Specification of carpel margin meristems and archesporial cells by the GRF-GIF duo is a prerequisite for the female and male reproductive competence of Arabidopsis.
Abstract
We investigated the biological roles of the Arabidopsis (Arabidopsis thaliana) GROWTH-REGULATING FACTOR (GRF) and GRF-INTERACTING FACTOR (GIF) transcriptional complex in the development of gynoecia and anthers. There are nine GRFs and three GIFs in Arabidopsis, and seven GRFs are posttranscriptionally silenced by microRNA396 (miR396). We found that overexpression of MIR396 in the gif1 gif2 double mutant background (gif1 gif2 35S:MIR396) resulted in neither ovary nor pollen. Histological and molecular marker-based analyses revealed that the mutant gynoecial primordia failed to develop carpel margin meristems and mature flowers lacked the ovary, consisting only of the stigma, style, and replum-like tissues. The mutant anther primordia were not able to form the pluripotent archesporial cells that produce pollen mother cells and microsporangia. Multiple combinations of GRF mutations also displayed the same phenotypes, indicating that the GRF-GIF duo is required for the formation of those meristematic and pluripotent cells. Most GRF proteins are localized and abundant in those cells. We also found that the weak gynoecial defects of pinoid-3 (pid-3) mutants were remarkably exacerbated by gif1 gif2 double mutations and 35S:MIR396, so that none of the gynoecia produced by gif1 gif2 pid-3 and 35S:MIR396 pid-3 developed ovaries at all. Moreover, gif1 gif2 double mutations and 35S:MIR396 also acted synergistically with 1-N-naphthylphthalamic acid in forming aberrant gynoecia. The results altogether suggest that the GRF-GIF duo regulates the meristematic and pluripotent competence of carpel margin meristems and the archesporial cell lineage and that this regulation is implemented in association with auxin action, ultimately conferring reproductive competence on Arabidopsis.
In flowering plants (angiosperms), normal development of the gynoecium and anther is essential for reproductive success, since they bear and house the egg and sperm cells, respectively. The gynoecium of Arabidopsis (Arabidopsis thaliana) consists of two carpels that are believed to arise congenitally fused along their margins from the center of the floral meristem (Ferrándiz et al., 1999, 2010; Supplemental Fig. S1). The fused margins correspond to the medial domain of the gynoecium, and, later, the two carpels become discernible by the formation of repla, which demarcate the two lateral ovary valves. The distal part of the ovary meets the style capped with the stigma, and its proximal part meets the gynophore. The gynoecium develops internal tissues, such as the ovule, septum, and transmitting tract (Ferrándiz et al., 1999, 2010; Supplemental Fig. S1). The ovule produces a functional megaspore, which performs gametogenesis to generate the embryo sac containing an egg cell. The septum compartmentalizes the ovary into two locules and forms the transmitting tract in the middle, which guides pollen tube growth. Importantly, all of these internal tissues are derived from a pair of tissues called carpel margin meristems (CMMs), which arise longitudinally from the adaxial medial portions of the gynoecial primordium at floral stage 7 and of which cells are, literally, proliferative and pluripotent. Therefore, the formation and maintenance of CMMs are critical for the female competence of Arabidopsis and angiosperms.
Gynoecium development in Arabidopsis has been studied over a couple of decades, elucidating a number of patterning and identity genes as well as regulatory networks (Reyes-Olalde et al., 2013). Meanwhile, only a few studies have focused specifically on CMM development. Nonetheless, it has been known that some members of the LEUNIG (LUG), SEUSS (SEU), and AINTEGUMENTA gene families and FILAMENTOUS FLOWER play pivotal roles in the development of CMMs and CMM-derived tissues (ovule, septum, transmitting tract; hereafter, CMM derivatives collectively). Mutational combinations of these genes resulted in a complete lack of CMM derivatives and repla, consequently producing medially split gynoecia only with ovary valves (Liu and Meyerowitz, 1995; Chen et al., 2000; Krizek et al., 2000; Franks et al., 2002; Azhakanandam et al., 2008). It has been proposed that these gene products exert an overlapping function in CMM development, probably forming a multimeric complex, although their detailed cellular and molecular mechanisms remain to be elucidated (Nole-Wilson and Krizek, 2006; Azhakanandam et al., 2008).
The Arabidopsis anther has a four-lobed structure. Each lobe internally develops a microsporangium that consists of three outer concentric parietal layers (i.e. endothecium, middle layer, and tapetum), and the microsporangium harbors pollen mother cells (PMCs; Sanders et al., 1999; Egger and Walbot, 2016; Supplemental Fig. S1). According to the lineage model, all of the layer cells and PMCs are formed through sequential mitotic divisions and differentiation of archesporial cells that arise in the L2 layer of the nascent anther primordium, thus implying that archesporial cells are, by nature, meristematic and pluripotent. PMCs perform sporogenesis and gametogenesis consecutively to generate pollen grains containing two sperm cells. As such, the establishment of archesporial lineage cells is of pivotal importance for male competence.
GROWTH-REGULATING FACTORs (GRFs) are plant-specific transcription factors and are conserved in all land plants, including mosses (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015). GRFs interact with GRF-INTERACTING FACTORs (GIFs) to form a transcriptional complex, which serves as a functional unit (Kim and Kende, 2004; Horiguchi et al., 2005). There are nine GRFs and three GIFs in Arabidopsis. They were first recognized by their growth-promoting activities in lateral organs: loss-of-function mutations of GRFs and GIFs resulted in small narrow leaves and petals (Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Lee et al., 2009). These studies demonstrated that most of the Arabidopsis GRF and GIF members examined were required, in a redundant manner, for the cell proliferation of lateral organs, thus determining their final size.
A plant microRNA, miR396, targets and cleaves GRF mRNAs, and it is highly conserved in all tracheophytes (Jones-Rhoades et al., 2006; Taylor et al., 2014). In Arabidopsis, miR396 is encoded by MIR396a and MIR396b and induces the cleavage of seven GRF mRNAs, with the exceptions of GRF5 and GRF6 transcripts that lack the target site (Jones-Rhoades et al., 2006; Liu et al., 2009; Rodriguez et al., 2010). Overexpression of MIR396 using the cauliflower mosaic virus 35S promoter (35S:MIR396a and 35S:MIR396b) caused remarkable reductions in the levels of target GRF mRNAs in leaves and floral organs (Liu et al., 2009; Rodriguez et al., 2010; Liang et al., 2014). The existence of miR396 not only revealed an additional regulatory layer in the control of GRF expression but also provided a genetic tool for overcoming the functional redundancy of GRFs.
Performing morphological and histological analyses of gif1 gif2 gif3 (briefly, gif1/2/3), we have shown that the GIF family plays an essential role in the development of Arabidopsis floral organs (Lee et al., 2014). As for gynoecial development, CMMs of the gif triple mutant partially lost their meristematic competence (hereafter, meristematicity) and pluripotency over time, so that the distal half of the gynoecium lacked CMM derivatives and the replum, leading to medially split gynoecia with ovary valves, each being topped by the partial stigma and style; the proximal half retained CMM activities, although poor, producing aberrant CMM derivatives and the replum. Ovules produced in the proximal half developed very limited integuments, and the functional megaspore lost its identity and, thus, ended in nucellar cells, consequently producing no embryo sac. As for anther development, archesporial lineage cells of the gif triple mutant, like CMM cells in the gynoecium, degenerated over time to become the same somatic cells as neighboring connective cells, producing neither microsporangia nor PMCs. Some of the archesporial cells survived to form pollen grains, which, however, were not released because of the lack of the dehiscence apparatus. These results indicated that GIF genes are required in a redundant manner for the formation and maintenance of the meristematicity of CMMs and archesporial lineage cells.
