Stomatal density can be increased in rice by expanding the expression domain of SHORTROOT, but contrary to theoretical predictions, enhanced stomatal numbers have no impact on photosynthetic capacity.
Abstract
The coordinated positioning of veins, mesophyll cells, and stomata across a leaf is crucial for efficient gas exchange and transpiration and, therefore, for overall function. In monocot leaves, stomatal cell files are positioned at the flanks of underlying longitudinal leaf veins, rather than directly above or below. This pattern suggests either that stomatal formation is inhibited in epidermal cells directly in contact with the vein or that specification is induced in cell files beyond the vein. The SHORTROOT pathway specifies distinct cell types around the vasculature in subepidermal layers of both root and shoots, with cell type identity determined by distance from the vein. To test whether the pathway has the potential to similarly pattern epidermal cell types, we expanded the expression domain of the rice (Oryza sativa ssp japonica) OsSHR2 gene, which we show is restricted to developing leaf veins, to include bundle sheath cells encircling the vein. In transgenic lines, which were generated using the orthologous ZmSHR1 gene to avoid potential silencing of OsSHR2, stomatal cell files were observed both in the normal position and in more distant positions from the vein. Contrary to theoretical predictions, and to phenotypes observed in eudicot leaves, the increase in stomatal density did not enhance photosynthetic capacity or increase mesophyll cell density. Collectively, these results suggest that the SHORTROOT pathway may coordinate the positioning of veins and stomata in monocot leaves and that distinct mechanisms may operate in monocot and eudicot leaves to coordinate stomatal patterning with the development of underlying mesophyll cells.
The coordinated differentiation of cell types within an organ is a crucial component of morphogenesis, necessary to ensure that the final form is appropriate for function. In this regard, photosynthetic function in plant leaves requires that chloroplast-containing cells in the middle leaf layers are interspersed with veins (to supply water and to redistribute metabolites) and are overlaid with stomatal pores through which carbon dioxide can enter the leaf. In grass leaves, cellular arrangements are defined by parallel longitudinal veins that extend from the base of the leaf sheath to the tip of the leaf blade, with short transverse veins interconnecting the longitudinal network (Sharman, 1942; Esau, 1943; Nelson and Dengler, 1997). This vascular framework underpins linear files of stomata in the epidermis, with each vein being flanked by one to three rows of stomata on both the medial and lateral sides (Stebbins and Shah, 1960). The genetic mechanisms that ensure optimal photosynthetic capacity in grass leaves, by coordinating the development of veins, photosynthetic cell types, and stomata, are not known.
Grass leaves develop basipetally such that cellular differentiation proceeds from the tip of the leaf to the base, with cell divisions gradually becoming restricted to the base of the leaf blade (Sharman, 1942; Sylvester et al., 1990). In maize (Zea mays), the procambial centers that will form leaf veins are specified in subepidermal cell layers before cell divisions become restricted to the leaf base (Sharman, 1942; Russell and Evert, 1985; Sylvester et al., 1990), but the epidermal cell files that will form stomata only become distinguishable after that stage, when increased mitotic activity leads to smaller cells than in adjacent epidermal cell files (Stebbins and Shah, 1960). Although divisions in stomatal cell files normally increase cell numbers within rows rather than contribute to adjacent cell files, cell lineage analysis in maize revealed that cells within a stomatal row are not necessarily clonally related (Hernandez et al., 1999). As such, positional signals must trigger the specification of stomatal cell fate. Because stomatal cell files are specified in positions flanking procambial centers, it is conceivable that positional signals emanating from the developing vein either inhibit stomatal specification in epidermal cells directly above and below the procambium or induce specification in epidermal cell files at the flanks of the procambial center.
In rice (Oryza sativa ssp japonica), maize, and the model grass Brachypodium distachyon, stomatal identity and differentiation appear to be regulated by the same group of basic helix-loop-helix (bHLH) transcription factors that operate in the well-established pathway that was discovered in the eudicot Arabidopsis (Arabidopsis thaliana; Liu et al., 2009; Raissig et al., 2016, 2017). However, orthologous genes have been adopted into different roles in the two patterning networks. In B. distachyon, the combined activity of the bHLH proteins SPEECHLESS (BdSPCH1 and BdSPCH2) and INDUCER OF CBF EXPRESSION1 (BdICE) leads to the specification of stomatal fate, with loss-of-function mutants in either BdSPCH1/BdSPCH2 or BdICE lacking the characteristically spaced files of small epidermal cells at the leaf base that are the first visible indication of stomatal specification (Raissig et al., 2016). In considering what might induce stomatal specification in those cell files, we reflected on known patterning mechanisms that operate around the vasculature. Of note is the SHORTROOT (SHR) pathway, which acts to radially pattern cells around veins in Arabidopsis (Levesque et al., 2006; Cui et al., 2014). AtSHR is expressed specifically in the vasculature, but the protein, which is a member of the GRAS transcription factor family, moves outward to surrounding cell layers, where it interacts with a second GRAS family member, SCARECROW (SCR). SHR-SCR interactions specify cells as endodermis in the root (Helariutta et al., 2000) or bundle sheath in the shoot (Wysocka-Diller et al., 2000; Cui et al., 2014). Given that stomatal patterning is coordinated with vascular patterning, and that both SHR and SCR are expressed during stomatal development in rice (Kamiya et al., 2003), we speculated that the SHR pathway might act to induce the bHLH pathway in epidermal cell files that flank leaf veins and, thus, to regulate the spacing and localization of stomatal rows.
Two SHR genes are present in the rice genome (OsSHR1 [Os07g39820] and OsSHR2 [Os03g31880]; Cui et al., 2007). In situ hybridization analyses demonstrated that OsSHR1 was expressed uniformly throughout the epidermis at the base of young leaf primordia, whereas the expression of OsSHR2 was undetectable in either shoots or roots (Kamiya et al., 2003). This difference in expression level also was seen in recent transcriptomic data sets, where OsSHR1 transcript levels were three times higher than those of OsSHR2 (van Campen et al., 2016). Notably, at the same stage that OsSHR1 transcripts accumulate uniformly in the epidermis, OsSCR1 transcripts are localized specifically in the files of small epidermal cells that will become stomatal rows (Kamiya et al., 2003). Later in development, once the asymmetric cell divisions associated with the specification of guard mother cells and subsidiary cells have occurred, both OsSHR1 and OsSCR1 transcripts are localized in individual stomatal complexes within stomatal cell files (Kamiya et al., 2003). This progressive spatial restriction is consistent with a role for OsSHR1-OsSCR1 interactions in self-refining domains of activity during stomatal development, as seen during radial root patterning of Arabidopsis (Helariutta et al., 2000). However, there is no evidence that OsSHR1 is expressed in the vasculature and, thus, no indication of a link between leaf veins and the specification of stomatal files in the epidermis. The expression domain of OsSHR2 has yet to be resolved.
To dissect the mechanisms that coordinate vein formation, stomatal patterning, and photosynthetic capacity in grass leaves, we first tested whether SHR activity can influence stomatal development. We show that OsSHR2 transcripts preferentially accumulate in developing vascular tissue of rice leaf primordia and, through a gain-of-function experiment, demonstrate that ectopic SHR activity in bundle sheath cells that surround leaf veins is sufficient to induce supernumerary stomatal rows in the epidermis. Despite theoretical predictions that the observed increase in stomatal density would improve photosynthetic capacity, no such gains were observed. Furthermore, in contrast to the phenotypes observed in Arabidopsis leaves with altered stomatal patterning (Dow et al., 2017), there was no increase in mesophyll cell density or in leaf thickness. These results illustrate the complexity of feedback mechanisms that coordinate the development of cell types in different layers of the leaf and point to distinct mechanisms operating in monocots and eudicots.
RESULTS
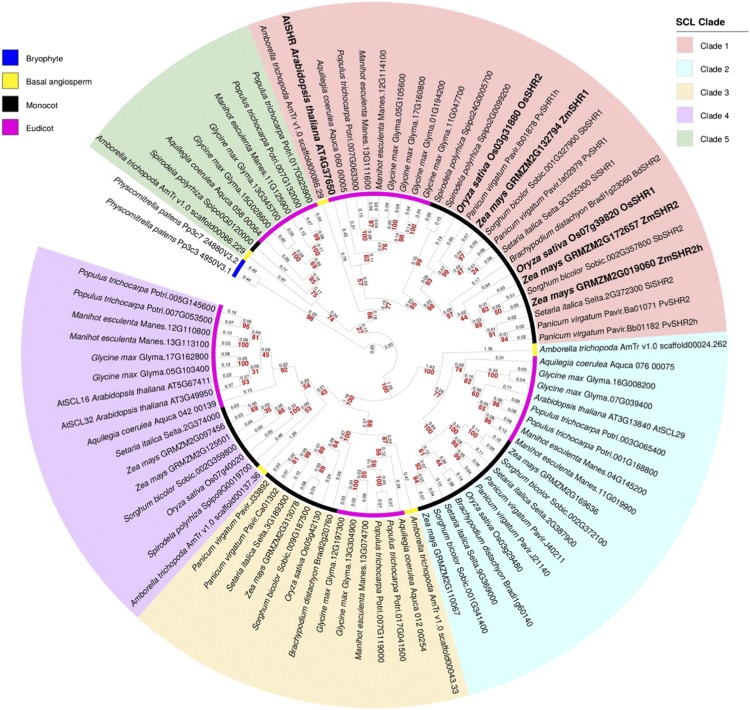
Duplicate SHR Genes in Grasses
Before testing the hypothesis that stomatal patterning in grass leaves is regulated by SHR activity in developing veins, the evolutionary relationships between AtSHR and related genes in the grasses were determined. Phylogenetic analysis was carried out with sequences from a broad range of land plant taxa, including five eudicot species: Arabidopsis, Glycine max, Manihot esculenta, Populus trichocarpa, and Aquilegia coerulea, the latter being a species that diverged early in the eudicot clade. Seven monocot species also were sampled: maize, rice, Sorghum bicolor, Setaria italica, Panicum virgatum, B. distachyon, and Spirodela polyrhiza. To improve tree resolution, sequences were included from Amborella trichopoda (an angiosperm that diverged before the dicot-monocot split), and the tree was rooted with the bryophyte Physcomitrella patens as the outgroup. Figure 1 reveals five angiosperm gene clades, each of which contains genes from monocots, eudicots, and A. trichopoda. Within the AtSHR clade (Fig. 1, clade 1), there is evidence of a gene duplication prior to speciation in the grasses and further within-species duplications in maize and P. virgatum that are consistent with known whole-genome duplications (Schnable et al., 2009; Okada et al., 2010). In maize, ZmSHR2 and its homeolog ZmSHR2h have both been retained in the genome, whereas only one copy of ZmSHR1 is present. The published nomenclature for these genes is somewhat confusing, because the ZmSHR1 sequence (GRMZM2G132794; Slewinski et al., 2014) is orthologous to OsSHR2 (Os03g31880), and the homeologs ZmSHR2 (GRMZM2G172657) and ZmSHR2h (GRMZM2G019060) are orthologous to OsSHR1 (Os07g39820). All five maize and rice SHR genes in clade 1 are equally related to AtSHR.
