The tangled1air9 double mutant had synthetic division plane orientation and root growth and cell file rotation defects that are rescued by full-length TAN1-YFP and only partially by TAN1-YFP deletion constructs.
Abstract
TANGLED1 (TAN1) and AUXIN-INDUCED-IN-ROOTS9 (AIR9) are microtubule-binding proteins that localize to the division site in plants. Their function in Arabidopsis (Arabidopsis thaliana) remained unclear because neither tan1 nor air9 single mutants have a strong phenotype. We show that tan1 air9 double mutants have a synthetic phenotype consisting of short, twisted roots with disordered cortical microtubule arrays that are hypersensitive to a microtubule-depolymerizing drug. The tan1 air9 double mutants have significant defects in division plane orientation due to failures in placing the new cell wall at the correct division site. Full-length TAN1 fused to yellow fluorescent protein, TAN1-YFP, and several deletion constructs were transformed into the double mutant to assess which regions of TAN1 are required for its function in root growth, root twisting, and division plane orientation. TAN1-YFP expressed in tan1 air9 significantly rescued the double mutant phenotype in all three respects. Interestingly, TAN1 missing the first 126 amino acids, TAN1-ΔI-YFP, failed to rescue the double mutant phenotype, while TAN1 missing a conserved middle region, TAN1-ΔII-YFP, significantly rescued the mutant phenotype in terms of root growth and division plane orientation but not root twisting. We use the tan1 air9 double mutant to discover new functions for TAN1 and AIR9 during phragmoplast guidance and root morphogenesis.
Plant cells are typically constrained by cell walls (Cosgrove, 2005) that are also connected via plasmodesmata (Brunkard and Zambryski, 2017), and they do not migrate relative to each other. In the absence of significant cell migration, the entire plant body must be built through elegant coordination between the division, expansion, and differentiation of cells. Therefore, division plane orientation, or the spatial control of cytokinesis, has important roles in plant development and growth (Pickett-Heaps et al., 1999).
The key steps of plant division plane orientation occur during interphase, G2, and mitosis. During interphase, the cortical microtubule array typically aligns perpendicular to the cell expansion axis (Baskin, 2001), and during G2 (Gunning and Wick, 1985), this promotes the formation of a land plant-specific microtubule and microfilament array called the preprophase band (PPB; Pickett-Heaps and Northcote, 1966). Although the location of the PPB accurately predicts the future division site (Gunning et al., 1978; Van Damme et al., 2007; Rasmussen et al., 2013; Lipka et al., 2014; Martinez et al., 2017), its role in division plane establishment has recently been called into question by mutants that do not form obvious PPBs but manage to produce relatively orderly roots. These plants have mutations in the tonneau1a locus (Zhang et al., 2016) and mutations in three related tonneau1 recruiting motif (trm) loci (Schaefer et al., 2017). Whether the PPB establishes the future division site or is the signpost of an earlier established cue for the future division site remains an active area of investigation. The PPB is a transient structure that forms in G2 and promotes proper spindle orientation (Chan et al., 2005; Ambrose and Cyr, 2008; Schaefer et al., 2017) but then disappears from the cortex. The PPB is thought to leave behind a division site marker that recruits the division machinery to the correct location (Rasmussen et al., 2013). The spindle separates the chromosomes in anaphase, and finally, the phragmoplast is formed during telophase. The phragmoplast is composed of opposing microtubule and microfilament disks, with the microtubule plus ends directed toward the middle of the cell (Jürgens, 2005; Lee and Liu, 2013; Smertenko et al., 2017). Vesicles containing cell wall materials are transported along the microtubule tracks toward the center of the phragmoplast, which specifies the location and growth axis of the newly forming cell wall, called the cell plate (Boruc and Van Damme, 2015). As the phragmoplast expands, microtubules at the center are disassembled (Smertenko et al., 2011; Murata et al., 2013). In wild-type cells, the phragmoplast expands toward the location demarcated previously by the PPB, and the cell plate fuses with the mother cell wall (Rasmussen et al., 2013). Finally, the phragmoplast is disassembled and the new cell wall is formed (Lee and Liu, 2013).
The tangled1 (tan1) mutant and its corresponding gene (GRMZM2G039113) were first identified as important for division plane orientation in maize (Zea mays), because mutants had misplaced cell walls (Smith et al., 1996, 2001) and cytoskeletal arrays (Cleary and Smith, 1998). However, the identification of AtTANGLED1 (TAN1 [At3g05330]; also called ATN, TAN, or AtTAN but referred to hereafter as TAN1) as the first positive marker of the division site in Arabidopsis (Arabidopsis thaliana; Walker et al., 2007) illuminated another aspect of its important role in division plane orientation. TAN1-YFP in both Arabidopsis (Walker et al., 2007) and maize (Martinez et al., 2017) localized to the division site when or after the PPB formed, until after disassembly of the phragmoplast. The maize tan1 mutant often failed to place the new cell wall in the location specified by the PPB, indicating significant defects in phragmoplast guidance to the division site (Martinez et al., 2017).
A deletion study in Arabidopsis defined regions of the TAN1 protein required for its localization to the division site. The first 126 amino acids defined a region of the protein (region I) that was both necessary and sufficient for colocalization with the PPB. Another, middle region (region II, defined by amino acids 126–222) was necessary and sufficient for TAN1 localization to the division site during telophase. Together, these two parts of the protein were sufficient for TAN1 localization from prophase until the end of cytokinesis (Rasmussen et al., 2011b). Unfortunately, functional analysis of the Arabidopsis TAN1 protein by transgenic rescue experiments has been hampered by a weak tan1 mutant phenotype. In maize, TAN1-YFP rescued the tan1 mutant phenotype and localized to the division site and also to mitotic microtubule arrays (Martinez et al., 2017).
AUXIN INDUCED IN ROOTS9 (AIR9 [AT2G34680]) was first identified as an auxin-induced mRNA in root cultures (Neuteboom et al., 1999). Later, the AIR9 protein was described as a microtubule-binding protein that localized to microtubules, including the PPB, and also to the cell plate insertion site when the phragmoplast reached the mother cell cortex (Buschmann et al., 2006). Although the mutant was originally thought to be lethal due to a linked mutation, later evidence indicated that air9 mutants grew similarly to wild-type plants and did not display obvious division plane or growth defects (Buschmann et al., 2015). AIR9 is conserved in plants and also is found in the eukaryotic parasitic trypanosomatid lineage (Buschmann et al., 2007). In Trypanosoma brucei, deletion of AIR9 altered cleavage furrow and nuclear positioning (May et al., 2012).
TAN1 and AIR9 localization at the division site hints that they may have important roles in division plane orientation, but single tan1 or air9 mutants have mild or no phenotypes, respectively. Here, we demonstrate that tan1 air9 double mutants have a synthetic (or synergistic) phenotype consisting of defects in root growth, division plane orientation, and differentiation zone root cell file rotation, suggesting both mitotic and interphase microtubule functions. We used the tan1 air9 double mutant as a tool to understand TAN1 function in Arabidopsis: full-length TAN1-YFP significantly rescued division plane defects, root length, and root twisting of the tan1 air9 mutant. A truncation study showed that the N-terminal region of TAN1 is indispensable for function, while a central region is required to buffer a previously unidentified interphase function of TAN1.
RESULTS
The tan1 air9 Double Mutants Show Synthetic Phenotypes in Root Growth and Patterning
In Arabidopsis, air9 mutants were crossed to tan1 mutants to determine whether tan1 could enhance the air9 mutant phenotype. One allele combination was generated in the Landsberg erecta (Ler) ecotype, while the other was generated in a mixed Wassilewskija (Ws) and Columbia (Col) background, as described in “Materials and Methods.” Both the Ler and Ws/Col single and double mutants had similar phenotypes, suggesting that the mutant combination of air9 with tan1 was responsible for the double mutant phenotype instead of an unlinked mutation. Because both allele combinations showed similar phenotypes, we show data for Ler ecotype mutants in the main figures and Ws/Col ecotype mutants in supplementary information. Adult tan1 air9 double mutant plants show an overall short phenotype compared with single mutants or Ler plants (Supplemental Fig. S1). While Ler and air9 and tan1 single mutants had minor or no defects in root cell patterning (Fig. 1, A–C; Supplemental Fig. S2, A–C), tan1 air9 double mutants had roots with disordered patterning (Fig. 1D; Supplemental Fig. S2D). The tan1 air9 double mutant roots were significantly shorter (Fig. 1E; Supplemental Fig. S2E) and wider when compared with Ler (Ler, 111.4 ± 2.2 µm, n = 9; tan1 air9, 123.9 ± 2.6 µm, n = 11; Student’s t test, P = 0.002; measured at the transition area between the meristematic and elongation zones [Wachsman et al., 2015] of 5-d-old roots). Several of these phenotypes are consistent with division plane defects described in detail in the next section.
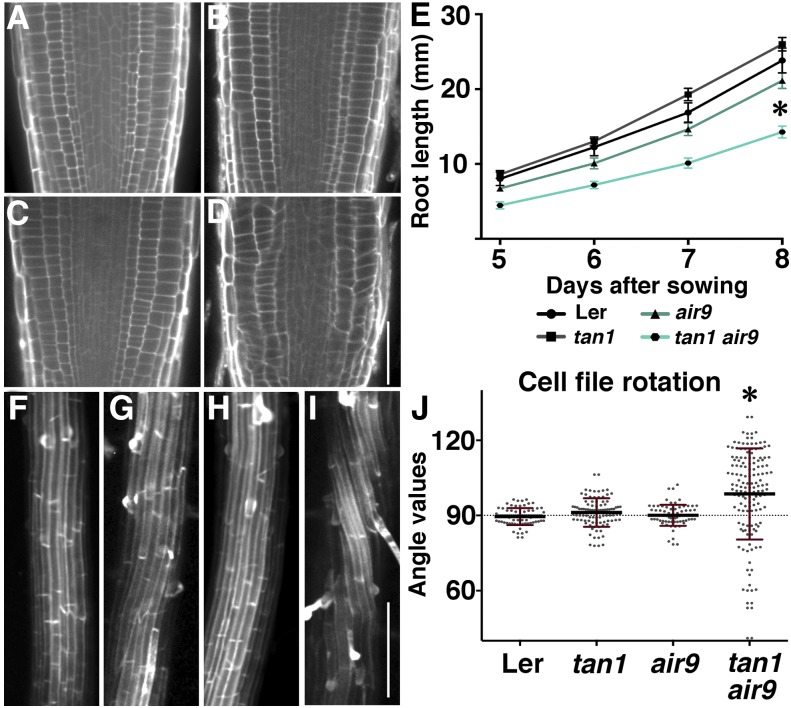
Figure 1.