We reckoned that the incomplete abolishment of CMM and archesporial lineage cells of the gif triple mutant might be due to the fact that the gif3 mutation was not null but hypomorphic (Lee et al., 2009), raising the possibility that, with a true null mutation of gif3, a complete abolishment of CMM and archesporial cells might be attained. However, no null mutation of gif3 is available at present. Given the tight coupling of the GRF-GIF duo in leaf growth, it is also conceivable that grf mutations, like gif, should cause similar aberrancies in floral development. It was reported that some strong overexpressors of MIR396a often produced single-carpel gynoecia (Liang et al., 2014; Pajoro et al., 2014). However, those studies did not resolve anatomical and histological details of how the down-regulation of miR396-targeted GRFs led to the phenotypes. Moreover, with regard to CMM and anther development, the role of the GRF family and its individual members has not been investigated yet. In this study, we aimed to test such notions by overexpressing MIR396b in the gif mutant background (gif 35S:MIR396b) and by constructing grf multiple mutants. Our data demonstrate that the GRF-GIF duo is crucial, on the one hand, for the meristematicity and pluripotency of CMM and archesporial cells and, on the other hand, for ovary identity. We also suggest that the duo functions in association with auxin action, since the pinoid-3 (pid-3) mutation and 1-N-naphthylphthalamic acid (NPA) aggravated these grf and gif phenotypes.
RESULTS
gif 35S:MIR396b and grf5 35S:MIR396b Gynoecia Fail to Develop CMMs and Ovary Valves
Scanning electron microscopy (SEM) analysis revealed that the wild-type Arabidopsis gynoecium begins to form CMMs longitudinally at its adaxial medial domain at the floral stage 7 (Fig. 1, A1 and A2). Along with the gynoecial primordium elongating, apical cells of both the medial and lateral domains differentiate to form the stigma with papillar cells and the style (Fig. 1, A3–A5). During the developmental period, the abaxial side of CMMs differentiates to the replum, culminating in a mature gynoecium, in which two carpels are recognized by the presence of two ovary valves and intervening repla (Fig. 1A6). To investigate the roles of the GRF-GIF duo in floral organ development, the gif1-1 gif2-1 (hereafter, gif1/2) double mutant was crossed with the 35S:MIR396b plant that had been shown to down-regulate the expression of target GRFs (GRF1–GRF4 and GRF7–GRF9; Liu et al., 2009; Rodriguez et al., 2010; for nomenclature and mutational nature, see Supplemental Table S1 and Supplemental Fig. S2). We found that the resulting gif1/2 35S:MIR396b mutant failed to form CMMs (Fig. 1, B1 and B2) but that the apical portion of the mutant gynoecium developed the stigma with papillar cells (Fig. 1, B3–B6). Strikingly, the preponderant majority (81%) of mutant gynoecia had no ovary valves at all and took a radial rod shape (Fig. 1B6; Table I). A minor proportion developed a pair of defective CMMs closely abutted and, thus, produced the gynoecium with a single ovary valve (Fig. 1, B11–B13). A similar proportion displayed split gynoecia, exposing ovules (Fig. 1, B18 and B19). Gynoecia with only a single, tiny valve also were sporadically observed (Fig. 1B14). In addition, various fusions between floral organs occurred in high proportions (Fig. 1, B2, B4, and B15–B17; Table I). The numbers of petals and stamens also were reduced significantly (Table I). In contrast, the parental plants, 35S:MIR396b and gif1/2, showed quasi-normal CMMs and no distinctive defects in those aspects of floral organ development (Fig. 1, C1–C4 and D1–D4, respectively), although 35S:MIR396b produced single-valved gynoecia, but only rarely (Fig. 1, C5 and C6; Table I). The same is true for the gif1 35S:MIR396b double mutant, but with weaker phenotypes than those of the triple mutant: fewer valveless gynoecia but more single-valved gynoecia and less reduction in the numbers of petals and stamens (Table I). Our previous study showed that, although the gif1/2/3 triple mutant developed ovary valves, it produced severely compromised CMMs, CMM derivatives, and repla as well as split gynoecia (Lee et al., 2014). Taken together, these results indicate that the GRF-GIF duo function was maximally impaired in gif1/2 35S:MIR396b and that the GRF-GIF duo is required not only for CMM development but also for ovary valve formation.
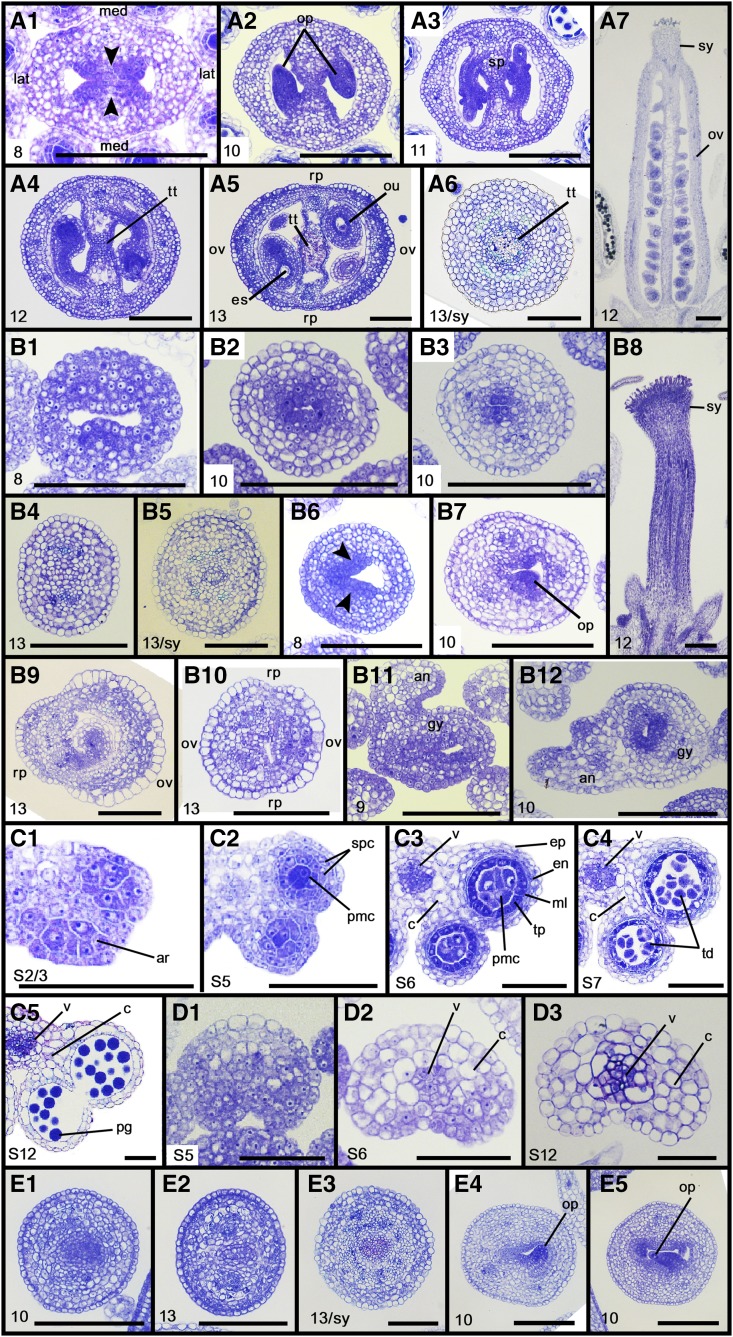
Figure 1.
