Cortical microtubule arrays in the Arabidopsis hypocotyl share a common underlying architecture that identifies their mode of construction.
Abstract
Microtubules at the plant cell cortex influence cell shape by patterning the deposition of cell wall materials. The elongated cells of the hypocotyl create a variety of microtubule array patterns with differing degrees of polymer coalignment and orientation to the cell’s growth axis. To gain insight into the mechanisms driving array organization, we investigated the underlying microtubule array architecture in light-grown epidermal cells with explicit reference to array pattern. We discovered that all nontransverse patterns share a common underlying array architecture, having a core unimodal peak of coaligned microtubules in a split bipolarized arrangement. The growing microtubule plus ends extend toward the cell’s apex and base with a region of antiparallel microtubule overlap at the cell’s midzone. This core coalignment continuously shifts between ±30° from the cell’s longitudinal growth axis, forming a continuum of longitudinal and oblique arrays. Transverse arrays exhibit the same unimodal core coalignment but form local domains of microtubules polymerizing in the same direction rather than a split bipolarized architecture. Quantitative imaging experiments and analysis of katanin mutants showed that the longitudinal arrays are created from microtubules originating on the outer periclinal cell face, pointing to a cell-directed, rather than self-organizing, mechanism for specifying the major array pattern classes in the hypocotyl cell.
The interphase microtubules at the plant cell cortex play a critical role in plant morphogenesis. Early experiments depolymerizing “spindle fibers” showed the curious property of changing plant cell shape. These observations led to a proposal that polymers at the cell cortex organized cell wall fibers on the other side of the plasma membrane to affect cell shape (Green, 1962). Early electron microscopy and immunocytochemistry provided images of cortical microtubule array patterns correlated with the growth habit of specific cell types (Hardham and Gunning, 1978; Hepler and Newcomb, 1964; Ledbetter, 1982; Lloyd et al., 1985; Shibaoka, 1974). Genetic analyses later showed that point mutations in tubulin genes led to cell morphology changes correlated with array pattern defects (Abe and Hashimoto, 2005; Ishida et al., 2007; Thitamadee et al., 2002). More recent investigations have shown that the cortical microtubule cytoskeleton provides a dynamic scaffold for both the targeting of cellulose-producing enzymes and for the orientation of cellulose deposition (Crowell et al., 2009; Desprez et al., 2007; Gutierrez et al., 2009; Paredez et al., 2006). Collectively, these observations provide compelling evidence that cortical microtubule pattern influences cellular morphogenesis by orienting the deposition of cell wall materials (Baskin, 2001; Cyr and Palevitz, 1995; Ehrhardt and Shaw, 2006; Emons et al., 2007; Lloyd, 2011; Sedbrook and Kaloriti, 2008).
How plant cells organize the microtubule cytoskeleton to specify cell morphology remains a central question for plant cell biology. The centrosome in animal cells gathers microtubule nucleation complexes to a central position in the cell resulting in a “radial” microtubule array pattern. The minus-ends remain anchored at the centrosome, with the dynamic microtubule plus ends radiating into the cytoplasm. Flowering plants do not have a centrosome or centralized microtubule-organizing center (Cyr and Palevitz, 1995). The microtubules nucleate from the same gamma-tubulin ring complexes (γ-TuRCs) found in animal cells, but the γ-TuRCs are not known to be clustered to specific sites in the cell (Liu et al., 1993; Murata et al., 2005; Nakamura et al., 2004). Hence, plant cortical arrays create a wide variety of patterns and organizational states with mixed polarities of microtubules (Ehrhardt and Shaw, 2006).
Axially growing hypocotyl cells are an important model for investigating the mechanisms driving microtubule array organization and the relationship of array pattern to cell morphogenesis. The cortical microtubule arrays on the outer periclinal cell face show a distribution of patterns, generally classed by the degree of microtubule coalignment and by the orientation of the alignment to the cell’s growth axis (Ehrhardt and Shaw, 2006; Vineyard et al., 2013). Dark-grown cells showing rapid expansion tend to have microtubules aligned transversely to the cell’s long axis (Crowell et al., 2011; Lindeboom et al., 2013a), with a high degree of coalignment (i.e. aligned to each other). This transverse pattern is hypothesized to create bands of cellulose around the cell’s short axis, restricting radial expansion and promoting axial growth (Baskin, 2001, 2005; Cosgrove, 1987). Light-grown hypocotyl cells grow more slowly and typically exhibit a variety of coaligned array patterns in transverse, oblique, and longitudinal orientations, with a separate class of “basket” patterned arrays having no obvious coalignment (Chan et al., 2007; Chan et al., 2010; Crowell et al., 2011; Dixit et al., 2006; Sambade et al., 2012; Takesue and Shibaoka, 1999; Vineyard et al., 2013; Yu et al., 2015). The role of nontransverse array patterns is more speculative, where the less-ordered arrays could be transitions between coaligned patterns or potentially important for creating more isotropic cell walls (Baskin, 2005; Chan et al., 2007, 2010, 2011; Emons et al., 2007; Gutierrez et al., 2009).
While array pattern and the information it carries for cell wall construction has been the focus of extensive study, the underlying architecture of the array remains poorly understood. Each array pattern is composed of both bundled and unbundled microtubules that have a position, orientation, and direction of polymerization on the cell face (i.e. array architecture). The individual microtubules that make up the pattern are constantly treadmilling (Shaw et al., 2003; Shaw and Lucas, 2011), requiring that a constant supply of new polymers be created to maintain the pattern. Investigations using an End Binding1-GFP (EB1-GFP) probe to visualize only the growing microtubule plus ends have shown groupings of microtubules polymerizing in the same direction on the outer cell face (Ambrose and Wasteneys, 2014; Chan et al., 2007, 2011; Dixit et al., 2006; Sambade et al., 2012; Vineyard et al., 2013). The microtubules have been characterized as either local cohorts, repatterning the array (Chan et al., 2007, 2010) or as a “bipolarity” of the entire array (Ambrose and Wasteneys, 2014; Sambade et al., 2012). These observations suggest a common origin or a cellular mechanism for orienting these microtubules. While the observations have not been reconciled with the array patterns observed in hypocotyl cells, they have been proposed to be important for creating alternating layers of cell wall material with different net orientations (Chan et al., 2010, 2011; Lloyd, 2011).
Patterning models have been proposed based on studies that identified specific properties and behaviors of individual microtubules (Ambrose et al., 2011; Chan et al., 2003; Dixit et al., 2006; Dixit and Cyr, 2004; Ehrhardt and Shaw, 2006; Lloyd and Chan, 2008; Mathur, 2006; Sambade et al., 2012; Wasteneys and Ambrose, 2009). In this view, microtubules given a set of polymerization parameters and rules for microtubule-microtubule interactions will, by the nature of those actions, self-organize into a specific pattern. Computational models have shown the plausibility of self-organizing mechanisms, often with a biasing feature in the cell geometry (Allard et al., 2010a, 2010b; Deinum et al., 2011; Eren et al., 2010; Hawkins et al., 2010; Sambade et al., 2012). Alternative views have suggested that array patterning occurs more similarly to the radial arrays in centrosomal systems, where the site and direction of microtubule nucleation is under explicit cellular control or via feedback from the cell wall (Ambrose et al., 2011; Bringmann et al., 2012; Burian et al., 2013; Heisler et al., 2010; Landrein and Hamant, 2013). The cortical microtubules in guard cells, for example, appear to be organized with polymerization coming from a site adjacent to the pore (Eisinger et al., 2012), suggesting that acentriolar plant cells could have mechanisms to assign the position and direction of microtubule nucleation.
Time lapse imaging has provided examples where array pattern changes gradually (Yuan et al., 1995) and, alternatively, where local groups of microtubules appear to abruptly alter or overwrite an existing microtubule pattern (Ambrose and Wasteneys, 2014; Chan et al., 2007, 2010, 2011; Dixit et al., 2006). The data are not well enough resolved to ask if the pattern changes arise from a state change in the microtubules (i.e. self-organization) or because of local cellular action on the microtubule array. Observations of transverse microtubule arrays in dark-grown plants transitioning to longitudinal patterns in the light were shown to involve KATANIN-dependent microtubule severing at points of microtubule crossover, followed by recovery of the new plus end through microtubule rescue (Lindeboom et al., 2013b). This mechanism of rapidly “amplifying” microtubules in a new direction provides a molecular mechanism for driving context-specific array transitions without substantial changes in de novo nucleation.
To date, no explicit molecular or phenomenological models have been proposed to account for the distribution of microtubule array patterns found in light-grown hypocotyl cells. As a means for understanding the mechanisms by which specific array patterns are created, we quantitatively examined how the individual microtubules are arranged within the four commonly observed array pattern classes. We discovered that all nontransverse array patterns share a common underlying array architecture, indicating a common mechanistic origin. Based on these findings, we used quantitative time-course studies, nucleation site markers, and a loss-of-function katanin p60 mutant to gain insight into how the nontransverse array patterns are created by the cell.
RESULTS
Cortical Array Patterns Maintain a Unimodal Core of Coaligned Microtubules
The information that a microtubule pattern confers to the cell wall, as a template for cellulose deposition, is generally related to the degree and direction of microtubule coalignment (Baskin, 2001, 2005; Green, 1962; Lloyd, 2011). Array pattern in hypocotyl cells has, therefore, generally been assigned based on a subjective visual judgment of the degree of array coalignment and the orientation of the array to the cell’s long (growth) axis. Visual classification has proven useful for describing the relative distribution of pattern types (Atkinson et al., 2014; Liu et al., 2013; Vineyard et al., 2013) but does not provide information about the microtubule polarities or the degree of polymer coalignment within the array. To quantitatively examine the array pattern and the arrangement of microtubules underlying the pattern we used 3D time lapse confocal microscopy to image an EB1-GFP probe (Mathur et al., 2003). EB1-GFP labels the growing plus ends of individual microtubules allowing for the assignment of microtubule position, orientation, and growth direction from short time lapsed sequences.
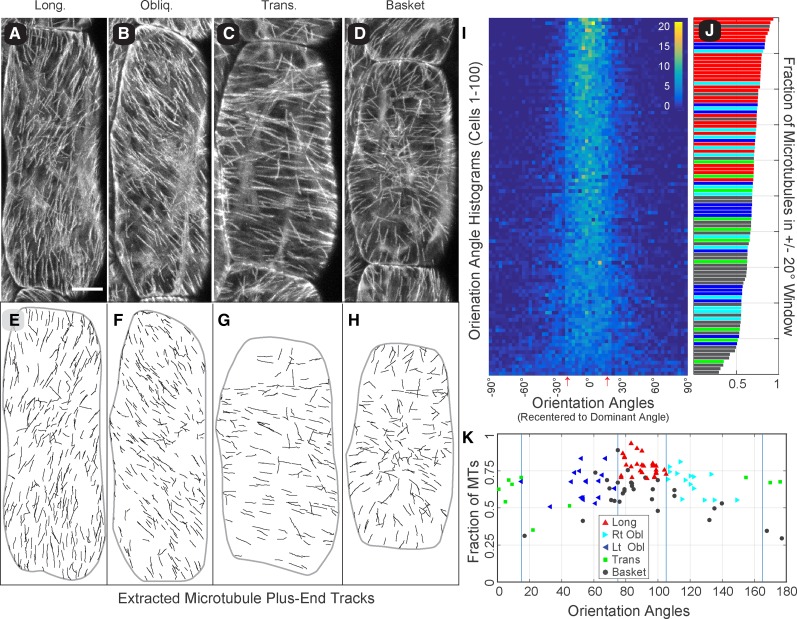
We began with a survey of 100 randomly selected hypocotyl cells from 6-d-old, light grown seedlings. Cells were imaged at 3 s intervals for 10 consecutive frames, using five to eight optical sections per interval. Array pattern was first assigned visually using “summation” images of the EB1-GFP time lapse data (Fig. 1, A–D). Cells were oriented 90° to the horizon with angles reading in the counterclockwise direction. We classified 70% of the arrays as coaligned and subdivided these cells by orientation as 10% transverse (0°–15°), 32% oblique (16°–74°; 17% left, 15% right), and 28% longitudinal (75°–90°). The remaining 30% of the arrays were classified as having no dominant microtubule coalignment (i.e. “basket” patterns), consistent with prior pattern assessments using GFP-tubulin probes (Atkinson et al., 2014; Le et al., 2005; Vineyard et al., 2013; Yu et al., 2015).
