Light and ethylene mediate hypocotyl cell elongation by coordinating the expression of microtubule-destabilizing protein MDP60 in Arabidopsis.
Abstract
Precise regulation of hypocotyl cell elongation is essential for plant growth and survival. Light suppresses hypocotyl elongation by degrading transcription factor phytochrome-interacting factor 3 (PIF3), whereas the phytohormone ethylene promotes hypocotyl elongation by activating PIF3. However, the underlying mechanisms regarding how these two pathways coordinate downstream effectors to mediate hypocotyl elongation are largely unclear. In this study, we identified the novel Microtubule-Destabilizing Protein 60 (MDP60), which plays a positive role in hypocotyl cell elongation in Arabidopsis (Arabidopsis thaliana); this effect is mediated through PIF3. Ethylene signaling up-regulates MDP60 expression via PIF3 binding to the MDP60 promoter. MDP60 loss-of-function mutants exhibit much shorter hypocotyls, whereas MDP60 overexpression significantly promotes hypocotyl cell elongation when grown in light compared to the control. MDP60 protein binds to microtubules in vitro and in vivo. The organization of cortical microtubules was significantly disrupted in mdp60 mutant cells and MDP60-overexpressing seedlings. These findings indicate that MDP60 is an important mediator of hypocotyl cell elongation. This study reveals a mechanism in which light and ethylene signaling coordinate MDP60 expression to modulate hypocotyl cell elongation by altering cortical microtubules in Arabidopsis.
Plants need to elaborately regulate growth to survive in response to multiple developmental and environmental cues, including modulating hypocotyl elongation. Hypocotyl elongation is significantly influenced by external and internal cues, such as light and phytohormones (Feng et al., 2008; Pierik et al., 2009). Light is a crucial negative regulator of hypocotyl elongation (Yang et al., 2009; Liu et al., 2013). Genetic evidence has shown that the mutants of many components in the light signaling pathway generally display defective hypocotyl elongation phenotypes, such as the loss-of-function mutant of the red light receptor PHYB, which exhibits greatly elongated hypocotyls (de Lucas et al., 2008; Kim et al., 2016). Light induces degradation of the basic helix-loop-helix proteins phytochrome interacting factors (PIFs), which act as transcription factors downstream of light signaling and positively regulate hypocotyl elongation (Huq and Quail, 2002; Kim et al., 2003). Monogenic PIF mutants, including pif3, pif4, and pif5, exhibit shorter light-grown hypocotyls than wild-type plants (Zhong et al., 2012), demonstrating that PIFs promote hypocotyl elongation in light by regulating downstream gene expression.
Phytohormone ethylene is a gaseous plant hormone that promotes light-grown hypocotyl elongation (Smalle et al., 1997; Zhong et al., 2012). Ethylene is detected by five membrane-bound receptors, and biological responses to ethylene are mediated by two plant-specific transcription factors: ETHYLENE-INSENSITIVE3/EIN3-LIKE1 (EIN3/EIL1; Guo and Ecker, 2003; Potuschak et al., 2003; Boutrot et al., 2010; Zhang et al., 2011). The ethylene-insensitive mutant ein2 displays shorter hypocotyls, whereas the constitutive ethylene response mutant ctr1 exhibits longer hypocotyls than wild-type seedlings grown in the light, demonstrating that ethylene signaling participates in promoting hypocotyl elongation in the light (Smalle et al., 1997; Alonso et al., 1999; Vandenbussche et al., 2007). Importantly, activation of PIF3 transcription, but not that of other PIFs, is essential for ethylene signaling-promoted light-grown hypocotyl elongation (Zhong et al., 2012). Thus, light and ethylene signaling coordinate hypocotyl elongation by elaborately regulating PIF3, although the downstream effectors of PIF3 that participate in hypocotyl regulatory pathways remain unclear.
Microtubules are involved in multiple key physiological processes in plants, including hypocotyl cell elongation. The organization of cortical microtubules is considered to be related to the growth statuses of hypocotyl cells, namely, that they are dominantly transversely oriented to the hypocotyl longitudinal growth axis in rapidly growing hypocotyl cells and longitudinally oriented when cell elongation stops (Li et al., 2011; Liu et al., 2013). Many environmental and developmental factors can alter cortical microtubule organization during hypocotyl cell elongation, including light, phytohormone ethylene, gibberellic acid, and brassinosteroid (BR; Shibaoka, 1993; Le et al., 2005; Wang et al., 2012; Sun et al., 2015; Lian et al., 2017). Alterations in the transcript and protein levels of many microtubule-associated proteins (MAPs) result in abnormal hypocotyl cell elongation by disrupting cortical microtubule organization in response to multiple cues. For example, overexpression of the Arabidopsis (Arabidopsis thaliana) microtubule-associated protein WDL3 dramatically inhibits hypocotyl cell elongation under light exposure by increasing the percentage of oblique and longitudinally oriented cortical microtubules (Liu et al., 2013). Thus, proper regulation of MAPs is important for plant cell growth in response to developmental and environmental signals, although the underlying mechanisms require further dissection. Recently, the microtubule-associated protein WDL5 was identified as a positive regulator in ethylene-inhibited etiolated hypocotyl elongation (Sun et al., 2015). Although WDL5 expression was also detected in light-grown seedlings, WDL5 did not affect light-grown hypocotyl elongation (Sun et al., 2015). This suggests that different regulatory pathways may require different MAPs to regulate cortical microtubules and thus mediate hypocotyl elongation. Identifying MAPs inolved in upstream signaling-mediated hypocotyl elongation will facilitate understanding underlying mechanisms of plant cell growth.
At3g01015 belongs to the targeting-proteins-for-Xklp2 (TPX2) family. In this study, we show that the At3g01015 gene product MICROTUBULE-DESTABILIZING PROTEIN60 (MDP60), which was named based on its molecular mass of ∼60 kD and its function in microtubule regulation, plays a positive role in hypocotyl cell elongation. Moreover, MDP60 acts as a downstream effector of light and ethylene signaling and is targeted by PIF3. Our study reveals a mechanism involved in light- and ethylene-coordinated MDP60 expression that ultimately mediates hypocotyl cell elongation by regulating cortical microtubule organization.