SEM analysis of floral organ phenotypes of gif1/2 35S:MIR396b, grf5 35S:MIR396b, and pid-14. A1 to A10, The wild type. B1 to B19, gif1/2 35S:MIR396b. C1 to C6, 35S:MIR396b. D1 to D4, gif1/2. E1 to E3, pid-14. Numbers in images indicate floral stages, as described by Smyth et al. (1990). Arrowheads, CMMs; asterisks, ovary valves; arrows, stamens fused to the gynoecium. ov, Ovary valve; rp, replum; sg, stigma; sy, style. Bars = 100 µm, except for A8, A10, B8, and B10 (10 µm).
Table I. Floral organ phenotypes of gif, grf5, and 35S:MIR396.
Flower organs at flower stage 12 from five different plants were examined. I, Normal double valves; II, no valves; III, single tiny valve; IV, single valve; V, split double valves; Se, sepal; Pe, petal; St, stamen; Ca, carpel. Values are means ± se (n = 200). Dashes denote 0%.
| Genotype | Percentage of Ovary Types |
Percentage of Organ Fusion |
No. of Organs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | Se + Pe | Pe + St | St + St | St + Ca | Se | Pe | St | |
| Wild type | 100 | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| gif1 | 100 | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| gif1/2 | 100 | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf5 | 100 | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| 35S:MIR396b | 98 | – | – | 2 | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| gif1 35S:MIR396b | – | 45 | 7 | 44 | 4 | 7 | 16 | 56 | 9 | 4.0 ± 0.0 | 3.1 ± 0.1 | 4.4 ± 0.2 |
| gif1/2 35S:MIR396b | – | 81 | 2 | 8 | 11 | 4 | 21 | 61 | 28 | 4.0 ± 0.0 | 2.3 ± 0.1 | 4.0 ± 0.2 |
| grf5 35S:MIR396b | 37 | 8 | – | 47 | 8 | 18 | – | – | 4 | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
We reasoned that, since GRF5 mRNAs are not targeted by miR396, the introduction of grf5 into the 35S:MIR396b plant should further reduce the levels of total GRF mRNAs and, thus, permit an evaluation of the influence of the GRF family alone, apart from the GIF family. Indeed, grf5 35S:MIR396b plants produced valveless (8%) and, mostly, single-valved (47%) gynoecia as well as normal, two-valved gynoecia (37%; Table I). The grf5 single mutant is indistinguishable from the wild type with regard to floral organ development. grf5 35S:MIR396a also showed similar phenotypes (Liang et al., 2014). These results indicate that a substantial reduction in the level of GRF mRNAs as a whole leads to the lack of CMMs and ovary valves.
The Mutant Gynoecial Rod Consists of Only Replum-Like Tissues at the Expense of Lateral Valves
Although gif1/2 35S:MIR396b gynoecia had the stigma with papillar cells, it was not obvious whether they had the style tissue and what the cellular identity of the gynoecial rod was. The epidermis of the wild-type style is distinguishable from that of ovary valves by crenelated deposits of wax and stomata (Sessions and Zambryski, 1995; Fig. 1, A7 and A8). The corresponding region of the mutant also displayed the same type of crenelated cells together with stomata (Fig. 1, B7 and B8), which is indicative of the style tissue. The epidermis of the wild-type ovary valve is composed of four to five cell clusters surrounding an immature stoma, while the epidermis of the wild-type replum comprises small rectangular cells with no stomata (Fig. 1, A9 and A10). In contrast, the corresponding region of the rod appeared to consist of only small rectangular cells, which were more like those of the replum than those of valves (Fig. 1, B9 and B10). It was noticeable, however, that the mutant epidermis contained sporadic stomata, as did the style tissue, raising the possibility that the rod epidermis may retain a modicum of valve identity.
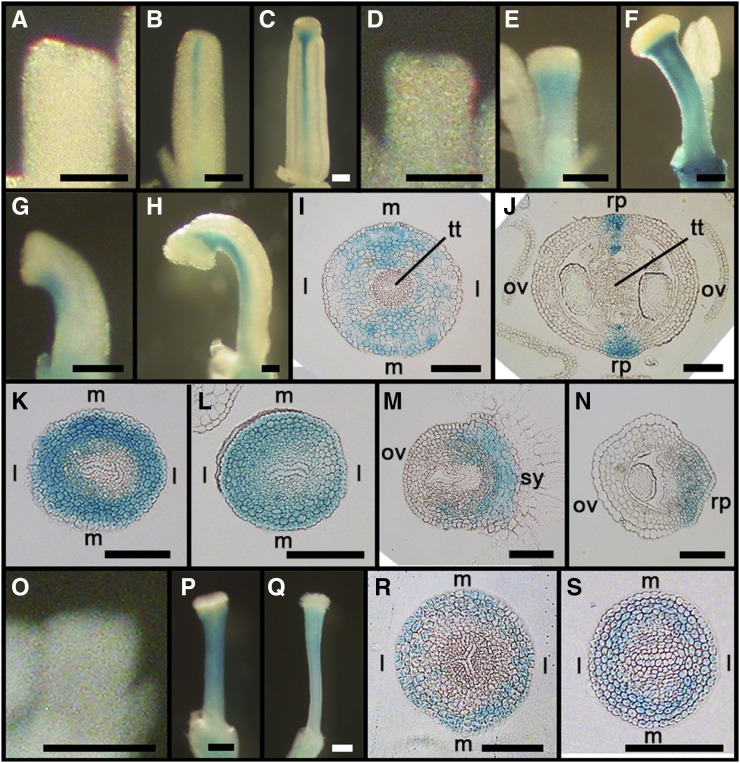
To clarify the identity of the mutant gynoecial rod, we constructed gif1 35S:MIR396b KNAT1:GUS-18 (briefly, KNAT1:GUS). The KNAT1:GUS marker was shown previously to express in the replum and style (Alonso-Cantabrana et al., 2007). In the wild type, the marker was not expressed in early primordial gynoecia but began to express exclusively in repla and, later, styles, as the tissues were developmentally established (Fig. 2, A–C). Cross-sectional images revealed that GUS signals were spread all over the stylar cells, denser in medial domains, but not in the central transmitting tract; in the ovary, the signals were restricted to the replum cells and medial vasculature (Fig. 2, I and J). In valveless gynoecia, the signals were not detected in early primordia, as in wild-type gynoecia, but were later detected all over the whole rod (Fig. 1, D–F). Similarly, in single-valved gynoecia, the signals were observed in the replum and style but not in the valve (Fig. 2, G and H). Cross-sectional images of valveless mutants displayed strong signals in the circular band of stylar cells and, notably, in all of the rod cells (Fig. 2, K and L). The single-valved mutant also showed a strong signal in the style and the expanded replum, but not in the valve (Fig. 2, M and N). grf5 35S:MIR396b KNAT1:GUS showed similar staining patterns (J.-H.J. and J.H.K., data not shown). SEM and KNAT1:GUS analyses indicate that the gynoecial rod consisted of only replum-like tissues and lost its mediolateral patterning of KNAT1:GUS expression.
Figure 2.
Expression patterns of KNAT1:GUS in gynoecia of gif1 35S:MIR396 KNAT1:GUS and pid-14 KNAT1:GUS. Whole-mount images are as follows. A to C, KNAT1:GUS. D to H, gif1 35S:MIR396b KNAT1:GUS. O to Q, pid-14 KNAT1:GUS. Cross-section images are as follows. I and J, Style and ovary regions of KNAT1:GUS, respectively. K and L, Style and ovary regions of valveless gynoecia of gif1 35S:MIR396b KNAT1:GUS, respectively. M and N, Style and ovary regions of single-valved gynoecia of gif1 35S:MIR396b KNAT1:GUS, respectively. R and S, Style and ovary regions of pid-14 KNAT1:GUS, respectively. l, Lateral domain; m, medial domain; ov, ovary valves; rp, replum; sy, style; tt, transmitting tract. Bars = 100 µm.