Figure 1.
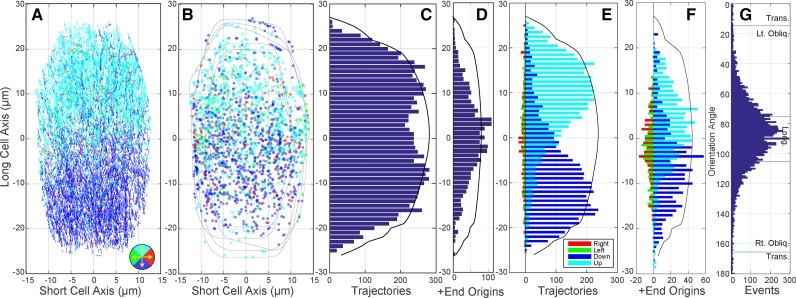
Cortical microtubule arrays in light-grown hypocotyl cells show a unimodal core of microtubule coalignment. Summation images of 3D time lapse confocal microscopy (5–8 optical sections, 3-s interval, 10 frames) showing the trajectory of growing microtubule plus ends labeled with EB1-GFP in 6-d-old Arabidopsis hypocotyl cells representing longitudinal (A), oblique (B), transverse (C), and basket (D) array patterns. Bar = 10 µm. EB1-GFP tracks extracted from each time lapse series (E–H) for quantitative evaluation of microtubule position, orientation, and growth direction. Microtubule coalignment was assayed in 100 randomly selected cells by creating histograms of the microtubule orientation angles displayed as a combined heat map (I). Each array histogram (row) is recentered on the dominant orientation angle (column 0°), and all arrays are sorted from most coaligned to least coaligned (I, top to bottom) using a ±20° sliding window. Color key indicates number of plus ends per 3° bin; red arrows indicate ±17° SD for cumulative data. The fraction of microtubules in the ±20° sliding window (J), for all 100 cells in (I), color coded for the visually determined array pattern (color key in K). The dominant microtubule orientation angle is plotted against the fraction of microtubule plus ends (MTs) found within a ±20° sliding window to evaluate the relationship between array pattern and the relative degree of microtubule coalignment (K). Markers represent the visually determined array pattern. Blue lines represent array classification boundaries.
To quantitatively examine the microtubule coalignment in these cells, we developed semiautomated software written for our imaging protocol to identify and associate individual EB1-GFP foci into short (>4 frames) tracks (Fig. 1, E–H; Supplemental Fig. S1, A and B; and see “Materials and Methods”). Since microtubule coalignment is based on the orientation of the microtubules, we converted the microtubule growth trajectory direction (0°–359°) to an orientation angle (0°–179°) with reference to the cell’s long axis (90°). A histogram of the orientation angles (1° bins) for each cell was then scanned using a sliding window to find the angular window range with the highest proportion of microtubule plus ends. We used the central angle from that window as the “dominant orientation angle” for the array and the fraction of growing microtubule plus ends within that window as a relative metric for the degree of microtubule coalignment. Surprisingly, we found that window size had little influence on the identification of a dominant orientation angle, where windows between 10° and 80° identified approximately the same dominant angle (±4°) for a given array (Supplemental Fig. S1C).
To investigate the microtubule coalignment, we identified the dominant orientation angle in all 100 cells using a ±20° window and ranked the cells by the fraction of microtubule plus ends (i.e. orientation angles) in that window range (Fig. 1, I and J). The histogram of orientation angles for each cell (bin = 3°) was recentered to the dominant orientation angle and converted to a heat map (Fig. 1I, left to right) ordered from most coaligned to least coaligned (Fig. 1I, top to bottom). The heat map showed that nearly all cells (92 of 100), including 23 of 30 classed as basket patterns, contained a core unimodal peak of microtubules. A cumulative histogram of recentered orientation angles indicated a core angular coalignment with a sd of ±17° (Supplemental Fig. S1D; Fig. 1I, red arrows). The core microtubule coalignment was independent of array orientation with respect to the cell’s axis. The fraction of microtubules found within the ±20° window, plotted as a bar graph (Fig. 1J), showed a mixing of different pattern classes (color code indicates visually assigned pattern) with a similar degree of coalignment (Supplemental Fig. S1E). We observed no clear examples of bimodal distributions where the eight arrays (seven baskets and one transverse) showing <50% of the microtubules in the ±20° window (Fig. 1J) had no distinguishable orientation bias in the histograms.
We plotted the dominant orientation angle for each array against the fraction of microtubules found in the ±20° window (Fig. 1K) and observed a 95% concurrence (67/70 cells) between the visual pattern classification and the position of the dominant orientation angle identified in the histograms for the coaligned arrays. The left- and right-oblique arrays were notably clustered toward a longitudinal orientation with relatively few shallow oblique patterns. The majority of arrays (20 of 30) that were classed as showing no dominant coalignment (i.e. basket) clustered with the longitudinal and steeply oblique arrays and (23 of 30) had >50% of the plus ends in the ±20° window. The mean fraction of microtubules in the ±20° window for basket patterns (0.58 ± 0.13) was approximately equal to the left-oblique (0.61 ± 0.11), right-oblique (0.66 ± 0.10), and transverse (0.61 ± 0.09) patterns, with longitudinal arrays (0.77 ± 0.06) significantly (P < 0.01) higher than all other classes. These data show that a majority of arrays subjectively classed as basket patterns had the same degree of array coalignment as coaligned arrays and clustered with the same orientation angles.
From these observations, using single microtubule tracks, we find that >90% of the hypocotyl cells at this developmental stage create a single unimodal core of microtubules that are coaligned within a 30° to 40° range, independent of orientation or array pattern.
The Core Coalignment of Microtubules Has a Split Bipolar Architecture
Prior studies have shown that cortical microtubules can appear as local cohorts, coordinated for the direction of polymerization (Ambrose and Wasteneys, 2014; Chan et al., 2007; Dixit et al., 2006; Sambade et al., 2012; Vineyard et al., 2013). How these local arrangements of treadmilling polymers relate to the array patterns in hypocotyl cells is not known. To examine the direction of polymerization, as a function of position on the outer cell face, we created microtubule trajectory maps with color-coded EB1-GFP trajectories based on 90° quadrants representing up, down, left, and right (Fig. 2, A–F).
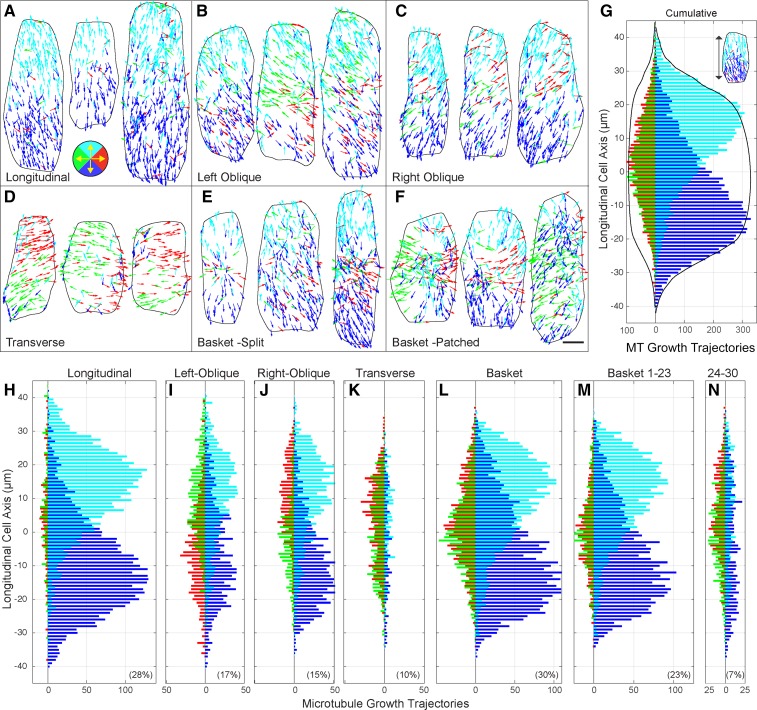
Figure 2.
Cortical microtubules exhibit a longitudinal split bipolar array architecture. Vector plots showing the color-coded growth trajectories of microtubule plus ends on the outer periclinal cell face for all array patterns; longitudinal (A), left-oblique (B), right-oblique (C), transverse (D), basket with split bipolarity (E), and basket with patched regions of organization (F). Color coding (key in A) for 90° quadrants representing up (cyan), down (blue), left (green), and right (red). Scale bar = 5 µm. Interleaved histograms showing the spatial distribution of microtubule trajectories as a function of the cell’s long axis in 90° quadrants (G–N). Cumulative histogram (G) of lateral (left side of ordinate) and longitudinal (right side of ordinate) trajectories (n = 24,369 tracks, 100 cells, 18 seedlings) showing spatial separation of apically and basally directed microtubule polymerization. Cumulative cell width (black lines) scaled for presentation. Trajectories for cells with longitudinal (H), left-oblique (I), and right-oblique (J) patterns exhibited a split bipolar arrangement not observed for transverse array patterns (K). Basket patterns (L) were separated by cells showing a split bipolarity (M) or only patches of organization (N). Percent of each pattern in the population in lower corner of frame.
The microtubule trajectory maps indicated a striking level of local coordination for the direction of microtubule polymerization. For longitudinal, oblique (left and right), and a majority of the basket-patterned cells, the apically polymerizing microtubule plus ends concentrated on the apical half of the cell face and basally polymerizing plus ends on the basal half (Fig. 2, A–C and E). Transverse coaligned patterns showed regions across the cell face where the microtubule plus ends grew in the same direction but did not show a consistent positional bias for microtubule growth direction or an obvious separation of lateral trajectories between left and right sides of the cell face (Fig. 2D). Basket arrays exhibiting >50% of the microtubules in a ±20° window exhibited the same spatial segregation of apically and basally directed plus ends (Fig. 2E). Examination of basket patterns with <50% of polymers in the window range had regions of the cell face with microtubules polymerizing in the same direction, but no consistent pattern was observed when comparing cells (Fig. 2F).