Root phenotypes of Ler, tan1, air9, and tan1 air9 double mutants. A to D, Root cell walls stained with propidium iodide of Ler (A), tan1 (B), air9 (C), and tan1 air9 (D) plants. E, Root length measurements from days 5 to 8 after sowing of Ler and mutant plants onto vertically oriented plates. The asterisk indicates that tan1 air9 roots were significantly shorter than others (P < 0.01). F to I, Maximum projections of 10 0.2-µm Z-stacks showing expansion to the differentiation zone of propidium iodide-stained roots of Ler (F), tan1 (G), air9 (H), and tan1 air9 (I). J, Root cell file rotation (twisting) measurements. Each dot represents an angle measured from the transverse wall to the long axis of the root. Angles greater than 90° are left twisting and angles less than 90° are right twisting. The asterisk indicates a significant difference in the distribution of tan1 air9 cell file rotation compared with others (P < 0.01). Bars = 50 µm (A–D) and 200 µm (F–I).
We examined the meristematic, elongation, and differentiation zones of tan1 air9 double mutant roots to determine how they contributed to root growth defects. Meristematic zones were identified by dividing cells, elongation zones were identified by cell lengths more than twice the size of meristematic cell lengths, and differentiation zones of roots were identified by the presence of root hairs or xylem (Wachsman et al., 2015). After both 5 and 7 d of growth, the meristematic zones of tan1 air9 roots were significantly shorter than in Ler roots (Supplemental Fig. S3, A, B, E, and H). Elongation zones also were statistically significantly shorter in the tan1 air9 double mutant compared with Ler controls at 5 and 7 d (Supplemental Fig. S3, A, B, E, and H). Next, we measured cell areas to estimate cell sizes in meristematic, elongation, and differentiation zones. Cell areas were similar in Ler and tan1 air9 roots (Supplemental Fig. S3, D and G). In addition, when we compared the ratio between the length and width of cells, there were small or no differences between Ler and tan1 air9 meristematic or elongation zone cell shapes. In contrast, differentiation zone root cells were significantly shorter and wider in the tan1 air9 double mutant (Supplemental Fig. S3, C and F). Together, these data indicated that the short-root phenotype was caused by smaller meristematic and elongation zones as well as by shorter, wider cells in the differentiation zone.
In addition to shorter, wider cells, the late differentiation zone of tan1 air9 roots showed increased cell file rotation or twisting compared with roots of single mutants or the wild type (Fig. 1, F–J; Supplemental Fig. S2, F–J). At least four roots of seedlings grown for 8 d on vertical plates from each genotype were used to measure the angle between the long axis of the root and the transverse cell wall. Typical transverse wall angles were 90° for cell files that do not twist. For Ler and single tan1 and air9 mutants, the cell file rotation angles were both similar to each other and close to the expected 90° value (Fig. 1J; n = 52, 72, and 54 with variance of 15.8, 24.3, and 31.1 and Wilcoxon signed rank test two-tailed P = 0.089, 0.366, and 0.301, respectively). In contrast, tan1 air9 double mutants had significant root cell file twisting, indicated by much higher variance and significant offset from 90° (Fig. 1J; n = 140, σ = 362.4, and P ≤ 0.0001). Angles above 90° indicate left-handed twisting, while angles below 90° indicate right-handed twisting. When cell file rotation was examined in straight (versus bent) sections of the root, the leftward-twisting tendency was more pronounced. Bent root sections tended toward right-handed twisting (Supplemental Fig. S4). Similarly, left root cell file twisting also was observed in the tan1 air9 double mutant in the Col/Ws ecotype, and the corresponding variance in cell file rotation angles was much higher than in single mutants or the wild type (Supplemental Fig. S2, F–J). These data indicate that TAN1 and AIR9 play a role in maintaining proper cell file orientation in roots and, therefore, suggest an interphase function.
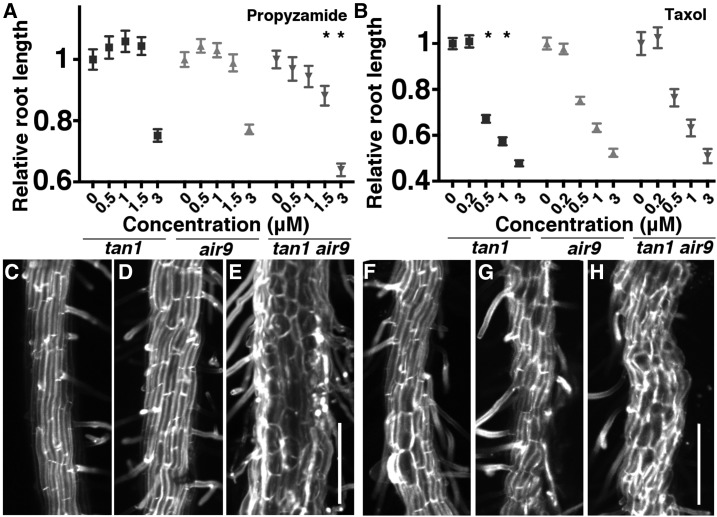
Based on the left-handed cell file rotation in the tan1 air9 double mutant and the short, wide root cells in the differentiation zone, we predicted that microtubule arrays would be destabilized in the tan1 air9 double mutant, similar to tubulin mutants (Ishida et al., 2007). We assessed microtubule array orientation using fixed, immunostained 7-d-old roots. In the elongation zone, microtubule organization was significantly more transverse in wild-type Ler cells (Supplemental Fig. S5A; n = 36 cells from eight plants) compared with tan1 air9 double mutant cells (Supplemental Fig. S5B; n = 38 cells from seven plants, Mann-Whitney P < 0.001). The microtubule arrays were not skewed in either direction in the double mutant but instead showed significantly more variability in their orientation (Supplemental Fig. S5C). To gain further insight into how TAN1 and AIR9 altered microtubule function, single and double mutant seedlings were grown for 4 d and then transferred to plates containing different concentrations of a microtubule-destabilizing drug, propyzamide, for 4 d. Each day, root lengths were measured. Graphs show root length normalized to dimethyl sulfoxide (DMSO) only (negative control) treated seedlings (day-8 graphs in Fig. 2A and days 5–7 in Supplemental Fig. S6). Overall, tan1 air9 mutant root growth was more sensitive to propyzamide than tan1 or air9 mutants. Not only was tan1 air9 root growth inhibited at a lower concentration (1.5 µm), but it grew proportionally more slowly in 3 µm propyzamide compared with either single mutant (Fig. 2A). In contrast, when tan1, air9, and tan1 air9 plants were treated with the microtubule-stabilizing drug taxol, only tan1 showed increased sensitivity to medium concentrations of taxol (0.5 and 1 µm). Increasing taxol concentrations caused reduced growth (Fig. 2B). In addition, although single tan1 (Fig. 2C) and air9 (Fig. 2D) mutants responded to 3 µm propyzamide with loss of anisotropy, the tan1 air9 plants had a more severe phenotype (Fig. 2E). Together, these data show that tan1 air9 mutants have disorganized microtubule arrays and that they are hypersensitive to propyzamide but do not respond differentially compared with air9 mutants to taxol (Fig. 2, B and F–H).
Figure 2.
Effects of propyzamide and taxol on tan1, air9, and tan1 air9 double mutant plants. A and B, Relative root length normalized by lengths of the 0 µm-treated roots of 8-d-old tan1, air9, and tan1 air9 plants grown on different concentrations of propyzamide (A) and taxol (B). Asterisks indicate significantly different responses to drug treatments (Kolmogorov–Smirnov test, P < 0.01; n > 24 plants per condition each day). C to H, Maximum projections of 10 (C, D, and F–H) and 30 (E) 1-µm Z-stacks of 8-d-old propidium iodide-stained differentiation zone roots of tan1 (C and F), air9 (D and G), and tan1 air9 (E and H) plants treated with 3 µm propyzamide (C–E) and 3 µm taxol (F–H). Bars = 200 µm.
Next, cell file rotation was assessed in the plants treated with propyzamide and taxol. After root growth assays were completed, the roots of plants treated with 0, 1, and 3 µm propyzamide or taxol were imaged and cell file rotation was measured as described earlier. Treatment with propyzamide or taxol increased cell file rotation, but neither single nor double mutants showed a differential response in either drug (Supplemental Fig. S7). Due to significant cell bulging, we were unable to accurately measure cell file rotation at 3 µm for propyzamide and taxol (Fig. 2, C–E).
