In Arabidopsis leaves, the catalytic C-terminal region of STARCH SYNTHASE 4 promotes starch granule initiation while its noncatalytic N-terminal region determines starch granules morphology.
Abstract
The formation of normal starch granules in Arabidopsis (Arabidopsis thaliana) leaf chloroplasts requires STARCH SYNTHASE 4 (SS4). In plants lacking SS4, chloroplasts typically produce only one round granule rather than multiple lenticular granules. The mechanisms by which SS4 determines granule number and morphology are not understood. The N-terminal region of SS4 is unique among SS isoforms and contains several long coiled-coil motifs, typically implicated in protein-protein interactions. The C-terminal region contains the catalytic glucosyltransferase domains, which are widely conserved in plant SS and bacterial glycogen synthase (GS) isoforms. We investigated the specific roles of the N- and C-terminal regions of SS4 by expressing truncated versions of SS4 and a fusion between the N-terminal region of SS4 and GS in the Arabidopsis ss4 mutant. Expression of the N-terminal region of SS4 alone did not alter the ss4 mutant phenotype. Expression of the C-terminal region of SS4 alone increased granule initiation but did not rescue their aberrant round morphology. Expression of a self-priming GS from Agrobacterium tumefaciens also increased the number of round granules. Remarkably, fusion of the N-terminal region of SS4 to A. tumefaciens GS restored the development of wild-type-like lenticular starch granules. Interestingly, the N-terminal region of SS4 alone or when fused to GS conferred a patchy subchloroplastic localization similar to that of the full-length SS4 protein. Considered together, these data suggest that, while the glucosyltransferase activity of SS4 is important for granule initiation, the N-terminal part of SS4 serves to establish the correct granule morphology by properly localizing this activity.
Leaf chloroplasts typically contain multiple lenticular starch granules that form between the thylakoid membranes. These semicrystalline, insoluble granules are composed of two glucose (Glc) polymers, 70% to 90% amylopectin and 10% to 30% amylose. Amylopectin is a large branched polymer, where Glc residues are linked via α-1,4-glucosidic bonds to form linear chains that are connected via α-1,6-bonds (branch points). The chains in amylopectin arrange in a distinct tree-like architecture in which unbranched chain segments cluster. Neighboring unbranched chain segments intertwine to form double helices that further align in layers to produce a semicrystalline matrix (Pérez and Bertoft, 2010; Buléon et al., 1998). The minor component, amylose, consists of lightly branched α-1,4-linked chains and is localized in the amorphous parts of the granule between the crystalline layers formed by amylopectin.
Starch is synthesized by three types of enzyme activity: starch synthases (SSs), starch branching enzymes, and isoamylase-type debranching enzymes (Myers et al., 2000; Zeeman et al., 2010; Pfister and Zeeman, 2016). SS transfers the glucosyl moiety from ADP-Glc to the nonreducing end of an acceptor glucan. Branching enzyme transfers part of a linear glucan chain to another chain, forming an α-1,6-linkage. Subsequently, isoamylase removes a fraction of the α-1,6-linkages and is thereby thought to facilitate the formation of the crystalline amylopectin layers (Ball et al., 1996).
Most plants contain multiple SS isoforms. These can be separated into six classes based on amino acid sequence: SS1, SS2, SS3, SS4, SS5, and the granule-bound starch synthase GBSS (Nougué et al., 2014). Insight into the roles of each class during starch synthesis has been obtained by studying mutant plants deficient in each isoform, as well as by characterizing the enzymes in vitro. The exception is SS5, which has not been characterized to date. According to these studies, SS1, SS2, and SS3 are important for establishing proper amylopectin structure: SS1 preferentially elongates newly placed branches to a length of around 8–10 Glc units, and SS2 further elongates these chains to around 13–18 Glc units (Delvallé et al., 2005; Fujita et al., 2006; Umemoto et al., 1999; Pfister et al., 2014). SS3 is proposed to synthesize long, cluster-spanning amylopectin chains (Fontaine et al., 1993; Fujita et al., 2007; Pfister and Zeeman, 2016). GBSS synthesizes amylose within the granular matrix (van de Wal et al., 1998; Tatge et al., 1999; Zeeman et al., 2002).
In contrast to the other SSs, SS4 does not have a major influence on the structure of amylopectin or on amylose synthesis (Roldán et al., 2007; Szydlowski et al., 2009), although in a heterologous yeast (Saccharomyces cerevisiae) system, it is capable of creating branched glucans together with the branching enzymes and isoamylase-type debranching enzyme (Pfister et al., 2016). Rather, it appears to play a critical role in starch granule initiation: the Arabidopsis (Arabidopsis thaliana) ss4 mutant usually has zero, one, or rarely two or three granules per chloroplast (Roldán et al., 2007; Malinova et al., 2017), instead of five to seven granules found in most wild-type chloroplasts (Crumpton-Taylor et al., 2012). The total starch content at dusk is lower in this mutant, yet this starch is incompletely metabolized at night. In young leaves, the phenotype is particularly severe, and most chloroplasts are starch free (Crumpton-Taylor et al., 2013). The ss4 mutant also accumulates very high levels of ADP-Glc, indicating that the remaining SS isoforms cannot utilize it when SS4 is absent, presumably due to the lack of glucan substrates (Crumpton-Taylor et al., 2013; Ragel et al., 2013). These findings are consistent with a role for SS4 upstream of the other SSs, that is, a role in granule initiation. Interestingly, the ss3ss4 double mutant is almost completely starch free, suggesting that SS3 is important for the less frequent initiations that still occur when SS4 is absent (Szydlowski et al., 2009; Seung et al., 2016). In addition to altering the number of starch granules, SS4 appears also to influence the granule’s size and shape: those from leaves of the Arabidopsis ss4 mutant are enlarged and near-spherical instead of lenticular (Roldán et al., 2007; Crumpton-Taylor et al., 2013).
The molecular features that give each SS its specific role are largely unknown. All share a common catalytic domain comprised of a glycosyltransferase 5 (GT5) and a glycosyltransferase 1 (GT1) subdomain (Pfister and Zeeman, 2016). This structure is conserved in prokaryotic glycogen synthases (GS), such as GlgA from Agrobacterium tumefaciens. In addition, most plant SS isoforms have N-terminal extensions of different lengths that, in some cases, have demonstrated functions. For instance, the carbohydrate binding modules in the N-terminal extensions of SS3 help confer a higher affinity for and activity on its glucan substrates (Senoura et al., 2007; Valdez et al., 2008).
Several SSs have been shown to engage in protein-protein interactions, often with other starch biosynthetic enzymes (Tetlow et al., 2008; Hennen-Bierwagen et al., 2008, 2009; Liu et al., 2009, 2012). For example, an external helix on the catalytic domain of GBSS enables it to engage in a coiled-coil-mediated interaction with PROTEIN TARGETING TO STARCH (PTST), which helps localize GBSS to the starch granule (Seung et al., 2015). Similarly, SS4 was recently shown to bind to PTST2, a protein related to PTST that also contains a carbohydrate-binding module and predicted coiled-coil motifs (Seung et al., 2017). This interaction appeared to involve the catalytic domain of SS4, although the exact interaction site between SS4 and PTST2 has not yet been identified. Like ss4 mutants, the ptst2 mutants have reduced numbers of starch granules, and this phenotype was exacerbated by loss of another homolog (PTST3; Seung et al., 2017). PTST2 was proposed to bind long malto-oligosaccharides with its carbohydrate-binding module and provide them as substrates to SS4 (Seung et al., 2017). This idea is broadly consistent with other observations that mutations affecting malto-oligosaccharide metabolism affect starch granule initiation (Malinova et al., 2017; Satoh et al., 2008; Seung et al., 2016).
The unique N-terminal region of SS4 is predicted to contain coiled-coil motifs (Leterrier et al., 2008), although this region is not needed for interaction with PTST2. However, fluorescent tagging showed that the N-terminal region confers a subchloroplastic localization to SS4, possibly at areas peripheral to starch granules (Gámez-Arjona et al., 2014). The N-terminal region of SS4 was also reported to enable its association with thylakoid membranes and to allow interaction with the thylakoid- and plastoglobule-bound fibrillin proteins FBN1a and FBN1b (Gámez-Arjona et al., 2014). It is unclear whether this a coiled-coil-mediated interaction. Further, the significance of this interaction remains to be established, since mutants deficient in both FBN1a and FBN1b have a normal starch phenotype.
Despite excellent progress in defining the role of SS4, the mechanisms by which it regulates the initiation of starch granules and controls their morphology are not fully understood. SS4 is presumed to determine the number of starch granules in a chloroplast by synthesizing specific amylopectin precursors (Roldán et al., 2007; Szydlowski et al., 2009), a role facilitated by its interaction with PTST2 (Seung et al., 2017). This emerging model contrasts with the priming of glycogen synthesis in other organisms. In metazoans and fungi, glycogen synthesis is primed by glycogenin proteins, which self-glucosylate to create primers that GS and branching enzymes can elaborate (Wilson et al., 2010; Ball et al., 2011). In prokaryotes, the priming process is less well studied. In A. tumefaciens, there is evidence that the GS can self-glucosylate when supplied only with ADP-Glc, thereby synthesizing glucan primers and fulfilling an analogous role to the eukaryotic glycogenins (Ugalde et al., 2003).
