed by compounds in insect saliva (Miles, 1999; Felton et al., 2014). Broadly speaking, effectors are proteins or other molecules produced by pathogens or insects that can alter host structures and functions (Hogenhout et al., 2009). Several insect effectors with diverse effects have been identified in aphids in recent years (Bos et al., 2010; Atamian et al., 2013; Rodriguez et al., 2014; Naessens et al., 2015). For example, expression of the aphid protein effector C002 in host plants increases the fecundity of green peach aphid, while another effector, Mp10, reduces aphid fecundity (Bos et al., 2010). Moreover, transient in planta expression of Mp10 activates jasmonic acid (JA) and salicylic acid (SA) signaling pathways (Rodriguez et al., 2014) and triggers chlorosis in Nicotiana benthamiana (Bos et al., 2010). Similarly, the expression of two candidate effectors, Me10 and Me23, from the potato aphid in host N. benthamiana plants increases aphid fecundity (Atamian et al., 2013), and MpMIF (a MIF cytokine secreted in aphid watery saliva during feeding) plays an important role in aphid survival and can affect both the SA and JA signaling pathways (Naessens et al., 2015). However, little is known about effectors from piercing-sucking herbivores other than aphids and their functions in host plants.
Plants have evolved sophisticated defense mechanisms to protect themselves from insect herbivores, most of which are initiated by the recognition of their saliva or oral secretions. The signals are transmitted within plants via transduction networks, including JA, ethylene, SA, and hypersensitive response (HR) pathways. Accordingly, infestation by piercing-sucking insects increases the production of JA, SA, and ethylene in rice (Oryza sativa; Yuan et al., 2005; Du et al., 2009; Hu et al., 2011). Key elements in these signaling pathways include MAPK cascades, which occur in all eukaryotes, are highly conserved, and modulate numerous cellular responses to diverse cues (Wu et al., 2007). These responses include complex defense responses against insects (Wu and Baldwin, 2010). For example, oral secretions from the chewing insect tobacco hornworm (Manduca sexta) induce MAPK-activated defense responses to herbivore attack in Nicotiana attenuata leaves (Wu et al., 2007). Similarly, aphid resistance conferred by the Mi-1 gene in tomato (Solanum lycopersicum) can be attenuated by virus-induced gene silencing (VIGS) of certain MAPKs and MAPK kinases (Li et al., 2006). MAPK cascades also play important roles in planthopper resistance gene-mediated immunity (Yuan et al., 2005). Mechanisms for resistance to phloem-feeding insects include the induction of forisome (sieve tube protein) dispersion, callose deposition, and thus, phloem plugging, which prevent insects from continuously ingesting phloem sap from plants (Will et al., 2007; Hao et al., 2008).
The brown planthopper (BPH), Nilaparvata lugens, is a severe herbivorous insect pest of rice that causes extensive yield losses and economic damage to rice both directly (by feeding) and indirectly (by transmitting viral diseases). During outbreaks, planthoppers can completely destroy crops, an effect called hopper burn (Backus et al., 2005). Like other piercing-sucking insects, BPHs secrete gelling and watery saliva. Recently, genomic tools such as proteomics and transcriptomics have been used to investigate BPH salivary glands and saliva at the molecular level (Konishi et al., 2009; Ji et al., 2013; Huang et al., 2016; Liu et al., 2016). Two secretary proteins that actively participate in salivary sheath formation were recently identified in BPHs (Huang et al., 2015, 2016). Furthermore, several salivary proteins that play important roles in interactions between BPH and rice were identified (Petrova and Smith, 2014; Ji et al., 2017; Ye et al., 2017). However, the functions of the majority of BPH-secreted proteins have not yet been experimentally determined. The biological roles of specific BPH salivary protein effectors in rice-BPH interactions remain poorly understood.
In an analysis of the BPH salivary gland transcriptome, we found a mucin-like protein gene highly expressed in BPH salivary glands. Mucins are a family of high-Mr, heavily glycosylated proteins that mostly constitute tandem repeats of identical or highly similar sequences rich in Ser, Thr, and Pro residues (Verma and Davidson, 1994). Mucin-like proteins are widely distributed in eukaryotes, bacteria, and viruses. Intestinal mucins and salivary gland mucins have been identified in insects. Intestinal mucin is a major protein constituent of the peritrophic membrane that facilitates the digestive process as well as protecting invertebrate digestive tracts from microbial infection (Wang and Granados, 1997). A mucin-like protein that was identified in the salivary glands of Anopheles gambiae through transcriptomic analysis might modulate parasite infectivity or help lubricate insect mouthparts (Francischetti et al., 2002). A mucin-like protein in the salivary proteome of BPH has been detected (Huang et al., 2016). However, the functions of mucin-like proteins in insects are largely unknown.
Here, we identified this N. lugens-secreted mucin-like protein (NlMLP) as an insect cell death-inducing protein involved in plant-insect interactions. NlMLP is required for salivary sheath formation and feeding of BPHs on their host plants. NlMLP induces defense responses in plant cells, including cell death, the expression of pathogen-responsive genes, and callose deposition. Finally, we found that the active part of NlMLP is located at its C-terminal region.
RESULTS
NlMLP Is Highly Expressed in N. lugens Salivary Glands and Secreted into Rice Tissues
Sequencing of a cDNA library produced from BPH salivary glands yielded 40,000,000 reads. After a series of assembly and alignment steps (Supplemental Methods S1), 13,969 unigenes were functionally annotated with gene descriptions. Assignment of Clusters of Orthologous Groups (COG) and Gene Ontology terms showed that the salivary gland proteins are involved in basic processes such as transcription and translation as well functions including binding, catalytic activity, and secretion (Supplemental Figs. S1 and S2).
Salivary proteins that are secreted outside of salivary gland cells to perform their functions should contain a secretory signal peptide; 399 unigenes in the BPH salivary gland transcriptome were predicted to encode proteins with signal peptides. Proteins with more than one predicted transmembrane domain, which are likely anchored in cell membranes of the salivary gland, were excluded. After these filtering steps, 256 potential secretory proteins were retained (Supplemental Table S1). A gene (CL865) showing high identity to the Laodelphax striatellus mucin-like protein was the most abundant in the transcriptome.
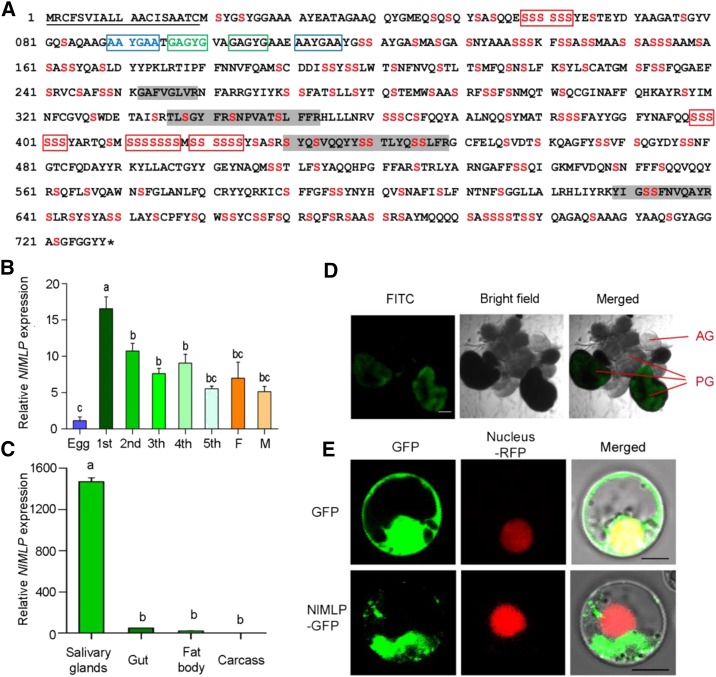
We obtained a full-length cDNA for this gene, which contains a 2,187-bp open reading frame and encodes a polypeptide of 728 amino acid residues (Fig. 1A). We named this gene NlMLP (accession no. KY348750). The first 19 amino acids constitute the signal peptide, with cleavage predicted between residues 19 and 20. NlMLP is rich in Ser (22.4%) residues, 36% of which are predicted to be potential mucin-type O-glycosylation sites. Some repeated amino acid sequences, a typical feature of mucin-like proteins (Verma and Davidson, 1994), were found. NlMLP protein has been detected in both gelling and watery saliva (Huang et al., 2016; Liu et al., 2016). To investigate the functions of NlMLP, we analyzed mRNA levels in BPHs at various developmental stages, including eggs, first to fifth instar BPHs, and female and male adults, via quantitative reverse transcription (qRT)-PCR. NlMLP expression was higher in insects at feeding stages (nymph or adult) than at the nonfeeding stage (egg; Fig. 1B). NlMLP transcripts were detected at higher levels in the salivary gland than in the gut, fat body, and remaining carcass (Fig. 1C). We also analyzed the expression of NlMLP in salivary glands by mRNA in situ hybridization. Hybridization signals were detected in the A-follicles of principal glands but not in the salivary ducts or accessory glands (Fig. 1D).
Figure 1.
Molecular characterization of NlMLP. A, Amino acid sequence of NlMLP. The solid underline indicates the signal peptide predicted by SignalP. The asterisk indicates the stop codon. The shaded amino acid residues indicate the peptides detected in BPH-infested rice tissue by liquid chromatography-tandem mass spectrometry analyses. Ser is shown in red. The repeat region is boxed. B and C, Expression patterns of NlMLP at different developmental stages (B) and in different tissues (C). Relative levels of NlMLP expression at the indicated developmental stages (1st to 5th, first to fifth instar; F, female adult; M, male adult) and in different tissues (salivary gland, gut, fat body, and remaining carcass) were normalized against β-actin gene expression, as determined by qRT-PCR. Data represent means ± se of three repeats; n = 30. Different letters above the bars indicate significant differences, as determined by Tukey’s honestly significant difference test (P < 0.05). D, mRNA in situ hybridization of NlMLP in salivary glands. Signals from anti-digoxigenin-fluorescein isothiocyanate (FITC; shown in green) bound to a digoxigenin-labeled antisense riboprobe used for hybridization to NIMLP transcripts were detected by confocal laser-scanning microscopy. AG, Accessory gland; PG, principal gland. Bar = 100 μm. E, NlMLP localizes to the cytoplasm of rice cells when transiently expressed. GFP or NlMLP-GFP fusion protein was expressed in rice protoplasts by polyethylene glycol-mediated transformation. Confocal laser-scanning microscopy was used to investigate fusion protein distribution 16 h after transformation. Bars = 5 μm.