Division Plane Maintenance Is Disrupted in tan1 air9 Double Mutant Cells
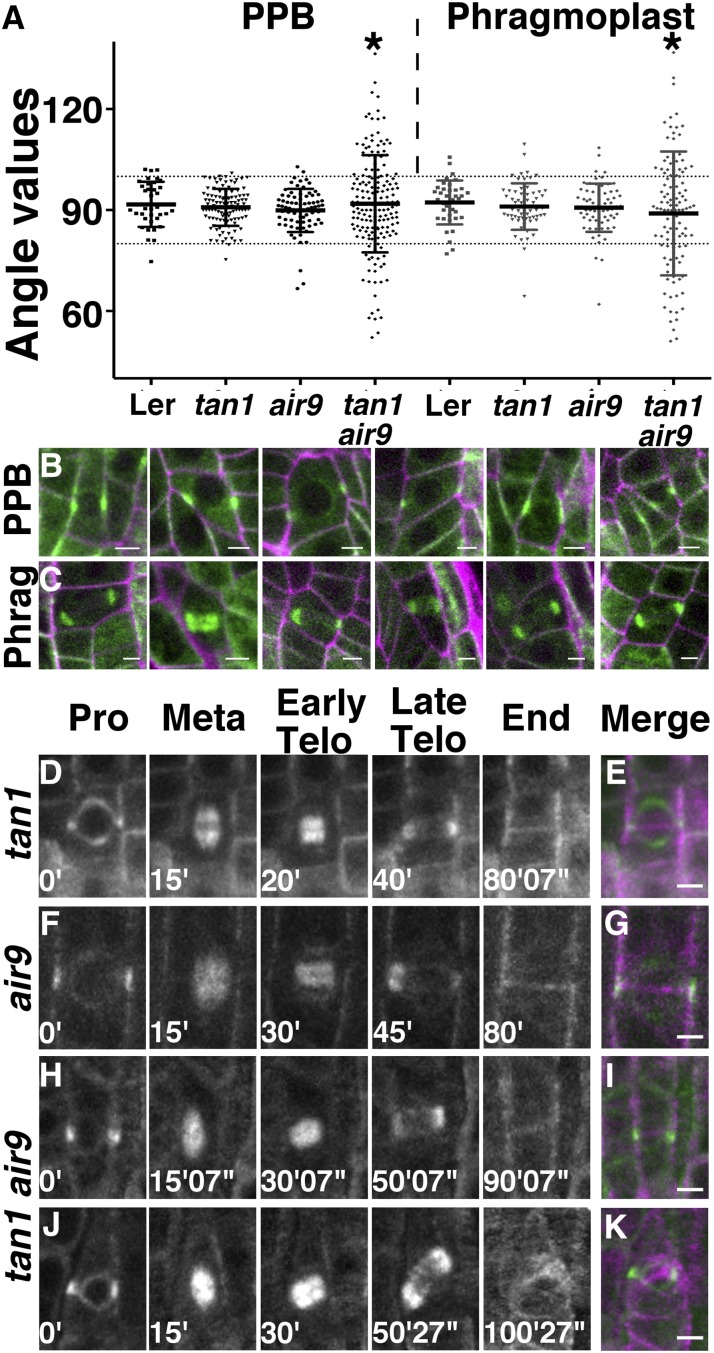
Mitotic microtubule array orientation was measured to determine whether tan1 air9 double mutants had division plane defects, as predicted from observed altered root cell patterning (Fig. 1D). For this purpose, single and double mutants were transformed with a live cell marker for microtubules, Cerulean fluorescent protein fused to ⍺-tubulin4 (CFP-TUBULIN; Kirik et al., 2007). Two independent CFP-TUBULIN transgenic lines in the single tan1 and air9 mutants and in the tan1 air9 double mutant were selected based on homogenous fluorescence in all cells and similar root growth compared with nontransformed sibling plants (Supplemental Fig. S8). Next, PPB orientation angle was measured by drawing the angle between the PPB and the lateral cell wall in cortex and epidermal root cells from seedlings grown for 5 to 7 d. In wild-type cells, epidermal and cortex root cells typically divide close to ∼90° compared with the long axis of the cell. In both tan1 and air9 single mutants, PPB angles were within 80° to 100° in more than 90% of cells (98.5% [n = 67] and 93.5% [n = 77], respectively), similar to the wild-type control (92.3% [n = 39]; Fig. 3A; Table I). In contrast, 38.8% of cells (n = 121) of the tan1 air9 double mutant had PPB angles outside 80° to 100°. The variance in PPB angles of single tan1 and air9 mutants was similar to the variance in the wild-type PPB angles of the wild-type control (σ = 44, 32, and 40 for the wild type, tan1, and air9, respectively) and not statistically significantly different (F test, P = 0.1258 and P = 0.7199 for tan1 and air9, respectively), whereas the variance for the double tan1 air9 mutant was ∼4.5-fold higher (σ = 216; F test, P < 0.0001). These measured angles and their relative frequencies were similar in at least one other independent CFP-TUBULIN transgenic line generated for each mutant background (Supplemental Table S1).
Figure 3.
PPB and phragmoplast angle measurements, micrographs, and time-lapse imaging of dividing cells expressing CFP-TUBULIN. A, PPB and phragmoplast angle orientation in dividing root cells. The angle was measured between the long axis of the cell wall and the orientation of the PPB or phragmoplast. The 80° and 100° angles are indicated by dotted lines. A line heterozygous for air9 and tan1 in the Ler background was used for Ler. Asterisks indicate statistically significant differences in distributions (F test, P < 0.0001). B and C, Merged confocal images showing CFP-TUBULIN (green) and propidium iodide (magenta) of tan1 air9 double mutant root cells. Images show PPBs (B) and phragmoplasts (C) with angles outside the 80° to 100° range. D to K, Time-lapse images of tan1 (D), air9 (F), oriented tan1 air9 (H), and misoriented tan1 air9 (J) division, showing the different phases of mitosis. Merged images of completed division starting with a PPB (green) and ending with a new cell wall (magenta) are shown for tan1 (E), air9 (G), properly oriented tan1 air9 division (I), and misoriented cell wall in a tan1 air9 division (K). Minutes and seconds are given in white at the bottom left sides of the time-lapse images. Bars = 5 µm.
Table I. Orientation of PPBs and phragmoplasts in Ler (tan1/+; air9/+), tan1, air9, and the tan1 air9 double mutant expressing CFP-TUBULIN (CT) to assess microtubule structure orientation.
The numbers in parentheses correspond to independent transformation events. Normal orientation is defined as division structure angles between 80° and 100°, while aberrant orientation is defined as angles outside this range. These data are plotted in Figure 2A.
| Parameter | Ler CT (#1) | tan1 CT (#9) | air9 CT (#1) | tan1 air9 CT (#1) |
|---|---|---|---|---|
| PPB | ||||
| Total (n) | 39 | 67 | 77 | 121 |
| Normal (%) | 92.3 | 98.5 | 93.5 | 61.2 |
| Aberrant (%) | 7.7 | 1.5 | 6.5 | 38.8 |
| Mean angle(°) | 91.7 | 90.7 | 89.9 | 91.5 |
| Variance | 43.7 | 31.7 | 40.4 | 215.8 |
| Phragmoplast | ||||
| Total (n) | 37 | 45 | 69 | 103 |
| Normal (%) | 86.5 | 86.7 | 89.9 | 49.5 |
| Aberrant (%) | 13.5 | 13.3 | 10.1 | 50.5 |
| Mean angle | 92.3 | 90.6 | 90.7 | 90.1 |
| Variance | 41.4 | 54.3 | 50.7 | 347.1 |
The aberrant PPB angles suggested two different hypotheses: (1) PPB placement is defective in the tan1 air9 double mutant, suggesting that TAN1 and AIR9 together have a G2 or interphase function vital for properly positioning the PPB; or (2) PPB angle reflects alterations in cell shape but not PPB placement defects per se. Close examination of tan1 air9 cells with PPB angles outside of 80° to 100° showed mostly PPBs in cells with nonrectangular prism shapes (representative examples are shown in Fig. 3B). Although the angles were not close to 90°, the PPBs appeared to divide the cell into two similarly sized volumes, possibly suggesting that the aberrant cell shape of the tan1 air9 double mutant, rather than incorrect placement of the PPB, may account for the majority of variability in PPB angle measurements.
To address whether there was a relationship between cell shape and PPB angle, we measured variance in cell edge angles as a measure of aberrant cell shape. As the cell shape is less rectangular in two dimensions, the cell edge angle variance increases dramatically, as reflected in the differences between Ler and tan1 air9 variances (Supplemental Fig. S9). When we plotted the absolute PPB angle (compared with the canonical 90° angle) versus the variance in cell edge angles, there was no detectable relationship either for Ler or the tan1 air9 double mutant (Supplemental Fig. S9). These data suggest that variability in cell edge angles does not predict alterations in PPB angle. This raises the possibility that tan1 air9 double mutants have defects in PPB placement per se rather than that cell shape alters PPB placement. These possibilities are not mutually exclusive, and further work is needed to directly answer this question.
Phragmoplast angle orientation, measured the same way as PPB angle orientation, was even more aberrant than PPB angle orientation in the tan1 air9 double mutant. Approximately 90% of cells in the single mutants tan1 (86.7%, n = 45) and air9 (89.9%, n = 69), as well as the wild-type control (86.5%, n = 37), had phragmoplast angles within 80° to 100° (Fig. 3A; Table I). No significant differences were observed between the variance observed for Ler (σ = 41.4) and tan1 (σ = 54.3; F test, P = 0.6399) or air9 (σ = 50.7; F test, P = 0.5409). However, 50.5% of tan1 air9 double mutant phragmoplasts were misoriented (outside 80°–100° [n = 103]; σ = 347.1; F test, P < 0.0001; Fig. 3, A and C; Table I). These results were similar in at least one other independent CFP-TUBULIN transgenic line generated for each mutant background (Supplemental Table S1). Cells of the tan1 air9 double mutant had a higher frequency of misoriented phragmoplasts than misoriented PPBs (Table I).
The tan1 air9 misoriented phragmoplasts observed might be due to aberrant cell shapes or misplaced PPBs but, additionally, might be due to failure in phragmoplast guidance back to the division site specified by the PPB, similar to tan1 mutants in maize (Martinez et al., 2017). Therefore, we performed time-lapse imaging experiments to determine whether the phragmoplast tracked back to the division site indicated by the PPB in the tan1 air9 double mutant. In eight and six time-lapse movies of tan1 and air9 mutants, respectively, the PPB and new cell wall had the same locations as shown in merged images in Figure 3, E and G. Correct orientation of new cell walls from time-lapse observation (Fig. 3, D–G) is consistent with the low amount of misoriented phragmoplasts reported previously (Walker et al., 2007). In contrast, when tan1 air9 double mutant cells were imaged from prophase until the end of cytokinesis, while 56.3% of divisions completed normally (Fig. 3, H and I), 43.7% had a phragmoplast guidance defect, because the location of the new cell wall did not overlap with the division site specified by the PPB (Fig. 3, J and K; n = 32). These data indicate that the tan1 air9 double mutant has significant phragmoplast guidance defects, which could account for many of the aberrant phragmoplast angles observed in the tan1 air9 double mutant. Together, these data show that TAN1 and AIR9 together are critical for phragmoplast guidance to the division site, similar to TAN1 in maize.
Full-Length TAN1-YFP and TAN1-ΔII-YFP Rescue the tan1 air9 Double Mutant, But TAN1-ΔI-YFP Does Not
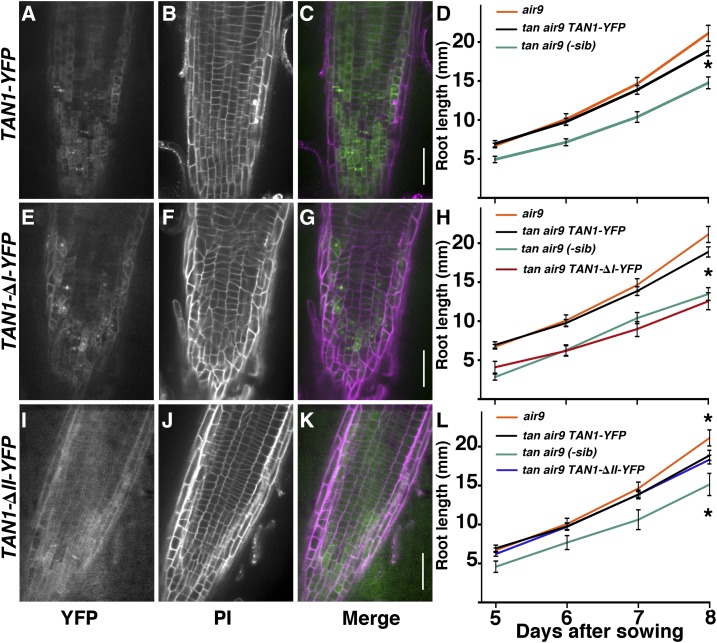
To assess whether TAN1-YFP could rescue the tan1 air9 double mutant, full-length Ler TAN1 coding sequence driven by the cauliflower mosaic virus 35S promoter (Walker et al., 2007) was transformed into the tan1 air9 double mutant. Three independent lines were selected, and their progeny were observed (Supplemental Fig. S10). Root cell patterning of double mutants expressing full-length TAN1-YFP looked similar to single mutants and wild-type roots, although they still had occasional division plane defects observed by propidium iodide staining (Fig. 4, A–C; compare Fig. 4B with Fig. 1, A–C; Supplemental Fig. S10) and quantified by measuring division structure angles below. Primary roots of tan1 air9 double mutants with TAN1-YFP were ∼30% longer at day 8 (n = 9) than nontransformed siblings (n = 8; Student’s t test, P = 0.001) and grew similarly to air9 mutants (n = 9; Student’s t test, P = 0.101; Fig. 4D), indicating rescue in terms of root length.