Previously, we showed that expression of A. tumefaciens GS in Arabidopsis ss3ss4 mutants could overcome the defect in starch granule initiation (Crumpton-Taylor et al., 2013), though granule morphology was still aberrant. To extend the understanding of how SS4 contributes to starch granule formation in vivo, we investigated the role of its N- and C-terminal regions by expressing truncated forms in the ss4 mutant background. We analyzed the localization of these truncated forms within the chloroplast and their capacity to complement the mutant phenotype. We further fused the N-terminal part of SS4 to A. tumefaciens GS and compared it to expression of GS alone. We show that the N-terminal region of SS4 alters the localization of GS and allows the formation of normal lenticular-shaped starch granules. We discuss the distinct functions of the catalytic C-terminal domains and noncatalytic N-terminal region in starch granule initiation and growth.
RESULTS
Generation of ss4 Plants Expressing A. tumefaciens GS or SS4 Variants
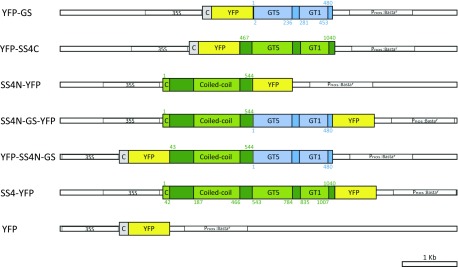
The A. tumefaciens GS was fused N terminally to the yellow fluorescent protein (YFP), targeted to the plastids by the transit peptide from the Arabidopsis Rubisco small subunit (At5g38430; amino acid residues 1–53), and expressed in the ss4-1 single mutant (Col-0 background; YFP-GS; Fig. 1). Since GS is homologous to the C-terminal half of SS4, we also designed a truncated SS4 construct consisting of its C-terminal region (amino acids 476–1040; including both GT1 and GT5 domains and a region immediately preceding them that is conserved in SS4 orthologs) that was N terminally fused to YFP (YFP-SS4C; Fig. 1). Further, we generated a construct containing the N-terminal half of SS4 (amino acids 1–544, also including the conserved region preceding the GT domains; SS4N-YFP) C terminally fused to YFP. We also made a chimeric construct where this N-terminal region of SS4 was fused to full-length A. tumefaciens GS and YFP (YFP-SS4N-GS or SS4N-GS-YFP, depending on the position of the YFP; Fig. 1). Finally, constructs encoding the full-length SS4 protein fused to YFP (SS4-YFP) and chloroplast-targeted YFP alone served as controls (Fig. 1). In all cases, constitutive expression was driven by the cauliflower mosaic virus 35S (CaMV 35S) promoter.
Figure 1.
Constructs used for the expression of GS- and SS4-derived proteins in ss4 mutants. Constructs encoding the full-length A. tumefaciens GS, the C-terminal region of Arabidopsis SS4 (SS4C), and the N-terminal region of Arabidopsis SS4 fused to GS (SS4N-GS) were cloned into vector pB7GWY2,0, modified to encode the Arabidopsis Rubisco small subunit B chloroplast transit peptide (C in gray; amino acids 1-53) upstream of YFP. Constructs encoding the N-terminal region of SS4 alone (SS4N), the N-terminal region linked to GS (SS4N-GS), and the full-length SS4 all contained the endogenous SS4 chloroplast transit peptide (C in green; amino acids 1-42) and were cloned into vector pB7WGY2,0, such that YFP was fused to their C termini. Constructs were driven by the CaMV 35S promoter (35S). Pnos:Bastar encoded a Basta resistance gene, driven by the A. tumefaciens nopaline synthase gene promoter. Blue bars and green bars indicate GS and SS4 regions, respectively. Both GS and the SS4C contain glycosyltransferase 5 (GT5) and glycosyltransferase 1 (GT1) domains. The N-terminal region of SS4 contains coiled-coil motifs. Protein domains were predicted by SMART (Letunic et al., 2015). Numbers indicated amino acid residues of the corresponding original full-length SS4 or GS protein sequences.
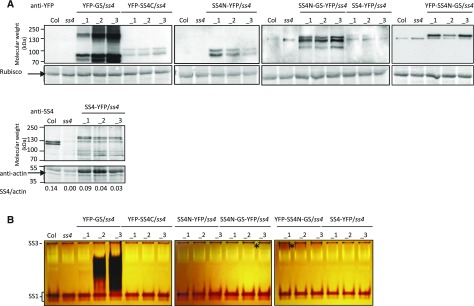
For each construct, we obtained numerous independent transgenic lines in the ss4 mutant background. We then selected three lines of each with different expression levels, judged by immunoblots on total protein extracts using either a monoclonal anti-YFP antibody or polyclonal antibodies against SS4 (Fig. 2A). Every plant line gave a band corresponding to the expected molecular mass of its mature introduced protein (i.e. lacking the chloroplast transit peptide). These bands were absent in protein extracts of the ss4 parent line and the wild type. There were additional, often closely migrating bands apparent in the transformed plant lines (Fig. 2A). It is possible that these additional bands reflect posttranslationally modified forms of the introduced protein (e.g. due to phosphorylation or proteolysis). In the case of YFP-GS, we observed additional slowly migrating bands, which possibly resulted from self-glucosylation of the protein.
Figure 2.
Detection of GS- and SS4-derived proteins in transformed ss4 plants. A, Soluble proteins were extracted from Arabidopsis ss4 mutant plants transformed with the constructs given in Figure 1. Equal amounts of protein from four plants of each line were pooled, separated by SDS-PAGE, and analyzed by immunoblotting with antibodies against YFP (top) or SS4 (bottom). The expected molecular weights of the mature introduced protein were: 80 kD for YFP-GS, 94 kD for YFP-SS4C, 85 kD for SS4N-YFP, 137 kD for SS4N-GS-YFP and YFP-SS4N-GS, and 141 kD for SS4-YFP. In some blots exposed for longer times, an endogenous 130-kD protein in the wild type and ss4 mutants cross reacted with the anti-YFP antibody. The amounts of SS4-YFP and endogenous SS4 are given as SS4/actin ratio below the anti-SS4 blot. B, For zymograms, equal amounts of soluble protein were separated on native-PAGE gels containing 0.3% (w/v) oyster glycogen. After electrophoresis, gels were incubated for 16 h in a solution containing 1 mm ADP-Glc. Dark bands corresponding to the endogenous starch synthase activities SS1 and SS3 were visible in all lines. Additional dark bands represent the introduced synthase activities. Asterisks mark areas where very faint additional synthase activities could be detected.
We investigated whether the chain-elongating activities of introduced enzymes were detectable on zymograms (native PAGE gels supplemented with glycogen as a substrate; Figure 2B). After incubating the gels in an ADP-Glc-containing medium, activity bands were visualized as dark bands by iodine staining, indicating elongation of glycogen chains. Bands corresponding to the endogenous starch synthases SS1 and SS3 (Wattebled et al., 2005; Zhang et al., 2005; Szydlowski et al., 2009) were detected in all plant extracts. In addition, a dark, smeared band appeared in lines expressing YFP-GS. Among these lines, YFP-GS_3 had the strongest GS activity and YFP-GS_1 had the weakest, consistent with the immunoblotting results (Fig. 2A). Faint bands were visible in some SS4N-GS-YFP-expressing and YFP-SS4N-GS-expressing lines. No additional activity bands were visible in lines expressing YFP-SS4C, SS4N-YFP, or SS4-YFP. However, these zymograms are evidently not able to detect all glucosyl transferase activities, since the endogenous SS2 and SS4 activities are not visible either (Szydlowski et al., 2009; Pfister et al., 2014). Multiple pale bands were also visible in the zymograms. These result from glucan hydrolytic activities that digest the glycogen substrates and could potentially mask the activities of comigrating glucan synthases.
Plant Growth and Leaf Starch Content Are Partially Restored by GS or the C-Terminal Region of SS4
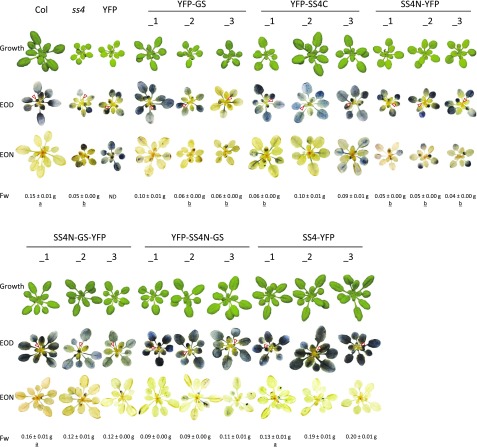
We evaluated our transformed plant lines for their growth phenotypes and for their leaf starch accumulation by iodine staining (Fig. 3). As previously reported, the ss4 mutant grew slowly and had pale and starch-free young leaves (Roldán et al., 2007; Crumpton-Taylor et al., 2013). The older leaves stained for starch at the end of day and, unlike the wild type, did not to fully degrade it during the subsequent night. Control ss4 plants expressing chloroplast-targeted YFP alone were indistinguishable from the ss4 parent.
Figure 3.