To confirm that NlMLP was secreted into rice tissue during feeding, we extracted proteins from the leaf sheaths of plants following BPH feeding and analyzed them by mass spectrometry. Four NlMLP peptides were detected in BPH-infested rice leaf sheaths but not in noninfested rice (Fig. 1A), indicating that NlMLP was secreted into the rice plants.
To determine the cellular localization of NlMLP in plant cells when transiently expressed, we conducted localization experiments using rice protoplasts and N. benthamiana leaf cells. When the NlMLP-GFP fusion protein was transiently expressed in rice protoplasts, GFP fluorescence was detected only in the cytoplasm, while control GFP fluorescence was detected in both the cytoplasm and the nucleus (Fig. 1E). When the NlMLP-YFP fusion protein was transiently expressed in N. benthamiana leaves via agroinfiltration, NlMLP localized to the cytoplasm (Supplemental Fig. S3).
NlMLP Is Required for the Feeding of BPHs on Rice Plants and for Insect Performance
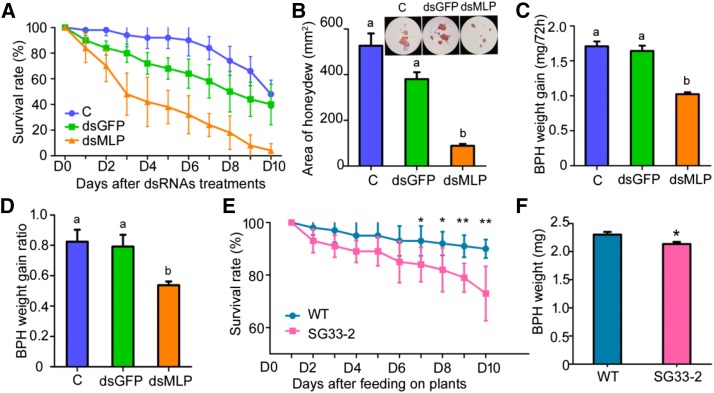
To elucidate the role of NlMLP in BPH, we synthesized double-stranded RNA (dsRNA) from NlMLP and injected it into fourth instar BPH nymphs to mediate RNA interference (RNAi). This treatment had a very strong silencing effect, reducing NlMLP transcript levels significantly (∼95%) on the first day after treatment compared with the levels in two control groups receiving either no injection or injection with double-stranded GFP (dsGFP; P < 0.001 for BPHs with no injection [C] and double-stranded MLP [dsMLP] from 1 to 6 d, P = 0.017 for dsGFP and dsMLP at 1 and 5 d, P = 0.016 for dsGFP and dsMLP at 2, 3, and 6 d, P = 0.015 for dsGFP and dsMLP at 4 d; Supplemental Fig. S4). The silencing was confirmed by RNA gel-blot analysis 2 d after injection (Supplemental Fig. S4). The treated BPH insects were allowed to feed on cv TN1 rice plants. The survival rate of BPHs harboring a silenced NlMLP gene was significantly lower (from 2 to 10 d following injection) than those of the two control groups (P < 0.001 for C and dsMLP and P = 0.046 for dsGFP and dsMLP at 2 d, P < 0.001 for C and dsMLP and P = 0.001 for dsGFP and dsMLP at 10 d; Fig. 2A). The cumulative mortality rate of BPHs injected with dsMLP, dsGFP, and C was 96%, 60%, and 52%, respectively, at 10 d after injection (Fig. 2A). BPHs subjected to the NlMLP RNAi treatment also excreted significantly less honeydew, a simple measurable indicator of BPH feeding activity, than the two control groups (P < 0.001 for C and dsMLP; P = 0.003 for dsGFP and dsMLP; Fig. 2B), as well as having smaller weight gain values (P = 0.011 for C and dsMLP, P = 0.031 for dsGFP and dsMLP; Fig. 2C) and weight gain ratios (P = 0.023 for C and dsMLP, P = 0.043 for dsGFP and dsMLP; Fig. 2D). Furthermore, silencing of NlMLP reduced BPH virulence. Rice plants died in 7 d after being infested by common BPH insects or BPH insects injected with dsGFP, while those plants infested by BPH insects injected with dsMLP still survived and grew normally (Supplemental Fig. S5). These results indicate that silencing the NlMLP gene significantly reduced the feeding and performance of BPHs on rice plants.
Figure 2.
Effects of NlMLP silencing on BPH feeding and performance. A, The survival rates of BPH insects after injection were monitored daily. The survival rates of BPHs injected with dsMLP were reduced significantly compared with those of BPHs with no injection and those injected with dsGFP after 2 d of treatment (Student’s t test). C, BPHs with no injection; dsGFP, BPHs injected with GFP-dsRNA; dsMLP, BPHs injected with NlMLP-dsRNA. The experiment was repeated five times with 10 BPHs per replicate. Data represent means ± sd of five repeats. B, Honeydew excretion by BPH insects on filter paper. The intensity of the honeydew color and area of the honeydew correspond to BPH feeding activity. The experiment was repeated three times with 10 filter papers per replicate. Data represent means ± se of three repeats. Different letters above the bars indicate significant differences, as determined by Tukey’s honestly significant difference test (P < 0.05). C and D, BPH weight gain (C) and BPH weight gain ratio (D) of BPHs after injection with dsGFP or dsMLP. Data represent means ± se of five independent experiments with 10 BPHs per replicate. Different letters above the bars indicate significant differences, as determined by Tukey’s honestly significant difference test (P < 0.05). E, Survival rates of BPH insects feeding on NlMLP-dsRNA-transgenic plants (SG33-2) and wild-type plants (WT). The experiment was repeated five times with 20 BPHs per replicate. Data represent means ± sd of five repeats. Asterisks above the columns indicate significant differences compared with wild-type plants (*, P < 0.05 and **, P < 0.01, Student's t test). F, BPH weight after feeding for 10 d on NlMLP-dsRNA-transgenic plants (SG33-2) and wild-type plants. The experiment was repeated five times with 10 BPHs per replicate. Data represent means ± se of five repeats. The asterisk above one column indicates a significant difference compared with wild-type plants (*, P < 0.05, Student’s t test).
Feeding on rice plants expressing dsRNAs was shown previously to trigger RNAi of a target gene in BPH (Zha et al., 2011). Therefore, we transformed BPH-susceptible rice plants with NlMLP-dsRNA and selected a T2 homozygous dsMLP-transgenic line expressing NlMLP-dsRNA via qRT-PCR analysis (Supplemental Fig. S6A). When second instar BPHs were fed on dsMLP-transgenic plants, their expression level of NlMLP was significantly (40%) lower than in BPHs fed on wild-type plants at 7 and 9 d after the start of exposure (P = 0.042 at 7 d, P = 0.045 at 9 d; Supplemental Fig. S6B). BPH survival and weight gain also were significantly lower in insects fed on dsMLP-transgenic plants than in those fed on wild-type plants from 7 to 10 d (P = 0.049 at 7 d, P = 0.046 at 8 d, P = 0.005 at 9 d, P = 0.008 at 10 d; Fig. 2E) and after 10 d (P = 0.023; Fig. 2F), respectively. These results clearly show that NlMLP protein is essential for BPH feeding and performance.
NlMLP Is Necessary for Salivary Sheath Formation
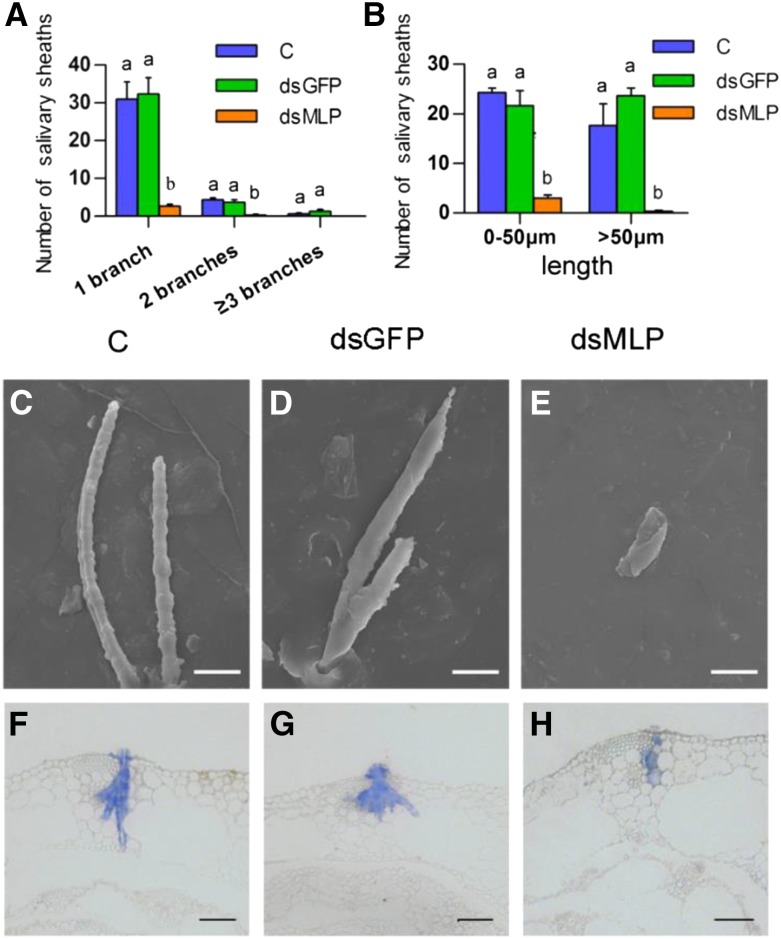
NlMLP was found in both gelling saliva and watery saliva (Huang et al., 2016). To further investigate the effects of NlMLP on feeding, we focused on salivary sheath formation. First, we fed BPHs on an artificial diet in Parafilm sachets for 2 d and analyzed their salivary sheaths by fluorescence microscopy and scanning electron microscopy observation. The fluorescence microscopy analysis revealed that BPHs subjected to the NlMLP RNAi treatment produced salivary sheaths that were significantly shorter and less branched than those produced by the control BPHs receiving either no injection or injection with dsGFP (Fig. 3, A and B). Moreover, the structure of the sheaths was incomplete or predominantly amorphous, or gelling saliva deposits at their stylet penetration sites were minimal, whereas those secreted by control BPHs had complete, typical structures (Fig. 3, C–E). Second, we collected stems from rice plants after BPH feeding and sectioned them to observe salivary sheath morphology in planta. Most salivary sheaths in rice stems produced by the control BPHs reached the phloem (Fig. 3, F and G), whereas most salivary sheaths produced by NlMLP-silenced BPHs were shorter and failed to reach the phloem, instead stopping in the rice epidermis or xylem (Fig. 3H). Together, these observations indicate that NlMLP is necessary for salivary sheath formation. Silencing of NlMLP in BPHs resulted in imperfect, short salivary sheaths, thus affecting phloem feeding.
