TsNAC1 improves abiotic stress resistance of Thelungiella halophila by regulating ion transport and retards vegetative growth by limiting the expansion of cells.
Abstract
NAC proteins constitute one of the largest families of plant-specific transcription factors, and a number of these proteins participate in the regulation of plant development and responses to abiotic stress. T. HALOPHILA STRESS RELATED NAC1 (TsNAC1), cloned from the halophyte Thellungiella halophila, is a NAC transcription factor gene, and its overexpression can improve abiotic stress resistance, especially in salt stress tolerance, in both T. halophila and Arabidopsis (Arabidopsis thaliana) and retard the growth of these plants. In this study, the transcriptional activation activity of TsNAC1 and RD26 from Arabidopsis was compared with the target genes’ promoter regions of TsNAC1 from T. halophila, and the results showed that the transcriptional activation activity of TsNAC1 was higher in tobacco (Nicotiana tabacum) and yeast. The target sequence of the promoter from the target genes also was identified, and TsNAC1 was shown to target the positive regulators of ion transportation, such as T. HALOPHILA H+-PPASE1, and the transcription factors MYB HYPOCOTYL ELONGATION-RELATED and HOMEOBOX12. In addition, TsNAC1 negatively regulates the expansion of cells, inhibits LIGHT-DEPENDENT SHORT HYPOCOTYLS1 and UDP-XYLOSYLTRANSFERASE2, and directly controls the expression of MULTICOPY SUPPRESSOR OF IRA14. Based on these results, we propose that TsNAC1 functions as an important upstream regulator of plant abiotic stress responses and vegetative growth.
NAC transcription factors include at least 107 members in Arabidopsis (Arabidopsis thaliana; Riechmann et al., 2000) and constitute one of the largest protein families in higher plants. NAC transcription factors have important roles in plant development (Guo et al., 2005; Petricka et al., 2012), cell apoptosis (Lee et al., 2014), abiotic stress tolerance (Tran et al., 2010), secondary cell wall formation (Yamaguchi and Demura, 2010), and endoplasmic reticulum stress resistance (Chi et al., 2017). NAC families have a conserved N terminus with a similar DNA-binding domain but highly diverse C termini, which do not contain any known protein domains (Ooka et al., 2003). Arabidopsis RESPONSIVE TO DESICCATION26 (RD26; AT4G27410) is a NAC transcription factor belonging to the ATAF subfamily, and it is markedly up-regulated by salt, drought, and abscisic acid (ABA) treatment. In addition, RD26 has the ability to positively regulate ABA signaling (Fujita et al., 2004; Tran et al., 2004). In cotton (Gossypium hirsutum), overexpression of the rice (Oryza sativa) NAC gene SNAC1 improved tolerance to drought and salt via enhanced root development and reduced transpiration rates (Liu et al., 2014). Recent studies also showed that, in maize (Zea mays), STRESS RELATED NAC1 (ZmSNAC1) worked as a stress-responsive transcription factor in positively modulating abiotic stress tolerance (Lu et al., 2012; Shiriga et al., 2014). However, the response of the downstream networks of these NAC transcription factors to abiotic stress have not been clarified.
Arabidopsis is a salt-sensitive plant and presents certain limitations in studies of salt and drought tolerance mechanisms. Thellungiella halophila (and Thellungiella salsuginea) is a close relative of Arabidopsis with a similar coding sequence. However, T. halophila does show differences at the metabolome level, including in the metabolism of Ala and the expression patterns of certain drought- or cold-responsive genes (Amtmann et al., 2005; Benina et al., 2013). T. halophila is a valuable model plant for the study of plant stress response mechanisms because it has a small genome and exhibits high tolerance to drought, low temperatures, and high salinity (Zhu, 2001; Inan et al., 2004).
VACUOLAR H+-PYROPHOSPHATASE (H+-PPASE) plays an important role in the abiotic stress tolerance of plants. Overexpression of V-PPASE1 (AVP1; AT1G15690) in Arabidopsis improved the drought and salt tolerance in this species (Gaxiola et al., 2001). The overexpression of T. HALOPHILA H+-PPASE1 (TsVP1) also enhanced salt and drought stress resistance in different species, such as cotton, maize, and tobacco (Nicotiana tabacum; Gao et al., 2006; Li et al., 2008; Lv et al., 2009). Although the expression of TsVP1 is apparently induced by salt stress, this is not the case for AVP1 in Arabidopsis. Furthermore, one 130-bp (−667 to −538) region was identified as the key sequence for the salt stress response from the promoter of TsVP1 in T. halophila (Sun et al., 2010).
Using the above-mentioned 130-bp region as a starting point, we identified T. HALOPHILA STRESS RELATED NAC1 (TsNAC1), which is an ortholog of RD26 in Arabidopsis. The overexpression of TsNAC1 substantially promoted the abiotic stress tolerance of plants and retarded the growth of T. halophila plants. We compared the differences between TsNAC1 and RD26 to determine the function of TsNAC1 in growth regulation and abiotic responses. A chromatin immunoprecipitation sequencing (ChIP-Seq) assay was used to reveal the downstream genes and regulation network. This study provides evidence that the growth and abiotic stress resistance responses of the halophyte T. halophila are regulated by the key transcription factor TsNAC1 in a dose-dependent manner.
RESULTS
TsNAC1 Is an Upstream Regulator of TsVP1
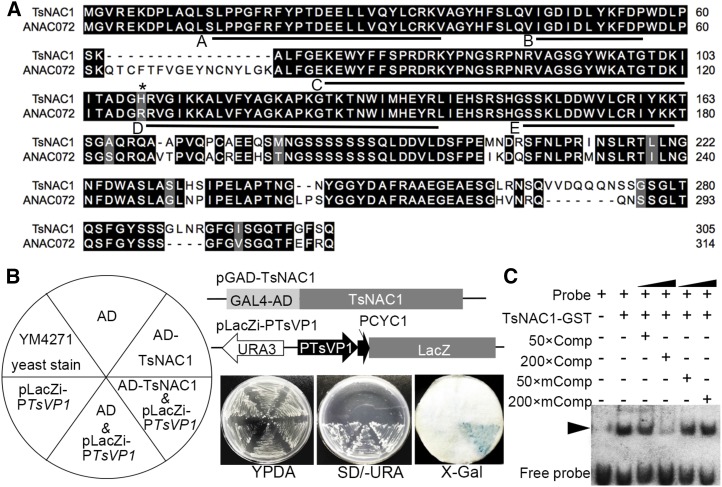
A plasmid containing a 3× tandemly repeated 530-bp DNA fragment of the TsVP1 promoter (nucleotides −667 to −138) was constructed and introduced into the yeast strain Y187. This strain was then transformed with the pGADT7-cDNA library comprising cDNAs from T. halophila plants exposed to a 12-h high-salt stress treatment. One target protein was identified from the strains grown on synthetic dextrose (SD)/-His/-Trp medium with 5 mm 3-aminotriazole. The candidate gene was cloned from T. halophila, and the BLAST results indicated that the target protein was TsNAC1, a member of the NAC superfamily. TsNAC1 has an open reading frame with a length of 918 bp, shares approximately 86% similarity with RD26 at the nucleotide level and 92% similarity at the amino acid level (Supplemental Data S1), and includes the A to E subdomains of the NAC DNA-binding domain (Fig. 1A). Evolutionary tree analysis indicated that TsNAC1 is the closest gene to RD26 (ANAC072) in T. halophila (Supplemental Fig. S1).
Figure 1.
The 130-bp key region of the promoter of TsVP1 is a direct binding target of TsNAC1. A, Multiple amino acid sequence alignment of TsNAC1 and its ortholog in Arabidopsis (RD26). Identical residues are shaded in black, and five subdomains (A–E) are indicated on the sequences. B, Transcriptional activity assay of TsNAC1 in yeast strain YM4271. TsNAC1 was cloned and fused with the GAL4 AD. The promoter of TsVP1 was cloned into pLacZi vector. Streaked transformants are shown on Yeast extract Peptone Dextrose medium with 0.003% Adenine hemisulfate (YPDA) and SD/-uracil medium. The activities of β-galactosidase were examined by X-gal staining. C, Electrophoretic mobility shifts assay (EMSA). Probes, TsNAC1-GST, competitors (130-bp unlabeled fragments), and mutated competitors at 50× and 200× molar excess were present (+) or absent (−) in each reaction. The protein-DNA complexes are marked by black arrowheads.
The coding region of TsNAC1 was fused to the GAL4 activation domain (AD), and the promoter of TsVP1 was cloned into the reporter plasmid pLacZi. Both of the plasmids were transformed into the yeast strain YM4271. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-gal) staining for β-galactosidase activity showed a blue color in strains with the promoter region of TsVP1 (Fig. 1B), indicating that TsNAC1 was bound to the promoter region to activate the transcription of TsVP1 in yeast.
The TsNAC1-GST fusion protein was expressed in Escherichia coli, and the 130-bp fragments of the TsVP1 promoter were labeled with digoxigenin-ddUTP for an EMSA. TsNAC1 was bound to the labeled probe, and the 200× molar excess unlabeled probe could competitively inhibit the combination (Fig. 1C). These results demonstrated that TsNAC1 could bind to the 130-bp sequence of the TsVP1 promoter in vitro.