Growth and starch staining phenotype of ss4 plants expressing GS- or SS4-derived proteins. Plants grown in soil for 25 d under a 12-h-day/12-h-night cycle were photographed at the end of the day (EOD). For iodine staining, rosettes harvested at the EOD and end of night (EON) were decolorized with ethanol and stained with Lugol’s solution. Col: wild-type plant. ss4: ss4-1 mutant plant. Plants designated _1, _2, and _3 are T2 individuals from independent transgenic lines, as indicated. The mean (± se) rosette fresh weight (Fw) for each line is given (n = 8-12; ND, not determined) and designated as “a” or “b” when not significantly different from the wild type or ss4 mutant, respectively (determined by ANOVA; P value < 0.05).
The YFP-GS-expressing plants had greener rosette leaves, and line 1 displayed significantly improved growth compared with ss4 mutants. Lines YFP-GS_1 and YFP-GS_2 also showed complementation in terms of starch accumulation, as both young and mature leaves stained for starch. Yet part of this starch remained after the night, resulting in patchy staining of the leaves. Similarly, transformants expressing the C-terminal region of SS4 (YFP-SS4C lines) grew better than ss4 mutants and accumulated starch in all rosette leaves. These plants also did not fully metabolize their starch during the night. Interestingly, expression of GS fused to the N-terminal region of SS4 (SS4N-GSYFP/YFP-SS4N-GS) resulted in improved growth and iodine-staining patterns that were very similar to those of the wild type and SS4-YFP control plants: all leaves were green, accumulated starch during the day, and degraded most of it in the subsequent night. In contrast, when the N-terminal region of SS4 was expressed alone (SS4N-YFP plants), the phenotype was indistinguishable from that of the ss4 mutant. Thus, GS or the C-terminal region of SS4 alone partially complement the ss4 mutant phenotype. The extent of complementation is improved by the presence of the N-terminal region of SS4, but the N-terminal region alone cannot complement the mutant.
The Influence of the N-Terminal Region of SS4 on Starch Metabolism
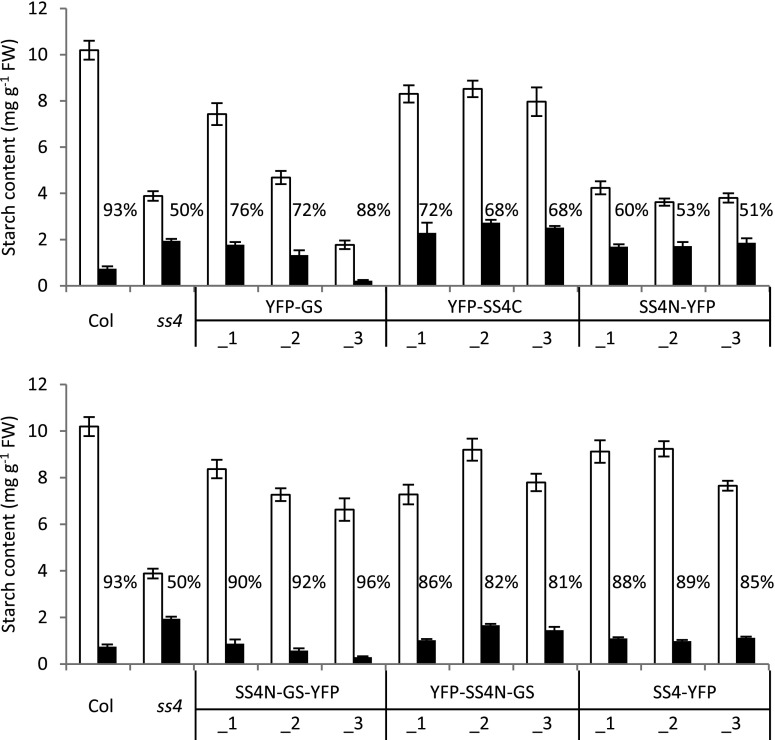
Iodine staining is a qualitative rather than quantitative indication of starch content. To quantify the starch content in our lines, we harvested whole rosettes at the end of a normal day and night, digested the starch enzymatically, and measured the released Glc (Fig. 4). As expected, ss4 plants accumulated less starch during the day than the wild type. They also degraded only one-half of their starch by the end of the night. These results were consistent with our observations from iodine staining (Fig. 3) and previous reports (Roldán et al., 2007; Crumpton-Taylor et al., 2013).
Figure 4.
Starch contents of ss4 plants expressing GS- or SS4-derived proteins. Plants grown for four weeks under 12-h-day/12-h-night cycles were harvested at the end of day (white bars) and end of night (black bars). The mean starch content (± se) for each line is given (n = 4 individual rosettes for Col, ss4, and SS4N-YFP; n = 6 for all other lines). For Col and for ss4, the data in the top and bottom panels are the same. The percentage of starch broken down during the night is indicated.
The YFP-GS lines varied considerably in starch content at the end of day, ranging from near-normal amounts of starch in YFP-GS_1 to starch levels even lower than in ss4 mutants in case of YFP-GS_3 (Fig. 4). Thus, starch content correlated inversely with the abundance and activity of GS (Fig. 2). Consistent with the iodine-staining (Fig. 3), the YFP-GS lines did not fully mobilize their starch during the night. The lines expressing YFP-SS4C reached starch levels close to those of the wild type, but also degraded only approximately 70% of the starch reserve during the night.
By contrast, SS4N-GS and SS4-YFP plants showed near-wild-type starch accumulation and breakdown, degrading around 80% to 90% of their starch by dawn (Fig. 4). Thus, having the N-terminal region of SS4 positively influenced starch turnover both when combined with GS or the C-terminal region of SS4. Yet, when expressed on its own, the N-terminal region of SS4 again showed no effect on the pattern of starch accumulation, as SS4N-YFP plants phenocopied the ss4 mutant (Fig. 4).
SS4 Domains Influence Starch Granule Shape and Numbers Independently
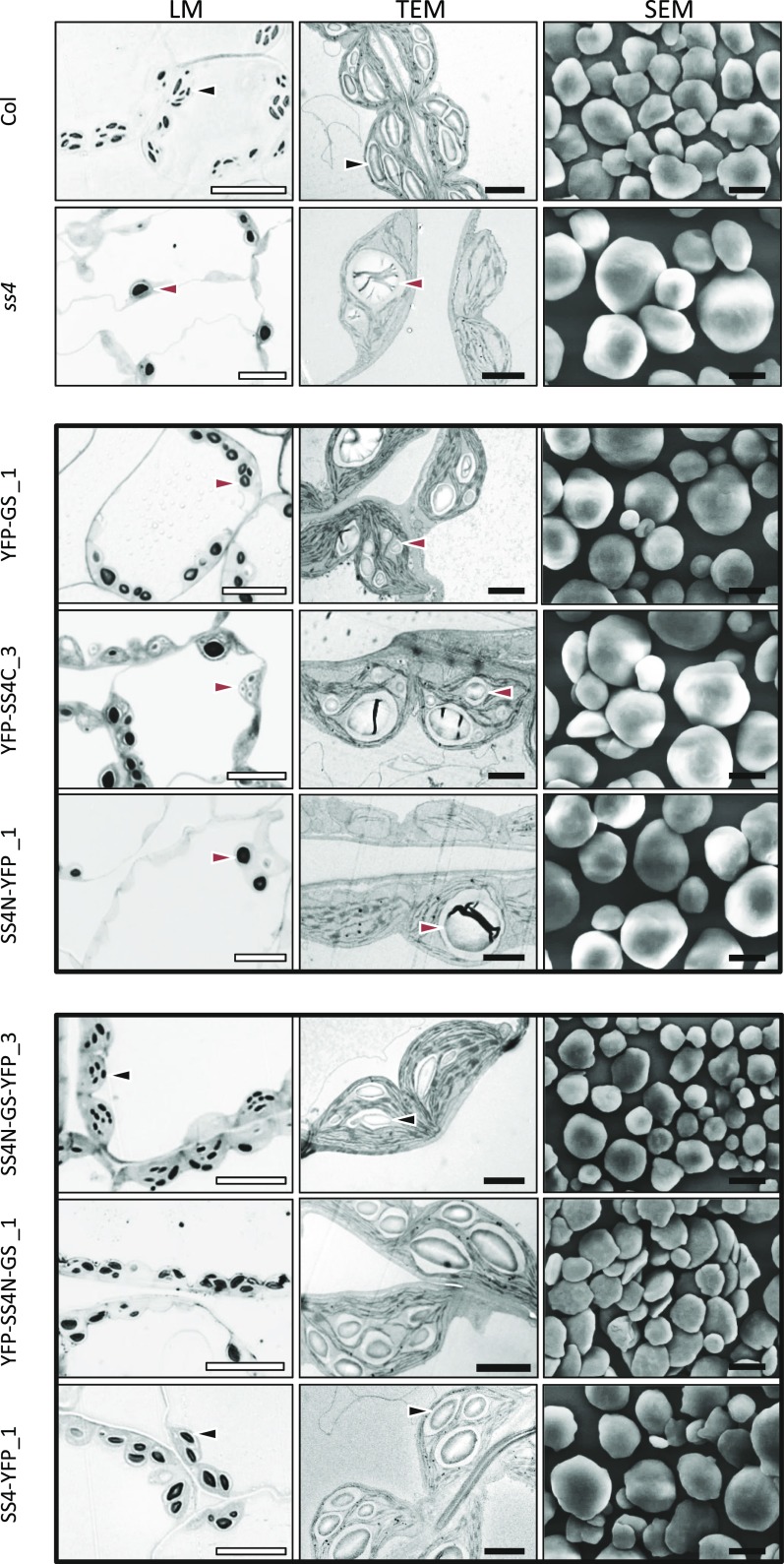
It was proposed that the reduced starch synthesis and degradation rates in ss4 mutants may be a consequence of its deposition as fewer, enlarged, and rounded granules when compared to the wild type (Roldán et al., 2007). Since a number of our complementation lines also still displayed impaired starch metabolism, we investigated the number and morphology of starch granules within their chloroplasts. Leaf number 7 (an expanding source leaf; Supplemental Figure S1) was harvested at the end of day, and samples from the middle of the leaf lamina were fixed and embedded in resin. We obtained light micrographs (LMs; Fig. 5; Supplemental Fig. S2) and transmission electron micrographs (TEMs; Fig. 5; Supplemental Fig. S3) of the leaf mesophyll tissue, where the majority of leaf starch is synthesized. To examine granule morphology in more detail, we also purified starch granules from whole rosettes and visualized them by scanning electron microscopy (Fig. 5; Supplemental Fig. S4).