Figure 3.
Effects of NlMLP silencing on salivary sheath formation. A and B, Distribution of branch number (A) and length (B) of salivary sheaths formed by BPHs on an artificial diet. BPH insects were fed with dietary Suc for 48 h, and the salivary sheaths were collected and counted using a fluorescence microscope. C, Noninjected BPHs; dsGFP, BPHs injected with GFP-dsRNA; dsMLP, BPHs injected with NlMLP-dsRNA. Data represent means ± se of three repeats. Different letters above the bars indicate significant differences, as determined by Tukey’s honestly significant difference test (P < 0.05). C to E, Scanning electron micrographs showing the morphology of salivary sheaths formed by BPHs on an artificial diet. Salivary sheaths from noninjected BPHs (C) show a complete structure. BPHs injected with GFP-dsRNA (D) formed similar types of sheaths. BPHs injected with NlMLP-dsRNA (E) produced abnormal salivary sheaths. Bars = 20 μm. F to H, Fluorescence microscopy images showing the morphology of salivary sheaths in rice plants. Rice plants were infested with BPH insects for 48 h and investigated by paraffin sectioning. Sheaths produced by noninjected BPHs (F), BPHs injected with GFP-dsRNA (G), and BPHs injected with NlMLP-dsRNA (H) are shown. Bars = 50 μm.
NlMLP Induces Plant Cell Death
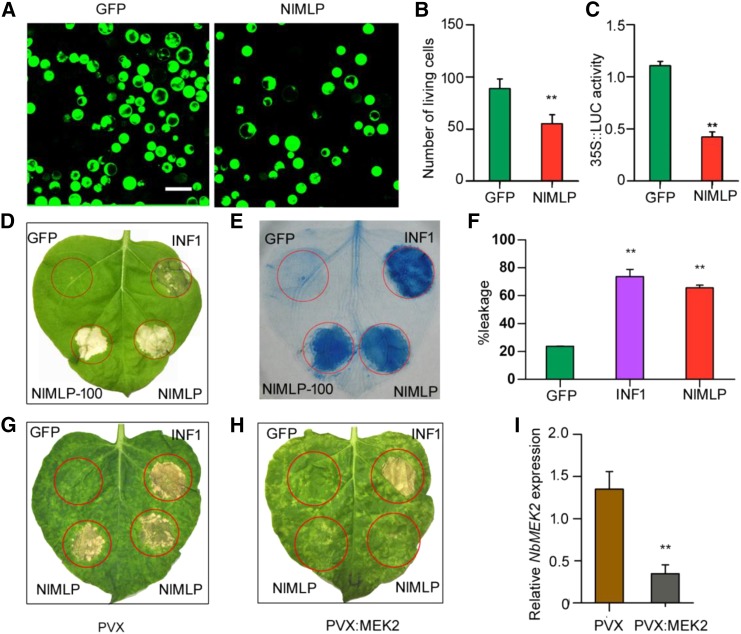
The above findings clearly show that NlMLP is secreted into rice tissues (Fig. 1A) and that it is localized to the cytoplasm of rice cells (Fig. 1E). To uncover the potential role of NlMLP in the host plant, we transiently expressed NlMLP without the signal peptide in rice protoplasts. Fluorescein diacetate (FDA) staining of the protoplasts showed that the cell viability of protoplasts expressing NlMLP was significantly lower than that of control protoplasts expressing GFP (P < 0.001; Fig. 4, A and B). We also coexpressed NlMLP together with the luciferase (LUC) gene in rice protoplasts. LUC activity was significantly lower in protoplasts coexpressing NlMLP compared with the control coexpressing GFP (P < 0.001; Fig. 4C). Immunoblotting confirmed that NlMLP and GFP were expressed properly in the rice protoplasts (Supplemental Fig. S8A). These observations indicate that NlMLP expression triggers cell death in rice protoplasts. To determine whether NlMLP-triggered cell death is affected by the presence of BPH-resistance genes in rice, we performed similar LUC assays after cotransfection of protoplasts with NlMLP and the genes Bph6, Bph9, and Bph14. The LUC activity was still significantly lower in the presence of NlMLP than in the GFP controls, regardless of the presence of resistance genes (P = 0.001 for Bph6, P = 0.005 for Bph9, P = 0.004 for Bph14; Supplemental Fig. S7). Therefore, NlMLP expression triggers cell death in rice protoplasts independently of these BPH-resistance genes.
Figure 4.
NlMLP causes cell death in rice protoplasts and N. benthamiana leaves. A, Images of FDA-stained viable rice protoplasts transformed with GFP or NlMLP. Living cells were visualized using confocal laser-scanning microscopy, and images were taken 20 h after transformation. GFP is a control protein that does not induce cell death. Bar = 25 μm. B, Number of viable rice protoplasts transformed with GFP or NlMLP determined after FDA staining. Average values and se were calculated from three independent experiments, and 10 randomly selected microscopy fields were counted per experiment. Asterisks above one column indicate a significant difference compared with GFP (**, P < 0.01, Student’s t test). C, LUC activity in rice protoplasts coexpressing LUC and NlMLP. Data represent means ± se of three independent experiments. Asterisks above one column indicate a significant difference compared with GFP (**, P < 0.01, Student’s t test). D and E, Leaves of N. benthamiana were infiltrated with A. tumefaciens carrying GFP, INF1, and NlMLP. The leaves were photographed 5 d after agroinfiltration (D), and the treated leaves were stained with Trypan Blue (E). INF1 is a control protein that induces cell death. GFP, INF1, and NlMLP were transiently expressed with the YFP-HA epitope tag; NlMLP-100, NlMLP with no epitope tag. F, Quantification of cell death by measuring electrolyte leakage in N. benthamiana leaves. Electrolyte leakage from the infiltrated leaf discs was measured as a percentage of leakage from boiled discs 4 d after agroinfiltration. Data represent means ± se of four repeats. Asterisks above the columns indicate significant differences compared with GFP (**, P < 0.01, Student’s t test). G and H, Silencing of NbMEK2 in N. benthamiana leaves compromises NlMLP-induced cell death but not INF1-induced cell death. GFP, INF1, and NlMLP were transiently expressed in N. benthamiana leaves expressing PVX vector (G) and leaves in which NbMEK2 had been silenced by VIGS (H). The leaves were photographed 5 d after agroinfiltration. I, NbMEK2 transcript abundance in silenced N. benthamiana leaves as measured by qRT-PCR. Data represent means ± se of three repeats. Asterisks above one column indicate a significant difference compared with GFP (**, P < 0.01, Student’s t test).
We further verified the ability of NlMLP to induce plant cell death by performing Agrobacterium tumefaciens-mediated expression of NlMLP in N. benthamiana leaves. INF1, an elicitin secreted by Phytophthora infestans that induces HR cell death in Nicotiana spp. plants (Derevnina et al., 2016), was used as a positive control, while GFP was used as a negative control. NlMLP, with or without a YFP-HA tag, triggered marked cell death (Fig. 4, D and E). Moreover, ion leakage was significantly higher from leaves expressing NlMLP or INF1 than from the GFP-expressing controls (P = 0.002 for GFP and INF1, P < 0.001 for GFP and NlMLP; Fig. 4F). Immunoblot analysis showed that GFP, INF1, and NlMLP proteins accumulated to comparable degrees in N. benthamiana leaves (Supplemental Fig. S8B). However, the cell death symptoms caused by INF1 and NlMLP were different. Leaves infiltrated with the INF1 strain appeared chlorotic in 4 d and became severely necrotic, accompanied by brown or black color, after 5 d. On leaves infiltrated with NlMLP, white or gray-white necrotic spots appeared in 4 d and became bigger around the infiltrated site as time went on (Supplemental Fig. S9). We further investigated the quantity of NlMLP required for the cell death symptoms. We set up different concentrations (OD600 = 0.005, 0.01, 0.02, 0.03, 0.04, 0.05, 0.08, 0.1, 0.2, and 0.3) of NlMLP strains to infect N. benthamiana leaves and found that cell death was caused by NlMLP strains in OD = 0.01 or over but was not when the OD was 0.005 (Supplemental Fig. S10A). Immunoblot analysis detected NlMLP protein in the leaves infected by strains in OD 0.01 or over but not in OD of 0.005 (Supplemental Fig. S10B).
To further characterize the physiological properties of the cell death induced by NlMLP, we examined the effects of treatments that inhibit various potential cell death-associated processes in rice protoplasts and N. benthamiana leaves. The application of LaCl3 blocked the induction of cell death by NlMLP, suggesting that the cell death process mediated by NlMLP is dependent on a calcium signaling pathway (Boudsocq et al., 2010; Table I). There was no difference in cell viability between protoplasts incubated in the light or dark, indicating that the cell death process induced by NlMLP is light independent (Asai et al., 2000). Bcl-xl is an antiapoptotic protein (Chen et al., 2012). The expression of Bcl-xl in N. benthamiana leaves suppressed cell death induced by subsequent NlMLP expression (Table I). Our results indicate that cell death induced by NlMLP shares some common properties with cell death induced by BAX and INF1. MAPK cascades play important roles in defense-related signal transduction (Yang et al., 2001). MEK2 is a MAPK kinase that acts upstream of SA-induced protein kinase and wounding-induced protein kinase and controls multiple defense responses to pathogen invasion (Yang et al., 2001). When we silenced MEK2 in N. benthamiana plants via VIGS (P = 0.006; Fig. 4I), NlMLP-triggered cell death was reduced significantly in MEK2-silenced plants (Fig. 4H) but not in control plants (Fig. 4G). However, the presence of INF1, which triggers cell death independently of MEK2 (Takahashi et al., 2007), clearly caused necrosis in MEK2-silenced plants (Fig. 4H). Therefore, NlMLP-triggered cell death is associated with MEK2-dependent MAPK cascades.