To quantitatively investigate this array architecture, we centered and oriented each cell by its perimeter and determined the polymerization direction of the microtubule plus ends as a function of the cell’s long axis. The starting positions of all EB1-GFP tracks from each pattern class were pooled into 1 µm linear bins where 90° quadrants were used to separate the growth directions. The data are presented as interleaved histograms with the same color coding as the trajectory maps, left/right (green/red) trajectories to the left of the ordinate and up/down (cyan/blue) trajectories to the right (Fig. 2, G–N). A cumulative histogram for all 100 cells (Fig. 2G) showed the principle architectural features of the cortical microtubule arrays in these light-grown, 6-d-old hypocotyl cells. The apically and basally polymerizing microtubules formed two populations (Figure 2, cyan/blue), distinctly offset (P ≤ 0.00001) or split by growth trajectory as a bipolar distribution across the cell face. This “longitudinal split bipolarity” was symmetric across the cell’s long axis and the apical and basal cell faces contributed no more than 1% (110/9,982 from apical and 64/8,596 from basal face) of the microtubules to the outer periclinal cell face, based on plus end counts from the apical/basal ends of the cell face. Transversely oriented microtubules were concentrated toward the middle of the cell but showed no left or right trajectory bias for the upper or lower cell face when examined over all pattern types. The accumulated cell width (Fig. 2, black traces) indicated that the tapering distribution at the cell’s apex and base arose because of different cell lengths being summed together rather than a decrease in plus end density. The cumulative distribution of microtubule plus end directions, normalized for cell length or plotted as distance from the respective cell edges (Supplemental Fig. S2, A–N), support these observations and show a beta distribution for the apically and basally polymerizing plus end distributions.
The longitudinal split bipolarity was a conserved architectural feature for all pattern classes, with the exception of transverse patterns and a subset of basket arrays (Fig. 2, H–N). The accumulated trajectories from visually classed longitudinal arrays showed a nearly mirrored distribution of oppositely growing plus ends between the upper and lower cell face (Fig. 2H). Left and right-oblique patterns shared the same split bipolar arrangement of longitudinal and steeply oblique microtubules (Fig. 2, I and J). Interestingly, the oblique arrays showed biases for the lateral (i.e. left side of ordinate) microtubules where the right-oblique arrays (Fig. 2J) were biased for rightward polymerizing microtubules on the upper half of the cell face and leftward polymerizing microtubules on the lower half of the cell face. The left-oblique array patterns (Fig. 2I) showed the opposite arrangement. This property in oblique-patterned cells corresponded to a coordinated leftward or rightward orientation of the core coaligned population of microtubules, placing some of the split bipolarity into the lateral 90° quadrants used for the directional analysis (Fig. 2, B and C).
Transverse array patterns showed no obvious spatial bias for microtubule growth direction as an average property of the arrays, relative to the long axis (Fig. 2K). The transversely oriented microtubules, growing in either direction, formed a relatively uniform distribution across the middle 75% of the cell face. We did not observe a split bipolar arrangement of the residual longitudinal polymers for the transverse pattern class, likely owing to the low number of counts for these microtubules. Transverse patterns showed some spatial bias for microtubule trajectories across the short axis of the cell when accumulated over all cells but did not exhibit the same split bipolarity observed for the longitudinal and oblique arrays (Supplemental Fig. S2, O–S).
Basket-patterned arrays exhibited approximately the same spatial distribution of microtubule trajectories as the cumulative histogram (Fig. 2L). Separating the 23 basket arrays with >50% of the microtubules coaligned in the sliding window (Fig. 2M) indicated that these cells (n = 6,067 tracks) exhibited the longitudinal split bipolar arrangement identified in the longitudinal and oblique classed arrays, but with a larger relative number of transverse microtubules appearing within a 20- to 30-µm region at the midzone. The remaining seven basket-patterned arrays (n = 2,064 tracks) did not show the core split bipolar organization of longitudinal microtubules and exhibited a broader distribution of transverse polymers across the cell face (Fig. 2N).
Observing that the split bipolar architecture of the core longitudinal coalignment was a consistent property of the nontransverse microtubule arrays, we asked if this array architecture was specific to the growth condition or developmental stage. Using the same imaging protocol, we examined 6-d-old plants grown under both 24-h light and 12-h light/dark cycles and light-grown seedlings on days one, three, and six after germination. In all cases, and independent of cell size, cell length, or aspect ratio, we observed the same split bipolar arrangement of longitudinally oriented microtubule trajectories relative to the cell’s long axis (Supplemental Fig. S3, A–I). Additionally, to determine if the longitudinal split bipolar architecture was specifically attributable to the EB1-GFP probe, we performed a time-based image subtraction of time lapse data from YFP-alpha-tubulin-5 (YFP-TuA5)-expressing cells (Lindeboom et al., 2013b) and observed the split bipolar architecture when tracking the microtubule ends across the cell face (Supplemental Fig. S3, J–N).
Gradual Transitions of Core Coalignment between Oblique and Longitudinal Patterns
Our analysis of single time points from light-grown seedlings at 6 d after germination indicated a highly structured microtubule array architecture with the majority of cells exhibiting a core of coaligned microtubules in a split bipolarized organization. Based on these observations, we wanted to determine how the cortical array transitions between the observed array patterns. We performed time-course studies on individual epidermal cells from the middle region of 6-d-old, light-grown hypocotyls. Microtubule treadmilling results in the apparent movement of polymers at 1 to 2 µm min−1, defined by the minus-end depolymerization rate (Shaw et al., 2003; Shaw and Lucas, 2011). We therefore imaged the arrays every 10 to 12 min for >2 h, using our previously implemented protocol (Fig. 3A), with the expectation that microtubules in any given 10 µm by 10 µm area will be “new” at each time interval. To quantify array changes over time, we tracked the EB1-GFP plus ends and used the sliding window (±20°) to define the dominant array orientation angle.
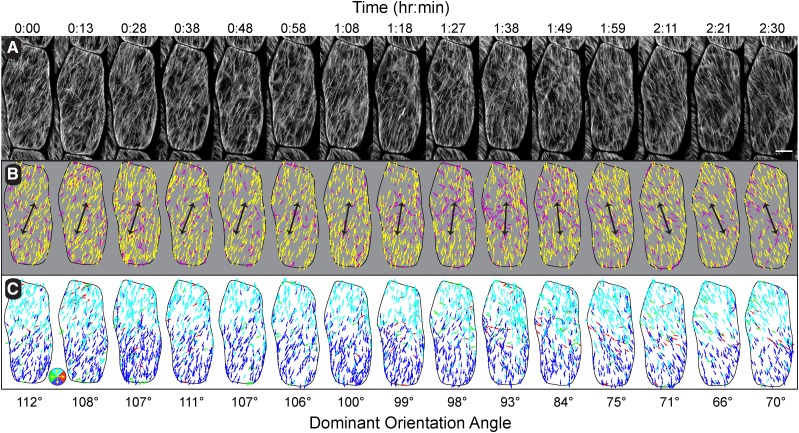
Figure 3.
Time course observations of array pattern changes. Summation images of EB1-GFP (10 frames at 3-s intervals) taken every 10 to 12 min for 2.5 h (A). Color-coding for the dominant array orientation angle (yellow) using a ±20° window (B) indicates a gradual shift in array pattern coordinated across the cell face. Black arrows indicate orientation angle in B. Trajectory maps of microtubule growth direction color-coded for up (cyan), down (blue), left (green), and right (red) in 90° quadrants indicating a sustained bias for the direction of microtubule polymerization across that long axis of the cell (C). Bar = 5 µm.
Orientation maps for a single representative cell over time showed the tracked microtubule plus ends within the ±20° window in yellow and microtubules outside of the dominant coalignment in magenta (Fig. 3B). Arrows on the orientation maps, indicating the dominant orientation angle, showed that the pattern shifts from right-oblique to longitudinal and then to left-oblique over the 2.5 h time course. The microtubule orientation was coordinated across the cell face, with polymers on the upper half appearing in a more leftward orientation and polymers on the lower half in a more rightward orientation. Microtubules appearing outside of the dominant coalignment did not show an obvious bias for position over this time course.
Evaluating the direction of microtubule polymerization (Fig. 3C) showed a clear maintenance and coordination of the split bipolar architecture while gradually transitioning between longitudinal and oblique patterns. While shifting orientation in a counterclockwise manner, new microtubules on the apical half of the cell face polymerized apically and more leftward, while polymers on the basal half of the cell face were directed basally and more rightward. The coordinated shift in pattern maintained an antiparallel region of microtubules across the entire width of the midzone and occurred without obvious reference to a centralized spot or hub on the cell face.
To quantitatively evaluate the time course data, we created histograms of the axial orientation angles for the cell at each time point, displayed as a heat map (Fig. 4A′). The core coalignment of microtubules shown in the heat map (Fig. 4A′) transitioned from right-oblique to longitudinal and to left-oblique (Fig. 3B) in a gradual manner, with the microtubule coalignment (i.e. peak width) remaining approximately constant in agreement with data from the fixed time point study (Fig. 1I). We additionally plotted the fraction of coaligned microtubules in a ±20° window over the dominant orientation angle (Fig. 4A′′). The relative degree of array coalignment changed over this time course (Fig. 4A′′) and covered the range previously defined in the fixed time point study for the majority of cells (Fig. 1K). We noted that as this cell transitioned from right- to left-oblique, the fraction of plus ends within the ±20° window remained in the range of the visually classed oblique and basket patterns from the fixed time point study and did not fall below 50%.
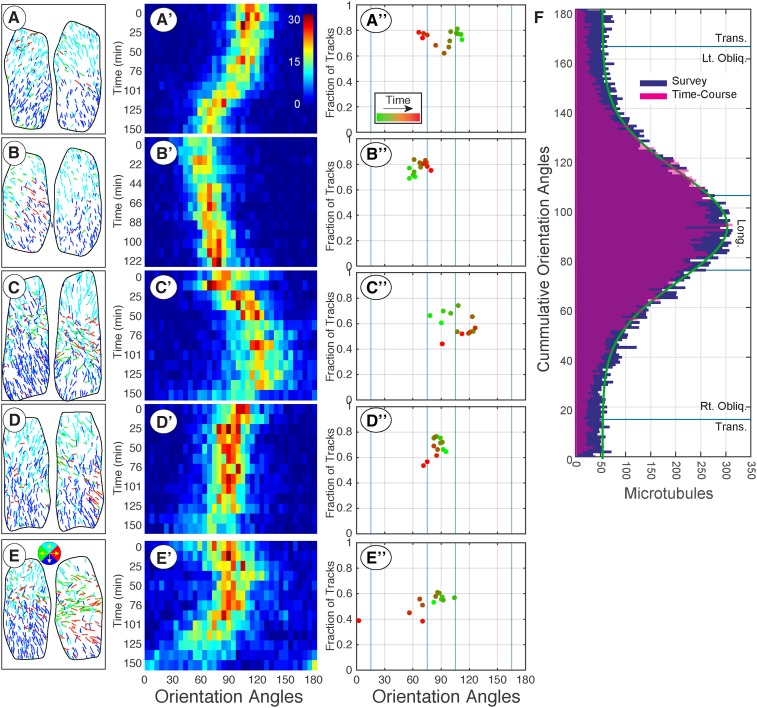
Figure 4.