RESULTS
MDP60 Promotes Hypocotyl Cell Elongation in the Light
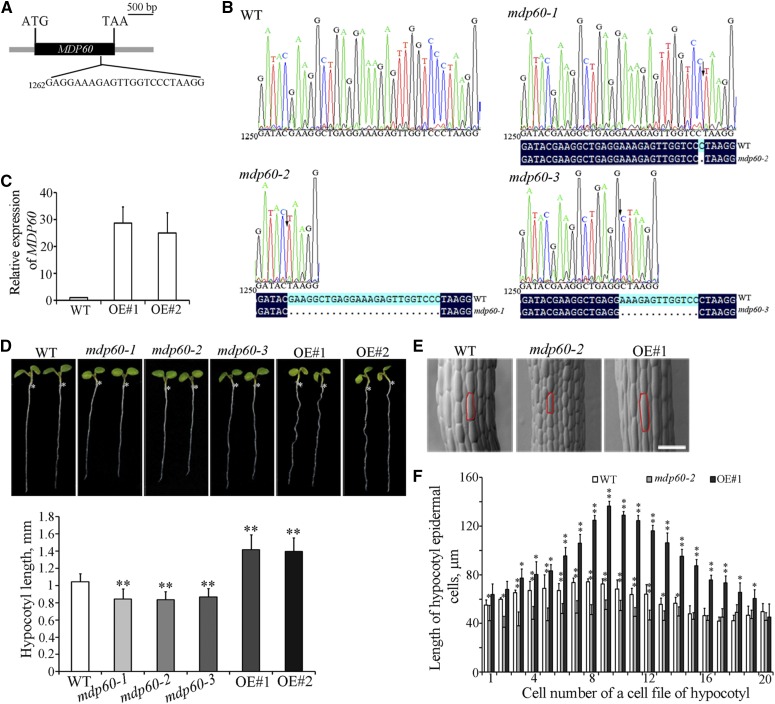
MDP60 belongs to the TPX2 protein family and MDP60 expression is obviously altered in the presence of 1-aminocyclopropane-1-carboxylic acid (ACC) and brassinolide phytohormones based on microarray data (Supplemental Fig. S1; Winter et al., 2007), suggesting a potential role of MDP60 in plant cell elongation. Because MDP60 knockdown or knockout T-DNA insertion lines are unavailable, MDP60-loss-of-function seedlings were generated using CRISPR/Cas9 technology to analyze the function of MDP60 in Arabidopsis (Wang et al., 2015). We obtained 32 T1 lines, three of which were homozygous. We sequenced the regions surrounding MDP60 gene target sites in three separate lines (mdp60-1, mdp60-2, and mdp60-3), showing loss of some nucleotide bases at different positions in the MDP60 sequence (Fig. 1, A and B). Additionally, of 22 MDP60-overexpressing wild-type (Arabidopsis Col-0 background) lines obtained, 12 exhibited the longer-hypocotyl phenotype in T3 generation, and line 2 (OE#1) and line 3 (OE#2) were selected for analysis. Quantitative real-time PCR showed that MDP60 transcription levels were considerably increased in two lines (Fig. 1C). Observation of 7-d-old light-grown mdp60-1, mdp60-2, and mdp60-3 seedlings revealed that hypocotyl length was demonstrably shorter than in wild-type seedlings. In contrast, hypocotyls were much longer in light-grown MDP60-overexpressing Arabidopsis than in wild-type plants (Fig. 1D). To confirm the hypocotyl phenotype in the mdp60 mutants, we also generated MDP60 RNA interference (RNAi) lines. We randomly selected three RNAi lines and found that MDP60 expression levels correlated with the short hypocotyl phenotype when seedlings were grown in the light (Supplemental Fig. S2, A and B), which is consistent with the phenotypes of mdp60 mutants.
Figure 1.
MDP60 is a positive regulator of light-grown hypocotyl cell elongation. A, Diagram of MDP60 showing the target sites for CRISPR/Cas9. The coding sequence and untranslated regions of MDP60 are indicated by a black box and gray boxes, respectively. B, The mdp60 mutants were generated by CRISPR/Cas9 technology. MDP60 gene mutation was evaluated by sequencing, and the mutant sites in MDP60 are indicated by arrows. C, Real-time PCR analysis of MDP60 transcripts in wild-type (WT) and MDP60-overexpressing (OE#1 and OE#2) seedlings. UBQ11 was used as a reference gene. D, The seedlings from mdp60 mutants show much shorter hypocotyls, whereas the hypocotyls are significantly longer in MDP60-overexpressing Arabidopsis when grown in the light for 7 d. The graph shows the average hypocotyl length measured from a minimum of 40 seedlings. E, Scanning electron microscopy images of hypocotyl epidermal cells from wild-type, mdp60-2 mutant, and MDP60-overexpressing seedlings grown in the light. Bar in E = 100 μm. F, Hypocotyl cell lengths of wild-type, mdp60-2 mutant, and MDP60-overexpressing seedlings were measured and calculated from at least 500 cells. t test, *P < 0.05, **P < 0.01. Error bars represent mean ± sd.
Because mdp60-1, mdp60-2, and mdp60-3 had comparable phenotypes, we were not surprised that the OE#1 and OE#2 lines also exhibited similar traits. Thus, we used mdp60-2 and OE#1 to perform subsequent analyses. No major differences in the cell shape of the hypocotyls were observed between wild-type, mdp60-2, and MDP60-overexpressing seedlings by scanning electronic microscopy (Fig. 1E). The numbers of cells in individual hypocotyl epidermal cell files in wild-type, mdp60-2, and MDP60-overexpressing seedlings were similar (∼20–22). However, the lengths of hypocotyl cells were decreased in mdp60-2 seedlings and significantly increased in MDP60-overexpressing Arabidopsis, particularly in the middle regions (Fig. 1F). Thus, MDP60 functions as a positive regulator of light-grown hypocotyl cell elongation.
Ethylene Signaling Mediates MDP60-Promoted Hypocotyl Cell Elongation
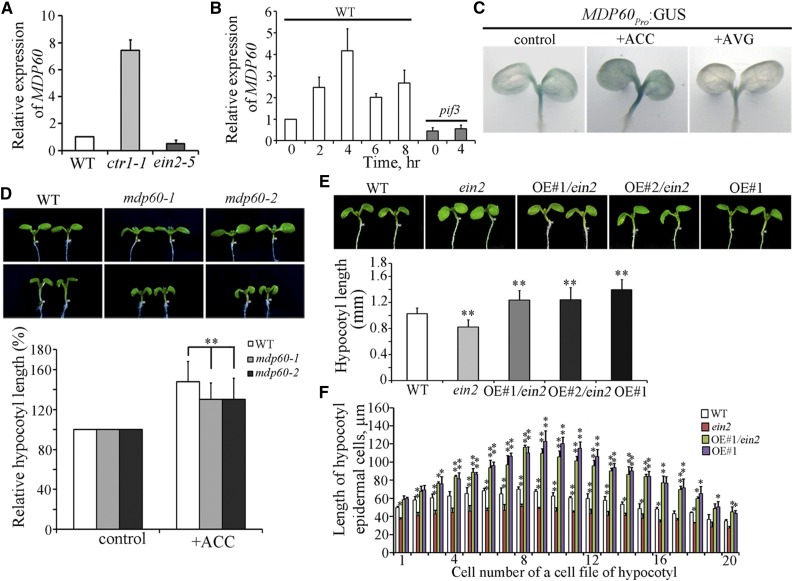
According to microarray data and the positive role of ethylene on light-grown hypocotyl elongation, we hypothesized that MDP60 participates in ethylene signaling-promoted hypocotyl elongation. We first determined whether and how ethylene regulates MDP60 expression. RNA was purified from 5-d-old light-grown seedlings from the constitutive ethylene response mutant ctr1-1 and ethylene-insensitive mutant ein2-5 and quantitative real-time PCR analyses were performed. MDP60 expression was much higher in the ctr1-1 mutant, but lower in the ein2-5 mutant compared to the wild type (Fig. 2A). Quantitative real-time PCR revealed that MDP60 expression is induced by ACC treatment, with peak levels detected 4 h after treatment (Fig. 2B, left panel), demonstrating that ethylene upregulates MDP60 expression. To further explore the exact role of ethylene signaling in hypocotyl MDP60 expression, transgenic lines expressing β-glucuronidase (GUS) driven by the native MDP60 promoter (MDP60pro:GUS) were generated and subsequently treated with ACC (a ethylene precursor) or AVG (aminoethoxyvinyl-Gly; an inhibitor of ethylene biosynthesis), respectively. Similar results were obtained for all 12 lines of MDP60pro:GUS transgenic seedlings investigated. GUS staining showed that MDP60 is mainly expressed in light-grown hypocotyls in MDP60pro:GUS transgenic seedlings (Fig. 2C, left panel), which is consistent with the physiological role of MDP60 in hypocotyls. In addition, GUS staining was significantly enhanced when the MDP60pro:GUS transgenic seedlings were transferred to the medium containing 10 μm ACC for 4 h (Fig. 2C, middle panel). However, GUS staining was obviously decreased when the MDP60pro:GUS transgenic seedlings were transferred to the medium containing 2 μm AVG for 4 h (Fig. 2C, right panel). These data indicate that ethylene signaling is critical for MDP60 transcription.
Figure 2.