Figure 4.
Root micrographs and root length measurements of tan1 air9 plants expressing full-length TAN1-YFP, TAN1-∆I-YFP, and TAN1-∆II-YFP. A to C, E to G, and I to K, Confocal images of root cell walls stained with propidium iodide (PI) of tan1 air9 plants expressing TAN1-YFP (A–C), TAN1-∆I-YFP (E–G), and TAN1-∆II-YFP (I–K) with YFP signal (A, E, and I), cell walls stained with PI (B, F, and J), and merged channels (C, G, and K). YFP signal is green and PI signal is magenta. Bars = 50 µm. D, H, and L, Root length measurements from day 5 to 8 of tan1 air9 plants expressing full-length TAN1-YFP (D), TAN1-∆I-YFP (H), and TAN1-∆II-YFP (L). Single mutant air9 and nontransformed sibling plants were used as controls. Asterisks indicate significant differences between root lengths by Student’s t test (P < 0.01).
Rescue experiments were performed using a construct missing a region of TAN1 that is critical for TAN1 localization at the division site during metaphase and telophase. TAN1-∆I-YFP, missing amino acids 3 to 126 of the TAN1 protein, localizes to the division site during preprophase but is not maintained at the division site after the PPB is disassembled (Rasmussen et al., 2011b). TAN1-∆I-YFP in the tan1 air9 double mutant phenocopied the double mutant in three independent lines in terms of root cell patterning (Fig. 4, E–G; Supplemental Fig. S10) and root growth (Fig. 4H), even though the fluorescence intensity at the division site during G2 and prophase was higher than that of full-length TAN1-YFP (Supplemental Fig. S11). These data suggest that region I of TAN1 is critical for TAN1 function in root cell patterning and overall root length.
Next, we assessed whether TAN1 missing a region critical for localization during prophase was important for TAN1 function. TAN1-∆II-YFP localizes to the division site during telophase but is absent during prophase (Rasmussen et al., 2011b). TAN1-∆II-YFP is missing amino acids 132 to 222 of the TAN protein. When TAN1-∆II-YFP was transformed into the double mutant, it significantly rescued the tan1 air9 double mutant phenotype. TAN1-∆II-YFP fluorescence intensities were similar to those of full-length TAN1-YFP, suggesting similar levels of expression (Supplemental Fig. S11). The roots of the double mutant tan1 air9 plants expressing TAN1-∆II-YFP had mostly normal root patterning (Fig. 4, I–K), similar to TAN1-YFP lines (compare Fig. 4J with 4B). Moreover, root length after 8 d of growth of tan1 air9 plants expressing TAN1-∆II-YFP was similar to that in full-length TAN1-YFP tan1 air9 plants (n = 14 and 12, respectively; P = 0.5309 at 8 d after sowing). This experiment was repeated three times with similar results. The root length of TAN1-∆II-YFP tan1 air9 was 30% longer than the root length of nontransformed siblings but still not as long as that of the air9 controls (Fig. 4L). This suggests that TAN1-∆II-YFP functioned as well as TAN1-YFP in overall root patterning and growth.
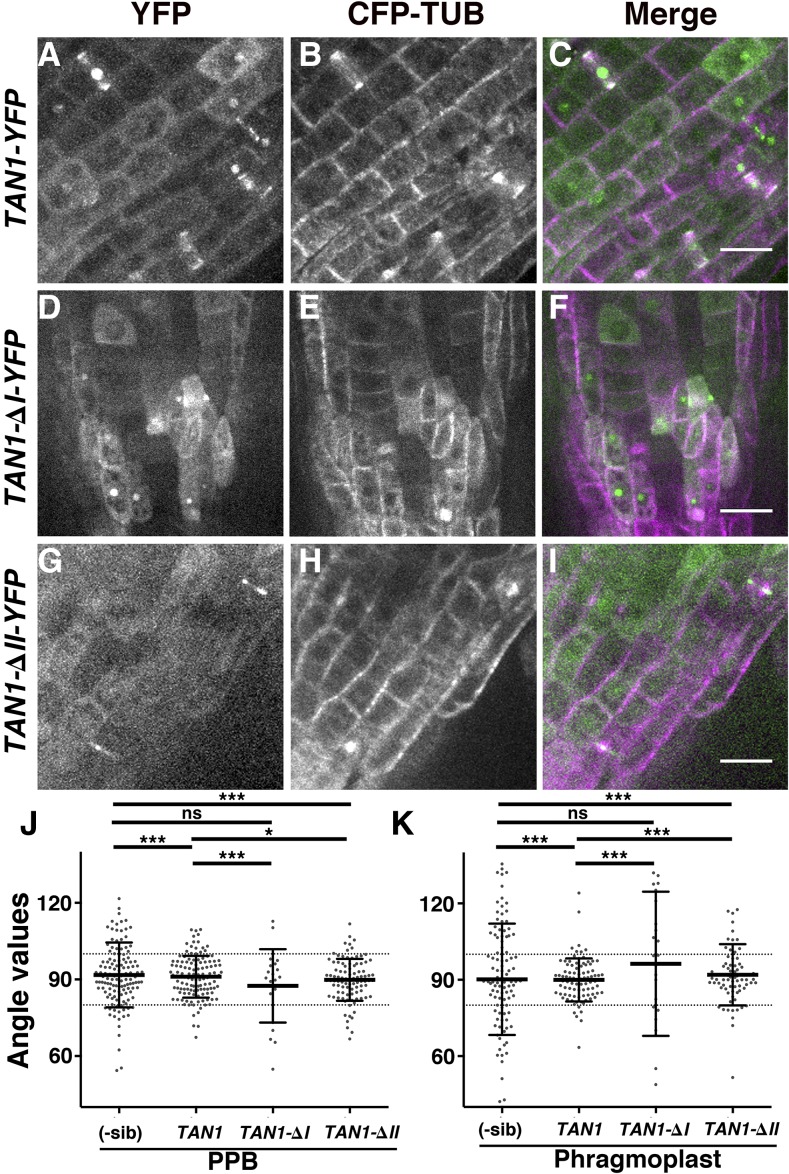
We tested the ability of full-length TAN1-YFP to rescue the PPB and phragmoplast positioning defects of the tan1 air9 double mutant. Double tan1 air9 mutants expressing TAN1-YFP and CFP-TUBULIN were generated by crossing. PPB angles of tan1 air9 expressing TAN1-YFP were within 80° to 100° of the long axis of the lateral wall 79% of the time, which results in a reduction of the variance (n = 124, σ = 66; Fig. 5, A–C and J; Table II), showing significant rescue compared with only 35.5% PPB angles within 80° to 100° in the nontransformed tan1 air9 sibling cells (n = 127, σ = 160.4; F test, P < 0.0001; Table II). However, although the rescue was evident, PPB angles were not completely restored to air9 single mutant PPB angles (compare Fig. 3A and 5, A–C; Tables I and II; F test, P = 0.02). Similarly, phragmoplast angles in double mutants expressing TAN1-YFP were significantly, but not fully, restored to air9 single mutant phragmoplast angles. Double mutants with TAN1-YFP had phragmoplast angles between 80° and 100° in 85.3% of cells (n = 89, σ = 70.7; Fig. 4, A–C and K; Table II). In nontransformed tan1 air9 double mutant siblings, only 41.2% of phragmoplast angles were between 80° and 100° (n = 102, σ = 474.3; P < 0.0001). However, TAN1-YFP tan1 air9 phragmoplasts were significantly more variable than air9 single mutant phragmoplast angles (P < 0.001 between air9 and TAN1-YFP tan1 air9). Therefore, the orientations of both PPBs and phragmoplasts were significantly but not perfectly restored in double mutant plants expressing TAN1-YFP. These results demonstrate that introduction of TAN1-YFP partially rescued the orientation of mitotic structures.
Figure 5.
PPB and phragmoplast orientation of tan1 air9 double mutants expressing full-length TAN1-YFP, TAN1-∆I-YFP, and TAN1-∆II-YFP. A to I, Confocal images of propidium iodide-stained roots of tan1 air9 plants with CFP-TUBULIN and TAN1-YFP (A–C), TAN1-∆I-YFP (D–F), and TAN1-∆II-YFP (G–I). Shown are YFP signal (A, D, and G), CFP-TUBULIN signal (B, E, and H), and merged images (C, F, and I). Bars = 20 µm. J and K, Orientation compared with the lateral cell walls of PPBs (J) and phragmoplasts (K) of tan1 air9 plants coexpressing CFP-TUBULIN and YFP-tagged versions of full-length TAN1, TAN1-∆I, and TAN1-∆II, as indicated. YFP-negative sibling plants (-sib) were used to measure the orientation of PPBs and phragmoplasts as a control. Each dot represents a measured angle. The F test was used to compare distributions. The single asterisk indicates P = 0.02, triple asterisks indicate P < 0.002, and ns indicates no significant difference detected.
Table II. Orientation of PPBs and phragmoplasts in tan1air9 mutants expressing TAN1-YFP, TAN1-ΔRI-YFP, and TAN1-ΔRII-YFP or the nontransformed siblings with CFP-TUBULIN (CT).