Figure 1.
Phylogenetic tree of SHR-like proteins. Maximum-likelihood inference was determined for phylogenetic relationships between SHR proteins in a basal angiosperm, six monocot, and five eudicot species, rooted with sequence from the bryophyte P. patens. Five distinct SCL clades are resolved. Branch lengths measured in number of amino acid substitutions per site are indicated in black, and bootstrap values for each node are shown in red. Homeologous relationships in monocot SHR proteins are indicated with h. Arabidopsis (AtSHR), rice (OsSHR1 and OsSHR2), and maize (ZmSHR1, ZmSHR2, and homeolog ZmSHR2h) proteins are highlighted in boldface.
OsSHR2 Is Expressed in Developing Leaf Veins at the Time When Stomata Are Specified in the Overlying Epidermis
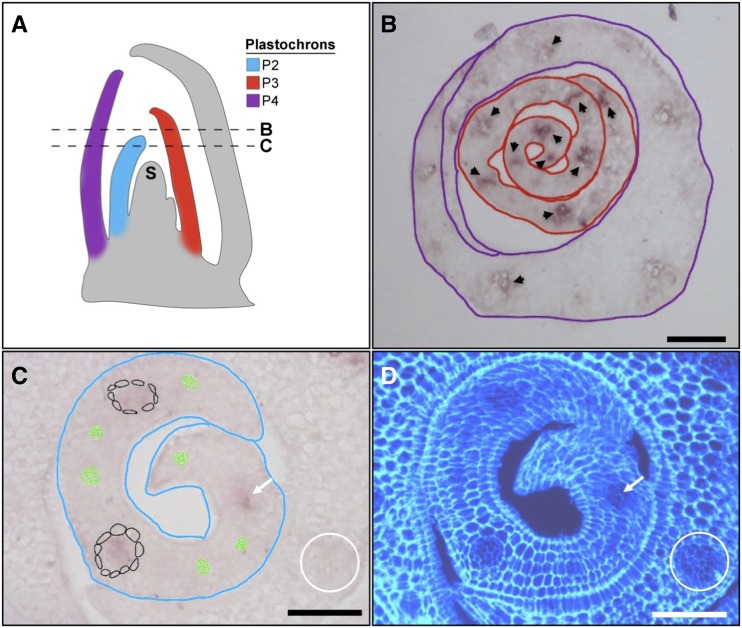
Although the SHR pathway is known to pattern cell types around both shoot and root vascular tissue in Arabidopsis, OsSHR1 is not expressed in developing leaf veins of rice (Kamiya et al., 2003). To determine the spatial expression domain of OsSHR2, a gene-specific fragment was hybridized to developing leaf primordia. Figure 2 shows that OsSHR2 transcripts accumulate in developing veins of plastochron P2 to P4 rice leaf primordia (where a plastochron is the time interval between the initiation of leaf primordia, and P1 is the youngest and closest to the shoot meristem from which the leaves are derived). In contrast to Arabidopsis, where AtSHR transcripts accumulate in procambium and persist throughout vascular development (Dhondt et al., 2010; Gardiner et al., 2011; Cui et al., 2014), OsSHR2 transcripts are detectable only in veins after the surrounding bundle sheath cells have differentiated, and then expression is relatively short-lived (Fig. 2C). Notably, the time window of OsSHR2 expression encompasses P3, which is when files of small cells that will become stomata are first visible in the epidermis (van Campen et al., 2016). The timing of OsSHR2 transcript accumulation in leaf veins, therefore, precludes a role in procambium formation but is consistent with a subsequent role in cellular differentiation, including the specification of stomatal cell files in the overlying epidermis.
Figure 2.
OsSHR2 transcripts accumulate in developing leaf veins. A, Schematic of a monocot shoot apex in longitudinal cross section showing the shoot apical meristem (S) and plastochron (P) leaf primordia. P2, P3, and P4 are color coded as indicated. Relative positions of the cross sections shown in B and C are indicated with dashed lines. B, Transverse cross section showing P3 (red outline) and P4 (purple outline) primordia hybridized in situ with a gene-specific fragment of OsSHR2. Transcripts are visible in developing veins of the P3 primordium (arrowheads) but are much less abundant by P4. C, Transverse cross section showing P2 primordium (blue outline) hybridized in situ with a gene-specific fragment of OsSHR2. Transcripts are detected in well-developed veins that are surrounded by bundle sheath cells (outlined in black or indicated by the white arrow) but not in procambium (outlined in green) or in older veins in the P3 primordium (white circle). D, Same section as in C counterstained with Calcofluor White and photographed under UV light to view cellular structure. Bars = 50 μm.
Ectopic Expression of ZmSHR1 Leads to the Formation of Extra Stomatal Rows in the Rice Epidermis
To determine whether SHR activity in subepidermal leaf layers has the potential to pattern stomata in the epidermis, transgenic rice lines were generated in which the temporal and spatial domains of SHR activity were expanded. Protein alignments gave no indication of substantial differences between rice and maize proteins in known functional domains (Supplemental Data Set S1); therefore, the orthologous ZmSHR1 gene was used as the transgene to avoid potential silencing of OsSHR2. ZmSHR1 was expressed either from the ZmSHR1 promoter or from a promoter that is preferentially active in bundle sheath cells surrounding the vasculature. The rationale behind this approach was that if endogenous OsSHR2 function in the developing vein specifies the position of stomatal rows on the flanks of the vein, expanding activity to cells encircling the vein should add extra rows of stomata in the epidermis.
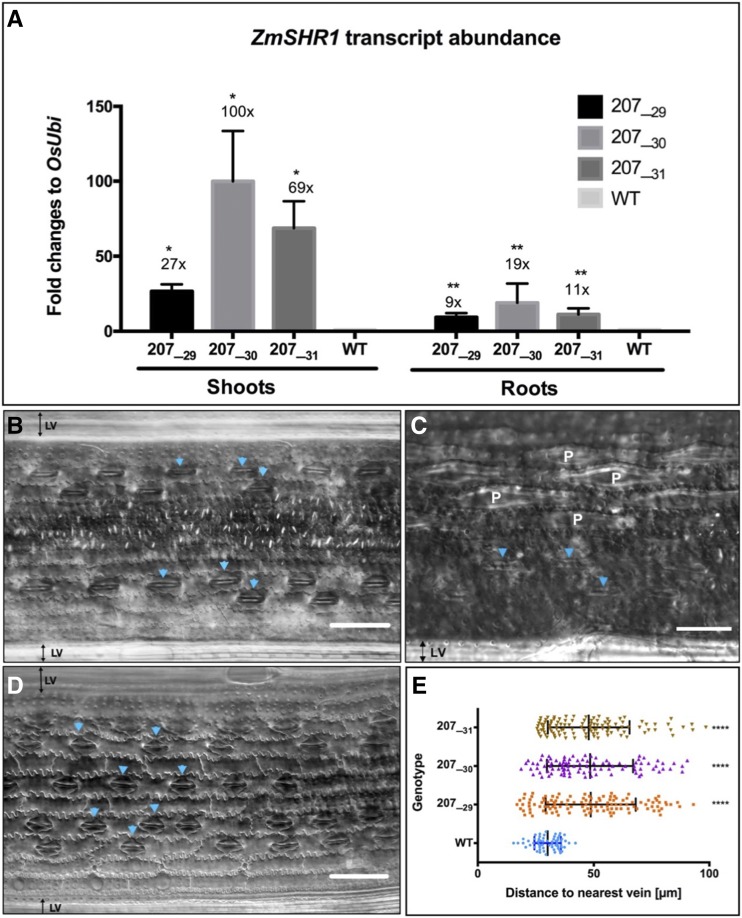
Transgenic lines were generated by fusing the coding sequence of ZmSHR1 either to the native maize promoter (ZmSHR1pro) or to the Zoysia japonica phosphoenolpyruvate carboxykinase promoter (ZjPCKpro) and transforming the constructs into the rice cv Kitaake. Although ZmSHR1 accumulates preferentially in bundle sheath cells of maize leaves (Tausta et al., 2014), the spatial domain of ZmSHR1pro activity in rice has not been determined, whereas ZjPCKpro has been shown previously to direct gene expression in veins and mature bundle sheath cells of rice leaves and in the vasculature of rice roots (Nomura et al., 2005). Transgenic lines were recovered from transformations with three different constructs (ZmSHR1pro:ZmSHR1, a translational reporter fusion [ZmSHR1pro:mTurquoise::ZmSHR1], and ZjPCKpro:ZmSHR1). In all three cases, supernumerary stomatal files were observed in the leaves of T0 regenerants (Supplemental Fig. S1), demonstrating that ectopic SHR activity is sufficient to perturb stomatal patterning in the rice epidermis. Due to very low fertility in ZmSHR1pro:ZmSHR1 and ZmSHR1pro:mTurquoise::ZmSHR1 lines and given the known fidelity of ZjPCKpro for bundle sheath preferential activity in rice (Nomura et al., 2005), the ZjPCKpro:ZmSHR1 line was advanced for further phenotypic analysis. Three T1 individuals (207_29, 207_30, and 207_31) were propagated into the T2 generation, and then homozygous T2 lines (207_29H, 207_30K, and 207_31J) that encompassed a range of transgene expression levels (from 27- to 100-fold above background in the shoot) were selected for all subsequent analyses. Figure 3A shows that ZmSHR1 transcripts accumulated in both shoot and root tissues of all three T2 lines, with levels relative to endogenous ubiquitin being highest in line 207_30 and levels in all lines being higher in shoots than in roots.
Figure 3.