Figure 5.
The N-terminal region of SS4 confers the discoid shape of starch granules. Left panels show light micrographs (LM) of toluidine blue-stained, semithin sections cut from fixed, embedded tissue from leaf number 7 of each line. Middle panels show TEM of ultrathin sections cut from the same samples. Right panels show scanning electron micrographs of starch granules extracted from whole rosettes of the given lines. The lines presented here are those showing the highest expression of the introduced construct, except for YFP-GS, where the line with lowest expression was chosen due to its relatively strong expression. Data from the remaining lines is presented in Supplemental Figures S2 to S4. White bars = 10 µm; black bars = 2 μm. Black arrow, lenticular starch granule; red arrow, spherical starch granule.
In sections from wild-type chloroplasts, we predominantly observed multiple ellipsoid starch granules, consistent with the flattened, lenticular shape of the granules seen by scanning electron micrograph (Fig. 5). In contrast, 80% of the sections of the ss4 mutant chloroplasts were devoid of starch (Supplemental Table S1). The minority of chloroplast sections that contained starch usually deposited it as one larger and rounded granule, as previously reported (Crumpton-Taylor et al., 2013; Roldán et al., 2007). It should be noted that chloroplast sections of approximately 500-750-nm thickness capture only a part of the chloroplast, meaning that the number of starch-containing chloroplasts will be slightly underestimated.
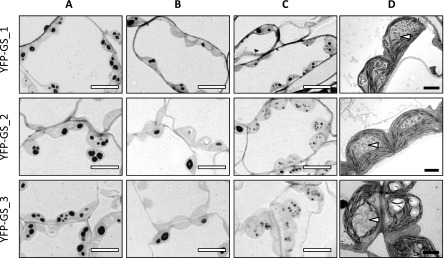
The lines expressing YFP-GS differed from the ss4 parental line in that they typically contained multiple rounded granules per chloroplast section (Fig. 5; Supplemental Figs. S2–S4), and only a few (10–20%) of the chloroplast sections were starch free (Supplemental Table S1). Interestingly, approximately 10% of the chloroplast sections contained supernumerary granules, which were tiny and of irregular shape (Fig. 6). The proportion of chloroplast sections containing these aberrant granules was highest in the line YFP-GS_3 (Supplemental Table S1), which expressed GS to the highest level (Fig. 2). In the YFP-SS4C lines, approximately 90% of chloroplast sections contained starch granules, similar to the YFP-GS lines. However, granules produced in the YFP-SS4C lines were larger and fewer in number than those from the YFP-GS lines. Interestingly, the granules still exhibited the rounded shape, characteristic of the ss4 mutant (Fig. 5; Supplemental Figs. S2–S4). No supernumerary granules were observed in the case of YFP-SS4C expression. Thus, while expression of either GS or SS4C in the ss4 background facilitated the formation of more starch granules, neither restored the normal lenticular morphology.
Figure 6.
Variation in granule morphology in chloroplasts of YFP-GS lines. Chloroplast sections of expanding source leaves from each YFP-GS line were analyzed by LM and TEM. A, LMs showing multiple granules in chloroplast sections. B, LMs showing zero, one or two granules in chloroplast sections. C, LMs showing supernumerary granules in chloroplast sections. D, TEM of chloroplasts with supernumerary granules. White bars = 10 µm; black bars = 2 µm. White arrows indicate supernumerary, irregular granules.
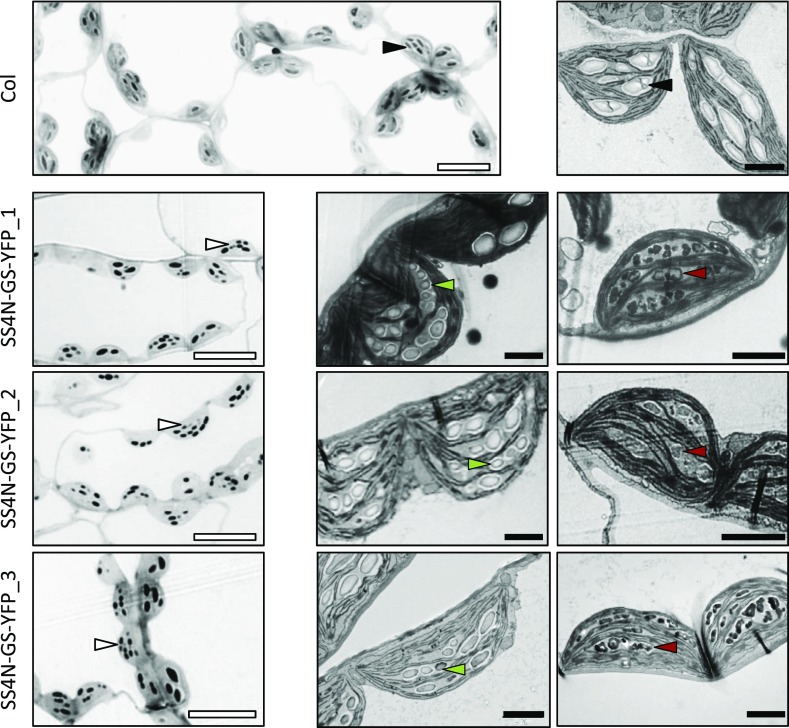
Expression of the SS4N-GS fusion constructs almost fully complemented the ss4 mutant phenotype: there were multiple granules per chloroplast section, and the granules had the wild-type-like shape despite being slightly smaller. As in the GS-YFP lines, these lines also contained some chloroplasts with supernumerary tiny granules within a single stromal pocket (observed in approximately 10–20% of the chloroplast sections). Often, these supernumerary granules had smooth surfaces, while in other sections they were irregular in appearance (Fig. 7; Supplemental Fig. S5).
Figure 7.
Variation in starch granule morphology in chloroplasts of SS4N-GS-YFP-expressing plants. LMs (left-hand panels) and TEMs (central and right-hand panels) of chloroplast sections from expanding source leaves of the wild type (Col) and each of the SS4N-GS-YFP-expressing lines. White bars = 10 µm; black bars = 2 µm. Black arrows, lenticular type granules; white arrows: supernumerary granules; green arrows, smooth supernumerary granules; red arrows, irregular supernumerary granules.
As expected, the three SS4-YFP lines also produced multiple wild-type-like granules per chloroplast section (Fig. 5; Supplemental Figs. S2–S4), even though they contained lower amounts of SS4 than the wild type (Fig. 2). In this case, no supernumerary granules were observed. Lines expressing SS4N-YFP had starch granules that were indistinguishable from those of the ss4 parent both in terms of morphology and frequency (Fig. 5; Supplemental Table S1), consistent with our other observations that the N-terminal region of SS4 by itself appears to have no effect on starch biosynthesis.
We investigated whether the differences in granule morphology were accompanied by changes in amylopectin structure. Therefore, we enzymatically debranched starch from each line and analyzed its chain-length distribution (CLD) by high performance anion-exchange chromatography with pulsed amperometric detection (Supplemental Fig. S6). This confirmed that the CLDs of starch from wild-type and ss4 mutant plants are very similar (Roldán et al., 2007). The CLDs of starches from our set of transformed lines were also all very similar to that of the wild type.
