Sucrose transporter expression in leaves is regulated at the transcriptional and post-transcriptional level to link changes in photosynthesis and sugar status with phloem loading and carbon export.
Abstract
Suc transporters (SUTs) play a key role in the allocation and partitioning of photosynthetically fixed carbon in plants. While a function could be assigned to many members of the SUT family, almost no information is available on their regulation. Here, the transcriptional regulation of SUTs in response to various environmental stimuli in the leaves of five dicots (Arabidopsis [Arabidopsis thaliana], soybean [Glycine max], potato [Solanum tuberosum], tomato [Solanum lycopersicum], and poplar [Populus spp.]) and four monocots (maize [Zea mays], rice [Oryza sativa], wheat [Triticum aestivum], and barley [Hordeum vulgare]) was investigated. Extensive data on expression of SUTs in relation to changes of environmental conditions were obtained through a global analysis of 168 transcriptomics data sets. Results were validated by quantitative PCR measurements and extended by the measurement of photosynthesis rate and phloem sugar content to draw insight on the correlation of SUT expression and sugar export from leaves. For the apoplasmic phloem loaders, a clear difference in transcriptional regulation in response to different environmental stimuli was observed. The consistent patterns of SUT expression under abiotic stress indicates which types of SUTs are involved in the regulation of leaf sugar status and in stress signaling. Furthermore, it is shown that down-regulation of phloem loading is likely to be caused by transcriptional regulation of SUTs, while up-regulation depends on post-transcriptional regulation. In poplar, expression of PtaSUT4 was found to consistently respond to environmental stimuli, suggesting a significant role in the regulation of sugar export from leaves in this passive symplasmic phloem loader.
In most herbaceous plants, including most crop plants, Suc transporters (SUTs) play a key role in the export of photosynthetically fixed carbon from leaves. In the phloem of minor veins, SUT proteins facilitate the uptake of Suc into the companion cells and sieve elements. Once in the sieve elements, Suc is transported by mass flow toward the carbon sink organs. SUTs are involved in other processes as well. There is evidence that at least in some species, SUTs reload Suc along the phloem path, or import Suc into sink organs, like fruit (Gould et al., 2012; Liesche et al., 2015). Furthermore, a function in signaling has been proposed repeatedly for several members of the SUT gene family, most prominently for SUT4 of potato (Solanum tuberosum), which was shown to play a major role in the shade avoidance response and influence the hormonal regulation of flowering and tuberization without making a major contribution to Suc transport itself (Chincinska et al., 2008, 2013).
SUTs are represented by a gene family usually consisting of between three and nine members, which can be grouped into three major classes according to sequence similarity, exon-intron structure, and function (Peng et al., 2014; Reinders et al., 2012). Type I SUTs, only present in eudicots, are necessary for essential functions such as phloem loading (Riesmeier et al., 1994; Gottwald et al., 2000) and normal pollen function (Sivitz et al., 2008). Group II is separated in the dicot-specific group IIA and the monocot-specific group IIB. Monocot species, such as maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare), were found to utilize type II SUTs for phloem loading (Aoki et al., 2004; Sivitz et al., 2005; Slewinski et al., 2009). While there has been controversy regarding the question if type II SUTs play the same role in rice (Oryza sativa) as they do in the other grasses, current evidence suggests that they do (Julius et al., 2017). A role in phloem unloading and Suc import into sink tissues has been assigned to several members of the type IIB SUTs (Kühn and Grof, 2010). All land plants contain type III SUTs. Some Type III SUTs are localized at the tonoplast, while others have been found at the plasma membrane (Chincinska et al., 2013). A function of tonoplast-localized SUTs in Suc storage and regulation of cytosolic Suc levels is evident (Endler et al., 2006; Payyavula et al., 2011). Contrastingly, the main function of plasma membrane-localized type III SUTs seem to be signaling (Chincinska et al., 2008, 2013).
Considering their importance in carbon partitioning and utilization, relatively little is known about the regulation of SUTs. A connection between SUT transcription and transport activity became evident through genetic manipulations of SUT expression (Bürkle et al., 1998; Slewinski et al., 2009; Wang et al., 2015). Several studies supported this link in nonmanipulated plants. For example, in potato SUT1 expression was shown to correlate with leaf Suc export rate (Chincinska et al., 2008) and in peach (Prunus persica), SUT2 expression correlates with Suc import into fruits (Vimolmangkang et al., 2016). However, a systematic analysis of which factors influence SUT expression is missing, even in the model plant Arabidopsis (Arabidopsis thaliana). Not knowing the extent to which plants rely on SUT regulation to control carbon partitioning is a major reason for our limited understanding of the control of this process (Ayre, 2011; Liesche et al., 2017).
Here, the question of how SUTs are regulated in response to various stimuli is addressed through a global analysis of publicly available gene expression data. How SUT expression correlates with phloem loading is then further investigated experimentally.
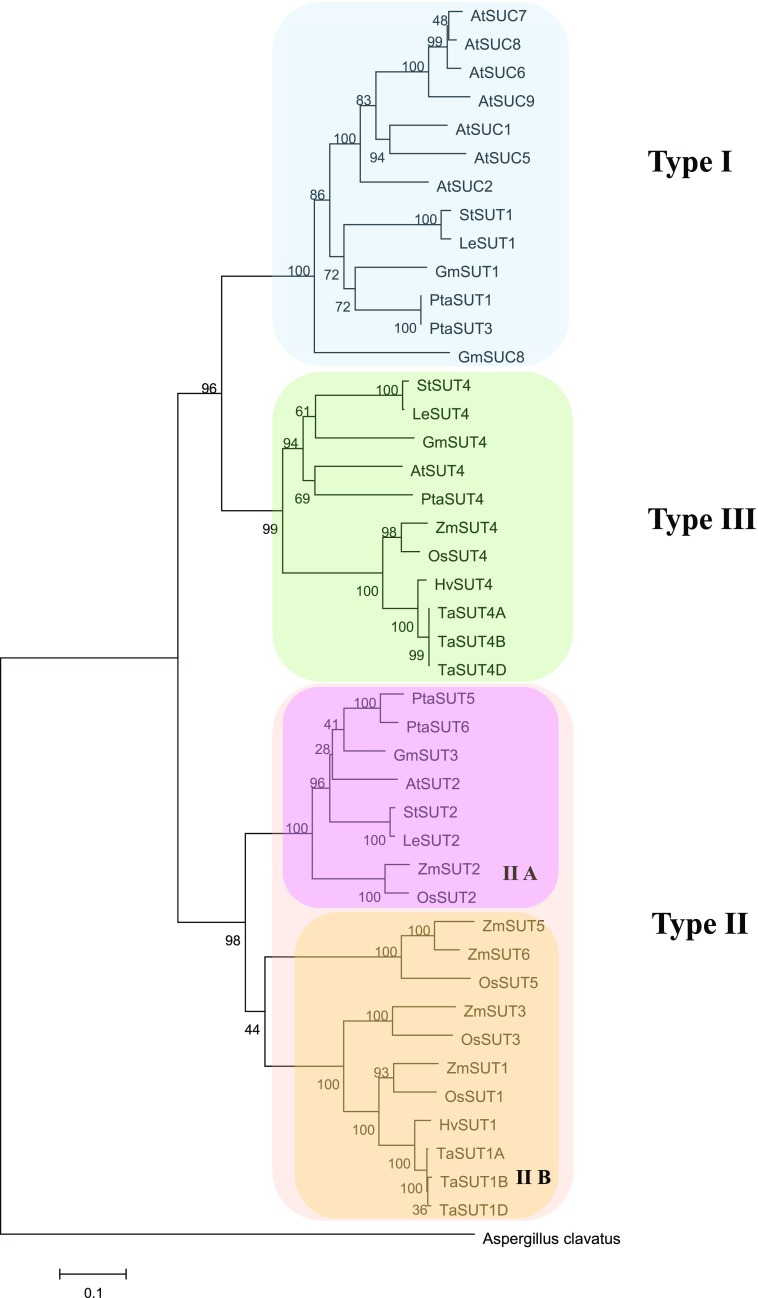
The analysis includes nine angiosperm species, four monocots, and five dicots, whose classification is shown in Figure 1. These species, except for poplar (Populus spp.), have been shown to load Suc into the phloem by an active apoplasmic mechanism. The sieve element companion cell complex (SECCC) is not connected to surrounding cells by plasmodesmata, allowing efficient import of Suc by one of the plasma membrane-localized SUTs of type I for dicots and type IIB for monocots. Poplar is a so-called passive symplasmic loader, a type mostly found in tree species (Davidson et al., 2011). Plasmodesmata enable diffusion along the whole prephloem pathway from mesophyll cells to the SECCC, and the high Suc concentration in the source phloem is thought to be the result of high Suc levels in the cytosol of all leaf cells (Rennie and Turgeon, 2009; Zhang et al., 2014). However, the altered pattern of carbohydrate partitioning in plants that lack the tonoplast-localized PtaSUT4 indicates that active Suc transport could still be relevant for poplar phloem loading (Payyavula et al., 2011; Liesche, 2017). The question if Suc transporters are involved in the regulation of carbohydrate export from leaves in passive loaders is a special focus in this study.
Figure 1.
Phylogenetic tree of plant Suc transporters based on protein sequence similarity. For the SUTs of the monocot species maize, wheat, rice, and barley, we follow Leach et al. (2017) in referring to all SUTs belonging to type IIA as SUT2 and to all SUTs belonging to type III as SUT4, even though different names can be found in online databases.
The investigation of changes in SUT expression focuses on parameters related to photosynthesis, such as light intensity and CO2 concentration, as well as different types of abiotic stress. Biotic stresses were not included as their effect is generally pathogen- and host-specific and therefore not suitable for a comparative analysis between species. Furthermore, the investigation is limited to source leaves, reflecting both the availability of data and this study’s focus on the regulation of phloem loading.
