Potyviruses antagonize NBR1-mediated antiviral autophagy that targets RNA granules containing the viral RNA silencing suppressor HCpro for degradation.
Abstract
Autophagy is a conserved intracellular degradation pathway and has emerged as a key mechanism of antiviral immunity in metazoans, including the selective elimination of viral components. In turn, some animal viruses are able to escape and modulate autophagy for enhanced pathogenicity. Whether host autophagic responses and viral countermeasures play similar roles in plant-virus interactions is not well understood. Here, we have identified selective autophagy as antiviral pathway during plant infection with turnip mosaic virus (TuMV), a positive-stranded RNA potyvirus. We show that the autophagy cargo receptor NBR1 suppresses viral accumulation by targeting the viral RNA silencing suppressor helper-component proteinase (HCpro), presumably in association with virus-induced RNA granules. Intriguingly, TuMV seems to antagonize NBR1-dependent autophagy during infection by the activity of distinct viral proteins, thereby limiting its antiviral capacity. We also found that NBR1-independent bulk autophagy prevents premature plant death, thus extending the lifespan of virus reservoirs and particle production. Together, our study highlights a conserved role of selective autophagy in antiviral immunity and suggests the evolvement of viral protein functions to inhibit autophagy processes, despite a potential trade-off in host survival.
Autophagy is a conserved intracellular transport and degradation pathway in which specialized double-membrane vesicles, so-called autophagosomes, deliver cytoplasmic material to lytic compartments (Klionsky and Codogno, 2013). Autophagosomes arise from expanding isolation membranes (or phagophores) that engulf autophagic substrates in a bulk or selective process, and subsequently fuse with the lysosome (in mammals) or vacuole (in yeast and plants) to release the sequestered cargo for breakdown by acid hydrolases. Initiation and completion of autophagy therefore relies on extensive membrane remodeling and trafficking events and involves the coordinated action of more than 30 autophagy-related proteins (ATGs; Mizushima et al., 2011; Lamb et al., 2013). Autophagy is constitutively active at basal levels to maintain cellular homeostasis, but can be substantially induced to promote cellular and organismal survival in various stress conditions including aging, starvation, and microbial infection (Boya et al., 2013; Klionsky and Codogno, 2013). While nonselective autophagy of cytoplasmic portions has been mainly implicated in nutrient remobilization and support of energy demand, selective autophagic mechanisms are increasingly recognized in the specific removal of superfluous or toxic cellular components (Reggiori et al., 2012). Multiple targets of selective autophagy have been identified, ranging from single and aggregated proteins to whole organelles and intracellular pathogens (Svenning and Johansen, 2013). Their recruitment to growing autophagosomes is mediated by cargo adaptor proteins like p62/SQSTM1 and NBR1 that interact with membrane-associated ATG8/LC3 through a conserved motif termed LC3-interacting region (LIR; Svenning and Johansen, 2013; Stolz et al., 2014; Zaffagnini and Martens, 2016). In addition, both proteins share an N-terminal PB1 domain for polymerization and a UBA domain at the C terminus that mediates the interaction with mono- and polyubiquitin and can be involved in cargo recognition (Svenning et al., 2011).
In metazoans, autophagy plays well-established roles in adaptive and innate immunity against viruses including the direct targeting of viral components, for example via p62/SQSTM1 (Shelly et al., 2009; Berryman et al., 2012; Judith et al., 2013). In turn, many viruses have evolved measures to antagonize antiviral autophagy or even hijack autophagic mechanisms for viral replication, subversion of immune responses, and promotion of cellular lifespan (Dong and Levine, 2013). In plants, autophagy has been linked to the regulation of defense hormone signaling and host cell death, and some pathogens were found to manipulate autophagy processes for enhanced virulence (Kabbage et al., 2013; Dagdas et al., 2016). Only very recently, functions of autophagy in virulent plant virus infection have begun to emerge. We could demonstrate that NBR1 mediates selective autophagic degradation of the viral capsid protein and particles of Cauliflower mosaic virus (CaMV) in a process referred to as xenophagy (Hafrén et al., 2017). In addition, the autophagy pathway was shown to contribute to antiviral immunity against geminiviruses (Haxim et al., 2017) and also appears to be co-opted by some viral proteins to remove plant defense components (Derrien et al., 2012; Cheng and Wang, 2016). Thus, autophagy may turn out to be similarly important in host-virus interactions in plants as observed in animals.
RNA silencing is regarded as the primary antiviral defense in plants, and successful viruses have evolved viral suppressors of RNA silencing (VSRs) to escape this resistance mechanism (Csorba et al., 2015). The absence or dysfunction of VSRs is sufficient to cause immunity to virus infection in plants, highlighting the importance of a functional RNA silencing pathway in host defense and VSR activity in viral counter-defense (Diaz-Pendon et al., 2007; Garcia-Ruiz et al., 2010). In general, RNA silencing involves the processing of dsRNA molecules by DICER-like proteins (DCLs) into virus-derived small RNAs, which are subsequently incorporated in the RNA-induced silencing complex (RISC) to guide the cleavage or translational inhibition of complementary viral RNAs. Emerging evidence indicates that VSRs are able to interfere with almost every step in the RNA silencing pathway. Intriguingly, the polerovirus VSR P0 was shown to target the key RISC component ARGONAUTE1 (AGO1) for degradation by autophagy mechanisms (Derrien et al., 2012). On the other hand, VSRs such as the potyviral helper-component proteinase (HCpro), cucumoviral 2b, and geminiviral bC1 themselves seem to be removed by autophagy as part of the plant’s counter-counterdefense (Nakahara et al., 2012; Haxim et al., 2017). Together, these findings support autophagy functions in antiviral RNA silencing in plants.
In this study, we investigated how bulk and NBR1-dependent selective autophagy pathways contribute to virus accumulation and disease development during turnip mosaic virus (TuMV) infection. TuMV is a positive-stranded RNA virus of the economically important genus Potyvirus (family Potyviridae), and a compatible pathogen of the model plant Arabidopsis (Arabidopsis thaliana). The cellular infection cycle of TuMV involves translation of a monocistronic viral RNA into at least 11 viral proteins, of which several participate in the assembly of membrane-associated viral replication complexes (VRCs; Revers and García, 2015). TuMV VRCs can concentrate, together with other membranous structures, ribosomes, electron-dense granules, and viral particles, in large perinuclear assemblies. Recently identified potyvirus-induced RNA granules (PGs) were found to locate in proximity to VRCs (Hafrén et al., 2015). Importantly, the viral silencing suppressor HCpro promotes and localizes to PGs together with several host proteins found in canonical cytoplasmic RNA granules like processing bodies (PBs) and stress granules (SGs) (Hafrén et al., 2015). Since the majority of PG components proved to be essential for the infection process, PG functions seem to be coupled to both viral translation and suppression of antiviral RNA silencing (Hafrén et al., 2015). Intriguingly, the unrelated geminivirus protein BV1 activates a PB component to weaken RNA silencing (Ye et al., 2015), suggesting a more general role of PBs in the control of antiviral RNA silencing in plants. However, similar to autophagy, the knowledge of RNA granule function during plant virus infection is still in its infancy compared to metazoan viruses (Tsai and Lloyd, 2014).
Here, we report that NBR1 specifically targets HCpro in the context of virus-induced RNA granules to suppress TuMV accumulation. The antiviral capacity of the NBR1-dependent autophagy pathway, however, seems to be antagonized by TuMV proteins during infection. Finally, we show that NBR1-independent autophagy prevents premature cell death and extends the time-span of virus production, suggesting a potential epidemiological trade-off between viral interference with autophagy and host survival.