The Mutant Carpels and Archesporial Cells Fail to Specify Their Meristematicity and Pluripotency
We performed histological analysis in order to investigate the cellular basis of the CMM and valve development. The early gynoecial primordium of the wild type develops a pair of CMMs at its adaxial medial sides (CMM cells are strongly stained by Toluidine Blue because of their dense cytoplasm; Fig. 3A1). It is evident that the gynoecium at this early stage takes on a bilateral symmetry, establishing mediolateral patterning. Later, CMMs grow inward to fuse together, developing the septum with the transmitting tract, which thus renders two carpels and locules separated (Fig. 3, A2–A5 and A7). The flanking regions of CMMs form ovule primordia that develop further to give rise to the embryo sac. Concomitantly, the abaxial medial sides differentiate into the replum; the lateral domains differentiate to ovary valves, of which epidermal cells are much larger than those of the replum; and the apical region of the gynoecium differentiates into the style with the transmitting tract, finally culminating in a mature gynoecium (Fig. 3, A5–A7).
Figure 3.
Histological analysis of gynoecia and anthers of gif1/2 35S:MIR396b and pid-14. A1 to A7, Wild-type gynoecia; A6, style. B1 to B12, gif1/2 35S:MIR396b gynoecia; B5, style. C1 to C5, Wild-type anthers. D1 to D3, gif1/2 35S:MIR396b anthers. E1 to E5, pid-14 gynoecia; E3, style. Numbers in images indicate floral and stamen stages, as described by Smyth et al. (1990) and Sanders et al. (1999). Arrowheads, CMMs. an, Anther; ar, archesporial cell; c, connective tissue; en, endothecium; ep, epidermis; es, embryo sac; gy, gynoecium; lat, lateral; med, medial; ml, middle layer; op, ovule primordium; ou, ovule; ov, ovary valve; pg, pollen grain; pmc, pollen mother cell; rp, replum; sp, septum; spc, secondary parietal cell; sy, style; td, tetrad; tp, tapetum; tt, transmitting tract; v, vasculature. Bars = 100 µm, except for C1 to D3 (50 µm).
In contrast, no typical CMMs were observed in gif1/2 35S:MIR396b gynoecia even at floral stage 8 (Fig. 3B1). At later stages, Toluidine Blue staining was strongly detected in several inner layers but soon restricted only to the innermost layer of cells and, finally, faded away (Fig. 3, B2–B4). It seems that these densely stained inner cells otherwise would have organized CMMs. Actually, however, they failed to take on meristematic and pluripotent properties, resulting in neither CMMs nor locules, and the gynoecium was simply filled in with cells (Fig. 3, B4 and B8). The gynoecium displayed no sign of ovary development either: the presumed ovary part resembled wholly the replum with regard to cell types and structural organization (Figs. 2 and 3B4). The mutant style also failed to develop such a typical transmitting tract as the wild type (compare Fig. 3, A6 and B5). Some mutant gynoecia developed two partial CMMs abutted at their flanks, from which ovule primordia occurred but were aborted (Fig. 3, B6, B7, and B9). In this case, the gynoecium apparently developed a single ovary valve with an expanded replum (Fig. 3B9). Occasionally, gynoecia in which a distinction between valves and repla was obscure were observed (Fig. 3B10). Congenital fusions between the gynoecium and anther occurred frequently (Fig. 3, B11 and B12). Taken together, our data clearly indicate that the GRF-GIF duo is absolutely required for carpel development by endowing the primordial cells of CMMs and ovary valves with meristematicity and pluripotency.
The wild-type anther formed a typical four-lobed structure, whereas the mutant anther usually formed a two-lobed structure (Fig. 1, A5 and B4). Histological analysis revealed the presence of the archesporial cells in the wild type and that they performed a series of cell divisions and differentiation events to form the microsporangium and PMCs (Fig. 3, C1–C3). Then, PMCs carried out sporogenesis and gametogenesis consecutively to produce pollen grains (Fig. 3, C4 and C5). Strikingly, however, the mutant anther primordium had no archesporial cells, thus producing no archesporial lineage cells, such as parietal cells and PMCs: mature anthers were simply filled with somatic connective cells (Fig. 3, D1–D3). These results clearly indicate that the GRF-GIF duo also is absolutely required for the specification and formation of archesporial cells.
GRF1 to GRF3 and GRF5 Are Critical for Carpel and Anther Development
We have hitherto employed 35S:MIR396b in order to investigate the role of the GRF family in floral organ development and to overcome the functional redundancy of its members. We next set out to dissect what portions of the influences exerted by 35S:MIR396b were attributable to individual members of the GRF family. To do that, we collected all available T-DNA insertion mutants of the Columbia accession, except grf2-0, which was in the Wassilewskija accession (Supplemental Table S1; Supplemental Fig. S2; grf1-3, grf2-0, grf3-1, grf4-1, grf4-2, grf5-2, grf7-1, grf8-1, and grf9-1 were used in this study), and constructed multiple mutants through a series of crosses and PCR-assisted genotyping (Table II). It should be noted that the grf1/2/3/5 quadruple mutant was established after five backcrosses of grf2-0 with grf1/3/5; grf1/2(+)/3/4-1/5 and grf1/2(+)/3/4-2/5 were established after crosses of the cleaned grf1/2/3/5 with grf1/3/4-1/5 and grf1/3/4-2/5, respectively. As a result, none of the single, double, or triple mutants, except grf1/3/5, displayed any visible defects with regard to floral organ development, and neither did some quadruple mutants (Table II; Supplemental Table S2). However, a substantial portion of grf1/3/5 triple mutants had single-valved gynoecia and showed slight aberrations in floral organ separation and numbers. Notably, these floral defects of grf1/3/5 were greatly enhanced by the addition of grf2: the majority of grf1/2/3/5 quadruple mutants had malformed gynoecia, mostly single valved and valveless, and displayed severe fusions between floral organs as well as significant reductions in numbers of petals and stamens. Even the presence of only a single copy of the grf2 allele enhanced the phenotypes, as observed in grf1/2(+)/3/4-1/5 and grf1/2(+)/3/4-2/5 (grf1/2/3/4-1/5 and grf1/2/3/4-2/5 homozygous for grf2 were embryo lethal). SEM and histological analyses confirmed that the valveless and single-valved gynoecia of grf1/2/3/5 displayed the same defects in forming CMMs, ovary valves, and archesporial cells as those of gif1/2 35S:MIR396b (J.-H.J. and J.H.K., data not shown). It should be noted, however, that the phenotypic enhancement was not obtained by the addition of other mutations: neither by such singles as grf4-1, grf4-2, grf7, grf8, and grf9 nor by such doubles as grf4-1 grf7, grf4-2 grf7, grf4-1 grf9, and grf4-2 grf9. Even the grf1/4-2/5/7/8 quintuple mutant was quite normal with regard to floral development. It also should be noted that the mutations used in this study are null or affect other developmental processes, such as leaf growth (Supplemental Table S1; Supplemental Fig. S2; Kim and Lee, 2006; Kim et al., 2012). These results indicate that GRF1 to GRF3 and GRF5 play, in a functionally redundant manner, a critical role in the development of CMMs, ovary valves, and archesporial cells, whereas GRF4 and GRF7 to GRF9 have little, if any, role. The role of GRF6 remains to be determined in the future because no mutant is available at present.
Table II. Floral organ phenotypes of grf multiple mutants.