RESULTS
Cross-Platform Comparison of Transcriptomics Data Provides Consistent Results on SUT Expression
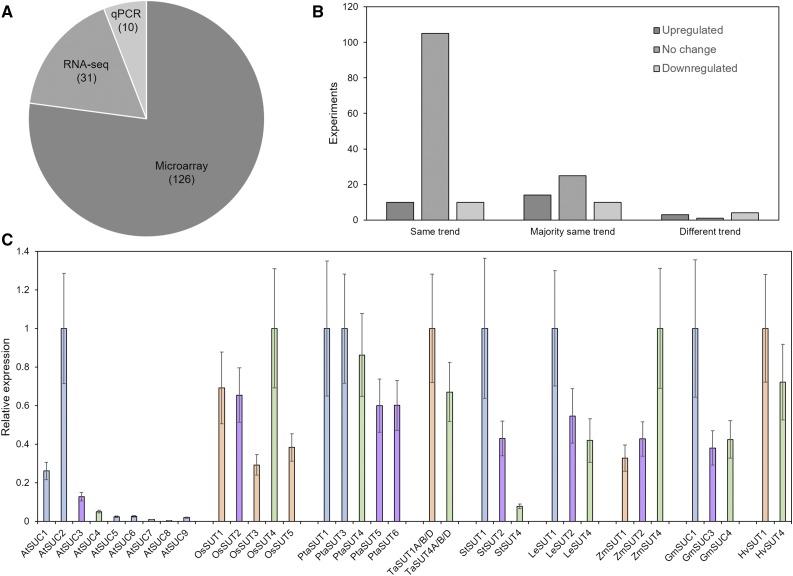
The comparative analysis of SUT expression is based on data, most of it publicly available, from 167 experiments (Supplemental Table S2). In these experiments, three different techniques were used to quantify gene expression: microarray, RNAseq, and quantitative PCR (qPCR). While microarray and RNAseq data sets include information for the whole transcriptome, qPCR data were acquired from studies that focus on SUT expression. The majority of data stems from microarray experiments, while only a limited number of RNAseq and qPCR experiments were analyzed (Fig. 2A), strictly reflecting the availability of data.
Figure 2.
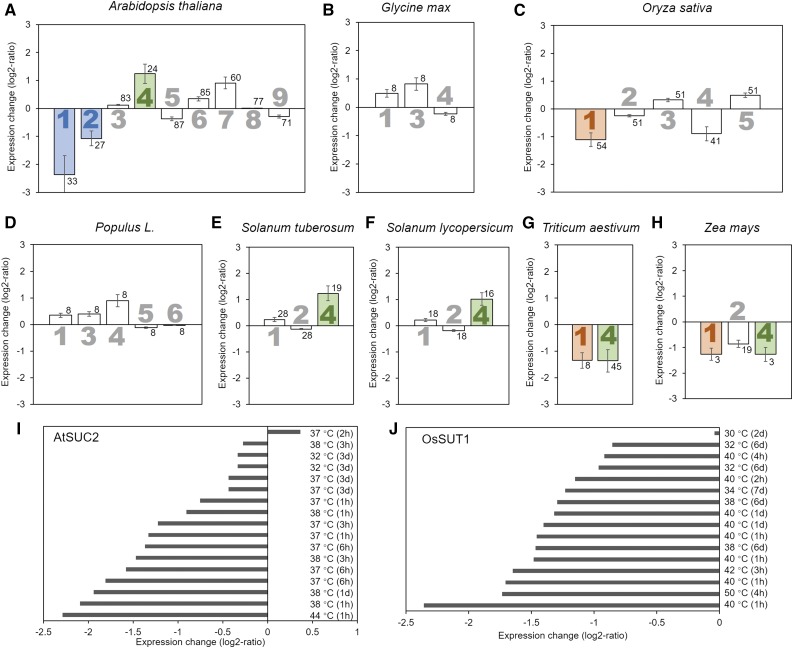
Sources and consistency of gene expression data used in this study. A, Percentages of the different techniques, that is microarray, RNAseq, and quantitative PCR, that were used to generate the 167 data sets used in this study. B, Consistency of the trend of biological replicates. C, Relative Suc transporter expression levels in mature leaves under control conditions according to all experiments considered in this study. Data were normalized to the highest expressing SUT for each species. Error bars indicate sd. The minimum number of biological replicates for each expression value in C was n = 38. Bar color indicates the type of Suc transporter (blue = type I, purple = type IIA, orange = type IIB, green = type III).
The combination of multiple data sets for similar experiments strengthens the reliability of the average values of the log2-fold change in gene expression, as these are based on a higher number of biological replicates (on average 30). This is especially important since the reliability of microarray data, which this study is mainly based on, has been doubted (Richard et al., 2014). Here, 71% of the average values for log2-fold change are based on biological replicates that all show the same trend (Fig. 2B). However, a further 28% of the values are supported by replicates that mostly show a similar trend, while only 1% of the values are based on contradictory data (Fig. 2B). Furthermore, we performed qPCR measurements of gene expression to compare with the averaged values yielded by the transcriptomics analysis.
The expression of SUT in leaves has been shown to differ markedly for different family members (Weise et al., 2000; Payyavula et al., 2011; Reuscher et al., 2014). The comparison of SUT expression levels under control conditions is in line with previous results, generally showing the SUT responsible for phloem loading as having the highest level of expression (Fig. 2C). While in Arabidopsis some of the SUTs are expressed at very low levels in relation to AtSUC2, the difference between phloem-loading SUT and other SUTs is less marked in the other species (Fig. 2C).
Photosynthetic Rate, Light Levels, and CO2 Concentration Have Limited Influence on SUT Expression
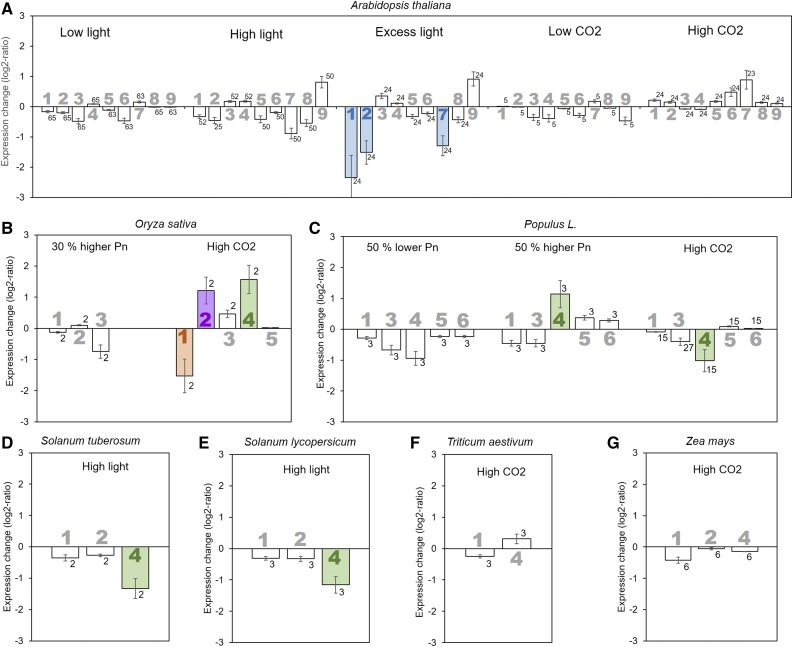
The export rate of carbohydrates from the leaf depends to a high degree on their production, that is photosynthetic activity, which is influenced by factors such as light and CO2 availability (Jiao and Grodzinski, 1996; Grodzinski et al., 1998; Amiard et al., 2005; Duan et al., 2014). Here, transfer of plants to an environment with high light (400–800 μmol photons m−2 s−1) or low light (<100 μmol photons m−2 s−1) was found to not significantly alter the expression of any SUT in species with apoplasmic phloem loading (Fig. 3, A, D, and E). The exception is SUT4 in the Solanaceae species tomato (Solanum lycopersicum) and potato, whose expression was reduced under high light conditions (Fig. 3, D and E). Only if Arabidopsis plants were subjected to excess light (1,300 μmol photons m−2 s−1), SUT expression changed, with a significant decrease for AtSUC1, AtSUC2, and AtSUC7 (Fig. 3A). This is likely due to the general detrimental effect on cell function of this condition (Jung et al., 2013). Like different light levels, exposure to increased (480–780 μg mL−1) or decreased (50–100 μg mL−1) atmospheric CO2 concentrations compared to ambient levels of 350 to 400 μg mL−1 did not significantly alter the expression level of the SUTs in apoplasmic loaders (Fig. 3), except for several rice SUTs. In rice, an increase in OsSUT2 and OsSUT4 expression and a decrease of OsSUT1 expression is observed in response to moderately increased CO2 levels (from 370 to 480 μg mL−1; Fig. 3B). However, these results are based on only two biological replicates and are inconsistent with the response of other grasses (Fig. 3).
Figure 3.
Influence of changes in light levels, atmospheric CO2 concentration, or photosynthetic rate (Pn) on Suc transporter gene expression. The average log2-fold change in expression compared to control conditions for the different Suc transporters (large numbers) is presented for Arabidopsis (A), rice (B), poplar (C), potato (D), tomato (E), wheat (F), and maize (G). Significant changes are highlighted by filled bars with color indicating the type of Suc transporter (blue = type I, purple = type IIA, orange = type IIB, green = type III). Error bars represent sd. Small numbers indicate number of biological replicates.
In contrast to the apoplasmic loaders, SUT expression in the passive symplasmic loader poplar changes with the photosynthetic rate and higher CO2 levels. Expression of the tonoplast-localized PtSUT4 is decreased when photosynthetic rate is reduced by 50% and significantly increased when photosynthetic rate is increased by 50% (Fig. 3C). When plants are transferred to a high CO2 environment (>550 or 720 μg mL−1), the SUT4 level significantly decreases (Fig. 3C). The results show that while in apoplasmic loaders SUT expression is generally not linked to photosynthesis, there is a connection in the passive symplasmic loader poplar. However, the up-regulation under high CO2 conditions indicates that additional factors influence expression.