Table I. Results of inhibition assays of cell death in rice protoplasts and N. benthamiana leaves.
ND, Not determined.
| Gene | Treatment |
|||||
|---|---|---|---|---|---|---|
| LaCl3a |
Darkb |
Bcl-xlc |
||||
| LUC Activity in RPd | Cell Death in NBLe | LUC Activity in RP | Cell Death in NBL | LUC Activity in RP | Cell Death in NBL | |
| GFP | 1.00 ± 0.19 | 0/8 | 1.00 ± 0.34 | 0/8 | ND | 0/8 |
| NlMLP | 0.74 ± 0.17 | 1/8 | 0.14 ± 0.02* | 8/8 | ND | 0/8 |
| BAX | 0.66 ± 0.21 | 3/8 | 0.09 ± 0.02* | 7/8 | ND | 0/8 |
| INF1 | 0.92 ± 0.34 | 3/8 | 0.38 ± 0.11* | 7/8 | ND | 6/8 |
LaCl3 was applied to protoplasts immediately after transfection at a final concentration of 1 mm. For agroinfiltration of N. benthamiana leaves, LaCl3 was added to resuspended A. tumefaciens cultures at a final concentration of 1 mm.
Rice protoplasts and N. benthamiana leaves were incubated in the dark for 30 min before transfection and maintained in the dark after transfection.
N. benthamiana leaves were preinfiltrated with A. tumefaciens cells harboring the Bcl-xl expression vector.
LUC activity in rice protoplasts. Data represent means ± se of three repeats. Asterisks indicate significant differences compared with the corresponding GFP (*, P < 0.05, Student’s t test).
Number of cell death sites/total number of infiltrated leaves of N. benthamiana.
NlMLP Triggers Plant Defense Responses
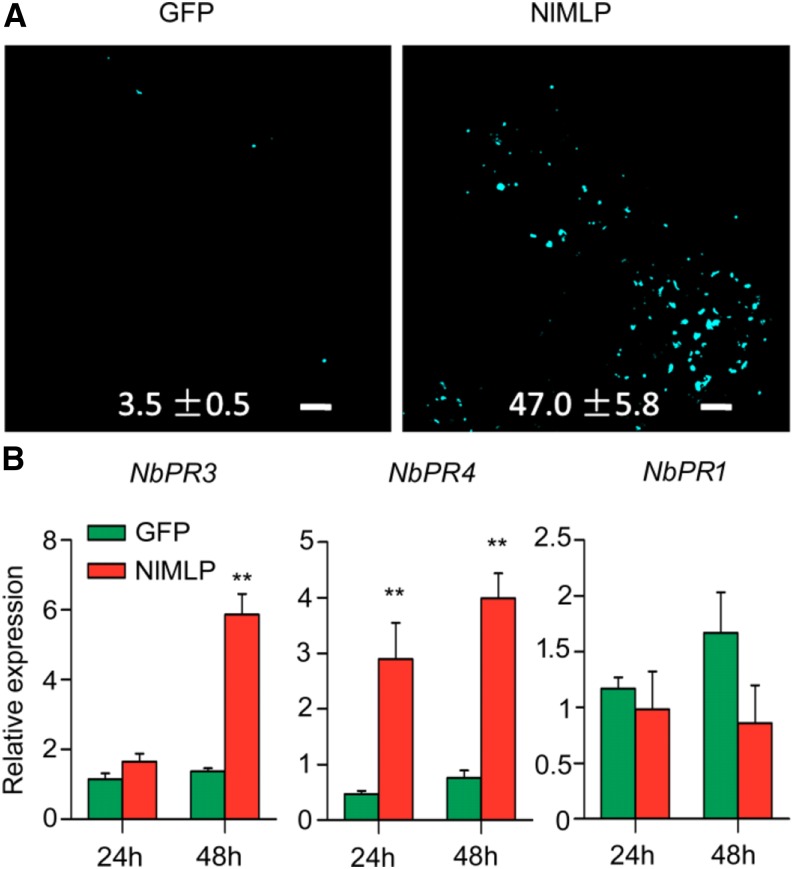
Callose deposition is used as a marker for plant basal defense responses and participates in plant defenses against phloem sap ingestion by insects (Hann and Rathjen, 2007; Hao et al., 2008). Thus, to determine whether NlMLP activates defense responses in planta, we expressed NlMLP in N. benthamiana leaves and investigated callose deposition by Aniline Blue staining. Many more callose spots were present in NlMLP-expressing leaves (47 per infiltration) than in GFP-expressing leaves (3.5 per infiltration; P = 0.001; Fig. 5A). Moreover, NlMLP induced transcriptional activation of the pathogen resistance (PR) genes NbPR3 (P = 0.143 at 24 h, P = 0.002 at 48 h) and NbPR4 (P = 0.002 at 24 h, P = 0.002 at 48 h), but not NbPR1 (P = 0.625 at 24 h, P = 0.18 at 48 h), within 48 h of infection (Fig. 5B). The up-regulation of genes encoding acidic NbPR1 protein is a characteristic feature of the activated SA-signaling pathway, while the induction of genes encoding basic NbPR3 and NbPR4 proteins is associated with JA-dependent defense responses (Zhang et al., 2012; Naessens et al., 2015). Thus, NlMLP appears to induce defense responses mediated by the JA signaling pathway, thereby promoting the production of PR proteins and the biosynthesis of cell wall-reinforcing callose.
Figure 5.
NlMLP affects plant defense responses during transfection. A, Aniline Blue staining of N. benthamiana leaves showing callose deposition spots in areas transfected with GFP or NlMLP. Photographs were taken 48 h after inoculation. Numbers indicate means ± se of the number of callose spots in four individual leaf discs. Bars = 100 mm. B, Expression of the defense-related genes NbPR3, NbPR4, and NbPR1 following transient expression of GFP and NlMLP in N. benthamiana leaves. Data represent means ± se of three repeats. Asterisks above the columns indicate significant differences compared with GFP (**, P < 0.01, Student’s t test).
The Functional Motif Is Located in the C Terminus of NlMLP
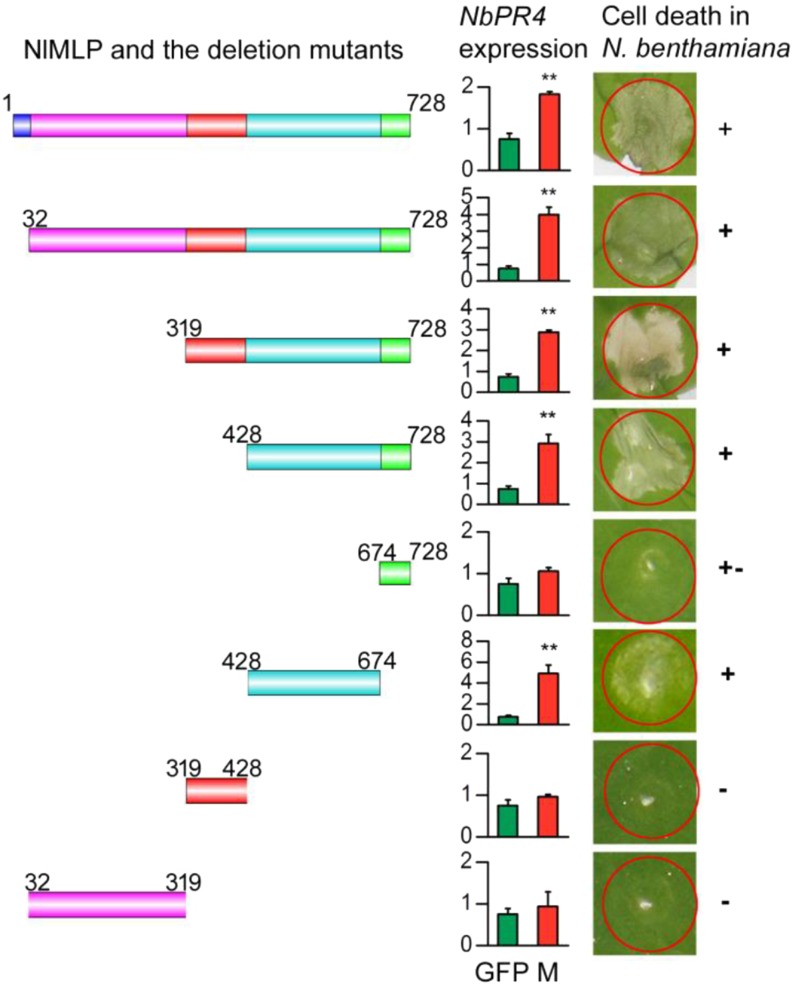
NlMLP showed no sequence similarity to any known cell death-inducing effector. To delineate the functional domains of NlMLP, we assayed the ability of N-terminal and C-terminal deletion mutant proteins to trigger cell death in N. benthamiana leaves (Fig. 6). The N-terminal deletion mutant M428-728 strongly triggered cell death, but the C-terminal deletion mutants M32-319 and M319-428 did not. Moreover, M428-674 triggered cell death less strongly than did M428-728, and the M674-728 mutant did not induce cell death at all. These findings suggest that the 428-674 amino acid fragment is required to trigger cell death and that the 674-728 amino acid fragment might promote this effect. We also found that the 428-674 fragment was required for the expression of NbPR4 (P = 0.001 for GFP and M1-728, P = 0.002 for GFP and M32-728, P = 0.008 for GFP and M319-728, P < 0.001 for GFP and M428-728, P = 0.129 for GFP and M674-728, P = 0.007 for GFP and M428-674, P = 0.209 for GFP and M319-428, P = 0.656 for GFP and M32-319; Fig. 6). Immunoblot analysis showed that the mutant proteins accumulated to comparable levels in N. benthamiana leaves (Supplemental Fig. S8C).