The numbers in parentheses refer to the transformation event number. Normal orientation is defined as angles between 80° and 100°, and aberrant orientation corresponds to angles outside this range. These data are plotted in Figure 4, J and K.
| Parameter | tan1 air9 CT (#1) TAN1-YFP (#1) | tan1 air9 CT (#1) TAN1-ΔRI-YFP (#3) | tan1 air9 CT (#1) TAN1-ΔRII-YFP (#4) | tan1 air9 CT(#1; Combined Data from All Nontransformed Siblings) |
|---|---|---|---|---|
| PPB | ||||
| Total (n) | 124 | 21 | 95 | 127 |
| Normal (%) | 79 | 66.6 | 82.1 | 64.6 |
| Aberrant (%) | 20.9 | 33.3 | 17.9 | 35.4 |
| Mean angle | 91.1 | 87.4 | 89.8 | 91.8 |
| Variance | 66.2 | 197 | 66.8 | 160.4 |
| Phragmoplast | ||||
| Total (n) | 89 | 26 | 76 | 102 |
| Normal (%) | 85.3 | 19.2 | 69.7 | 41.2 |
| Aberrant (%) | 14.6 | 80.7 | 30.2 | 58.8 |
| Mean angle | 89.9 | 90.8 | 91.9 | 91.8 |
| Variance | 70.7 | 1076 | 142.7 | 474.3 |
To determine whether TAN region I was necessary for PPB and phragmoplast orientation, we measured the angles of tan1 air9 double mutants expressing TAN1-∆I-YFP and CFP-TUBULIN (Fig. 5, D–F, J, and K; Tables I and II). Unlike TAN1-YFP, TAN1-∆I-YFP did not rescue either PPB orientation or phragmoplast orientation. Only 66.6% of PPB angles (n = 21, σ = 197; Fig. 4J; Table I) and 19.2% of phragmoplast angles (n = 26, σ = 1,076; Fig. 4K; Table II) of TAN1-∆I-YFP tan1 air9 plants were within 80° to 100°. No statistically significant differences between PPB and phragmoplast angles were observed between TAN1-∆I-YFP tan1 air9 and nontransformed siblings (F test, P = 0.4088 and P = 0.0807, respectively). Accordingly, PPB (F test, P < 0.0001) and phragmoplast (F test, P < 0.0001) angle values in TAN1-∆I-YFP tan1 air9 plants were statistically different from the ones observed in TAN1-YFP tan1 air9 plants. These data indicate that region I is critical for TAN1 function for PPB and phragmoplast angle orientation as well as root growth and cell shape described earlier.
Finally, PPB and phragmoplast angle values were measured in double mutant tan1 air9 plants expressing TAN1-∆II-YFP and CFP-TUBULIN to determine whether TAN1-∆II-YFP could rescue the double mutant division plane orientation phenotype (Fig. 5, G–K; Tables I and II). TAN1-∆II-YFP expression in the tan1 air9 double mutant significantly restored PPB and phragmoplast orientation compared with double mutant siblings. For PPB orientation, 82.1% of TAN1-∆II-YFP tan1 air9 PPB angles were between 80° and 100° (n = 95, σ = 66.8; Fig. 4J; Table I), significantly closer to air9 single mutants than 64.6% of nontransformed siblings (F test, P < 0.0001). For phragmoplast orientation, 69.7% of TAN1-∆II-YFP tan1 air9 phragmoplast angles were between 80° and 100° (n = 76, σ = 142.7; Fig. 5K; Table II) compared with 41.2% of nontransformed siblings (F test, P < 0.0001). Although both PPB and phragmoplast orientation was significantly restored, TAN1-∆II-YFP tan1 air9 PPB and phragmoplast angles were less normally distributed compared with air9 single mutants (F test, P = 0.02 and P < 0.0001, respectively). Moreover, when PPB and phragmoplasts angles of TAN1-YFP tan1 air9 were compared with TAN1-∆II-YFP tan1 air9, PPB angles were indistinguishable (F test, P = 0.9446) but phragmoplast angles of TAN1-∆II-YFP did not rescue as well as TAN1-YFP (F test, P < 0.002).
All together, these data indicate that region I is critical for TAN1 function in PPB and phragmoplast orientation, root patterning, and growth. Region II, however, plays a minor role in PPB and phragmoplast orientation and is dispensable for root growth and root cell patterning. After 4 weeks of growth, TAN1-YFP tan1 air9 and TAN1-∆II-YFP tan1 air9 plants grew similarly to air9 mutants, while TAN1-∆I-YFP tan1 air9 plants had short stature similar to the tan1 air9 double mutant (Supplemental Fig. S12).
Regions I and II of TAN1 Are Important for Proper Root Cell File Rotation
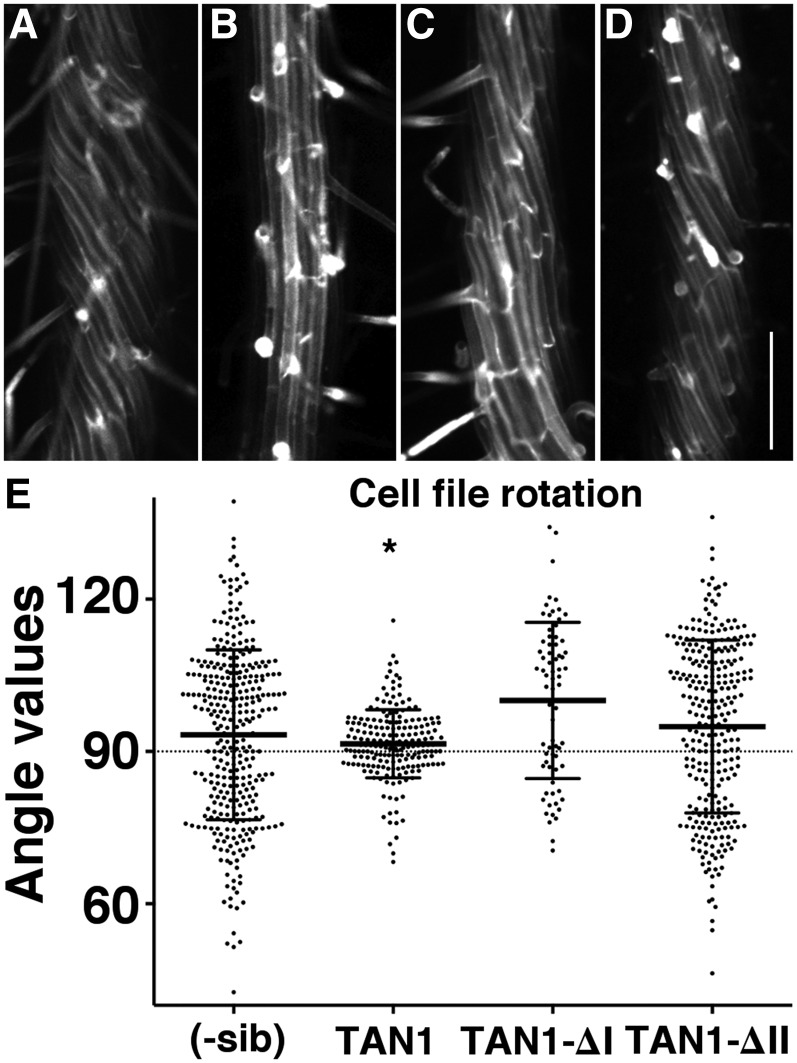
TAN1 localization in dividing cells (Walker et al., 2007; Rasmussen et al., 2011b; Martinez et al., 2017) and periodic mRNA expression patterns similar to M phase-expressed transcripts (Menges and Murray, 2002) strongly suggested that it only functions during mitosis and cytokinesis. However, TAN1-YFP driven by its own promoter has not been well characterized in Arabidopsis. We measured fluorescence intensities of TAN1-YFP driven by its native promoter in the root. As expected, TAN1-YFP localized to the division site, but it also accumulated in the cytoplasm of nondividing cells in the meristematic zone (Supplemental Fig. S13). There was no evidence that TAN1-YFP colocalized with microtubules in nondividing cells. When the fluorescence of native promoter TAN1-YFP was measured in elongation and differentiation zone cells, there was no fluorescence observed above background (Supplemental Fig. S13C). Although we did not observe TAN1-YFP above background, TAN1 may perform some function in these cells due to disorganized microtubule arrays in the elongation zone and twisted differentiation zone root cell files in tan1 air9 plants (Fig. 1, I and J; Supplemental Fig. S2, I and J). These data suggest that TAN1 may perform a function during interphase. We assessed whether TAN1-YFP could rescue the cell file rotation of tan1 air9 double mutants (Fig. 6, B and E). TAN1-YFP in tan1 air9 double mutants rescued the cell file rotation phenotype. The variance in root file angles was 44.7 for TAN1-YFP tan1 air9 (n = 204) and 31.1 for air9 single mutants (n = 42; Levene’s test, P = 0.36) but markedly different from that of nontransformed siblings (σ = 282, n = 330; Levene’s test, P < 0.0001; Fig. 6, B and E). Neither TAN1-∆I-YFP nor TAN1-∆II-YFP could rescue the root cell file twisting. The variance of cell file rotation of both TAN1-∆I-YFP and TAN1-∆II-YFP tan1 air9 (n = 72, σ = 233 and n = 283, σ = 289, respectively) was similar to that of nontransformed siblings (σ = 282; Levene’s test, P = 0.37 and P = 0.74, respectively; Fig. 6, C–E). This result indicates that the first 222 amino acids of TAN1 (regions I and II; Rasmussen et al., 2011b) are important for maintaining proper cell file rotation in differentiation zone root cells when AIR9 function is compromised. Interestingly, even though TAN1-∆II-YFP in tan1 air9 double mutants had a significant defect in cell file rotation, the rate of early root growth (days 5–8) was rescued to air9 levels.
Figure 6.
Differentiation zones of tan1 air9 roots expressing full-length TAN1-YFP, TAN1-∆I-YFP, and TAN1-∆II-YFP. A to D, Maximum projections of 10 Z-stacks of 0.2 µm of propidium iodide-stained roots of tan1 air9 plants (A) and tan1 air9 plants expressing TAN1-YFP (B), TAN1-∆I-YFP (C), and TAN1-∆II-YFP (D). Bar = 200 µm. E, Cell file rotation angles of double mutant tan1 air9 plants expressing TAN1-YFP, TAN1-∆I-YFP, or TAN1-∆II-YFP and the corresponding nontransformed sibling plants (-sib). Angles greater than 90° are left twisting and angles less than 90° are right twisting. The asterisk indicates a significantly smaller variance (P < 0.001 by Levene’s test; n = 330).