Ethylene upregulates MDP60 expression to promote hypocotyl cell elongation. A, MDP60 expression was determined using quantitative real-time PCR with RNA purified from wild-type, ctr1-1, or ein2-5 5-d-old light-grown seedlings. Error bars represent ± sd (n = 3). B, Quantitative real-time PCR analysis of MDP60 RNA levels in 5-d-old wild-type and pif3 mutant seedlings treated with 10 μm ACC for the indicated times. UBQ11 was used as a reference gene. Error bars represent mean ± sd (n = 3). C, GUS staining of MDP60pro:GUS transgenic lines in the absence or presence of ACC and AVG. D, Wild-type and mdp60 mutant seedlings were grown on half-strength Murashige and Skoog medium supplemented with or without ACC in the light for 7 d. The graph shows the relative hypocotyl length measured from at least 66 seedlings per sample grown on the medium supplemented with 0 and 10 μm ACC under light growth conditions. Three independent experiments were performed with similar results, each with three biological repeats. t test, **P < 0.01, error bars represent the mean ± se, n = 3. E, MDP60 transgenic mutant ein2-5 (OE#1/ein2 and OE#2/ein2) seedlings had much longer hypocotyls than wild-type seedlings but were similar in hypocotyl length to MDP60 transgenic wild-type (OE#1) seedlings when grown in the light for 7 d. The graph shows the average hypocotyl length measured from at least 41 seedlings per sample (**P < 0.01, t test). Error bars indicate the mean ± sd. F, Lengths of hypocotyl cells from wild-type, ein2-5, and MDP60 transgenic ein2-5 mutants (OE#1/ein2) and wild-type seedlings (OE#1) grown in the light for 7 d. t test, *P < 0.05, **P < 0.01. Error bars represent mean ± sd.
Wild-type, mdp60-1, and mdp60-2 seedlings were then cultured on medium containing ACC. Hypocotyl length in 7-d-old light-grown seedlings was substantially increased in the presence of ACC in wild-type controls, but not mdp60-1 or mdp60-2 mutants (Fig. 2D), indicating that MDP60 loss-of-function mutants are much less sensitive to ACC during light-grown hypocotyl elongation than the wild type. Furthermore, MDP60 was overexpressed in the ein2-5 mutant background to investigate whether increasing MDP60 expression could recover the shorter hypocotyls of the ein2-5 mutant. Quantitative real-time PCR showed that MDP60 transcription levels were considerably enhanced in lines (ein2-5 mutant background) overexpressing MDP60 (OE#1/ein2 and OE#2/ein2; Supplemental Figure S3A). Observation of 7-d-old light-grown seedlings showed that the hypocotyl lengths were dramatically increased in the MDP60-overexpressing ein2-5 mutants, which is similar to the hypocotyl lengths of MDP60-overexpressing wild-type seedlings (Fig. 2E). Scanning electron microscopy revealed that hypocotyl cell lengths were increased in ein2-5 mutants when MDP60 overexpression was enhanced (Fig. 2F). Thus, these observations demonstrate that MDP60 plays a positive role in ethylene-promoted hypocotyl cell elongation.
Light Signaling Negatively Regulates MDP60 Expression through PIF3
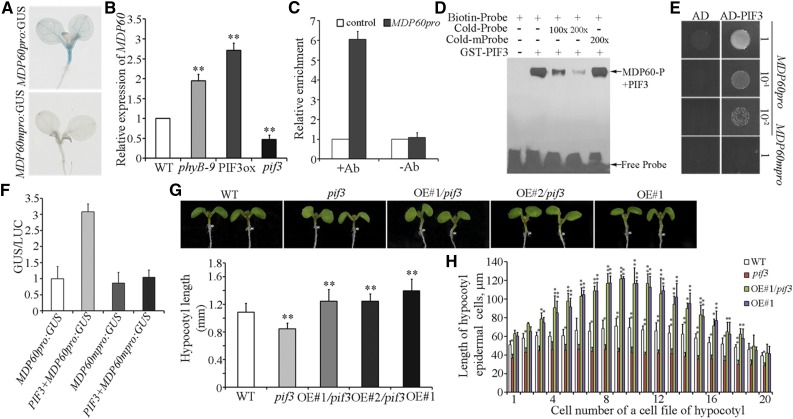
To understand the underlying mechanisms of MDP60 expression regulation during light-grown hypocotyl elongation, we sequenced the MDP60 promoter, identifying a typical G-box motif (located −910 to −916 upstream of the transcription start site), which is present within the promoters of many light-regulated genes. We next assessed the importance of this motif for MDP60 expression by mutating G-box (5′-CACGTG-3′ to 5′-ACTACA-3′) to generate MDP60pro:GUS transgenic lines (MDP60mpro:GUS). GUS staining showed no obvious signal in MDP60mpro:GUS transgenic seedlings compared to MDP60pro:GUS transgenic seedlings (Fig. 3A), demonstrating that the G-box in the MDP60 promoter sequence is essential for MDP60 expression.
Figure 3.
Light downregulates MDP60 through PIF3 during hypocotyl cell elongation. A, GUS staining of MDP60pro:GUS and MDP60mpro:GUS transgenic seedlings. B, MDP60 expression as determined using quantitative real-time PCR with RNA purified from wild-type, phyB-9, PIF3-overexpressing (PIF3ox), or pif3 mutant light-grown seedlings. Error bars represent ± sd (n = 3). C, ChIP-qRT-PCR assay of PIF3 binding to MDP60 promoters in vivo. Chromatin from light-grown PIF3pro:PIF3-YFP transgenic seedlings was immunoprecipitated with an anti-GFP antibody, and the amount of indicated DNA in the immune complex was determined by qRT-PCR. DNA precipitated without addition of the antibody (-Ab) as a negative control. At least three independent experiments were performed with similar results. Data are the mean values of three replicates ± sd from one experiment. D, EMSA assay for PIF3 binding to MDP60 promoters. Each biotin-labeled DNA fragment was incubated with the GST-PIF3 protein. Competition for labeled promoter sequences was performed by adding an excess of unlabeled wild-type or mutated probes. The arrow indicates bands caused by PIF3 binding to the P fragment in the MDP60 promoter. E, Y1H analysis using P fragments containing a wild-type G-box (MDP60pro) and mutated G-box (MDP60mpro) as bait and PIF3 as prey. Representative growth status of yeast cells is shown on SD/-UHL agar media with 3-amino-1,2,4-triazole from triplicate independent trials. Numbers on the right side of each photograph indicate relative densities of the cells. F, Transient expression of PIF3 and MDP60pro:GUS or MDP60mpro:GUS in N. benthamiana leaves. Each data bar represents the mean ± sd (n = 3). G, The MDP60 transgenic pif3 mutant (OE#1/pif3 and OE#2/pif3) had much longer hypocotyls than wild-type seedlings but had similar hypocotyl length to MDP60 transgenic wild-type (OE#1) seedlings grown in the light for 7 d. The graph shows the average hypocotyl length measured from at least 43 seedlings per sample. (**P < 0.01, t test). Error bars indicate mean ± sd. H, Lengths of hypocotyl cells from wild-type, pif3, MDP60 transgenic pif3 mutant (OE#1/pif3), and wild-type (OE#1) seedlings grown in the light for 7 d. t test, *P < 0.05, **P < 0.01. Error bars represent the mean ± sd.
It is well established that the transcription factor PIF3 positively mediates hypocotyl elongation via the light signaling pathway and also acts as a crucial downstream regulator participating in ethylene signaling-promoted light-grown hypocotyl elongation (Zhong et al., 2012; Yu and Huang, 2017). Thus, we hypothesized that PIF3 links the light and ethylene signaling pathways and may play a critical role in regulating MDP60 expression. We first interrogated MDP60 expression in mutants of several components of the light signaling pathway. Real-time PCR showed that MDP60 expression was obviously increased in the phyB-9 mutant and PIF3-overexpressing seedlings, but significantly decreased in the pif3 mutant (Fig. 3B). Next, we investigated whether ethylene upregulates MDP60 expression through PIF3. We found no obvious change in MDP60 expression in the pif3 mutant after ACC treatment for 4 h compared to vehicle treatment (Fig. 2B, right panel). These data demonstrate that light and ethylene signaling coordinate MDP60 expression through PIF3.