Flower organs at flower stage 12 from five different plants were examined. I, Normal double valves; II, no valves; III, single tiny valve; IV, single valve; V*, reduced double valves; Se, sepal; Pe, petal; St, stamen; Ca, carpel. Values are means ± se (n = 200). Dashes denote 0%.
| Genotype | Percentage of Ovary Types |
Percentage of Organ Fusion |
No. of Organs |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V* | Se + Se | Se + Pe | Pe + St | St + St | St + Ca | Se | Pe | St | |
| Wild type | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf1/3/4-2 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf1/3/5 | 91 | – | – | 8 | 1 | 1 | – | – | 3 | 1 | 3.9 ± 0.1 | 3.7 ± 0.1 | 5.7 ± 0.1 |
| grf1/5/7 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf4-2/5/7 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf5/7/8 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf1/2/3/5a | 21 | 13 | 6 | 51 | 9 | 7 | 14 | 64 | 14 | 37 | 4.1 ± 0.1 | 2.3 ± 0.1 | 4.8 ± 0.1 |
| grf1/3/4-1/5 | 87 | – | – | 13 | – | 1 | – | 2 | 2 | – | 3.9 ± 0.0 | 3.9 ± 0.0 | 5.7 ± 0.1 |
| grf1/3/4-2/5 | 89 | – | – | 11 | – | – | – | – | 3 | 2 | 4.0 ± 0.0 | 3.9 ± 0.0 | 5.7 ± 0.1 |
| grf1/3/5/7 | 93 | – | – | 7 | – | – | – | – | 1 | 1 | 4.0 ± 0.0 | 3.8 ± 0.0 | 5.8 ± 0.0 |
| grf1/5/7/8 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf4-1/5/7/8 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf4-2/5/7/8 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| grf1/2(+)/3/4-1/5b | 65 | – | – | 35 | – | – | 1 | 9 | 7 | – | 4.2 ± 0.0 | 3.3 ± 0.0 | 5.7 ± 0.0 |
| grf1/2(+)/3/4-2/5b | 74 | – | – | 25 | – | – | – | 19 | 5 | 1 | 4.1 ± 0.0 | 3.5 ± 0.0 | 5.7 ± 0.0 |
| grf1/3/4-1/5/7 | 89 | 11 | 1 | 1 | 4 | 4.0 ± 0.0 | 3.8 ± 0.1 | 5.8 ± 0.1 | |||||
| grf1/3/4-2/5/7 | 86 | 12 | 2 | 2 | 4 | 3.9 ± 0.0 | 3.8 ± 0.1 | 5.7 ± 0.1 | |||||
| grf1/3/4-1/5/9 | 90 | 6 | 4 | 3 | 1 | 1 | 4.0 ± 0.0 | 3.8 ± 0.1 | 5.7 ± 0.1 | ||||
| grf1/3/4-2/5/9 | 89 | 11 | 1 | 7 | 2 | 3.9 ± 0.0 | 3.7 ± 0.1 | 5.6 ± 0.1 | |||||
| grf1/4-2/5/7/8 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
The mutant was constructed through five back-crosses of grf2 in the Wassilewskija background with grf1/3/5 in Columbia.
2(+) indicates that the quintuple mutants are heterozygous for grf2, and they were obtained by crossing the genetically cleaned grf1/2/3/5 mutant with grf1/3/4-1/5 and grf1/3/4-2/5.
Localization Patterns and Abundance of GRF:GUS Proteins in Floral Organs
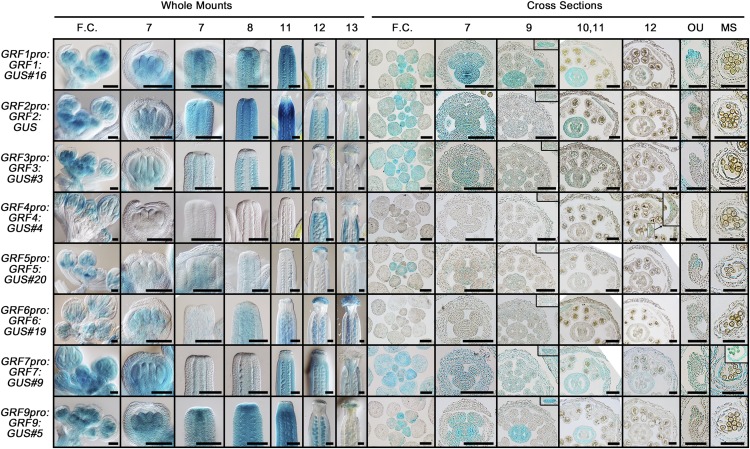
To analyze the expression patterns of GRFs, we adopted a translational reporter system in which a genomic fragment containing the approximately 2-kb promoter region, exons, and introns of each GRF member was fused to the GUS coding sequence. GRF8 was excluded in this analysis because of the obscure annotation of its genomic structure, and GRF2pro:GRF2:GUS was provided by Dr. Javier Palatnik (Rodriguez et al., 2010). Most GRF members are actively expressed in floral organs (Fig. 4). Cross-sectional images of flower clusters revealed that most GRF members were highly expressed in floral meristems (Fig. 4). GRF1 to GRF3 and GRF5 to GRF9 were detected in the whole primordia of gynoecia and anthers from the incipient stage to stage 7. The gynoecial signals were maintained until stage 11, after which the signals became weaker or faded away, being restricted to stigmatic tissues, ovules, or valves. The anther signals also were maintained for a while, finally being restricted to the tapetum; expression of most GRF members was detected in petal primordia as well. The expression pattern of GRF4 was atypical in that it was expressed neither in floral meristems nor in incipient floral organs but later in sepals and the tapetum as well as in endocarpic tissues of ovary valves.
Figure 4.
Localization patterns and abundance of GRF:GUS fusion proteins in floral organs. Representative lines (hashtag numbers) of the T2 generation were subjected to the GUS staining assay. F.C., Flower cluster; OU, ovule; MS, microsporangium. Numbers on top indicate floral stages. Insets of cross-sectional images at stages 11 and 12 show petal primordia and an enlarged part of an ovary valve, respectively. Bars = 100 µm, except for the last two columns (50 µm).
gif1/2 and 35S:MIR396 Enhance the Weak Phenotypes of pid-3 Gynoecia
Interestingly, the gynoecial phenotypes of gif 35S:MIR396 and grf multiple mutants are virtually identical to those of auxin-related mutants. Mutations in polar auxin transport, such as pin-formed1 and pid (Okada et al., 1991; Bennett et al., 1995; Huang et al., 2010), and mutations in auxin biosynthetic genes, such as yuc1 yuc4 and wei8 tar2 (Cheng et al., 2006; Stepanova et al., 2008), produced a gynoecial rod without CMM derivatives and ovary valves. Treatment with the polar auxin transport inhibitor NPA (Nemhauser et al., 2000; Larsson et al., 2014) also caused similar defects. For a detailed illustration of the phenotypic similarity, we selected and analyzed a null mutant allele of PID, pid-14. pid-14 mutants developed almost no ovaries: most of them were valveless and single valved (Table III). In addition, floral organs were frequently fused; the number of stamens was reduced, whereas the number of petals was increased. These are all characteristic phenotypes of pid mutants (Bennett et al., 1995; Huang et al., 2010). The pid-14 gynoecium, like gif 35S:MIR396b, failed to develop CMMs and ovary valves (Fig. 1, E1–E3) and was filled in with cells, leaving no locules and consisting of replum-like tissues (Fig. 3, E1–E3). Some gynoecia sporadically developed ovule primordia, which never culminated in mature ovules (Fig. 3, E4 and E5). The pid-14 KNAT1:GUS gynoecium showed the same staining patterns as gif 35S:MIR396b KNAT1:GUS, indicating that the pid-14 gynoecial rod consisted of only expanded medial replum tissues with neither CMMs nor lateral valves (Fig. 2, O–S).