DISCUSSION
Although tan1 (Walker et al., 2007) and air9 (Buschmann et al., 2015) single mutants have a minor or no phenotype, respectively, in Arabidopsis, their combination in the tan1 air9 double mutant produced plants with severe division plane orientation and growth defects. The roots of tan1 air9 double mutants were both short and twisted, and adult plants were smaller compared with wild-type and single mutant plants. Similar phenotypes were observed in two independent double mutant combinations. Furthermore, TAN1-YFP rescued a large part of the tan1 air9 double mutant phenotype. Together, these data imply that previously identified tan1 mutant alleles (tan-csh in Ler and tan-mad in Col) represent either partial or total loss-of-function mutants, even though they produce tan1 transcripts (Walker et al., 2007). The mutant alleles of AIR9 used in this work consist of a large deletion of its 5′ region (air9-31; Ler ecotype) and a T-DNA insertion that results in defective AIR9 transcripts (air9-5; Col ecotype; Buschmann et al., 2015), which both likely result in a complete loss of AIR9 function.
The strong mutual enhancement of phenotype observed in tan1 air9 double mutants suggests a synthetic genetic interaction. This indicates that, in the wild type, AIR9 and TAN1 functions buffer each other. Buffering can occur when two separate genetic pathways converge to produce a common outcome. Alternately, buffering can occur when genes act in a single process or pathway, for example when the proteins function in a common protein complex (Hartman et al., 2001). Intriguingly, both TAN1 and AIR9 have only one copy in the Arabidopsis genome, and the two proteins share no common domain architecture, although they both bind microtubules (Smith et al., 2001; Buschmann et al., 2006). Direct protein interaction was not observed by yeast two-hybrid analysis (data not shown). It is possible that TAN1 and AIR9 function together in a common protein complex or that they may act in parallel or redundant pathways.
The tan1 air9 Double Mutant Has Defects in Division Plane Orientation
The tan1 air9 double mutants had a significant defect in phragmoplast guidance to the division site, with ∼50% of new cell walls placed outside the location specified by the PPB. These data show that TAN1 and AIR9 promote proper phragmoplast guidance to the division site, indicating that they are both required to execute division plane orientation in Arabidopsis. Importantly, to our knowledge, our results provide the first direct evidence that AIR9 plays a functional role in the plant division plane alignment. Indeed, the significant phragmoplast guidance defects and short stature of the tan1 air9 double mutant are similar to those of the tan1 mutant in maize (Smith et al., 1996, 2001; Martinez et al., 2017) or the Arabidopsis double mutant in the two related phragmoplast-orienting kinesins (POKs), pok1 pok2 (Müller et al., 2006; Lipka et al., 2014). The division site-localized POK1 and POK2 proteins are required to maintain TAN1 at the division site during telophase (Lipka et al., 2014), possibly via direct interaction with the first ∼100 amino acids of TAN (Rasmussen et al., 2011b). TAN1 localizes to the division site throughout mitosis (Walker et al., 2007), whereas AIR9 accumulates at the division site during prophase and then at the division site when the phragmoplast reaches the cell cortex (Buschmann et al., 2006). The partial colocalization of TAN1 and AIR9 might indicate that both proteins may work together, perhaps in a complex, to promote proper phragmoplast guidance to the division site after the PPB is disassembled. Whether this is due to microtubule-binding capability or another function remains unclear.
In addition to phragmoplast guidance defects, PPB angles were widely divergent from 90° in tan1 air9 double mutants, suggesting a possible defect in division plane establishment. It is also possible that aberrant PPB angles reflect significant alterations in cell shape rather than an actual defect in PPB placement. This interpretation also may account for the misoriented PPBs in pok1 pok2 double mutants (Müller et al., 2006), maize tan1 mutants (Cleary and Smith, 1998), and sabre and clasp mutants (Pietra et al., 2013), because their cell shapes also are irregular. There is a pressing need to assess whether mutants with misplaced PPBs are correctly or incorrectly positioned according to the geometry of the cell. Here, we used cell edge angle measurements as a proxy for describing cell shape. While useful in determining if the cell shape is aberrant, it cannot be used to determine whether PPB placement is accurate for the specific shape of the cell. Observations of symmetric cell division suggest that the new cell wall forms according to the minimization of its surface area, similar to soap films (Errera, 1888; Flanders et al., 1990; Lloyd, 1991; Besson and Dumais, 2014). Future research will assess whether PPB placement in tan1 air9 double mutants is correctly or incorrectly positioned using a geometry-based model.
Many mutants with PPB placement defects lack proteins required for asymmetric divisions (Abrash and Bergmann, 2009; Rasmussen et al., 2011a; Kajala et al., 2014; Shao and Dong, 2016). Yet other mutants lack PPBs altogether (Camilleri et al., 2002; Azimzadeh et al., 2008; Wright et al., 2009; Spinner et al., 2010, 2013; Kirik et al., 2012). Many mutants with cells that never formed PPBs also have significant defects in cortical microtubule organization and cell expansion, making it difficult to separate the interphase function from the G2 or mitotic function (Camilleri et al., 2002; Azimzadeh et al., 2008; Wright et al., 2009; Spinner et al., 2010, 2013; Kirik et al., 2012). Two interesting exceptions of mutants lacking obvious PPBs have recently been identified: tonneau1a (Zhang et al., 2016) and a triple mutant in three related trm loci (Schaefer et al., 2017). One hypothesis is that the mostly normal cortical microtubule array without a PPB is sufficient to direct the formation of a properly placed new cell wall. In support of this, PPB narrowing is not required for final cell wall placement (Marcus et al., 2005). Also, the cortical microtubule array realigns toward the future PPB location directly after DNA synthesis (Gunning and Sammut, 1990). An alternative hypothesis is that the PPB, although not localized prominently in the tonneau1a or trm mutant, still may assemble well enough to recruit some division plane factors and, therefore, promote proper division plane orientation.
Potential Interphase Function of TAN1 and AIR9
A likely interphase function of TAN1 and AIR9 was identified by significant cell file rotation in differentiation zone roots in the tan1 air9 double mutant. This result is surprising, because TAN1 transcripts do not accumulate in nondividing tissues (Walker et al., 2007), nor is the protein obvious in nondividing cells (Walker et al., 2007; Martinez et al., 2017). Its transcript is coexpressed with mitosis-specific transcripts (Menges and Murray, 2002). TAN1-YFP expressed from its native promoter is observed in interphase cells in the meristematic zone but does not colocalize with cortical microtubule arrays. Native promoter-driven TAN1-YFP is not expressed above background in elongation or differentiation zones of roots, where the cell file rotation, cell shape, and microtubule arrays are altered (Supplemental Fig. S13). In contrast, AIR9 transcripts accumulate across many cell types, including those that do not divide (Winter et al., 2007). Although the underlying causes of cell file rotation implicate circumnutation, gravity, and mechanosensing responses, they are not completely understood (Migliaccio and Piconese, 2001; Weizbauer et al., 2011; Roy and Bassham, 2014). Several mutants that alter microtubule dynamics also have cell file rotation defects. Right-handed twisting occurs in tortifolia1/spiral2 and spiral1 mutants, which lack microtubule-binding proteins (Furutani et al., 2000; Buschmann et al., 2004; Sedbrook et al., 2004; Galva et al., 2014). Left-handed twisting occurs in Ler plants treated with the microtubule-destabilizing drug propyzamide (Nakamura et al., 2004) and in mutants such as microtubule organization1 (Whittington et al., 2001; Sugimoto et al., 2003), lefty1, and lefty2 (Thitamadee et al., 2002). In addition to mutations that alter microtubule dynamics, sometimes mutants that modulate actin dynamics, ethylene levels, or auxin transport also have altered cell file rotation patterns (Buer et al., 2003; Yuen et al., 2003; Millar et al., 2011; Roy and Bassham, 2014). Although cell file rotation occurred in the tan1 air9 mutants, the roots themselves did not show obvious root skewing in classical tilted agar experiments. Even though root skewing can be a powerful assay for detecting twisting mutants, not all twisters show dramatic root skewing. Right-handed tortifolia2 mutants show little overall root skewing (Buschmann et al., 2009). This may have to do with the extent to which root elongation is affected. Similar to tan1 air9 mutants, tortifolia2 mutants have roots with severely inhibited growth rates.
Cell file rotation defects of the tan1 air9 double mutant differentiation zone suggest destabilized cortical microtubule arrays. Indeed, disorganized, but not oblique, microtubule arrays were observed in the elongation zone of tan1 air9 roots. Similar results were observed in the spr1/sku6 mutant, in which microtubule arrays were not skewed but cell file rotation still occurred (Sedbrook et al., 2004). In addition, tan1 air9 double mutants were hypersensitive to the microtubule-destabilizing drug propyzamide in terms of root growth and morphology. Together, these results suggest that TAN1 and AIR9 have a combined interphase function in stabilizing cortical microtubule arrays.
TAN1-YFP Rescued the Mutant Phenotype, While TAN1-ΔII-YFP Partially Rescued It and TAN1-ΔI-YFP Did Not Rescue It
The full-length TAN1-YFP coding sequence, driven by the constitutively active cauliflower mosaic virus 35S promoter, significantly rescued the tan1 air9 double mutant in root growth, division plane orientation, and cell file rotation. Although TAN1-YFP did not fully restore double mutants to wild-type or air9 single mutant phenotypes, our data show that TAN1-YFP is indeed functional. TAN1-YFP tan1 air9 was used as a benchmark to compare rescue using deletion constructs expressed in the double mutant. It was not surprising that TAN1-ΔI-YFP failed to rescue the tan1 air9 double mutant, because this region of the protein is highly conserved across the land plant lineage. Intriguingly, region I of TAN1 localizes to the division site during telophase, but not during prophase or metaphase, and interacts directly with the kinesin POK1 (Rasmussen et al., 2011b). Both POK1 and POK2 are required for TAN localization during telophase (Walker et al., 2007; Lipka et al., 2014). POK1 and POK2 also are required for the localization of two closely related proteins called PHGAP1 and PHGAP2 to the division site, but only after the PPB disassembles. These putative small GTPase-activating proteins (ROP-GAPs, or pleckstrin homology [PH] GAPs) play a role in division plane orientation (Stöckle et al., 2016). Although it is tempting to speculate that the division site accumulation of TAN during telophase is critical for its function, we cannot rule out other functions of region I, such as interaction with other proteins. We attempted to address the role of TAN1 during telophase by fusing a cyclin B destruction box onto TAN1-YFP (D-TAN1-YFP; Martinez et al., 2017) to remove TAN1 by proteosome-mediated degradation during anaphase, as used to eliminate other proteins (Krupnova et al., 2009; Van Damme et al., 2011). Although the destruction box reduced TAN1 accumulation during telophase, it did not eliminate TAN1 at the division site. The majority of D-TAN-YFP transformants rescued the tan1 mutant phenotype; therefore, the specific TAN1 function during telophase is still unknown (Martinez et al., 2017).