We next sought to determine the regulatory relationship between PIF3 and MDP60 expression. Chromatin immunoprecipitation (ChIP) was performed to determine whether PIF3 protein binds to the MDP60 promoter in vivo. A PIF3-YFP fusion protein was expressed using a PIF3 native promoter (Al-Sady et al., 2006) and immunoprecipitated using an antibody recognizing the GFP tag. Quantitative real-time PCR revealed that the chromatin immunoprecipitating with the anti-GFP antibody was enriched in fragment P (located −837 to −997 upstream of the transcription start site) in the MDP60 promoter, but not in the control in which DNA was precipitated without the anti-GFP antibody (Fig. 3C). Furthermore, electrophoretic mobility shift assays (EMSAs) showed that the GST-PIF3 fusion protein binds to fragment P of the MDP60 promoter in vitro. This binding was abolished by addition of increasing amounts of unlabeled P probes (Fig. 3D), indicating that PIF3 can directly bind to the MDP60 promoter in vitro. We then confirmed binding by yeast one-hybrid analysis, showing that PIF3 targets the MDP60 promoter sequence containing a wild-type G box sequence, but not a mutated G-box sequence (Fig. 3E), demonstrating that the MDP60 is a PIF3-targeted gene. Finally, we performed transient expression analysis to investigate how PIF3 regulates MDP60 expression. MDP60pro:GUS or MDP60mpro:GUS was cotransformed with 35S:PIF3 into Nicotiana benthamiana leaves. As shown in Figure 3F, PIF3 significantly increases MDP60 transcription through a target G-box in the MDP60 promoter sequence. Collectively, these results demonstrate that PIF3 targets and upregulates MDP60 expression.
As our data showed that MDP60 is directly upregulated by PIF3, we hypothesized that MDP60 overexpression in a pif3 mutant background could attenuate its short hypocotyls. Real-time PCR showed that MDP60 transcription levels were considerably enhanced in lines (pif3-3 mutant background) overexpressing MDP60 (OE#1/pif3 and OE#2/pif3; Supplemental Fig. S3B). Hypocotyl length in MDP60-overexpressing pif3 mutants was dramatically increased in 7-d-old light-grown seedlings, which is similar to that observed in MDP60-overexpressing wild-type seedlings (Fig. 3G). Scanning electron microscopy revealed that the hypocotyl cell lengths of pif3 mutants were increased when MDP60 overexpression was enhanced (Fig. 3H). Thus, based on these findings, we conclude that MDP60 that acts as a downstream effector of PIF3 during light signaling-mediated hypocotyl elongation.
MDP60 Directly Binds to Microtubules
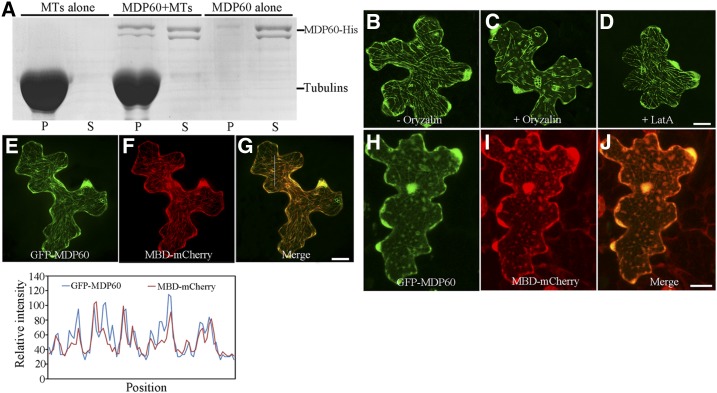
Many proteins containing a TPX2 domain, such as proteins of the WVD2/WDL family and MDP40, exhibited microtubule localization in Arabidopsis cells (Perrin et al., 2007; Wang et al., 2012), suggesting that MDP60 is a putative microtubule-associated protein in Arabidopsis. To test this, a cosedimentation assay was performed. SDS-PAGE analysis revealed that a MDP60-His fusion protein purified from Escherichia coli directly bound to microtubules in vitro (Fig. 4A). Cellular localization of MDP60 was investigated in Arabidopsis cells. Transient assay showed that GFP-MDP60 exhibited filamentous localization in pavement cells (Fig. 4B), which was disrupted by treatment with the microtubule-disrupting reagent oryzalin (Fig. 4C), but were nearly intact in the presence of LatA (a reagent that depolymerizes actin filaments; Fig. 4D), suggesting that this localization is related to microtubules, but not F-actin. To confirm this result, GFP-MDP60 and MBD-mCherry (an mCherry-tagged microtubule binding domain of MAP4) were transiently coexpressed in Arabidopsis pavement cells. Confocal microscopy showed that the GFP-MDP60 green fluorescent signal overlapped with the MBD-mCherry red fluorescent signal (Fig. 4, E–G); this colocalization was able to be disrupted by oryzalin (Fig. 4, H–J), confirming that MDP60 colocalized with microtubules. These data demonstrate that MDP60 colocalizes with microtubules in vitro and in the cells.
Figure 4.
MDP60 directly binds to microtubules. A, MDP60-His was cosedimented with paclitaxel-stabilized microtubules. MDP60-His was most abundant in the supernatant (S) in the absence of microtubules, but cosedimented with microtubules into pellets (P). B, GFP-MDP60 was transiently expressed in Arabidopsis pavement cells where it formed filamentous structures. The filamentous pattern of GFP-MDP60 was disrupted when cells were treated with 10 μm oryzalin for 30 min (C) but was unaffected when treated with 100 nm LatA for 30 min (D). E to G, Colocalization of transiently expressed GFP-MDP60 and MBD-mCherry. Plot of a line scan drawn in G showing a strong correlation between spatial localization of GFP-MDP60 and MBD-mCherry. H to J, The localization could be disrupted via oryzalin. Bars in D, G, and J = 20 μm.
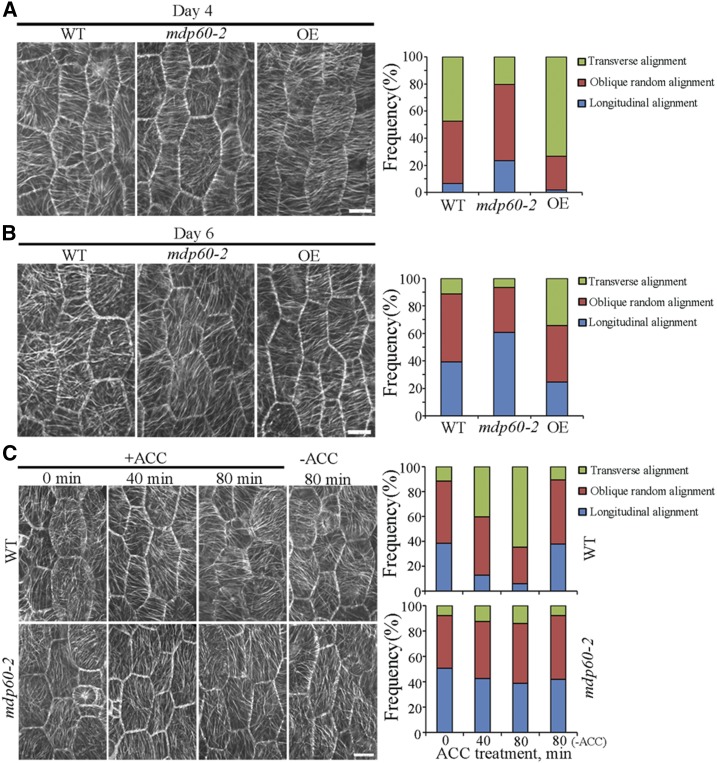
Cortical Microtubule Organization Is Obviously Altered in Epidermal Cells of MDP60 Transgenic Seedlings
Since the organization of cortical microtubules is related to the elongation status of hypocotyl epidermal cells and since MDP60 binds to microtubules, we hypothesized that MDP60 regulates cortical microtubule organization during hypocotyl cell elongation. To test this possibility, we analyzed cortical microtubules in epidermal cells from the hypocotyl middle region of wild-type, MDP60-overexpressing, and mutant mdp60-2 Arabidopsis seedlings carrying YFP-tagged tubulin. Cells were examined at days 4 and 6 of light exposure. We observed generally transverse and oblique parallel arrays of cortical microtubules oriented relative to the longitudinal hypocotyl growth axis in epidermal cells from wild-type hypocotyls. In contrast, cortical microtubules were randomly, obliquely, or longitudinally oriented in most mdp60-2 cells, while we observed mainly transverse cortical microtubule arrays in MDP60-overexpressing hypocotyl cells at day 4 (Fig. 5A). Cortical microtubules were dominantly oriented in a longitudinal manner in wild-type and mutant mdp60-2 hypocotyl cells, whereas oblique and transverse cortical microtubules were detected in most MDP60-overexpressing Arabidopsis cells at day 6 (Fig. 5B). Thus, the alteration of cortical microtubule organization observed in MDP60-overexpressing seedlings is consistent with the significant increase in hypocotyl cell elongation caused by increased MDP60 expression.