Expression Profile of TsNAC1
To evaluate the expression pattern of TsNAC1 in T. halophila, real-time reverse transcription (RT)-PCR was performed with diverse organ samples collected from embryogenesis to senescence. The results suggested that the senescent organs, including senescent cauline leaves, rosette leaves, and roots, always had higher expression levels than the other stages. Parts of the floral organs, such as sepals and petals, also exhibited a clear expression pattern (Supplemental Fig. S2A). These data were consistent with the public microarray data for RD26 from the e-FP browser (Schmid et al., 2005).
Wild-type plants of T. halophila were sprayed and watered with 100 µm ABA or methyl jasmonate (MeJA). The roots were sensitive to ABA and showed a 600-fold expression peak after 24 h of treatment, whereas the shoots were sensitive to MeJA and showed a 720-fold expression peak after 24 h of treatment. The expression of TsNAC1 also was induced by cold, drought, and active oxygen stress in the shoots and by drought and salt stress in the roots (Supplemental Fig. S2B).
Overexpression of TsNAC1 Retards the Growth of T. halophila
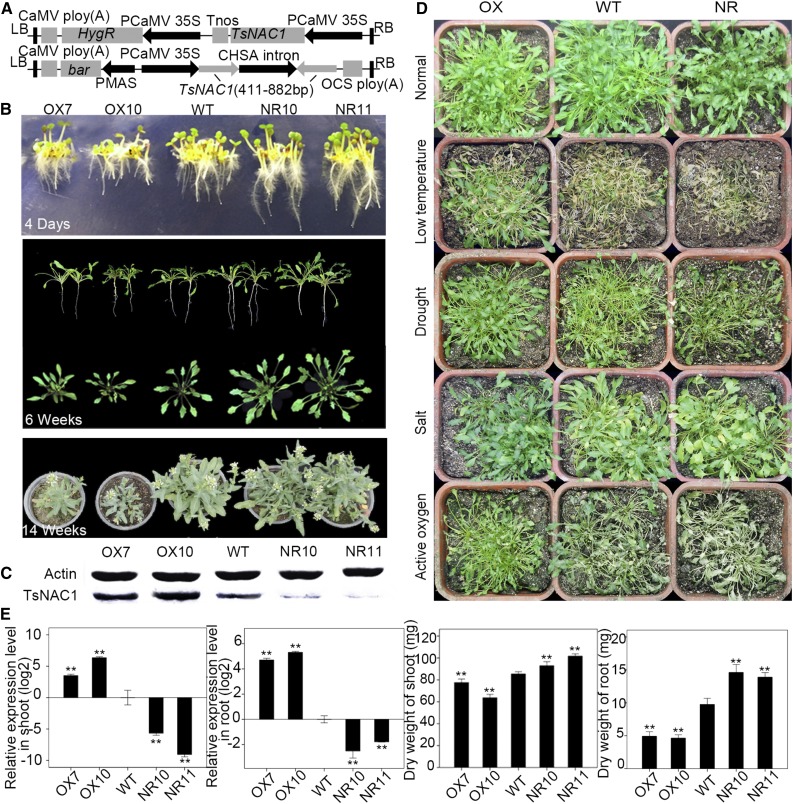
Transgenic plants overexpressing TsNAC1 (OX; 35S:TsNAC1) and TsNAC1 RNA interference (RNAi) plants (NR; harboring hairpin-shaped TsNAC1 mRNA fragments [411–882 bp] driven by the cauliflower mosaic virus 35S promoter) were generated from T. halophila, and Arabidopsis overexpressing TsNAC1 plants (OXA; 35S:TsNAC1) also were generated (Fig. 2A). Western-blot analyses indicated that the protein levels of these independent lines were consistent with the transcription levels of TsNAC1 (Fig. 2C). The overexpressing lines OX7 and OX10 and the RNAi lines NR10 and NR11 of T. halophila were selected to determine the effects of TsNAC1 transcript levels on growth and abiotic stress responses. Seeds were germinated on one-half-strength Murashige and Skoog-agar medium, and seeds of the overexpressing lines OX7 and OX10 germinated later than the wild-type and RNAi seeds (Fig. 2B). After 6 weeks of growth, the OX7 and OX10 plants were much smaller than the wild-type plants (Fig. 2B), and the dry weights of the shoots and roots were substantially lighter (Fig. 2E). The dry weight of 6-week-old plants from the T. halophila OX lines was reduced by 10.1% to 25.4% in the shoots and by 49.5% to 52.5% in the roots compared with those of the wild type (Fig. 2E). In contrast, the NR10 and NR11 plants had larger leaves and stronger roots (Fig. 2B). The biomass and growth rates were inversely related to the expression levels of TsNAC1 (Fig. 2E). Taken together, these results led to the conclusion that TsNAC1 retarded plant growth.
Figure 2.
TsNAC1 regulated the growth of T. halophila. A, Diagrams of the plant overexpression vector and RNAi vector. B, Phenotypes of TsNAC1 transgenic plants. Shown are seedlings grown on Murashige and Skoog-agar for 4 d after sowing and plants grown in soil for 6 weeks and 14 weeks (after vernalization treatment for 5 weeks). C, Western-blot results of transgenic lines and the wild type (WT). D, Phenotypes of OX, wild-type, and NR T. halophila plants after 48 h of treatment at −4°C with 3,500 lx, after 1 week of drought stress with an 18% PEG6000 solution, after keeping the 600 mm NaCl concentration of culture medium for 2 weeks, and after 36 h of active oxygen stress with 0.1 mL of 3 mm paraquat solution for every pot. E, TsNAC1 expression levels in OX7, OX10, NR10, NR11, and the wild type (the expression was normalized with Actin), and the dry weight of shoots and roots. Bars represent means ± sd, with three biological replicates in the experiment and five plants for each repeat. Significant differences by Student’s t test are indicated with asterisks: **, P < 0.01.
Overexpressing TsNAC1 Improves Abiotic Stress Resistance of Plants
MeJA-responsive and ABA-responsive cis-acting regulatory elements are observed in the promoter sequences of TsNAC1 and RD26, and the TsNAC1 promoter has an anaerobic induction element and certain cold- and dehydration-responsive elements, including the TGA element, ARE, and TC-rich repeats (Supplemental Table S1). To identify whether TsNAC1 expression enhances plant tolerance to drought, high salinity, low temperatures, and reactive oxygen species (ROS) stress in transgenic T. halophila, the seeds from the transgenic lines were germinated in a medium of peat and vermiculite (at a 2:1 ratio) and the seedlings were grown for 6 weeks under normal conditions. Under the low-temperature stress (−4°C) treatment (Fig. 2D), the overexpressing plants had a higher survival rate after 48 h of treatment (Supplemental Fig. S3A). After 6 weeks of growth, plants were watered with 18% (w/v) PEG6000 solution for 1 week (Fig. 2D), and overexpressing plants exhibited higher tolerance to drought conditions than wild-type or RNAi transgenic plants (Supplemental Fig. S3B). To examine salt resistance, the plants were thoroughly watered with an 800 mm NaCl solution and then watered with 10 mL of a 400 mm NaCl solution every 3 d (Fig. 2D). After 14 d of exposure, the transgenic RNAi and wild-type plants developed necrotic leaves and certain senescent leaves died, whereas the TsNAC1 overexpression plants displayed the same characteristics as untreated plants (Supplemental Fig. S3C). Uniform seedlings of transgenic and wild-type plants were sprayed with 0.1 mL of 3 mm paraquat per pot. After a 36-h treatment, the leaves of OX lines all showed greater resistance and higher survival probabilities than the wild-type or NR plants (Supplemental Fig. S3D). These results suggested that overexpressing TsNAC1 significantly improved plant abiotic resistance.
