A tonoplast sugar transporter ClTST2 is regulated by a sugar-induced WRKY transcription factor SUSIWM1.
Abstract
How sugar transporters regulate sugar accumulation in fruits is poorly understood and particularly so for species storing high-concentration Suc. Accumulation of soluble sugars in watermelon (Citrullus lanatus) fruit, a major quality trait, had been selected during domestication. Still, the molecular mechanisms controlling this quantitative trait are unknown. We resequenced 96 recombinant inbred lines, derived from crossing sweet and unsweet accessions, to narrow down the size of a previously described sugar content quantitative trait locus, which contains a putative Tonoplast Sugar Transporter gene (ClTST2). Molecular and biochemical analyses indicated that ClTST2 encodes a vacuolar membrane protein, whose expression is associated with tonoplast uptake and accumulation of sugars in watermelon fruit flesh cells. We measured fruit sugar content and resequenced the genomic region surrounding ClTST2 in 400 watermelon accessions and associated the most sugar-related significant single-nucleotide polymorphisms (SNPs) to the ClTST2 promoter. Large-scale population analyses strongly suggest increased expression of ClTST2 as a major molecular event in watermelon domestication associated with a selection sweep around the ClTST2 promoter. Further molecular analyses explored the binding of a sugar-induced transcription factor (SUSIWM1) to a sugar-responsive cis‐element within the ClTST2 promoter, which contains the quantitative trait locus (QTL) causal SNP. The functional characterization of ClTST2 and its expression regulation by SUSIWM1 provide novel tools to increase sugar sink potency in watermelon and possibly in other vegetable and fruit crops.
Carbohydrates are the most important source of energy for human diet, and sugar partitioning is fundamental to plant growth and development. The disaccharide Suc and the monsaccharides Glc and Fru are accumulated to comparably high levels in plants (Goldschmidt and Huber, 1992). Suc is synthesized de novo in source organs (leaves) and transported through the phloem to sink tissues (fruits, roots, and stems). Fruits of watermelon (Citrullus lanatus; Yativ et al., 2010) and melon (Cucumis melo; Dai et al., 2011), roots of sugar beet (Beta vulgaris; Giaquinta et al., 1979), or stems of sugar cane (Saccharum officinarum; Hawker and Hatch, 1965) are heterotrophic organs, accumulating high concentrations of soluble sugars. When sugars and amino acid residues are at high concentrations in the cell cytoplasm, they may undergo nonenzymatic glycosylation (also referred to as glycation), resulting in damage to functional and structural proteins (Yamauchi et al., 2002). The transport of sugars into the plant cell vacuoles is devised to avoid nonenzymatic glycosylation of proteins while maintaining healthy cell function (Slewinski, 2011). The vacuoles vary in size and can occupy up to 90% of plant cell volume and play a critical role in regulating nutrients, salt, calcium, energy, and cellular pH levels during cell growth and division and during biotic and abiotic stress (Martinoia et al., 2007).
TST genes associated with vacuole transport have been characterized and cloned from Arabidopsis (Arabidopsis thaliana) (Wingenter et al., 2010; Cho et al., 2010), berry, apple, and litchi (Afoufa-Bastien et al., 2010; Li et al., 2012; Wang et al., 2015) fruit development. However, TSTs and the mechanisms regulating their expression have not been studied in detail in plant species that produce fruits consumed by humans. The published TSTs are based on T-DNA insertion lines (Wingenter et al., 2010) and transcriptome or proteome studies (Jung et al., 2015). Moreover, no TST sequence variation has been associated with fruit sugar accumulation or genetically mapped based on accumulating sugars variation. Consequently, no TST gene-based genetic marker has been developed for molecular breeding. Until recently, our knowledge and understanding of TST genes was mainly limited to the TST gene family of the model species Arabidopsis and our understanding of the molecular mechanism that drives Suc import capacity across the tonoplast of high-Suc-accumulating plant organs had been limited (Hedrich et al., 2015). Recent breakthrough studies characterized the gene that uploads Suc to the vacuoles of the sugar beet taproots (Jung et al., 2015; Dohm et al., 2014). Unlike the Arabidopsis genome that contains three TST genes (TST1, 2, and 3), the sugar beet genome contains four TST orthologs, with a duplicated TST2 (Dohm et al., 2014). The duplicated TST genes were later termed BvTST2.1 and BvTST2.2. The BvTST2.1 gene is highly and specifically expressed in the sugar beet taproots, in correlation with sugar accumulation. The BvTST2.1 protein, which shows transmembrane Suc transportation activity against large concentration gradients, is specifically highly abundant in the sugar beet taproot (Jung et al., 2015).