SUT Expression Responds to Abiotic Stress
Exposure to abiotic stress can have significant effects on carbohydrate partitioning, including the export of carbohydrates from leaves, as shown for drought stress (Hummel et al., 2010; Durand et al., 2016), salt stress (Hasegawa et al., 2000), and heat stress (Berry and Bjorkman, 1980).
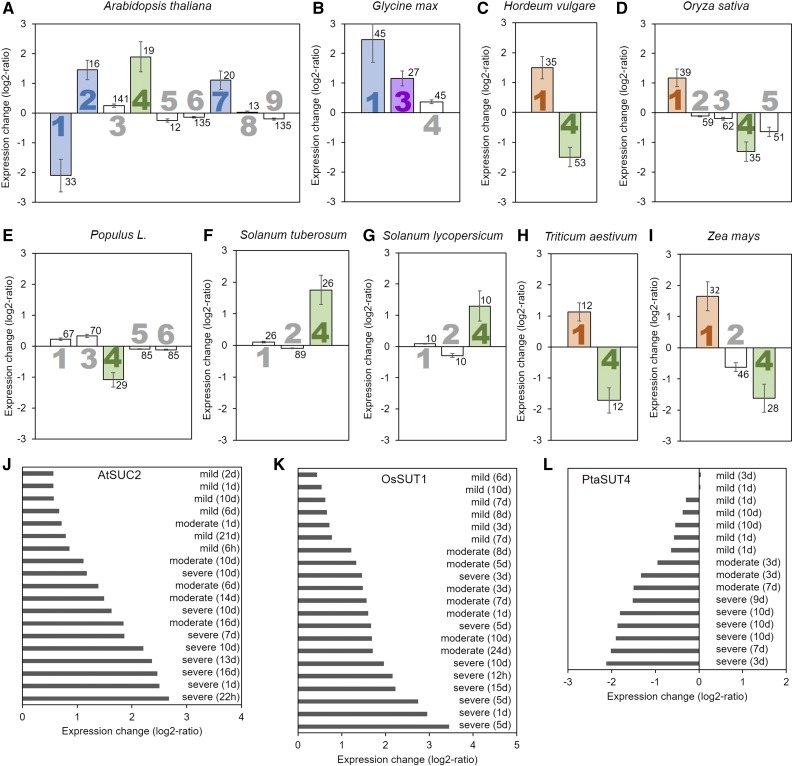
Drought stress, ranging from mild to severe levels, causes significant changes in the expression of SUT genes in all species analyzed here (Fig. 4). In the apoplasmic loaders Arabidopsis, soybean (Glycine max), barley (Hordeum vulgare), rice (Oryza sativa), wheat (Triticum aestivum), and maize (Zea mays), the SUT responsible for phloem loading is up-regulated, but not in tomato or potato (Fig. 4). Of the other SUTs, especially the type III SUTs show a change in expression when subjected to drought stress. In the apoplasmic loading dicots, SUT4 is up-regulated, although not significantly in soybean (Fig. 4, A, B, F, and G). In the grasses, SUT4 is down-regulated (Fig. 4, C, D, H, and I). In the passive symplasmic loader poplar, PtSUT4 is down-regulated (Fig. 4E), confirming its involvement in the drought response indicated by the study of knock-out plants (Frost et al., 2012; Xue et al., 2016). Also notable is the strong reduction in expression of Arabidopsis AtSUC1 (Figure 4A), a type I SUT not directly involved in phloem loading, which was also down-regulated in response to drought stress.
Figure 4.
Influence of drought stress on Suc transporter expression. The average log2-fold change in expression compared to control conditions for the different Suc transporters (large numbers) is presented for Arabidopsis (A), soybean (B), barley (C), rice (D), poplar (E), potato (F), tomato (G), wheat (H), and maize (I). Perturbation plots of Arabidopsis AtSUC2 (J), rice OsSUT1 (K), and poplar PtaSUT4 (L). Significant changes are highlighted by filled bars with color indicating the type of Suc transporter (blue = type I, purple = type IIA, orange = type IIB, green = type III). Error bars represent sd. Small numbers indicate number of biological replicates.
The response to salt stress, triggered by treatment with 100 to 300 mm NaCl, strongly resembles the response to drought stress (Supplemental Fig. S3). Expression of the same SUTs responds with significant changes. The only difference is found in barley and maize, where HvSUT4 and ZmSUT4 are not down-regulated under salt stress, as observed under drought stress. Furthermore, up-regulation of wheat TaSUT1 is not significant under salt stress (Supplemental Fig. S3).
The pattern of SUT expression in plants subjected to heat stress differs from that for drought and salt stress. Arabidopsis AtSUC2 is down-regulated under heat stress (around 15°C increase) but up-regulated under drought/salt (Fig. 5). Similarly, in the monocots rice, wheat, and maize, the phloem-loading SUT1 is down-regulated under heat stress (7–15°C increase; Fig. 5) instead of up-regulated under drought and salt stress. In contrast, in the symplasmic loader poplar, PtaSUT4 is up-regulated under heat stress (17°C increase; Fig. 5D) instead of down-regulated under drought stress (Fig. 4E). Interestingly, the response in the Solanaceae, tomato and potato, is the same for all three stresses, with only SUT4 expression significantly increased (Fig. 5, E and F).
Figure 5.
Influence of heat stress on Suc transporter expression. The average log2-fold change in expression compared to control conditions for the different Suc transporters (large numbers) is presented for Arabidopsis (A), soybean (B), rice (C), poplar (D), potato (E), tomato (F), wheat (G), and maize (H). Perturbation plots of Arabidopsis AtSUC2 (I) and rice OsSUT1 (J). Significant changes are highlighted by filled bars with color indicating the type of Suc transporter (blue = type I, purple = type IIA, orange = type IIB, green = type III). Error bars represent sd. Small numbers indicate number of biological replicates.
Correlation of SUT Expression and Phloem Loading
While the analysis of transcriptomics data reveals patterns in the regulation of SUTs, no definitive conclusions regarding its effect on phloem loading can be drawn without concomitant measurements of expression and export on the same plants. We subjected two dicots, Arabidopsis and tomato, and two monocots, maize and wheat, to the experimental stimuli discussed above and determined the photosynthesis rate, whole-leaf sugar levels, SUT expression levels, and Suc concentration in phloem exudate.
As samples gained through EDTA-facilitated exudation are prone to contamination (Liu et al., 2012), we performed several tests to verify that the measured Suc levels correspond to the concentration in the leaf phloem. Exudation profiles were compared for a leaf blade with petiole, leaf blade with minimal petiole, and petiole only. All samples contained similar levels of Glc and Fru, but Suc content was several times higher when the leaf blade was present compared to the petiole alone (Supplemental Fig. S4). This suggests that the exudate mostly stems from the leaf blade and indicates that hexoses can be considered as contamination stemming from the injured cells of the petiole. This is supported by the fact that most hexoses are exuded during the first hour, whereas the Suc content continually increases with the time of exudation (Supplemental Fig. S4). Furthermore, composition and concentrations of exudates was found to differ markedly from that of whole leaf sap (Figs. 6 and 7), corroborating earlier studies that found exudate to be representative of phloem sap (King and Zeevert 1974; Guelette et al., 2012).
Figure 6.
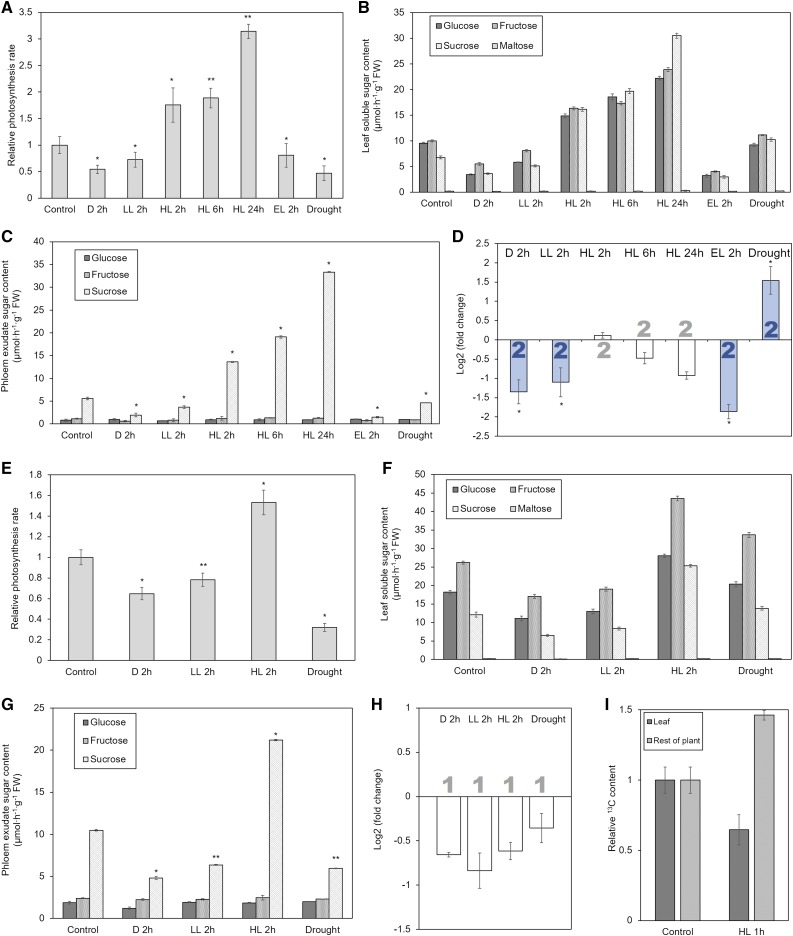
Comparison of photosynthesis rate, sugar content in leaf extract, sugar content in phloem exudate, carbon export, and Suc transporter gene expression in Arabidopsis (A–D) and tomato (E–I). All parameters were measured at the same time on comparable leaves of the same plants, except for the measurement of carbon export (I). This was performed on plants at the same growth stage as those for the other experiments by applying a pulse of 13CO2 to a source leaf, transfer to experimental conditions for 1 h, and determination of the 13C content of the leaf and the rest of the plant. Significance of differences to control values is indicated by *P < 0.05 and **P < 0.01. Error bars indicate sd. Significant changes of gene expression are highlighted by filled bars with color indicating the type of Suc transporter (blue = type I). Average values were obtained from at least three biological replicates. Abbreviations: D, dark; LL, low light; HL, high light.