Exploration of the Target Genes of TsNAC1
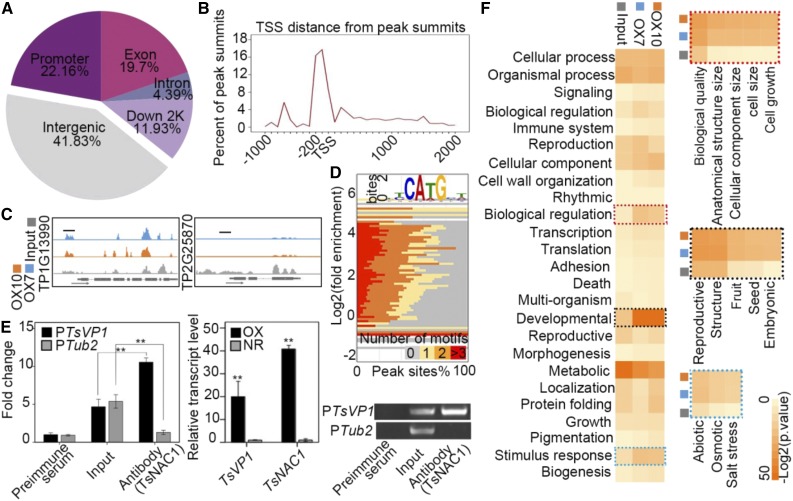
To investigate how the overexpression of TsNAC1 retarded plant vegetative development, the downstream target genes of TsNAC1 were identified by ChIP-Seq using overexpressing lines OX7 and OX10 and the TsNAC1 polyclonal antibody, the specificity of which was tested by western blot, with Columbia-0 (Col-0) and OXA leaves as materials and with the β-Actin antibody as the contrast (Supplemental Fig. S4). Each sequencing read was mapped to the T. halophila genome (http://thellungiella.org/) to identify its position using the ultrafast Bowtie aligner (Langmead et al., 2009). The ChIP-Seq data included 14.2 million mapped reads, with 7.8-fold of genome coverage on average. After excluding the peaks in intergenic regions, MACS software (Zhang et al., 2008) detected 1,868 and 2,454 genes associated with a gene model with P < 0.05 for each of the ChIP-Seq data sets. A total of 892 genes were highly enriched in both sets, and 22.16% of these gene peaks were located in the promoter region (a fragment from −2,000 bp to ATG; Fig. 3A). Additionally, except for the peaks in the intergenic region, the read distribution was close to the transcription start site at −200 to +100 bp (Fig. 3B). TsNAC1 is an upstream regulator of TsVP1. The ChIP-Seq signals of TP1G13990 (TsVP1) and TP2G25870 (Tub2), which were used as the positive and negative controls, respectively, were visualized on the genome browser (Fig. 3C). Based on ChIP-Seq, motifs around the binding peaks located at the promoter regions and a short and ungapped CA(T/A)G sequence were investigated (Fig. 3D). The ChIP samples were quantified via qPCR using primers specific to the promoters of TsVP1 and Tub2. As shown in Figure 3E, the promoter region of TsVP1 was effectively enriched, and TsVP1 was expressed at higher levels in OX lines than in the NR lines. These data demonstrated that TsNAC1 targeted the promoter of TsVP1 and that the ChIP-Seq results were valid.
Figure 3.
ChIP-Seq assay of TsNAC1. A, Distribution of TsNAC1 transcription factor-binding sites. B, Identified peak distance from transcription start sites (TSS) for TsNAC1. The peaks were highly enriched from −200 to +100 bp to the transcription start sites. C, ChIP-Seq signals of TP1G13990 (TsVP1) and TP2G25870 (Tub2) on the genome browser. The tag counts were normalized in each bin according to the total number of reads. Short black lines mark regions used for the ChIP-quantitative PCR (qPCR) assay. D, Binding situation of TsNAC1 quantified by the percentage occurrence of the CA(T/A)G motif (x axis) plotted against the log2 fold enrichment of immunoprecipitation samples compared with the input sample (y axis), with the color of each square mapped to the number of indicated peak motifs. E, Anti-TsNAC1 ChIP-qPCR validation and transcript detection of TsVP1 and TsNAC1. Samples immunized by preimmune serum were used as the negative control in the ChIP-qPCR assay. Bars represent means ± sd, with three biological replicates in the experiment and five plants for each repeat. Significant differences by Student’s t test are indicated with asterisks: **, P < 0.01. F, GO analysis results of the input sample data and data for two immunoprecipitation samples (OX7 and OX10). The dotted rectangles indicate significant and consistent GO enrichment and the hierarchical classification results of biological regulation, developmental processes, and responses to stimuli.
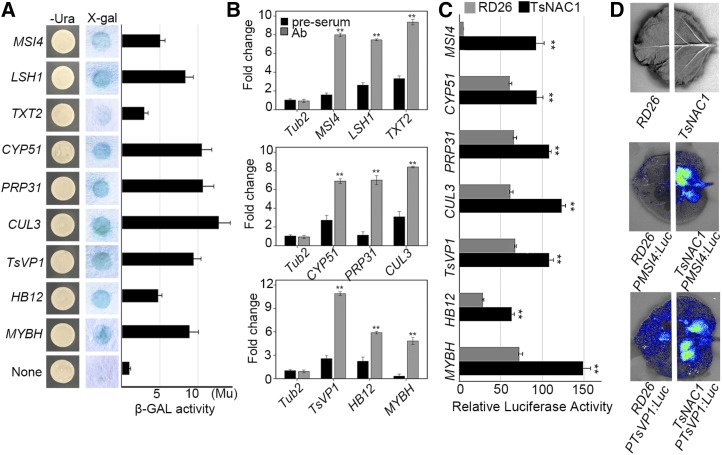
To further explore the biological processes regulated by TsNAC1, the binding target genes were classified into Gene Ontology (GO) groups. The GO analysis results for 892 coenriched genes showed a significant and coincident enrichment for biological regulation (GO:0065007), developmental processes (GO:0032502), and responses to stimuli (GO:0050896) compared with the input sample data (Fig. 3F). The next hierarchical classification of these genes primarily involved biological processes, including cell growth (GO:0016049), embryonic development ending in seed dormancy (GO:0009793), and response to salt stress (GO:0009651; Fig. 3F). Genes with fold enrichment greater than 20 (normalized by the input sample) and peaks located in the promoter region of genes from these three groups were selected to test for the expression levels in transgenic plants, including the levels of TsVP1. Seven genes were up-regulated [log2 (OX/NR) > 2] and two genes were down-regulated [log2 (OX/NR) < −1] by TsNAC1 (Table I). The promoter regions for these nine genes were recombined into a pLacZi vector for one-hybrid yeast system assays of the YM4271 strain. X-gal staining showed that these regions were targeted by TsNAC1 in yeast (Fig. 4A).
Table I. Candidate target genes of TsNAC1 identified by ChIP-Seq.
| TGI No.a | AGI No.b | Motifc (Y/N) | Aliasd | Enrichment Folde | Relative Expression Levelf |
|---|---|---|---|---|---|
| Tp3g32900 | AT2G19520 | Y | MSI4 | 35.71 | 3.93 |
| Tp2g20180 | AT5G28490 | Y | LSH1 | 25.51 | −2.18 |
| Tp6g02410 | AT4G02500 | Y | TXT2 | 31.06 | −1.92 |
| Tp1g10290 | AT1G11680 | Y | CYP51 | 59.52 | 3.46 |
| Tp2g04490 | AT1G60170 | Y | PRP31 | 44.64 | 3.01 |
| Tp1g21950 | AT1G26830 | Y | CUL3 | 41.21 | 2.96 |
| Tp1g13990 | AT1G15690 | Y | TsVP1 | 29.76 | 3.43 |
| Tp5g01460 | AT3G61890 | Y | HB12 | 39.68 | 2.57 |
| Tp2g12110 | AT5G47390 | Y | MYBH | 29.76 | 2.18 |
TGI, T. halophila Genome Initiative. bAGI, the ortholog of T. halophila in the Arabidopsis Genome Initiative. cMotif, whether with CAT(A)G in the promoter region of target genes in T. halophila. dAlias, the ortholog of target genes’ short names in Arabidopsis, except for TsVP1, which has been reported. eEnrichment fold, which was normalized by the input sample data. fRelative expression level, log2 (relative expression of the target gene in OXA lines against that in wild-type plants, normalized with the expression level of Tub2), referenced to the results of real-time RT-PCR.
Figure 4.
Validation of candidate downstream genes. A, Downstream target promoter fragments were connected into pLacZi and transformed into the pAD-GAL4-TsNAC1 YM4271 strain. β-Galactosidase (β-GAL) activities were validated by X-gal staining. β-Galactosidase activity was measured with o-nitrophenyl-β-d-galactopyranoside (ONPG) as substrate. B, Binding of TsNAC1 to the promoter of downstream genes as determined by ChIP-PCR. Six-week-old wild-type rosette leaves were used as materials and immunoprecipitated with TsNAC1 antibody and protein A agarose beads. Negative control reactions were performed in parallel using preimmune serum. β-Galactosidase units (Mu) were determined as follows: 1,000 × OD574/(t × V × OD600), where t is elapsed incubation time (min) and V is volume of culture (mL). C and D, Transient expression of the P35S:TsNAC1 or P35S:RD26 construct with promoter:Luc reporter constructs in T. halophila protoplasts (C) and tobacco leaves (D). Representative bioluminescence images are shown. Bars in B and C represent means ± sd, with three biological replicates in the experiment. Significant differences by Student’s t test are indicated with asterisks: **, P < 0.01.
TsNAC1-Regulated Genes with the CA(T/A)G Element in the Promoter
Motif discovery with the DNA fragments identified via ChIP-Seq around the binding peaks was performed using MEME-ChIP (Bailey et al., 2015), and 197 DNA sequences (length between 67 and 434, average length of 151.1) located at the promoter regions and a short and ungapped CA(T/A)G motif with E < 8.5e-018 were investigated (Fig. 3D). The distribution of the motif was visualized using a histogram that showed the percentage occurrence of the CA(T/A)G motif (x axis) plotted against the log2 fold enrichment of immunoprecipitation samples compared with the input sample (y axis), with the color of each square mapped to the number of indicated peak motifs (Fig. 3D). In general, peaks had one or more motifs at log2 > 0, and the proportion of peaks containing at least one motif increased along with the log2 fold enrichment of TsNAC1 binding.