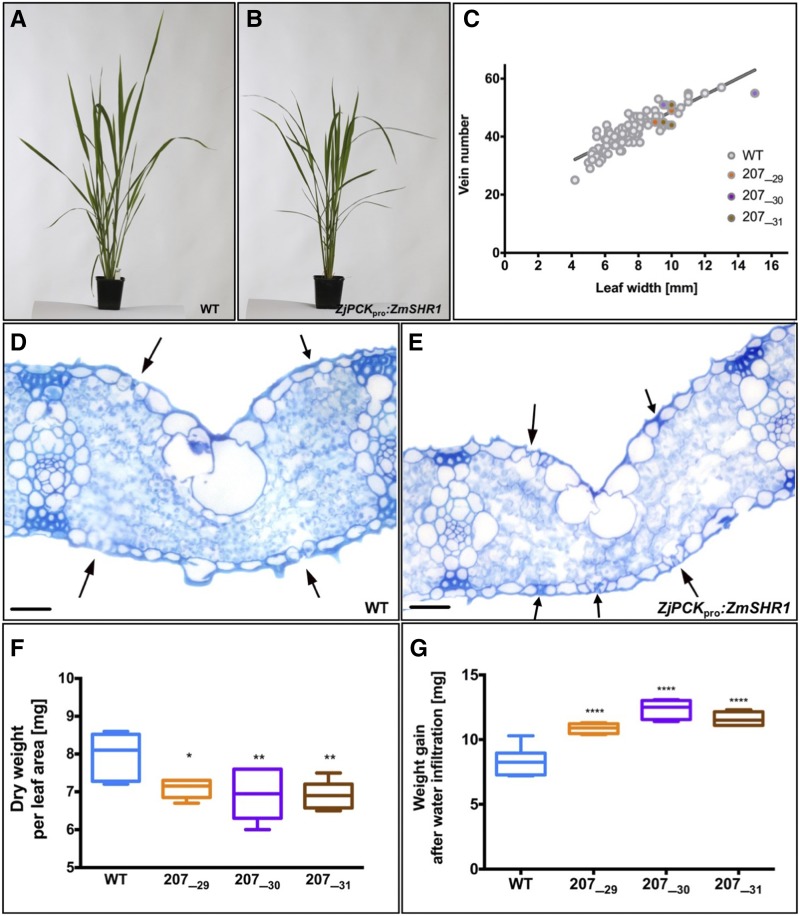
Ectopic ZmSHR1 expression perturbs stomatal patterning in rice. A, Bar charts representing ZmSHR1 transcript abundance in shoots and roots of wild-type (WT) cv Kitaake plus three T2 transgenic lines (207_29H, 207_30K, and 207_31J). Expression was quantified by quantitative reverse transcription-PCR. Transcript levels were normalized to endogenous rice ubiquitin (OsUbi) transcripts and are presented as fold changes above background (wild-type levels are set to 1). Values represent means of two biological replicates, and error bars represent se. *, P < 0.02 and **, P < 0.004 as determined by a two-tailed Student’s t test. B to D, Paradermal views of the abaxial epidermis of cleared wild-type (B) and ZjPCKpro:ZmSHR1 (D) leaves visualized by DIC microscopy. A magnified view (C) illustrates papillae cells (denoted as P) in the wild type. Arrowheads point to stomata. LV, Lateral vein. Bars = 50 µm (B and D) and 75 µm (C). E, Box plot representing measured distances between stomata and the nearest vein in leaves of the wild type and three ZjPCKpro:ZmSHR1 T2 lines. Each data point represents a single stomatal complex. P values were determined by one-way ANOVA where transgenics were individually compared with the wild type: ****, P = 0.0001.
To determine the extent to which the expression of the ZjPCKpro:ZmSHR1 transgene perturbs stomatal patterning, both adaxial and abaxial epidermal layers of transgenic and wild-type plants were cleared and analyzed by differential interference contrast (DIC) microscopy. In wild-type leaves, stomata are organized in one or two linear cell files close to both large and small veins. Given the distance between veins, this pattern leads to three or four stomatal cell files plus at least three or four nonstomatal files between each vein pair (Fig. 3, B and C). In leaves of the ZjPCKpro:ZmSHR1 plants, stomatal rows were more evenly distributed between veins in both the adaxial and abaxial (Fig. 3D) epidermis. In the abaxial epidermis, the extra stomatal files replaced the epidermal papillae that are usually present on the wild-type epidermis (Fig. 3, B–D). To quantify the observed changes, the distance between individual stomata and the nearest vein was measured. Figure 3E illustrates how stomatal position varies from 20 to 100 μm from a vein in the transgenic lines, as opposed to a range of just 20 to 40 μm in the wild type.
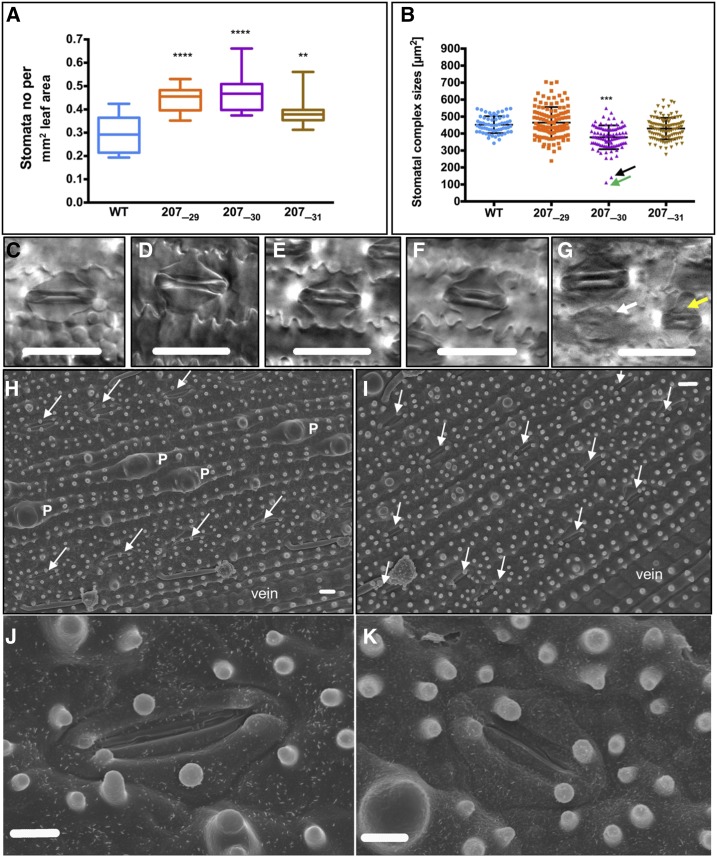
To assess whether the ectopic positioning of stomata also reflects an increase in overall stomatal number across the leaf, stomatal density (stomata per mm2) was measured. Figure 4A shows that stomatal density was increased significantly in all three ZjPCKpro:ZmSHR1 lines as compared with the wild type. Because stomatal density has been shown previously to be negatively correlated with stomatal size (Ohsumi et al., 2007; Franks et al., 2009; Franks and Beerling, 2009), measurements of stomatal size also were taken. A significant decrease in size was observed only for line 207_30 (Fig. 4, B and G), the line that had the highest levels of transgene expression (Fig. 3A) and the highest stomatal density (Fig. 4A). Other than the size, stomatal anatomy in all three transgenic lines was essentially indistinguishable from that of the wild type (Fig. 4, C–F and H–K). No consistent difference in the number of stomata per row could be detected. Together, these results demonstrate that the additional stomatal rows result from the formation of supernumerary stomata in the epidermis and that, to some extent, this increase in number is associated with a decrease in stomatal size.
Figure 4.
Increased stomatal density is not associated with altered stomatal anatomy. A, Box plot showing stomata number measured per mm2 of leaf area of the wild type (WT) and three ZjPCKpro:ZmSHR1 T2 lines. B, Box plot of stomatal complex sizes calculated from stomatal height multiplied by stomatal length. Error bars = 50 µm. All P values were determined by one-way ANOVA where transgenics were individually compared with the wild type: ****, P = 0.0001; ***, P < 0.001 and **, P < 0.01. C to F, Representative examples of stomatal morphology in the wild type (C), 207_29 (D), 207_30 (E), and 207_31 (F) lines photographed by DIC microscopy. G, Morphology of the smallest complexes in line 207_30. The white arrow corresponds to the data point indicated by the black arrow in B, and the yellow arrow corresponds to the green arrow in B. H and I, Positive imprints of the abaxial epidermis of wild-type (H) and ZjPCKpro:ZmSHR1 line 207_31 (I) leaves visualized by scanning electron microscopy. Papillae cells are indicated as P, arrows point to stomata, and veins are indicated. J and K, Representative examples of stomatal morphology in wild-type (J) and 207_31 (K) lines photographed by scanning electron microscopy. Bars = 30 µm (C–G), 10 µm (H and I), and 5 µm (J and K).
The Development of Supernumerary Stomata Has No Significant Impact on General Shoot Morphology or Overall Leaf Anatomy
To determine whether the presence of extra stomata on the leaf surface had any impact on general shoot development in rice, overall plant morphology was examined. Plant stature, tiller number, growth rate, and flowering time were similar in both wild-type (Fig. 5A) and ZjPCKpro:ZmSHR1 transgenic (Fig. 5B) lines, suggesting that neither ectopic ZmSHR1 activity nor the presence of supernumerary stomatal files caused any gross alterations in shoot anatomy. To confirm this observation, particularly given that loss-of-function mutations in shr1 in maize are reported to perturb vein spacing and the specification of bundle sheath cells (Slewinski et al., 2014), a range of leaf anatomy traits also were quantified. Table I shows measurements of leaf width, leaf length, total number of longitudinal veins, bundle sheath cell size, cell number, and distance between veins for four individuals of each T2 line and the wild type. ANOVA revealed no significant differences between wild-type and transgenic lines for any of these traits. In addition, the ratio of leaf width to vein number was normal in transgenic lines (Fig. 5C). Histological examination of leaf sections similarly demonstrated normal patterning of subepidermal layers (Fig. 5, D and E), with no significant difference observed between wild-type and transgenic lines in mesophyll cell size, mesophyll cell number between veins, or leaf thickness (Table II). In lines 207_30 and 207_31, a significant difference was observed in the number of cell layers present between abaxial and adaxial epidermal layers. However, given equivalent leaf thickness in the wild type and all three transgenic lines, the reduced number of cell layers must have been compensated for by subtle differences in cell size, cell number, and orientation (Table II). Notably, lines 207_30 and 207_31 did not have significantly higher stomatal densities than line 207_29 (Fig. 4A), but they did have much higher transgene expression levels (Fig. 3A). As such, the effect on mesophyll cell layers could be a direct consequence of ectopic SHR activity rather than an effect of stomatal density. Extra stomata in all of the transgenic lines were generally associated with extra substomatal air spaces, as determined by loss of leaf weight after drying and gain of leaf weight after vacuum infiltration of water (Byott, 1976). When compared with the wild type, transgenic leaves were clearly lighter after drying (Fig. 5F) and heavier after water infiltration (Fig. 5G), suggesting the presence of more air spaces in leaves of the transgenic lines. Together, these data suggest that ZmSHR1-mediated induction of supernumerary stomata in rice does not perturb general shoot morphology or the overall leaf anatomy.