Intriguingly, TAN1-ΔII-YFP, which eliminates highly conserved amino acids 132 to 222, rescues the tan1 air9 mutant phenotype almost as well as full-length TAN1-YFP. Root growth and patterning and division plane orientation were almost completely rescued, although there was no apparent rescue of the cell file rotation phenotype. This separation between putative interphase function and mitotic function suggests that region II plays a more significant role during interphase. Previous results supported the hypothesis that TAN1 plays distinct roles in microtubule-dependent processes and division plane orientation via the separation of mitotic progression delays and division plane defects of the tan1 mutant in maize. An additional unexpected result is that TAN1-ΔII-YFP tan1 air9 roots grew as well as TAN1-YFP tan1 air9 roots, despite having a cell file rotation defect. However, other mutants with defects in cell file rotation can have root growth equivalent to wild-type plants (Roy and Bassham, 2017). Analysis of partially or fully rescued lines will drive forward the understanding of TAN1 and AIR9 function.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Single mutants in the TAN1 locus (AT3G05330), csh-tan (Ler) and tan-madison (Col/Ws; Walker et al., 2007), and in the AIR9 locus (AT2G34680), air9-31 (Ler) and air9-5 (Col/Ws; Buschmann et al., 2015), were described previously. Double mutants were generated by crossing lines carrying single mutations. Double homozygous mutants were selected by observation and then verified using PCR-based markers. For in vitro growth of Arabidopsis (Arabidopsis thaliana) seedlings, seeds were sown on one-half-strength Murashige and Skoog (MS) medium (MP Biomedicals; Murashige and Skoog, 1962) containing 0.5 g L−1 MES (Fisher Scientific), pH 5.7, and 0.8% (w/v) agar (Fisher Scientific). Seeds were placed at 4°C for 2 to 3 d. After stratification, plants were grown vertically in a growth chamber (Percival) with 16/8-h light/dark cycles and temperature set to 22°C. For root length experiments, transgenic T2 lines expressing TAN-YFP, TAN-∆I-YFP, or TAN-∆II-YFP, with or without CFP-TUBULIN, were stratified and grown vertically on one-half-strength MS plates solidified with 0.8% agar. After 4 d of growth, root lengths were measured every 24 h from day 5 to 8. After measuring, seedlings were screened by microscopy for YFP or CFP signal and identified as a positive plant or nontransformed sibling. Root length measurements were made in FIJI and plotted in Prism (GraphPad).
Confocal Microscopy
Images were taken on an inverted Nikon Ti with motorized stage (ASI Piezo) and spinning-disk confocal microscope (Yokogawa W1) run with Micromanager software (micromanager.org) and built by Solamere Technology. Solid-state lasers (Obis) and emission filters (Chroma Technology) used were as follows: excitation, 445; emission, 480/40 (for CFP-TUBULIN); excitation, 561; emission, 620/60 (for propidium iodide); and excitation, 514; emission, 540/30 (for TAN1-YFP and other YFP translational fusions). 40× or 60× water-immersion objectives were used with perfluorocarbon immersion liquid (RIAAA-678; Cargille) with 1.15 and 1.2 numerical aperture, respectively. The 10× objective has 0.45 numerical aperture. Root images shown in Supplemental Figure S3 were taken with a Leica SP8 and were assembled by stitching micrographs together using the ImageJ plugin Stitching (Preibisch et al., 2009).
Generation of Transgenic Lines
Arabidopsis flower buds were used for Agrobacterium tumefaciens-mediated transformation as described (Clough and Bent, 1998). Transgenic plants expressing YFP-tagged TAN1 versions were selected on 15 µg mL−1 glufosinate (Finale; Bayer) with 0.1% (v/v) Tween (Fisher Scientific) and either screened directly by microscopy or transferred to soil and selfed. Transgenic lines expressing CFP-TUBULIN were selected on 20 µg mL−1 hygromycin (Fisher Scientific). Lines coexpressing TAN1-YFP, TAN1-∆I-YFP, and TAN1-∆II-YFP with CFP-TUBULIN were generated by crossing tan1 air9 double mutants expressing the transgenes and screening the subsequent progeny by microscopy.
Plasmid Construction
TAN1-YFP, TAN-∆I-YFP, and TAN-∆II-YFP coding sequences were subcloned by EcoRI/BamHI double digestion from plasmids described previously (Rasmussen et al., 2011b) into pEZT-CL vector (a generous gift from David Ehrhardt, Carnegie Institute, Stanford University; https://deepgreen.dpb.carnegiescience.edu/cell%20imaging%20site%20/html/vectors.html) for selection with glufosinate (Finale; Bayer). The vector containing CFP-TUBULIN was described previously (Kirik et al., 2007).
Analysis of Mutant Phenotypes
For imaging root cell walls, 5- to 8-d-old seedlings were stained with a solution of 10 µm propidium iodide for 1 to 3 min and then washed with water before imaging via confocal microscopy using a 10× objective. To measure PPB and phragmoplast angles, seedlings expressing CFP-TUBULIN were stained with propidium iodide and imaged by confocal microscopy. Angles were measured between the lateral wall and PPBs or phragmoplasts in FIJI (FIJI Is Just ImageJ; http://fiji.sc/). For analysis of cell file rotation, roots were stained with propidium iodide, and differentiation zone epidermal root cells, defined by the presence of root hairs, were imaged. Measurements of cell lengths and widths, angles, and meristematic, elongation, and differentiation zones were made using FIJI (http://fiji.sc/). Prism (GraphPad) was used to perform statistical analyses and to make graphs. For normally distributed data, F tests were used to compare normally distributed variances and Student’s t test was used to compare differences. For nonnormally distributed data (Fig. 5), Levene’s test was performed online (http://scistatcalc.blogspot.co.uk).
Immunostaining
Ler and tan1 air9 double mutant plants were grown vertically on one-half-strength MS plates in a growth chamber at 22°C with a 16/8-h light/dark cycle for 7 d. They were fixed and processed for immunofluorescence microscopy using 1 µg mL−1 monoclonal anti-α-tubulin B-5-1-2 antibody (Life Technologies; 32-2500) followed by 2 µg mL−1 Alexa-568 goat anti-mouse antibody (Thermo Fisher; A-11004) as described previously (Sugimoto et al., 2000). Microtubule angles were measured using the ImageJ plugin AngleJ (Günther et al., 2015).
Treatment with Propyzamide and Taxol
Arabidopsis seeds were sterilized with chlorine gas for 2 h at room temperature (∼21°C). Seeds were plated on 0.8% agar plates with 0.05% DMSO and 0.5 g L−1 MES, pH 5.7, and placed at 4°C for 2 d. Plates were then moved to a growth chamber at 22°C with a 16/8-h light/dark cycle for 4 d. After 4 d, four plates of seedlings (n = 23–45) were transferred onto new plates with 0.05% DMSO and 0, 0.5, 1, 1.5, or 3 µm propyzamide and returned to the growth chamber. The length of the roots was marked every 24 h for 4 d. The plates were then scanned, and the root lengths were measured using FIJI. Then, the mean and se were calculated for each condition, normalized to 0 µm for each day, and plotted as percentages. Taxol treatments were performed the same way, but the concentrations used were 0, 0.2, 0.5, 1, and 3 µm.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ631804.1 (tangled1), DQ291137.1 (air9).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overall growth of 4-week-old Ler, tan1, air9, and tan1 air9 double mutants in the Ler background and overall phenotypes of 4-week-old Col/Ws and tan1 air9 double mutant in the Col/Ws background.
Supplemental Figure S2. Root phenotypes of Col, tan1, air9, and tan1 air9 double mutants.
Supplemental Figure S3. Root phenotype of tan1 air9 double mutants.
Supplemental Figure S4. Comparison of cell file twisting of straight and bent (or waving) root segments.
Supplemental Figure S5. Analysis of microtubule orientation in elongation zone cells of Ler and the tan1 air9 double mutant.
Supplemental Figure S6. Effects of propyzamide and taxol on root length of tan1, air9, and double tan1 air9 plants.
Supplemental Figure S7. Effects of propyzamide and taxol on root cell file rotation of tan1, air9, and double tan1 air9 plants.
Supplemental Figure S8. Growth and roots of mutant lines expressing CFP-TUBULIN A to D.
Supplemental Figure S9. Comparison between cell edge angle variance and PPB angle.
Supplemental Figure S10. Root cell images from independent transgenic lines expressing YFP-tagged versions of TAN1, TAN1-ΔRI, and TAN1-ΔRII.
Supplemental Figure S11. Fluorescence intensity measurements of independently transformed lines containing different versions of YFP-tagged TAN1 at the division site in tan1 air9 double mutants taken with identical imaging conditions.
Supplemental Figure S12. Photograph of 4-week-old plants.
Supplemental Figure S13. TAN1-YFP expressed from its native promoter.
Supplemental Table S1. Orientation of PPBs and phragmoplasts in tan1, air9, and tan1 air9 double mutants expressing CFP-TUBULIN.
Acknowledgments
Thanks to Sean Cutler (University of California, Riverside [UCR]) for use of the growth chamber and scanner and Danielle Garceau (UCR) for analysis of AIR9 gene expression patterns. Thanks to Jaimie Van Norman (UCR) for use of the confocal microscope and suggestions on identifying zones and analyzing roots. Thanks to Rasmussen laboratory members for critical reading of the article.
Footnotes
C.G.R. gratefully acknowledges funding from NSF-MCB #1505848, #1716972 and USDA-NIFA-CA-R-BPS-5108-H. V.H.M. was supported by an American Society for Plant Biology Summer Research for Undergraduates (ASPB-SURF) 2017 fellowship.
Articles can be viewed without a subscription.