Quantitative evaluation of time course data shows gradual and sustained shifting between oblique and longitudinal array patterns. Time course data for five cells imaged at 10- to 12-min intervals for >2 h Microtubule trajectory maps for the first and last time point of each series (A–E) with color coding for up, down, left, and right (key in E). The distribution of microtubule orientation angles (5° bins) for each time point assembled into a heat map (A′–E′) with time running from top to bottom. The heat maps show a gradual shifting of the dominant polymer orientation without a substantial broadening of the microtubule coalignment. The dominant orientation angle was plotted against the fraction of coaligned microtubules using a ±20° window for each cell and color coded green-to-red (A′′–E′′) to show the temporal sequence. The cumulative distribution of all microtubule orientation angles from the 100 cell fixed time point data (blue) plotted with the cumulative distribution of all orientation angles from the five time course experiments (F). Blue lines represent array classification boundaries and green trace represents Gaussian fit to fixed time point data set.
Time course data sets for four additional cells, including the first and last trajectory maps (Fig. 4, B–E), showed similar properties. The core coalignment continuously and gradually reoriented in leftward (Fig. 4, A and D) or rightward (Fig. 4, B andC) directions over the 2.5-h time course with evidence of switching between directions (Fig. 4E). The average change in angle was 6.4° ± 4.9° per 10- to 12-min interval (n = 5 cells, 63 time points) with a mean axial orientation angle of 86° ± 21° (n = 5 cells, 68 time points). While the arrays continuously reoriented, we did not observe array patterns to “rotate” past ±30° from longitudinal into shallow oblique or transverse patterns. We observed only one instance of the array becoming transverse (Fig. 4E; 150 min) in 13 h of total recorded time, and this case did not involve a continuous rotation of the core microtubule coalignment. Arrays shifting between 60° and 120° (i.e. ±30° from longitudinal at 90°) did not change orientation at a constant rate, though we estimate 90 to 120 min for a complete 60° reorientation. We found no cases where the array orientation remained temporally fixed for more than two frames (12 min).
The time-course data showed that the array patterns gradually and continuously change between steeply oblique and longitudinal patterns, maintaining the longitudinal split bipolar architecture over several hours. We hypothesized that this action would potentially explain the distribution of steeply oblique and longitudinal patterns, including the majority (23/30) of basket patterns, observed in the fixed time-point study (Fig. 1K). To test this hypothesis, we compared the accumulated orientation angles from our fixed time-point study (n = 24,369 tracks, 100 cells, 18 seedlings) to the accumulated orientation angles from the five time-course studies (n = 5 cells, 68 time points, 20,255 trajectories; Fig. 4F). The data from the fixed time point study was dominated by the longitudinal and steeply oblique patterns where the angles formed a single peak, centered at 90° (longitudinal), that was normally distributed above a baseline value (µ = 90°, σ = 21°, offset = 51; Figure 1K, green trace). The tails (0°–35° and 145°–180°) formed a uniform distribution (Supplemental Fig. S1, E and F). The polymers were 34.08% longitudinal, 31.59% left-oblique, 26.24% right-oblique, and 8.18% transverse. The time-course orientation angles formed a nearly identical distribution, with a peak centered at 87.4° and a Gaussian distribution through the tails (σ = 23°) owing to near absence of transverse array patterns. The polymers were 37.10% longitudinal, 35.04% left-oblique, 24.32% right-oblique, and 3.46% transverse. Thus, the central core of coaligned, bipolarly directed arrays gradually and constantly shifts orientation between ±30° from longitudinal to form the majority of nontransverse patterns at this developmental stage.
Longitudinal Patterns Are Created by Microtubules Originating on the Outer Cell Face
Collectively, our data show that the outer periclinal face of these light-grown hypocotyl cells forms patterns from two principle architectures. Between 80% and 90% of the arrays exhibited an apical/basal split bipolar arrangement, with the remaining arrays forming transverse patterns or potential transition states with local regions of polymers growing in the same direction. Because so few microtubules were observed to enter the longitudinal arrays from the apical or basal end walls (i.e. <1%), we hypothesized that the majority of the longitudinal microtubules originated from positions on the outer periclinal cell face. Prior work has shown that microtubules are nucleated from γ-TuRCs on the outer periclinal cell face (Liu et al., 1994; Murata et al., 2005), but microtubules can also be contributed from the nuclear surface (Ambrose and Wasteneys, 2014), the lateral anticlinal cell faces (Sambade et al., 2012; Vineyard et al., 2013), and through amplification of polymers using a KATANIN-based severing mechanism (Lindeboom et al., 2013b).
To determine the origin of the microtubules that are giving rise to the longitudinal and steeply oblique arrays, we extended our imaging protocol to 100 frames at 3-s intervals (Fig. 5A; n = 5 cells × 100 frames each, 10,351 trajectories) and identified, by hand, the position and direction of all newly created EB1-GFP-labeled microtubule ends (Fig. 5B; n = 2,139 origins). These data represent the number, position, and direction of the combined microtubule nucleation events, microtubule rescue events, and any microtubules coming from the cell’s lateral or axial side faces. We then asked if the events we counted could plausibly account for the steady-state longitudinal array pattern on the outer periclinal cell face.
Figure 5.
Newly appearing EB1-GFP foci are at a higher density at the cell’s midzone. Trajectory map (A) for microtubule plus ends in five longitudinally oriented arrays imaged for 5 min at 3-s intervals (n = 500 frames, 5 cells, 11,512 trajectories). Color coding represents 90° bins with up (cyan), down (blue), left (green), and right (red) per legend. The positions of all newly appearing EB1-GFP foci (i.e. microtubule contributions from nucleation, rescue, and internal cell positions) were identified by hand (n = 2,139 events) and plotted with the same color coding for direction (B). Black traces represent cell perimeters. Histograms representing the total trajectory count (C) and nascent EB1-GFP events (D) as a function of the cells’ long axes (bin = 1 µm). The accumulated cell width, scaled for appearance (C, black trace), indicates a relatively uniform trajectory distribution with a depression at the midzone. Interleaved histograms representing the direction of mapped trajectories (E) and nascent EB1-GFP events (F) as a function of the cells’ long axis (bin = 1 µm) indicate a split bipolar arrangement of longitudinal events for both populations. Lateral events plotted to the left of the ordinate and longitudinal events to the right for E and F with black traces indicating the accumulated cell width. The accumulated orientation angles for the five cells (G) showing a symmetric, peaked distribution centered at longitudinal (µ = 87° ± 26°).
We counted 270 ± 70 EB1-GFP foci per cell face over an average projected area of 907 ± 228 µm2, yielding a density of 0.29 ± 0.03 µm−2 or about 30 growing plus ends per 100 µm2 (n = 5 cells, 5 min each, 20 time points per min). Assuming that 65% to 75% of the microtubule plus ends were in a growth phase (i.e. EB1-GFP labeled) at steady state (Shaw et al., 2003; Shaw and Lucas, 2011; Shaw and Vineyard, 2014), we estimated a total average population of 360 microtubule ends (40 per 100 µm2) with ∼90 microtubules (10 per 100 µm2) in a shortening or pause phase not labeled with EB1-GFP. New EB1-GFP foci appeared at the cell cortex at a rate of 85.56 ± 15.15 min−1 normalized by area to 0.096 ± 0.013 µm−2 min−1 or 10 events per 100 µm2. Using prior measurements of 3.7 ± 0.94 microtubule nucleation events per 100 µm2 min−1 of cell face area (Nakamura et al., 2010), we inferred that the remaining 5 to 7 nascent EB1-GFP events were either rescue events, consistent with prior estimates of rescue in the range of 0.01 to 0.02 min−1, 50 to 100 s (Shaw et al., 2003; Shaw and Lucas, 2011), or microtubules arriving from the lateral side walls. These measurements for newly appearing EB1-GFP foci, therefore, are well within the range to account for the microtubules observed on the outer periclinal cell face.
To determine if the newly appearing EB1-GFP events occurred in a uniform spatial distribution, we compared the trajectory map for all EB1-GFP tracks (Fig. 5, A and C) with the mapped the positions of all newly appearing EB1-GFP foci (Fig. 5, B and D). The overall distribution of tracked plus ends formed a weakly bimodal distribution along the long axis of the cell (Fig. 5C), where the nascent EB1-GFP events formed a peaked distribution centered at the cell’s midzone (Fig. 5D). Comparing the direction of microtubule growth, taken in 90° quadrants (Fig. 5, E and F), both measurements produced a spatially distinct separation of apically and basally directed plus ends along the cell’s long axis. The newly appearing EB1-GFP foci formed relatively symmetric distributions of apically and basally directed polymers, offset from the cells’ midpoint, where the bulk of these foci preceded (i.e. were closer to the cell’s midpoint) the larger population of growing plus ends.
The newly appearing plus ends included a larger proportion of lateral orientations (20%; n = 420 of 2,139) than we observed for all tracked trajectories (7%; n = 728 of 10,351). These nascent lateral tracks were concentrated near the midzone and not at the apex or base of the cell face (Fig. 5F), accounting for 10% to 15% of the overall midzone bias in the density distribution (Fig. 5D). We observed that up to 8% (n = 174 of 2,139) of the nascent EB1-GFP foci were close enough to the cell periphery (≤0.5 µm) and moving inward from the sidewalls to be contributed from lateral or apical/basal cell faces. We found no evidence for short-lived (i.e. less than five frames) nascent microtubules near the apex or base of the cell face in the hand-tracked data that would suggest a mechanism of selective microtubule depolymerization (Oda and Fukuda, 2012) contributing to the split bipolarity (n = 25 min at 3 s intervals). The accumulated orientation angles for the tracked plus ends in all five cells (Fig. 5G) had a mean orientation of 87° with SD ±26°, similar to both the fixed time-point and time-course data sets.
We investigated the possibility that some fraction of the new EB1-GFP foci at the midzone arose from microtubules arriving from the interior of these highly vacuolated cells, initiating on the nuclear surface (Ambrose and Wasteneys, 2014). We imaged double transgenic lines, expressing GFP-tubulin or EB1-GFP in combination with an mCherry-Histone2b protein, and observed the nucleus near the midzone in nearly all cases. However, 3D image reconstructions revealed that the nucleus was resident at the inner periclinal cell face (n = 506 of 507 cells) and was not contributing microtubules directly to the outer periclinal cell face (Supplemental Fig. S4).
Gamma-Tubulin Complexes Are Uniformly Distributed across the Outer Cell Face
We hypothesized that the split bipolar arrangement of longitudinal microtubules occurs, in part, because of the absence of microtubules entering the array from the apical and basal cell faces. However, the higher fraction of nascent EB1-GFP labeled plus ends appearing at the cell’s midzone suggested that the split bipolarity could also be driven by a concentration of nucleation events around the cell’s midzone. To address this hypothesis, we asked if the γ-TuRCs showed a spatial bias similar to the accumulation of newly appearing EB1-GFP foci (Fig. 5D). Nucleation of microtubules from gamma-tubulin complex GCP2-GFP- and GCP3-GFP-labeled γ-TuRCs has previously been shown to strictly correlate with residency times at the cell cortex (Nakamura et al., 2010). When nucleation was not observed, γ-TuRC complexes persisted for an average of only 7.4 s and rarely remained beyond 24 s. γ-TuRCs exhibiting microtubule nucleation persisted over a broad range of times between 24 and 104 s, related to microtubule release, with a reported average of 58.9 s (Nakamura et al., 2010).