Figure 7.
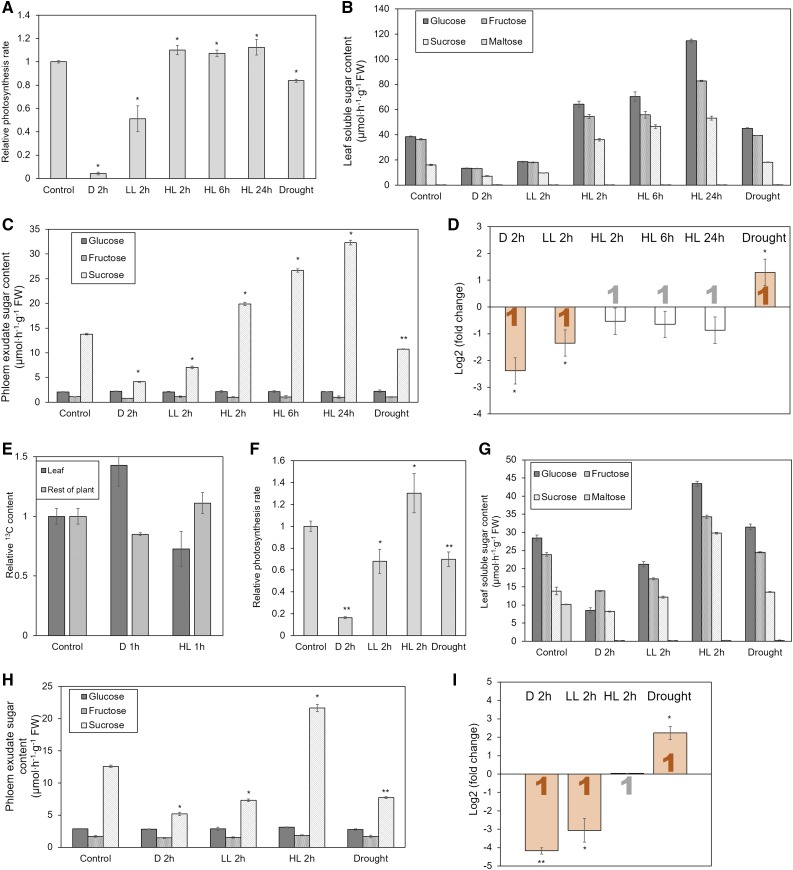
Comparison of photosynthesis rate, sugar content in leaf extract, sugar content in phloem exudate, carbon export, and Suc transporter gene expression in maize (A–E) and wheat (F–I). All parameters were measured at the same time on comparable leaves of the same plants, except for the measurement of carbon export (E). This was performed on plants at the same growth stage as those for the other experiments by applying a pulse of 13CO2 to a source leaf, transfer to experimental conditions for 1 h, and determination of the 13C content of the leaf and the rest of the plant. Significance of differences to control values is indicated by *P < 0.05 and **P < 0.01. Error bars indicate sd. Significant changes of gene expression are highlighted by filled bars with color indicating the type of Suc transporter (orange = type IIB). Average values were obtained from at least three biological replicates, except of the measurement of carbon export (E), which is based on two replicates. D, dark; LL, low light; HL, high light.
The expression of cell wall invertases, which could potentially alter sugar ratios during the exudation period, does not significantly differ between the various environmental conditions (Supplemental Fig. S4). Furthermore, starch levels do not change significantly during short incubation periods (2 h) or the period of exudation (5 h), as tested in tomato, showing that only soluble sugar levels are relevant in the present context of short-term regulation of phloem loading (Supplemental Fig. S4). Importantly, to complement measurements on exudate, we assessed phloem loading in tomato and maize using isotopic carbon. Plants were transferred to the respective experimental conditions after pulse-labeling a leaf with 13CO2. The observed differences in the amount of isotopic carbon that is exported agreed with the differences in exudate sugar levels measured by EDTA-facilitated exudation (Figs. 6 and 7).
When plants were exposed to different light conditions, photosynthesis rate and Suc concentration in leaves and exudate scaled with the light level in Arabidopsis and tomato (Fig. 6) as well as in maize and wheat (Fig. 7), unless it reached excessive levels (Fig. 6A). Exposure to low light did lead to down-regulation of the phloem-loading SUTs in Arabidopsis, maize, and wheat, which contrasts with the transcriptomics results presented above (Fig. 3). This discrepancy might be due to the variety of experimental conditions combined in each averaged value derived from the transcriptomics data, as some conditions might not be severe enough to cause a significant effect. Tomato LeSUT1 was also down-regulated, although the change of 84% was not considered significant. The lower expression of the phloem-loading SUTs coincided with lower Suc concentration in the phloem exudate (Figs. 6 and 7). The same response, in some cases with higher intensity, could be observed when plants were transferred to dark conditions (Figs. 6 and 7).
In accordance with the transcriptomics results presented above, SUT expression measurement by qPCR did not show up-regulation under high light conditions in these four apoplasmic loaders (Figs. 6 and 7). When testing different exposure times in Arabidopsis, we found that even 24-h exposure to high light conditions, which leads to continuously increasing levels of Glc, Fru, and Suc in the leaf and similarly increasing Suc levels in the phloem exudate, does not affect the expression of AtSUC2 (Fig. 6, A–D). Similar results were obtained for maize, where longer incubation led to increased levels of leaf sugars and Suc in the exudate, without significant change in expression of ZmSUT1 (Fig. 7, A–D).
Other SUTs of all four species that were tested showed the same results as obtained from the analysis of transcriptomics data, except AtSUC5, which was strongly down-regulated under low light (Supplemental Fig. S5). Notable is the pattern of tomato SUT4, which was found to be up-regulated under low light and down-regulated under high light, exactly the opposite from its homologs in the other three species (Supplemental Fig S5).
Exposure to moderate drought stress caused considerable changes in SUT expression (Figs. 6 and 7; Supplemental Fig. S5), which matched with the results from transcriptomics analysis (Fig. 4). The SUTs responsible for phloem loading were up-regulated in Arabidopsis, maize, and wheat, but not tomato, where expression was unchanged (Figs. 6 and 7). Photosynthesis rate and Suc content in whole-leaf extract and phloem exudate decreased in all four species, although in maize not significantly (Figs. 6 and 7).
Correlation of Tomato LeSUT1 Expression with Protein Abundance
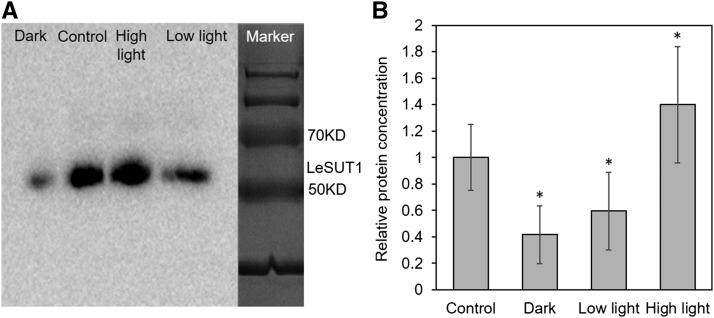
To test how SUT gene expression corresponds to protein abundance, we performed western blots using protein extracts of the same leaf material as for qPCR and an antibody against tomato LeSUT1 with proven binding specificity (Hackel et al., 2006). Even though expression of LeSUT1 did not change significantly in response to exposure to high light, low light, or dark conditions, protein abundance showed strong differences. The band for LeSUT1 protein collected from high light samples was about 40% stronger as from plants grown under control conditions, and LeSUT1 abundance was significantly reduced under low light and dark conditions (Fig. 8). These changes in protein abundance mirror the change in Suc content in phloem exudate observed in the same plants (Fig. 6G).
Figure 8.
Protein abundance of tomato LeSUT1 in response to different light levels. A, Representative western blot showing the LeSUT1-specific band at 56 kD. B, Average intensity of LeSUT1-specific bands measured in four experiments. Plant material was the same as used for qPCR (Fig. 6F). Significance of differences to control values is indicated by *P < 0.05.
DISCUSSION
SUT Expression Does Not Always Correspond to Phloem Loading
In species with apoplasmic phloem loading, it is generally assumed that the SUT-facilitated pumping of Suc into the SECCC directly influences source strength (Ainsworth and Bush, 2011). Reduced expression or knock-out of the phloem-loading SUT leads to Suc accumulation, while overexpression can lead to increased source strength (Dasgupta et al., 2014). The notion of SUT expression regulating phloem loading is corroborated here, but only for down-regulation in response to reduced light levels. High light and other stimuli that enhance photosynthesis, leaf sugar levels, and phloem loading did not influence the expression of the phloem-loading SUTs in the eight apoplasmic loading species analyzed here. Therefore, a direct link between SUT expression and phloem loading rate does not seem to exist for the up-regulation of phloem loading under these conditions. This contrasts with studies that artificially up-regulate a relevant SUT through overexpression, which, in some cases, did result in increased phloem loading (Dasgupta et al., 2014; Wang et al., 2015).