Figure 5.
The cortical microtubule array is greatly altered in hypocotyl epidermal cells of MDP60 transgenic seedlings. A and B, Cortical microtubules in epidermal cells from the middle regions of hypocotyls from wild-type (WT), mdp60-2 mutant, and MDP60 transgenic (OE) seedlings in a YFP-tubulin background were observed by confocal microscopy after growth in the light for 4 or 6 d. The graphs show the frequencies of different microtubule orientation patterns in light-grown hypocotyl epidermal cells from wild-type, mdp60-2, and OE seedlings (n > 65 cells). Bars in A and B = 10 μm. C, Wild-type and mdp60-2 hypocotyls in a YFP-tubulin background were transferred to medium containing 0 or 10 μm ACC for 0, 40, and 80 min. Cortical microtubules from the middle regions of hypocotyl epidermal cells were observed. The graphs show the frequencies of microtubule orientation patterns in the middle regions of wild-type and mdp60-2 hypocotyl epidermal cells (n > 65 cells). Bar in C = 10 μm.
Ethylene is demonstrated to participate in the cortical microtubule reorganization during etiolated hypocotyl cell elongation (Sun et al., 2015; Ma et al., 2016). Because MDP60 plays a positive role in ethylene-promoted hypocotyl elongation, we analyzed the effects of MDP60 in regulating cortical microtubule organization in response to ethylene. Wild-type and mdp60-2 seedlings were grown for 6 d in the light and then transferred to the medium containing 10 μm ACC. Cortical microtubules were mostly longitudinally oriented to the hypocotyl growth axis in the middle regions of hypocotyls in wild-type and mdp60-2 mutants, whereas almost 40% of transverse cortical microtubules appeared in the hypocotyl cells of wild-type seedlings after treatment with ACC for 40 min. In contrast, most of the cells in mdp60-2 hypocotyls had random, oblique, or longitudinal microtubule arrays. Increasing the duration of treatment induced predominantly transverse cortical microtubules in wild-type, but not mdp60-2, cells (Fig. 5C), indicating that cortical microtubule reorganization in response to ACC treatment was partially hindered in mdp60-2 cells. This demonstrates that the microtubule reorganization from a longitudinal to a transverse orientation in mdp60 mutant cells is less sensitive to ethylene.
MDP60 Is a Microtubule-Destabilizing Protein
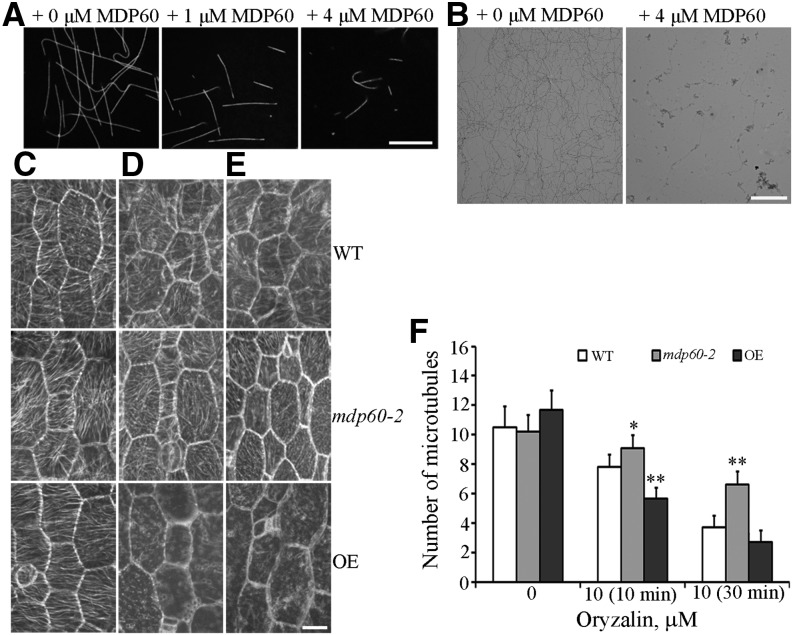
Given that MDP60 colocalizes and alters cortical microtubule organization, we further investigated the molecular basis for MDP60-mediated microtubule organization during hypocotyl elongation. Microtubules polymerized from rhodamine-labeled tubulin incubated with or without MDP60 were observed using confocal microscopy, revealing much shorter microtubules in the presence of 1 and 4 μm MDP60 than in the absence of MDP60 (Fig. 6A). This result was further confirmed using negative-stain electron microscopy (Fig. 6B), suggesting that MDP60 destabilizes microtubules in vitro.
Figure 6.
MDP60 is a microtubule destabilizer. A, Fluorescent images of microtubules polymerized in 20 μm rhodamine-labeled tubulin solution in the presence of 0, 1, and 4 μm MDP60. Bar in A = 10 μm. B, Negative stain electron micrographs of A. Bar in B = 5 μm Cortical microtubules were observed in epidermal cells in the middle regions of hypocotyls from light-grown wild-type (WT), mdp60-2, and MDP60-overexpressing (OE) seedlings after treatment with 0 μm oryzalin (C), 10 μm oryzalin for 10 min (D), and 10 μm oryzalin for 30 min (E). Bar in E = 10 μm. F, Quantification of cortical microtubules in hypocotyl epidermal cells of wild-type, mdp60-2, and OE seedlings using ImageJ software (n > 30 cells for each sample). The y axis represents the number of cortical microtubules that crossed a fixed line (∼10 μm) perpendicular to the orientation of the majority of cortical microtubules in the cell. *P < 0.05, **P < 0.01, t test. Error bars represent mean ± sd.
The microtubule-destabilizing activity of MDP60 was further interrogated in hypocotyl cells using the microtubule-disrupting drug oryzalin. To quantify the effects of oryzalin on the stability of cortical microtubules in wild-type, mdp60-2, and MDP60-overexpressing cells, we estimated the densities of microtubules in hypocotyl epidermal cells as previously reported (Li et al., 2011; Sun et al., 2015). Paired Student’s t tests were employed to identify significant differences. Cortical microtubule densities in wild-type, mdp60-2, and MDP60-overexpressing cells were not obviously different before treatment but were significantly different after treatment (Fig. 6F). Most cortical microtubules were disrupted in MDP60-overexpressing epidermal cells after treatment with 10 μm oryzalin for 10 min. However, microtubules in wild-type and mutant mdp60-2 cells were largely unaffected (Fig. 6, C, D, and F). Increasing oryzalin treatment to 30 min resulted in disruption of the majority of cortical microtubules in wild-type and MDP60-overexpressing cells, whereas many intact cortical microtubules were observed in mutant mdp60-2 cells (Fig. 6, E and F). Thus, the sensitivity of microtubules to oryzalin treatment was significantly enhanced when MDP60 expression was increased. These results confirm that MDP60 functions as a microtubule destabilizer.
DISCUSSION
Understanding how upstream signals crosstalk to regulate cortical microtubules in multiple conditions is essential to elucidate the mechanisms underlying plant growth. In this study, we demonstrate that the microtubule-destabilizing protein MDP60 acts as a downstream effector of light and ethylene signaling to control hypocotyl cell elongation.