Table III. Floral organ phenotypes of gif, 35S:MIR396, and pid.
Flower organs at flower stage 12 from five different plants were examined. I, Normal double valves; II, no valves; III, single, tiny valve; IV, single valve; V*, reduced double valves; Se, sepal; Pe, petal; St, stamen; Ca, carpel. Values are means ± se (n = 200). Dashes denote 0%.
| Genotype | Percentage of Ovary Types |
Percentage of Organ Fusion |
No. of Organs |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V* | Se + Se | Pe + Pe | Pe + St | St + St | St + Ca | Se | Pe | St | |
| Wild type | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| gif1 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| gif1/2 | 100 | – | – | – | – | – | – | – | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| 35S:MIR396b | 96 | – | – | 4 | – | – | – | 2 | – | – | 4.0 ± 0.0 | 4.0 ± 0.0 | 6.0 ± 0.0 |
| pid-3 | 46 | 2 | 8 | 25 | 19 | 10 | 8 | – | 8 | 5 | 4.2 ± 0.1 | 4.3 ± 0.1 | 4.8 ± 0.2 |
| pid-14 | – | 67 | 11 | 11 | 11 | 11 | 44 | – | 22 | 13 | 3.8 ± 0.2 | 5.7 ± 0.3 | 2.2 ± 0.3 |
| gif1 pid-3 | 25 | 4 | 2 | 50 | 19 | 17 | 12 | 4 | 13 | 2 | 4.7 ± 0.1 | 4.9 ± 0.1 | 4.1 ± 0.2 |
| gif1/2 pid-3 | – | 100 | – | – | – | 30 | 45 | 8 | – | – | 4.6 ± 0.1 | 6.5 ± 0.2 | 1.3 ± 0.1 |
| 35S:MIR396b pid-3 | – | 81 | 3 | 16 | – | 9 | 12 | 3 | 5 | – | 4.7 ± 0.1 | 5.9 ± 0.3 | 1.7 ± 0.4 |
To investigate the genetic interaction between the GRF-GIF duo and PID, we chose the pid-3 mutant that harbored a hypomorphic weak allele (Bennett et al., 1995). About half of the pid-3 mutants produced normal, two-valved gynoecia, whereas the other half developed aberrant gynoecia, mostly single valved and reduced, two valved (Table III). However, the addition of the gif1 and gif1/2 mutations synergistically enhanced pid-3 phenotypes, so that gif1/2 pid-3 gynoecia were all valveless. In addition, all gif1/2 pid-3 mutants had pin-formed inflorescence primary stems (11 of 11 plants), whereas gif 1/2 and pid-3 had 0% (zero of 17) and 19% (four of 21), respectively, indicating that the synergism also was manifested in the development of inflorescence stems. Similarly, almost all of the 35S:MIR396b pid-3 gynoecia are valveless. The numbers of petals and stamens in the gif/2 pid-3 and 35S:MIR396b pid-3 mutants also were affected severely, being close to those of pid-14. When wild-type flower clusters were treated with NPA, they occasionally produced single-valved and reduced, two-valved gynoecia (Table IV; Nemhauser et al., 2000). When gif1/2 double mutants and 35:MIR396b plants were treated with NPA, the proportions of malformed gynoecia were increased significantly, compared with those of the NPA-treated wild type, indicating that gif1/2 and 35S:MIR396b gynoecia are hypersensitive to NPA treatment.
Table IV. Gynoecial phenotypes of gif1/2 and 35S:MIR396b treated with NPA.
Flower organs at flower stage 12 from five different plants were examined. I, Normal double valves; II, no valves; III, single, tiny valve; IV, single valve; V*, reduced double valves. Dashes denote 0%.
| Genotype | Percentage of Ovary Types |
No. | ||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V* | ||
| Mock wild type | 100.0 | – | – | – | – | 169 |
| NPA wild type | 96.5 | – | 0.4 | 2.2 | 1.3 | 228 |
| Mock gif1/2 | 100.0 | – | – | – | – | 120 |
| NPA gif1/2 | 79.5 | 4.0 | 1.5 | 9.5 | 5.5 | 200 |
| Mock 35S:MIR396b | 98.3 | – | – | 1.2 | 0.6 | 171 |
| NPA 35S:MIR396b | 73.0 | 7.0 | 1.9 | 12.1 | 6.1 | 215 |
DISCUSSION
The GRF-GIF Duo Endows Carpel Tissues and Archesporial Cells with Meristematicity and Pluripotency
Our previous phenotypic analysis of gif1/2/3 triple mutants demonstrated that the GIF family plays essential roles in the formation and maintenance of CMMs, their derivatives, and archesporial lineage cells (Lee et al., 2014). Yet, CMMs and archesporial cells were not completely compromised by the gif triple mutations. Moreover, it should be noted that two carpels, manifested by the presence of valves, were formed in the triple mutant, although aberrant and split. In addition, it has not been investigated to date whether GRFs play a role in floral development and exert a partnership with GIFs, as they do in other biological processes (Kim and Tsukaya, 2015). Although a role of the GRF family in carpel development has been suggested from 35S:MIR396 phenotypes (Liang et al., 2014), no detailed studies of the roles of 35S:MIR396 and GRFs per se have been reported. In this study, we showed that simultaneous knockout and/or down-regulation of both GRF and GIF families by gif 35S:MIR396 led not only to a complete abolishment of CMMs and archesporial cells but also to no ovary valves (Figs. 1 and 3). Analysis of grf multiple mutants also revealed that, among nine GRF members, GRF1 to GRF3 and GRF5 exerted dominant roles (Table II). These results indicate that both GRF and GIF play essential roles in determining the identities of CMMs, ovary valves, and archesporial cells. Histological analysis led us to propose that the cellular function of the duo is to confer meristematicity and pluripotency on carpel tissues of the gynoecium (CMMs, their derivatives, and valves) as well as on the archesporial cells and their derivatives, so that these cells are capable of proliferating and giving rise to various cell types (Fig. 3). We also suggest the GRF-GIF duo as one of the unknown carpel factors that, despite a long pursuit, have remained cryptic due to their functional redundancy (Wynn et al., 2011). Judging from microarray and in situ hybridization analyses, those authors proposed that GRF5 might be one of these cryptic factors. The localization patterns and abundance of GRF:GUS proteins largely account for the floral defects of gif 35S:MIR396 and grf multiple mutants and also are in good agreement with those of GIF:GUS proteins and GIF mRNAs (Lee et al., 2014), with those of GFP-fused GRF2, GRF5, and GRF8 proteins (Pajoro et al., 2014), and with the GRF promoter activities (Liang et al., 2014). In addition, the in situ localization patterns of GRF5 transcripts were virtually identical to those of GRF5:GUS fusion proteins (Fig. 4; Wynn et al., 2011). In conclusion, it is obvious that the GRF-GIF duo is absolutely required for the reproductive competence of both the female and male organs of Arabidopsis.
We found that, in spite of the abundances of GRF7 and GRF9 in almost all floral organs, their mutations seldom contributed to floral defects (Fig. 4; Table II). Liang et al. (2014) showed that overexpression of miR396-resistant versions of GRF7 and GRF9 nullified the effect of 35S:MIR396a on gynoecial development, suggesting that these GRFs may be equivalent to other GRFs, at least at the protein level. Alternatively, GRF7 may not be involved directly in the regulation of the meristematicity and pluripotency of floral primordia, for it was reported previously that GRF7 acted as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A (Kim et al., 2012).