RESULTS
Autophagy Promotes Plant Fitness during TuMV Infection
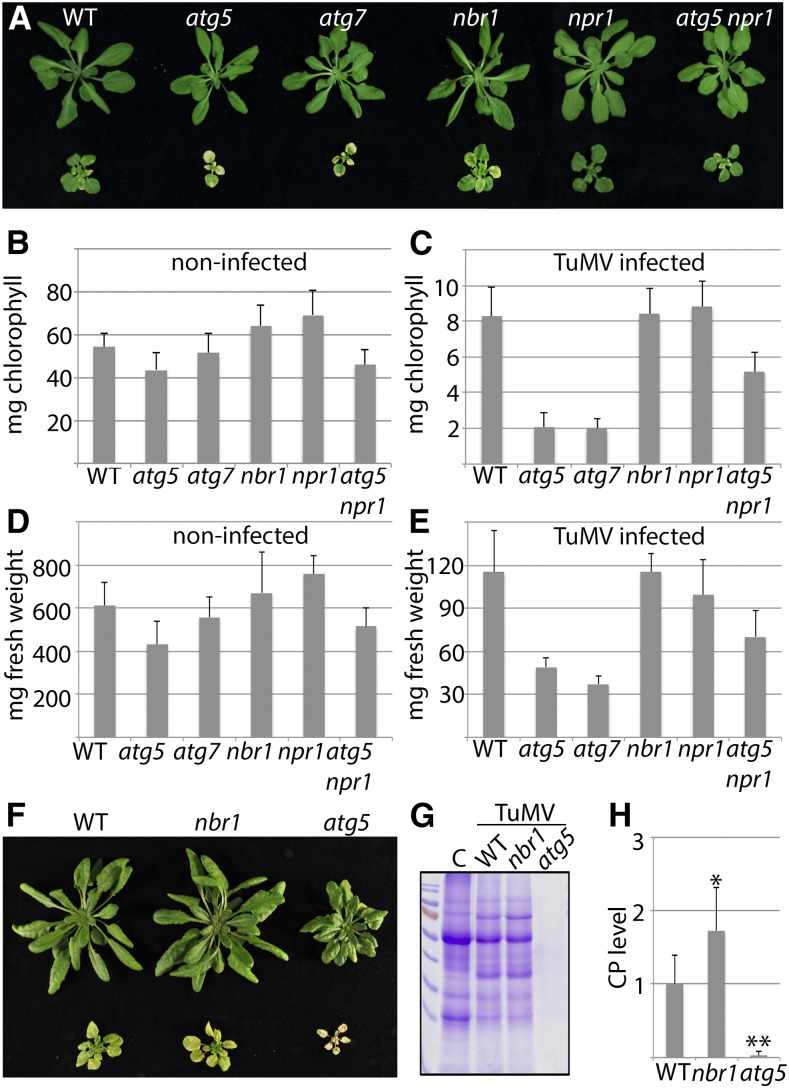
To evaluate whether autophagy plays a role in disease development during TuMV infection, we first examined symptom severity in Arabidopsis loss-of-function mutants of the core autophagy genes ATG5 and ATG7 and the autophagy cargo receptor NBR1. Twenty-one days after inoculation (DAI) with TuMV, we observed stronger disease symptoms including severe stunting, accelerated leaf senescence, and tissue necrosis in atg5 and atg7 compared to Col-0 wild-type and nbr1 plants (Fig. 1A). The pronounced disease phenotype was supported by the drastically increased reduction in total chlorophyll content and fresh weight for infected atg5 and atg7 plants in comparison to wild type and nbr1 (Fig. 1, B–E).
Figure 1.
Autophagy promotes plant fitness during TuMV infection. A, Virus-induced symptoms in wild-type, atg5, atg7, nbr1, npr1, and atg5 npr1 plants at 21 DAI with TuMV (bottom) compared to noninfected controls (top). B and C, Total chlorophyll content of infected (B) and noninfected (C) plants at 21 DAI (n = 8 individual plants). D and E, Fresh weight of infected (D) and noninfected (E) plants at 21 DAI (n = 8 individual plants). F, Virus-induced symptoms in wild-type, nbr1, and atg5 plants at 28 DAI (bottom) compared to noninfected controls (top). G, Total protein analysis by SDS-PAGE and Coomassie Brilliant Blue staining in TuMV-infected wild-type, nbr1, and atg5 plants at 28 DAI compared to noninfected wild-type control (C). H, Direct ELISA detecting viral CP levels in wild-type, nbr1, and atg5 at 28 DAI (n = 6 individual plants). All values represent means ± sd. Statistical significance (*P < 0.05; **P < 0.01) was revealed by student’s t test (compared with wild type).
Autophagy-deficient mutants such as atg5 are known to display enhanced aging- and pathogen-induced senescence in association with inappropriate accumulation of ubiquitinated proteins and enhanced ER stress (Munch et al., 2014). This phenotype is promoted by elevated levels of salicylic acid (SA) and repressed by loss of the SA response regulator NONEXPRESSOR OF PR1 (NPR1) in the atg5 npr1 double mutant (Yoshimoto et al., 2009; Munch et al., 2014). Since potyvirus infection has previously been linked to ER stress and the unfolded protein response (UPR; Luan et al., 2016; Verchot, 2016), we tested whether the disease outcome in atg mutants is accelerated by NPR1. We found that TuMV-induced symptom development was clearly attenuated in atg5 npr1 compared to atg5, but still significantly stronger than in wild-type or npr1 plants (Fig. 1).
Together, these results indicate that the increased disease phenotype triggered by autophagy deficiency is partly coupled to SA-dependent NPR1 stress responses, and that autophagy promotes plant fitness and dampens symptom severity during TuMV infection independently of NBR1. Intriguingly, symptom analysis later in infection revealed complete collapse and death of atg5 plants (Fig. 1F), which was supported by the dramatic loss of protein content compared to wild-type and nbr1 plants (Fig. 1G). ELISA analysis of viral capsid protein (CP) further showed that, in contrast to wild type and nbr1, TuMV was hardly detectable in atg5 plants at this late infection stage, suggesting that autophagy-mediated plant fitness is essential for virus survival (Fig. 1, G–H).
NBR1-Dependent Autophagy Suppresses TuMV Infection
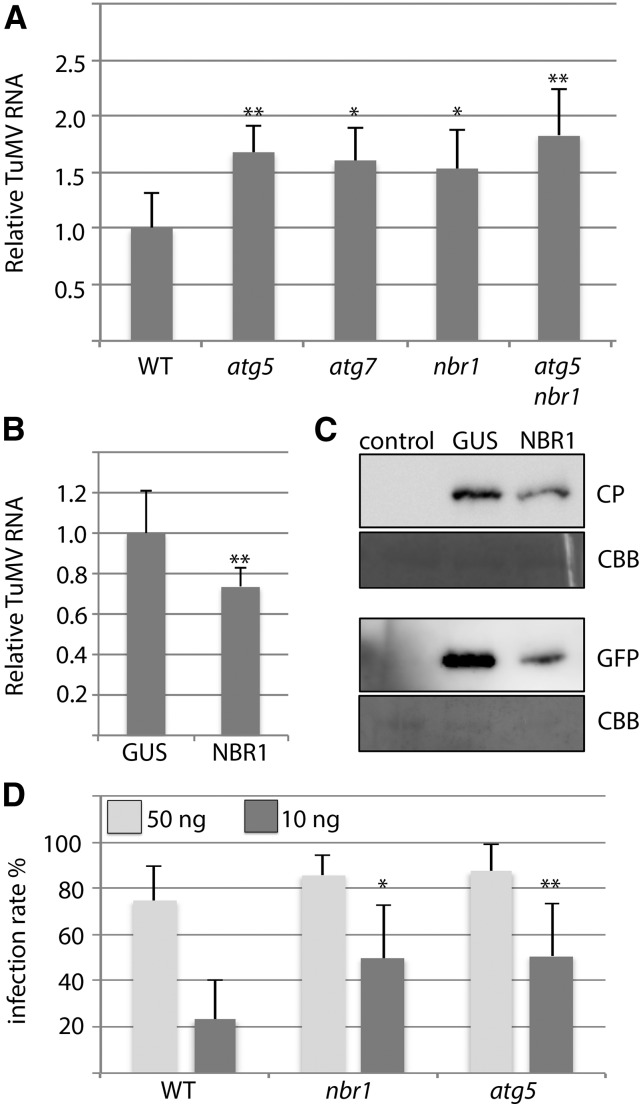
Since CP determination in nbr1 indicated significantly enhanced TuMV levels compared to wild type (Fig. 1H), we sought to investigate the impact of NBR1 and autophagy on virus accumulation in more detail. We first examined TuMV RNA levels in wild-type, atg5, atg7, and nbr1 plants at different time points of infection. Importantly, all autophagy-deficient mutants displayed increased viral RNA accumulation relative to wild type at 12 DAI, but the effect occurred with a slightly delayed kinetics in nbr1 compared to atg5 and atg7 plants (Supplemental Fig. S1). To test whether NBR1 suppressed viral titers by autophagy-dependent functions, we used an atg5 nbr1 double mutant and compared TuMV RNA levels to the single mutants. We did not detect a significant additive effect of nbr1 and atg5 mutations on virus accumulation, indicating that NBR1 restricts TuMV infection through the autophagy pathway (Fig. 2, A and B). We then verified the NBR1-mediated impact on TuMV accumulation by transient overexpression of NBR1 in Nicotiana benthamiana leaves. Agroinfiltration of an NBR1 expression construct together with an infectious TuMV clone decreased both viral RNA and protein levels in comparison to the coexpressed GUS control (Fig. 2C), further supporting the antiviral activity of NBR1. Finally, we found that nbr1 and atg5 mutant plants displayed significantly enhanced infection rates upon mechanical inoculation with reduced concentration of isolated particles (Fig. 2D), suggesting that NBR1-dependent autophagy suppresses the establishment of TuMV infection.
Figure 2.