Figure 5.
Expression of ZmSHR1 in rice does not perturb overall shoot morphology or leaf anatomy. A and B, Representative phenotypes of 6-week-old wild-type (WT; A) and transgenic (B) soil-grown plants. C, Regression plot showing vein number versus leaf width for four individuals from three T2 lines. Gray dots represent wild-type measurements from Table I combined with measurements of wild-type leaves at various developmental stages (Supplemental Data Set S2. D and E), Transverse sections of penultimate leaves from representative wild-type (D) and ZjPCKpro:ZmSHR1 (E) plants. Arrows indicate stomata. Bars = 500 µm. F and G, Dry weight (F) and weight gain after water infiltration (G) for leaves of the wild type and three ZjPCKpro:ZmSHR1 T2 lines. All P values were determined by one-way ANOVA where transgenics were individually compared with the wild type: ****, P = 0.0001; **, P < 0.01; and *, P < 0.1.
Table I. Quantification of leaf anatomy traits.
Measurements were taken from the sixth fully expanded adult leaf on the first tiller for four individuals from each line. For distance and cell number between veins, measurements were taken from three or four pairs of intermediate veins for each leaf. Bundle sheath cell size measurements were taken from six cells in each leaf. Values represent means ± se. Significance was tested using a one-way ANOVA (P < 0.05). There were no significant differences between the three transgenic lines and the wild type (WT) for any of the measurements.
| Line | Leaf Width | Leaf Length | Total No. of Veins | Average Distances between Veins (n = 3) | Average Cell No. between Veins (n = 3–4) | Average Bundle Sheath Cell Size (n = 6) | Vein No. per mm of Leaf Area |
|---|---|---|---|---|---|---|---|
| mm | µm | µm | |||||
| WT1a | 11 | 465 | 52 | 212 ± 6.7 | 6.5 ± 0.6 | 9.9 ± 1.7 | 4.7 |
| WT1b | 11 | 435 | 54 | 185 ± 2.9 | 6.3 ± 0.6 | 10.2 ± 3.1 | 4.7 |
| WT1c | 12 | 550 | 55 | 219 ± 4.2 | 6.7 ± 0.6 | 9.8 ± 2.8 | 4.6 |
| WT1d | 13 | 565 | 57 | 213 ± 43.2 | 6 ± 0 | 10 ± 2.4 | 4.4 |
| WT total | 11.9 ± 0.4 | 503.8 ± 31.8 | 54.5 ± 1.0 | 207.3 ± 7.6 | 6.4 ± 0.1 | 10.0 ± 0.1 | 4.6 ± 0.1 |
| 207_29-1a | 10 | 455 | 44 | 150 ± 8.4 | 8.4 ± 0.5 | 8.6 ± 0.2 | 4.4 |
| 207_29-1b | 11 | 478 | 55 | 203 ± 6.3 | 5.2 ± 0.5 | 11.2 ± 3.8 | 5.0 |
| 207_29-1c | 10 | 490 | 49 | 194 ± 8.3 | 6.8 ± 0.5 | 8.3 ± 1.6 | 4.9 |
| 207_29-1d | 9 | 405 | 45 | 199 ± 16.2 | 6.25 ± 0.5 | 9.2 ± 0.9 | 5.0 |
| 207_29 total | 10.0 ± 0.4 | 457.0 ± 18.8 | 48.4 ± 2.5 | 186.4 ± 12.4 | 6.7 ± 0.7 | 9.3 ± 0.7 | 4.8 ± 0.1 |
| 207_30-1a | 10 | 512 | 44 | 215 ± 13.3 | 7.4 ± 1.1 | 8.7 ± 0.3 | 4.4 |
| 207_30-1b | 15 | 595 | 55 | 202 ± 4.2 | 7.3 ± 0.6 | 9.0 ± 1.7 | 3.7 |
| 207_30-1c | 9.5 | 435 | 51 | 194 ± 3.5 | 7.3 ± 0.6 | 10.4 ± 1.7 | 5.4 |
| 207_30-1d | 10 | 443 | 51 | 205 ± 8.1 | 7.6 ± 0.6 | 8 ± 2.2 | 5.1 |
| 207_30 total | 11.1 ± 1.3 | 496.3 ± 37.2 | 50.3 ± 2.3 | 204.1 ± 4.4 | 7.4 ± 0.1 | 9.0 ± 0.5 | 4.6 ± 0.4 |
| 207_31-1a | 10 | 420 | 44 | 187 ± 16.6 | 8 ± 1.0 | 9.4 ± 0.5 | 4.4 |
| 207_31-1b | 9.5 | 579 | 45 | 219 ± 2.5 | 8.6 ± 0.5 | 8.6 ± 1.7 | 4.7 |
| 207_31-1c | 11 | 560 | 52 | 211 ± 5.1 | 7.5 ± 0.5 | 10.1 ± 2.2 | 4.7 |
| 207_31-1d | 10 | 513 | 51 | 180 ± 10.4 | 6.5 ± 0.5 | 9.0 ± 1.9 | 5.1 |
| 207_31 total | 10.1 ± 0.3 | 518 ± 35.5 | 48.0 ± 2.0 | 199.1 ± 9.3 | 7.7 ± 0.4 | 9.2 ± 0.3 | 4.7 ± 0.1 |
Table II. Quantification of mesophyll cell size and layers.
Measurements were taken from hand-cut cross sections of fully expanded adult leaves of different leaf widths. Leaf thickness represents the mean of three measurements taken at three points across the mediolateral axis: at a vein, directly adjacent to a vein, and midway between two veins. To cover the normal range seen in the wild type (WT), leaves with widths between 9 and 13 mm were chosen for measurements. Mesophyll cell sizes represent means of six cell size measurements in one leaf. For cell number between veins, measurements were taken between three or four pairs of intermediate veins for each leaf. Values represent means ± se. Significance was tested using a one-way ANOVA. There was a significantly lower number of mesophyll cell layers between line 20730 and the wild type (***, P < 0.0005) and between line 20731 and the wild type (*, P < 0.05). Leaf thickness, mesophyll cell size, and mesophyll cell number between veins were not significantly different between transgenic lines and the wild type.
| Line | Leaf Width | Leaf Thickness | No. of Mesophyll Cell Layers | Mesophyll Cell Size | Mesophyll Cell No. between Veins |
|---|---|---|---|---|---|
| mm | µm | µm2 | |||
| WT1a | 11 | 91.7 ± 4.3 | 6 | 240.5 ± 35.3 | 8–9 |
| WT1b | 11 | 94.3 ± 5.0 | 5 | 186. 33 ± 45.8 | 8–9 |
| WT1c | 11 | 88.3 ± 2.8 | 5 | 172.7 ± 46 | 8 |
| WT2a | 12 | 88.7 ± 8.8 | 6 | 218.3 ± 20.5 | 7–8 |
| WT2b | 12 | 91 ± 1.0 | 6 | 205.7 ± 30.2 | 7–9 |
| WT2c | 12 | 82 ± 2.6 | 5 | 210.5 ± 25.1 | 7 |
| WT3a | 13 | 85.6 ± 5.6 | 6 | 251.8 ± 33.9 | 7–9 |
| WT3b | 13 | 82.6 ± 2.6 | 5 | 241.8 ± 32.9 | 7–9 |
| WT3c | 13 | 76 ± 5.1 | 5 | 195.3 ± 30.1 | 7–9 |
| WT4a | 10 | 79.7 ± 4.9 | 4 | 205.2 ± 12.4 | 8 |
| WT4b | 10 | 80.7 ± 4.1 | 4 | 229.8 ± 30.9 | 7 |
| WT4c | 10 | 80 ± 5.1 | 3 | 190.7 ± 20.2 | 8 |
| WT5a | 9 | 77.5 ± 4.3 | 4 | 205.8 ± 23.3 | 8 |
| WT5b | 9 | 82 ± 6.0 | 4 | 197.5 ± 37 | 8 |
| WT5c | 9 | 69.3 ± 9.2 | 3 | 172.8 ± 37.2 | 7 |
| WT total | 83.3 ± 1.7 | 4.7 ± 0.3 | 205.3 ± 6.1 | 7–9 | |
| 207_29-1a | 9 | 71.7 ± 6.0 | 5 | 131.2 ± 34.3 | 11 |
| 207_29-1b | 9 | 67 ± 4.0 | 4 | 186.5 ± 43.2 | 8 |
| 207_29-1c | 9 | 78 ± 2.1 | 5 | 229.7 ± 63.7 | 7 |
| 207_29-2a | 10 | 91 ± 7.5 | 4 | 233.3 ± 58.8 | 7 |
| 207_29-2b | 10 | 77 ± 7.1 | 4 | 191.2 ± 16.5 | 7 |
| 207_29-2c | 10 | 74.3 ± 9.7 | 4 | 185.7 ± 41 | 9 |
| 207_29-3a | 12 | 84.7 ± 12.3 | 4 | 224.2 ± 29.9 | 8–9 |
| 207_29-3b | 12 | 90.3 ± 11.8 | 4 | 200.3 ± 40.3 | 8 |
| 207_29-3c | 12 | 84.7 ± 12.9 | 3 | 209 ± 27.9 | 7 |
| 207_29 total | 79.9 ± 2.8 | 4.1 ± 0.2 | 199 ± 10.4 | 7–11 | |
| 207_30-1a | 10 | 78 ± 4.2 | 3 | 261 ± 44.9 | 6–8 |
| 207_30-1b | 10 | 81.3 ± 3.7 | 3 | 188.8 ± 26.3 | 8 |