The occurrence of increased phloem loading in the absence of transcriptional up-regulation of SUTs means that other factors influence phloem loading, that the existing capacity of SUTs is large enough to increase loading without additional proteins, or that SUT activity is not regulated at the transcriptional, but the posttranscriptional level. Indeed, experiments in potato have demonstrated the potential for the regulation of SUT activity through differential intracellular localization of SUT1 (Liesche et al., 2010), as well as through dimerization (Krügel et al., 2008) and mRNA stability (He et al., 2008). The existence of posttranslational control of SUT activity has also been shown in Arabidopsis, where the amount of AtSUC2 protein was found to not correlate with Suc transport rate (Sakr et al., 1997). In addition to protein localization, also the proton motive force might influence SUT activity. Constitutive overexpression of a proton-pumping pyrophosphatase in Arabidopsis leaf companion cells has recently been shown to increase phloem loading (Khadilkar et al., 2016). Here, the amount of tomato LeSUT1 protein was found to be increased in response to high light conditions, despite unchanged gene expression. This indicates a regulatory step at the posttranscriptional level, which could be a change in mRNA translation efficiency or a change in protein turnover rate. Furthermore, it suggests that the existing capacity of SUTs is insufficient to increase phloem loading rate in tomato, instead requiring additional SUT synthesis.
SUT Regulation Is Part of the Abiotic Stress Response
Understanding the plant’s response to abiotic stresses is a major focus for plant biologists today. There is increasing evidence that different types of SUTs are involved in the response to abiotic stress (Sivitz et al., 2008; Frost et al., 2012; Gong et al., 2015; Jia et al., 2015). Based on the extensive transcriptomics data available for drought, salt, and heat stress, these data are extended here by the identification of patterns in the response of SUT expression. In contrast to nonstressed conditions, when phloem loading rate and SUT expression seems to be a function of leaf sugar status (Vaughn et al., 2002, and results presented above), more complex patterns were observed under stress, probably reflecting the changes in water status and sink activity. Drought and salt stress were found to have the same effect on SUT expression in all species, which is not surprising considering the overlap of plant responses to osmotic stresses such as drought, and salinity shown in genomics studies (Kreps et al., 2002; Buchanan et al., 2005). The common pattern shows that both require the same adaptations regarding the transport of sugars.
Similarity was also found in the change of SUT expression in response to drought and salt stress among all four tested monocot species. In all cases, SUT1 was up-regulated while SUT4 was down-regulated. Interestingly, this pattern did not depend on the severity of the applied stress. Monocot SUT4s belong to type III and are tonoplast localized (Endler et al., 2006; Eom et al., 2011; Leach et al., 2017). Down-regulation of the tonoplastic SUTs can be expected to increase Suc storage in the vacuole, thereby increasing the osmotic potential of the leaf cells. That this can affect phloem loading has recently been shown in maize (Leach et al., 2017). Up-regulation of SUT1 might be necessary to maintain phloem transport despite the decrease in Suc availability caused by increased sequestration of Suc to the vacuole, increased production of osmoprotective sugars such as raffinose, and decreased Suc production. This is consistent with the observed capacity of root growth under severe drought stress, at least in the lines with higher stress tolerance (Zheng et al., 2010), and could also apply to Arabidopsis where a similar pattern was observed. In contrast to drought and salt stress, SUT1 was found to be significantly down-regulated under heat stress in the tested monocots, while SUT4 expression showed the same response. This result matches the observed strong reduction of growth in all tissues observed in heat-stressed grasses (Kotak et al., 2007; Frey et al., 2015).
Dicots do not show a universal response like the monocots. While in Arabidopsis and soybean the phloem-loading SUTs are up-regulated, they do not show any significant change in the Solanaceae potato and tomato. Although the potato and tomato varieties used here, like most commercially grown cultivars, are drought-sensitive (Pmk and Ledent, 2001; Sprenger et al., 2016), Suc content in phloem exudate is only moderately reduced, making this observation even more puzzling. In these species, only expression of SUT4 changed significantly in response to the different stresses. Type III SUTs of Solanaceae are located at the plasma membrane in contrast to Type III SUTs in all other species, which are tonoplast localized (Chincinska et al., 2013). These SUT4s have been previously linked to light and hormone signaling (Chincinska et al., 2008). Our data demonstrates that Solanaceae SUT4 are involved in the response to a broad range of environmental stimuli, including drought, salt, and heat stress, and different light conditions. Considering the relatively low expression level in leaves (Chincinska et al., 2008), a direct involvement in Suc transport seems unlikely, even though potato SUT4 was shown to be a high-capacity transporter. Our data agree with the idea of SUT4 as posttranslational, negative regulator of SUT1. This would be possible, since both are coexpressed in the same cells and were shown to be able to interact (Reinders et al., 2002).
The Arabidopsis homolog to the Solanaceae SUT4, AtSUC4, is also up-regulated under all three types of stress, although considering the different intracellular localization, its role must be a different one. Indeed, AtSUC4-facilitated Suc exchange with the vacuole has been described as essential for salt stress tolerance (Gong et al., 2015). Another SUT whose expression was found to consistently respond to abiotic stress is AtSUC1. It has been previously implicated in ABA signaling (Hoth et al., 2010) and sugar signaling (Sivitz et al., 2008) in roots and pollen. In contrast to those tissues, AtSUC1 expression in mature leaves is very low, and its domain of expression is most likely limited to developing trichomes (Sivitz et al., 2007). The down-regulation of AtSUC1 could mean that Suc partitioning is stopped under various stress conditions, including water, salt, heat, and light stress. That AtSUC9, which has also been linked to abiotic stress resistance (Jia et al., 2015), does not show changes here is not surprising considering that its domain of action does not include leaves.
Poplar SUT4 Might Regulate Phloem Loading
How passive symplasmic phloem loaders regulate carbohydrate transport from leaves is an open question. In an unregulated system, carbohydrate export from leaves would be directly coupled to their production. Since this is not the case, mechanisms that either modulate the cytosolic Suc concentration or the capacity for transport into the phloem must exist (Liesche et al., 2017). In poplar, down-regulation of the vacuolar Suc transporter PtaSUT4 via RNAi decreases source strength and leads to Suc accumulation in the leaf as well as lower photosynthetic activity (Payyavula et al., 2011; Frost et al., 2012). It can be hypothesized that PtaSUT4 activity regulates the cytosolic concentration of Suc in leaf mesophyll cells by controlling the exchange with the vacuole, thereby influencing the Suc gradient along the prephloem pathway that determines the rate of phloem loading (Liesche, 2017). This notion of PtaSUT4 as an important link between Suc production, storage, and export is confirmed here by showing that its expression scales with photosynthetic rate. If the PtaSUT4 level remained constant under low photosynthesis conditions, then the vacuolar Suc concentration would be continuously reduced as the Suc import is expected to follow the concentration potential between cytosol and vacuole, which potentially affects nighttime supply.
PtaSUT4 could be a central factor for the adaptation of the leaf carbon balance in response to environmental stimuli, as its expression was found to change in response to abiotic stress and long-term and short-term elevated CO2 levels. In the responses to drought and salt stress, down-regulation of PtaSUT4 coincides with reduced Suc export from the leaf, indicated by the growth inhibition observed in these trees (Xue et al., 2016). PtaSUT4 is also down-regulated under elevated CO2 even though export rates are higher. This is likely due to the strongly induced carbon assimilation rate that allows for increased storage as well as increased growth as reported from similar experiments (Wullschleger et al., 1992; Zhao et al., 2011). These data point to PtaSUT4 as an important factor for determining this pattern of carbon allocation. The reduced expression presumably leads to increased Suc storage in the vacuole and, thereby, a lowering of the concentration gradient along the prephloem pathway.
The data highlights the potential importance of PtaSUT4 in poplar phloem loading. This could constitute the general mode of how symplasmic phloem loaders regulate carbon export from leaves, but data from additional species are needed for confirmation.
CONCLUSIONS
The results presented here show an important role of transcriptional regulation of SUTs in the stress response in source leaves, especially of the phloem-loading SUTs and the vacuolar SUTs. They also point at a potential role in signaling, for example of the Solanaceae SUT4. Furthermore, it could be shown that transcriptional as well as posttranscriptional mechanisms are used to regulate leaf sugar export in the analyzed monocot and dicot species with apoplasmic phloem loading. In the passive loaders, represented by poplar, results indicate participation of vacuolar SUTs in the regulation of leaf sugar export.
MATERIALS AND METHODS
Assembly of a Phylogenetic Tree
All 44 SUT protein sequences were obtained from GenBank (Benson et al., 2013) for 9 angiosperm species, the dicots Arabidopsis (Arabidopsis thaliana), poplar (Populus spp.), tomato (Solanum lycopersicum), potato (Solanum tuberosum), soybean (Glycine max), the monocots rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), maize (Zea mays), and the fungus Aspergillus clavatus, which was used as the outgroup. All sequence data were converted to FASTA format. Multiple protein sequence alignments were generated with Clustal W in MEGA6 (Tamura et al., 2013), and the variable length N- and C-terminal regions of the alignment were removed. Phylogenetic analysis was done using the Neighbor-joining method in MEGA6 with 1,000 bootstrap replicates. Trees were also visualized using the MEGA6 software. For monocot SUTs, we follow Leach et al. (2017) in naming all SUTs belonging to type IIA SUT2 and all belonging to type III SUT4.
Collection of Gene Expression Data
All expression data were collected from online sources, except for one experiment for which data were directly provided by the corresponding author. The microarray data and RNAseq data were collected from Gene Expression Omnibus (Edgar et al., 2002) and ArrayExpress (Kolesnikov et al., 2015). The qPCR data were extracted from research articles with sources listed in Supplemental Table S2. Only data from experiments conducted on mature leaves were collected. Experiments were selected which provided comparative information for gene expression under control conditions and under the influence of one of the following factors: increased or decreased photosynthetic rate, increased or decreased light intensity, increased or decreased atmospheric CO2 concentration, different times of day, drought stress, salt stress, and heat stress.
Analysis of Microarray Data
The identifier of the microarray probes corresponding to the SUT genes was found through the gene annotation information for the relevant platforms, which was downloaded from the Web site of the supplier of the array. For genes, which may have more than one probe, the one that showed the highest expression values and clearest differences between different conditions were selected. The values for SUT expression were extracted from each biological replicate of every series related to the parameters mentioned above. Only the processed data that have been normalized were used for the subsequent analysis. The average values, sd, the log2 values, and linear values were all calculated. Only the variables with log2 values >1 or below −1 were considered significantly different. Furthermore, the difference is described as upregulated with a log2 value >1 and downregulated with a log2 value below −1.