The modern sweet-dessert watermelon is the result of many years of cultivation and selection for watermelon with desirable qualities, appealing for human consumption. Because of its sweetness and attractive flesh color and flavor, watermelon is among the most-consumed fresh fruits throughout the world. On the other hand, watermelons collected in the center of origin of Citrullus spp. in Africa (Chomicki and Renner, 2015), Citrullus lanatus var citroides and Citrullus colocynthis, are often unsweet or bitter (2–3 Brix) and have hard white, green, or yellow flesh (Liu et al., 2013). The many years of domestication and breeding selection have led to marked increase in fruit quality of watermelon cultivars (10–12 Brix). However, there is no sufficient information about the mechanisms and evolution of genes controlling the transport and accumulation of sugars in cell vacuoles of the sweet watermelons versus unsweet watermelon fruits.
Recently, a single-nucleotide polymorphism (SNP)-based genetic linkage map was constructed using a population of 96 recombinant inbred lines (RIL) derived from a cross between the high-sugar-content Chinese watermelon line 97103 (Citrullus lanatus var lanatus) and the unsweet accession of Citrullus lanatus var citroides U.S. Plant Introduction (PI) 296341-FR (Sandlin et al., 2012; Ren et al., 2014). The 96 RIL families were analyzed for fruit sugar content, and a QTL “QBRX2-1(or Qfru2-3)” with a large effect (R2 = ∼20%) associated with sugar accumulation in watermelon fruit was mapped on chromosome 2. However, the QBRX2-1 QTL covers a large chromosomal interval of a 2.9 Mega-base (Mb). A fine mapping analysis approach is needed to further expedite and locate the gene(s) controlling sugar accumulation in watermelon.
In this study, we used genome sequencing of 96 RILs to construct an ultra-high-density SNP linkage bin map, combined with different biochemical and genetic approaches, to identify the causal gene underlying QBRX2-1 QTL interval and elucidate major molecular event that might be associated with the evolution and domestication of sweet-dessert watermelon.
RESULTS
Mapping and Association Study of QBRX2-1, a Sugar Content QTL in Watermelon
To map candidate genes in the previously identified QBRX2-1 (or Qfru2-3) QTL interval on chromosome 2 (Sandlin et al., 2012; Ren et al., 2014), we resequenced all 96 RILs derived from 97103 × PI296341-FR (Supplemental Fig. S1). Using segregation data of 0.948 million SNPs and a bin-mapping strategy that combines the cosegregating SNP markers between two contiguous block borders into a bin, we constructed an ultra-high-density SNP map (Supplemental Fig. S2). The average physical length of a recombination bin was 138.1 kb, ranging from 3.0 kb to 970 kb, resulting in a nearly saturated map suitable for mapping genes.
The 96 RILs were phenotyped for Brix, Fru, Glc, and Suc content in three sequential years. The heritability (h2) of sugar content traits was relatively high (>85%), except for Suc (65%; Supplemental Table S1). Significant (P < 0.001) genotype by environment interactions for traits were also observed (Supplemental Table S1). Bin-based QTL analysis narrowed down this QTL from published 2.9 Mb to a physical region of 797.4 kb. The mapping procedure anchored the ∼70-kb scaffold 6994, which was not associated to any chromosome in the watermelon genome assembly version 1 (Guo et al., 2013) to chromosome 2, between positions 17,657,266 to 18,454,759 (Fig. 1, A and B), explaining ∼20% of the total phenotypic variation.