Figure 6.
Deletion analysis of NlMLP. Left column, Schematic diagrams of NlMLP and the deletion mutants. Middle column, Relative expression of NbPR4 in N. benthamiana leaves agroinfiltrated with GFP or NlMLP deletion mutants, as determined by qRT-PCR. Data represent means ± se of three repeats. M, NlMLP or the deletion mutants. Asterisks above the columns indicate significant differences compared with GFP (**, P < 0.01, Student’s t test). Right column, Cell death lesions in N. benthamiana leaves expressing the NlMLP deletion mutants. +, +–, and – indicate obvious cell death, weak cell death, and no cell death, respectively. Each experiment was repeated at least three times with similar results.
DISCUSSION
To our knowledge, this is the first report of a planthopper salivary protein that plays important roles in feeding and interactions with the host plant. We identified an N. lugens mucin-like protein, NlMLP, that is highly expressed in the salivary glands of BPHs and secreted into rice tissue during BPH feeding. NlMLP is necessary for the probing of rice plants by BPHs to obtain phloem sap for insect survival. BPH feeding was inhibited and insect performance was reduced significantly when NlMLP expression was knocked down (Fig. 2). Furthermore, the salivary sheaths produced by NlMLP-silenced BPHs were shorter and less branched than those of control BPHs fed on both an artificial diet and rice tissue. The sheaths had incomplete structures and were predominantly amorphous.
Mucin-like proteins have been identified in various organisms; some mucins are involved in controlling mineralization (Boskey, 2003) and are associated with processes including nacre formation in mollusks (Marin et al., 2000), calcification in echinoderms (Boskey, 2003), and bone formation in vertebrates (Midura and Hascall, 1996). Based on our results here, mucin appears to be an essential component of the BPH salivary sheath. Notably, NlMLP is rich in Ser, which provides attachment sites for carbohydrate chains that participate in the formation of large extracellular aggregates (Korayem et al., 2004), thus functioning in the formation of salivary sheaths, which support stylet movements and exploration of the host plant tissue. Therefore, reductions in the levels of NlMLP may prevent the construction of complete sheaths. The immediate consequence of this reduction in NlMLP-RNAi insects is that salivary sheaths formed in rice plants do not reach into the sieve tube, which likely accounts for the reduction in phloem feeding and the performance of these insects on rice plants.
In aphids, the salivary protein SHP contributes to the solidification of gelling saliva and sheath formation, partly through the formation of disulfide cystine bonds (Carolan et al., 2009; Will and Vilcinskas, 2015). Mucins can form disulfide-dependent soluble dimers and multimeric insoluble gels through the cross linking of Cys residues (Axelsson et al., 1998). Cys residues in NlMLP might behave in a similar fashion and strongly contribute to the formation of the polymeric matrix during sheath hardening via the formation of intermolecular disulfide bonds. NlMLP also might function in the growth and development of BPHs. Silencing of NlMLP affected insect development, which, in turn, reduced salivary sheath formation, feeding, and performance.
Plants usually detect molecules emitted by parasites to trigger defense responses. When BPHs feed on phloem sap, their saliva is secreted into rice tissue, which contains bioactive components involved in inducing the expression of defense genes. We demonstrated that NlMLP is one such bioactive component. Mucin-like proteins also have been detected in fungal pathogens. The surfaces of many parasites, including the protozoan parasite Trypanosoma cruzi (Buscaglia et al., 2006), the fish pathogen Trypanosoma carassii (Lischke et al., 2000), and the potato pathogen Phytophthora infestans (Görnhardt et al., 2000), are covered in mucins, which participate in interactions with host cells during the invasion process (Buscaglia et al., 2006; Larousse et al., 2014). Similarly, NlMLP is highly expressed in salivary glands and is secreted into the plant. We found that NlMLP expression triggered cell death in rice protoplasts and N. benthamiana leaves (Fig. 4) as well as plant immunity responses, including the induction of PR gene expression and callose synthesis in leaves (Fig. 5). NlMLP molecules on the surface of the salivary sheath, representing an essential component of this structure, can be detected as a signal of BPH feeding by host cells and evoke defense responses. NlMLP in watery saliva may play the same role as well. Our results indicate that the functional portion of NlMLP is located at its C terminus. The presence of the 428-674 amino acid fragment was sufficient to trigger cell death and induce the expression of NbPR4. Mucin-like proteins also are detected in other piercing-sucking insects such as leafhopper (Hattori et al., 2015) and several mosquito species (Das et al., 2010). The wide taxonomic range of hosts in which NlMLP can trigger cell death suggests that NlMLP may be recognized by a conserved protein found in many plants. The plant receptor that recognizes NlMLP remains to be identified.
The defense reaction elicited by NlMLP shares common features with immune responses shown by well-known effectors and pathogen-associated molecular patterns. NlMLP might be an elicitor that is involved in the pattern-triggered immunity process. It may be recognized by plant PRRs, which trigger plant defensive responses. NlMLP triggers cell death, a common phenomenon in effector-triggered immune responses. Ca2+ is a well-known secondary signal in eukaryotes. The early defense response to BPH in rice involves Ca2+ influx, which is a common early plant response triggered by insect feeding (Hao et al., 2008; Hogenhout and Bos, 2011; Bonaventure, 2012). Our results indicate that the cell death process induced by NlMLP is dependent on Ca2+ influx. Moreover, cell death induced by NlMLP is not dependent on light and is suppressed by the antiapoptotic protein Bcl-xl. NlMLP-triggered cell death requires the MEK2 MAP kinase signal transduction pathway. MEK2 is a common component in MAP kinase pathways required for HR followed by certain Avr-R interactions in tomato and tobacco (Nicotiana tabacum; Morris, 2001; del Pozo et al., 2004; King et al., 2014). NbMEK2 also is required for cell death triggered by the Phytophthora sojae RxLR effector Avh241 in N. benthamiana (Yu et al., 2012). Given the broad range of plants responding to NlMLP and the dependence of these responses on the MEK2 pathway, we speculate that plants respond to NlMLP via a conserved upstream component of plant signaling pathways. The expression of NlMLP induced the JA pathway marker genes NbPR3 and NbPR4. NbPR3 is a chitinase gene associated with JA-dependent defenses that is induced by elicitors (Naessens et al., 2015), while NbPR4 encodes a hevein-like chitinase that also is associated primarily with JA responses (Kiba et al., 2014). These findings suggest that defense responses triggered by NlMLP are associated with the JA signaling pathway. NlMLP expression in N. benthamiana leaves also induced callose deposition. Callose deposition is a useful mechanism conferring plant immunity to insects and pathogens (Luna et al., 2011). In the interaction of rice and BPH, callose deposition on the sieve plates occludes sieve tubes, thereby directly inhibiting continuous feeding by BPHs (Hao et al., 2008).
Nevertheless, BPH can successfully attack most rice cultivars and even results in hopper burn. This success is thought to be because of the evolved ability of BPH to suppress or counteract rice defense responses triggered by NlMLP and other elicitors (Walling, 2008; Kaloshian and Walling, 2016). Hemipteran saliva is a complex mixture of biomolecules with potential roles in overcoming plant immune responses and enables hemipterans to manipulate host responses to their advantage (Miles, 1999; Kaloshian and Walling, 2016). One example is that BPH can decompose the deposited callose and unplug sieve tube occlusions by activating β-1,3-glucanase genes and, thereby, facilitate the continuous phloem feeding in rice plants (Hao et al., 2008). To date, some effectors, including C002, Me10, Me23, Mp1, Mp2, and Mp55, have been identified by assays of in planta overexpression and RNAi in aphids. These effectors contribute to aphid survival and suppress host defense (Mutti et al., 2008; Bos et al., 2010; Atamian et al., 2013; Pitino and Hogenhout, 2013; Elzinga et al., 2014). The effectors may target any component in pattern-triggered immunity or the host cell trafficking pathway (Derevnina et al., 2016; Kaloshian and Walling, 2016; Rodriguez et al., 2017). It is likely that potent effectors in BPH also target such cell processes in rice and enable BPH to successfully feed on rice plants, which induces cascade reactions termed the BPH-feeding cascade and results in the death of susceptible rice varieties (Cheng et al., 2013). In contrast, in resistant rice, the resistance gene induces a strong defense response that inhibits insect feeding, growth, and development and enables the plant to grow normally.
In summary, our results indicate that NlMLP secreted by BPHs into rice plants plays dual roles as a component in feeding sheath formation and activating plant defense responses. The defense responses induced by NlMLP in plant cells are related to Ca2+ mobilization and the MEK2 MAP kinase and JA signaling pathways. Further studies are needed to identify the target of NlMLP in rice cells and BPH effectors that suppress rice defense responses to further explore the interaction mechanisms between plants and planthoppers.
MATERIALS AND METHODS
Plant Materials and Insects
Wild-type and transgenic rice (Oryza sativa) plants were grown in the experimental fields at Wuhan University Institute of Genetics under routine management practices. BPH (Nilaparvata lugens) biotype 1 insect populations were reared on 1-month-old plants of the susceptible rice cv TN1 under controlled environmental conditions (26°C ± 0.5°C, 16-h-light/8-h-dark cycle).
Transcriptome Analysis of BPH Salivary Glands
BPH adult females were dipped in 70% (v/v) ethanol for several seconds and washed in 0.85% (w/v) NaCl solution. Salivary glands were dissected in phosphate-buffered saline solution (pH 7.4). This was accomplished by pulling the head off of the thorax with forceps and carefully removing the salivary glands that emerged from the distal region of the severed head. A total of 2,000 salivary glands were dissected and directly dipped into 300 µL of RNAiso Plus (Takara) for RNA preparation. The poly(A) RNA was isolated, broken into smaller fragments (200–700 bp), and reverse transcribed to synthesize cDNA for Illumina HiSeq 2000 sequencing. After stringent quality assessment and data filtering, 40,000,000 reads were selected as high-quality reads (after removing adapters and low-quality regions) for further analysis. A set of 12,668,039 high-quality reads with an average length of 400 bp was assembled, resulting in 59,510 contigs. The contigs were assembled into 31,645 unigenes, consisting of 3,960 clusters and 27,685 singletons. All unigene sequences were aligned by BLASTx to protein sequences in databases, including non-redundant, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes, and COG, followed by sequences in the nonredundant National Center for Biotechnology Information protein database (with E < 10−5 in both cases).