Analysis of RNAseq Data
For each data set, low-quality reads were filtered by Trimmomatic (Bolger et al., 2014), with the following parameters [LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 MINLEN:20]. Retained reads were mapped to SUT genes in the corresponding species using TopHat2 (v2.1.0) (Kim et al., 2013) with the parameters [Arabidopsis: -i 20 -I 5000 -g 5 -p 30; other species: -i 20 -I 10000 -g 5 -p 30]. The expression abundance of each gene was estimated in terms of reads per kilobase of transcript per million mapped reads (RPKM) using the formula: RPKM = (109 * C)/(N * L), where C represents the number of high-quality reads mapped to the gene, N is the total number of reads in the library, and L means length of the gene. The number of reads that mapped to the gene was counted by BEDtools multicov (Quinlan and Hall, 2010) with default parameters. For some experiments, data were extracted through Genevestigator (Nebion AG).
Plant Growth and Environmental Stimuli
All plants were grown in growth chambers. The Arabidopsis (cv Col-0 ecotype) seeds were sterilized in 75% alcohol for 2 min and 10% (v/v) sodium hypochlorite for 10 min, washed 5 times with sterilized water, and imbibed for 3 d in the dark at 4°C. The 1/2 Murashige and Skoog (MS) solid medium was prepared with 1% (w/v) Suc and 1% (w/v) agar at pH 5.8. The seeds were grown on 1/2 MS medium plates for about 5 d (22°C, 24 h dark) until germinated and then transferred to a growth chamber on moist soil in pots kept at 16-h-light/8-h-dark period, 90 μmol photons m−2 s−1, with temperature at 22°C. The maize (cv inbred line B73) and tomato (cv Micro-Tom) seeds were sterilized in sterilized water for 30 min at 30°C and 10% Na3PO4 for 20 min and then soaked in sterilized water for about 6 h. The seeds were imbibed on a wet towel at 27°C in the dark for about 3 d and then transferred to a growth chamber on moist soil in pots kept at 16-h-light/8-h-dark period, about 300 μmol photons m−2 s−1 with a temperature of 26°C during the day and 24°C at night. The wheat (cv Chinese spring) seeds were imbibed on moist filter paper with sterilized water for about 2 d at room temperature until germinated and then transferred to a growth chamber on moist soil in pots kept at a 16-h-light/8-h-dark period, 300 μmol photons m−2 s−1 with temperature about 12°C.
The plants were fully irrigated by watering until the start of the drought stress. The 3-week-old Arabidopsis seedlings, the V3 leaf stage (about 2–3 weeks) maize seedlings, the 2-week-old tomato seedlings, and approximately 5-week-old wheat seedlings were subjected to drought stress by withholding water for several days until a level of moderate drought was reached (soil relative moisture content <40%). The control plants were still watered. The leaf relative water content (RWC) was also used to determine the degree of drought stress. For determination of RWC, fresh leaves were removed and then immediately the fresh weight measured. The leaves were then incubated in distilled water for 12 h at 4°C in the dark, blotted, dried, and then the turgid weight measured. At last, the leaves were subjected to oven drying at 105°C for at least 24 h, and then the dry weight (DW) measured. The RWC was calculated by the equation: RWC (%)= [(fresh weight − DW)/(turgid weight − DW)] * 100, and for moderate drought, the RWC value should be about 60%. After treatment, the leaves were harvested from drought-stressed and control plants, quickly frozen in liquid nitrogen, and stored at −80°C for the isolation of RNA for qPCR.
For the different light intensity stimuli, leaves from the top of the plants (stage described above) were subjected to different light intensities for 2 h: excess light (900 µmol photons m−2 s−1) for Arabidopsis; high light (400 µmol photons m−2 s−1) for Arabidopsis; high light (700 µmol photons m−2 s−1) for maize, wheat, and tomato; low light (40 µmol photons m−2 s−1) for Arabidopsis; low light (100 µmol photons m−2 s−1) for tomato, maize, and wheat; and darkness (0 µmol photons m−2 s−1) for Arabidopsis, maize, wheat, and tomato. For Arabidopsis and maize, 6 and 24 h under high light stimuli were also tested. After treatment, the leaves were harvested from different light-treated and control plants, quickly frozen in liquid nitrogen, and stored at −80°C for the isolation of RNA for qPCR.
Measurement of Photosynthesis, Leaf Sugar Levels, and Sugar Export
The photosynthesis measurement was conducted using a Li-6400 portable photosynthesis measuring system (LI-COR, Inc.) with a 6-cm2 leaf chamber under control conditions and after subjection to different stimuli (described above) on plant leaves. The photosynthetically active radiation was kept at 1,000 µmol photons m−2 s−1 for Arabidopsis and 1,500 µmol photons m−2 s−1 for tomato, maize, and wheat. The reference CO2 concentration was kept at about 400 μmol CO2 mol−1 air (μg mL−1). Each measurement took 60 s to complete. Measurements were conducted during the day at 9:00 am to 12 am. To assure the homogeneity, especially for the light intensity experiment, measurements were always performed on a nonshaded leaf from the top of the plants. The values of three measurement repetitions were averaged. For Arabidopsis and wheat, a single leaf would not fill the whole measurement chamber. Therefore, an image was taken from which the leaf area was calculated using ImageJ (Schindelin et al., 2012). Photosynthetic parameters were then recalculated for the actual leaf area inside the chamber.
Phloem exudates were obtained using the EDTA-facilitated exudation method (Tetyuk et al., 2013) and adapted for all the plants tested in this experiment. For Arabidopsis, about 20 leaves from the control and plants subjected to different stimuli were cut with a razor blade at the base of the petiole, close to the center of the rosette, and immediately placed in petri dishes containing 20 mm EDTA solution. Then, petioles were recut at the base and the leaves immediately put into 1.5-mL tubes containing 20 mm EDTA solution. After 1-h incubation in wet paper towels in the dark, leaves were gently removed from the tubes and washed thoroughly using distilled water to remove all EDTA. Leaves were then transferred into new 1.5-mL tubes containing sterilized water and further incubated in wet paper towel in the dark to collect the exudates. A time course experiment was performed with sampling times of 0, 0.5, 1, 2, 3, and 5 h. The results showed that the amount of the sugars present in the exudates increased with time, and the 5-h time point provided reproducible amounts of sugars using the EDTA facilitated exudation method. Therefore, 5-h incubation was chosen for the following experiments. After 5-h collection, the phloem exudates were frozen in liquid nitrogen and stored at −80°C. To evaluate the possibility that the sap may come from the petiole instead of the leaf blade, the petiole only and the leaf blade with just a bit of petiole were used to collect the exudates as a control, which was then compared to the whole leaf samples.
For tomato, the exudates were collected using the same method as Arabidopsis, but only about five leaves were used. For maize and wheat, we followed the method described by Yesbergenova-Cuny et al. (Yesbergenova-Cuny et al., 2016). Leaves were cut off at their base, close to the stem, and recut under the exudation buffer (10 mm HEPES, 10 mm EDTA, adjusted to pH 7.8 with NaOH). After 1-h incubation in wet paper towels in dark, leaves were removed and washed thoroughly using distilled water. Then, leaves were transferred into new 10-mL tubes containing sterilized water and further incubated in wet paper towels in the dark. A time course experiment was also performed as described for Arabidopsis above. After 4-h collection, phloem exudates were frozen in liquid nitrogen and stored at −80°C.
The leaf sugars were extracted using the ethanol method. The leaves of Arabidopsis, tomato, maize, and wheat under control conditions were ground into powder using liquid nitrogen and then dissolved in 80% ethanol and incubated at 80°C in a water bath for 30 min. The supernatant was taken off after centrifuging at 12,000 x g for 20 min and the ethanol evaporated by incubation at 90°C in a water bath. The powder was suspended in 5 mL pure water and filtered using a 0.45-µm microaperture filter membrane, immediately frozen in liquid nitrogen, and stored at −80°C.
To quantify the sugars (Glc, Fru, maltose, Suc) in the collected phloem exudates, the exudate was diluted about 1:5 and subjected to anion chromatography using an ion chromatograph equipped with integral pulse amperometric detection (ICS-5000+, ThermoFisher). The CarboPac PA 10 column (4 x 250 mm) was used as separation column, and mixtures of pure water and 200 mm NaOH solution in different ratios were used as mobile phase in the gradient elution. The flow rate was 0.5 mL ·min−1. In the amperometric detection, the gold electrode was used as working electrode and the Ag/AgCl electrode was used as reference electrode. The percentage of 200 mm NaOH during the gradient eluting procedure changed from 0 to 20% in during the first 14 min, from 20 to 60% in the following 4 min, from 60 to 100% in the following 12 min, and from 100 to 80% in the following 15 min. The other part of the solution consisted of pure water. The mixture of four sugars (Glc, Fru, Suc, and maltose) was used to make the standard curves. The relative concentrations of the different sugars were analyzed quantitatively by the peak area normalizing method implemented in Chromeleon 7 software (ThermoFisher).
Quantification of Gene Expression by qPCR
Total RNA from all the leaf samples was extracted using Trizol (Invitrogen). The following changes were made to the standard procedure recommended by the manufacturer. After homogenization of 100 mg material in liquid nitrogen and addition of Trizol, samples were incubated for 10 min in an ice bath. After addition of chloroform, samples were shaken vigorously for 15 s and incubated in the ice bath for 15 min. All reagents used were cooled to 4°C. The quantity and quality of RNA was assessed spectroscopically (Nanodrop ND-2000) and by agarose gel electrophoresis. The first-strand cDNA was synthesized from 1 µg of total RNA with the PrimeScript RT reagent kit (Perfect Real Time, TaKaRa) according to the manufacturer’s instructions. The cDNA synthesis reaction conditions for the SYBR Green analysis method were 37°C for 15 min and 85°C for 5 s. The transcript levels of various genes (see primer sequences in Supplemental Table S1) were quantified by qPCR using the CFX Connect Real Time System (Bio-Rad) with TransStart Tip Green qPCR SuperMix (Transgen) according to the manufacturer’s instructions. The qPCR experiments were performed on two independent biological samples and on four technical replicates, and the suitable Cq values for these replicates were averaged. Expression levels were normalized to reference gene (Arabidopsis: AtActin12 and AtUBQ10, tomato: LeGAPDH and LeUBI3, maize: ZmGAPDH and ZmActin2, wheat: TaActin7 and TaGAPDH) expression levels. Quantification of mRNA expression level was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). For normalizing the expression level of each mRNA, fold change values and the se were calculated. To evaluate the significance of differences, P values were calculated using Student's t test method implemented in the SPSS software (IBM).