It had been demonstrated that TsNAC1 could recognize the 130-bp key region of the promoter of TsVP1 in vivo and vitro. To identify the specific binding site, a serious 5′ deletion fragment was amplified by PCR from −667 to −388 (280 bp). M1 was the full length of 280 bp (Supplemental Data S2), M2 was 10 bp shorter than M1, and by such analogy, M14 lost the full length of the 130-bp key fragment. We incubated the TsNAC1-GST with both a moderately labeled 130-bp key fragment and excess unlabeled 5′ deletion fragments, and the results of the gel retardation assay validated that the combination between TsNAC1 and M13/M14 was weaker than M1 to M12, and the 20-bp fragment GAATATACCATGGATAAGCA was the core recognition sequence (Supplemental Fig. S5B). The promoter of TsVP1 was able to be recognized by TsNAC1, with two CA(T/A)G motifs (−545 and −400 to ATG) in this region (Supplemental Table S3). To identify the specific binding site via EMSA, TsNAC1 was bound to these sites, and when excess competitors (unlabeled sequences) were added, the binding signals decreased remarkably while the mutant competitors (unlabeled mutated sequences) could not (Supplemental Fig. S5A). The one-hybrid yeast system results using series substitutions as reporters indicated that CA(T/A)G is the core DNA-binding motif of TsNAC1 (Supplemental Fig. S5C). These results suggested that TsNAC1 could recognize CA(T/A)G and, thus, regulate the downstream network.
Multiple Cell Expansion Regulators Are Targeted by TsNAC1
LIGHT-DEPENDENT SHORT HYPOCOTYLS1 (LSH1) and UDP-XYLOSYLTRANSFERASE2 (TXT2) exerted positive regulation on cell expansion by acting as transcription factors (Iyer and Aravind, 2012) or maintaining the structure of cell walls (Wang et al., 2014). MULTICOPY SUPPRESSOR OF IRA1 4 (MSI4) negatively regulated the vegetative biomass by silencing the expression of FLOWERING LOCUS C (FLC; Morel et al., 2009). The ChIP-Seq and ChIP-PCR analyses (Fig. 4B) indicated that these loci were the TsNAC1 target genes. Quantitative RT-PCR results indicated that the expression of MSI4 was up-regulated but that of TXT2 and LSH1 was down-regulated (Table I) by TsNAC1 in the OX lines. Taken together, these data suggest that the role of LSH1, TXT2, and MSI4 in retarding cell expansion is directly regulated by TsNAC1. TsVP1, which was shown previously to play an important role in cell expansion, was a target gene of TsNAC1 and was up-regulated in the OX lines but down-regulated in the NR lines.
TsNAC1 Plays a Crucial Role in Plant Tolerance to Salt Stress
An analysis of the TsNAC1 ChIP-Seq data identified a number of positive regulators of salt stress tolerance, including TsVP1, HOMEOBOX12 (HB12), and MYB HYPOCOTYL ELONGATION-RELATED (MYBH), as candidate TsNAC1 target genes. TsNAC1 has been shown to bind to the 130-bp sequence of the TsVP1 promoter in vivo and in vitro. Two peaks were identified at 450 to 600 bp (29.76 relative fold enrichment) and 300 to 450 bp (14.29 relative fold enrichment) upstream of the ATG of TsVP1 (Fig. 3C). Binding of TsNAC1 to two other genes also was identified by the one-hybrid yeast assay (Fig. 4A) and by ChIP-PCR assay (Fig. 4B). The expression levels of TsVP1, HB12, and MYBH were all up-regulated by TsNAC1 (Table I). The results confirmed that TsNAC1 positively regulates the expression of TsVP1, HB12, and MYBH in plant salt stress tolerance.
TsNAC1 Shows Higher Transcriptional Activation Activity Than RD26
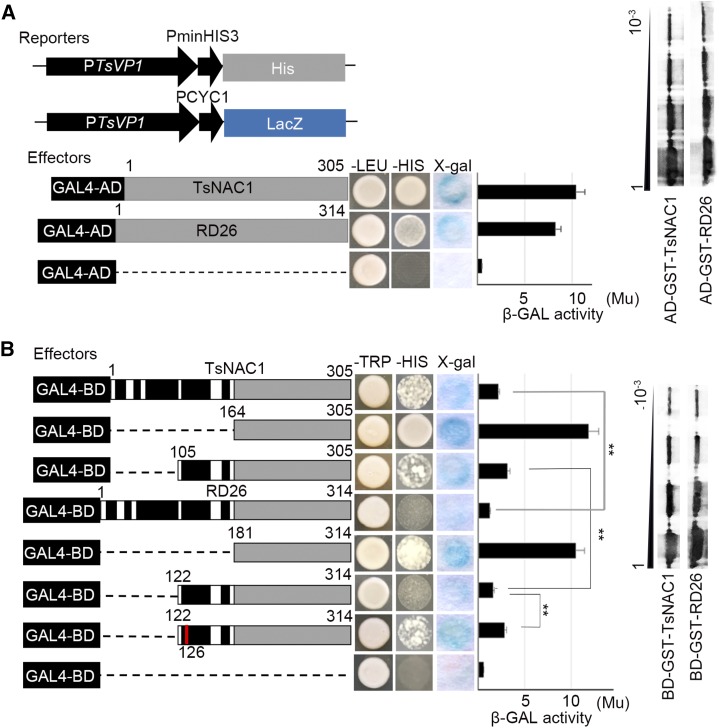
Two one-hybrid yeast systems were constructed to test for differences between the binding domain and AD of RD26 and TsNAC1. First, open reading frames were cloned by translational fusion with the yeast GAL4 AD, and the TsVP1 promoter was linked to pLacZi or pHis2.1 as reporter. The cotransformed YM4271 strains were cultured on SD/-His, and β-galactosidase activity was examined to verify their DNA-binding ability. Although TsNAC1 and RD26 were able to bind to the TsVP1 promoter, X-gal staining demonstrated that the binding ability of RD26 was slightly weaker than that of TsNAC1 (Fig. 5A).
Figure 5.
Assays of transcription factor activity in the yeast system. A, Binding activity of TsNAC1 and RD26 to the promoter of TsVP1. TsNAC1 and RD26 were cloned and fused with the yeast GAL4 AD. The promoter of TsVP1 was linked into pHis2.1 and pLacZi vectors as reporters. B, Truncation analysis of the transcriptional activation activity of TsNAC1 and RD26. Numbers on the top indicate the positions of amino acids. Subdomains A to E of the NAC DNA-binding domains are colored in black, the ADs are colored in gray, and the mutation of the amino acid is colored in red. The C-terminal AD (164–305 and 181–314), the AD with the repression domain (105–305 and 122–314), and the full-length transcription factor domain (1–305 and 1–314) were cloned and fused with the yeast GAL4 binding domain (BD). Transformants were screened on SD/-Leu or SD/-Trp and SD/-His, and the β-galactosidase (β-GAL) activities were validated by X-gal staining and quantified by ONPG assay. Western blot was executed with the GST antibody. Bars represent means ± sd, with three biological replicates in the experiment. Significant differences by Student’s t test are indicated with asterisks: **, P < 0.01.
RD26 was reported as having a repression domain (122–141) that suppresses the AD (Hao et al., 2010). The full-length RD26 (1–314), a C-terminal AD (181–314), and the AD with the repression domain (122–314) were each recombined into a pBD vector and then transformed in the YRG-2 strain. The strains containing only the AD (181–314) grew well on the selection SD/-His medium and showed high β-galactosidase activity, and the other two stains were stunted on SD/-His medium and showed much lower β-galactosidase activity. The same TsNAC1 deletion strains (1–305, 105–305, and 164–305) also were examined following training on SD/-His medium with an ONPG assay, and the region from amino acid 105 to 163 of TsNAC1 was found to suppress the transcriptional activation of AD (164–305; Fig. 5B).
The 164 to 305 region of TsNAC1 exerted limited higher transcriptional activity compared with the 181 to 314 region of RD26, although the β-galactosidase activity of the strains with the TsNAC1 105 to 305 region was twice as great as that of the strains with the RD26 122 to 314 region. These results suggest that the repression ability of the TsNAC1 105 to 163 region was lower than that of the RD26 122 to 180 region. Alignments with ClustalW indicated that the presence of Arg at RD26-126 was the only difference between the two protein sequences (Fig. 1A). A point mutation fragment was constructed in pBD, and the results of the transcriptional activity assay supported our hypothesis that the mutation from Arg to His weakened the repression ability (Fig. 5B). Overall, significant differences were not observed in the binding ability of RD26 and the TsNAC1 target to the promoter of TsVP1, although the transcript activation activity was much lower than that of TsNAC1 in yeast.
Referenced to the results of ChIP-Seq and the one-hybrid yeast system, seven genes (including TsVP1) were up-targeted by TsNAC1. And except for MSI4, the promoters of these genes contained the target motifs of RD26. To demonstrate whether TsNAC1 has higher transcript activation activity in vivo, the promoter regions of these seven genes were connected into pGreenII 0800-Luc (with PMSI4:Luc as the negative control) and cotransformed with 35S:TsNAC1 or 35S:RD26 into the protoplasts of T. halophila (Fig. 4C). Injection of the transformed Agrobacterium tumefaciens GV3101 pGreenII 0800-PTsVP1:Luc and 35S:TsNAC1 into the leaves of tobacco in pairs also showed higher luciferase signal than that of 35S:RD26 (Fig. 4D). All these results demonstrated that TsNAC1 had higher transcript activity than RD26 in vivo.