Possible Molecular Genetic Mechanisms by Which the GRF-GIF Duo Acts
Detailed molecular mechanisms by which the GRF-GIF duo specifies meristematicity and pluripotency of those floral primordia remain to be elucidated in the future. However, recent studies provided clues for inferring several possible mechanisms. First, a series of tandem affinity-purification experiments using Arabidopsis and maize (Zea mays) leaves revealed that GIF1 (ANGUSTIFOLIA3 [AN3]) and GRF proteins were associated with SWI/SNF complexes, suggesting that the GIF1/AN3 transcription cofactor recruits SWI/SNF complexes to render cis-elements of target genes exposed to GRF transcription factors and, thus, that the GRF-GIF duo may act as a key transcriptional complex in transcriptional networks involved in leaf organ growth (Debernardi et al., 2014; Vercruyssen et al., 2014; Nelissen et al., 2015). This notion may hold up for the developmental processes of floral organs as well, since GIF1/AN3 was found to be associated with the promoters of a number of genes and the enhanced expression of some of them, including HECATE1 (HEC1; Vercruyssen et al., 2014). HEC1 is known to be necessary for carpel fusion as well as the development of the transmitting tract and stigma (Schuster et al., 2015). Second, the tandem affinity-purification experiments also revealed that GIF1/AN3 was copurified with SEU, SEU-LIKE1 (SLK1), SLK2, and LEUNIG_ HOMOLOG (LUH; Nelissen et al., 2015). SEU and SLKs are transcriptional adaptors that form a complex with LUG and regulate CMM development (Azhakanandam et al., 2008; Stahle et al., 2009; Bao et al., 2010). LUH is a transcription corepressor belonging to the same family that LUG does (Lee and Golz, 2012). Both LUG and LUH interact with SEU to regulate CMM development in a functionally redundant manner (Sridhar et al., 2004, 2006; Sitaraman et al., 2008; Bao et al., 2010). It was proposed that SEU and SLKs, probably interacting physically with LUG and LUH, sustain the meristematicity of CMM tissues (Stahle et al., 2009; Bao et al., 2010). Therefore, it is tempting to speculate that the GRF-GIF duo may regulate CMM development in association with multimeric complexes consisting of members of the SEU and LUG families. Third, a chromatin immunoprecipitation-coupled sequencing study revealed that GRFs were potential direct targets of SEPALLATA3 (SEP3) and APETALA1 (AP1) transcription factors (Kaufmann et al., 2009; Pajoro et al., 2014). In fact, the floral phenotypes of a weak mutant allele, ap1-3, resemble those of 35S:MIR396a and pANT:MIR396a: ap1-3 often displayed petal-stamen mosaic structures and a reduction in carpel number (Mandel et al., 1992; Bowman et al., 1993). These results suggest that GRFs may function redundantly downstream of SEP3 and AP1 to regulate the patterning and differentiation of floral organs. Finally, the molecular action of GRFs may be associated with CUP-SHAPED COTYLEDON (CUC) transcription factors, since grf mutations interacted genetically with cuc mutations, resulting in severe fusions of cotyledons and floral organs (Lee et al., 2015), and since GRF1 proteins interacted physically with CUC1 and CUC2 proteins in vitro (Jae Og Jeon and J.H.K., data not shown). We showed that gif 35S:MIR396 and grf multiple mutants displayed severe intraorgan and interorgan fusions (Figs. 1 and 3). It has been reported that CUC1 and CUC2 redundantly regulate CMM formation (Kamiuchi et al., 2014).
As for the anther, a host of genes have been known to be involved in its development (Ma, 2005; Egger and Walbot, 2016). Among them, only a few genes function in specifying archesporial cell fate and/or delineating archesporial and somatic cells. To date, the SPOROCYTELESS/NOZZLE (SPL/NZZ) and BARELY ANY MERISTEM (BAM1 and BAM2) genes are known to play pivotal roles in the specification processes, acting as positive and negative regulators, respectively (Schiefthaler et al., 1999; Yang et al., 1999; Hord et al., 2006). SPL/NZZ and BAMs encode a MADS transcription factor and Leu-rich repeat-receptor-like kinases, respectively. However, little is known about how these proteins cause the specification of archesporial cells in cellular and molecular terms. The spl mutant anther consists of only somatic connective cells, producing no archesporial cells, as did gif1/2 35S:MIR396, indicating that the GRF-GIF duo is a novel positive regulator in specifying archesporial cells. This notion is in line with the fact that most GRFs and GIFs are actively expressed in the anther primordium (Fig. 4; Lee et al., 2014). Therefore, it is conceivable that future studies on the genetic and molecular interaction between the GRF-GIF duo and SPL/NZZ would shed light on the cellular and molecular processes involved in the specification processes.
The GRF-GIF Duo May Be Tightly Associated with the Auxin Signaling Network
Lack of the GRF-GIF duo led to aberrant gynoecia with neither CMMs nor ovary valves but only with expanded abaxial medial tissues, repla, at the expense of valves (Figs. 1–3), which is indicative of defects in the establishment of mediolateral patterning. These are typical phenotypes caused by mutations of auxin-related genes, as mentioned earlier, and are shown in the detailed histological analyses of pid-14 in this study. It has been proposed that auxin flows occurring in the lateral domains of the gynoecial primordium exert a dual action: promoting the outgrowth of the lateral domains but preventing them from obtaining medial domain identity (Larsson et al., 2014; Marsch-Martínez and de Folter, 2016). This concept may explain why mutations of auxin-related genes and NPA treatment result in a lack of lateral carpel tissues and the expansion of medial repla. In this regard, the gynoecial phenotypes of gif 35S:MIR396b also may be a manifestation of failures in promoting the outgrowth of the lateral domains and in preventing them from obtaining medial domain identity. Indeed, gif mutations and 35S:MIR396b synergistically enhanced those weak gynoecial defects of pid-3 and NPA-treated plants (Tables III and IV). It is noteworthy that GIF1/AN3 activates HEC1 expression and that HEC1 activates the expression of PIN1 and PIN3 in the gynoecium (Vercruyssen et al., 2014; Schuster et al., 2015). Furthermore, HEC1 expression is induced by auxin, and SEU and SLKs may facilitate the auxin response to support organ development from meristematic tissues (Bao et al., 2010; Schuster et al., 2015). Taken together, it is conceivable that the action mode of the GRF-GIF duo may be tightly associated with the auxin signaling network. The fact that gif1/2 greatly enhanced the pin-formed stem phenotype of pid-3 mutants is in line with this notion. Elucidation of the convergence points between the action modes of the GRF-GIF duo and auxin should be an important focus in future researches.
CONCLUSION
Our data demonstrate a novel and pivotal role of the GRF-GIF duo in carpel and anther development, drawing attention to its importance in the reproductive competence of Arabidopsis and angiosperms. The genetic interaction between GRF-GIF and PID provides substantial evidence that the action mode of the GRF-GIF duo may be associated with that of auxin. This study also uncovered a part of the unknown factors that should be involved in CMM development, as suggested by Wynn et al. (2011), and provides testable hypotheses with regard to the duo’s action, such as the involvement in chromatin remodeling and genetic interactions with floral identity genes and CUCs. This should help decipher how plants regulate the meristematicity and pluripotency of CMM and archesporial cells.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) Columbia-0 plants were used, and all mutants and transgenic plants were of the same accession, except grf2. Seeds were sown on autoclaved wet soil (Mix5; Sunshine), stratified at 4°C for 3 d, and transferred to a growth room at 23°C under a photoperiod of 16 h of light/8 h of darkness. 35S:MIR396b and GRF2pro:GRF2:GUS (Rodriguez et al., 2010), pid-3 (Bennett et al., 1995), pid-14 (Huang et al., 2010), and KNAT1:GUS-18 (Alonso-Cantabrana et al., 2007) have been described before. For nomenclature and detailed description of grf and gif mutants, see Supplemental Table S1 and Supplemental Figure S2. All mutants were confirmed by PCR-assisted genotyping and phenotyping (for primer sequence information, see Supplemental Table S3). For NPA treatments, flower clusters were sprayed in the morning and afternoon with a 100 µm NPA solution (Sigma-Aldrich) containing 0.01% (v/v) Silwet L-77 and 0.1% (v/v) dimethyl sulfoxide, as described by Nemhauser et al. (2000). Mock treatments were performed only with 0.01% (v/v) Silwet L-77 and 0.1% (v/v) dimethyl sulfoxide.