SignalP 3.0 was used to predict the presence of signal peptides. To predict transmembrane domains, each amino acid sequence with a signal peptide was submitted to the TMHMM Server version 2.0.
Cloning and Sequencing of NlMLP cDNA
The cDNA sequences of NlMLP were obtained from salivary gland transcriptomes of BPHs. To obtain the full-length sequence of each truncated sequence from BPHs, 5′ and 3′ RACE amplification was performed using the 5′-Full RACE Kit and 3′-Full RACE Core Set version 2.0 (Takara) following the manufacturer’s instructions. For 5′ RACE, gene-specific primers were designed based on sequencing data, and an external reverse primer (5′-RACE-EP) and a nested primer (5′-RACE-NP) were used. For 3′ RACE, the cDNA was then amplified by nested PCR with the external forward primer (3′-RACE-EP) and the nested forward primer (3′-RACE-NP; Supplemental Table S2) with La Taq DNA polymerase (Takara). Purified PCR products were ligated to the pMD18-T simple vector (Takara), and positive colonies were sequenced.
Proteomics Analysis of Rice Leaf Sheaths
The cv TN1 rice plants were grown in pots (8 cm in diameter and 15 cm in height). At the fifth leaf stage, second and third instar BPH nymphs were released onto the plants at 20 insects per plant. The leaf sheaths of BPH-infested plants and noninfested control plants were collected after 72 h, immediately frozen in liquid nitrogen, and stored at −80°C. Protein extraction and digestion, iTRAQ labeling, strong cation-exchange chromatography fractionation, and liquid chromatography-tandem mass spectrometry proteomic analysis were performed according to the manufacturer’s protocols using the iTRAQ Reagent 8 Plex Kit (SCIEX). A TripleTOF 5600 (AB SCIEX), fitted with a NanoSpray III source (AB SCIEX) and a pulled quartz tip as the emitter (New Objectives), was used for tandem mass spectrometry. Protein identification was performed using Mascot 2.3.02 (Matrix Science) against the transcriptomic database of N. lugens salivary glands (containing 18,099 protein-coding sequences).
Expression Analysis of NlMLP
The temporal and spatial expression patterns of NlMLP were investigated by qRT-PCR as follows. For spatial expression pattern analysis, salivary glands, guts, fat bodies, and carcasses (i.e. samples consisting of all parts remaining after the removal of the salivary gland, gut, and fat body) were dissected from BPH adults using a stereomicroscope. Total RNA was isolated from each type of tissue using RNAiso Plus (Takara). For temporal expression pattern analysis, total RNA was isolated from BPH at various developmental stages, including eggs, first to fifth instar nymphs, female adults, and male adults. First-strand NlMLP cDNA was obtained from all samples by reverse transcription using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara) according to the manufacturer’s instructions, followed by amplification by qRT-PCR using the Bio-Rad CFX-96 Real-Time PCR system with the iTaq Universal SYBR Green Supermix Kit (Bio-Rad) and gene-specific primers qNlMLP-F and qNlMLP-R (Supplemental Table S2). As an endogenous control to normalize expression levels with average threshold cycle numbers, a partial fragment of the BPH actin gene was amplified with primers qNlActin-F and qNlActin-R (Supplemental Table S2). A relative quantitative method (ΔΔCt) was applied to evaluate the variation in expression among samples.
Fluorescence in Situ Hybridization of BPH Salivary Glands
The expression patterns of NlMLP within the salivary glands were examined by in situ mRNA localization and confocal microscopy as follows. BPHs were anesthetized on ice, and their salivary glands were dissected and fixed in 4% (v/v) formaldehyde in PTw (phosphate-buffered saline + 0.1% (v/v) Tween) overnight at 4°C. The fixed glands were treated with 5 mg mL−1 proteinase K for 10 min and subjected to various hybridization steps (Villarroel et al., 2016). Briefly, the glands were prehybridized in hybridization buffer for 1 h at 52°C, incubated in fresh hybridization buffer and probe overnight at 52°C, and subjected to six 25-min washes at 52°C with wash buffer. After washing at room temperature with PTw, the glands were incubated for 3 to 5 h at 4°C in the dark in a 1:200 dilution of antidigoxigenin fluorescein (Fab fragments; Roche) in PTw. The samples were then washed with PTw for 20 min and mounted in 70% (v/v) glycerol in PTw. Fluorescence images were obtained using an Olympus Fluoview FV1000 confocal laser-scanning microscope.
dsRNA Synthesis and Injection to Trigger RNAi in Insects
The DNA template for dsMLP synthesis was obtained using primer pair dsMLP-F and dsMLP-R. dsMLP was synthesized using the MEGAscript T7 High Yield Transcription Kit (Ambion) according to the manufacturer’s instructions. The amplified NlMLP fragment (500 bp) was confirmed by sequencing. The primers dsGFP-F and dsGFP-R, which were designed based on the GFP gene template, were used to synthesize dsGFP as a negative control in the RNAi experiments. The dsRNA was purified by phenol chloroform extraction and resuspended in nuclease-free water at a concentration of 5 µg µL−1.
Fourth instar nymphs were anesthetized with carbon dioxide for approximately 20 s until they were unconscious and placed on 2% (w/v) agarose plates with their abdomens facing up. A 46-nL volume of dsMLP or dsGFP was injected into each insect at the junction between the prothorax and mesothorax using a microprocessor-controlled Nanoliter 2010 injector (World Precision Instruments). The injected BPHs were reared on rice cv TN1 plants.
Bioassay of BPHs after dsRNA Injection
The survival rates of BPHs injected with dsGFP or dsMLP and the noninjected controls were determined as follows. Pots (diameter, 7 cm; height, 9 cm), each containing a single 1-month-old cv TN1 rice plant grown and maintained as described above, were individually covered with plastic cages into which 10 injected BPH nymphs were released. The number of surviving BPH nymphs on each plant was recorded daily for 10 d. The experiment was repeated five times.
BPH growth rates were analyzed using newly emerged brachypterous females (within 1 d of emergence), Parafilm sachets (2 × 2.5 cm), and 1-month-old cv TN1 rice plants grown and maintained as described. After weighing the insects and sachets, the insects were placed into the sachets, which were then attached to the plants (with one BPH per sachet and one sachet per plant). After 72 h, each insect and Parafilm sachet was reweighed, and the changes in weight of the BPH and sachet were defined as BPH weight gain and honeydew weight, respectively. The BPH weight gain ratio was calculated as the change in weight relative to the initial weight. The experiment was repeated 10 times per group, and the experiments were conducted five times.
Honeydew Excretion on Filter Paper
Honeydew excretion on filter paper was measured as described previously (Du et al., 2009). One-month-old rice cv TN1 plants grown in pots (diameter, 7 cm; height, 9 cm; one plant per pot) as described were covered with an inverted transparent plastic cup placed over filter paper resting on a plastic petri dish. Two days after injection, the BPHs were placed into the chamber. After 2 d of feeding, the filter paper was collected and treated with 0.1% (w/v) ninhydrin in acetone solution. After 30 min of oven drying at 60°C, the honeydew stains appeared violet or purple due to their amino acid content. The area of the ninhydrin-positive deposits was measured with ImageJ software. The experiment was repeated three times.
Development of dsMLP-Transgenic Rice and the BPH Bioassay
To constitutively express NlMLP-dsRNA in rice, a 500-bp template fragment (the same as the target sequence used for microinjection) and stuffer sequence fragment (a PDK intron) were used to generate a hairpin RNAi construct as described previously (Zha et al., 2011). The construct was inserted into binary vector pCXUN (accession no. FJ905215) under the control of the plant ubiquitin promoter. The construct was transformed into rice cv Kasalath (an indica rice variety susceptible to BPH and amenable to gene transformation) using an Agrobacterium tumefaciens-mediated method. Integration of the foreign DNA in T0 plants was confirmed by PCR and DNA gel-blot analysis. The plants were cultivated, and T2 seeds were collected for the bioassay.
The survival rates of BPHs on transgenic and wild-type plants (cv Kasalath) were determined by releasing 20 second instar nymphs onto each plant. The number of surviving BPHs was recorded daily for 10 d. The experiment was repeated five times. NlMLP expression in BPHs was evaluated by qRT-PCR. BPH weight was measured on a microbalance after 10 d of feeding.
Observation of Salivary Sheaths on Parafilm and in Planta
To observe the morphology of the salivary sheaths, BPHs were fed on an artificial diet (D-97) consisting of amino acids, vitamins, inorganic salts, and Suc (Fu et al., 2001) using plastic cylinders (4 cm long and 2.5 cm in diameter) as feeding chambers. Sachets formed from two layers of stretched Parafilm M (∼4 times the original area), each containing 200-µL portions of D-97, were placed at one end of the chamber. The opposite end of the chamber was covered with a piece of nylon mesh after the test insects had been introduced. Insects could feed by piercing through the inner Parafilm layer of the diet sachets, leaving salivary sheaths in the sachets. To assess the effects of dsRNA injection on salivary sheaths, 2 d after injection, nymphs were individually placed in the chambers and allowed to feed for 48 h. The inner Parafilm layers were then placed on a microscope slide, and the salivary sheaths were counted using a BX51 light microscope (Olympus). Sets of 10 dsNlMLP-injected, dsGFP-injected, and noninjected control BPHs were used per experiment, and the experiment was repeated three times.
To obtain samples for scanning electron microscopy, the inner Parafilm layer was placed on a microscope slide and salivary sheaths were identified with a light microscope (Olympus BX51). Regions of interest were labeled, and the Parafilm was cut with a scalpel. The salivary sheaths from the Parafilm were attached to a sample holder, coated with gold, and observed with an S-3400N scanning electron microscope (Hitachi). Five replicates were prepared per treatment, and 20 randomly chosen salivary sheaths were observed.