Tracing of Isotopic Carbon
Maize and tomato plants were cultivated as described above. A leaf chamber for labeling with 13CO2 was made by sealing a plastic bag with tape. The 13CO2 gas was generated through the reaction of 50 mg of NaH13CO3 (99 atom% 13C, Sigma-Aldrich) with 100 μL of saturated citric acid, both of which were injected into the leaf chamber and mixed with the help of a syringe. After incubation for 30 min, the bags were taken off and plants were either left under control conditions or transferred to high light or dark conditions. The labeled leaves and the rest of the plant, including most of the roots, were separately harvested after a chase period of 1 h. Samples were then dried in an oven at 75°C for about 5 d. The samples were ground to a fine powder in a mortar, which was then sifted through a 0.2-mm mesh sieve. The content of 13C and total C of the samples were analyzed using an isotope ratio mass spectrometer (Delta V Advantage and Elemental analyzer, Thermo Fisher Scientific, Inc.) by the company Huake Stable Isotope Laboratory, Shenzhen. Two plants and two replications were used for each light intensity, and relative 13C values of the leaves and rest of the plant were calculated.
Quantification of Protein by Western Blot
Tomato leaves from plants exposed to different light intensities as described above, as well as from the control group, were ground to powder using the high throughput tissue grinder (SCIENTZ-48) in liquid nitrogen. Then, 2 mL extraction buffer (50 mm NaH2PO4, adjusted to pH 8.0 with NaOH, 200 mm NaCl, 1 mm EDTA, 0.6% polyvinylpyrrolidone-30, 5 mm ascorbic acid, 2 mm dithiothreitol, 50 mm sodium pyrophosphate, 25 mm sodium fluoride, 1 mm sodium molybdate, 1 mm 1,10- phenanthroline, 5 mm β- sodium glycerin phosphate, and 0.5 mm phenylmethylsulfonyl fluoride, dissolved in ethanol, 1× protease inhibitor cocktail), was added to about 1 g leaf powder, vortexed thoroughly until well mixed, and then centrifuged at 10 000g for 15 min (4°C). The supernatant was transferred to a new tube and recentrifuged at 10,000 g for 15 min (4°C) to remove the precipitated particles that might influence ultracentrifugation. Then the supernatant was transferred to a centrifuge tube and centrifuged at 100,000 g for 1 h (4°C). The supernatant was removed and discarded and the precipitate resuspended using the resuspension buffer (2 mm EGTA, 2 mm EDTA, 100 mm MOPs, adjusted to pH 7.0 with NaOH, 1 mm dithiothreitol, 0.5 mm sodium pyrophosphate, 0.5 mm sodium fluoride, 0.5 mm sodium molybdate, 0.5 mm phenylmethylsulfonyl fluoride, dissolved in ethanol, 10% glycerol, and 1×protease inhibitor cocktail). The protein was quantified by Easy II Protein Quantitative kit (BCA, TransGen Biotech) and the spectrophotometer Infinite M200Pro (Tecan) according to the manufacturer’s protocol with bovine serum albumin as standard. After protein quantification, the proteins were frozen in liquid nitrogen and stored at −80°C for electrophoresis and western-blot analysis.
For SDS-PAGE, 5% stacking and 12% separating gel were used. To investigate the difference in LeSUT1 abundance between different light intensity, samples of 80 μg were supplemented with trace bromophenol blue and denaturated at 100°C for 5 min. After that, proteins were resolved under constant voltage of 80 V for the stacking gel for 30 min and constant voltage of 120 V for the separating gel for 1.5 h in a DYY-6D Protein equipment (Liuyi Beijing) until bromophenol blue reached the bottom of the gel. Semidry electrotransfer was used for the membrane transfer, and eight filter sheets to fit the measurement of the gel (8.5 x 5.5 cm) and one polyvinylidene fluoride (PVDF) membrane with the same dimensions were also used. After wetting the filter paper in transfer buffer and wetting the PVDF membrane in methanol, a transfer sandwich was created as follows: four filter papers, PVDF, gel, four filter papers (from the positive pole to the negative pole). Semidry electrotransfer was performed in JY-ZY3 semidry cell (JUNYI) at room temperature, applying constant current of 70 mA for 85 min until the protein transferred from gel to the PVDF membrane. The membrane was washed with TBST for 5 min and 3 times, then blocked with 5% skim milk in TBST for 1.5 h at room temperature, and then LeSUT1-specific antirabbit antibody (diluted 1,000-fold in 5% skim milk in TBST) was added and incubated for 1.5 h at room temperature on a shaker. After incubation, the membrane was washed with TBST for 5 min and 3 times. Secondary antibody (Goat anti-Rabbit IgG, HRP conjugated, diluted 5,000-fold in 5% skim milk in TBST) was added and incubated for 2 h at room temperature on a shaker. The membrane was washed with TBST for 5 min and 3 times. Bands on the membrane were then detected using the EasySee western-blot kit (TransGen Biotech) according to the manufacturer’s protocol. The band of the target LeSUT1 gene was quantified using the ImageJ software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtSUC1 (AT1G71880), AtSUC2 (AT1G22710), AtSUC3 (AT2G02860), AtSUC4 (AT1G09960), AtSUC5 (AT1G71890), AtSUC6 (AT5G43610), AtSUC7 (AT1G66570), AtSUC8 (AT2G14670), AtSUC9 (AT5G06170), CWINV2 (AT3G52600), CWINV4 (AT2G36190), CWINV5 (AT3G13784), GmSUC1 (AJ563364), GmSUC3 (BQ452560), GmSUC4 (BI784806), HvSUT1 (AM055812), HvSUT4 (AJ272308), LeSUT1 (AB845639), LeSUT2 (AF166498), LeSUT4 (AF176950), OsSUT1 (D87819), OsSUT2 (AY137242), OsSUT3 (AB071809), OsSUT4 (HQ875341), OsSUT5 (AB091674), PtaSUT1 (POPTR_0013s11950), PtaSUT3 (POPTR_0019s11560g), PtaSUT4 (POPTR_0002s10710g), PtaSUT5 (XM_002311560), PtaSUT6 (POPTR_0010s10370), TaSUT1 (AF408842), TaSUT4 (BJ308154), StSUT1 (X69165), StSUT2 (AF166498), StSUT4 (AF176950), ZmSUT1 (AB008464), ZmSUT2 (AY639018), ZmSUT4 (AY581895).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Sequences of gene-specific primers used for qPCR.
Supplemental Table S2. Transcriptomics data sets analyzed in this study.
Supplemental Figure S3. Influence of salt stress on Suc transporter expression.
Supplemental Figure S4. Control experiments for the EDTA-facilitated exudation of phloem sap.
Supplemental Figure S5. Change of Suc transporter expression in response to various stimuli.
Acknowledgments
Antibodies against tomato SUT1 were kindly provided by Dr. Christina Kühn, Humboldt University Berlin. The authors declare no conflict of interest.
Footnotes
Articles can be viewed without a subscription.