We examined the spatial distribution and residency times of γ-TuRCs on the outer periclinal cell face of GCP2 mutant seedlings, complemented with a GCP2-3xGFP transgene, and expressing mCherry-TuA5 to mark the microtubules (Lindeboom et al., 2013b). Images were taken at 6-s intervals using spinning-disk confocal microscopy (Fig. 6), accruing 15 to 18 frames before photobleaching precluded identification of individual foci. γ-TuRC markers were identified at the cell cortex in each image frame, and the number of consecutive time intervals within a 0.5-µm spatial range, and with a one-frame gap allowance, was recorded for each complex (n = 10 cells). The distribution of residency times for identified γ-TuRC foci showed a roughly exponential decrease, beyond the first time interval (Fig. 6A; n = 10 cells), comparable to previously reported distributions (Nakamura et al., 2010). We found 579 γ-TuRC foci persisting for >24 s for 10 cells over 16.5 min of imaging time on the cell cortex. Given a projected cell-face area of 995 ± 292 µm2, we observed 3.72 ± 1.13 γ-TuRC complexes per 100 µm2 min−1 with residency times predicted to yield nucleation events (>24 s), in excellent agreement with prior findings of 3.7 ± 0.94 per 100 µm2 min−1 (Nakamura et al., 2010).
Figure 6.
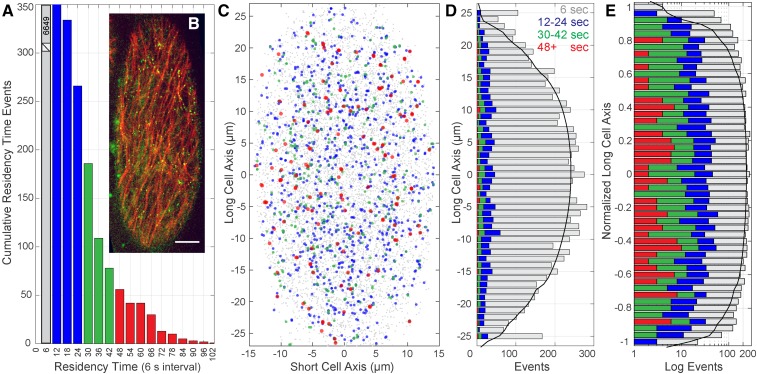
Gamma-tubulin complexes are distributed uniformly across the outer periclinal cell face. The distribution of residency times (A) for GCP2-GFP foci at the cell cortex of Arabidopsis hypocotyl cells (B) imaged every 6 s for >90 s using mCherry-TuA5 to label microtubules. Scale bar = 5 µm in (B); green is GCP2-GFP and red is mCherry-TuA5. Combined positions of all GCP2-GFP localized foci from 10 cells (n = 8,179 foci, 990 s total time) color coded for residency times (C) according to A. Histogram of the accumulated GCP2-GFP foci as a function of position on the cells’ long axis (D) color coded as in A (1-µm bins). Histogram of GCP2-GFP residency times as a function of normalized cell length (E) for 10 cells with events on a log scale to compare distributions of less frequent events. Cumulative totals for 6 s (6,649), 12 to 24 s (951), 30 to 42 s (429), and >48 s (150) for 10 cells with an average projected outer face surface area of 995 ± 292 µm2.
We examined the spatial distribution of γ-TuRCs by plotting the combined positions from 10 cells and color coding each γ-TuRC position for residency time (Fig. 6C). We observed no general pattern or concentration of γ-TuRC complexes across the cell face. Plotting the distribution of γ-TuRC positions along the cell’s long axis (n = 10 cells, 8,179 events) as a stacked bar graph indicating residency time (Fig. 6D), we found no evidence to support our hypothesis that γ-TuRC localization accounts for the increased fraction of nascent EB1-GFP foci at the cell’s midzone. Normalizing the data for cell length and plotting the number of γ-TuRCs on a log scale to visualize the distribution of infrequent events (Fig. 6E), we observed that γ-TuRCs persisting beyond 24 s were evenly distributed across the longitudinal axis, suggesting that γ-TuRC-based nucleation is not concentrated at the midzone in these hypocotyl cells.
The Longitudinal Split Bipolarity Forms Independently of KATININ Severing Activity
The microtubule severing protein, KATANIN p60, plays an outsized role in creating and maintaining the coaligned microtubule arrays in plants (Bouquin et al., 2003; Burk et al., 2001; Burk and Ye, 2002; Nakamura et al., 2010; Stoppin-Mellet et al., 2002, 2003, 2006; Zhang et al., 2013). In addition to other ascribed functions, KATANIN severs microtubules at crossover sites to increase microtubule number through an amplification mechanism (Lindeboom et al., 2013b) and releases microtubules from γ-TuRC-related nucleation sites to initiate polymer treadmilling (Nakamura et al., 2010). Our observations provide evidence that the dominant class of longitudinal, split bipolar arrays is created and maintained by microtubules that are continually made on the outer periclinal cell face. To determine if an amplification mechanism is required for the longitudinal split bipolarity and to assess the role of microtubule treadmilling in creating or maintaining the dominant array class, we crossed the EB1-GFP plus end marker into the katanin p60 null mutant and assessed the array architecture by imaging for 100 frames at 3-s intervals (Fig. 7) for comparison to wild-type plants (Fig. 5).
Figure 7.
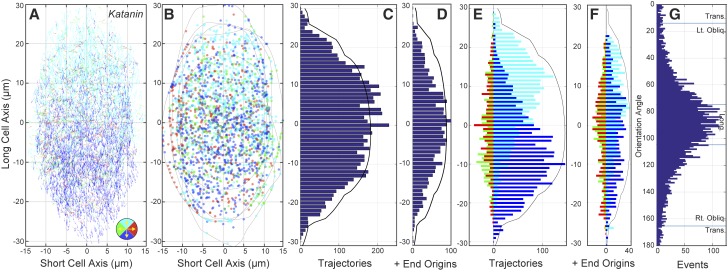
The katanin p60 mutant maintains a split bipolar array architecture. Trajectory map (A) for microtubule plus ends in five longitudinally oriented arrays from katanin p60 mutant seedlings imaged for 5 min at 3 s intervals (n = 500 frames, 5 cells, 6810 trajectories). Color coding represents 90° bins with up (cyan), down (blue), left (green), and right (red) per legend. The position of all newly appearing EB1-GFP foci (n = 2,506 events) were plotted with the same color coding for direction (B). Black traces represent cell perimeters. Histograms representing the total trajectory count (C) and nascent EB1-GFP events (D) as a function of the cells’ long axes (bin = 1 µm). Interleaved histograms representing the direction of mapped trajectories (E) and nascent EB1-GFP events (F) as a function of the cells’ long axis (bin = 1 µm) show a split bipolar arrangement of longitudinal events for both populations. The accumulated orientation angles for the five cells (G) showing a symmetric, peaked distribution centered at longitudinal (µ = 90° ± 33°).
We imaged five cells having an average projected area of 976 ± 209 µm2 and counted 214 ± 30 EB1-GFP foci per cell face, yielding a density of 0.22 ± 0.03 µm−2, or about 22 growing plus ends per 100 µm2 (n = 5 cells, 5 min each, 20 time points per min). These data indicated that while the projected mean cell-face area was slightly (8%) larger, the katanin mutant had ∼25% fewer (22 versus 30 EB1-GFP foci per 100 µm2) growing plus ends per unit area than wild type. New EB1-GFP foci appeared at the cell cortex at an average rate of 100 ± 6.6 min−1, normalized to 0.10 ± 0.07 µm−2 min−1 by area or 10 events per 100 µm2 min−1, equivalent to the wild-type measurement. We observed 345 new EB1-GFP events within 0.5 µm of the cell perimeter, with 288 polymerizing inward, suggesting that the higher ratio of nascent EB1-GFP events to total plus end density is partly accounted for by a slightly higher number of plus ends extending over the cell edges from the anticlinal sidewalls (11% katanin versus 7% wild type) in the katanin mutant.
To assess the spatial distribution and direction of plus end growth, we created trajectory maps from consecutive 10-frame sequences in the 100-frame time lapse data sets (Fig. 7A; n = 5 cells, 6,810 trajectories). We then plotted the newly appearing EB1-GFP foci in each 5-min series (Fig. 7B; n = 2506 origins) and found that the distributions were remarkably similar to that of wild-type cells (Fig. 5, A and B), showing a distinct spatial separation of the apically and basally polymerizing plus ends along the longitudinal axis of the cells. The arrays in the katanin mutant showed a uniform distribution of tracked plus ends over the cell’s long axis (Fig. 7C), where the distribution of newly appearing EB1-GFP foci formed a more peaked distribution, centered at the midzone (Fig. 7D).
Microtubules in the katanin mutant exhibited a strong apical/basal orientation bias and an unambiguous longitudinal split bipolar arrangement on the cell face (Fig. 7E). The nascent EB1-GFP foci (Fig. 7F) polymerizing in an apical or basal direction were spatially separated into distinct populations on the cell face (P < 0.001) but were visibly less resolved into independent populations when compared to wild-type cells (Fig. 5F). A cumulative histogram of the EB1-GFP orientations (Fig. 7G) showed the same Gaussian-shaped peak observed for wild-type cells (Fig. 5G), with a larger sd (±32.7° versus ±26°), indicating that KATANIN p60 has an impact on microtubule orientation in these cells but is not specifically required for setting the overall orientation of the longitudinal split bipolar array.
DISCUSSION
The Outer Face of the Hypocotyl Cell Forms Two Cortical Array Pattern Classes
The cortical microtubule cytoskeleton plays an influential role in plant cell morphogenesis by templating the deposition of cellulose into the cell wall. A substantial body of work shows that the cortical microtubules act as sites for cellulose synthase insertion into the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009) and guide the direction of synthase movement (Paredez et al., 2006). How the microtubule array patterns are specified by the cell and how the newly deposited cellulose acts to affect cell shape remain less clear (Ivakov et al., 2017; Liu et al., 2016; Lloyd, 2011; Saffer et al., 2017; Shaw, 2013). We examined the cortical array patterns in 6-d-old hypocotyl cells and found a dynamic yet highly structured underlying architecture. The majority of cells formed a continuum of longitudinal and steeply oblique patterns with a split bipolar arrangement of polymers, while a minority formed transversely coaligned patterns with patches of microtubules polymerizing in the same direction. We argue from our quantitative observations that the epidermal hypocotyl cells, at this developmental stage, form two principle microtubule array pattern classes on the outer periclinal cell face, differentiated by polymer orientation to the cell axis and the arrangement of microtubule polarity within the arrays.
The majority of cells at this developmental stage form a coaligned microtubule array pattern that constantly reorients over a 60° range, centered on the longitudinal axis of the cell. The array orientation is dynamic and nonrandom when taken over a time scale of hours. A unimodal core of coaligned microtubules (±17°) is retained while the array changes orientation, ultimately forming a broader Gaussian distribution of orientation angles (±30° from longitudinal) on a 90 to 120 min time scale. The defining feature of this pattern class is the split bipolarized arrangement of microtubules, where the dynamic plus ends extend toward the cell’s apex or base and the less dynamic minus ends form a region of antiparallel overlap around the cell’s midzone. This archetypal feature was discovered to underpin the longitudinal, right-oblique, left-oblique, and a majority (23/30) of the visually defined basket patterns, suggesting a common mechanism for creating and maintaining these apically/basally oriented arrays. Subsequent time lapse experiments confirmed that these arrays are contiguous in their construction. We, therefore, argue that for the purposes of cellulose patterning and for understanding the mechanisms creating the array patterns, these arrays constitute a singular pattern class.