The dsRNA-treated BPHs were reared on 1-month-old cv TN1 rice seedlings for 48 h, and the leaf sheaths were collected and used for paraffin sectioning. One-month-old rice cv TN1 plants (grown and maintained as described) were infested with a set of 10 dsMLP-injected, dsGFP-injected, or noninjected BPHs. The leaf sheaths produced by the insects were collected, fixed in 4% (v/v) paraformaldehyde, dehydrated, embedded in paraffin, and cut into 10-mm-thick sections with a microtome. The sections were mounted on microscope slides and stained with 0.25% (w/v) Coomassie Blue staining solution for 10 min. The slides were dewaxed, rehydrated, examined, and photographed using a light microscope.
Viability Assay Using Rice Protoplasts
Rice cv 9311 (an indica rice variety susceptible to BPH) protoplasts were isolated from 10-d-old plants as described previously (Zhang et al., 2011). The cell viability and LUC assays were conducted using rice protoplasts as described (Zhao et al., 2016). Briefly, for the cell viability assay, protoplasts were transfected with the indicated plasmids for 20 h and stained with 220 μg mL−1 FDA. For the LUC assays, protoplasts were cotransfected with the reporter Renilla LUC gene and another construct carrying GFP or NlMLP. LUC activity was measured 40 h after transfection using a LUC assay system (Promega).
A. tumefaciens Infiltration Assays of Nicotiana benthamiana Leaves
A sequence of interest was amplified by PCR. The PCR products and the PUC19 vector were both digested by NotI and AscI and bound by DNA ligase to create entry vectors. Using LR Clonase reaction enzyme mix (Invitrogen), the target sequence was recombined into the destination vector pEarleyGate 101 (with the YFP-HA epitope tag) or pEarleyGate 100 (with no epitope tag). Constructs were introduced into A. tumefaciens strain GV3101 by electroporation. Recombinant strains were cultured in Luria-Bertani medium supplemented with 50 µg mL−1 kanamycin and 50 μg mL−1 rifampicin, harvested, washed three times in infiltration medium (10 mm MES, pH 5.6, 10 mm MgCl2, and 150 µm acetosyringone), and resuspended in infiltration medium to an OD600 of 0.3. The resuspended recombinant strains were incubated for 2 h at room temperature and infiltrated into the leaves of 4- to 6-week-old N. benthamiana plants (grown in a greenhouse at 25°C/20°C under a 16-h-light/8-h-dark cycle) through a nick created using a needleless syringe. Symptom development in N. benthamiana was monitored visually 3 to 8 d after infiltration. To test the difference of cell death symptoms caused by INF1 and NlMLP, 12 pieces of N. benthamiana leaves were injected. Symptom development in N. benthamiana was monitored visually every day after infiltration. Four independent biological replicates were conducted and got similar results.
To test the induction of plant defense-related gene expression by NlMLP, RNA was extracted from N. benthamiana leaves 48 h after infiltration. SYBR Green qRT-PCR assays were performed to determine gene expression levels as described previously. Three independent biological replicates were conducted for each experiment.
Agroinfiltrated N. benthamiana leaves were harvested at 48 h post inoculation. Total protein extracts were prepared by grinding 400 mg of leaf tissue in 1 mL of extraction buffer (20 mm Tris-Cl, 1% (w/v) SDS, and 5 mm EDTA) in the presence of 10 mm DTT. The samples were shaken on a vortex for 30 s and centrifuged at 800g for 10 min at 4°C. The resulting supernatant was used for immunoblot analysis.
Cell death was assayed by measuring ion leakage from leaf discs. Four leaf discs of equal dimension (10 mm) were placed into 5 mL of distilled water. The first set was incubated at room temperature overnight, and its conductivity (C1) was recorded using a conductivity meter (FiveGO-FG3). The second set was boiled, and its conductivity (C2) was recorded after cooling. Relative electrolyte leakage [(C1/C2) × 100] was calculated. The experiments were carried out four times.
Callose deposition in infiltrated leaf discs 48 h after infiltration was visualized after Aniline Blue staining following published methods (Naessens et al., 2015). Briefly, dissected leaf discs were destained by successive washes in 70%, 95%, and 100% (v/v) ethanol for 2 h per wash, washed twice with distilled, deionized water, and incubated in Aniline Blue solution (70 mm KH2PO4 and 0.05% [w/v] Aniline Blue, pH 9) for 1 h. Leaf discs were mounted in 80% (v/v) glycerol and observed with a fluorescence microscope (FV1000; Olympus). After collecting callose images, fluorescence in the images was quantified with ImageJ software. Each experiment was repeated three times independently.
VIGS of MEK2 in N. benthamiana Plants
Silencing of NbMEK2 in N. benthamiana was performed by Potato virus X (PVX) VIGS as described by Sharma et al. (2003). The NbMEK2 sequence (267 bp from the 3′ terminus) was inserted into the PVX vector in the antisense direction to generate PVX:MEK2. The construct containing the insert and the empty vector PVX were transformed into A. tumefaciens strain GV3101. Bacterial suspensions were applied to the undersides of N. benthamiana seedlings (∼20 d) using a 1-mL needleless syringe. The plants exhibited mild mosaic symptoms 3 to 4 weeks after inoculation. MEK2 gene expression was measured by qRT-PCR, and plants in which silencing was established were subjected to further analysis.
Cell Death Inhibition Assays
The effects of the calcium channel inhibitor LaCl3 and the human antiapoptotic gene Bcl-xl on cell death (under dark conditions) were determined as described previously (Chen et al., 2013). For the rice protoplast transient expression assay and agroinfiltration into N. benthamiana leaves, rice protoplasts or A. tumefaciens cultures were resuspended in LaCl3 at a final concentration of 1 mm. Transient expression assays were performed under dark conditions to determine whether cell death induced by NlMLP is light dependent. Isolated protoplasts and N. benthamiana plants were incubated in the dark for 30 min before transfection and maintained in the dark after transfection. Bcl-xl was cloned into binary vector pGR107 and introduced into A. tumefaciens strain GV3101. NlMLP or the cell death-inducing gene BAX or INF1 was expressed in N. benthamiana leaves 24 h after introduction of the Bcl-xl expression vector or the empty control vector via agroinfiltration.
Statistical Analysis
Data were compared using Student’s t test or Tukey’s honestly significant difference test using PASW Statistics version 18.0.
Accession Numbers
NlMLP sequence data from this article can be found in the EMBL/GenBank data libraries under accession number KY348750. Binary vector pCXUN sequence data from this article can be found in the EMBL/GenBank data libraries under accession number FJ905215.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. COG classification of sequences in the BPH salivary gland transcriptome.
Supplemental Figure S2. Gene Ontology classification of sequences in the BPH salivary gland transcriptome.
Supplemental Figure S3. NlMLP is localized to the cytoplasm in plant cells.
Supplemental Figure S4. Confirmation of the effect of RNAi on NlMLP expression in BPHs.
Supplemental Figure S5. Virulence of BPHs on rice plants.
Supplemental Figure S6. Relative expression of NlMLP in transgenic rice and BPH insects.
Supplemental Figure S7. NlMLP induces cell death in rice irrespective of the presence of BPH-resistance genes (Bph6, Bph9, and Bph14).
Supplemental Figure S8. Immunoblot analysis of proteins in rice protoplasts and N. benthamiana.
Supplemental Figure S9. Symptoms of cell death caused by INF1 and NlMLP in N. benthamiana leaves.
Supplemental Figure S10. Quantitative estimate of NlMLP required for plant symptoms.
Supplemental Table S1. List of PCR primers used in this study.
Supplemental Table S2. List of qRT-PCR primers used in this study.
Supplemental Methods S1. Transcriptome analysis of BPH salivary glands.
Supplemental Methods S2. Gene silencing analysis by qRT-PCR.
Supplemental Methods S3. Gene silencing analysis by RNA gel blotting.
Supplemental Methods S4. Virulence of BPHs on rice plants.
Supplemental Methods S5. Determining the quantity of NlMLP required for the cell death symptoms.
Footnotes
This work was supported by grants from National Program on Research and Development of Transgenic Plants Grants (2016ZX08009003-001-008), National Natural Science Foundation of China (31630063, 31230060, 31401732), and National Key Research and Development Program (2016YFD0100600, 2016YFD0100900).
Articles can be viewed without a subscription.