The N-Terminal Region of SS4 Influences the Subchloroplastic Localization
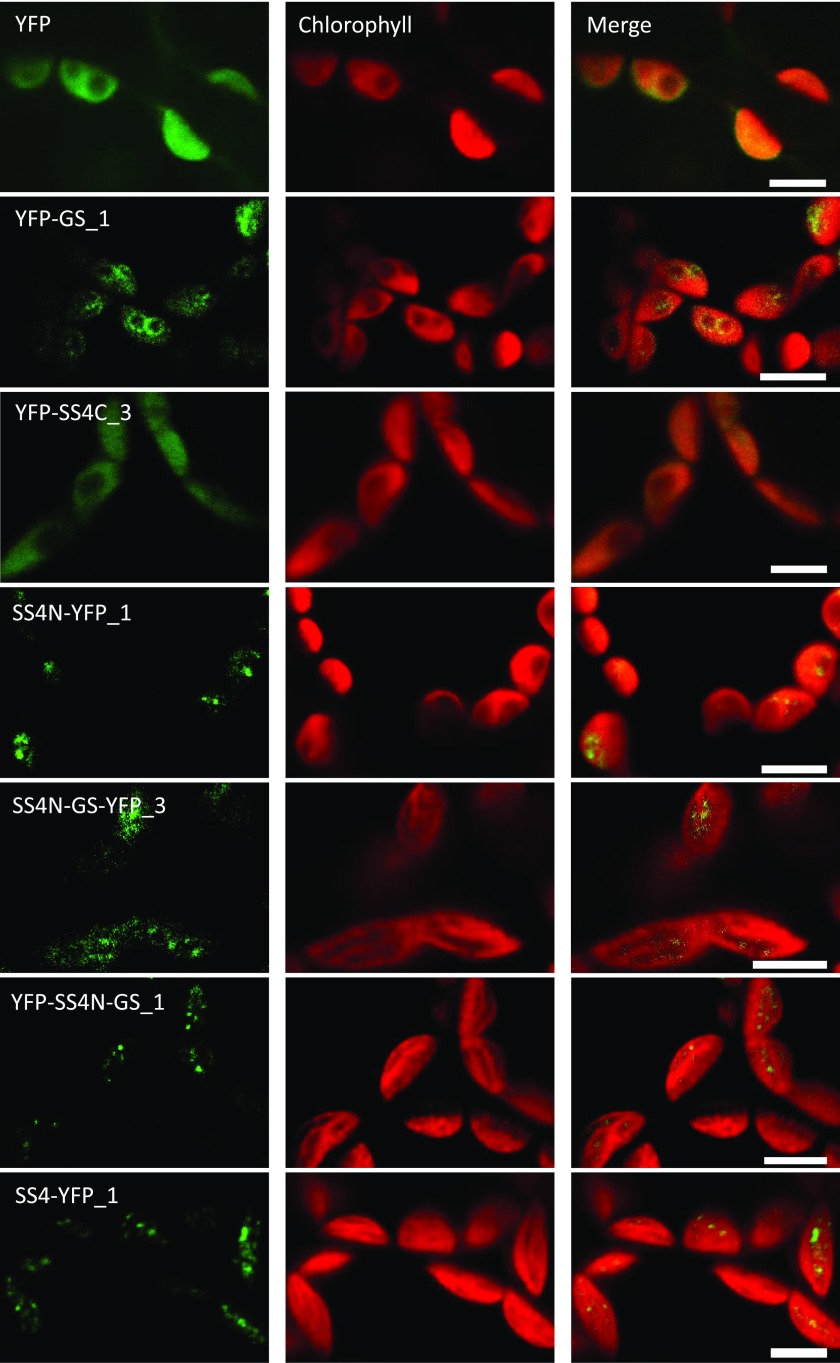
Previous work indicated that SS4 is not homogeneously distributed within the chloroplast stroma, but rather localizes to discrete spots at the edges of starch granules. Furthermore, this localization was suggested to be dependent on its N-terminal region (Gámez-Arjona et al., 2014; Szydlowski et al., 2009). To investigate the localization of our expressed proteins, we imaged YFP fluorescence in Arabidopsis leaves during the light period by confocal laser scanning microscopy (Fig. 8). As expected, free YFP was uniformly distributed within the chloroplast stroma (nonfluorescent patches typically reflect the presence of starch granules). This localization was similar in the case of YFP-SS4C, which also gave a uniform stromal YFP fluorescence. This result differs from that described by Gámez-Arjona et al. (2014), where the C-terminal region of SS4 appeared as a single dot at one end of the chloroplast. This is most likely due to differences between stable expression in transformed Arabidopsis lines used here versus transient overexpression Nicotiana benthamiana used previously.
Figure 8.
Subchloroplastic localization of YFP-tagged SS4 or GS proteins in mesophyll cell chloroplasts from mature Arabidopsis leaves. Confocal fluorescence micrographs showing free YFP and the YFP-tagged proteins (as indicated) are presented in the left-hand panels. The corresponding chlorophyll fluorescence is given in the central panels and the merged images in the right-hand panels. Bars = 5 µm.
Interestingly, YFP-GS appeared as ring-like structures surrounding the rounded starch granules, indicating that it preferentially associates with the granule surface. When the N-terminal part of SS4 was fused to GS, however, it did not uniformly surround the granules anymore, but rather appeared to localize to discrete areas at the periphery of starch granules. This distribution was similar to that of full-length SS4-YFP and the N-terminal region of SS4 alone (Fig. 8). Thus, the N-terminal region of SS4 appears both necessary and sufficient to confer the distinct subchloroplastic localization typical of SS4.
DISCUSSION
This work provides significant new insight into the function of SS4 in starch granule formation. On the one hand, the catalytic C-terminal region of SS4 promotes granule formation, presumably via the catalytic activity of its glucosyltransferase domains, a role that can be substituted by A. tumefaciens GS. On the other hand, the N-terminal region of SS4 confers a specific subchloroplastic localization, which is critical in determining granule morphology. It is remarkable that this N-terminal region of SS4 similarly guides the activity A. tumefaciens GS, which is then able to substitute surprisingly well for SS4.
Agrobacterium GS Increases Granule Number in the ss4 Mutant
The data presented here support our previous work indicating that A. tumefaciens GS promotes the initiation of starch granules in the absence of SS4 (Crumpton-Taylor et al., 2013). In the ss4 parental line, 80% of the chloroplast sections examined had zero granules, with the remainder having one or at most two granules. Most chloroplasts in the GS-expressing lines contained numerous small, round granules. Very few chloroplasts were granule free, while some contained supernumerary granules (Figs. 5 and 6; Supplemental Table S1). GS could function to increase the number of granules either by increasing the total amount of glucan-elongation activity, such that it increases the frequency of stochastic granule initiations. Alternatively (or in addition), GS could promote granule initiation via its self-priming ability (Ugalde et al., 2003), providing a substrate for the other starch biosynthetic enzymes. Consistent with this idea, immunoblotting and zymogram analyses revealed multiple and/or smeared bands for the YFP-GS protein and its activity, respectively (Fig. 2). Such behavior on SDS- and native-PAGE gels was proposed to be due to self-glucosylation, which changes the protein’s molecular mass and properties, thereby affecting its migration (Ugalde et al., 2003).
Among the GS-expressing lines, granule numbers per chloroplast appeared to correlate positively with GS protein/activity. However, starch metabolism was far from normal in these lines; granule morphology was still aberrant, as in the ss4 mutant, and the extent of starch metabolism during the diel cycle was highly variable. Surprisingly, starch content at the end of the day was negatively correlated with GS protein/activity: YFP-GS_1 had low GS activities and comparable amounts of starch to the wild type, while YFP-GS_3 had the largest amount of GS activity but the lowest amount of starch (Figs. 2 and 4). The reason for this is unclear. One possibility is that, despite increasing the number of granule initiations, excessive amounts of GS somehow interfere with the activities of the plant’s endogenous enzymes, restricting the rate of starch biosynthesis. YFP-GS had a distinct localization, surrounding the starch granules (Fig. 8). This is somewhat reminiscent of the subcellular localization of the native GS (EcGS or GlgA) in Escherichia coli cells, which associates with glycogen particles at the cell poles (Wilson et al., 2010). Association of GS with the starch granule surface might crowd out the other chain elongating, branching and debranching enzymes necessary for starch synthesis. Interestingly, a somewhat analogous phenotype was observed in transgenic potato tubers expressing EcGS; these produced less starch, which was more branched, and accumulated more soluble sugars (Shewmaker et al., 1994). However, in our lines expressing YFP-GS, chain-length distributions revealed no such alterations in the structure of the remaining starch (Supplemental Fig. S6), and no polyglucans were measured in the water-soluble fraction (data not shown). An alternative explanation for the low starch content in the presence of high GS could be that glucan degradation proceeds simultaneously with synthesis. If glucans made by abundant GS together with the other starch biosynthetic enzymes are unable or slow to crystallize, they may be subject to hydrolysis. Such situations have been reported in Arabidopsis (e.g. in the isa1 isoamylase-deficient mutants or the ss2ss3 double mutant; Delatte et al., 2005; Pfister et al., 2014), but again correlated with a measurable change in the structure of the starch (Supplemental Fig. S6). Interestingly, when GS was expressed in the ss3ss4 mutant background, starch structure was altered, having more short (d.p. 5–7) and fewer intermediate (d.p. 10–15) chains than wild-type starch (Crumpton-Taylor et al., 2013). We suggest that when GS represents a greater fraction of the total synthase activity, its preference for producing short like chains typical of glycogen becomes more apparent.
Despite variation in the amount of starch synthesized, the proportion of starch degraded at night was increased in all three YFP-GS-expressing lines relative to ss4 (Fig. 4). Low starch turnover in ss4 plants was proposed to result from having fewer, large starch granules; this changes the surface area available for both synthesis during the day and degradation at night (Roldán et al., 2007). Clearly, the numerous small, round granules made in the presence of YFP-GS have a greater surface area on which starch metabolic enzymes can act, compared with the few large, round granules in ss4 plants (Fig. 5, Fig. 6). Thus, expression of YFP-GS may help proper starch turnover indirectly by increasing the granule number. However, whether the granule surface area is indeed a limiting factor for synthesis and/or degradation remains to be tested.