| 207_30-1c | 10 | 85 ± 5.7 | 3 | 220 ± 48.5 | 7 |
| 207_30-2a | 11 | 79 ± 3.1 | 3 | 200.4 ± 30.6 | 6 |
| 207_30-2b | 11 | 88.3 ± 7.5 | 3 | 210.5 ± 51.8 | 7 |
| 207_30-2c | 11 | 78.7 ± 9.9 | 3 | 251.3 ± 39.8 | 6 |
| 207_30 total | 81.7 ± 1.7 | 3 ± 0*** | 222 ± 11.7 | 6–8 | |
| 207_31-1a | 10 | 80 ± 9.1 | 4 | 158 ± 40.3 | 9 |
| 207_31-1b | 10 | 81.7 ± 4.1 | 4 | 227.3 ± 51.7 | 7 |
| 207_31-1c | 10 | 81.7 ± 3.5 | 3 | 147 ± 10.9 | 8 |
| 207_31-2a | 11.5 | 84.3 ± 3.2 | 4 | 271.7 ± 27.8 | 8 |
| 207_31-2b | 11.5 | 79.7 ± 3.9 | 4 | 230.3 ± 22.5 | 7 |
| 207_31-2c | 11.5 | 74.3 ± 5.2 | 3 | 198.2 ± 28.8 | 8 |
| 207_31 total | 80.3 ± 1.4 | 3.6 ± 0.2* | 205.4 ± 19.3 | 7–9 |
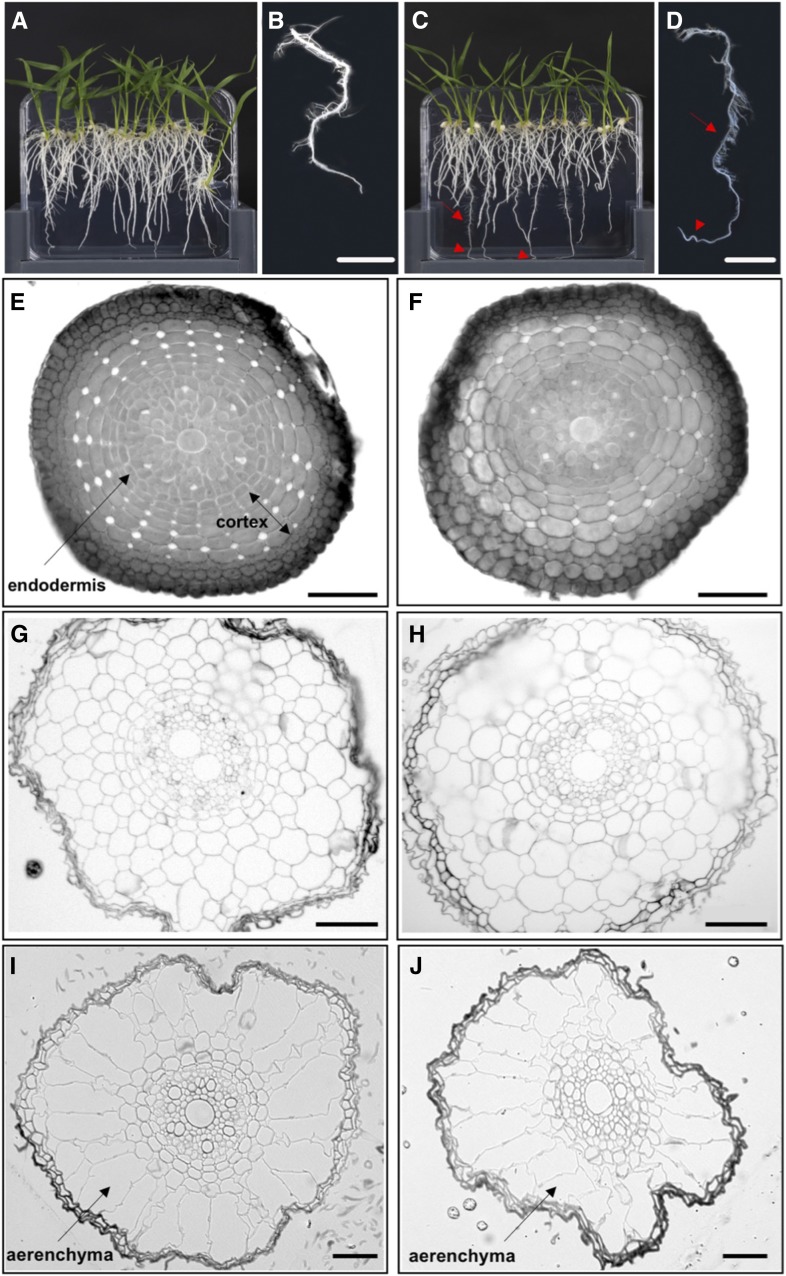
Root Architecture Is Modified in Rice Lines That Express ZmSHR1
In all flowering plant species examined thus far, loss of SHR function in the root perturbs radial patterning of endodermal and cortical cells around the vasculature and leads to shorter roots overall. Consistent with this observation, gain of SHR function in ZjPCKpro:ZmSHR1 transgenic rice plants led to longer roots with finer tips and an increased number of lateral roots compared with the wild type (Fig. 6, A–D). The formation of extra lateral roots is analogous to the role of AtSHR in lateral root formation in Arabidopsis (Lucas et al., 2011). However, the long-root phenotype contrasts with previous gain-of-function experiments in which the constitutive expression of AtSHR or OsSHR2 led to short roots with perturbed radial patterning that was manifest by the presence of extra cortical cell layers (Henry et al., 2017). Transverse sections through zones of wild-type roots (Fig. 6, E, G, and I) and ZjPCKpro:ZmSHR1 transgenic roots (Fig. 6, F, H, and J) revealed that the central vasculature, endodermal layer, and number of cortical layers were the same in both. Collectively, these data demonstrate that ZjPCKpro-driven expression of ZmSHR1 in rice leads to root elongation and the production of more lateral roots, but these perturbations are not accompanied by patterning defects and do not impact shoot development in any obvious way.
Figure 6.
ZjPCKpro:ZmSHR1 transgenic seedlings develop longer roots than the wild type. A to D, Two-week-old wild-type (A and B) and ZjPCKpro:ZmSHR1 (C and D) plants grown on one-half-strength Murashige and Skoog plates, with magnified images of detached wild-type (B) and ZjPCKpro:ZmSHR1 (D) roots. Red arrows indicate fine lateral roots, and red arrowheads point to elongated root tips. E to J, Transverse sections through the root apex (E and F), elongation zone (G and H), and maturation zone (I and J) of 2-week-old wild-type (E, G, and I) and ZjPCKpro:ZmSHR1 (F, H, and J) roots. Bars = 1 cm (B and D) and 50 µm (E–J).
Increased Stomatal Density Predicts a Higher Photosynthetic Capacity in ZjPCKpro:ZmSHR1 Lines, But the Predicted Gains Are Not Realized
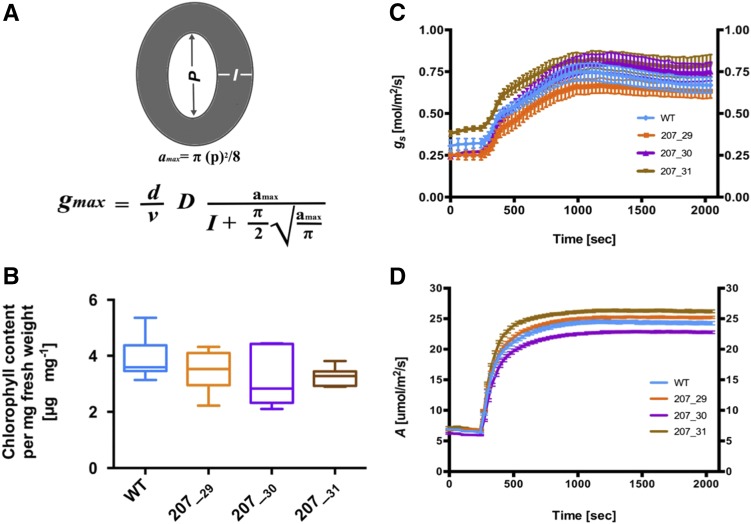
Because stomatal density and the size of the stomatal pore determine stomatal conductance to CO2 and water, and thus strongly correlate with photosynthetic capacity (Fischer et al., 1998; Franks et al., 2009; Franks and Beerling, 2009), it has been hypothesized that increased stomatal density could increase photosynthetic capacity and, hence, crop yield (Jones, 1977, 1987). However, such an approach has had little success in conventional breeding programs. The increased stomatal density observed in transgenic ZjPCKpro:ZmSHR1 lines enabled the impact of varying stomatal numbers to be evaluated both theoretically and experimentally. Measurements of stomatal density and of pore length (µm) and depth (µm; equivalent to guard cell width) can be used to calculate the theoretical maximum rate of photosynthesis (Fig. 7A; Brown and Escombe, 1900; Franks and Farquhar, 2001). Therefore, pore length and depth were measured in leaves of wild-type and ZjPCKpro:ZmSHR1 transgenic lines (Table III). These values were then averaged and used to calculate the theoretical maximum rate of stomatal conductance to water vapor (gmax; Fig. 7A) and, from that, the theoretical maximum stomatal conductance to CO2 (gcmax [by dividing gmax values by 1.6 to account for the differences in molecular diffusion between water vapor and CO2]; Farquhar and Sharkey, 1982). Table III shows that predicted gmax values of ZjPCKpro:ZmSHR1 lines are 25% to 60% higher than those in the wild type and that gcmax values are 20% to 40% higher than those in the wild type. Together, these data suggest the potential for a higher stomatal conductance and, therefore, greater photosynthetic capacity in ZjPCKpro:ZmSHR1 lines as compared with the wild type.
Figure 7.
Increased stomatal number in ZjPCKpro:ZmSHR1 lines does not lead to significant increases in chlorophyll, photosynthetic assimilation rates, or stomatal conductivity. A, Schematic representation of calculations used to determine photosynthetic capacity. Pore length (P; µm), pore depth (I; µm), and stomatal density (D) measurements were used to calculate gmax, which is an integrated measurement of D and the stomatal geometry embedded in a physical gas-diffusion model. P was used to first calculate the mean maximum stomatal pore area (amax; µm2). gmax was then calculated from amax, D, and I. The equation constants are π, approximated to 3.142, d represents the diffusivity of water (24.6 × 10−6 m2 s−1 at 25°C), and ν represents the molar volume of air (24.4 × 10−3 m3 mol−1 at 25°C and 101.3 kPa). B, Box plot representing chlorophyll content in each line (n = 6). C, Temporal response of net gs to an increase in irradiance from 100 to 1,000 µmol m−2 s−1. Data points are means ± se (n = 5–7). D, Temporal response of net A to an increase in irradiance from 100 to 1,000 µmol m−2 s−1. Data points are means ± se (n = 5–7). gs and A were both measured under ambient CO2 levels of 400 µL L−1. Steady-state levels of gs were achieved within 30 to 60 min from an initial PPFD of 100 µmol m−2 s−1. WT, Wild type.
Table III. Stomatal pore measurements.