NBR1-mediated autophagy suppresses TuMV infection. A, TuMV RNA levels determined by RT-qPCR in systemic leaves of atg5, atg7, nbr1, and atg5 nbr1 relative to wild-type plants at 10 DAI. Values represent means ± sd (n = 4 biological replicates) relative to wild type and were normalized with PP2A as internal reference. B, RT-qPCR analysis of TuMV RNA levels in response to transient over-expression of NBR1 in N. benthamiana leaves at 5 DAI relative to GUS expression (control). Values represent mean ± sd (n = 4) and were normalized to PP2a. C, Immunoblot analysis of viral protein levels (GFP and CP) in samples of NBR1 over-expressing N. benthamiana leaves used in B. Coomassie Brilliant Blue staining of the membrane verified comparable protein loading. D, Mechanical inoculation of wild-type, nbr1, and atg5 plants with 50 or 10 ng of isolated TuMV particles. The infection rates were based on symptom appearance in systemic leaves at 21 DAI and are given as mean ± sd (n = 7 independent inoculations of 18 plants per genotype). Statistical significance (**P < 0.01) was revealed by Student’s t test.
NBR1, ATG8a, and Ubiquitin Accumulate in Large Aggregate-Like Structures during Infection
The observed altered disease development and TuMV accumulation in autophagy-deficient mutants prompted us to analyze whether autophagy levels are changed during TuMV infection. We first monitored expression of the autophagic markers ATG8a and NBR1 (Zhou et al., 2013) and found that transcript and protein levels of both genes were highly increased upon TuMV challenge compared to the noninfected control, suggesting a virus-triggered autophagy response (Fig. 3, A and B). However, since the cargo adaptor NBR1 itself is a substrate of the autophagy pathway (Svenning et al., 2011), the drastic accumulation of NBR1 protein might indicate reduced degradation and thus perturbed autophagic flux during TuMV infection (Svenning et al., 2011; Munch et al., 2014, 2015). Notably, we did not observe similar accumulation of NBR1 during previous CaMV infections despite comparably elevated transcript levels (Hafrén et al., 2017). To analyze further the significance of the NBR1 degradation defects, we compared NBR1 protein levels in infected wild-type plants to those in atg5 and atg7 mutants, which are completely blocked in NBR1 delivery and turnover in the vacuole (Svenning et al., 2011; Munch et al., 2014). NBR1 accumulation was almost as strong in wild-type as in atg5 and atg7 plants during TuMV infection, supporting the notion that TuMV substantially reduces NBR1 flux (Fig. 3C). In addition, we assayed whether increased levels of NBR1 correlated with the amount of poly-ubiquitinated proteins, which are known targets of NBR1-mediated autophagic degradation (Svenning et al., 2011; Zhou et al., 2013). While all genotypes showed a marked accumulation of ubiquitinated proteins in response to TuMV, the effect was more pronounced in atg5, atg7, and nbr1 compared to wild-type plants (Fig. 3C). These results suggested that NBR1 flux is compromised during TuMV infection.
Figure 3.
TuMV infection induces large aggregate-like structures containing NBR1, ATG8, and ubiquitin. A, RT-qPCR analysis of ATG8a and NBR1 transcript levels in TuMV infected wild-type plants at 10 DAI compared to noninfected plants. Values represent mean ± sd (n = 3) relative to noninfected control and were normalized to UBQ9 and PP2a. Statistical significance (**P < 0.01) was revealed by Student’s t test. B, Immunoblot analysis of ATG8a and NBR1 protein levels in TuMV-infected and control plants at 10 DAI. Two biological replicates were analyzed and accumulation of the cell cycle protein CDC2 served as reference. C, Immunoblot analysis of NBR1 and ubiquitin (Ub) in noninfected (-) and infected (+) wild-type, atg5, atg7, and nbr1 plants. Ponceau S staining (PS) of the membrane was used as loading control. D, Localization of stably coexpressed NBR1-RFP and GFP-ATG8a in TuMV-infected transgenic Col-0 plants at 14 DAI. The images are Z-stack projections and scale bar = 20 μm. E, Localization of GFP-ubiquitin and NBR1-RFP in stably expressing transgenic Col-0 plants. Images represent single confocal plains. Scale bar = 20 μm. F, The number of GFP-ATG8a puncta was quantified from TuMV-infected and healthy Col-0 that stably expressed GFP-ATG8a at 14 DAI. Puncta were calculated from single plains of individual Z-stacks (n = 7 per condition) using ImageJ. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles and data points are plotted as white circles. G, Immunoblot analysis of NBR1 accumulation and GFP-ATG8a processing in stably expressing Col-0 plants in the absence (control) or presence of TuMV, using anti-NBR1 and anti-GFP antibodies. PS of the membrane served as loading control. The non-GFP tagged TuMV-2 isolate was used in D, E, F and G. H, Association of RFP-tagged NBR1 mutants K11A, W661A I664A, and ΔUBA2 in comparison to NBR1 wild type with virus factories (arrow) upon coexpression with TuMV-GFP in N. benthamiana. Scale bar = 10 μm. N indicates nucleus. Images represent single confocal planes.
We then analyzed the subcellular localization of NBR1 in response to TuMV challenge using transgenic plants expressing NBR1-RFP under its own promoter. In contrast to the even distribution in the vacuole of noninfected plants, NBR1-RFP accumulated to high levels and localized in large cytoplasmic aggregate-like structures during systemic infection (Supplemental Fig. S2). Imaging of GFP-ATG8a and NBR1-RFP coexpressing transgenic plants revealed a large number of GFP-ATG8a structures that partially accumulated in NBR1 aggregates during infection (Fig. 3D). Similar to NBR1 aggregates, the GFP-ATG8a-labeled punctate structures were mainly observed in response to infection (Supplemental Fig. S3). Furthermore, we also found that NBR1-RFP aggregates were colabeled with GFP-ubiquitin in coexpressing transgenic plants (Fig. 3E). Quantification of GFP-ATG8a-labeled puncta underscored their strong accumulation during TuMV infection (Fig. 3F). To test whether NBR1-independent autophagy processes are similarly impacted during TuMV infection, we applied the established GFP-ATG8 processing assay to monitor “bulk” autophagy flux (Chung et al., 2010). Interestingly, this analysis revealed an increased accumulation of free GFP, derived from autophagic turnover of GFP-ATG8 in the vacuole, in TuMV- compared to non-infected plants (Fig. 3G). Together, these findings suggest that the autophagy pathway is induced but NBR1 flux substantially reduced during TuMV infection. Notably, the observed free GFP generation from GFP-ATG8a (Fig. 3G) as well as the enhanced ubiquitin (Fig. 3C) and viral RNA (Fig. 2A) accumulation in atg5, atg7, and nbr1 mutants compared to wild type support the notion that autophagy and NBR1 flux remain, at least to some extent, functional during TuMV infection.
To further dissect which NBR1 domains are important for the TuMV-induced NBR1 localization, we analyzed previously described NBR1 mutant variants (Svenning et al., 2011) in the transient N. benthamiana system. These mutants carried a point mutation (K11A) in the PB1 domain, a double-point mutation (W661A/I664A) in the LIR-domain, or a deletion in the UBA2 domain (ΔUBA2). In marked contrast to wild-type NBR1, all mutants lost almost completely the ability to localize to the TuMV-induced perinuclear aggregate structure (Fig. 3H), indicating that self-oligomerization as well as ATG8- and ubiquitin-binding of NBR1 are required for infection-triggered redistribution. These results reinforced that NBR1 functions are coupled to autophagy and are in line with the colocalization of NBR1, ATG8a, and ubiquitin during TuMV infection.
NBR1 Targets RNA Granules Containing the Viral RNA Silencing Suppressor HCpro
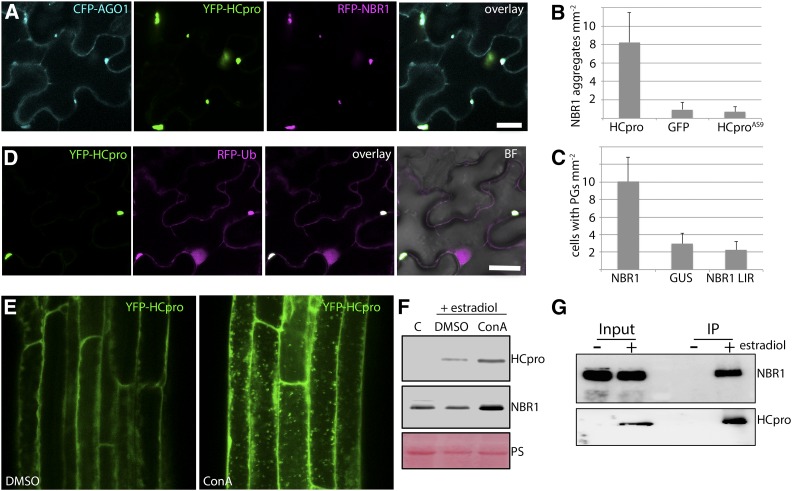
HCpro functions as potyviral suppressor of RNA silencing and was previously proposed to undergo autophagic degradation based on its partial stabilization by the autophagy inhibitor 3-MA (Nakahara et al., 2012). As HCpro turnover would be a plausible explanation for how autophagy suppresses TuMV infection, we addressed if HCpro is a direct target of NBR1. Intriguingly, transient expression of RFP-NBR1 and YFP-HCpro revealed colocalization in granule-like cytoplasmic structures (Supplemental Fig. S4), resembling those recently described as PGs (Hafrén et al., 2015). PGs were so far shown to be induced by the silencing competent HCpro and to contain RNA regulating proteins AGO1, ribosomal P0, eIF(iso)4E, UBP1, VCS, and DCP1, in addition to viral RNA. Consistent with this, we observed localization of AGO1 to HCpro and NBR1 colabeled foci (Fig. 4A) and also confirmed that expression of HCpro but not of the silencing suppression-defective HCpro mutant AS9 (HCproAS9) increased the frequency of NBR1 aggregates compared to the GUS control (Fig. 4B). Furthermore, we found that overexpression of NBR1, but not the NBR1 LIR mutant, increased the frequency of PGs (Fig. 4C), indicating that NBR1 promotes PG assembly in an ATG8-binding dependent manner.