Light and Ethylene Coordinate Hypocotyl Cell Elongation through MDP60
As important components of the light signaling pathway, PIF transcription factors play crucial roles in promoting hypocotyl elongation (Leivar et al., 2008; Leivar and Quail, 2011; Zhong et al., 2014). It is well known that PIF protein levels are regulated by the 26S proteasome in response to light signals (Castillon et al., 2007). PIF proteins are more stable in the dark than in the light, which favors seedlings rapidly elongating their etiolated hypocotyls upward in search of the soil surface. Our results indicate that MDP60 functions as a positive regulator of hypocotyl elongation in the light, which is mediated by PIF3; thus, we hypothesized that it may be an important factor facilitating seedling growth toward the soil surface. Thus, we further analyzed the expression of MDP60 in the dark. Interestingly, no obvious MDP60 expression was detected in etiolated hypocotyls (Supplemental Fig. S4A, left panel), suggesting that MDP60 may be not involved in rapid etiolated hypocotyl elongation in the dark. In agreement with these results, both the pif3 mutant and PIF3-overexpressing seedlings exhibited similar etiolated hypocotyl lengths as wild-type seedlings (Zhong et al., 2012), although the exact course of this phenomenon remains unclear. Considering our data, we propose that PIF3 may be nonfunctional in etiolated hypocotyl elongation and acts as a crucial regulator for MDP60 expression. Thus, our data demonstrated that MDP60 expression levels are positively regulated by PIF3 (which, in turn, is known to be negatively regulated by light) and that the MDP60 promoter is a direct target of PIF3.
Ethylene inhibits etiolated hypocotyl elongation in Arabidopsis seedlings in the dark, but promotes hypocotyl elongation in the light. Our results indicate that ethylene signaling is critical for MDP60 expression in light via PIF3-mediated regulation. Thus, we interrogated whether ethylene also plays a role in MDP60 expression in the dark. GUS staining indicated no obvious MDP60 expression in etiolated hypocotyls when treated with ACC (Supplemental Fig. S4A, right panel). Furthermore, etiolated hypocotyl elongation was similar between mdp60 mutants and wild-type seedlings when grown in medium containing ACC (Supplemental Fig. S4B). Thus, these data suggest that MDP60 may not participate in ethylene-inhibited etiolated hypocotyl elongation in the dark.
Furthermore, we found that hypocotyl length in MDP60-overexpressing wild-type seedlings was shorter than in ctr1-1 mutant, PIF3-overepxressing, and phyB-9 mutant seedlings when grown in the light for 7 d (Supplemental Fig. S5A). In addition, hypocotyl length in mdp60-2 mutant seedlings was longer than ein2-5, pif3 mutant, and PHYB-overexpressing seedlings (Supplemental Fig. S5B). A previous study indicated that PIF3 is an essential component required for ethylene-induced light-grown hypocotyl elongation (Zhong et al., 2012). Combined with the EMSA and ChIP-qPCR assay, our data thus support the hypothesis that MDP60 acts downstream of PIF3 and plays a central role in ethylene-induced light-grown hypocotyl elongation and also functions as a negative regulator involved in light-suppressed hypocotyl elongation. Thus, light and ethylene signaling elaborately regulate MDP60 expression for proper hypocotyl elongation. Increasing evidence points to PIF3 playing a central role in promoting hypocotyl elongation under multiple abiotic and environmental conditions. For example, one study has shown thermoperiodic control of hypocotyl elongation through ethylene signaling-mediated regulation of PIF3 activity (Bours et al., 2015). Future studies are needed to investigate whether and how MDP60 functions in these processes.
MAPs Are Required to Mediate Hypocotyl Elongation in Different Regulatory Pathways
Several MAPs have exhibited their effects on hypocotyl cell elongation in response to environmental and developmental cues. For example, the microtubule plus-end tracking protein SPRIAL1 (SPR1) promotes dark-grown hypocotyl cell elongation, whereas the microtubule-stabilizing protein WDL3 suppresses hypocotyl cell elongation in the light (Nakajima et al., 2004, 2006; Liu et al., 2013; Lian et al., 2017). In this study, MDP60 participates in light and ethylene crosstalk-mediated hypocotyl elongation, suggesting that some MAPs may common participants in diverse regulatory pathways mediating plant growth under multiple growth conditions.
Transcript regulation is a mechanism that controls the activity of microtubule-associated proteins during hypocotyl elongation. For example, SPR1 transcripts were detected in etiolated hypocotyls, but not light-grown hypocotyls, suggesting that light affects SPR1 expression, which regulates SPR1-mediated control of hypocotyl growth in response to light (Nakajima et al., 2004, 2006). However, the upstream transcription factors that regulate SPR1 expression in response to light remain unknown. Arabidopsis MDP40 is directly targeted by BRASSINAZOLE-RESISTANT1, a key transcription factor in the BR signaling pathway, and participates in BR-induced etiolated hypocotyl cell elongation (Wang et al., 2012). Here, we report that MDP60 is directly targeted by PIF3 during hypocotyl cell elongation. This suggests that regulating MAP expression through transcription factors alters cortical microtubule organization and is important for plant cell responses to diverse cues.
Microtubule-Destabilizing Proteins Participate in Ethylene-Promoted Hypocotyl Cell Elongation
Genetic evidence has shown that increasing or decreasing the expression of microtubule destabilizers or stabilizers results in abnormal hypocotyl elongation by altering cortical microtubule organization (Wang et al., 2007; Liu et al., 2013). Previous studies have shown that destabilizing microtubules partially rescued ethylene-inhibited stem elongation in pea (Pisum sativum) seedlings and that WDL5, which functions as a microtubule stabilizer, plays a positive role in ethylene-inhibited etiolated hypocotyl elongation in Arabidopsis (Steen and Chadwick, 1981; Sun et al., 2015), suggesting that microtubule-stabilizing activity is involved in ethylene-inhibited etiolated hypocotyl elongation. In this study, we showed that the microtubule-destabilizing protein MDP60 plays a positive role in ethylene-induced hypocotyl elongation. This demonstrates that regulation of microtubule stability by various MAPs may be important for ethylene-mediated hypocotyl elongation in response to signals.
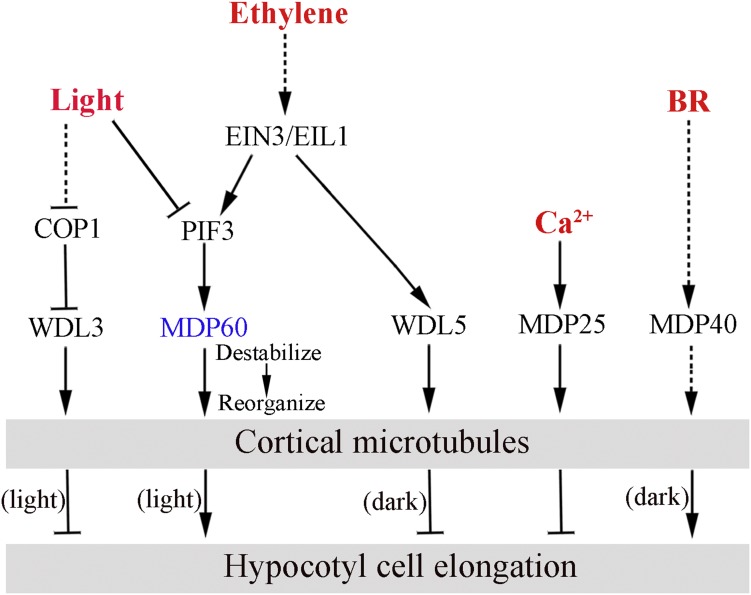
Combined with previous studies involving microtubule-associated protein WDL3, WDL5, MDP25, and MDP40 (Li et al., 2011; Wang et al., 2012; Liu et al., 2013; Sun et al., 2015; Lian et al., 2017), this study provides a strong outline of upstream signaling regulating cortical microtubules in hypocotyl cell elongation (Fig. 7). WDL3 suppresses hypocotyl cell elongation and is regulated by an E3 ubiquitin ligase COP1 in response to light. Ethylene and BR regulate WDL5 and MDP40 expression to mediate etiolated hypocotyl cell elongation through altering cortical microtubule organization. Ca2+ regulates the activity of MDP25 on cortical microtubules to inhibit hypocotyl cell elongation. In this study, we showed that light and ethylene signaling play opposite roles in controlling MDP60 expression through PIF3, and MDP60 acts on cortical microtubules via its microtubule destabilizing activity to maintain a transverse orientation, which ultimately promotes hypocotyl cell elongation. Future studies should aim at characterizing additional MAPs in this regulatory network and further address the underlying mechanisms of microtubules in signaling crosstalk in response to multiple developmental and environmental cues.