DISCUSSION
Abiotic stress, such as high-salt stress, has a considerable impact on the quality and yield of agricultural products. Thus, the mechanisms of the abiotic stress response must be investigated at the molecular level. T. halophila is a halophytic plant with excellent salt and drought resistance; therefore, it is an interesting system in which to study the mechanisms of the abiotic stress response. Here, we explored the abiotic stress resistance of TsNAC1 overexpression lines to identify target genes for further investigation.
TsNAC1 Participates in Plant Responses to Multiple Abiotic Stressors
Previous studies of Arabidopsis showed that RD26 is an important regulator of abiotic stress that is involved in the ABA-dependent dehydrate stress-signaling pathway (Fujita et al., 2004; Tran et al., 2004). TsNAC1 is highly expressed in mature tissues (Supplemental Fig. S1A), which is similar to the expression pattern of RD26 (Schmid et al., 2005). However, the expression of TsNAC1 was induced by salt stress, drought, and ABA treatments as well as by low temperatures, ROS stress, and MeJA treatments (Supplemental Fig. S1B), which is inconsistent with the findings for RD26. The promoter sequences of both TsNAC1 and RD26 have light-responsive, MeJA-responsive, and ABA-responsive cis-acting regulatory elements. However, the TsNAC1 promoter also had auxin-responsive, anaerobically induced, and cold- and dehydration-responsive elements (Supplemental Table S1), and these findings are consistent with the expression pattern (Supplemental Fig. S1B). Moreover, the overexpression lines of T. halophila showed slower growth and better survivability after exposure to low temperature, drought stress, high-salt stress, and ROS stress, whereas RNAi lines showed more rapid growth and were sensitive to abiotic stresses. Several regulator genes involved in the response to abiotic stress were positively regulated by TsNAC1. For example, the expression of DIACYLGLYCEROL KINASE2, ASPARTATE AMINOTRANSFERASE5 (AAT3), and CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR1 is induced by low temperature to improve plant freezing tolerance (Lee et al., 2005; Goulas et al., 2006; Doherty et al., 2009), and their expression was increased in the OX lines (Supplemental Table S2). ACONITASE3, POLLEN-PISTIL INCOMPATIBILITY2, and NAD-DEPENDENT MALIC ENZYME2 (NAD-ME2) are induced by oxidative stress and protect plants by increasing the stability in the molar ratio of metal ions (Moeder et al., 2007; Tan et al., 2010), and the expression of these genes was up-regulated by TsNAC1, as shown by the real-time RT-PCR results for the OX lines against the wild-type plants (Supplemental Table S2). These data suggest that TsNAC1 also plays a role in the control of ROS levels, which indicates that the expression level of TsNAC1 was positively correlated with the resistance to abiotic stresses of the plant and that TsNAC1 is a crucial regulator for multiple abiotic stress responses in T. halophila.
TsNAC1 Involvement in the Regulation of Cell Ion Transport
TsNAC1 has been shown to play an important role in salt stress. Moreover, TsNAC1 targets the promoter regions of TsVP1, HB12, and MYBH, and the expression levels of these genes were reduced in the NR lines. The homologous analysis identified two H+-PPases in T. halophila, including TsVP1, which is a crucial proton transporter protein. Utilizing the energy from the hydrolysis of inorganic pyrophosphate, TsVP1 cooperates with H+-ATPase (Silva and Gerós, 2009) to transfer H+ into a vacuole or the extracellular environment to improve salinity and drought stress tolerance (Gao et al., 2006; Bao et al., 2009; Kumar et al., 2014). Prior studies have shown that AVP1 could not respond to salt stress and was not positively regulated by RD26 (Sun et al., 2010). These differences between TsNAC1 and RD26 contribute to differences in salt tolerance between Arabidopsis and T. halophila.
HB12 is in the ABA-induced homeodomain-Leu zipper subfamily I, and it specifically functions in Na+ ion homeostasis by regulating the expression of PMR2A (an Na+/Li+ translocating P-type ATPase) in yeast and responds to salt and drought stresses (Shin et al., 2004; Valdés et al., 2012). AtHB-12, the ortholog of HB12, is the target gene of ANAC019 (Hickman et al., 2013), and its T-DNA insertion mutant exhibits longer inflorescence stems via the negative regulation of the expression of GA 20-oxidase (Son et al., 2010). In addition to HB12, the expression of MYBH, another transcription factor involved in salt and drought stresses, also is controlled by TsNAC1. MYBH in Arabidopsis responds to salt and drought stresses (Rasheed et al., 2016) and also plays a crucial role in auxin accumulation by up-regulating the expression of PHYTOCHROME-INTERACTING FACTOR4 (Kwon et al., 2013). MYB families, such MYB2, MYB21, MYB108, MYB112, MYB116, etc., are the target genes of RD26 (Hickman et al., 2013), and they are involved in salt and dehydration tolerance (Yang et al., 2012; Wang et al., 2015). The RD26 target motif also occurs in the promoters of the MYBH and HB12 genes in Arabidopsis, which suggests that these promoters are involved in the salt stress response. Previous studies of rice showed that the K+ TRANSPORTER (OsHKT1) reduces Na+ accumulation as a response to salt stress and showed that it is the downstream gene of OsMYBc (Zhang et al., 2015). Mutant hkt1-1 in Arabidopsis is sensitive to NaCl and exhibits greater root growth than the wild type. The OsHKT1 ortholog in T. halophila is induced dramatically by salt stress (Ali et al., 2013), which suggests that MYBH might be involved in the transport of K+ by regulating the expression of HKT1. These results show that TsNAC1 improves the salt and drought tolerance of plants by directly or indirectly regulating the transport of ions to maintain the proton gradient across the cell membrane and also provides energy for secondary active transport, such as the transport of auxin, which is consistent with observations in Arabidopsis (Li et al., 2005; Yang et al., 2014).
TsNAC1 Retards Growth by Restricting Cell Expansion and Organ Size
The homozygotes of the T-DNA insertion mutants of RD26 (N1, SALK_072276; N2, SALK_083756) were highly sensitive to high-salt conditions (C.L., Q. Sun, B. Li, Z.L., Z.P., and J.Z., unpublished data), although significant differences in organ size were not observed compared with Col-0 (Fujita et al., 2004). A recent study indicated that transgenic 35S:RD26 plants displayed a stunted growth phenotype (Ye et al., 2017). When TsNAC1 derived by the cauliflower mosaic virus 35S promoter was introduced into N1 and N2, the plants showed a smaller organ size than Col-0 but bigger organs than the TsNAC1 transgenic plants derived from Col-0, and the lines also had significantly improved salt tolerance (unpublished data). We calculated the cell size of vascular tissue in the root maturation zone 5 d after sowing, and the cell size of the NR lines was increased by 20% and that of the OX lines was reduced by 27.5% compared with the wild type (Supplemental Fig. S6).
In Arabidopsis, AVP1 participates in auxin transport and accumulates in the root (Li et al., 2005). And TsVP1, which is the ortholog of AVP1 in T. halophila, is up-regulated in TsNAC1 overexpression lines and also regulates plant growth. Moreover, the overexpression of AVP1 increases the shoot biomass of plants (Li et al., 2005; Schilling et al., 2014), and the overexpression of TsVP1 in tobacco, cotton, and maize also enhances plant biomass (Gao et al., 2006; Li et al., 2008; Lv et al., 2009). Thus, crucial regulators must retard growth and reduce organ size in TsNAC1 overexpression lines. Plants from TsNAC1 overexpression lines showed slower growth and smaller organs than plants that overexpressed TsVP1. This result suggests that the effect of TsVP1 on growth in TsNAC1 overexpression lines was overwhelmed by the other target genes regulated by TsNAC1.
The organ size of plants is related to cellular processes, such as cell proliferation and expansion (Horváth et al., 2006; Du et al., 2014; Cheng et al., 2016). The expression of MSI4 was up-regulated in TsNAC1 OX lines, and Arabidopsis MSI4 mutants showed a severalfold increase in vegetative biomass and a 1.3-fold expansion in cell size (Morel et al., 2009). These results suggest that MSI4 is a negative regulator of cell expansion. The ortholog of MSI4 in Arabidopsis has been shown to associate with HISTONE DEACETYLASE6 and LYSINE SPECIFIC DEMETHYLASE1 to repress FLC (Ausín et al., 2004; Yu et al., 2016), and flc-3 mutants flower later than wild-type plants (Shindo et al., 2006) and present phenotypes similar to that of OX lines (Supplemental Fig. S7). In addition to the up-regulation of MSI4, two positive regulators of cell expansion, LSH1 and TXT2, are negatively regulated by TsNAC1. ALOG family proteins act as the key developmental regulators of shoot organs (Takeda et al., 2011) and seeds (Zhao et al., 2004). LSH1 functions with several other transcription factors to promote unidimensional cell enlargement (Zhao et al., 2004; Iyer and Aravind, 2012). The development of primary cell walls plays a crucial role in plant growth (Cosgrove, 2005) and morphogenesis (Schopfer, 2006). Xyloglucan in the primary cell wall of dicotyledonous plants is a restraining factor on wall extensibility (Keegstra et al., 1973; Hayashi, 2003; Park and Cosgrove, 2015). The biosynthesis of xyloglucan is catalyzed by the ortholog of TXT2, a GT34 family member (Vuttipongchaikij et al., 2012; Zabotina et al., 2012), in Arabidopsis (Cavalier et al., 2008). Ye et al. (2017) discovered that TCH4 and EXPL2 were down-regulated in 35S:RD26 plants to retard plant growth. RT-PCR results indicated that TXT2, which is located in the same subfamily with TCH4, was suppressed 1.92-fold [log (OX/wild type)] (Table I), but EXPA6, which is located in the same subfamily with EXPL2 and with a 6.19 relative fold enrichment in its promoter region, was suppressed less than 0.23-fold (data not shown) in OX lines. Overall, variations in the expression levels of MSI4, LSH1, and TXT2 induced by TsNAC1 lead to the retardation of plant growth by retarding cell expansion.