Mean stomatal measurements D, P, and I (see Fig. 7A) were used to calculate the maximum theoretical rate of gmax and gcmax in the wild type and three independent transgenic lines. Values represent means ± se of stomatal measurements taken from the middle of the penultimate leaf of the first tiller. Asterisks indicate significant differences from the wild type as determined by a one-way ANOVA test: **, P > 0.01; ***, P > 0.001; and ****, P > 0.0001.
| Genotype | D (n = 12) | P (n = 40–48) | I (n = 40–48) | gmax | gcmax |
|---|---|---|---|---|---|
| mm−2 | µm | mol water m−2 s−1 | mol CO2 m−2 s−1 | ||
| Wild type | 0.29 ± 0.1 | 13.11 ± 0.3 | 2.53 ± 0.10 | 1.03 | 0.65 |
| 207_29 | 0.46 ± 0.1**** | 11.22 ± 0.2**** | 1.89 ± 0.04**** | 1.62 | 1.01 |
| 207_30 | 0.48 ± 0.1**** | 11.00 ± 0.2**** | 1.99 ± 0.10**** | 1.68 | 1.05 |
| 207_31 | 0.39 ± 0.1** | 10.90 ± 0.2**** | 2.11 ± 0.10*** | 1.28 | 0.80 |
To investigate whether the theoretically predicted increase in photosynthetic capacity was realized in ZjPCKpro:ZmSHR1 transgenic lines, leaf chlorophyll content was quantified, and gas-exchange experiments were carried out to measure both stomatal conductivity to water vapor (gs) and photosynthetic carbon assimilation rates (A). No significant difference in chlorophyll content was observed between wild-type and transgenic lines (Fig. 7B), suggesting that mesophyll cell chloroplast capacity is normal in the transgenic lines. For gas-exchange experiments, a light intensity of 1,000 µmol m−2 s−1 (photosynthetic photon flux density [PPFD]) was first identified as the lowest PPFD at which the saturation point of CO2 assimilation was reached in one representative ZjPCKpro:ZmSHR1 line and the wild type (Supplemental Fig. S2). To resolve the impact of stomatal distribution on stomatal opening and photosynthetic induction, net gas exchange was measured in leaves during a low (100 µmol m−2 s−1) to high (1,000 µmol m−2 s−1) light transition. There was a rapid and immediate increase in gs (Fig. 7C) and A (Fig. 7D) in response to high light in all measured lines. However, despite the increased gcmax values (Table III), the three transgenic lines showed no increase in either gs (steady-state values or rate of induction; Fig. 7C) or A (Fig. 7D). These results indicate that alterations to stomatal density are physiologically buffered to protect the leaf from water loss and, presumably, to coordinate capacity between veins, photosynthetic cell types, and stomata.
DISCUSSION
The SHR pathway has been characterized primarily in Arabidopsis, where a single gene encodes a mobile protein that moves outward from its expression domain in the vasculature to specify cell fate in surrounding cell layers (Helariutta et al., 2000; Nakajima et al., 2001; Gallagher et al., 2004; Levesque et al., 2006; Cui et al., 2007). Phylogenetic analysis revealed that the presence of a single SHR gene is typical of eudicot genomes, except where within-species genome duplications have occurred (Fig. 1). This contrasts with grasses, where duplicated SHR1 and SHR2 genes are present in all species examined except B. distachyon, where only one member of the orthogroup has been retained (Fig. 1). Transgenic Arabidopsis lines that express OsSHR1, OsSHR2, or BdSHR in the expression domain of AtSHR exhibit supernumerary cortical cell layers in the roots of a wild-type background and a phenotype in the loss-of-function Atshr background that indicates rescue of the mutant phenotype in addition to the supernumerary cortical cell layers (Wu et al., 2014). Together with data showing that all three of the monocot proteins are mobile in Arabidopsis (Wu et al., 2014), these observations suggest that the role of the SHR pathway in patterning cell layers around the root vasculature is conserved in Arabidopsis, rice, and B. distachyon, with the monocot proteins able to move farther from the vasculature than AtSHR.
As in roots, the SHR pathway regulates radial patterning around vascular tissue in the hypocotyl of Arabidopsis (Fukaki et al., 1998; Tasaka et al., 1999), but roles in shoot development have not been well described in monocots, with the only functional insight provided by the Zmshr1 loss-of-function mutant in maize, in which vascular patterning in the leaf is perturbed (Slewinski et al., 2014). Vascular perturbations in Zmshr1 plants are subtle, however, as might be expected given the potential for redundancy with ZmSHR2 and ZmSHR2h. The transgenic rice lines reported here express ZmSHR1 in the bundle sheath cells and mature veins of the leaf in addition to expression of the endogenous ortholog OsSHR2 in developing leaf veins. Despite the spatial and temporal expansion of the expression domain, no defects in vascular patterning were observed, and the specification of cell types around the veins was normal (Fig. 5; Table I). As such, ZmSHR1 expression in bundle sheath cells is not sufficient to alter vascular patterning in rice. There are three possible explanations for this observation. Most likely, by the time the transgene is expressed, the developmental time window for procambium initiation in neighboring cells may have passed. Alternatively, downstream targets and/or interaction partners of ZmSHR1 may not be recognized in rice. Or ZmSHR1 plays no role in leaf venation patterning per se. Although OsSHR2 is expressed in developing veins, the timing of expression (Fig. 2) is consistent with the suggestion that the ZmSHR1/OsSHR2 orthologs are not involved in the initiation of new veins in the leaf but, instead, play a role in specifying cell types either within or around the developing vascular bundle.
Although subepidermal patterning was normal in rice leaves expressing ZmSHR1, stomatal patterning was perturbed (Figs. 3 and 4; Table III). Specifically, expanding the effective expression domain of OsSHR2, through transgenic manipulation of the maize ortholog ZmSHR1, led to the formation of additional stomatal rows at a greater distance from the vein than in wild-type leaves. The normally consistent position of stomatal cell files on the flanks of veins in monocot leaves has been proposed previously to result either from an inhibitory signal transmitted from the developing vein to directly overlying epidermal cells or from an inductive signal transmitted to epidermal cells at a specific distance from the vein (Hernandez et al., 1999). The stomatal phenotype reported here supports an inductive role for the SHR pathway, either through direct action of the mobile SHR protein or indirectly via SHR target molecules (but does not preclude the possibility of an antagonistic inhibitory signal also acting directly over the veins). In an analogous mechanism to that observed in the radial patterning of roots, limited movement of the SHR protein from the site of vascular expression could specify stomatal cell files in the epidermis at the flanks of the vein. In this scenario, expansion of the SHR expression domain to bundle sheath cells surrounding the vein would lead to extra stomatal rows positioned farther away from the vein than normal.
A number of genes act downstream of SHR during root development, with SCR being the best characterized (Helariutta et al., 2000; Nakajima et al., 2001; Levesque et al., 2006). The distinct expression pattern of OsSCR during guard cell and subsidiary cell formation in the rice epidermis predicts a further role for SCR in stomatal formation (Kamiya et al., 2003). However, ectopic expression of ZmSCR1 in rice does not perturb stomatal density or patterning, at least when expressed constitutively and at high levels (Wang et al., 2017). Therefore, stomatal specification must be mediated by interactions with alternative downstream targets, the most likely candidates being homologs of BdSPCH and BdICE, which have been shown to act in the earliest stages of specification in B. distachyon (Raissig et al., 2016). The role of OsSCR may be limited to later stages in the process, where asymmetric cell divisions are needed to form the stomatal complex (Kamiya et al., 2003). Clearly, further analyses are needed to determine exactly how the SHR pathway integrates with the canonical bHLH pathway that regulates stomatal differentiation.
Many aspects of the mechanisms that coordinate stomatal patterning with leaf venation and leaf gas exchange remain unresolved, particularly in crop plants. In part, this paucity of information is because appropriate lines for phenotypic analyses have been unavailable. The transgenic rice lines generated in this work allowed an initial investigation into the interrelationship between stomatal patterning and photosynthetic capacity in a monocot. Our results showed that, despite predicted increases in photosynthetic capacity through increased gmax (Table III), actual rates of gs and photosynthesis were indistinguishable between the transgenic lines and the wild type (Fig. 7). Studies with stomatal density and distribution (sdd1-1) mutants in Arabidopsis also showed that stomatal density cannot be correlated directly with gs, because increased numbers can be compensated for by decreased functional stomatal aperture (Büssis et al., 2006; Lawson and Blatt, 2014). This compensation mechanism appears to be environmentally dependent, in that no difference in gs or assimilation rates were detected when the wild type, sdd-1 mutants with increased stomatal density, and SDD1 overexpression lines with a 40% reduction in stomatal number were compared under light conditions of 180 µmol m−2 s−1 PPFD (Berger and Altmann, 2000; Von Groll et al., 2002). When measured at high light, however, the increase in stomatal density in sdd-1 mutants translated into 30% higher photosynthetic rates in Arabidopsis (Schlüter et al., 2003). Future analyses of ZjPCKpro:ZmSHR1 leaves under different light intensities may reveal that a similar compensatory mechanism operates in grasses to balance the tradeoff between carbon gain and water loss via transpiration. A second compensatory mechanism also operates in Arabidopsis, whereby altered stomatal patterning is accompanied by altered development of underlying mesophyll cells, ensuring that both gas exchange and water use efficiency are optimized across the leaf (Dow et al., 2017). Given that increased stomatal density did not lead to increased mesophyll cell density in the rice leaves examined here, it is possible that different compensatory mechanisms operate in eudicot and monocot leaves to coordinate gas exchange, carbon fixation, and water loss.
CONCLUSION
Collectively, the work reported here has revealed that the spatial expression domain of ZmSHR1/OsSHR2 orthologs in the subepidermal layers of rice leaves influences the positioning of stomatal cell files in the overlying epidermis. By expanding the vascular-specific expression domain to include bundle sheath cells surrounding the vein at later stages of development, extra stomatal rows were formed. The resulting increased stomatal density did not increase mesophyll cell density in underlying tissue and did not enhance photosynthetic efficiency under the limited conditions tested, but the transgenic lines provide a vehicle for more rigorous testing of the relationship between stomatal density and photosynthetic capacity in the future.
MATERIALS AND METHODS
Plant Growth Conditions
Rice (Oryza sativa ssp japonica) ‘Kitaake’ lines were grown in soil (John Innes Compost No. 2) in a transgenic greenhouse in either Oxford, United Kingdom, or Düsseldorf, Germany. Day/night temperature was maintained at 28°C/22°C ± 3°C with a diurnal light regime of 16 h of light (supplemented to ∼300 μmol m−2 s−1) and 8 h of dark. For fixation prior to in situ hybridization, seedlings were grown in water-saturated medium-grade vermiculite (Sinclair) for 17 d in a glasshouse at 28°C (day) and 23°C (night) with up to 16 h of supplemental light (100 μmol m−2 s−1) per day.