Figure 7.
Working model of upstream signaling-mediated hypocotyl cell elongation through diverse MAP regulation of cortical microtubules. Arrows show positive regulation, and bar ends show inhibitory action. WDL3 suppresses hypocotyl cell elongation in the light, whereas WDL5 and MDP40 mediate hypocotyl cell elongation in the dark. Ca2+ regulates activity of MDP25 to inhibit hypocotyl cell elongation. In this study, we showed that light and ethylene coordinate MDP60 expression through PIF3, and MDP60 alters cortical microtubule organization via microtubule-destabilizing activity, which promotes hypocotyl cell elongation.
METHODS
Plant Materials and Growth Conditions
All plant materials used in this study were from the Arabidopsis (Arabidopsis thaliana) Columbia (Col) ecotype background. Seeds were sterilized and placed on half-strength Murashige and Skoog medium (Sigma-Aldrich) with 0.8% agar and 1% Suc. For hypocotyl measurements, plates were placed at 22°C in the light for 7 d after stratification at 4°C for 2 d. Mutants ein2-5 (Alonso et al., 1999), ctr1-1(Kieber et al., 1993), pif3-3 (Monte et al., 2004), phyB-9 (Reed et al., 2000), PHYB-GFP (Zheng et al., 2013), and 35S:Tubulin5A-YFP transgenic plants (Kirik et al., 2012) were used in this study.
Isolation of MDP60 cDNA Clones from Arabidopsis
The full-length MDP60 cDNA sequence was amplified using RT-PCR. Primers used to amplify MDP60 were 5′-CCATGGGAATGGAGTCGACGAATTTGA-3′ and 5′-GCGGCCGCGAGGTCTTGTTGGTATAAGAG-3′. MDP60-His-tagged fusion proteins were expressed and purified according to the manufacturer’s protocols. Protein concentration was determined using a Bio-Rad protein assay kit. Protein samples were analyzed by SDS-PAGE.
Analysis of the MDP60 Promoter:GUS Activity
A DNA fragment of the MDP60 promoter containing 1689 bp upstream of the translation start site was amplified by PCR. Primers used for amplification were 5′-GTCGACTCTTATTTTGTGAATGTTCAGGCTG-3′and 5′-GGATCCGTCTTTTCGAGTGAGTGAAGGGA-3′. The sequence was then cloned into the pCAMBIA1391 vector (Invitrogen), and the resulting construct was transformed into Arabidopsis plants using Agrobacterium tumefaciens (strain GV3101) using the Arabidopsis floral dip method (Clough and Bent, 1998). Thirty-seven independent transgenic lines were obtained, and homozygous seedlings were used for histochemical localization of GUS activity in hypocotyl cells. The GUS staining procedure was performed as previously described by Wang et al. (2007).
Generation of MDP60 Overexpressing and RNAi Arabidopsis Lines
To stably express MDP60-MYC, full-length MDP60 cDNA was amplified by PCR and subcloned into the pBI221 vector (Invitrogen), which expressed MDP60 under the control of the 35S promoter and a nopaline synthase terminator. MYC was amplified and inserted at the C terminus of MDP60. The cDNAs encoding MDP60 and MYC were then amplified and subcloned into the pCAMBIA1300 expression vector and transformed into wild-type (Columbia ecotype) Arabidopsis plants with A. tumefaciens (strain GV3101). Transgenic homozygous Arabidopsis lines from T3 generation were used.
To prepare stable MDP60 RNAi Arabidopsis lines, a MDP60 RNAi vector was generated by amplifying 200- or 200 bp MDP60 coding sequences in the sense and antisense orientations and inserting them into the pFGC5941 vector. Primers used for amplification of MDP60 RNAi sequences were 5′-GGATCCTGGAGGATGGAAATGAACAAGTC-3′ and 5′-CCCGGGTAGAAGTTACCTTCGAAAGTGGGAA-3′, and 5′-CCATGGTGGAGGATGGAAATGAACAAGTC-3′ and 5′-CTCGAGTAGAAGTTACCTTCGAAAGTGGGAA-3′.
Generation of mdp60 Using CRISPR/Cas9 Technology
Ribozyme-based CRISPR/Cas9 technology was described by Wang et al. (2015). Wild-type Arabidopsis plants, Col-0 ecotype, were transformed with the CRISPR construct by floral dipping. mdp60 plants were identified at the T2 stage.
Transient Expression Assays in Nicotiana benthamiana
MDP60pro:GUS or MDP60mpro:GUS with pCAMBIA1391 and 35S:PIF3 constructs were transformed into A. tumefaciens (GV3101). A. tumefaciens cells were harvested by centrifugation and suspended in a solution containing 10 mm MES, pH 5.6, 10 mm MgCl2, and 200 mm acetosyringone to an optical density (600 nm) of 0.7. Cells were incubated at room temperature for 4 h and used to infiltrate N. benthamiana leaves using a needle-free syringe. GUS activity of the infiltrated leaves was quantitatively assessed.
Yeast One-Hybrid Assay
PIF3 full-length coding sequences were amplified by PCR and cloned into the NdeI-BamHI sites of the pGAD7 vector (Clontech). Fragments of MDP60 promoters were amplified and cloned into AflII-XhoI sites of the PHIS1 vector (Clontech). AD-fused protein and LacZ reporter plasmids were cotransformed into the AH109 yeast strain. Yeast transformation and growth were performed based on the Yeast Protocols Handbook (Clontech).
Microtubule Cosedimentation Assay
Porcine brain tubulins were purified using a method previously published by Castoldi and Popov (2003) and used for sedimentation assays. Tubulin assembly and cosedimentation of microtubules with MDP60-His fusion proteins were performed as described by Mao et al. (2005) and Li et al. (2011). Purified proteins were centrifuged at 150,000g at 4°C for 20 min before use. Prepolymerized, paclitaxel-stabilized microtubules (5 μm) were incubated with 4 μm MDP60 fusion proteins in PEM buffer (1 mm MgCl2, 1 mm EGTA, and 100 mm PIPES-KOH, pH 6.9) plus 20 μm paclitaxel at room temperature for 20 min. After centrifugation at 100,000g for 20 min, the supernatant and pellets were subjected to SDS-PAGE.
Electron microscopy was performed as described previously (Mao et al., 2005), using a JEM-1230 transmission electron microscope (JEOL Co.).
PCR Analysis
Quantitative real-time PCR analysis was performed to assess MDP60 transcript levels in wild-type, MDP60-overexpressing, ein2-5, phyB-9, PIF3-overexpressing, and pif3 seedlings. Total RNA was isolated using TRIzol reagent (Invitrogen) from 5-d-old seedlings grown in the light. For quantitative real-time PCR, an ABI 7500 real-time PCR system (Applied Biosystems) was used according to the manufacturer’s instructions. Primers used for subsequent detection of MDP60 expression were 5′-ATGAGCCAGCCTAAGTGTTC-3′ and 5′-GAGGCATGAGGTTGTTCTAGTC-3′. UBQ11 was used as an internal control (5′-GCAGATTTTCGTTAAAACC-3′ and 5′-CCAAAGTTCTGCCGTCC-3′). Three biological replicates and two to three technical replicates (for each biological replicate) were used for each treatment. The average and sd were calculated from the biological replicates.