The C-terminal Region of SS4 Alone Promotes Granule Initiation in ss4 Mutants
The C-terminal region of SS4 is homologous to the full-length GS from A. tumefaciens and the phenotypes of plants transformed with the SS4C or the GS construct were similar in several respects. First, expression of both constructs partially restored starch accumulation, with over 80% of the chloroplasts producing multiple starch granules (Fig. 5; Supplemental Figs. S2 and S3). Second, in both cases a greater fraction of the starch could be degraded at night than in ss4 (Fig. 4). Third, neither construct altered amylopectin structure compared to the wild type (and ss4 parental line; Supplemental Fig. S6). However, also in both instances the starch granules still had the rounded ss4-like morphology (Fig. 5; Supplemental Fig. S4), demonstrating that neither YFP-SS4C nor YFP-GS could rescue all aspects of the ss4 mutant phenotype.
Despite the similarities between the YFP-SS4C and the YFP-GS lines, the phenotypes were not identical. The YFP-SS4C lines had fewer starch granules per chloroplast than the YFP-GS lines, and we did not observe supernumerary granules (Supplemental Table S1). This might be due to the relatively low amount of YFP-SS4C protein expressed (Fig. 2A). YFP-SS4C activity could also not readily be detected using zymograms, even though Raynaud et al. (2016) reported that a similarly truncated form of SS4, when recombinantly expressed in E. coli, was measurable this way. Indeed, the phenotypes of the YFP-SS4C lines were most similar to that of YFP-GS_1, that is, the line expressing the lowest amounts of YFP-GS. It is possible that higher amounts of YFP-SS4C would have led to more granules and lower starch contents, as seen with high levels of YFP-GS expression.
However, there were also qualitative differences between the SS4C and GS proteins that could be relevant to the distinct phenotypes observed. First, immunoblots of the YFP-SS4C protein did not yield multiple, slow-migrating bands as they did for YFP-GS (Fig. 2A). This is presumably because SS4C does not have self-priming activity (Szydlowski et al., 2009) and rather relies on interaction with the recently described PTST2 and/or PTST3 proteins to deliver its substrates (Seung et al., 2017). Second, our YFP fluorescence data revealed that SS4C was uniformly distributed in chloroplasts, unlike the granule-surface localization of GS (Fig. 8), suggesting a lower inherent affinity to glucans. Third, it is possible that the glucan elongating activity and specificity of YFP-SS4C differs from that of YFP-GS. Indeed, if glycogen is a poor substrate for SS4C, this might help explain why we did not detect its activity in glycogen-containing native gels.
The N-Terminal Domain of SS4 Determines the Morphology of Starch Granules
Our data indicate that the N-terminal region of SS4 has a very specific and important role in starch granule biosynthesis. It contains predicted coiled-coil domains between amino acids 204 and 466 (see Pfister and Zeeman, 2016 and references therein). This sequence is conserved in SS4 orthologs but is absent in the other SS isoforms, although some of them have distinct predicted coiled-coil motifs of their own (Pfister and Zeeman, 2016). The N-terminal region of SS4 had a patchy subchloroplastic localization similar to that of the full-length SS4 protein (Fig. 8), suggesting that this region determines the protein’s localization. However, it did not rescue the growth, starch synthesis, and turnover, or granule number and morphology phenotypes of ss4 (Figs. 3–5; Supplemental Figs. S2–S4). These data show that the N-terminal region of SS4 alone, while correctly localized, is not functional without the catalytic C-terminal domains.
Remarkably, both the function and localization of A. tumefaciens GS could be changed by fusing the N-terminal region of SS4 to it. Expressing the chimeric protein (SS4N-GSYFP or YFP-SS4N-GS) in ss4 plants complemented most aspects of the ss4 phenotype: the plants displayed near-normal growth rates (Fig. 3), starch levels, and nighttime starch degradation (Fig. 4). Strikingly, SS4N-GS expression not only increased granule numbers but also resulted in wild-type-like lenticular starch granules (Fig. 5; Supplemental Figs. S2–S4). SS4N-GS furthermore showed a subchloroplastic localization that resembled that of SS4 rather than that or GS (Fig. 8). Nevertheless, we still observed subtle differences to the wild type. For example, there was a weak negative correlation between SS4N-GS abundance and starch synthesis: The line that contained the highest activity of SS4N-GS also produced the lowest amount of starch among the three lines and vice versa (e.g. SS4N-GS-YFP_3 versus SS4N-GS-YFP_1; Figs. 2 and 4). This correlation was reminiscent of what was observed in lines expressing just GS (discussed above). Moreover, the SS4N-GS-expressing lines had noticeably more starch granules than the SS4-complementation lines (Fig. 5; Supplemental Figs. S2–S3) and sometimes produced supernumerary smooth-surfaced tiny granules within single stromal pockets of chloroplasts, something not seen in the wild type or in SS4-YFP lines (Fig. 7; Supplemental Fig. S5).
The Functions of SS4
Granule initiation probably requires more than just priming of polysaccharide synthesis: It may require that the precrystalline glucan is not subject to degradation (e.g. by α-amylase or isoamylase; Seung et al., 2016; Pfister et al., 2016) and also that the polysaccharide can be further elaborated and assembled with other glucan molecules to provide the basis for a semicrystalline starch granule. Potentially, these processes are mediated by protein scaffolds that localize SS4 at distinct subplastidial spots. Such a protein scaffold may also serve to guide the biosynthetic machinery after initiation has occurred, directing the pattern of synthesis. Based on our and other results, we propose that the domain structure of SS4 serves distinct important roles in these aspects of starch granule initiation and growth.
The C-terminal domains confer the catalytic activity for glucan chain elongation by SS4 (Szydlowski et al., 2009), which may be particularly important during the early steps of granule initiation. The C-terminal region also appears to confer the capacity to interact with PTST2, which provides it with long malto-oligosaccharide substrates (Seung et al., 2017). A conserved domain immediately adjacent to the C-terminal glucosyltransferase domains allows SS4 dimerization, which was suggested to be required for activity (Raynaud et al., 2016). Dimerization may promote the elongation of long malto-oligosaccharides in proximity to one another. This could increase the likelihood of glucan interactions that must precede the formation of the higher order, semicrystalline structures observed in starch granules. In line with these hypotheses, recombinant barley (Hordeum vulgare) SS4 exhibits significant glucan-elongating activity only on linear malto-oligosaccharides, but not on branched polysaccharides (Cuesta-Seijo et al., 2016). Although the in vivo substrate affinity by SS4 may be modulated by PTST2, it is tempting to speculate that this combination of catalytic specificity, interaction with PTST2, and self-dimerization renders SS4 capable of granule initiation.
Notably, barley isoforms of SS1, SS2, SS3, and GBSS did not display such a preference for malto-oligosaccharides, and were equally or more active on branched polysaccharides as on linear oligosaccharides (Cuesta-Seijo et al., 2016). Phylogenetic analyses of starch synthases suggest that SS1 and SS2 are derived from the duplication of an ancient cyanobacterial GBSS gene, while SS3 and SS4 belong to a clade that either derived from the host or a chlamydial pathogen (Ball et al., 2011). This difference in origin could mean that SS1 and SS2 do not share the catalytic capacities required for priming with SS4. Indeed, when expressed in yeast, Arabidopsis SS1 and, to a lesser extent, SS2 were less efficient in glucan synthesis than SS3 and SS4 when the yeast’s endogenous glycogenins were absent (Pfister et al., 2016). It therefore appears unlikely that the catalytic regions of SS1 or SS2 alone or fused to the N-terminal region of SS4 would be capable of granule initiation in the ss4 mutant background.
The N-terminal region positions SS4 into specific subchloroplastic domains. Thereby, it may help define not only where a granule forms, but also the way in which the granule, once initiated, continues to grow, resulting in its characteristic lenticular shape. The nature of these subchloroplastic domains and how the N-terminal region localizes to them remain unclear. It seems likely that the N-terminal region exerts its function by interacting with other proteins (i.e. the protein scaffold hypothesized above). Two candidate interactors of the N-terminal region of SS4, the thylakoid-associated fibrillin proteins FBN1a and FBN1b, have already been identified. Yet, these alone cannot explain the localization and function of SS4, since lenticular granules are still made when both fibrillins are abolished (Gámez-Arjona et al., 2014). Further studies will be needed to identify additional factors and to build a detailed model of the cell biological context for starch granule biosynthesis.
Finally, the experiments described here also further highlight the apparent link between granule numbers/morphology and diel starch turnover. The ability of the C-terminal region of SS4 or full-length GS to increase granule numbers, and of the attached N-terminal region of SS4 to further restore their normal morphology, correlated with increases in starch amount and its diel turnover. This is important, since starch metabolism is central to the plant’s ability to control its overall carbon budget and to maintain efficient growth (Stitt and Zeeman, 2012). However, it is important to note that in other, recently described genotypes, this link seems to be broken: plants deficient in or overexpressing the SS4-interacting protein PTST2 have fewer or more numerous granules than the wild type, respectively, yet still maintain near-normal diel starch turnover (Seung et al., 2017).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type Col-0, T-DNA insertion mutant ss4-1 (GABI-Kat_290D11), and T2 generation of ss4-1 plants transformed with each of YFP-GS, YFP-SS4C, SS4N-GS-YFP, YFP-SS4N-GS, and SS4-YFP were used. Seeds were sown on soil and placed for 2 d at 4°C in the dark and were grown in a Percival growth chamber (CLF Plant Climatics) under a 12-h-day/12-h-night regime. The temperature was 20°C, relative humidity was 70%, and the light intensity was 150 µmol quanta m−2 s−1.