Transverse microtubule patterns represented only a minority of the arrays at this developmental time point, consistent with prior studies of these cells when grown in the light (Atkinson et al., 2014; Vineyard et al., 2013). Transverse patterns exhibited the same degree of core coalignment as the other array types, but in contrast to the other pattern class, they lacked a distinct, split bipolar organization of microtubules separating leftward- and rightward-directed microtubules. The arrays had broad regions of microtubule plus ends that were polymerizing in the same direction on the upper or lower half of the cell face or as part of the left or right side of the cell face. However, unlike the longitudinal split bipolar arrangement, we found no evidence that we could predict where a transverse region would show coordinated polymerization on the cell face. The regions showed no consistent spatial pattern between cells. We found that a minority of the basket-patterned arrays (7/30) in our fixed time point study also exhibited local regions or “cohorts” of co-oriented microtubules (Chan et al., 2010, 2011), but these regions lacked a dominant polymer orientation across the cell face. We propose that transverse arrays, therefore, differ in their architecture from the longitudinal patterns and constitute a second pattern class in these cells.
Our quantitative assessment showed that microtubule arrays that were visually classified as having no dominant array coalignment, variously termed “basket,” “random,” “mixed,” or “variably coaligned” in the literature (Crowell et al., 2011; Kirik et al., 2012; Le et al., 2005; Liu et al., 2013; Ma et al., 2016; Peaucelle et al., 2015; Sambade et al., 2012; Sun et al., 2015; Vineyard et al., 2013; Yu et al., 2015), typically (23/30 cells) had an equivalent degree of microtubule coalignment to visually classed coaligned arrays. We surmise that the subjective visual inspection does not discriminate well between bundled and unbundled microtubules, overweighting the sparse transverse polymers we observed in the quantitative assessments and underweighting the bundled polymers in more longitudinal orientations. The minority of basket-patterned cells (7/30) that did not have a dominant coalignment still retained regions of local organization, suggesting they represent a transition state into or out of a transverse pattern. We propose that the visually assigned “basket” or “random” designation constitutes a valuable qualitative class when describing a population of arrays but does not denote a specific pattern class with explicit relevance to cellulose deposition.
Self-Organization and Local Control of Array Patterning
Our quantitative observations of wild type and katanin mutant seedlings provide critical insight into the mechanisms by which the longitudinal array patterns are created and maintained. Models for array patterning have come chiefly from computational simulations emphasizing self-organization (Allard et al., 2010b; Deinum et al., 2011; Eren et al., 2010) or from mutant studies (Ambrose et al., 2011; Lindeboom et al., 2013b; Sambade et al., 2012). Time course studies showing pattern changes have mainly focused on transitions into a transverse or a longitudinal coalignment and not on the broader spectrum of patterns observed in steady-state cells (Ambrose and Wasteneys, 2014; Chan et al., 2007; Dixit et al., 2006; Sambade et al., 2012; Vineyard et al., 2013). The live-cell work showing steady-state arrays has highlighted the local regions or patches of spatially coordinated polymers that are proposed to travel around the cell, overwriting or altering the existing patterns to produce new cortical arrangements (Chan et al., 2007, 2011).
Our observations, integrating array architecture with pattern, show that these steady state microtubule arrays form and maintain a unimodal core of coaligned microtubules on the outer periclinal cell face, independent of array orientation. This central architectural feature is consistent with predictions from simulation studies suggesting that coalignment is an inescapable consequence of angle-dependent bundling and biases associated with the nucleation of microtubules from the side of existing microtubules (Deinum et al., 2011; Eren et al., 2012). Consistent with this view, we observed that the gradual transitions between oblique and longitudinal array patterns occurred without obvious dissolution or dispersion of the array coalignment. Hence, we view the near ubiquitous unimodal core coalignment as evidence that self-organizing properties have a substantial general effect on array organization.
A defining feature of the core coalignment in the majority of these arrays is the split bipolar arrangement of the microtubules. Our study, examining a broad survey of cells, short-interval time lapse, long-interval time courses, a developmental series, and multiple fluorescent probes, indicates that this architectural feature is a fundamental property of the hypocotyl periclinal cell face and not an occasional or incidental feature that might arise by chance. We propose that at least part of the split bipolar architecture arises because of the near absence of microtubules entering the outer periclinal cell face from the anticlinal apical or basal cell faces, in agreement with computational simulations (Sambade et al., 2012). We found no evidence in our hand-tracked data for short-lived EB1-GFP foci near the edges of the cell face that might have revealed selective destabilization of “wrong-way” microtubules contributing to the spatial bias for microtubule growth trajectory (Oda and Fukuda, 2012). Collectively, these data indicate that when the cortical array is oriented in a steeply oblique or longitudinal pattern, the absence of microtubules coming from the endwalls will result in a natural bias for microtubule polymerization in the direction of the respective endwall.
Prior computational simulations of bipolarity have assumed that nucleation occurs more frequently at the midzone, implying a second cell-directed mechanism contributing to the split bipolarity (Sambade et al., 2012). Using direct counts from cells and several assumptions from the literature, we presented a numerical model showing that the microtubules comprising the longitudinal split bipolar array can be accounted for by microtubules nucleated on the outer cell face. We subsequently found that the γ-TuRCs on the cell face formed no obvious spatial pattern and that the residency times expected to yield nucleation were in agreement with prior measurements (Nakamura et al., 2010). These observations suggest that concentrating nucleation at the cell’s midzone, and away from the apical and basal cell face regions, is not contributing to the split bipolarity we observed. We did find that the frequency of new EB1-GFP events (representing nucleation, rescue, and microtubules from the side walls), was highest toward the middle of cell. The most parsimonious explanation for this observation is a higher local rescue frequency occurring in the midzone, where we anticipate a higher level of antiparallel microtubule overlap. A higher rescue frequency for apically and basally oriented polymers could contribute to the split bipolarity and the bias for longitudinal polymers. Alternatively, the nascent EB1-GFP events could correspond to mechanisms for microtubule nucleation outside of the conventional γ-TuRC driven assembly (Petry et al., 2013; Schatz et al., 2003).
We proposed that the general degree of microtubule coalignment and a substantial part of the split bipolarity can be explained by self-organizing mechanisms. However, we find no evidence for self-organization in the continuous production of microtubules in a longitudinal or steeply oblique orientation. Our data supports a model where the microtubules giving rise to the dominant longitudinal pattern class are nucleated on the outer periclinal cell face with a longitudinal orientation bias. The split bipolarity itself indicates that the longitudinal orientation is not being provided by microtubules coming from the anticlinal endwalls or by polymers continually treadmilling around the longitudinal circumference of the cell. We effectively ruled out direct contributions from the nucleus (Ambrose and Wasteneys, 2014), and our counting of both new EB1-GFP foci and γ-TuRC residency times supports the case for producing a steady-state population of treadmilling microtubules on the outer cell face. We additionally showed that the katanin mutant, which is ostensibly blocked for polymer treadmilling and other self-organizing behaviors (Nakamura et al., 2010; Zhang et al., 2013), still creates microtubules on the outer cell face in a predominantly longitudinal orientation with a split bipolarity. Finally, the absence of microtubule “amplification” or “pruning” mechanisms in the katanin mutant (Lindeboom et al., 2013b; Zhang et al., 2013) restricts the available mechanisms for creating the longitudinal pattern to microtubule nucleation on the outer periclinal cell face.
Based on our observations, we predict that a molecular mechanism, resident on the outer periclinal cell face, promotes nucleation of longitudinally oriented microtubules to specify the dominant apical/basal pattern class in these hypocotyl cells. Once initiated, the absence of microtubules coming from apical or basal endwalls, coupled with some amount of antiparallel nucleation across the cell face, would account for the maintenance of the split bipolar architecture. The apically and basally polymerizing microtubule plus ends each form highly skewed beta distributions across the long axis of the cell (Supplemental Fig. S2, B and C). These microtubules treadmill from their respective nucleation sites into the apical or basal cell faces, creating a constant antiparallel flux of polymer mass. We propose that antiparallel nucleation, recently documented in cotyledons (Yagi et al., 2017), is required to prevent the apically and basally oriented halves of the array from effectively treadmilling past one another and clearing the cell face of longitudinal polymers. Parallel branched nucleation (Chan et al., 2009) could further contribute to both the accumulation of oriented plus ends nearest to the endwalls and to the constant reorientation of the arrays. In sum, we propose from our observations that a cell-directed mechanism, rather than strict self-organization, specifies the initial longitudinal orientation of the microtubules and that antiparallel microtubule nucleation across the cell face is required to maintain the observed split bipolar architecture.
Implications for Cellulose Deposition and the Secretory System
The hypocotyl plays a critical role in early plant development, extending the cotyledons into the light, carefully balancing axial extension with radial expansion (Vandenbussche et al., 2005). At 6 d postgermination, when the seedlings in this study were examined, the cells are still competent for rapid axial extension (Vineyard et al., 2013). However, 6-d-old, light-grown seedlings do not show the relatively high, sustained growth rates observed in dark-grown plants or 1- to 2-d-old seedlings. The appearance of coaligned regions or cohorts of cortical microtubules moving around these cells was previously used as evidence that these nontransverse microtubule arrays could be making layers of cellulose with alternating or offsetting orientations (Chan et al., 2007; Lloyd and Chan, 2008). While multilamellar cell walls have been observed in hypocotyls, neither the mechanism of their construction nor the effect on cell morphogenesis has been directly determined (Chan et al., 2007, 2010; Refrégier et al., 2004).
We found that the majority of cells at this developmental stage were constantly and gradually shifting the main array orientation over a 60° range, centered at longitudinal. We found no cells with array patterns that rotated beyond ±30° of longitudinal to form shallow oblique patterns in 13 h of time course observation and only one instance of reorganization to a transverse coalignment. Additionally, we found that the accumulated distribution of microtubule orientations, from both the fixed time point survey and the time course data, formed a nearly identical Gaussian distribution, centered at longitudinal. Based on these observations, we predict that cellulose is being deposited in a predominantly longitudinal orientation on the outer cell face, with an offset to either side of longitudinal.
The proposal that these arrays generate independent layers of cellulose with offsetting orientations depends upon both the rate of cellulose deposition and the rate of array reorientation. Based on reported cellulose synthase velocities of 0.3 µm min−1 and persistence times measured between 3 and 10 min (Chen et al., 2010; Crowell et al., 2009; Gutierrez et al., 2009; Paredez et al., 2006), we would expect these cells to produce discrete cellulose fibers that are oriented with the average angular dispersion of the microtubule coalignment (±17°) over the persistence time of the synthase complex activity. However, if the cortical array gradually reoriented from right-oblique to left-oblique (60° to 120°) over a 2-h period (e.g. Fig. 3), we would expect the cell wall to show a continuous change in net orientation over a 60° angular domain that mirrors the reorientation of the core array coalignment. The issue of whether this mechanism creates a poly-lamellate wall would then depend upon the number of active cellulose synthase complexes over that time. The observation that the steady-state action of the cortical array appears to be dynamic, rather than static, may help to explain why many mutants in microtubule-associated proteins do not lead to dramatic changes in array pattern per se but do lead to effects on cell expansion (Lucas et al., 2011).