Generation of Transformation Constructs
ZmSHR1 (GRMZM2G132794) coding sequence was amplified from genomic DNA by PCR using Phusion High-Fidelity Polymerase (Thermo Scientific) and Gateway-compatible primers (MS5F, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGTGGGTTAGATGGACACACTGTTTAGA-3′, and MS5R, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCATGGCCTCCACGCGCTCGT-3′). The amplified sequence was subcloned into the Gateway donor vector pDONR207 in a BP reaction. The resultant entry clones were sequenced, and the target sequences were then cloned in an LR reaction downstream of Zoysia japonica PCKpro (Nomura et al., 2005) into a destination vector modified from pVec8-GFP (Kim and Dolan, 2016) or into the binary destination vector pSC210. pSC210 vector was created by Sarah Covshoff and kindly gifted to us by Julian Hibberd (University of Cambridge). ZmSHR1pro:mTurquoise::ZmSHR1 and ZmSHR1pro:ZmSHR1 constructs were generated using the Golden Gate cloning system (Weber et al., 2011) with synthesized promoter and coding sequence modules.
Rice Transformation
Callus induced from mature rice seeds was used for the transformation of cv Kitaake with Agrobacterium tumefaciens strain EHA105 carrying the construct of interest. Callus induction, transformant selection, and seedling regeneration were performed at 32°C under continuous light according to a protocol modified from Toki et al. (2006; available to download from https://langdalelab.files.wordpress.com/2015/07/kitaake_transformation_2015.pdf). Hygromycin-resistant T0 seedlings confirmed positive for transgene presence by PCR screening (see below) were transplanted into soil in 0.73-L pots. T1 lines with single transgene insertion events were selected on the basis of a 3:1 segregation pattern on hygromycin-containing selective medium. Homozygous plants were identified by quantification of transgene copy number in hygromycin-resistant T1 plants, using the gCount service of iDNA genetics, and propagated into the T2 generation. Homozygous T2 lines 207_29H, 207_30K, and 207_31J, which represented a single transgene insertion event, were used for all phenotypic analyses except those shown in Supplemental Figure S1.
DNA and RNA Extraction
Genomic DNA was isolated from cv Kitaake lines using a modified 2% (v/v) cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980), and rice leaf RNA was extracted using TRIzol reagent.
Genomic PCR Screening
Regenerated T0 plants and T1 seedlings (2–3 weeks after germination) were subjected to genomic PCR using primers specific to the cloning vector pSC210 (pSC210-F, 5′-CTGCTGCTGCTGCTCTCTC-3′, located in ZjPCKpro, and pSC1/2/310R, 5′-AAGACCGGCAACAGGATTC-3′, located in the nos terminator). PCR amplification was performed in a total reaction volume of 20 µL containing 9.2 µL of 2xGoTaq mix (Promega) and 1 m betaine. PCR conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 30 s, 58°C for 40 s, and 72°C for 2.5 min; and 72°C for 5 min. All PCR products were detected by agarose gel electrophoresis using standard protocols (Sambrook, 1982).
Real-Time Quantitative Reverse Transcription-PCR
Gene expression analysis was carried out using RNA extracted from fully expanded fourth leaf tissue. Total RNA was treated with TURBO DNAse (Thermo Scientific), and cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Specific primers were used to amplify genes of interest (qRT5F, 5′-ATGAAGGTGTTCACCGAGGG-3′, and qRT5R, 5′-TCGTTGCTCGCCTTGGG-3′) or an endogenous UBIQUITIN gene (Os03g13170; pW65F, 5′-CCAGGACAAGATGATCTGCC-3′, and pW65R, 5′-AAGAAGCTGAAGCATCCAGC-3′). Reactions were carried out using KAPA SYBR FAST Master Mix (2×) universal (KAPA Biosystems) in a 20-µL PCR volume. Transcript amplification and detection was performed on the 7500 Fast Real-Time PCR System (Applied Biosystems) using the following thermal profile: 95°C for 5 min; followed by 45 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Three technical replicates were carried out per sample, and transcript abundance was normalized to the endogenous rice UBIQUITIN gene. Relative expression fold changes were calculated using the Pfaffl method (Pfaffl, 2001).
In Situ Hybridization
Shoot apices were harvested on ice, fixed in 4% (w/v) paraformaldehyde overnight, and then dehydrated using a graded ethanol series before storage in 70% (v/v) ethanol at 4°C. Following wax embedding in Paraplast Plus (Sigma-Aldrich), 8-μm-thick mounted sections were pretreated as described by Javelle et al. (2011) with the following modifications: the 30-s 10% (v/v) ethanol and 1-min water washes were replaced with a 2-min 0.85% (w/v) sodium chloride wash, and the proteinase K incubation time was increased to 30 min. The slides were hybridized to a 141-bp digoxigenin-labeled RNA probe specific to OsSHR2. The probe sequence was PCR amplified using primers appended with T3 and T7 RNA polymerase-binding sites (5′-AATTAACCCTCACTAAAGGGAGATCCCCTTTGCATCCAT-3′ and 5′-TAATACGACTCACTATAGGGCTGGCTACCACCATACAGCA-3′) and then labeled according to Langdale (1994), but with the following reaction mix: 1× RNA polymerase buffer, 0.5 mm ATP, 0.5 mm GTP, 0.5 mm CTP, 0.1 mm digoxigenin-UTP, 0.8 units μL−1 RNA polymerase (all Thermo Scientific), and 1.6 units μL−1 RNAseOUT (Invitrogen). The reaction was incubated for 1.5 h at 37°C and terminated by 10 min of incubation at 37°C with 3 volumes of DNase solution (10 mm Tris-HCl, pH 7.5, 10 mm magnesium chloride, 50 mm sodium chloride, 2.5 μg μL−1 tRNA, and 0.02 units μL−1 DNase I). Hybridization was carried out at 55°C essentially according to Langdale (1994) with posthybridization washes and visualization as described by Javelle et al. (2011) using 0.05× SSC buffer. After dehydration through ethanol and counterstaining for 5 min with 0.1% (w/v) Calcofluor White (Fluorescent Brightener 28; Sigma-Aldrich), slides were mounted in Entellan (Merck Millipore) and imaged under bright-field and UV light using a Leica DMR8 microscope, QImaging MicroPublisher (QImaging) camera, and Image-Pro Insight (MediaCybernetics) software.
Histological Analysis of Root Tissue
Roots were fixed in 3:1 ethanol:acetic acid for 30 min, dehydrated in an ethanol series, and embedded in Araldite epoxy resin (prepared from four single embedding components of Durcupan ACM A,B,C,D; Sigma-Aldrich). Transverse sections of 10 µm were cut with a glass knife microtome, stained with a drop of Toluidine staining solution (1% (w/v) Toluidine Blue in 1% (w/v) borate) for 20 s, and rinsed with distilled water. Dried slides were then mounted with DePeX medium (Serva) for microscopic analysis.
Scanning Electron Microscopy
Negative imprints of the abaxial leaf surface were taken from the middle of fully expanded adult leaves (11–12 mm diameter) using ESPE Express two-component ultra-light body dental wax (3M). Silicone imprints were dried for 5 min, covered with epoxy resin HP E25KL (HP Textiles), and dried overnight at room temperature to produce positive imprints for scanning electron microscopy. Resin imprints were coated with gold particles in an argon sputter coater and visualized with a Zeiss Leo 1430 VP scanning electron microscope.
Phenotypic Analysis of Leaf Anatomy
Anatomical measurements were carried out on the expanded sixth leaf of the first tiller. Leaf width and length were first measured, and then fresh segments were hand cut from the middle of the leaf blade. Sections were exposed to UV light and then viewed and photographed with a Leica DMRB microscope or a Zeiss Axio Imager M2m. The cell number between a pair of intermediate veins positioned between the second and third lateral veins from the leaf margin, as well as interveinal distances of three pairs of veins and cell size of six bundle sheath cells, were recorded for each sample. Two to five leaf samples were quantified for each line. Least-squares regression lines of leaf width versus vein number were plotted for the cv Kitaake wild type and ZjPCKpro:ZmSHR1 data sets using GraphPad Prism 6.
Quantification of Stomatal Phenotypes
The leaf clearing method used was adapted from Dow et al. (2014). Penultimate leaves were fixed and cleared with 7:1 ethanol:acetic acid solution overnight, softened for 30 min in 1 m potassium hydroxide, rinsed twice with water, and then mounted on microscope slides using a chloral hydrate-glycerol mounting solution (40 g of chloral hydrate in 16 g of glycerol). The abaxial epidermis was analyzed by differential interference contrast microscopy with Zeiss Axio ImagerM2m. Stomatal density (mm−2) was counted between two lateral veins in three different areas of four individual leaves per line. Stomatal sizes defined as stomatal height multiplied by stomatal width (µm2) were measured and averaged from 40 to 48 stomata counted from four to five leaves per individual line using ImageJ software (Abràmoff et al., 2004), available at https://imagej.nih.gov/ij/.
Leaf Drying and Weight-Gain Experiments
Leaf discs were punched from freshly detached leaves of equivalent size and then dried at room temperature for 3 d. Dry weights were determined for three pooled leaf discs per leaf, from six leaves in total. For weight-gain experiments, four leaf discs were punched from six freshly detached leaves and weighed. The discs were then immediately vacuum infiltrated with distilled water, blotted dry on filter paper, and then weighed once more to determine weight gain.
Quantification of Chlorophyll
Chlorophyll was extracted from freshly detached leaves of equivalent sizes as described previously (Lichtenthaler and Wellburn, 1983; Porra et al., 1989). Three leaf discs (10 mm each in diameter) were punched from the middle of mature fully expanded leaves and ground in liquid nitrogen. Chlorophyll was extracted by incubating the ground powder for 20 min in 80% (v/v) acetone in 50 mm Tris-HCl, pH 7.8. The absorption properties of 1 mL of extract were measured with an Eppendorf Biospectrometer, and chlorophyll content was calculated as follows: chlorophyll a (µg mL−1) = 12.21 (A663) − 2.81 (A646); chlorophyll b (µg mL−1) = 20.13 (A646) − 5.03 (A663); total chlorophyll content = 17.76 (A646) + 7.43 (A663).
Leaf Gas-Exchange Measurements
A and gs were measured on fully expanded leaves of 5- to 6-week-old plants using infrared gas analysis (Li-Cor 6400XT, Portable Photosynthesis System; LICOR Biosciences). Leaves were first equilibrated at a PPFD of 100 µmol m−2 s−1 until both A and gs reached steady state, this being defined as a less than 10% change in rate during a 10-min period (after 30–60 min). Once steady state was reached, A and gs were recorded for an initial period of 5 min at 100 µmol m−2 s−1 every 60 s. PPFD was then increased to 1,000 µmol m−2 s−1 for 30 min, while A and gs were measured every 30 s. The leaf cuvette was maintained at 400 µmol mol−1 CO2 concentration, leaf temperature of 27°C ± 2°C, and a vapor pressure deficit (VPD) of 1.3 ± 0.3 kPa. All measurements were completed before 3 pm to avoid any unwanted diurnal or circadian effects on photosynthesis.