References
- Ainsworth EA, Bush DR (2011) Carbohydrate export from the leaf: a highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol 155: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard V, Mueh KE, Demmig-Adams B, Ebbert V, Turgeon R, Adams WW III (2005) Anatomical and photosynthetic acclimation to the light environment in species with differing mechanisms of phloem loading. Proc Natl Acad Sci USA 102: 12968–12973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Scofield GN, Wang X-D, Patrick JW, Offler CE, Furbank RT (2004) Expression and localisation analysis of the wheat sucrose transporter TaSUT1 in vegetative tissues. Planta 219: 176–184 [DOI] [PubMed] [Google Scholar]
- Ayre BG. (2011) Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol Plant 4: 377–394 [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41: D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J, Bjorkman O (1980) Photosynthetic response and adaptation to temperature in higher plants. Annu Rev Plant Physiol 31: 491–543 [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, Weers BD, Klein RR, Pratt LH, Cordonnier-Pratt M-M, Klein PE, et al. (2005) Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol 58: 699–720 [DOI] [PubMed] [Google Scholar]
- Bürkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB (1998) The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol 118: 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincinska I, Gier K, Krügel U, Liesche J, He H, Grimm B, Harren FJ, Cristescu SM, Kühn C (2013) Photoperiodic regulation of the sucrose transporter StSUT4 affects the expression of circadian-regulated genes and ethylene production. Front Plant Sci 4: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincinska IA, Liesche J, Krügel U, Michalska J, Geigenberger P, Grimm B, Kühn C (2008) Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol 146: 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta K, Khadilkar AS, Sulpice R, Pant B, Scheible W-R, Fisahn J, Stitt M, Ayre BG (2014) Expression of sucrose transporter cDNAs specifically in companion cells enhances phloem loading and long-distance transport of sucrose but leads to an inhibition of growth and the perception of a phosphate limitation. Plant Physiol 165: 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A, Keller F, Turgeon R (2011) Phloem loading, plant growth form, and climate. Protoplasma 248: 153–163 [DOI] [PubMed] [Google Scholar]
- Duan Z, Homma A, Kobayashi M, Nagata N, Kaneko Y, Fujiki Y, Nishida I (2014) Photoassimilation, assimilate translocation and plasmodesmal biogenesis in the source leaves of Arabidopsis thaliana grown under an increased atmospheric CO2 concentration. Plant Cell Physiol 55: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M, Porcheron B, Hennion N, Maurousset L, Lemoine R, Pourtau N (2016) Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol 170: 1460–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom J-S, Cho J-I, Reinders A, Lee S-W, Yoo Y, Tuan PQ, Choi S-B, Bang G, Park Y-I, Cho M-H et al. (2011) Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol 157: 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey FP, Urbany C, Hüttel B, Reinhardt R, Stich B (2015) Genome-wide expression profiling and phenotypic evaluation of European maize inbreds at seedling stage in response to heat stress. BMC Genomics 16: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost CJ, Nyamdari B, Tsai C-J, Harding SA (2012) The tonoplast-localized sucrose transporter in Populus (PtaSUT4) regulates whole-plant water relations, responses to water stress, and photosynthesis. PLoS One 7: e44467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Liu M, Zhang L, Ruan Y, Ding R, Ji Y, Zhang N, Zhang S, Farmer J, Wang C (2015) Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiol Plant 153: 119–136 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould N, Thorpe MR, Pritchard J, Christeller JT, Williams LE, Roeb G, Schurr U, Minchin PE (2012) AtSUC2 has a role for sucrose retrieval along the phloem pathway: evidence from carbon-11 tracer studies. Plant Sci 188-189: 97–101 [DOI] [PubMed] [Google Scholar]
- Grodzinski B, Jiao J, Leonardos ED (1998) Estimating photosynthesis and concurrent export rates in C3 and C4 species at ambient and elevated CO21,2. Plant Physiol 117: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelette BS, Benning UF, Hoffmann-Benning S (2012) Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J Exp Bot 63: 3603–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel A, Schauer N, Carrari F, Fernie AR, Grimm B, Kühn C (2006) Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J 45: 180–192 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- He H, Chincinska I, Hackel A, Grimm B, Kühn C (2008) Phloem mobility and stability of sucrose transporter transcripts. Open Plant Sci J 2: 15–26 [Google Scholar]
- Hoth S, Niedermeier M, Feuerstein A, Hornig J, Sauer N (2010) An ABA-responsive element in the AtSUC1 promoter is involved in the regulation of AtSUC1 expression. Planta 232: 911–923 [DOI] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteillé M, Stitt M, et al. (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Zhang L, Wu D, Liu S, Gong X, Cui Z, Cui N, Cao H, Rao L, Wang C (2015) Sucrose transporter AtSUC9 Mediated by a low sucrose level is involved in Arabidopsis abiotic stress resistance by regulating sucrose distribution and ABA accumulation. Plant Cell Physiol 56: 1574–1587 [DOI] [PubMed] [Google Scholar]
- Jiao J, Grodzinski B (1996) The effect of leaf temperature and photorespiratory conditions on export of sugars during steady-state photosynthesis in Salvia splendens. Plant Physiol 111: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius BT, Leach KA, Tran TM, Mertz RA, Braun DM (2017) Sugar transporters in plants: new insights and discoveries. Plant Cell Physiol 58: 1442–1460 [DOI] [PubMed] [Google Scholar]
- Jung H-S, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J (2013) Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci USA 110: 14474–14479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadilkar AS, Yadav UP, Salazar C, Shulaev V, Paez-Valencia J, Pizzio GA, Gaxiola RA, Ayre BG (2016) Constitutive and companion cell-specific overexpression of AVP1, encoding a proton-pumping pyrophosphatase, enhances biomass accumulation, phloem loading and long-distance transport. Plant Physiol 170: 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Zeevaart JA (1974) Enhancement of Phloem exudation from cut petioles by chelating agents. Plant Physiol 53: 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, et al. (2015) ArrayExpress update--simplifying data submissions. Nucleic Acids Res 43: D1113–D1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf K-D (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Krügel U, Veenhoff LM, Langbein J, Wiederhold E, Liesche J, Friedrich T, Grimm B, Martinoia E, Poolman B, Kühn C (2008) Transport and sorting of the solanum tuberosum sucrose transporter SUT1 is affected by posttranslational modification. Plant Cell 20: 2497–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang H-S, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Grof CP (2010) Sucrose transporters of higher plants. Curr Opin Plant Biol 13: 288–298 [DOI] [PubMed] [Google Scholar]
- Leach KA, Tran TM, Slewinski TL, Meeley RB, Braun DM (2017) Sucrose transporter2 contributes to maize growth, development, and crop yield. J Integr Plant Biol 59: 390–408 [DOI] [PubMed] [Google Scholar]
- Liesche J. (2017) Sucrose transporters and plasmodesmal regulation in passive phloem loading. J Integr Plant Biol 59: 311–321 [DOI] [PubMed] [Google Scholar]
- Liesche J, He H-X, Grimm B, Schulz A, Kühn C (2010) Recycling of Solanum sucrose transporters expressed in yeast, tobacco, and in mature phloem sieve elements. Mol Plant 3: 1064–1074 [DOI] [PubMed] [Google Scholar]
- Liesche J, Pace MR, Xu Q, Li Y, Chen S (2017) Height-related scaling of phloem anatomy and the evolution of sieve element end wall types in woody plants. New Phytol 214: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesche J, Windt C, Bohr T, Schulz A, Jensen KH (2015) Slower phloem transport in gymnosperm trees can be attributed to higher sieve element resistance. Tree Physiol 35: 376–386 [DOI] [PubMed] [Google Scholar]
- Liu DD, Chao WM, Turgeon R (2012) Transport of sucrose, not hexose, in the phloem. J Exp Bot 63: 4315–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Payyavula RS, Tay KH, Tsai CJ, Harding SA (2011) The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J 65: 757–770 [DOI] [PubMed] [Google Scholar]
- Peng D, Gu X, Xue L-J, Leebens-Mack JH, Tsai C-J (2014) Bayesian phylogeny of sucrose transporters: ancient origins, differential expansion and convergent evolution in monocots and dicots. Front Plant Sci 5: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pmk D, Ledent JF (2001) Effects of moderate drought conditions on green leaf number, stem height, leaf length and tuber yield of potato cultivars. Eur J Agron 14: 31–41 [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Schulze W, Kühn C, Barker L, Schulz A, Ward JM, Frommer WB (2002) Protein-protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14: 1567–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Sivitz AB, Ward JM (2012) Evolution of plant sucrose uptake transporters. Front Plant Sci 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Turgeon R (2009) A comprehensive picture of phloem loading strategies. Proc Natl Acad Sci USA 106: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuscher S, Akiyama M, Yasuda T, Makino H, Aoki K, Shibata D, Shiratake K (2014) The sugar transporter inventory of tomato: genome-wide identification and expression analysis. Plant Cell Physiol 55: 1123–1141 [DOI] [PubMed] [Google Scholar]
- Richard AC, Lyons PA, Peters JE, Biasci D, Flint SM, Lee JC, McKinney EF, Siegel RM, Smith KG (2014) Comparison of gene expression microarray data with count-based RNA measurements informs microarray interpretation. BMC Genomics 15: 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB (1994) Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J 13: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakr S, Noubahni M, Bourbouloux A, Riesmeier J, Frommer WB, Sauer N, Delrot S (1997) Cutting, ageing and expression of plant membrane transporters. Biochim Biophys Acta 1330: 207–216 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Johnson ME, Krentz AD, Grof CP, Perroux JM, Ward JM (2007) Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol 143: 188–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM (2005) Analysis of the transport activity of barley sucrose transporter HvSUT1. Plant Cell Physiol 46: 1666–1673 [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM (2008) Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol 147: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski TL, Meeley R, Braun DM (2009) Sucrose transporter1 functions in phloem loading in maize leaves. J Exp Bot 60: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger H, Kurowsky C, Horn R, Erban A, Seddig S, Rudack K, Fischer A, Walther D, Zuther E, Köhl K, et al. (2016) The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ 39: 2370–2389 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetyuk O, Benning UF, Hoffmann-Benning S (2013) Collection and analysis of Arabidopsis phloem exudates using the EDTA-facilitated Method. J Vis Exp 80: e51111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99: 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimolmangkang S, Zheng H, Peng Q, Jiang Q, Wang H, Fang T, Liao L, Wang L, He H, Han Y (2016) Assessment of sugar components and genes involved in the regulation of sucrose accumulation in peach fruit. J Agric Food Chem 64: 6723–6729 [DOI] [PubMed] [Google Scholar]
- Wang L, Lu Q, Wen X, Lu C (2015) Enhanced sucrose loading improves rice yield by increasing grain size. Plant Physiol 169: 2848–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger SD, Norby RJ, Hendrix DL (1992) Carbon exchange rates, chlorophyll content, and carbohydrate status of two forest tree species exposed to carbon dioxide enrichment. Tree Physiol 10: 21–31 [DOI] [PubMed] [Google Scholar]
- Xue L-J, Frost CJ, Tsai C-J, Harding SA (2016) Drought response transcriptomes are altered in poplar with reduced tonoplast sucrose transporter expression. Sci Rep 6: 33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesbergenova-Cuny Z, Dinant S, Martin-Magniette M-L, Quilleré I, Armengaud P, Monfalet P, Lea PJ, Hirel B (2016) Genetic variability of the phloem sap metabolite content of maize (Zea mays L.) during the kernel-filling period. Plant Sci 252: 347–357 [DOI] [PubMed] [Google Scholar]
- Zhang C, Han L, Slewinski TL, Sun J, Zhang J, Wang Z-Y, Turgeon R (2014) Symplastic phloem loading in poplar. Plant Physiol 166: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Xu X, Zhang Y, Korpelainen H, Li C (2011) Nitrogen deposition limits photosynthetic response to elevated CO2 differentially in a dioecious species. Oecologia 165: 41–54 [DOI] [PubMed] [Google Scholar]
- Zheng J, Fu J, Gou M, Huai J, Liu Y, Jian M, Huang Q, Guo X, Dong Z, Wang H, et al. (2010) Genome-wide transcriptome analysis of two maize inbred lines under drought stress. Plant Mol Biol 72: 407–421 [DOI] [PubMed] [Google Scholar]