References
- Abrash EB, Bergmann DC (2009) Asymmetric cell divisions: a view from plant development. Dev Cell 16: 783–796 [DOI] [PubMed] [Google Scholar]
- Ambrose JC, Cyr R (2008) Mitotic spindle organization by the preprophase band. Mol Plant 1: 950–960 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Nacry P, Christodoulidou A, Drevensek S, Camilleri C, Amiour N, Parcy F, Pastuglia M, Bouchez D (2008) Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20: 2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2001) On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Besson S, Dumais J (2014) Stochasticity in the symmetric division of plant cells: when the exceptions are the rule. Front Plant Sci 5: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van Damme D (2015) Endomembrane trafficking overarching cell plate formation. Curr Opin Plant Biol 28: 92–98 [DOI] [PubMed] [Google Scholar]
- Brunkard JO, Zambryski PC (2017) Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Curr Opin Plant Biol 35: 76–83 [DOI] [PubMed] [Google Scholar]
- Buer CS, Wasteneys GO, Masle J (2003) Ethylene modulates root-wave responses in Arabidopsis. Plant Physiol 132: 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW (2006) Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr Biol 16: 1938–1943 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Dols J, Kopischke S, Peña EJ, Andrade-Navarro MA, Heinlein M, Szymanski DB, Zachgo S, Doonan JH, Lloyd CW (2015) Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4-FERM domain. J Cell Sci 128: 2033–2046 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Fabri CO, Hauptmann M, Hutzler P, Laux T, Lloyd CW, Schäffner AR (2004) Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr Biol 14: 1515–1521 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Hauptmann M, Niessing D, Lloyd CW, Schäffner AR (2009) Helical growth of the Arabidopsis mutant tortifolia2 does not depend on cell division patterns but involves handed twisting of isolated cells. Plant Cell 21: 2090–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H, Sanchez-Pulido L, Andrade-Navarro MA, Lloyd CW (2007) Homologues of Arabidopsis microtubule-associated AIR9 in trypanosomatid parasites: hints on evolution and function. Plant Signal Behav 2: 296–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D (2002) The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C (2005) Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell 17: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AL, Smith LG (1998) The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 10: 1875–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Errera L. (1888) Über Zellformen und Siefenblasen. Bot Centralblatt 34: 395–399 [Google Scholar]
- Flanders DJ, Rawlins DJ, Shaw PJ, Lloyd CW (1990) Nucleus-associated microtubules help determine the division plane of plant epidermal cells: avoidance of four-way junctions and the role of cell geometry. J Cell Biol 110: 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masukawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T (2000) The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127: 4443–4453 [DOI] [PubMed] [Google Scholar]
- Galva C, Kirik V, Lindeboom JJ, Kaloriti D, Rancour DM, Hussey PJ, Bednarek SY, Ehrhardt DW, Sedbrook JC (2014) The microtubule plus-end tracking proteins SPR1 and EB1b interact to maintain polar cell elongation and directional organ growth in Arabidopsis. Plant Cell 26: 4409–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning B, Sammut M (1990) Rearrangements of microtubules involved in establishing cell division planes start immediately after DNA synthesis and are completed just before mitosis. Plant Cell 2: 1273–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning BE, Wick SM (1985) Preprophase bands, phragmoplasts, and spatial control of cytokinesis. J Cell Sci Suppl 2: 157–179 [DOI] [PubMed] [Google Scholar]
- Gunning BES, Hardham AR, Hughes JE (1978) Evidence for initiation of microtubules in discrete regions of the cell cortex in Azolla root-tip cells, and an hypothesis on the development of cortical arrays of microtubules. Planta 134: 161–179 [DOI] [PubMed] [Google Scholar]
- Günther MI, Günther M, Schneiders M, Rupp R, Blesch A (2015) AngleJ: A new tool for the automated measurement of neurite growth orientation in tissue sections. J Neurosci Methods 251: 143–150 [DOI] [PubMed] [Google Scholar]
- Hartman JL IV, Garvik B, Hartwell L (2001) Principles for the buffering of genetic variation. Science 291: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kaneko Y, Iwano M, Hashimoto T (2007) Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 8544–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. (2005) Cytokinesis in higher plants. Annu Rev Plant Biol 56: 281–299 [DOI] [PubMed] [Google Scholar]
- Kajala K, Ramakrishna P, Fisher A, Bergmann DC, De Smet I, Sozzani R, Weijers D, Brady SM (2014) Omics and modelling approaches for understanding regulation of asymmetric cell divisions in Arabidopsis and other angiosperm plants. Ann Bot 113: 1083–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V (2012) TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Herrmann U, Parupalli C, Sedbrook JC, Ehrhardt DW, Hülskamp M (2007) CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J Cell Sci 120: 4416–4425 [DOI] [PubMed] [Google Scholar]
- Krupnova T, Sasabe M, Ghebreghiorghis L, Gruber CW, Hamada T, Dehmel V, Strompen G, Stierhof YD, Lukowitz W, Kemmerling B, et al. (2009) Microtubule-associated kinase-like protein RUNKEL needed [corrected] for cell plate expansion in Arabidopsis cytokinesis. Curr Biol 19: 518–523 [DOI] [PubMed] [Google Scholar]
- Lee YRJ, Liu B (2013) The rise and fall of the phragmoplast microtubule array. Curr Opin Plant Biol 16: 757–763 [DOI] [PubMed] [Google Scholar]
- Lipka E, Gadeyne A, Stöckle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Müller S (2014) The Phragmoplast-Orienting Kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26: 2617–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CW. (1991) How does the cytoskeleton read the laws of geometry in aligning the division plane of plant cells? Development (Suppl 1) 113: 55–651684934 [Google Scholar]
- Marcus AI, Dixit R, Cyr RJ (2005) Narrowing of the preprophase microtubule band is not required for cell division plane determination in cultured plant cells. Protoplasma 226: 169–174 [DOI] [PubMed] [Google Scholar]
- Martinez P, Luo A, Sylvester A, Rasmussen CG (2017) Proper division plane orientation and mitotic progression together allow normal growth of maize. Proc Natl Acad Sci USA 114: 2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May SF, Peacock L, Almeida Costa CIC, Gibson WC, Tetley L, Robinson DR, Hammarton TC (2012) The Trypanosoma brucei AIR9-like protein is cytoskeleton-associated and is required for nucleus positioning and accurate cleavage furrow placement. Mol Microbiol 84: 77–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Murray JAH (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Migliaccio F, Piconese S (2001) Spiralizations and tropisms in Arabidopsis roots. Trends Plant Sci 6: 561–565 [DOI] [PubMed] [Google Scholar]
- Millar KDL, Johnson CM, Edelmann RE, Kiss JZ (2011) An endogenous growth pattern of roots is revealed in seedlings grown in microgravity. Astrobiology 11: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Han S, Smith LG (2006) Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr Biol 16: 888–894 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, Machida Y, Hasebe M (2013) Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun 4: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Neuteboom LW, Ng JM, Kuyper M, Clijdesdale OR, Hooykaas PJ, van der Zaal BJ (1999) Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol Biol 39: 273–287 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD, Gunning BE, Brown RC, Lemmon BE, Cleary AL (1999) The cytoplast concept in dividing plant cells: cytoplasmic domains and the evolution of spatially organized cell. Am J Bot 86: 153–172 [PubMed] [Google Scholar]
- Pickett-Heaps JD, Northcote DH (1966) Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J Cell Sci 1: 109–120 [DOI] [PubMed] [Google Scholar]
- Pietra S, Gustavsson A, Kiefer C, Kalmbach L, Hörstedt P, Ikeda Y, Stepanova AN, Alonso JM, Grebe M (2013) Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat Commun 4: 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P (2009) Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen CG, Humphries JA, Smith LG (2011a) Determination of symmetric and asymmetric division planes in plant cells. Annu Rev Plant Biol 62: 387–409 [DOI] [PubMed] [Google Scholar]
- Rasmussen CG, Sun B, Smith LG (2011b) Tangled localization at the cortical division site of plant cells occurs by several mechanisms. J Cell Sci 124: 270–279 [DOI] [PubMed] [Google Scholar]
- Rasmussen CG, Wright AJ, Müller S (2013) The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J 75: 258–269 [DOI] [PubMed] [Google Scholar]
- Roy R, Bassham DC (2014) Root growth movements: waving and skewing. Plant Sci 221-222: 42–47 [DOI] [PubMed] [Google Scholar]
- Roy R, Bassham DC (2017) TNO1, a TGN-localized SNARE-interacting protein, modulates root skewing in Arabidopsis thaliana. BMC Plant Biol 17: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E, Belcram K, Uyttewaal M, Duroc Y, Goussot M, Legland D, Laruelle E, de Tauzia-Moreau ML, Pastuglia M, Bouchez D (2017) The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356: 186–189 [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible WR, Somerville CR (2004) The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Dong J (2016) Polarity in plant asymmetric cell division: division orientation and cell fate differentiation. Dev Biol 419: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Assaad F, Baluška F, Bezanilla M, Buschmann H, Drakakaki G, Hauser M-T, Janson M, Mineyuki Y, Moore I, et al. (2017) Plant cytokinesis: Terminology for structures and processes. Trends Cell Biol 12: 885–894 [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Piette B, Hussey PJ (2011) The origin of phragmoplast asymmetry. Curr Biol 21: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Smith LG, Gerttula SM, Han S, Levy J (2001) Tangled1: a microtubule binding protein required for the spatial control of cytokinesis in maize. J Cell Biol 152: 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Hake S, Sylvester AW (1996) The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122: 481–489 [DOI] [PubMed] [Google Scholar]
- Spinner L, Gadeyne A, Belcram K, Goussot M, Moison M, Duroc Y, Eeckhout D, De Winne N, Schaefer E, Van De Slijke E, et al. (2013) A protein phosphatase 2A complex spatially controls plant cell division. Nat Commun 4: 1863. [DOI] [PubMed] [Google Scholar]
- Spinner L, Pastuglia M, Belcram K, Pegoraro M, Goussot M, Bouchez D, Schaefer DG (2010) The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 137: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Stöckle D, Herrmann A, Lipka E, Lauster T, Gavidia R, Zimmermann S, Müller S (2016) Putative RopGAPs impact division plane selection and interact with kinesin-12 POK1. Nat Plants 2: 16120. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Himmelspach R, Williamson RE, Wasteneys GO (2003) Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15: 1414–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO (2000) New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T (2002) Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E (2011) Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23: 4013–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Vanstraelen M, Geelen D (2007) Cortical division zone establishment in plant cells. Trends Plant Sci 12: 458–464 [DOI] [PubMed] [Google Scholar]
- Wachsman G, Sparks EE, Benfey PN (2015) Genes and networks regulating root anatomy and architecture. New Phytol 208: 26–38 [DOI] [PubMed] [Google Scholar]
- Walker KL, Müller S, Moss D, Ehrhardt DW, Smith LG (2007) Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr Biol 17: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizbauer R, Peters WS, Schulz B (2011) Geometric constraints and the anatomical interpretation of twisted plant organ phenotypes. Front Plant Sci 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AJ, Gallagher K, Smith LG (2009) discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21: 234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CYL, Pearlman RS, Silo-Suh L, Hilson P, Carroll KL, Masson PH (2003) WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiol 131: 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iakovidis M, Costa S (2016) Control of patterns of symmetric cell division in the epidermal and cortical tissues of the Arabidopsis root. Development 143: 978–982 [DOI] [PMC free article] [PubMed] [Google Scholar]