The plant cortical microtubule array is largely composed of bundled polymers, made in both parallel and antiparallel organizations (Lucas et al., 2011; Shaw and Lucas, 2011). The substantial amount of antiparallel microtubule organization has led to questions about how the cell could use the inherent polarity of the microtubule lattice to distribute cargo directionally within the cell. We propose that the longitudinal split bipolar arrangement of the microtubules may have implications for how plant cells use that structural polarity. Based on the average displacement of the cortical microtubules, we expect an enrichment of proteins that bridge antiparallel microtubule bundles (e.g. MAP65 family) at the cell’s midzone (Lucas et al., 2011). We further predict that trafficking of materials on microtubules could be biased toward the cell’s apex and base (Ambrose and Wasteneys, 2014), through plus-end-directed motors, and biased toward the cell’s midzone through minus-end-directed motors. In this context, this “midzone-out” polymer arrangement appears to be opposite to the arrangement of the plant phragmoplast (Murata et al., 2013) and bears a striking similarity to the radial arrays formed from centrosomes in animals and fungi.
METHODS AND MATERIALS
Plant Materials
Seed was sterilized with a 19:1 (v/v) solution of 87.5% ethanol:30% H2O2 on filter paper and dried in a sterile hood before sowing on 1% agar (Sigma-Aldrich) plates containing 0.5× Murashige and Skoog medium (Murashige and Skoog Basal Salt Mixture; Sigma-Aldrich). Plates were wrapped in metal foil and kept at 4°C for 1 to 3 d to synchronize germination. The foil was removed and plates were oriented vertically under continuous light at 22°C for 6 d prior to imaging. Wild-type Arabidopsis (Arabidopsis thaliana; Columbia-0) plants expressed a 35S-EB1b-GFP transgene for visualizing polymerizing microtubule plus ends (Mathur et al., 2003). The katanin p60 null mutant (Arabidopsis Biological Resource Center; SAIL_343_D12, CS816005) contains a T-DNA in the fifth exon of KATANIN (AT1G80350) and was crossed to plants expressing 35S-EB1b-GFP, with F3 seedlings stably expressing 35S-EB1b-GFP used to visualize polymerizing microtubule plus ends. γ-TuRC complexes were visualized along with cortical microtubules in plants expressing GCP2-3xGFP and mChy-TUA5 transgenes in homozygous gcp2-1 null background (Lindeboom et al., 2013b).
Confocal Microscopy
Hypocotyl cells were imaged using a Leica SP8 confocal laser-scanning microscope with an HC PL APO CS2 40× 1.1 NA water immersion objective lens or a Nikon A1 laser-scanning confocal microscope with a Plan Fluor 40× 1.3 NA oil immersion objective lens. GFP-based probes were excited using a 488-nm laser line, and excitation was collected using the spectrophotometric unit with hybrid detectors for the Leica SP8 and the photomultiplier tube for the Nikon A1.
Time course experiments were performed on seedlings transferred from agar plates to microscope slides containing 0.5 mL of 0.5× Murashige and Skoog medium. Seedlings were partially sealed into the liquid environment with a cover glass and silicon grease (Dow-Corning). For microtubule plus-end tracking, 3D time lapse imaging was performed with the Leica SP8 confocal laser-scanning microscope. Axial optical sections of the rounded outer periclinal face of hypocotyl cells expressing EB1-GFP were taken at 0.4 to 0.6 µm steps to create a 3D image volume that extended down to the borders of adjacent cells, typically five to eight axial sections. The process was repeated 10 times at 3- to 4-s intervals to create the primary data set for plus-end identification and tracking. Attention was given to insuring that the cell face was not in contact with the cover glass at any time. Two-dimensional projections were created by converting proprietary Leica files to TIFF format and subsequently creating a maximum projection using ImageJ (NIH, Wayne Rasband).
For microtubule array pattern distributions, we imaged hypocotyl cells expressing GFP-TUB1 to a minimum depth of 10 µm with the Nikon A1 laser-scanning confocal microscope. Hypocotyls were imaged as two to four tiled sections beginning with the root/hypocotyl junction and progressing to the apical meristem. Optical sections for reconstructed image projections were taken at 0.5- to 0.8-µm steps. Two-dimensional projections were created by converting proprietary Nikon files to TIFF format and subsequently creating a maximum projection using ImageJ (NIH, Wayne Rasband). Images of entire hypocotyls were stitched by hand using Photoshop (Adobe Software).
For GCP2-3xGFP imaging, an Olympus inverted microscope was used with a 1.3 NA 100× silicon oil objective lens. The microscope used a Yokagawa W1 spinning-disk confocal scanning unit with 50 µm pinholes and motorized switching between green and red emission windows to a Hamamatsu Flash 4 V2.4 sCMOS camera.
Image Processing and Data Analysis
MATLAB (The Mathworks) scripts were developed and implemented as a series of graphical user interfaces for assessment of EB1-GFP position, tracking, and for computing summary statistics and figures. A general workflow was established for analyzing imported 10-frame time lapse EB1-GFP data (Supplemental Fig. S1A).
(1) Cell Perimeter
Users hand-selected cell boundary points that were connected and interpolated to a 1-pixel distance to form the cell perimeter. Perimeter data, together with a micrometer-verified user-input pixels-per-micrometer conversion factor, were used for determining projected cell-face area and for establishing cell orientation. Cells were imaged with the apical and basal cell faces in known positions for cell orientation during processing.
(2) Identification
Users identified >6 EB1-GFP foci for input into an optimization routine that identified two sigma values for Gaussian functions that maximize local signal intensity over background at these positions after subtracting the Gaussian filtered images. Users then set an optimized threshold value and the EB1-GFP foci were reported as transparent marks overlain on the original time lapse images. All image sequences were subsequently reviewed and hand edited for errors. The positions and 3 × 3 mean intensities of each position were reported for assessment of polymerizing microtubule plus-end position and total density.
(3) Trajectory Mapping
The noise associated with streaming cytoplasm in the projected 3D data sets, and the bundled nature of the plant cortical microtubules precluded use of commercial or publicly available scripts for EB1-GFP tracking (Applegate et al., 2011; Yang, 2014). Based on work from the Danuser and Jaqaman Laboratories (Applegate et al., 2011), a generalized optimization algorithm was developed for discovering consecutive positions of EB1-GFP foci having the same velocity and angular trajectory. For each 10-frame image sequence, users hand tracked >3 trajectories that were assessed for input parameters. The script then identified all possible image-to-image “tracks” for all identified foci in each frame. The algorithm iteratively computed tracks of >4 time intervals with the most consistent image-to-image velocity and least deviation from a straight line trajectory. Gap filling introduced false tracks into high-density arrays and was not used. Potentially overlapping foci were used once unless hand edited to create a track. A user input “sensitivity” value set a lower bound on deviation from the two criteria (i.e. weighted sum of deviations in velocity and three-point curvature in the trajectory). The algorithm was run on the first 5 frames of the 10-frame time lapse and mapped all possible trajectories to the 10th frame in each case. The final output from the algorithm was a matrix of values showing the linked positions of each “tracked” plus end. The positions were overlain onto the raw image series for hand editing as above. The algorithm is dependent upon image data at 2- to 4-s intervals where EB1-GFP foci are adequately separated for sequential identification. Empirical tests indicated 65% to 80% of the total EB1-GFP foci were captured into tracks, independent of cell-face area, where the >4 frame limitation ensured robust identification of trajectories. The number of tracks per cell generally followed the average density of EB1-GFP foci (Supplemental Fig. S1B). Randomizing the image order results in ∼5% of the EB1-GFP foci being captured into tracks using the parameters set for the correctly ordered series. Review and hand editing of each sequence effectively produced an output exactly concurrent with hand-tracked data.
(4) Display and Analysis
Centering and rotating each data set to the cell-growth axis was performed using the cell perimeter to create a geometric center (mean of x and y) and a rotation matrix for orientation angle (from Eigenvector of largest Eigenvalue, using single value decomposition of positive definite matrix from centered perimeter values). Trajectory angle was calculated from the mean slope of each track (minimum of five points) and related to the long axis of the cell. Trajectory maps were created using the initial position of each track in the 10-frame time lapse and drawing a vector (arrow) to signify the direction and magnitude of polymerization rate.
Histograms of trajectory as a function of position on the long or short axis of the cell were created by binning all initial trajectory positions in 1-µm increments from all cells and partitioning into 90° up, down, left, and right quadrants for interleaved bar graphs. The accumulated width of all cells was calculated by collecting all cell width measures from all perimeter traces per 1-µm linear dimension and scaling to relative units for display against the histograms. Normalization for cell length and width were accomplished by creating a unit length for cells and extracting plus-end position as a fraction of the unit position (Supplemental Fig. S2). A beta function was fit to the normalized distribution of apically and basally directed plus ends using fitting tools in MATLAB. Statistical tests for the likelihood that the apically and basally directed populations constituted a separated spatial population were accomplished with a two-tailed Student’s t test in all cases.
A moving-window method was developed for estimating the dominant axial orientation angle of the microtubule trajectories and classifying array pattern. Trajectory angles were converted to axial orientation angles by centering and vertically orienting the cell by the perimeter trace and then subtracting 180° from values >180°. The new value represents the orientation of the microtubule relative to the horizon as it would be used for pattern determination, without respect to polymer polarity. The distribution of orientation angles binned to 1° units was padded, creating a continuous histogram distribution from −25° to 205°. A range of angle values was selected as a moving “window,” where the number of orientation angles within the range was summed before incrementing the position of the window by one degree and repeating the summation across the entire padded distribution. The number of orientation angles in each window was divided by the total number of orientation angles such that the window with the maximum fraction of all orientation angles tracked in a given cell could be identified. Window sizes ranging from ±5° to 45° were tested where the sensitivity to window size was minimal between 15° and 25° (Supplemental Fig. S1C).
The GCP2-GFP foci were identified using the same methods as outlined for EB1-GFP foci. Residency time was scored as the number of frames (6-s intervals) having a spot within 0.5 µm of the prior position and having a one-frame gap allowance for missed identifications. The number of events per minute beyond 24-s residency time was determined by dividing the sum of all events with residency times >24 s by the total imaging time per cell, minus one interval, and calculating the average for 10 cells.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Methods validation and microtubule co-orientation.
Supplemental Figure S2. Normalized distributions of plus-end trajectories.
Supplemental Figure S3. Split bipolar architecture observed in other developmental contexts and using YFP-TuA5 probe.
Supplemental Figure S4. Cell nuclei are resident at the inner periclinal cell face and do not directly contribute microtubules to the cortical array on the outer cell face.
Acknowledgments
The authors wish to thank the members of the Shaw Lab and the Hangarter lab for helpful comments. The work would not have been possible without Dr. James Powers and the IU Light Microscopy Imaging Center.
Footnotes
A.E. and S.L.S. wish to thank the National Science Foundation for support, including MCB:1157982 and MCB:1615907 to S.L.S. A.E, was supported during part of the project by the Quantitative Chemical Biology grant (NIH:T32 GM109825).