References
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM (2000) Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12: 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Chaudhary R, Cin VD, Bao E, Girke T, Kaloshian I (2013) In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol Plant Microbe Interact 26: 67–74 [DOI] [PubMed] [Google Scholar]
- Axelsson MA, Asker N, Hansson GC (1998) O-Glycosylated MUC2 monomer and dimer from LS 174T cells are water-soluble, whereas larger MUC2 species formed early during biosynthesis are insoluble and contain nonreducible intermolecular bonds. J Biol Chem 273: 18864–18870 [DOI] [PubMed] [Google Scholar]
- Backus EA, Serrano MS, Ranger CM (2005) Mechanisms of hopperburn: an overview of insect taxonomy, behavior, and physiology. Annu Rev Entomol 50: 125–151 [DOI] [PubMed] [Google Scholar]
- Bonaventure G. (2012) Perception of insect feeding by plants. Plant Biol (Stuttg) 14: 872–880 [DOI] [PubMed] [Google Scholar]
- Bos JI, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskey AL. (2003) Biomineralization: an overview. Connect Tissue Res (Suppl 1) 44: 5–9 [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia CA, Campo VA, Frasch AC, Di Noia JM (2006) Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nat Rev Microbiol 4: 229–236 [DOI] [PubMed] [Google Scholar]
- Carolan JC, Fitzroy CI, Ashton PD, Douglas AE, Wilkinson TL (2009) The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9: 2457–2467 [DOI] [PubMed] [Google Scholar]
- Chen H, Stout MJ, Qian Q, Chen F (2012) Genetic, molecular and genomic basis of rice defense against insects. Crit Rev Plant Sci 31: 74–91 [Google Scholar]
- Chen S, Songkumarn P, Venu RC, Gowda M, Bellizzi M, Hu J, Liu W, Ebbole D, Meyers B, Mitchell T, et al. (2013) Identification and characterization of in planta-expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol Plant Microbe Interact 26: 191–202 [DOI] [PubMed] [Google Scholar]
- Cheng X, Zhu L, He G (2013) Towards understanding of molecular interactions between rice and the brown planthopper. Mol Plant 6: 621–634 [DOI] [PubMed] [Google Scholar]
- Das S, Radtke A, Choi YJ, Mendes AM, Valenzuela JG, Dimopoulos G (2010) Transcriptomic and functional analysis of the Anopheles gambiae salivary gland in relation to blood feeding. BMC Genomics 11: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB (2004) MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derevnina L, Dagdas YF, De la Concepcion JC, Bialas A, Kellner R, Petre B, Domazakis E, Du J, Wu CH, Lin X, et al. (2016) Nine things to know about elicitins. New Phytol 212: 888–895 [DOI] [PubMed] [Google Scholar]
- Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, et al. (2009) Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci USA 106: 22163–22168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact 27: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Chung SH, Hernandez MGE, Louis J, Peiffer M, Tian D (2014) Herbivore oral secretions are the first line of protection against plant-induced defences. Annu Plant Rev 47: 37–76 [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM (2002) Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J Exp Biol 205: 2429–2451 [DOI] [PubMed] [Google Scholar]
- Fu Q, Zhang ZT, Hu C, Lai FX, Sun ZX (2001) A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stal) (Homoptera: Delphacidae). Appl Entomol Zool (Jpn) 36: 111–116 [Google Scholar]
- Görnhardt B, Rouhara I, Schmelzer E (2000) Cyst germination proteins of the potato pathogen Phytophthora infestans share homology with human mucins. Mol Plant Microbe Interact 13: 32–42 [DOI] [PubMed] [Google Scholar]
- Hann DR, Rathjen JP (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J 49: 607–618 [DOI] [PubMed] [Google Scholar]
- Hao P, Liu C, Wang Y, Chen R, Tang M, Du B, Zhu L, He G (2008) Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol 146: 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Komatsu S, Noda H, Matsumoto Y (2015) Proteome analysis of watery saliva secreted by green rice leafhopper, Nephotettix cincticeps. PLoS ONE 10: e0123671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout SA, Bos JIB (2011) Effector proteins that modulate plant-insect interactions. Curr Opin Plant Biol 14: 422–428 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22: 115–122 [DOI] [PubMed] [Google Scholar]
- Hu J, Zhou J, Peng X, Xu H, Liu C, Du B, Yuan H, Zhu L, He G (2011) The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol 156: 856–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HJ, Liu CW, Cai YF, Zhang MZ, Bao YY, Zhang CX (2015) A salivary sheath protein essential for the interaction of the brown planthopper with rice plants. Insect Biochem Mol Biol 66: 77–87 [DOI] [PubMed] [Google Scholar]
- Huang HJ, Liu CW, Huang XH, Zhou X, Zhuo JC, Zhang CX, Bao YY (2016) Screening and functional analyses of Nilaparvata lugens salivary proteome. J Proteome Res 15: 1883–1896 [DOI] [PubMed] [Google Scholar]
- Ji R, Ye W, Chen H, Zeng J, Li H, Yu H, Li J, Lou Y (2017) A salivary endo-β-1,4-glucanase acts as an effector that enables the brown planthopper to feed on rice. Plant Physiol 173: 1920–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji R, Yu H, Fu Q, Chen H, Ye W, Li S, Lou Y (2013) Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence. PLoS ONE 8: e79612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloshian I, Walling LL (2016) Plant immunity: connecting the dots between microbial and hemipteran immune responses. In Czosnek H, Ghanim M, eds, Management of Insect Pests to Agriculture. Springer International Publishing, Switzerland, Cham, pp 217–243 [Google Scholar]
- Kiba A, Galis I, Hojo Y, Ohnishi K, Yoshioka H, Hikichi Y (2014) SEC14 phospholipid transfer protein is involved in lipid signaling-mediated plant immune responses in Nicotiana benthamiana. PLoS ONE 9: e98150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SR, McLellan H, Boevink PC, Armstrong MR, Bukharova T, Sukarta O, Win J, Kamoun S, Birch PR, Banfield MJ (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK ε to suppress plant immune signaling. Plant Cell 26: 1345–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Noda H, Tamura Y, Hattori M (2009) Proteomic analysis of the salivary glands of the rice brown planthopper, Nilaparvata lugens (Stal) (Homoptera: Delphacidae). Appl Entomol Zool (Jpn) 44: 525–534 [Google Scholar]
- Korayem AM, Fabbri M, Takahashi K, Scherfer C, Lindgren M, Schmidt O, Ueda R, Dushay MS, Theopold U (2004) A Drosophila salivary gland mucin is also expressed in immune tissues: evidence for a function in coagulation and the entrapment of bacteria. Insect Biochem Mol Biol 34: 1297–1304 [DOI] [PubMed] [Google Scholar]
- Larousse M, Govetto B, Séassau A, Etienne C, Industri B, Theodorakopoulos N, Deleury E, Ponchet M, Panabières F, Galiana E (2014) Characterization of PPMUCL1/2/3, three members of a new oomycete-specific mucin-like protein family residing in Phytophthora parasitica biofilm. Protist 165: 275–292 [DOI] [PubMed] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, Kaloshian I (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol Plant Microbe Interact 19: 655–664 [DOI] [PubMed] [Google Scholar]
- Lischke A, Klein C, Stierhof YD, Hempel M, Mehlert A, Almeida IC, Ferguson MA, Overath P (2000) Isolation and characterization of glycosylphosphatidylinositol-anchored, mucin-like surface glycoproteins from bloodstream forms of the freshwater-fish parasite Trypanosoma carassii. Biochem J 345: 693–700 [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhou H, Zhao J, Hua H, He Y (2016) Identification of the secreted watery saliva proteins of the rice brown planthopper, Nilaparvata lugens (Stål) by transcriptome and shotgun LC-MS/MS approach. J Insect Physiol 89: 60–69 [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Marin F, Corstjens P, de Gaulejac B, de Vrind-De Jong E, Westbroek P (2000) Mucins and molluscan calcification: molecular characterization of mucoperlin, a novel mucin-like protein from the nacreous shell layer of the fan mussel Pinna nobilis (Bivalvia, Pteriomorphia). J Biol Chem 275: 20667–20675 [DOI] [PubMed] [Google Scholar]
- Midura RJ, Hascall VC (1996) Bone sialoprotein: a mucin in disguise? Glycobiology 6: 677–681 [DOI] [PubMed] [Google Scholar]
- Miles PW. (1999) Aphid saliva. Biol Rev Camb Philos Soc 74: 41–85 [Google Scholar]
- Morris PC. (2001) MAP kinase signal transduction pathways in plants. New Phytol 151: 67–89 [DOI] [PubMed] [Google Scholar]
- Mutti NS, Louis J, Pappan LK, Pappan K, Begum K, Chen MS, Park Y, Dittmer N, Marshall J, Reese JC, et al. (2008) A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci USA 105: 9965–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naessens E, Dubreuil G, Giordanengo P, Baron OL, Minet-Kebdani N, Keller H, Coustau C (2015) A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr Biol 25: 1898–1903 [DOI] [PubMed] [Google Scholar]
- Petrova A, Smith CM (2014) Immunodetection of a brown planthopper (Nilaparvata lugens Stål) salivary catalase-like protein into tissues of rice, Oryza sativa. Insect Mol Biol 23: 13–25 [DOI] [PubMed] [Google Scholar]
- Pitino M, Hogenhout SA (2013) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol Plant Microbe Interact 26: 130–139 [DOI] [PubMed] [Google Scholar]
- Reymond P. (2013) Perception, signaling and molecular basis of oviposition-mediated plant responses. Planta 238: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PA, Escudero-Martinez C, Bos JI (2017) An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol 173: 1892–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PA, Stam R, Warbroek T, Bos JIB (2014) Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol Plant Microbe Interact 27: 30–39 [DOI] [PubMed] [Google Scholar]
- Sharma PC, Ito A, Shimizu T, Terauchi R, Kamoun S, Saitoh H (2003) Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol Genet Genomics 269: 583–591 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Nasir KH, Ito A, Kanzaki H, Matsumura H, Saitoh H, Fujisawa S, Kamoun S, Terauchi R (2007) A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii. Plant J 49: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Verma M, Davidson EA (1994) Mucin genes: structure, expression and regulation. Glycoconj J 11: 172–179 [DOI] [PubMed] [Google Scholar]
- Villarroel CA, Jonckheere W, Alba JM, Glas JJ, Dermauw W, Haring MA, Van Leeuwen T, Schuurink RC, Kant MR (2016) Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J 86: 119–131 [DOI] [PubMed] [Google Scholar]
- Walling LL. (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19: 195–216 [DOI] [PubMed] [Google Scholar]
- Walling LL. (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Granados RR (1997) An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci USA 94: 6977–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJ (2007) Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA 104: 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Vilcinskas A (2015) The structural sheath protein of aphids is required for phloem feeding. Insect Biochem Mol Biol 57: 34–40 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Yu H, Jian Y, Zeng J, Ji R, Chen H, Lou Y (2017) A salivary EF-hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in rice. Sci Rep 7: 40498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tang J, Wang Q, Ye W, Tao K, Duan S, Lu C, Yang X, Dong S, Zheng X, et al. (2012) The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol 196: 247–260 [DOI] [PubMed] [Google Scholar]
- Yuan H, Chen X, Zhu L, He G (2005) Identification of genes responsive to brown planthopper Nilaparvata lugens Stal (Homoptera: Delphacidae) feeding in rice. Planta 221: 105–112 [DOI] [PubMed] [Google Scholar]
- Zha W, Peng X, Chen R, Du B, Zhu L, He G (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS ONE 6: e20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Y, Lu W, Meng F, Wu CA, Guo X (2012) Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J Exp Bot 63: 3935–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Huang J, Wang Z, Jing S, Wang Y, Ouyang Y, Cai B, Xin XF, Liu X, Zhang C, et al. (2016) Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc Natl Acad Sci USA 113: 12850–12855 [DOI] [PMC free article] [PubMed] [Google Scholar]