Figure 4.
NBR1 associates with and mediates autophagic degradation of HCpro. A, Colocalization of CFP-AGO1, YFP-HCpro, and RFP-NBR1 upon transient coexpression in N. benthamiana leaves. Single confocal planes are shown together with the overlay. Scale bar = 20 μm. B, Frequency of NBR1 aggregates upon coexpression of RFP-NBR1 with HCpro, GFP, or HCproAS9 in N. benthamiana leaves. Values represent means ± sd (n = 16 independent leaf areas of 1 mm2). C, Frequency of cells with HCpro-labeled PGs upon coexpression of YFP-HCpro with control GUS, NBR1, and NBR1 (W661A, W664A) LIR mutant in N. benthamiana. Values represent means ± sd (n = 16 independent leaf areas of 1 mm2). D, Colocalization of YFP-HCpro and RFP-ubiquitin (Ub) upon coexpression in N. benthamiana 3 d after infiltration. Scale bar = 20 μm. E, Localization of YFP-HCpro in the presence or absence of ConA in estradiol-inducible transgenic Col-0 plants. β-estradiol (5 μm) induction was done together with DMSO or 0.5 μm ConA, followed by confocal imaging of the roots. YFP-labeled puncta detectable upon ConA treatment indicate HCpro accumulation in the vacuole. F, HCpro and NBR1 protein accumulation in estradiol-induced transgenic seedlings in the presence of DMSO or ConA. Immunoblot analysis was done in parallel with E and used anti-GFP antibody to monitor HCpro expression and anti-NBR1 antibody for detection of native NBR1 levels. Noninduced transgenic seedlings served as control and PS was used to indicate equal loading. G, Immunoprecipitation (IP) of estradiol-induced YFP-HCpro reveals specific association with NBR1. Immunoblots of input and IP samples from non- (-) or estradiol- (+) induced seedlings were probed with anti-GFP and anti-NBR1 antibodies as in F. Both induced and control seedlings were treated with 0.5 μm ConA to prevent autophagic degradation.
Similar to the association of NBR1 with perinuclear TuMV viroplasm (Fig. 3H), its colocalization with HCpro in PGs was highly decreased by mutations in the PB1, LIR, and UBA2 domains (Supplemental Fig. S4). Moreover, PGs were clearly labeled by ubiquitin as revealed by coexpression of YFP-HCpro and RFP-ubiquitin in N. benthamiana leaves (Fig. 4D). We therefore speculate that ubiquitination attracts NBR1 to PGs via the UBA2 domain and involves NBR1 self-oligomerization. In addition, the requirement of ATG8 binding suggested autophagosomal targeting of PGs. To test if HCpro undergoes autophagic degradation, we used a transgenic line expressing YFP-HCpro under control of an estradiol-inducible promoter and applied Concanamycin A (ConA) to block vacuolar proteolysis. Following HCpro expression, we observed multiple YFP-labeled structures reminiscent of autophagic bodies in root vacuoles of ConA-treated seedlings compared to the control (Fig. 4E). In addition, the ConA-stabilized HCpro structures were frequently colabeled with NBR1-RFP (Supplemental Fig. S5), and we also detected ConA-mediated stabilization of both NBR1 and HCpro at the protein level (Fig. 4F). Finally, NBR1 was identified in immunoprecipitates of induced HCpro protein (Fig. 4G).
Together, these results reinforced the link between HCpro and NBR1 and supported that HCpro is targeted for vacuolar degradation via NBR1-mediated autophagy.
NBR1 Suppresses TuMV Infection by Targeting HCpro
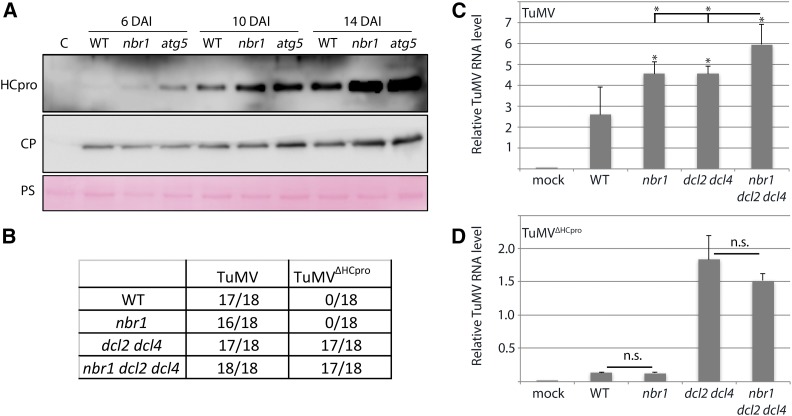
To address whether HCpro is degraded via NBR1-mediated autophagy during TuMV infection, we followed accumulation of both HCpro and CP over time in wild-type, nbr1, and atg5 plants (Fig. 5A). While CP accumulated to slightly higher levels in atg5 at 10 DAI and in nbr1 and atg5 at 14 DAI compared to wild-type plants, it remained rather unchanged in both mutants at the early time point (6 DAI). In contrast, increased accumulation of HCpro was more pronounced and occurred earlier than that of CP in nbr1 and atg5 plants relative to wild type, supporting that HCpro is degraded by NBR1-mediated autophagy during infection.
Figure 5.
NBR1 suppresses TuMV infection by targeting HCpro. A, Immunoblot analysis of HCpro and CP accumulation in wild-type, nbr1, and atg5 plants at 6, 10, and 14 DAI with TuMV. PS was used as loading control. B, Infectivity of TuMV and TuMVΔHCpro in wild-type, nbr1, dcl2 dcl4, and dcl2 dcl4 nbr1 plants after Agrobacterium-mediated inoculation. Appearance of typical TuMV disease symptoms in systemic tissue was used to score infections at 14 DAI, and the numbers of infected plants of 18 inoculated plants per treatment are indicated. C and D, Viral RNA levels determined by RT-qPCR in systemic leaves of TuMV- (C) and TuMVΔHCpro- (D) infected wild-type, nbr1, dcl2 dcl4, and dcl2 dcl4 nbr1 plants. Values represent means ± sd (n = 4) of viral RNA relative to the internal reference (PP2a). Statistical significance (*P < 0.05) was revealed by Student’s t test. n.s. = not significant.