EMSA
EMSA was performed according to Zhang et al. (2012). Briefly, recombinant GST-PIF3 protein was purified from Escherichia coli according to the manufacturer’s instructions. Biotin-labeled DNA fragments were synthesized and used as probes, and biotin-unlabeled DNA fragments of the same sequences were used as competitors. Nucleotide sequences of the double-stranded oligonucleotides for MDP60 P (which fragment containing G-box motif) were 5′-TCGAGGGGACACGTGAACGGTGAG-3′ and 5′-CTCACCGTTCACGTGTCCCCTCGA-3′. Primers were labeled using the Biotin 5′ End DNA labeling kit (Pierce). Standard reaction mixtures (20 μL) for EMSA contained 1 μg purified proteins, 2 μL biotin-labeled annealed oligo nucleotides, 2 μL 10× binding buffer (100 mm Tris, 500 mm KCl, and 10 mm DTT, pH 7.5), 1 μL 50% glycerol, 1 μL 1% Nonidet P-40, 1 μL 1 m KCl, 1 μL 100 mm MgCl2, 1 μL 200 mm EDTA, 1 μL 1 mg/mL poly(dI-dC), and 10 μL ultrapure water. Reactions were incubated at room temperature (25°C) for 30 min and loaded onto a 6% native polyacrylamide gel in TBE buffer (45 mm Tris, 45 mm boric acid, and 1 mm EDTA, pH 8.3). The gel was sandwiched and transferred to an N+ nylon membrane (Millipore) in 0.5× TBE buffer at 380 mA at 4°C for 60 min. Detection of biotin-labeled DNA by chemiluminescence was performed based on the instructions provided in the Light Shift Chemiluminescent EMSA kit (Pierce).
ChIP
Seven-day-old light-grown seedlings expressing PIF3-YFP under a PIF3 native promoter were used (Al-Sady et al., 2006). ChIP was performed as previously described (Johnson et al., 2002) using an anti-GFP monoclonal antibody (Sigma-Aldrich) for immunoprecipitation. Equal quantities of starting plant material and ChIP reagents were used for the real-time PCR reaction. Primers used to detect the PIF3 target MDP60 promoter were P (5′-GACGTTTGCCTTAGCATCATCA-3′ and 5′-TCTGATTACTGATTATCGTGGAAAA-3′) and actin2 as a control (5′-GGTAACATTGTGCTCAGTGGTGG-3′ and 5′-AACGACCTTAATCTTCATGCTGC-3′). ChIP experiments were performed independently three times.
Ballistics-Mediated Transient Expression in Leaf Epidermal Cells
Subcellular localization of GFP-MDP60 and cortical microtubules was visualized using transiently expressed 35S:GFP-MDP60 and 35S:MBD-mCherry constructs in Arabidopsis (Col ecotype) leaf epidermal cells. Experiments were performed as previously described by Fu et al. (2002). One microgram of 35S:GFP-MDP60 and 1 μg of 35S:MBD-mCherry DNA were used for particle bombardment. Six to eight hours after bombardment, GFP and mCherry signals were detected using a Zeiss LSM 510 META confocal microscope. Filamentous structures containing GFP-MDP60 in leaf epidermal cells were visualized after treatment with 10 μm oryzalin and 100 nm LatA for 30 min.
Accession Numbers
Sequence data can be found in the Arabidopsis Genome Initiative under accession number At3g01015 (MDP60).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MDP60 Expression Varies in Seedlings Treated with Different Hormones.
Supplemental Figure S2. MDP60 Expression Is Associated with the Hypocotyl Phenotype in MDP60 RNAi Lines.
Supplemental Figure S3. MDP60 Expression Is Increased in MDP60 Transgenic ein2 and pif3 Mutant Seedlings.
Supplemental Figure S4. MDP60 May Not Be Involved in Ethylene-Inhibited Etiolated Hypocotyl Cell Elongation in the Dark.
Supplemental Figure S5. MDP60 Acts Downstream of PIF3 in Ethylene-Stimulated and Light-Suppressed Hypocotyl Elongation.
Acknowledgments
We thank Dr. Shuhua Yang (China Agricultural University) and Dr. Suiwen Hou (Lanzhou University) for generously providing the ethylene-related Arabidopsis mutant and PHYB-GFP transgenic seeds.
Footnotes
Articles can be viewed without a subscription.
T.M. designed the project; Q.M., X.W., and J.S. performed specific experiments and analyzed the data; T.M. wrote, revised, and edited the manuscript; all authors read and approved the final manuscript.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bours R, Kohlen W, Bouwmeester HJ, van der Krol A (2015) Thermoperiodic control of hypocotyl elongation depends on auxin-induced ethylene signaling that controls downstream PHYTOCHROME INTERACTING FACTOR3 activity. Plant Physiol 167: 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Castoldi M, Popov AV (2003) Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32: 83–88 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim J, Song K, Park E, Kim K, Bae G, Choi G (2016) Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously. Plant Cell 28: 2770–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V (2012) TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, De Cnodder T, Van Der Straeten D, Verbelen JP (2005) Cell elongation and microtubule behaviour in the Arabidopsis hypocotyl: responses to ethylene and auxin. J Plant Growth Regul 24: 166–178 [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Qin T, Zhang Y, Liu X, Sun J, Zhou Y, Zhu L, Zhang Z, Yuan M, Mao T (2011) MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23: 4411–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian N, Liu X, Wang X, Zhou Y, Li H, Li J, Mao T (2017) COP1 mediates dark-specific degradation of microtubule-associated protein WDL3 in regulating Arabidopsis hypocotyl elongation. Proc Natl Acad Sci USA 114: 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Qin T, Ma Q, Sun J, Liu Z, Yuan M, Mao T (2013) Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell 25: 1740–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Sun J, Mao T (2016) Microtubule bundling plays a role in ethylene-mediated cortical microtubule reorientation in etiolated Arabidopsis hypocotyls. J Cell Sci 129: 2043–2051 [DOI] [PubMed] [Google Scholar]
- Mao T, Jin L, Li H, Liu B, Yuan M (2005) Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol 138: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T (2004) SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Kawamura T, Hashimoto T (2006) Role of the SPIRAL1 gene family in anisotropic growth of Arabidopsis thaliana. Plant Cell Physiol 47: 513–522 [DOI] [PubMed] [Google Scholar]
- Perrin RM, Wang Y, Yuen CY, Will J, Masson PH (2007) WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J 49: 961–971 [DOI] [PubMed] [Google Scholar]
- Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LA (2009) Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol 149: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, Elumalai RP, Millar AJ (2000) Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol 122: 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaoka H. (1993) Regulation by gibberellins of the orientation of cortical microtubules in plant cells. Aust J Plant Physiol 20: 461–470 [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten DV (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen DA, Chadwick AV (1981) Ethylene effects in pea stem tissue: Evidence of microtubule mediation. Plant Physiol 67: 460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ma Q, Mao T (2015) Ethylene regulates the Arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiol 169: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Vancompernolle B, Rieu I, Ahmad M, Phillips A, Moritz T, Hedden P, Van Der Straeten D (2007) Ethylene-induced Arabidopsis hypocotyl elongation is dependent on but not mediated by gibberellins. J Exp Bot 58: 4269–4281 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T (2012) Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24: 4012–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M (2007) Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Jang IC, Henriques R, Chua NH (2009) FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Huang R (2017) Integration of ethylene and light signaling affects hypocotyl growth in Arabidopsis. Front Plant Sci 8: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xu Y, Guo S, Zhu J, Huan Q, Liu H, Wang L, Luo G, Wang X, Chong K (2012) Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Z, Quan R, Li G, Wang R, Huang R (2011) An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol 157: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wu S, Zhai H, Zhou P, Song M, Su L, Xi Y, Li Z, Cai Y, Meng F, et al. (2013) Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H (2012) A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW (2014) Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc Natl Acad Sci USA 111: 3913–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]