Cloning and Isolation of Transgenic Lines
The full-length coding sequence of GS from Agrobacterium tumefaciens (Atu4075) was amplified from genomic DNA using GS-fw and GS-rev primers, given in Supplemental Table S2. The PCR product was cloned into the pDONR221 vector and was further cloned into the pB7WGY2 vector (modified to contain a chloroplast transit peptide [rCT: amino acids 1-53 of At5g38430] upstream and in frame with YFP) via Gateway recombination cloning technology (Invitrogen). The resulting construct was named YFP-GS (Fig. 1). The same YFP-GS construct as used in Crumpton-Taylor et al. (2013) was introduced into ss4 mutant plants, yielding the two independent lines YFP-GS_1 and YFP-GS_3. The third independent line, YFP-GS_2, was generated by crossing the ss4 mutant with one of our previously generated YFP-GS lines in the ss3 ss4 background (GS-5-3; Crumpton-Taylor et al., 2013) and selecting for the homozygous YFP-GS, ss4, and wild-type SS3 alleles. Similarly, the C-terminal region of Arabidopsis SS4 (SS4C; amino acid residues 467 to 1040) was amplified from the pDONR-AtSS4 plasmid (Roldán et al., 2007) using SS4C-fw and SS4C-rev primers (Supplemental Table S2). The amplified SS4C fragment was cloned into rCT containing pB7WGY2,0 vector and the resulting construct was named YFP-SS4C (Fig. 1). The N-terminal domain of SS4 (SS4N; amino acid residues 1 to 544) was amplified with attB sites using SS4-fw, SS4N-GS-rev primers. The SS4NΔctp-fw and SS4N-GS-rev primers were used to amplify the N-terminal domain of SS4 without its predicted chloroplastic transit peptide (amino acid residues 43 to 544). The pDONR-AtSS4 plasmids were used as template. The amplicons of SS4N and GS were cleaved and ligated via Bcl1 restriction sites, cloned into pB7YWG2,0 and into the rCT-containing pB7WGY2,0 vectors. The resulting constructs were named SS4N-GS-YFP and YFP-SS4N-GS (Fig. 1). The full-length cDNA of SS4 was cloned into pB7WGY2,0 via the Gateway LR reaction, and named SS4-YFP (Fig. 1).
We transformed each expression construct (YFP-GS, YFP-SS4C, SS4N-GS-YFP, YFP-SS4N-GS, and SS4-YFP) into ss4-1 mutants. The T1 seeds from each transformation were harvested and sown on soil. Multiple T1 transgenic plants for each transformation were isolated by Basta resistance screening. Two or three independent lines for each transformation were selected for further analysis, based on different abundances and fluorescence appearance of transformed proteins. T2 plants carrying the ss4-1 allele and transformed constructs were identified by PCR-based genotyping using primers shown in Supplemental Table S2.
Immunoblotting and Zymograms
Whole rosettes from single 25-d-old plants were harvested at the end of day, weighed, frozen in liquid N2, and ground into fine powder. Then 10 to 20 mg of the frozen powder from each plant was mixed in the extraction solution by the ratio of 100 µL extraction solution /10 mg frozen powder, containing 100 mM MOPS, pH 7.0, 1 mM EDTA, 5 mM DTT, glycerol, and protease inhibitors (Complete EDTA-free from Roche) with 0.5% Triton-X, 10% (v/v). After 5 min at 16,000g centrifugation, soluble protein extracts were isolated, and concentrations were quantified by using BCA protein assay kit (Sigma). Equal abundance of soluble protein extract from each sample of each line was pooled and used for immunoblotting and Zymogram. Then 20 µg pooled soluble protein extracts from each line were mixed with an equal volume of SDS loading solution (50 mm Tris, pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 5 mm dithiothreitol), boiled 5 min, and loaded to 10% SDS PAGEs. After electrophoresis at 10V cm-1 for 1 h at room temperature, proteins were transferred to polyvinylidene difluoride membranes, which were probed with monoclonal antibodies against YFP (Clontec JL-08) or SS4. Antibodies against beta-actin (from Sigma) were added for quantitative internal controls. Two-color IRDye secondary antibodies and Odyssey Infrared Imaging System (Odyssey CLx) were then used to visualize signals for transformed proteins (Green) and beta-actin (Red).
Abundances of detected proteins were quantified by using Image Studio Lite software. The remained homogenate was centrifuged for 5 min at 16,000g at 4°C. Soluble proteins in the supernatant were quantified using Bio-Red Protein Assay. For zymograms, 50 µg protein extracts from each line were loaded onto 7.5% native PAGE gels containing 0.3% oyster glycogen, following the procedures described in Crumpton-Taylor et al., 2013.
Starch Measurement
Whole rosettes of 25-d-old plants from each line were harvested at the end of day and at the end of night, decolorized in hot 70% (v/v) ethanol, stained with Lugol’s solution (Sigma-Aldrich), and stored in cold water overnight to remove excess iodine from rosettes.
To quantify starch content, whole rosettes of 25-d-old plants of Col, ss4, and each of the transformed lines were harvested at the end of day and end of night. Plants were weighed, frozen in liquid N2, ground to a powder, and further homogenized in 2 mL of 1.12 m perchloric acid (HClO4). The homogenates were subjected to centrifugation (3,000g, 10 min, 20°C) to separate the insoluble and soluble fractions. The insoluble fraction contained starch and was washed once with cold water, four times with 70% (v/v) ethanol, and resuspended in 1 mL water. Starch was quantified in the insoluble fraction as described in Hostettler et al. (2011).
Confocal Microscopy
A 1-cm2 leaf piece from a 30-d-old plant was harvested at the middle of day and observed by Zeiss LSM 780 confocal laser scanning with a 40x water immersion objective. YFP-fused proteins were excited by a 488/514 nm argon laser and detected at 520 to 570 nm. Chlorophyll auto-fluorescence was excited by a 594-nm helium neon laser and detected at 620 to 700 nm.
Isolation and Structural Analysis of Starch Granules
Whole rosettes of 5-week-old plants from Col, ss4, and each of the transformed lines were harvested after an 11-h photoperiod, frozen in liquid N2, ground to a powder, homogenized in starch extraction medium (50 mm Tris-HCl [pH 8], 0.2 mm EDTA, and 0.5% [v/v] Triton X-100), and filtered through a series of nylon nets (100-µm, 30-µm, and 15-µm pore sizes). Starch granules were separated from the filtrate by centrifugation (3,000g, 20 min, at 20°C) over a 95% (v/v) Percoll (Sigma-Aldrich) cushion. Starch pellets were washed three times with 0.5% (w/v) SDS and four times with water to remove the remaining SDS. The pellets were dried for 16 h in vacuo. Half a milligram of pure starch granules was boiled for 20 min, cooled at room temperature, debranched for 3 h by incubation in a solution containing 10 mm sodium acetate pH 4.8, 600 U isoamylase (from Pseudomonas sp.; Sigma), and 1 U pullulanase (M1, from Klebsiella planticola, Megazyme). Linear chains were analyzed by high performance anionexchange chromatography with pulsed amperometric detection (HPAEC-PAD) as described in Streb et al. (2008).
Light Microscopy and Electron Microscopy
Leaf pieces (4 mm2) were cut from three young rosette leaves from three 30-d-old plants and fixed for 16 h in 2% (v/v) glutaraldehyde and 0.05 m sodium cacodylate, pH 7.4, at 4°C. The leaf pieces were washed three times with 0.1 m sodium cacodylate, pH 7.4, and incubated overnight in 1% (w/v) osmium tetroxide with 0.1 m sodium cacodylate, pH 7.4 at 4°C. Sequentially, pieces were washed three times with ice-cold 0.1 m sodium cacodylate, pH 7.4, and once with water. Samples were dehydrated in a series of aqueous ethanol solutions from 50% (v/v) to 100% ethanol and once with 100% acetone. The leaf pieces were incubated sequentially for 2 h in 25% (v/v) and 50% epoxy resin (Spurr’s; Agar Scientific, in acetone), 16 h in 75% epoxy resin, and 7 h in 100% epoxy resin. Embedding was completed by incubation for 48 h with fresh 100% epoxy resin at 70°C. Semithin sections were cut with a Diatome 45° diamond knife. Then 500-nm-thick sections were placed on glass slides, stained for 10 min with toluidine blue, washed with water, and dried. Light micrographs were taken using Zeiss Axio Imager Z2 light microscope with a 100x oil immersion objective. For transmission electron microscopy, sections of 70-nm thickness were placed on formvar carbon-coated copper grids, stained with 2% (w/v) uranyl acetate and Reynold’s lead citrate and imaged with a FEI Morgagni 268 transmission electron microscope. Pictures are representative for the analysis of sections from two individual plants per genotype. For scanning electron microscopy, pure starch granules were coated with a 3-nm platinum layer using Balzers MED010 coating device and visualized by Leo 1530 Gemini microscope (Zeiss).