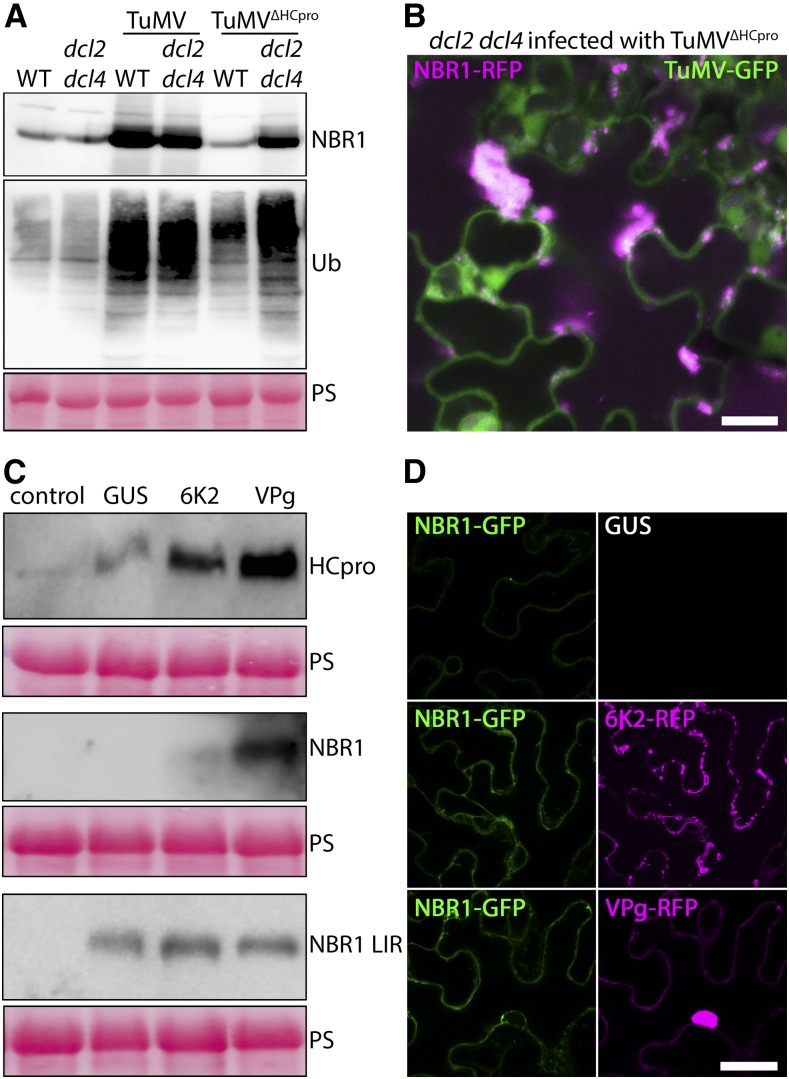
To assess whether NBR1 restricts TuMV infection in an HCpro-dependent manner, we then constructed a TuMV mutant devoid of HCpro (TuMVΔHCpro). TuMVΔHCpro did not cause any symptoms in wild type and nbr1, but remained virulent in dcl2 dcl4 plants (Fig. 5B). Infection with the TuMV wild-type strain resulted in a similar elevation of viral RNA levels in dcl2 dcl4 and nbr1 relative to wild-type plants and an additional increase in the dcl2 dcl4 nbr1 triple mutant (Fig. 5C), suggesting that NBR1-mediated suppression of TuMV infection is still operating in the dcl2 dcl4 background. In the case of TuMVΔHCpro, viral RNA accumulated to low levels in both wild-type and nbr1 plants but, as expected, was substantially increased in dcl2 dcl4 plants (Fig. 5D). RT-PCR analysis verified the absence of HCpro in TuMVΔHCpro infected tissue (Supplemental Fig. S6). Importantly, TuMVΔHCpro RNA did not accumulate to higher levels in nbr1 compared to wild type or in dcl2 dcl4 nbr1 compared to dcl2 dcl4 plants. Together, these results indicate that NBR1 limits infection via HCpro independently of DCL2- and DCL4-mediated processes. Moreover, TuMVΔHCpro RNA levels were significantly reduced in dcl2 dcl4 compared to wild-type TuMV, further supporting a proviral role of HCpro in the dcl2 dcl4 background and thus explaining the additive viral RNA accumulation in dcl2 dcl4 nbr1.
Viral VPg and 6K2 Proteins Block NBR1 and HCpro Degradation
We then aimed to identify the cause of NBR1 accumulation during TuMV infection. Notably, NBR1 and poly-ubiquitinated proteins accumulated also to high levels during TuMVΔHCpro infection of dcl2 dcl4 plants (Fig. 6A), essentially uncoupling HCpro from the response. This notion was further strengthened by the abundant aggregation of NBR1-RFP in dcl2 dcl4 upon TuMVΔHCpro infection (Fig. 6B). Previous work showed that the viral 6K2 protein interacts with the ER and early secretory pathway and induces distinct types of ER-derived vesicles required for VF formation (Wei and Wang, 2008; Wei et al., 2010). Because the 6K2-related 3A protein of enteroviruses was shown to colocalize with ATG8/LC3 upon ectopic expression (Klein and Jackson, 2011), we hypothesized that 6K2 might interfere with the autophagy pathway to block NBR1 flux. Additionally, the potyviral protein VPg was shown to promote viral RNA accumulation and translation in an HCpro- and PG-dependent manner (Hafrén et al., 2015). Thus, we addressed whether these proteins can affect NBR1 and HCpro amounts. We found that transient expression of VPg, and to a lesser extent also 6K2, increased the level of coexpressed YFP-HCpro and GFP-NBR1 (Fig. 6C). GFP-NBR1 remained below detection limit in the GUS control. Importantly, neither VPg nor 6K2 caused accumulation of the ATG8 binding-deficient GFP-NBR1 LIR mutant that is no longer degraded by autophagy (Svenning et al., 2011; Fig. 6C), suggesting that accumulation of NBR1 was caused through VPg/6K2-mediated inhibition of autophagy.
Figure 6.
Viral VPg and 6K2 proteins block NBR1 and HCpro degradation. A, Western-blot analysis of NBR1 and ubiquitin (Ub) accumulation in TuMV- and TuMVΔHCpro-infected wild-type and dcl2 dcl4 compared to the noninfected plants. PS was used as loading control. B, Localization of stably expressed NBR1-RFP in TuMVΔHCpro-infected transgenic dcl2 dcl4 plants at 14 DAI. The images are Z-stack projections and scale bar = 20 μm. C, Western-blot detection of GFP-NBR1, YFP-HCpro, and GFP-NBR1 LIR mutant after transient coexpression with GUS, 6K2-RFP, or VPg-RFP in N. benthamiana at 3 d after infiltration. PS was used as loading control. D, Confocal analysis of NBR1-GFP coexpressed with GUS, 6K2-RFP, or VPg-RFP in N. benthamiana at 3 d after infiltration. Scale bar = 20 μm.
Microscopy analysis further supported that GFP-NBR1 accumulated to higher levels upon coexpression of 6K2-RFP and VPg-RFP compared to the GUS control (Fig. 6D). However, GFP-NBR1 remained evenly distributed when coexpressed with 6K2 and VPg, indicating that these proteins alone are not sufficient to induce the NBR1-labeled aggregates observed during TuMV infection. Together, the identification of viral proteins that interfere with the NBR1-mediated degradation of HCpro reveals a high complexity in the interaction of TuMV with autophagic processes. However, the virus-induced inhibitory effect is not complete, because NBR1 still degrades HCpro to some extent during infection and reduces viral RNA accumulation via HCpro (Fig. 5).
NBR1-Mediated Autophagy Suppresses Infection of Another Potyvirus
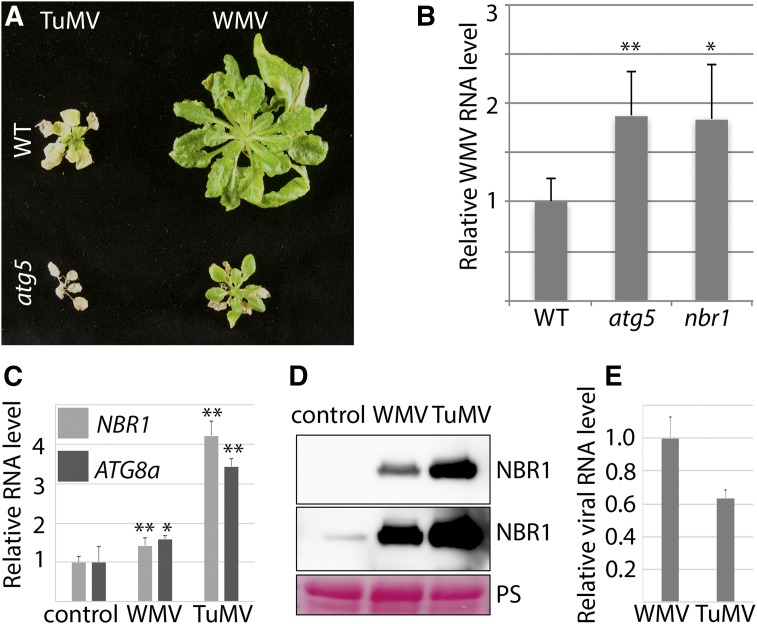
Finally, we addressed whether the effects of NBR1 and autophagy also applies to other potyviruses and thus analyzed the response to watermelon mosaic virus (WMV). Symptom severity during WMV infection was much stronger in atg5 compared to wild type, but overall milder than for TuMV (Fig. 7A). Similar to TuMV, atg5 plants developed tissue death, but its progression was considerably slower upon WMV challenge. We also found that WMV RNA accumulated to higher levels in nbr1 and atg5 plants compared to wild type (Fig. 7B). In contrast to TuMV, both NBR1 and ATG8a transcript levels were largely unaffected by WMV infection (Fig. 7C). Nevertheless, the NBR1 protein level was strongly elevated also for WMV (Fig. 7D), reinforcing that both potyviruses reduce NBR1 flux. Because viral RNAs of TuMV and WMV accumulated to comparable levels (Fig. 7E), the infection strength per se does not seem to account for the differences in symptom severity and NBR1/ATG8a transcription. We conclude that both the antiviral NBR1 response and autophagy-mediated promotion of plant survival applies also to WMV and possibly potyviruses in general.
Figure 7.