Protein Alignment and Phylogenetic Analysis
BLASTP (protein) and BLASTN (nucleotide) search algorithms were used on proteomes and genomes from 13 angiosperm and one bryophyte species obtained from the Phytozome version 10 database (Goodstein et al., 2012). Protein and genomic sequences that were significantly similar (threshold, E < 1 × 10−3) to the amino acid and transcript sequences from ZmSHR1 were identified. The amino acid sequences were aligned using MergeAlign software (Collingridge and Kelly, 2012; Supplemental Data Set S1. A) maximum-likelihood method was used to infer the phylogenetic tree using RAxML software (Stamatakis, 2014); 100 rapid bootstrap analysis runs were used to produce a best-scoring maximum-likelihood tree (the best-fitting protein substitution model used for inference was selected automatically by the software). The phylogenetic tree was visualized using Interactive Tree of Life software (Letunic and Bork, 2016).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Three independent T0 lines exhibit extra stomatal rows when ZmSHR1 is ectopically expressed.
Supplemental Figure S2. Light saturation curve of 207_30 versus the wild type.
Supplemental Data Set S1. Multiple sequence alignment of SHR-like proteins.
Supplemental Data Set S2. Data used for regression analysis of vein number versus leaf width.
Acknowledgments
We thank Sayuri Ando, Zara Lewis, and Julie Bull for technical support; Peng Wang, Dana Vlad, Tom Hughes, and Tracy Lawson for invaluable discussions throughout the course of the work; Sarah Covshoff for providing plasmid pSC210 to generate the promoter construct; Dana Vlad for making the Golden Gate constructs; Jane Knerova and Julian Hibberd for preliminary gas-exchange measurements; and Steve Kelly, Kim Gallagher, and Tom Hughes for comments on the article.
Footnotes
This work was funded by a grant (C4 Rice) from the Bill & Melinda Gates Foundation to the International Rice Research Institute (2012–2015; OPPGD1394) and the University of Oxford (2015–2019; OPP1129902). Additional awards contributed support for M.L.S. (EU FP7 Award 3to4 and CEPLAS) and O.V.S. (University of Oxford Clarendon and Somerville College Scholarships). B.J.W. is supported by an Alexander von Humboldt postdoctoral fellowship.
[CC-BY]Article free via Creative Commons CC-BY 4.0 license.
References
- Abràmoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–41 [Google Scholar]
- Berger D, Altmann T (2000) A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Brown HT, Escombe F (1900) Static diffusion of gases and liquids in relation to the assimilation of carbon and translocation in plants. Proc R Soc Lond 67: 124–128 [Google Scholar]
- Büssis D, Von Groll U, Fisahn J, Altmann T (2006) Stomatal aperture can compensate altered stomatal density in Arabidopsis thaliana at growth light conditions. Funct Plant Biol 33: 1037–1043 [DOI] [PubMed] [Google Scholar]
- Byott GS. (1976) Leaf air space systems in C3 and C4 species. New Phytol 76: 295–299 [Google Scholar]
- Collingridge PW, Kelly S (2012) MergeAlign: improving multiple sequence alignment performance by dynamic reconstruction of consensus multiple sequence alignments. BMC Bioinformatics 13: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Kong D, Liu X, Hao Y (2014) SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J 78: 319–327 [DOI] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Dhondt S, Coppens F, De Winter F, Swarup K, Merks RMH, Inzé D, Bennett MJ, Beemster GTS (2010) SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiol 154: 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow GJ, Bergmann DC, Berry JA (2014) An integrated model of stomatal development and leaf physiology. New Phytol 201: 1218–1226 [DOI] [PubMed] [Google Scholar]
- Dow GJ, Berry JA, Bergmann DC (2017) Disruption of stomatal lineage signaling or transcriptional regulators has differential effects on mesophyll development, but maintains coordination of gas exchange. New Phytol 216: 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. (1943) Ontogeny of the vascular bundle in Zea mays. Hilgardia 15: 327–368 [Google Scholar]
- Farquhar GD, Sharkey TD (1982) Stomatal conductance and photosynthesis. Annu Rev Plant Physiol 33: 317–345 [Google Scholar]
- Fischer RA, Rees D, Sayre KD, Lu ZM, Condon AG, Saavedra AL (1998) Wheat yield progress associated with higher stomatal conductance and photosynthetic rate, and cooler canopies. Crop Sci 38: 1467 [Google Scholar]
- Franks PJ, Beerling DJ (2009) CO2-forced evolution of plant gas exchange capacity and water-use efficiency over the Phanerozoic. Geobiology 7: 227–236 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Drake PL, Beerling DJ (2009) Plasticity in maximum stomatal conductance constrained by negative correlation between stomatal size and density: an analysis using Eucalyptus globulus. Plant Cell Environ 32: 1737–1748 [DOI] [PubMed] [Google Scholar]
- Franks PJ, Farquhar GD (2001) The effect of exogenous abscisic acid on stomatal development, stomatal mechanics, and leaf gas exchange in Tradescantia virginiana. Plant Physiol 125: 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M (1998) Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14: 425–430 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN (2004) Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Gardiner J, Donner TJ, Scarpella E (2011) Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev Dyn 240: 261–270 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Henry S, Dievart A, Divol F, Pauluzzi G, Meynard D, Swarup R, Wu S, Gallagher KL, Périn C (2017) SHR overexpression induces the formation of supernumerary cell layers with cortex cell identity in rice. Dev Biol 425: 1–7 [DOI] [PubMed] [Google Scholar]
- Hernandez ML, Passas HJ, Smith LG (1999) Clonal analysis of epidermal patterning during maize leaf development. Dev Biol 216: 646–658 [DOI] [PubMed] [Google Scholar]
- Javelle M, Marco CF, Timmermans M (2011) In situ hybridization for the precise localization of transcripts in plants. J Vis Exp e3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HG. (1977) Transpiration in barley lines with differing stomatal frequencies. J Exp Bot 28: 162–168 [Google Scholar]
- Jones HG. (1987) Breeding for stomatal characters. In Zeiger E, Farquhar G, Cowan I, eds, Stomatal Function. Stanford University Press, Palo Alto, CA, pp 431–443 [Google Scholar]
- Kamiya N, Itoh J, Morikami A, Nagato Y, Matsuoka M (2003) The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J 36: 45–54 [DOI] [PubMed] [Google Scholar]
- Kim CM, Dolan L (2016) ROOT HAIR DEFECTIVE SIX-LIKE class I genes promote root hair development in the grass Brachypodium distachyon. PLoS Genet 12: e1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. (1994) In situ hybridization. In Freeling M, Walbot V, eds, The Maize Handbook. Springer, New York, NY, pp 165–179 [Google Scholar]
- Lawson T, Blatt MR (2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol 164: 1556–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11: 591–592 [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC (2009) Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nelson T, Dengler N (1997) Leaf vascular pattern formation. Plant Cell 9: 1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Higuchi T, Ishida Y, Ohta S, Komari T, Imaizumi N, Miyao-Tokutomi M, Matsuoka M, Tajima S (2005) Differential expression pattern of C4 bundle sheath expression genes in rice, a C3 plant. Plant Cell Physiol 46: 754–761 [DOI] [PubMed] [Google Scholar]
- Ohsumi A, Kanemura T, Homma K, Horie T, Shiraiwa T (2007) Genotypic variation of stomatal conductance in relation to stomatal density and length in rice (Oryza sativa L.). Plant Prod Sci 10: 322–328 [Google Scholar]
- Okada M, Lanzatella C, Saha MC, Bouton J, Wu R, Tobias CM (2010) Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing and multilocus interactions. Genetics 185: 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 45e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Raissig MT, Abrash E, Bettadapur A, Vogel JP, Bergmann DC (2016) Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc Natl Acad Sci USA 113: 8326–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Matos JL, Gil MXA, Kornfeld A, Bettadapur A, Abrash E, Allison HR, Badgley G, Vogel JP, Berry JA, et al. (2017) Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355: 1215–1218 [DOI] [PubMed] [Google Scholar]
- Russell SH, Evert RF (1985) Leaf vasculature in Zea mays L. Planta 164: 448–458 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T, Rich EF (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schlüter U, Muschak M, Berger D, Altmann T (2003) Photosynthetic performance of an Arabidopsis mutant with elevated stomatal density (sdd1-1) under different light regimes. J Exp Bot 54: 867–874 [DOI] [PubMed] [Google Scholar]
- Sharman B. (1942) Developmental anatomy of the shoot of Zea mays L. Ann Bot 6: 245–284 [Google Scholar]
- Slewinski TL, Anderson AA, Price S, Withee JR, Gallagher K, Turgeon R (2014) Short-root1 plays a role in the development of vascular tissue and Kranz anatomy in maize leaves. Mol Plant 7: 1388–1392 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL, Shah SS (1960) Developmental studies of cell differentiation in the epidermis of monocotyledons. Dev Biol 2: 477–500 [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M (1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Tasaka M, Kato T, Fukaki H (1999) The endodermis and shoot gravitropism. Trends Plant Sci 4: 103–107 [DOI] [PubMed] [Google Scholar]
- Tausta SL, Li P, Si Y, Gandotra N, Liu P, Sun Q, Brutnell TP, Nelson T (2014) Developmental dynamics of Kranz cell transcriptional specificity in maize leaf reveals early onset of C4-related processes. J Exp Bot 65: 3543–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- van Campen JC, Yaapar MN, Narawatthana S, Lehmeier C, Wanchana S, Thakur V, Chater C, Kelly S, Rolfe SA, Quick WP, et al. (2016) Combined chlorophyll fluorescence and transcriptomic analysis identifies the P3/P4 transition as a key stage in rice leaf photosynthetic development. Plant Physiol 170: 1655–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Groll U, Berger D, Altmann T (2002) The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Karki S, Biswal AK, Lin HC, Dionora MJ, Rizal G, Yin X, Schuler ML, Hughes T, Fouracre JP, et al. (2017) Candidate regulators of early leaf development in maize perturb hormone signalling and secondary cell wall formation when constitutively expressed in rice. Sci Rep 7: 4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lee CM, Hayashi T, Price S, Divol F, Henry S, Pauluzzi G, Perin C, Gallagher KL (2014) A plausible mechanism, based upon Short-Root movement, for regulating the number of cortex cell layers in roots. Proc Natl Acad Sci USA 111: 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]