Quantification of Floral Organs
To determine the numbers of floral organs, we dissected and examined flowers at stage 12 from both the primary and secondary stems. Flower samples of pid-3, pid-14, gif1/2 pid-3, and 35S:MIR396b pid-3 were mostly from secondary branches, because their pin-formed primary stems had a limited capacity to produce flowers, only up to a few.
SEM Analysis
Flower clusters were prepared as described by Lee et al. (2014), and photographs of floral organs were obtained using SEM (S-4300 and EDX-350; Hitachi).
GUS Assay
The GUS staining procedure was performed according to Lee et al. (2014), and photographs were obtained using a light microscope (Eclipse NI-U; Nikon).
Histological Analysis
Flower clusters were fixed in 50% (v/v) ethanol, 10% (v/v) formaldehyde, and 5% (v/v) acetic acid at 4°C overnight and dehydrated using an ethanol series (50%, 70%, 80%, 90%, 95%, and 100%). The tissues were then embedded using the Technovit 7100 resin kit according to the manufacturer’s instructions (Heraeaus Kulzer). Tissue blocks were sectioned 4 μm in thickness by a microtome (Leica RM2125RT) and stained with 0.1% (w/v) Toluidine Blue O (Sigma-Aldrich).
Construction of GRFpro:GRF:GUS
GRFpro:GRF:GUS constructs were prepared using the In-Fusion Advantage PCR cloning kit (Clontech) according to the manufacturer’s instructions. In brief, genomic DNA from wild-type plants was amplified by PCR using primer pairs (Supplemental Table S3). Amplified DNA fragments included the promoter region (∼2 kb in length), exons, and introns, except the stop codon and 3′ untranslated region. In-Fusion enzymes joined the resulting PCR products and pBI101.1 vectors linearized with HindIII and BamHI to be in frame with GUS. These recombinant plasmids were confirmed by sequencing and introduced into Arabidopsis plants by the Agrobacterium tumefaciens-mediated transformation method (Clough and Bent, 1998). Dozens of independent T1 plants for each construct were selected on Murashige and Skoog agar plates (0.5× Murashige and Skoog salts, 1% (w/v) Suc, 0.8% (w/v) phytoagar, and 50 μg mL−1 kanamycin). Flower clusters of T2 plants were subjected to the GUS staining procedure. All of the transgenic lines for each construct showed similar staining patterns, and a typical pattern was presented.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Structural and developmental features of the Arabidopsis gynoecium and anther.
Supplemental Figure S2. Schematic representation of gene structures and T-DNA insertions.
Supplemental Table S1. Nomenclature of grf and gif mutant alleles.
Supplemental Table S2. Floral phenotypes of grf mutant alleles.
Supplemental Table S3. Primer sequences for PCR amplification.
Acknowledgments
We thank Dr. Javier F. Palatnik (Instituto de Biologia Molecular y Celular de Rosario) for the transgenic plants, 35S:MIR396b and GRF2pro:GRF2:GUS; Dr. David Smyth (Monash University) for pid-3; and Dr. Antonio Marinez-Laborda (Universidad Miguel Hernández) for KNAT1:GUS-18. We also thank the Arabidopsis Biological Resource Center (Ohio State University) for grf and pid-14 mutant seeds.
Footnotes
This work was supported by the Korea Research Foundation Grants (2015R1D1A1A01059934).
Articles can be viewed without a subscription.
References
- Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrándiz C, Martínez-Laborda A (2007) Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134: 2663–2671 [DOI] [PubMed] [Google Scholar]
- Azhakanandam S, Nole-Wilson S, Bao F, Franks RG (2008) SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol 146: 1165–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F, Azhakanandam S, Franks RG (2010) SEUSS and SEUSS-LIKE transcriptional adaptors regulate floral and embryonic development in Arabidopsis. Plant Physiol 152: 821–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8: 505–520 [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Chen C, Wang S, Huang H (2000) LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26: 42–54 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inzé D, Rodriguez RE, Palatnik JF (2014) Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J 79: 413–426 [DOI] [PubMed] [Google Scholar]
- Egger RL, Walbot V (2016) A framework for evaluating developmental defects at the cellular level: an example from ten maize anther mutants using morphological and molecular data. Dev Biol 419: 26–40 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Fourquin C, Prunet N, Scutt CP, Sundberg E, Trehin C, Vialette-Guiraud AC (2010) Carpel development. Adv Bot Res 55: 1–73 [Google Scholar]
- Ferrándiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z (2002) SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263 [DOI] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H (2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galván-Ampudia CS, Offringa R (2010) Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kamiuchi Y, Yamamoto K, Furutani M, Tasaka M, Aida M (2014) The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front Plant Sci 5: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Kim JH, Tsukaya H (2015) Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot 66: 6093–6107 [DOI] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Kidokoro S, Maruyama K, Nakajima J, Nakashima K, Mitsuda N, Takiguchi Y, Ohme-Takagi M, Kondou Y, et al. (2012) Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24: 3393–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Roberts CJ, Claes AR, Franks RG, Sundberg E (2014) Polar auxin transport is essential for medial versus lateral tissue specification and vascular-mediated valve outgrowth in Arabidopsis gynoecia. Plant Physiol 166: 1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Jeon JO, Lee MM, Kim JH (2015) Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ seperation. Plant Signal Behav 10: e98807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH (2009) The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol 151: 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Wynn AN, Franks RG, Hwang YS, Lim J, Kim JH (2014) The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev Biol 386: 12–24 [DOI] [PubMed] [Google Scholar]
- Lee JE, Golz JF (2012) Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal Behav 7: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu D (2014) Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol 164: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D (2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Marsch-Martínez N, de Folter S (2016) Hormonal control of the development of the gynoecium. Curr Opin Plant Biol 29: 104–114 [DOI] [PubMed] [Google Scholar]
- Nelissen H, Eeckhout D, Demuynck K, Persiau G, Walton A, van Bel M, Vervoort M, Candaele J, De Block J, Aesaert S, et al. (2015) Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC (2000) Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888 [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA (2006) AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol 141: 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B (2015) Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Mol Plant 8: 998–1010 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. (2014) Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol 15: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zuñiga-Mayo VM, Chávez Montes RA, Marsch-Martínez N, de Folter S (2013) Inside the gynoecium: at the carpel margin. Trends Plant Sci 18: 644–655 [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C, Gaillochet C, Lohmann JU (2015) Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142: 3343–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC (1995) Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121: 1519–1532 [DOI] [PubMed] [Google Scholar]
- Sitaraman J, Bui M, Liu Z (2008) LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol 147: 672–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101: 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Stahle MI, Kuehlich J, Staron L, von Arnim AG, Golz JF (2009) YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Taylor RS, Tarver JE, Hiscock SJ, Donoghue PCJ (2014) Evolutionary history of plant microRNAs. Trends Plant Sci 19: 175–182 [DOI] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn AN, Rueschhoff EE, Franks RG (2011) Transcriptomic characterization of a synergistic genetic interaction during carpel margin meristem development in Arabidopsis thaliana. PLoS ONE 6: e26231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]