NBR1-mediated autophagy suppresses infection of watermelon mosaic virus (WMV). A, Virus-induced symptoms in wild-type (top) and atg5 (bottom) plants at 40 DAI with TuMV (left) and WMV (right). B, Accumulation of WMV RNA determined by RT-qPCR in systemic leaves of wild-type, nbr1, and atg5 plants at 20 DAI. Values represent means ± sd (n = 4) relative to wild type, and PP2a was used as internal reference. Statistical significance (*P < 0.05, **P < 0.01) was revealed by Student’s t test (compared with wild type). C, RT-qPCR analysis of ATG8a and NBR1 transcript levels in WMV- and TuMV-infected wild-type plants at 20 DAI compared to noninfected plants. Values represent mean ± sd (n = 4) relative to noninfected control and were normalized to UBQ9 and PP2a. Statistical significance (*P < 0.05, **P < 0.01) was revealed by Student’s t test (compared with control). D, NBR1 protein accumulation determined by western-blot analysis at 20 DAI in WMV- and TuMV-infected tissue. Top, shorter exposure; bottom, longer exposure. PS was used as loading control. E, RT-qPCR analysis of viral RNA levels performed in parallel with transcript (C) and protein (D) analysis. Values represent mean ± sd (n = 4) relative to noninfected control and were normalized to UBQ9 and PP2a.
DISCUSSION
A wide range of virus infections in animals has been shown to involve autophagy functions. These include roles in immune signaling, maintenance of cellular homeostasis and viability, provision of membranes for virus replication, as well as degradation of viral components in a process termed xenophagy or virophagy (Dong and Levine, 2013). The frequent capacity of animal viruses to suppress, evade, and adopt the autophagy pathway for successful pathogenesis conveys its potential antiviral roles and coevolution with infections. A similar relevance of autophagy processes for plant-virus interactions has just recently begun to emerge (Cheng and Wang, 2016; Hafrén et al., 2017; Haxim et al., 2017; Li et al., 2017).
In this study, we reveal the complex interplay of plant autophagy with TuMV, a positive-stranded RNA virus belonging to the economically important picorna-like potyviruses. Our findings suggest that (1) the autophagy cargo receptor NBR1 suppresses infection by targeting potyvirus-induced RNA granules including the viral silencing suppressor HCpro for degradation, (2) the antiviral capacity of NBR1-dependent autophagy is antagonized by viral proteins during infection, and (3) NBR1-independent bulk autophagy promotes plant fitness to the benefit of virus production and potyvirus epidemiology. We propose that the crosstalk and functions of autophagy in defense and counter-defense reflects on the coevolutionary arms race between plants and potyviruses (Fig. 8).
Figure 8.
A hypothetical model of autophagy functions in the coevolutionary arms race between potyviruses and plants. (1) Viral RNA replication triggers antiviral RNA silencing. (2) Components that benefit infection accumulate in potyvirus-induced RNA granules (Hafrén et al., 2015), including viral RNA, the stress granule components UBP1 and eIF4E, the processing body components AGO1, DCP1, and VCS, ribosomal P0, as well as the viral silencing suppressor HCpro. The composition of PGs suggests a close link to RNA quality control and RNA silencing, and many PG components were shown to be required for potyvirus infection and translation (Eskelin et al., 2011; Hafrén et al., 2013, 2015). (3) PGs are ubiquitinated and targeted by NBR1 in an attempt to eliminate proviral PG complexes via autophagic degradation. (4) This antiviral process is counteracted by a yet unidentified mechanism impairing NBR1 flux, which is mediated by the viral proteins VPg and 6K2. From the coevolutionary perspective, we propose that that this crosstalk reflects on an arms race between pathogen and host, involving successive establishment of defense and counter-defense strategies.
We identified NBR1-dependent selective autophagy to contribute to the suppression of TuMV infection. Importantly, plant infection with a TuMV mutant devoid of HCpro (TuMVΔHCpro) was no longer suppressed by NBR1, pinpointing HCpro-dependent processes as the essential antiviral autophagy target. NBR1 is regarded as a functional hybrid of the animal autophagy cargo receptors NBR1 and p62/SQSTM1 (Svenning et al., 2011; Judith et al., 2013), of which the latter has been shown to participate in antiviral immunity by mediating the degradation of capsid proteins and particles of several animal viruses (Shelly et al., 2009; Orvedahl et al., 2010, 2011; Berryman et al., 2012; Judith et al., 2013). In plants, however, specific NBR1 targets remained largely unknown beyond a general role in delivering poly-ubiquitinated substrates to autophagy (Svenning et al., 2011; Zhou et al., 2013). Only recently, we found that NBR1-mediated targeting of nonassembled capsid protein and particles is an integral part of the antiviral immune response against CaMV infection (Hafrén et al., 2017).
Previous work suggested that the host protein rgs-CaM promotes autophagic degradation of both HCpro and the unrelated VSR 2b of cucumoviruses in N. benthamiana (Nakahara et al., 2012). Consistent with targeting by the autophagy pathway, HCpro was also found to accumulate in the vacuole during TuMV infection of N. benthamiana (Wan et al., 2015). Interestingly, 2b and HCpro associate closely with AGO1 (Zhang et al., 2006; Hafrén et al., 2015), and autophagic degradation of AGO1 was reported to be triggered by the VSR P0 of poleroviruses or defects in microRNA pathways (Derrien et al., 2012). We found that NBR1 colocalized with HCpro and AGO1 in PGs, suggesting that these complexes are targeted by NBR1. Our results clearly indicate that NBR1 self-aggregation as well as ATG8- and ubiquitin-binding are essential for NBR1 localization to PGs, and thus reveal a different targeting mechanism compared to the ubiquitin-independent association of NBR1 with the viral CP of CaMV (Hafrén et al., 2017). Accordingly, we show that PGs are ubiquitinated structures that increase in number upon overexpression of NBR1. Similarly, transient expression of the PG components AGO1, ribosomal protein P0, and UBP1, but not DCP1, promoted PG assembly (Hafrén et al., 2015). However, in striking contrast to the antiviral activity of NBR1, other PG components have proviral functions during potyvirus infection including the essential susceptibility factor eIF4E/eIF(iso)4E (Eskelin et al., 2011; Hafrén et al., 2013, 2015; Moury et al., 2014). It is interesting that NBR1 requires a functional LIR domain to induce PGs formation. One intriguing possibility is that the microscopically visible PG structures arise from NBR1-dependent clustering of smaller “PG” ribonucleoprotein complexes in connection to autophagosome biogenesis. The alternative scenario that NBR1 would have a negative impact on PG degradation, for example by saturating ATG8-binding, is not supported by the observed reduction of viral titers upon NBR1 overexpression. Although the mechanisms of NBR1-driven PG assembly require further investigation, we propose that NBR1 mediates degradation of these potyvirus-induced RNA granules. PGs were recently characterized as infection-specific unconventional RNA granules, which share hallmark proteins of the usually distinct structures of PBs and SGs. PGs contain also viral RNA and HCpro and are, in contrast to PBs and SGs, insensitive to cycloheximide (Hafrén et al., 2015). A selective form of autophagy termed granulophagy was previously described in yeast, where CDC48 functions as an autophagic receptor protein for SG and PB degradation (Buchan et al., 2013). Intriguingly, autophagy-deficient yeast was reported to accumulate RNA granules colabeled by both PB and SG markers. This finding shows that PB and SG markers can coaggregate in RNA granules destined for the granulophagy pathway, and thus provides a potential explanation for why these markers colocalize in PGs (Hafrén et al., 2015). Granulophagy was also observed in mammalian cells (Seguin et al., 2014), and our results support the existence of this selective autophagy pathway in plants.
RNA silencing represents a major innate immune response by which plants defend themselves against virus infections. Its importance is highlighted by the parallel and universal evolution of RNA silencing suppressors (VRS) in unrelated plant viruses to counteract RNA silencing (Csorba et al., 2015). Given that potyviruses depend on HCpro or the complementation by unrelated VSRs for successful infection (Maliogka et al., 2012), NBR1-mediated degradation of HCpro could have devastating consequences. Nonetheless, viral titers appeared only modestly repressed by NBR1. Our data indicate that NBR1 flux is suppressed as NBR1 aggregates as well as ubiquitin and ATG8a-labeled puncta accumulate in large cytoplasmic clusters, thus limiting the antiviral capacity of NBR1. We found that in particular the potyviral proteins VPg and, yet to a lesser extent, 6K2 antagonize NBR1-mediated autophagic turnover of PG-associated HCpro. It is of most interest that VPg has been shown to increase viral RNA stability and gene expression in a mechanism that is fully dependent on HCpro and other PG components (Eskelin et al., 2011; Hafrén et al., 2013, 2015). Since high concentration of VPg increased viral translation and RNA accumulation while reducing the number of PGs, a functional interdependence between viral RNA localization to PGs and translation was proposed (Hafrén et al., 2015). According to our current hypothesis that PGs are closely linked to NBR1-mediated antiviral autophagy, we speculate that the capacity of VPg to reduce autophagic degradation contributes to the promotion of viral RNA translation associated with PG components. While the mechanism behind this response remains to be identified, it appears strikingly different from our previous observations during CaMV infection. In this case, the majority of virus-induced ATG8a and NBR1 structures became visible only after blocking vacuolar degradation with ConA, indicative of high autophagic flux rates (Hafrén et al., 2017).