TsNAC1 Regulates Embryonic Development
Although the OX transgenic lines had smaller organ size, the fruit set was not lower than that of the wild-type and NR lines. Several positive regulators involved in embryo development are regulated by TsNAC1. Campesterol is the precursor in the synthesis of brassinosteroids (Hartmann, 1998) and affects postembryonic growth in higher plants (Clouse and Sasse, 1998). CYP51 is involved in the biosynthesis of campesterol, and cyp51 mutants are always embryo lethal (O’Brien et al., 2005) or seedling lethal (Kim et al., 2005) via the generation of ROS and ethylene (Kim et al., 2010). In the OX lines, CYP51 exhibits up-regulated expression, which could enhance the biosynthesis of campesterol, which is beneficial for embryonic development. One member of the Cullin family, CUL3, associates with RING-FINGER PROTEIN and BROAD COMPLEX domain proteins and works as an E3 ligase (Figueroa et al., 2005), which affects embryo pattern formation at the heart stage (Thomann et al., 2005). In T. halophila, the expression of CUL3 is up-regulated by TsNAC1 overexpression, which could regulate embryonic development. The ortholog of PRP31 in Arabidopsis encodes a splicing factor and interacts with STABILIZED1 and ZINC-FINGER AND OCRE DOMAIN-CONTAINING PROTEIN1 to regulate embryonic development by regulating the expression of RNA HELICASE22 (Kanai et al., 2013) and TRIGALACTOSYLDIACYLGLYCEROL1 (Xu et al., 2005) under abiotic stress treatments, such as cold stress (Du et al., 2015). In the T. halophila OX lines, the expression of PRP31 was up-regulated by TsNAC1 overexpression, which suggests that PRP31 or TsNAC1 plays a role in abiotic stress resistance by improving embryonic development.
Stress Tolerance Coupled to Growth Retardation
Prior studies showed that plants or tissues with lesser cell size contribute to plant abiotic stress tolerance, such as moisture resistance (Cutler et al., 1977; Bradford and Hsiao, 1982). Under abiotic stress, one way to improve the tolerance of plants is to construct a new ion homeostasis via active transport, such as improving the salt stress tolerance of plants by overexpressing H+-PPase AVP1 (Li et al., 2005). In this study, TsNAC1 improved the salt stress tolerance of plants by enhancing the expression of TsVP1, HB12, MYBH, etc., to regulate the transformation of ion, and it retarded plant growth via restricting cell expansion by affecting the expression level of some plant growth genes, such as MSI4, LSH1, and TXT2 (Fig. 6). With the advantage of lesser volume, smaller cells wasted less energy on the active transportation of ion to maintain osmotic pressure, and more energy could be used to power metabolic reactions to improve the abiotic stress resistance of plants. Recently, Ye et al. (2017) reported that RD26 mediates cross talk between drought and brassinosteroid (BR) signaling. When overexpressed, the BES1 target gene RD26 can inhibit BR-regulated growth. Global gene expression studies suggest that RD26 can act antagonistically to BR to regulate the expression of a subset of BES1-regulated genes, thereby inhibiting BR function. In this study, we found that the overexpressed TsNAC1 increased the expression of CYP51 that took part in the biosynthesis of campesterol, the precursor of BR, but the nexus of TsNAC1 function and BR signaling should mandate further investigations. It is concluded that TsNAC1 acts as an upstream regulator of plant abiotic stress resistance and vegetative growth functions directly on the promoters of MYBH, HB12, TsVP1, NAD-ME2, AAT3, PRP31, CYP51, MSI4, LSH1, TXT2, etc., upon increased or reduced expression and coordinates both abiotic stress resistance and growth retardation by reducing cell expansion and growth rate.
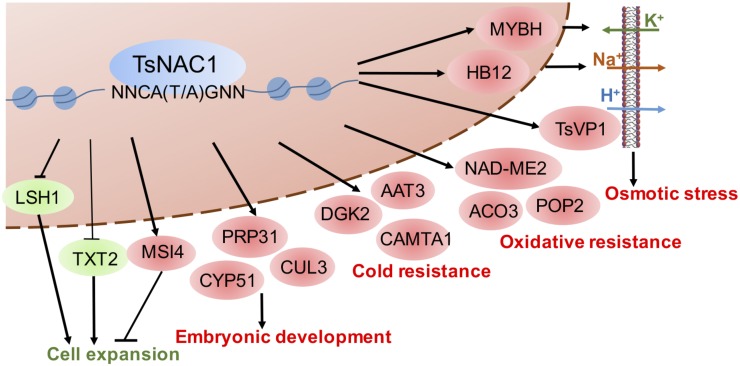
Figure 6.
Proposed model for the regulation of plant development and stress responses by TsNAC1. TsNAC1 functions as an upstream regulator of plant osmotic stress (regulates ion transportation), oxidative stress, and cold stress responses and coordinates both vegetative growth (retards cell expansion) and reproductive growth.
Comparison between RD26 and TsNAC1
The binding activity to the promoter, the candidate motif, and the downstream genes of TsNAC1 and RD26 were compared in this study. As shown in Figure 5, a higher transcriptional activation activity of TsNAC1 was found compared with RD26. In this study, the CA(T/A)G sequence was verified as the core DNA-binding motif for TsNAC1 (Supplemental Fig. S5), while RD26 recognizes and binds to regions containing a CACG consensus sequence (Tran et al., 2004). TsNAC1 preferred CATG to CAAG. The TsVP1 promoter sequence (−667 to −138 bp) used for the binding activity assay contained both of these motifs, which explains why the binding activity assay showed a similar level. A comparison of promoter sequences of major TsNAC1-targeted genes in T. halophila and their ortholog genes in Arabidopsis (Supplemental Table S3) showed that the distribution of motifs was different between species. Compared with the RNA sequencing results with the RD26 OX lines as materials from Ye et al. (2017), in the 892 downstream target candidate genes of TsNAC1, 135 genes were up-regulated in RD26 OX lines, and the main biological process was enriched in response to salt stress (including the orthologs of HB12 and MYBH) and the regulation of transcription (Table I; Supplemental Fig. S8). A total of 258 genes were down-regulated and enriched in the plant developmental process (including the orthologs of LSH1, TXT2, TCH4, and EXPA6, which play roles in restricting cell expansion) and nitrogen metabolism (Table I; Supplemental Fig. S8). It was worth noting that the expression of AVP1 is not regulated in RD26 OX lines. All these findings were consistent with the RT-PCR results of our study (Table I). The other 499 genes were enriched in postembryonic development, ion transport, response to cold, and oxidative stress. These findings may provide evidence to clarify the similarities and differences between the OX lines of TsNAC1 and RD26 in plant development and stress resistance.
The promoter and the coding sequence regions of AVP1 and TsVP1 have approximately 75% similarity. Five RD26-targeted motifs are observed in the AVP1 promoter, whereas three TsNAC1-targeted motifs are observed in the TsVP1 promoter (Supplemental Table S3). Both motifs and promoters are differential, which might have caused the discrepancy in response to salt stress. In the T. halophila OX lines, the TsNAC1 target genes presented altered expression levels, with MSI4, TsVP1, etc., showing up-regulated expression and LSH1 and TXT2 showing down-regulated expression. These data suggest that there may be different regulation patterns of TsNAC1 based on the distribution of the target motifs. Additional work is required to determine whether polymorphisms of the regulatory mechanisms occur. TsNAC1 appears to play similar but not identical roles in T. halophila and Arabidopsis for the regulation of plant growth and responses to abiotic stressors.
CONCLUSION
Plants from TsNAC1 overexpression lines showed higher survival rates under abiotic stress and grew slowly under normal conditions. The ChIP-Seq data offer evidence that the overexpression of TsNAC1 up-regulates the genes involved in the response to abiotic stresses and ion transport, thereby improving stress resistance, and negatively regulates the genes involved in plant vegetative growth, thereby reducing cell expansion (Fig. 6). TsNAC1 regulates target genes with the CA(T/A)G consensus sequence, which differs from the results for RD26. In the model extremophile plant T. halophila, TsNAC1 is a crucial upstream regulator for abiotic stress resistance and cell expansion.