References
- Abe T, Hashimoto T (2005) Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J 43: 191–204 [DOI] [PubMed] [Google Scholar]
- Allard JF, Ambrose JC, Wasteneys GO, Cytrynbaum EN (2010a) A mechanochemical model explains interactions between cortical microtubules in plants. Biophys J 99: 1082–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard JF, Wasteneys GO, Cytrynbaum EN (2010b) Mechanisms of self-organization of cortical microtubules in plants revealed by computational simulations. Mol Biol Cell 21: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO (2011) A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat Commun 2: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Wasteneys GO (2014) Microtubule initiation from the nuclear surface controls cortical microtubule growth polarity and orientation in Arabidopsis thaliana. Plant Cell Physiol 55: 1636–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G (2011) plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol 176: 168–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S, Kirik A, Kirik V (2014) Microtubule array reorientation in response to hormones does not involve changes in microtubule nucleation modes at the periclinal cell surface. J Exp Bot 65: 5867–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2001) On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Bouquin T, Mattsson O, Naested H, Foster R, Mundy J (2003) The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J Cell Sci 116: 791–801 [DOI] [PubMed] [Google Scholar]
- Bringmann M, Landrein B, Schudoma C, Hamant O, Hauser MT, Persson S (2012) Cracking the elusive alignment hypothesis: the microtubule-cellulose synthase nexus unraveled. Trends Plant Sci 17: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian A, Ludynia M, Uyttewaal M, Traas J, Boudaoud A, Hamant O, Kwiatkowska D (2013) A correlative microscopy approach relates microtubule behaviour, local organ geometry, and cell growth at the Arabidopsis shoot apical meristem. J Exp Bot 64: 5753–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Ye ZH (2002) Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14: 2145–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C (2007) Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat Cell Biol 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chan J, Calder GM, Doonan JH, Lloyd CW (2003) EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat Cell Biol 5: 967–971 [DOI] [PubMed] [Google Scholar]
- Chan J, Crowell E, Eder M, Calder G, Bunnewell S, Findlay K, Vernhettes S, Höfte H, Lloyd C (2010) The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J Cell Sci 123: 3490–3495 [DOI] [PubMed] [Google Scholar]
- Chan J, Eder M, Crowell EF, Hampson J, Calder G, Lloyd C (2011) Microtubules and CESA tracks at the inner epidermal wall align independently of those on the outer wall of light-grown Arabidopsis hypocotyls.J Cell Sci 124: 1088–1094 [DOI] [PubMed] [Google Scholar]
- Chan J, Sambade A, Calder G, Lloyd C (2009) Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21: 2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR (2010) Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci USA 107: 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (1987) Wall relaxation and the driving forces for cell expansive growth. Plant Physiol 84: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Timpano H, Desprez T, Franssen-Verheijen T, Emons A-M, Höfte H, Vernhettes S (2011) Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell 23: 2592–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA (1995) Organization of cortical microtubules in plant cells. Curr Opin Cell Biol 7: 65–71 [DOI] [PubMed] [Google Scholar]
- Deinum EE, Tindemans SH, Mulder BM (2011) Taking directions: The role of microtubule-bound nucleation in the self-organization of the plant cortical array. Phys Biol 8: 056002. [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Chang E, Cyr R (2006) Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol Biol Cell 17: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Cyr R (2004) Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Shaw SL (2006) Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Eisinger WR, Kirik V, Lewis C, Ehrhardt DW, Briggs WR (2012) Quantitative changes in microtubule distribution correlate with guard cell function in Arabidopsis. Mol Plant 5: 716–725 [DOI] [PubMed] [Google Scholar]
- Emons AMC, Höfte H, Mulder BM (2007) Microtubules and cellulose microfibrils: How intimate is their relationship? Trends Plant Sci 12: 279–281 [DOI] [PubMed] [Google Scholar]
- Eren EC, Dixit R, Gautam N (2010) A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol Biol Cell 21: 2674–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren EC, Gautam N, Dixit R (2012) Computer simulation and mathematical models of the noncentrosomal plant cortical microtubule cytoskeleton. Cytoskeleton 69: 144–154 [DOI] [PubMed] [Google Scholar]
- Green PB. (1962) Mechanism for plant cellular morphogenesis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hardham AR, Gunning BE (1978) Structure of cortical microtubule arrays in plant cells. J Cell Biol 77: 14–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RJ, Tindemans SH, Mulder BM (2010) Model for the orientational ordering of the plant microtubule cortical array. Phys Rev E Stat Nonlin Soft Matter Phys 82: 011911. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM (2010) Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Newcomb EH (1964) Microtubules + fibrils in cytoplasm of coleus cells undergoing secondary wall deposition. J Cell Biol 20: 529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kaneko Y, Iwano M, Hashimoto T (2007) Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 8544–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivakov A, Flis A, Apelt F, Fünfgeld M, Scherer U, Stitt M, Kragler F, Vissenberg K, Persson S, Suslov D (2017) Cellulose synthesis and cell expansion are regulated by different mechanisms in growing Arabidopsis hypocotyls. Plant Cell 29: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V (2012) TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Hamant O (2013) How mechanical stress controls microtubule behavior and morphogenesis in plants: History, experiments and revisited theories. Plant J 75: 324–338 [DOI] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, De Cnodder T, Van Der Straeten D, Verbelen JP (2005) Cell elongation and microtubule behavior in the Arabidopsis hypocotyl: Responses to ethylene and auxin. J Plant Growth Regul 24: 166–178 [Google Scholar]
- Ledbetter MC. (1982) The role of microtubules in plant-cell wall growth. Recent Adv Phytochem 16: 125–150 [Google Scholar]
- Lindeboom JJ, Lioutas A, Deinum EE, Tindemans SH, Ehrhardt DW, Emons AM, Vos JW, Mulder BM (2013a) Cortical microtubule arrays are initiated from a nonrandom prepattern driven by atypical microtubule initiation. Plant Physiol 161: 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom JJ, Nakamura M, Hibbel A, Shundyak K, Gutierrez R, Ketelaar T, Emons AM, Mulder BM, Kirik V, Ehrhardt DW (2013b) A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 342: 1245533. [DOI] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP (1994) gamma-Tubulin in Arabidopsis: Gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 6: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Marc J, Joshi HC, Palevitz BA (1993) A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J Cell Sci 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Liu X, Qin T, Ma Q, Sun J, Liu Z, Yuan M, Mao T (2013) Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell 25: 1740–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Schneider R, Kesten C, Zhang Y, Somssich M, Zhang Y, Fernie AR, Persson S (2016) Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev Cell 38: 305–315 [DOI] [PubMed] [Google Scholar]
- Lloyd C. (2011) Dynamic microtubules and the texture of plant cell walls. Int Rev Cell Mol Biol 287: 287–329 [DOI] [PubMed] [Google Scholar]
- Lloyd C, Chan J (2008) The parallel lives of microtubules and cellulose microfibrils. Curr Opin Plant Biol 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Lloyd CW, Clayton L, Dawson PJ, Doonan JH, Hulme JS, Roberts IN, Wells B (1985) The cytoskeleton underlying side walls and cross walls in plants: molecules and macromolecular assemblies. J Cell Sci Suppl 2: 143–155 [DOI] [PubMed] [Google Scholar]
- Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL (2011) Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell 23: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Sun J, Mao T (2016) Microtubule bundling plays a role in ethylene-mediated cortical microtubule reorientation in etiolated Arabidopsis hypocotyls. J Cell Sci 129: 2043–2051 [DOI] [PubMed] [Google Scholar]
- Mathur J. (2006) Local interactions shape plant cells. Curr Opin Cell Biol 18: 40–46 [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Srinivas BP, Hülskamp M (2003) A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr Biol 13: 1991–1997 [DOI] [PubMed] [Google Scholar]
- Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, Machida Y, Hasebe M (2013) Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun 4: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M (2005) Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ehrhardt DW, Hashimoto T (2010) Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat Cell Biol 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T (2004) Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2012) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD (2013) Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refrégier G, Pelletier S, Jaillard D, Höfte H (2004) Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol 135: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer AM, Carpita NC, Irish VF (2017) Rhamnose-containing cell wall polymers suppress helical plant growth independently of microtubule orientation. Curr Biol 27: 2248–2259.e4 [DOI] [PubMed] [Google Scholar]
- Sambade A, Pratap A, Buschmann H, Morris RJ, Lloyd C (2012) The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell 24: 192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE (2003) Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J 22: 2060–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Kaloriti D (2008) Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci 13: 303–310 [DOI] [PubMed] [Google Scholar]
- Shaw SL. (2013) Reorganization of the plant cortical microtubule array. Curr Opin Plant Biol 16: 693–697 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW (2003) Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Lucas J (2011) Intrabundle microtubule dynamics in the Arabidopsis cortical array. Cytoskeleton 68: 56–67 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Vineyard L. 2014. Cortical microtubule array organization and plant cell morphogenesis. In Fukada T., ed, Plant Cell Wall Patterning and Cell Shape. Wiley, Hoboken, NJ, pp. 97–126. [Google Scholar]
- Shibaoka H. (1974) Involvement of wall microtubules in gibberellin promotion and kinetin inhibition of stem elongation. Plant Cell Physiol 15: 255–263 [Google Scholar]
- Stoppin-Mellet V, Gaillard J, Vantard M (2002) Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem J 365: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V, Gaillard J, Vantard M (2003) Plant katanin, a microtubule severing protein. Cell Biol Int 27: 279. [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet V, Gaillard J, Vantard M (2006) Katanin’s severing activity favors bundling of cortical microtubules in plants. Plant J 46: 1009–1017 [DOI] [PubMed] [Google Scholar]
- Sun J, Ma Q, Mao T (2015) Ethylene regulates the Arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiol 169: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takesue K, Shibaoka H (1999) Auxin-induced longitudinal-to-transverse reorientation of cortical microtubules in nonelongating epidermal cells of azuki bean epicotyls. Protoplasma 206: 27–30 [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T (2002) Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Verbelen JP, Van Der Straeten D (2005) Of light and length: Regulation of hypocotyl growth in Arabidopsis. BioEssays 27: 275–284 [DOI] [PubMed] [Google Scholar]
- Vineyard L, Elliott A, Dhingra S, Lucas JR, Shaw SL (2013) Progressive transverse microtubule array organization in hormone-induced Arabidopsis hypocotyl cells. Plant Cell 25: 662–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO, Ambrose JC (2009) Spatial organization of plant cortical microtubules: Close encounters of the 2D kind. Trends Cell Biol 19: 62–71 [DOI] [PubMed] [Google Scholar]
- Yagi N, Matsunaga S, Hashimoto T (2017) Insights into cortical microtubule nucleation and dynamics in Arabidopsis leaf cells. J Cell Sci 0: 1–7 [DOI] [PubMed] [Google Scholar]
- Yang G. (2014) Image-based computational tracking and analysis of spindle protein dynamics. Methods Mol Biol 1136: 57–78 [DOI] [PubMed] [Google Scholar]
- Yu J, Qiu H, Liu X, Wang M, Gao Y, Chory J, Tao Y (2015) Characterization of tub4(P287L), a β-tubulin mutant, revealed new aspects of microtubule regulation in shade. J Integr Plant Biol 57: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Warn RM, Shaw PJ, Lloyd CW (1995) Dynamic microtubules under the radial and outer tangential walls of microinjected pea epidermal cells observed by computer reconstruction. Plant J 7: 17–23 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Fishel E, Bertroche T, Dixit R (2013) Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr Biol 23: 2191–2195 [DOI] [PubMed] [Google Scholar]