Interestingly, autophagic membrane formation is induced by several enteroviruses, including poliovirus (PV), human rhinovirus (HRV), and coxsackievirus B3 (CVB3; Klein and Jackson, 2011), which could be analogous to the proliferation of GFP-ATG8a structures observed during the TuMV response. PV and HRV are believed to trigger the autophagy pathway and benefit from the generated membranes in replication. CVB3 does not share this feature with PV and HRV and autophagy was proposed to act as an antiviral mechanism against CVB3 instead. Most intriguingly, CVB3-induced autophagosomes also cluster in arge perinuclear structures termed megasomes along with p62/SQSTM1 accumulation (Kemball et al., 2010). Together, autophagy plays an antiviral role against CVB3 and TuMV and both induce autophagosome clustering along with NBR1/p62 accumulation, suggesting similarity in autophagy modulation between these picorna-like viruses.
In addition to antiviral NBR1-dependent autophagy, we show that bulk autophagy limits symptom development and prevents premature plant death. By increasing the biomass and survival of infected plants, NBR1-independent autophagy supports virus production and enhances potyvirus epidemiology. This apparent proviral role could involve attenuation of SA-dependent defense responses connected to activation of the unfolded protein response during infection (Zhang et al., 2015), as autophagy-deficient plants display hypersensitivity to SA during ER stress (Munch et al., 2014). Similarly, disruption of autophagy increased host mortality after infection with an avirulent strain of Tobacco mosaic virus (Liu et al., 2005), virulent strains of CaMV (Hafrén et al., 2017), and several animal viruses (Dong and Levine, 2013). Notably, interference with autophagy to disrupt NBR1 flux may also impact the proviral effects of bulk autophagy, thus implying a potential trade-off between suppression of antiviral autophagy and host survival.
In conclusion, we propose a model that integrates autophagy in the potyviral coevolutionary arms race with the plant host (Fig. 8). Briefly, RNA production during TuMV replication provokes the onset of RNA silencing, which is the primary line of defense and countered by the VSR HCpro including co-option of host components associated with PGs. PGs are, however, ubiquitinated and destined for NBR1-dependent autophagic degradation. This plant counter-counter-defense suppresses infection, but potyviral proteins VPg and 6K2 are able to impair NBR1 flux and HCpro degradation by a yet unidentified mechanism, thereby allowing potyviruses to largely evade antiviral autophagy.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type plants were Arabidopsis (Arabidopsis thaliana) ecotype Col-0. Loss-of-function mutants atg5-1, atg7-2, nbr1-2, npr1-1, atg5-1 npr1-1, and dcl2-1 dcl4-2, as well as the mRFP-ATG8a transgenic line, have been described previously (Deleris et al., 2006; Hofius et al., 2009; Yoshimoto et al., 2009; Zhou et al., 2013; Munch et al., 2015). Arabidopsis plants were grown on soil for infection experiments under short-day conditions (8/16-h-light/-dark cycles) in a growth cabinet, and N. benthamiana plants were cultivated for transient expression assays under long-day conditions (16/8-h-light/-dark cycles) in a growth room at 150 μE/m2s, 21°C and 70% relative humidity, respectively. Sterile plants were cultivated in vitro on half-strength Murashige and Skoog (MS) medium with a 16-h photoperiod at 150 μE/m2s and 21°C.
Plasmid Construction and Generation of Transgenic Lines
For subcellular localization, a genomic fragment of NBR1 containing 2 kb of the predicted promoter and the coding region without stop codon was amplified (see Supplemental Table S1 for primer sequences), cloned into pENTR/D-TOPO, and subsequently recombined into pAUL11 (Lyska et al., 2013) to add 3′-end sequences encoding Strep(S)-III and 3xHA tags. The NBR1 fragment containing the 3xHA and SIII extension was reamplified by PCR, cloned into pENTR/D-TOPO, and recombined into pGWB459 and pGWB559 (Nakagawa et al., 2007). The resulting pNBR1:NBR1-SIII-3xHA-tagRFP constructs were used for transformation of Col-0 wild-type and dcl2 dcl4 plants. The pENTR clones containing the coding sequences of NBR1 wild-type, K11A, UBA2, and LIR domain mutants were described previously (Svenning et al., 2011) and used for recombination into the binary vector pUBN-DEST-mRFP (Grefen et al., 2010) and pGWB606 (Nakagawa et al., 2007) for transient expressions. VPg and 6K2 of TuMV were cloned into pENTR/D-TOPO and further recombined into pGWB454 for transient expression of mRFP-fusions. HCpro was deleted from the TuMV genome by PCR mutagenesis on the NcoI/AgeI fragment of pCB-TuMV-GFP (accession EF028235) to create TuMVΔHCpro. HCpro of TuMV was cloned into pENTR/D-TOPO and further recombined into pGWB442 for transient expression of YFP-HCpro. The YFP-HCpro fusion was amplified by PCR, cloned into pENTR/D-TOPO, recombined into pMDC160, and transformed to the Col-0 XVE driver line (Brand et al., 2006) to generate estradiol-inducible YFP-HCpro transgenic plants. The CFP-AGO1 and YFP-HCproAS9 expression constructs were described previously (Hafrén et al., 2015). Ubiquitin was amplified by PCR, cloned into pENTR/D-TOPO, and recombined into pUBN-DEST-GFP to obtain GFP-Ub. All binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101 for transient expression of N. benthamiana at the four- to six-leaf stage or stable Arabidopsis transformation using the floral dip method (Clough and Bent, 1998). Transgenic lines coexpressing two markers were obtained by crossing and F1 plants that were used for the experiments.
Transient Expression in N. benthamiana by Agrobacterium
Agrobacteria were grown on liquid LB-media with antibiotics and harvested by centrifugation at 3,000g for 5 min. Bacteria were washed once in ddH2O; resuspended in 10 mm MES pH 5.7, 10 mm MgCl2, and 150 μm acetosyringone; and incubated for 2 h standing on the bench. Optical densities at 600 nm were used to adjust the amount of bacteria to be equivalent between different cultures before mixing and infiltration to N. benthamiana leaves using a needleless syringe.
TuMV Inoculation, ELISA, and RNA Quantification
Viral particles purified from TuMV isolate 2 (Richter et al., 1994; pathotype 4, typed by Jenner and Walsh, HRI Wellesbourne, UK), TuMV-GFP (Garcia-Ruiz et al., 2010) or WMV-GFP-infected N. benthamiana plants were mechanically rubbed on the first true leaves of 3-week-old Arabidopsis plants using a sponge and spatula with carborundum as abrasive. For susceptibility testing, only a single leaf per plant was inoculated with different amounts of particles, and infected plants were identified by visual inspection of typical disease symptoms and the presence of TuMV-expressed GFP in upper, noninoculated leaves 3 weeks after inoculation. In experiments including TuMVΔHCpro-GFP, all viruses including TuMV-GFP were agroinoculated.
Direct ELISA was done essentially as described (Hafrén et al., 2017). Briefly, TuMV-infected individual wild-type, nbr1, and atg5 plants were harvested and assayed for the relative amount of viral CP using anti-CP antibodies (n = 6 individual plants per genotype). A serial dilution of the same infected plant extracts served as standard curve for quantitation and verified that quantitative detection was in the linear range.
For RT-qPCR, plants were sampled in biological replicates, each containing three individual plants from which inoculated leaves were removed. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen), and on-column DNA digestion was performed with DNase I (Qiagen). First-strand cDNA was synthesized from 1 μg of total RNA using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR analysis was done with Maxima SYBR Green/Fluorescein qPCR Master Mix (Thermo Fisher Scientific) using the CFX Connect Real-Time PCR detection system (Bio-Rad) with gene-specific primers listed in Supplemental Table S1. Normalization was done using PP2A (AT1G69960) and UBQ9 (AT5G37640).
Confocal Microscopy
Live cell images were acquired from abaxial leaf epidermal cells using a Zeiss LSM 780 microscope. Excitation/detection parameters for GFP and RFP were 488 nm/490 to 552 nm and 561 nm/569 to 652 nm, respectively, and the sequential scanning mode was used for covisualization of both fluorophores. Inhibitor treatment was carried out by syringe-infiltration of (mature leaves) or incubation (seedlings) in 0.5 μm concanamycin A (ConA) in 1/2 Murashige and Skoog medium 14 h before confocal analysis. Confocal images were processed with ZEN (version 2011) and Image J (version 1.48v) software. Quantification of GFP-ATG8a puncta was done on Z-stacks comprised of 10 plains and acquired at 2-μm intervals from the epidermal cell layer. Single plain images (100 μm x 100 μm) were converted to eight-bit grayscale and then counted for GFP-ATG8a puncta either manually or by the Particle Analyzer function of ImageJ.
Immunoprecipitation
Ten-d-old XVE:YFP-HCpro transgenic seedlings were incubated on liquid 1/2 Murashige and Skoog containing 0.5 μm ConA either with or without 20 μm estradiol for inducing YFP-HCpro expression. Seedlings were homogenized in 5 mL buffer [100 mm Tris pH8, 150 mm NaCl, 1% Triton X-100 supplemented with protease inhibitor cocktail (Roche)] per gram of fresh weight, centrifuged for 5 min with 4000g at 4°C, and filtered through two layers of miracloth. The resulting lysates were subjected to GFP-based immunoprecipitation using the anti-GFP microbeads (mMACS GFP Isolation Kit; Miltenyi Biotec) according to the manufacturer manual.