MATERIALS AND METHODS
One-Hybrid Yeast Screening of Thellungiella halophila cDNA Libraries
Six-week-old T. halophila wild plants, exposed to a 12-h high-salt stress treatment, were used to construct the cDNA library. The extraction of mRNA was performed with the Oligotex mRNA Mini Kit (Qiagen). Long-distance PCR (LD-PCR) was executed by the PCR cDNA Synthesis Kit (SMART). All the fragments were linked to the vector pGADT7-AD and transformed into yeast strain Y187 (Invitrogen) following the Yeast Protocols Handbook (Clontech). The transformed stains were cultured on SD/-Leu medium. Yeast plasmid was extracted and amplified with the T7 Sequencing Primer and the 3′ AD Sequencing Primer on plasmid; most fragments were from 700 to 2,000 bp. Considering the stability of the specific binding between the protein and the DNA sequence, 3× tandem-repeated 530-bp DNA fragments of the TsVP1 promoter (−667 to −138 bp) were cloned into pHis2.1 as bait. The bait vector was transformed into the cDNA library and cultured on SD/-His/-Trp with 5 mm 3-aminotriazole. The plasmid of the surviving stains was extracted and amplified with the T7 Sequencing Primer and the 3′ AD Sequencing Primer. The amplicons were transformed into Escherichia coil and sequenced at BioSune.
Binding Active Assay of TsNAC1 and RD26 in the Yeast System
The coding sequences of TsNAC1 and RD26 were obtained by PCR, and the products were digested by EcoRI/SmaI and BamHI/SacI, then connected into the pGADT7-AD vector, which containing the GAL4 AD. The promoters of TsVP1 and candidate target genes were cloned into the EcoRI/KpnI sites of pLacZi or the EcoRI/SpeI sites of pHis2.1. Plasmids were transformed in the yeast strain YM4271 (Invitrogen) in pairs. The β-galactosidase activities were examined by X-gal staining and measured as described in the Yeast Protocols Handbook (Clontech) using ONPG as the substrate.
Vector Construction and Transformation
The full-length cDNA of TsNAC1 was ligated into the vector pCAMBI1300 that contains hygromycin as the selective marker. The specific fragment (418–882 bp) of TsNAC1 was reorganized into the vector pFGC-5941 to interfere with the expression of TsNAC1, and bar was used as the selective marker. Vectors were introduced into T. halophila upon Agrobacterium tumefaciens GV3101-mediated transformation by the floral dip method. T. halophila seeds were planted in peat:vermiculite (2:1) and grown at 24°C under long-day conditions (16 h of light and 8 h of dark) with a relative humidity of 50% for 6 weeks. The plants were transformed to 2°C to 4°C for 5 weeks of vernalization and then put back to 24°C to enter the mature period.
Polyclonal Antibody of TsNAC1
First, the full-length TsNAC1 was cloned and connected into pGEX-4T-1, and then the integrated plasmid was transformed into BL21 stain. The prokaryotic expression strain was cultivated, and the expression of protein NAC-GST was induced as described in “Molecular Cloning: A Laboratory Manual III” (Sambrock and Russel, 2001). Purification of the soluble NAC-GST protein was performed by the Chelating Sepharose Fast Flow (GE Healthcare). Preparation of the polyclonal antibody was accomplished by Abmart. The titer of the antibody was over 5 × 104 IU.
EMSA
Probes were generated using the DIG Gel Shift Kit (Roche). The mutated competitor of the 130-bp key region in the promoter of TsVP1 was the sequence between PT7 and PT8 (−667 to −538 bp), which region was reported with no salt response (Sun et al., 2010) and sited beside the 130-bp key region.
Plant Performance under Different Treatments in the Greenhouse
T. halophila seeds were planted in peat:vermiculite (2:1) and grown at 24°C under long-day conditions (16 h of light and 8 h of dark) with a relative humidity of 50%. Different treatments were performed after 6 weeks. Uniform seedlings of transgenic plants and the wild type were sprayed with 0.1 mL of 3 mmol L−1 paraquat for every pot to mimic the active oxygen stress treatment. In the low-temperature stress treatment experiments, transgenic plants were transferred into the other culturing room at −4°C and with greater than 3,500 lx light intensity for 48 h. These plants were watered with 50 mL of 18% PEG6000 solution for every pot to mimic the drought stress and with an additional 10 mL every 2 d. These transgenic lines were watered with an 800 mm NaCl solution and then watered with 10 mL of a 400 mm NaCl solution every 3 d to keep the 600 mm NaCl concentration of culture medium for 2 weeks. The hormone treatment was performed by spraying and watering the 100 μm ABA or 100 μm MeJA, 1 mL for each pot.
Plant RNA Extraction and Real-Time RT-PCR
Total RNA was extracted by Trizol (Takara) reagent. An amount of 500 ng of total RNA was used for inverse transcription with the Transcriptor First Strand cDNA Synthesis Kit (Takara). The cDNA was diluted 10 times and then used as the template for real-time RT-PCR. The RT-PCR system was as follows: SYBR Premix Ex Taq (2×), 5 μL; PCR Forward Primer (10 μm), 0.2 μL; PCR Reverse Primer (10 μm), 0.2 μL; cDNA template, 1 μL, with distilled deionized water added up to 10 μL. The PCR conditions were as follows: incubate for 3 min at 95°C, followed by a total of 40 cycles of 15 s at 95°C, 30 s at 58°C, and 30 s at 72°C.
Luciferase Assay
Promoter fragments in the pGreenII 0800:Luc vector were cotransformed with the pSoup helper plasmid into A. tumefaciens GV3101 and injected together with 35S:TsNAC1 or 35S:RD26 into tobacco (Nicotiana tabacum) leaves. The isolation of T. halophila protoplast and transformation were performed according to the protocol described previously (Yoo et al., 2007; Wu et al., 2009). Cells were harvested 18 h posttransfection, and luciferase activity (Luc/Rluc) was measured after cell lysis using the Double-Luciferase Reporter Assay Kit (TransDetect).
ChIP and ChIP-qPCR
The details of ChIP were as described by Kaufmann et al. (2010). The sequencing platform was Illumina HiSeq 4000. Aligner and parameter were Clean Parameter soap_mm_gz -p 4 -v 5 -s 35 -m 0 -x 600. Peak caller and parameter were Peak Calling Parameter macs14 -g 411831487 -p 1e-5 -w–space 50 -m 10,30. Independent lines OX7 and OX10 were used for the ChIP-qPCR assay, and all samples were diluted to 10 ng µL−1 and reacted with the following system: SYBR Premix Ex Taq (2×), 5 μL; PCR Forward Primer (10 μm), 0.2 μL; PCR Reverse Primer (10 μm), 0.2 μL; cDNA template, 1 μL, with distilled deionized water added up to 10 μL.
The primers used to amplify the enriched region of the target genes are listed in Supplemental Table S4. All the target genes of TsNAC1 are listed in Supplemental Table S5. The row data of ChIP-Seq were uploaded to the data-sharing platform figshare: https://figshare.com/projects/Exploration_of_the_target_genes_of_TsNAC1/25135
Transcript Activation Activity Assay
The C-terminal AD (164–305), the AD with the repression domain (105–305), and the full-length transcription factor domain (1–305) of TsNAC1 were connected into the EcoRI and PstI sites of pGBKT7-BD. The C-terminal AD (181–314), the AD with the repression domain (122–314), the full-length transcription factor domain (1–314), and the point mutant (R126H) of RD26 also were inserted and connected into the BamHI and PstI sites of pGBKT7-BD vector. Plasmids were transformed in the yeast reporter strain YRG-2 (Agilent Stratagene). The β-galactosidase activities were examined by X-gal staining and measured by ONPG assay.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers TsNAC1(GenBank: AK352535.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Evolutionary relationships of TsNAC1.
Supplemental Figure S2. Expression pattern analysis of TsNAC1 in wild-type plants of T. halophila.
Supplemental Figure S3. Abiotic stress treatment comparison between the OX, wild-type, and NR lines.
Supplemental Figure S4. Validity of the TsNAC1 antibody.
Supplemental Figure S5. Determination of the core binding motif.
Supplemental Figure S6. Comparison of growth parameters in root 5 d after sowing.
Supplemental Figure S7. Differences of phenotype and flowering time among the different lines.
Supplemental Figure S8. Biological process GO analysis.
Supplemental Table S1. Different sites between the promoters of TsNAC1 and RD26.
Supplemental Table S2. Candidate target genes of TsNAC1 on other abiotic stresses.
Supplemental Table S3. Distribution of motifs in the promoters of target genes.
Supplemental Table S4. Primers used in this study.
Supplemental Table S5. Candidate target genes of TsNAC1 in T. halophila.
Supplemental Data S1. TsNAC1 peptide sequence.
Supplemental Data S2. Promoter sequence of TsVP1.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for Arabidopsis mutants.
Footnotes
This work was supported by the National Natural Science Foundation of China [grant no. 31571674 to J.Z.] and National Major Projects for Genetically Modified Organisms Breeding in China [grant no. 2016ZX08003004-003 to J.Z.].