Accession Numbers
Arabidopsis Genome Initiative gene code for Arabidopsis STARCH SYNTHASE 4 (SS4) is At4g18240. A. tumefaciens genome annotation for GS is Atu4075.
Supplemental Material
The following supplemental materials are available.
Supplemental Figure S1. The rosette leaf chosen for light and transmission electron microscopy.
Supplemental Figure S2. Light micrographs of leaf sections of three independent lines transformed with each construct encoding GS- or SS4-derived proteins.
Supplemental Figure S3. Transmission electron micrographs of mesophyll cell chloroplasts of three independent lines transformed with each construct encoding GS- or SS4-derived proteins.
Supplemental Figure S4. Scanning electron micrographs of starch granules from three independent lines transformed with each construct encoding GS- or SS4-derived proteins.
Supplemental Figure S5. Transmission electron micrographs showing supernumerary granules in chloroplasts of YFP-SS4N-GS lines.
Supplemental Figure S6. Chain length distribution of starch from the wild type (Col), ss4, and ss4 plants transformed with each construct encoding GS- or SS4-derived proteins.
Supplemental Table S1. Distribution of starch granules in chloroplast sections of the wild type (Col), ss4, and ss4 plants lines transformed with each construct encoding GS- or SS4-derived proteins.
Supplemental Table S2. Primers sequences used for ss4 mutant genotyping and for the amplification of coding sequences of A. tumefaciens GS and Arabidopsis SS4.
Acknowledgments
We thank Ángel Mérida for seeds of ss4-1 and for great suggestions during the course of this work. We thank the staff at ScopeM (ETH Zurich) for advice on electron microscopy. We thank Andrea Ruckle for assistance with plant cultivation. We thank Michaela Fischer-Stettler for technical assistance with operation of HPAEC-PAD Dionex. We thank David Seung for reading and providing insightful suggestions during manuscript preparation.
Footnotes
This work was financially supported by the Swiss National Science Foundation (grant no. 31003A–153144/1 to S.C.Z.) and by ETH Zurich.
Articles can be viewed without a subscription.
References
- Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C (2011) The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J Exp Bot 62: 1775–1801 [DOI] [PubMed] [Google Scholar]
- Ball S, Guan HP, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J (1996) From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell 86: 349–352 [DOI] [PubMed] [Google Scholar]
- Buléon A, Colonna P, Planchot V, Ball S (1998) Starch granules: structure and biosynthesis. Int J Biol Macromol 23: 85–112 [DOI] [PubMed] [Google Scholar]
- Crumpton-Taylor M, Grandison S, Png KMY, Bushby AJ, Smith AM (2012) Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol 158: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpton-Taylor M, Pike M, Lu K-J, Hylton CM, Feil R, Eicke S, Lunn JE, Zeeman SC, Smith AM (2013) Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol 200: 1064–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Seijo JA, Nielsen MM, Ruzanski C, Krucewicz K, Beeren SR, Rydhal MG, Yoshimura Y, Striebeck A, Motawia MS, Willats WGT, et al. (2016) In vitro biochemical characterization of all barley endosperm starch synthases. Front Plant Sci 6: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830 [DOI] [PubMed] [Google Scholar]
- Delvallé D, Dumez S, Wattebled F, Roldán I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, et al. (2005) Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J 43: 398–412 [DOI] [PubMed] [Google Scholar]
- Fontaine T, D’Hulst C, Maddelein M-L, Routier F, Pépin TM, Decq A, Wieruszeski JM, Delrue B, Van den Koornhuyse N, Bossu JP, et al. (1993) Toward an understanding of the biogenesis of the starch granule. Evidence that Chlamydomonas soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem 268: 16223–16230 [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park J-H, Jane J-L, et al. (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144: 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámez-Arjona FM, Raynaud S, Ragel P, Mérida A (2014) Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J 80: 305–316 [DOI] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149: 1541–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Liu F, Marsh RS, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM (2008) Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol 146: 1892–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettler C, Kölling K, Santelia D, Streb S, Kötting O, Zeeman SC (2011) Analysis of starch metabolism in chloroplasts. Methods Mol Biol 775: 387–410 [DOI] [PubMed] [Google Scholar]
- Leterrier M, Holappa LD, Broglie KE, Beckles DM (2008) Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol 8: 98–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43: D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Makhmoudova A, Lee EA, Wait R, Emes MJ, Tetlow IJ (2009) The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J Exp Bot 60: 4423–4440 [DOI] [PubMed] [Google Scholar]
- Liu F, Romanova N, Lee EA, Ahmed R, Evans M, Gilbert EP, Morell MK, Emes MJ, Tetlow IJ (2012) Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. Biochem J 448: 373–387 [DOI] [PubMed] [Google Scholar]
- Malinova I, Alseekh S, Feil R, Fernie AR, Baumann O, Schöttler MA, Lunn JE, Fettke J (2017) Starch synthase 4 and plastidal phosphorylase differentially affect starch granule number and morphology. Plant Physiol 174: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougué O, Corbi J, Ball SG, Manicacci D, Tenaillon MI (2014) Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway. BMC Evol Biol 14: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez S, Bertoft E (2010) The molecular structures of starch components and their contribution to the architecture of starch granules: A comprehensive review. Starke 62: 389–420 [Google Scholar]
- Pfister B, Lu K-J, Eicke S, Feil R, Lunn JE, Streb S, Zeeman SC (2014) Genetic evidence that chain length and branch point distributions are linked determinants of starch granule formation in Arabidopsis. Plant Physiol 165: 1457–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister B, Sánchez-Ferrer A, Diaz A, Lu K, Otto C, Holler M, Shaik FR, Meier F, Mezzenga R, Zeeman SC (2016) Recreating the synthesis of starch granules in yeast. eLife 5: 1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister B, Zeeman SC (2016) Formation of starch in plant cells. Cell Mol Life Sci 73: 2781–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragel P, Streb S, Feil R, Sahrawy M, Annunziata MG, Lunn JE, Zeeman S, Mérida Á (2013) Loss of starch granule initiation has a deleterious effect on the growth of arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol 163: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud S, Ragel P, Rojas T, Mérida Á (2016) The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. J Biol Chem 291: 10759–10771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D’Hulst C, Mérida A (2007) The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J 49: 492–504 [DOI] [PubMed] [Google Scholar]
- Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, Okita TW, Kaneko N, Fujita N, Yoshida M, et al. (2008) Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20: 1833–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoura T, Asao A, Takashima Y, Isono N, Hamada S, Ito H, Matsui H (2007) Enzymatic characterization of starch synthase III from kidney bean (Phaseolus vulgaris L.). FEBS J 274: 4550–4560 [DOI] [PubMed] [Google Scholar]
- Seung D, Boudet J, Monroe J, Schreier TB, David LC, Abt M, Lu KJ, Zanella M, Zeeman SC (2017) Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves. Plant Cell 29: 1657–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D, Lu K-J, Stettler M, Streb S, Zeeman SC (2016) Degradation of glucan primers in the absence of starch synthase 4 disrupts starch granule initiation in Arabidopsis. J Biol Chem 291: 20718–20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D, Soyk S, Coiro M, Maier BA, Eicke S, Zeeman SC (2015) PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker CK, Boyer CD, Wiesenborn DP, Thompson DB, Boersig MR, Oakes JV, Stalker DM (1994) Expression of Escherichia coli glycogen synthase in the tubers of transgenic potatoes (Solanum tuberosum) results in a highly branched starch. Plant Physiol 104: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC (2012) Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol 15: 282–292 [DOI] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman SC (2008) Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20: 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero M, Muñoz FJ, Ovecka M, Bahaji A, Planchot V, et al. (2009) Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 21: 2443–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatge H, Marshall J, Martin C, Edwards EA, Smith AM (1999) Evidence that amylose synthesis occurs within the matrix of the starch granule in potato tubers. Plant Cell Environ 22: 543–550 [Google Scholar]
- Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, Bresolin NS, Wait R, Morell MK, Emes MJ (2008) Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol 146: 1878–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde JE, Parodi AJ, Ugalde RA (2003) De novo synthesis of bacterial glycogen: Agrobacterium tumefaciens glycogen synthase is involved in glucan initiation and elongation. Proc Natl Acad Sci USA 100: 10659–10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T, Nakamura Y, Satoh H, Terashima K (1999) Differences in amylopectin structure between two rice varieties in relation to the effects of temperature. Starke 2–3: 58–62 [Google Scholar]
- Valdez HA, Busi MV, Wayllace NZ, Parisi G, Ugalde RA, Gomez-Casati DF (2008) Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry 47: 3026–3032 [DOI] [PubMed] [Google Scholar]
- van de Wal M, D’Hulst C, Vincken JP, Buléon A, Visser R, Ball S (1998) Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J Biol Chem 273: 22232–22240 [DOI] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al. (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Roach PJ, Montero M, Baroja-Fernández E, Muñoz FJ, Eydallin G, Viale AM, Pozueta-Romero J (2010) Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev 34: 952–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2002) The priming of amylose synthesis in Arabidopsis leaves. Plant Physiol 128: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]