Immunoblot Analysis
Proteins were extracted in 100 mm Tris pH 7.5 with 2% SDS, boiled for 5 min in Laemmli sample buffer, and cleared by centrifugation. The protein extracts were then separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Amersham, GE Healthcare), blocked with 5% skimmed milk in PBS, and incubated with primary antibodies anti-NBR1 (Svenning et al., 2011), anti-ATG8a (Yoshimoto et al., 2004), anti-CP (gift of F. Rabenstein), anti-HCpro (Kasschau et al., 2003), anti-Ub (Agrisera), and anti-GFP (Clonetech) using 1:2000 dilution in PBS 0.1% Tween 20 and secondary horseradish peroxidase-conjugated antibodies 1:5000 in PBS 0.1% Tween 20 (Amersham, GE Healthcare). The immunoreaction was developed using the ECL Prime kit (Amersham, GE Healthcare) and detected in a LAS-3000 Luminescent Image Analyzer (Fujifilm, Fuji Photo Film).
Data Analysis and Presentation
Data are presented as mean ± sd and statistical significance was analyzed by Student’s t test with P values < 0.05 denoted * and P values < 0.01 denoted **. Significance levels with P values < 0.001 or < 0.0001 were not specifically indicated. The number of replicates is given in the respective figure legends (n).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NBR1 (AT4G24690), ATG5 (AT5G17290), ATG7 (AT5G45900), NPR1 (AT1G64280), ATG8a (AT4G21980), DCL2 (AT3G03300), DCL4 (AT5G20320), and Ubiquitin/UBQ11 (AT4G05050).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NBR1-mediated autophagy suppresses TuMV infection.
Supplemental Figure S2. NBR1-RFP aggregates in TuMV-infected tissue.
Supplemental Figure S3. GFP-ATG8a and NBR1-RFP in noninfected tissue.
Supplemental Figure S4. PB1, LIR, and UBA domains recruit NBR1 to large PGs.
Supplemental Figure S5. Colocalization of YFP-HCpro and NBR1-RFP in roots upon ConA treatment.
Supplemental Figure S6. RT-PCR analysis detecting HCpro and VPg cistrons in TuMV and TuMVΔHCpro inoculated plants.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank John Mundy and Morten Petersen (University of Copenhagen; Denmark) for their support during the initiation of this project; Jim Carrington (Donald Danforth Plant Science Center, Michigan) for providing the TuMV-GFP and TuMV-GFP-AS9 clones as well as the anti-HCpro antibody; Cécile Desbiez (INRA, Monfavet Cedex, France) for the infectious WMV cDNA clone; Kristiina Mäkinen (University of Helsinki, Finland) for the CFP-AGO1 plasmid; Peter Brodersen (University of Copenhagen, Denmark) for dcl2 dcl4 seeds, and Frank Rabenstein (JKI Quedlinburg, Germany) for the TuMV-2 isolate and antiserum.
References
- Berryman S, Brooks E, Burman A, Hawes P, Roberts R, Netherton C, Monaghan P, Whelband M, Cottam E, Elazar Z, et al. (2012) Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J Virol 86: 12940–12953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Reggiori F, Codogno P (2013) Emerging regulation and functions of autophagy. Nat Cell Biol 15: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand L, Hörler M, Nüesch E, Vassalli S, Barrell P, Yang W, Jefferson RA, Grossniklaus U, Curtis MD (2006) A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol 141: 1194–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153: 1461–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang A (2016) The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J Virol 91: e01478–e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyán J (2015) viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479-480: 85–103 [DOI] [PubMed] [Google Scholar]
- Dagdas YF, Belhaj K, Maqbool A, Chaparro-Garcia A, Pandey P, Petre B, Tabassum N, Cruz-Mireles N, Hughes RK, Sklenar J, et al. (2016) An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5: e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313: 68–71 [DOI] [PubMed] [Google Scholar]
- Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci USA 109: 15942–15946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW (2007) Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Levine B (2013) Autophagy and viruses: adversaries or allies? J Innate Immun 5: 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelin K, Hafrén A, Rantalainen KI, Mäkinen K (2011) Potyviral VPg enhances viral RNA translation and inhibits reporter mRNA translation in planta. J Virol 85: 9210–9221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Hafrén A, Eskelin K, Mäkinen K (2013) Ribosomal protein P0 promotes Potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J Virol 87: 4302–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A, Lõhmus A, Mäkinen K (2015) Formation of Potato Virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog 11: e1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci USA 114: E2026–E2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxim Y, Ismayil A, Jia Q, Wang Y, Zheng X, Chen T, Qian L, Liu N, Wang Y, Han S, et al. (2017) Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6: e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Judith D, Mostowy S, Bourai M, Gangneux N, Lelek M, Lucas-Hourani M, Cayet N, Jacob Y, Prévost MC, Pierre P, et al. (2013) Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep 14: 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M, Williams B, Dickman MB (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog 9: e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL (2010) Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol 84: 12110–12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KA, Jackson WT (2011) Picornavirus subversion of the autophagy pathway. Viruses 3: 1549–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P (2013) The mechanism and physiological function of macroautophagy. J Innate Immun 5: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA (2013) The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 14: 759–774 [DOI] [PubMed] [Google Scholar]
- Li F, Zhao N, Li Z, Xu X, Wang Y, Yang X, Liu SS, Wang A, Zhou X (2017) A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog 13: e1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Luan H, Shine MB, Cui X, Chen X, Ma N, Kachroo P, Zhi H, Kachroo A (2016) The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol 172: 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyska D, Engelmann K, Meierhoff K, Westhoff P (2013) pAUL: a gateway-based vector system for adaptive expression and flexible tagging of proteins in Arabidopsis. PLoS One 8: e53787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliogka VI, Calvo M, Carbonell A, García JA, Valli A (2012) Heterologous RNA-silencing suppressors from both plant- and animal-infecting viruses support plum pox virus infection. J Gen Virol 93: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132 [DOI] [PubMed] [Google Scholar]
- Moury B, Charron C, Janzac B, Simon V, Gallois JL, Palloix A, Caranta C (2014) Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked protein (VPg): a game of mirrors impacting resistance spectrum and durability. Infect Genet Evol 27: 472–480 [DOI] [PubMed] [Google Scholar]
- Munch D, Rodriguez E, Bressendorff S, Park OK, Hofius D, Petersen M (2014) Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D, Teh OK, Malinovsky FG, Liu Q, Vetukuri RR, El Kasmi F, Brodersen P, Hara-Nishimura I, Dangl JL, Petersen M, et al. (2015) Retromer contributes to immunity-associated cell death in Arabidopsis. Plant Cell 27: 463–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakahara KS, Masuta C, Yamada S, Shimura H, Kashihara Y, Wada TS, Meguro A, Goto K, Tadamura K, Sueda K, et al. (2012) Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci USA 109: 10113–10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, MacPherson S, Sumpter R Jr, Tallóczy Z, Zou Z, Levine B (2010) Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. (2011) Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Komatsu M, Finley K, Simonsen A (2012) Autophagy: more than a nonselective pathway. Int J Cell Biol 2012: 219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers F, García JA (2015) Molecular biology of potyviruses. Adv Virus Res 92: 101–199 [DOI] [PubMed] [Google Scholar]
- Richter J, Proll E, Rabenstein F, Stanarius A (1994) Serological Detection of Members of the Potyviridae with Polyclonal Antisera. J Phytopathol 142: 11–18 [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, Carra S (2014) Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21: 1838–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly S, Lukinova N, Bambina S, Berman A, Cherry S (2009) Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30: 588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16: 495–501 [DOI] [PubMed] [Google Scholar]
- Svenning S, Johansen T (2013) Selective autophagy. Essays Biochem 55: 79–92 [DOI] [PubMed] [Google Scholar]
- Svenning S, Lamark T, Krause K, Johansen T (2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Lloyd RE (2014) Cytoplasmic RNA Granules and Viral Infection. Annu Rev Virol 1: 147–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J. (2016) How does the stressed out ER find relief during virus infection? Curr Opin Virol 17: 74–79 [DOI] [PubMed] [Google Scholar]
- Wan J, Basu K, Mui J, Vali H, Zheng H, Laliberté JF (2015) Ultrastructural characterization of turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J Virol 89: 12441–12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Huang TS, McNeil J, Laliberté JF, Hong J, Nelson RS, Wang A (2010) Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J Virol 84: 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Wang A (2008) Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J Virol 82: 12252–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Yang J, Sun Y, Zhao P, Gao S, Jung C, Qu J, Fang R, Chua NH (2015) Geminivirus activates ASYMMETRIC LEAVES 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLoS Pathog 11: e1005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Martens S (2016) Mechanisms of Selective Autophagy. J Mol Biol 428(9 Pt A): 1714–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen H, Brandizzi F, Verchot J, Wang A (2015) The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet 11: e1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z (2013) NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]