References
- Ali A, Cheol Park H, Aman R, Ali Z, Yun DJ (2013) Role of HKT1 in Thellungiella salsuginea, a model extremophile plant. Plant Signal Behav 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Bohnert HJ, Bressan RA (2005) Abiotic stress and plant genome evolution: search for new models. Plant Physiol 138: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME Suite. Nucleic Acids Res 43: W39–W49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao AK, Wang SM, Wu GQ, Xi JJ, Zhang JL, Wang CM (2009) Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci 176: 232–240 [Google Scholar]
- Benina M, Obata T, Mehterov N, Ivanov I, Petrov V, Toneva V, Fernie AR, Gechev TS (2013) Comparative metabolic profiling of Haberlea rhodopensis, Thellungiella halophyla, and Arabidopsis thaliana exposed to low temperature. Front Plant Sci 4: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford KJ, Hsiao TC (1982) Physiological Responses to Moderate Water Stress. Springer, Berlin [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Chen X, Zhu J, Huang H (2016) Overexpression of a Hevea brasiliensis ErbB-3 Binding protein 1 gene increases drought tolerance and organ size in Arabidopsis. Front Plant Sci 7: 1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Melencion SM, Alinapon CV, Kim MJ, Lee ES, Paeng SK, Park JH, Nawkar GM, Jung YJ, Chae HB, et al. (2017) The membrane-tethered NAC transcription factor, AtNTL7, contributes to ER-stress resistance in Arabidopsis. Biochem Biophys Res Commun 488: 641–647 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cutler JM, Rains DW, Loomis RS (1977) The importance of cell size in the water relations of plants. Physiol Plant 40: 255–260 [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du JL, Zhang SW, Huang HW, Cai T, Li L, Chen S, He XJ (2015) The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol Plant 8: 1053–1068 [DOI] [PubMed] [Google Scholar]
- Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, et al. (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Gao F, Gao Q, Duan X, Yue G, Yang A, Zhang J (2006) Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot 57: 3259–3270 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardeström P, Schröder W, Hurry V (2006) The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J 47: 720–734 [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Hartmann MA. (1998) Plant sterols and the membrane environment. Trends Plant Sci 3: 170–175 [Google Scholar]
- Hayashi T. (2003) Xyloglucans in the primary cell wall. Annu Rev Plant Biol 40: 139–168 [Google Scholar]
- Hickman R, Hill C, Penfold CA, Breeze E, Bowden L, Moore JD, Zhang P, Jackson A, Cooke E, Bewicke-Copley F, et al. (2013) A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J 75: 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, et al. (2004) Salt cress: a halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L (2012) ALOG domains: provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol Direct 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Hayashi M, Kondo M, Nishimura M (2013) The plastidic DEAD-box RNA helicase 22, HS3, is essential for plastid functions both in seed development and in seedling growth. Plant Cell Physiol 54: 1431–1440 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Moiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Talmadge KW, Bauer WD, Albersheim P (1973) The structure of plant cell walls. III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol 51: 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Lee H, Oh CJ, Lee HY, Eum HL, Kim HS, Hong YP, Lee Y, Choe S, An CS, et al. (2010) Postembryonic seedling lethality in the sterol-deficient Arabidopsis cyp51A2 mutant is partially mediated by the composite action of ethylene and reactive oxygen species. Plant Physiol 152: 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann KA, Feyereisen R (2005) Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol 138: 2033–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar T, Uzma, Khan MR, Abbas Z, Ali GM (2014) Genetic improvement of sugarcane for drought and salinity stress tolerance using Arabidopsis vacuolar pyrophosphatase (AVP1) gene. Mol Biotechnol 56: 199–209 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Kim JH, Nguyen HN, Jikumaru Y, Kamiya Y, Hong SW, Lee H (2013) A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J Exp Bot 64: 3911–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee HJ, Huh SU, Paek KH, Ha JH, Park CM (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227: 76–83 [DOI] [PubMed] [Google Scholar]
- Li B, Wei A, Song C, Li N, Zhang J (2008) Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnol J 6: 146–159 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Liu G, Li X, Jin S, Liu X, Zhu L, Nie Y, Zhang X (2014) Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 9: e86895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Ying S, Zhang DF, Shi YS, Song YC, Wang TY, Li Y (2012) A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep 31: 1701–1711 [DOI] [PubMed] [Google Scholar]
- Lv SL, Lian LJ, Tao PL, Li ZX, Zhang KW, Zhang JR (2009) Overexpression of Thellungiella halophila H+-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta 229: 899–910 [DOI] [PubMed] [Google Scholar]
- Moeder W, Del Pozo O, Navarre DA, Martin GB, Klessig DF (2007) Aconitase plays a role in regulating resistance to oxidative stress and cell death in Arabidopsis and Nicotiana benthamiana. Plant Mol Biol 63: 273–287 [DOI] [PubMed] [Google Scholar]
- Morel P, Tréhin C, Breuil-Broyer S, Negrutiu I (2009) Altering FVE/MSI4 results in a substantial increase of biomass in Arabidopsis: the functional analysis of an ontogenesis accelerator. Mol Breed 23: 239–257 [Google Scholar]
- O’Brien M, Chantha SC, Rahier A, Matton DP (2005) Lipid signaling in plants: cloning and expression analysis of the obtusifoliol 14α-demethylase from Solanum chacoense Bitt., a pollination- and fertilization-induced gene with both obtusifoliol and lanosterol demethylase activity. Plant Physiol 139: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56: 180–194 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed S, Bashir K, Matsui A, Tanaka M, Seki M (2016) Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Front Plant Sci 7: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Sambrock J, Russel DW (2001) Molecular Cloning: A Laboratory Manual (3rd edition). Immunology 49: 895–909 [Google Scholar]
- Schilling RK, Marschner P, Shavrukov Y, Berger B, Tester M, Roy SJ, Plett DC (2014) Expression of the Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) improves the shoot biomass of transgenic barley and increases grain yield in a saline field. Plant Biotechnol J 12: 378–386 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schopfer P. (2006) Biomechanics of plant growth. Am J Bot 93: 1415–1425 [DOI] [PubMed] [Google Scholar]
- Shin D, Koo YD, Lee J, Lee HJ, Baek D, Lee S, Cheon CI, Kwak SS, Lee SY, Yun DJ (2004) Athb-12, a homeobox-leucine zipper domain protein from Arabidopsis thaliana, increases salt tolerance in yeast by regulating sodium exclusion. Biochem Biophys Res Commun 323: 534–540 [DOI] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C (2006) Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiriga K, Sharma R, Kumar K, Yadav SK, Hossain F, Thirunavukkarasu N (2014) Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2: 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P, Gerós H (2009) Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signal Behav 4: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O, Hur YS, Kim YK, Lee HJ, Kim S, Kim MR, Nam KH, Lee MS, Kim BY, Park J, et al. (2010) ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol 51: 1537–1547 [DOI] [PubMed] [Google Scholar]
- Sun Q, Gao F, Zhao L, Li K, Zhang J (2010) Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol 10: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Hanano K, Kariya A, Shimizu S, Zhao L, Matsui M, Tasaka M, Aida M (2011) CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J 66: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Tan YF, O’Toole N, Taylor NL, Millar AH (2010) Divalent metal ions in plant mitochondria and their role in interactions with proteins and oxidative stress-induced damage to respiratory function. Plant Physiol 152: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U, Genschik P (2005) Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J 43: 437–448 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K (2010) Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1: 32–39 [DOI] [PubMed] [Google Scholar]
- Valdés AE, Overnäs E, Johansson H, Rada-Iglesias A, Engström P (2012) The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol Biol 80: 405–418 [DOI] [PubMed] [Google Scholar]
- Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ (2012) Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol 195: 585–595 [DOI] [PubMed] [Google Scholar]
- Wang C, Li S, Ng S, Zhang B, Zhou Y, Whelan J, Wu P, Shou H (2014) Mutation in xyloglucan 6-xylosytransferase results in abnormal root hair development in Oryza sativa. J Exp Bot 65: 4149–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W (2015) The rice High-Affinity Potassium Transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol 168: 1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Froehlich JE, Awai K, Benning C (2005) Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Demura T (2010) Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol 27: 237–242 [Google Scholar]
- Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63: 2541–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhang X, Gaxiola RA, Xu G, Peer WA, Murphy AS (2014) Over-expression of the Arabidopsis proton-pyrophosphatase AVP1 enhances transplant survival, root mass, and fruit development under limiting phosphorus conditions. J Exp Bot 65: 3045–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Liu S, Tang B, Chen J, Xie Z, Nolan TM, Jiang H, Guo H, Lin HY, Li L, et al. (2017) RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat Commun 8: 14573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu CW, Chang KY, Wu K (2016) Genome-wide analysis of gene regulatory networks of the FVE-HDA6-FLD complex in Arabidopsis. Front Plant Sci 7: 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG (2012) Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol 159: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang R, Jing W, Xiao L, Jin Y, Shen L (2015) The OsHKT1;1 transporter is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol 168: 1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Nakazawa M, Takase T, Manabe K, Kobayashi M, Seki M, Shinozaki K, Matsui M (2004) Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J 37: 694–706 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]