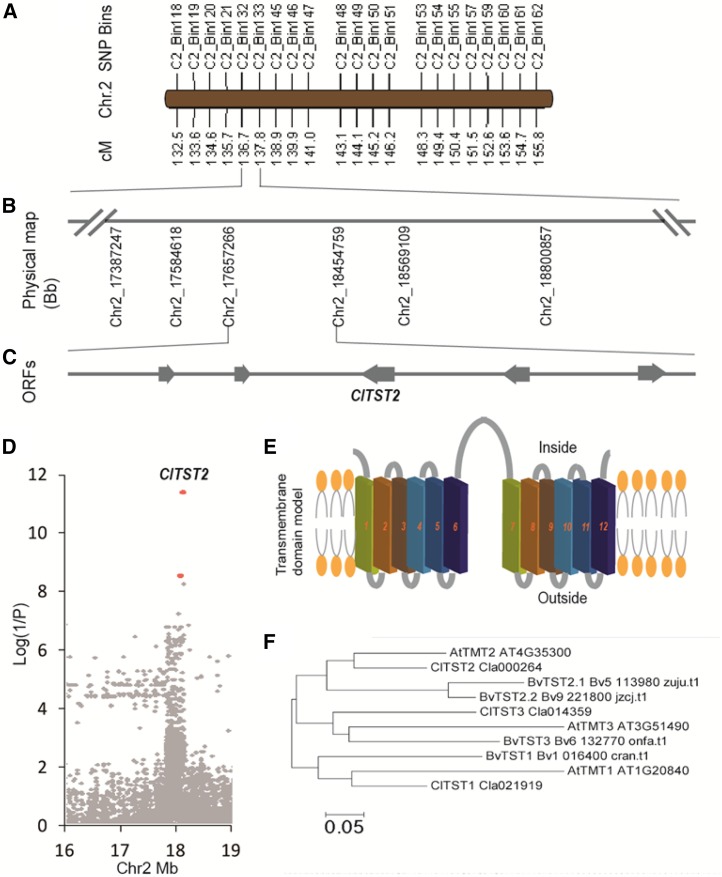
Figure 1.
Mapping, transmembrane domain modeling, and phylogenetic analysis of the ClTST2 gene. A, SNP bin map of the sugar content QTL (QBRX2-1). B, Mapping of QBRX2-1 to a physical region of 797.4 kb on chromosome 2 between nucleotides 17,657,266 and 18,454,759 in the watermelon genome. C, Five expressed genes (Supplemental Table S2) localized in the region, one of which is ClTST2 (Cla000264). D, Association study of sugar content, based on 21,249 SNPs identified in ClTST2 region (Chr2 16–19 Mb) by genomic resequencing of this region in 400 watermelon accessions. The most significant SNPs, marked with red dots, are located in the promoter of ClTST2. E, Transmembrane helix modeling for the ClTST2 protein showing 12 transmembrane domains. F, A neighbor-joining phylogenetic tree of Arabidopsis (At), sugar beet (Bv, B. vulgaris), and watermelon (Cl, C. lanatus) ClTST1, ClTST2, and ClTST3.
This 797.4 kb QTL region harbors only five fruit expressed open reading frames (Supplemental Table S2) as determined by RNA-seq transcriptome analysis of the parental lines (Fig. 1C; Guo et al., 2015). To identify the causal gene underlying this sugar QTL, we performed an association study on 400 worldwide collected representative watermelon accessions, by using 3 Mb (Chr2, from 16–19 Mb) genomic resequencing data of the region that covers the 797.4 kb QTL. The most significant SNPs, out of 21,249 SNPs identified in this region, are located on the Cla000264 promoter (Fig. 1D). Cla000264 annotated as a membrane sugar transporter was highly expressed, while the other four expressed genes are functionally irrelevant (Supplemental Table S2). These facts made Cla000264 the putative causal gene for QBRX2-1 sugar content QTL. Cla000264 shares about 78% amino acid identity and 88% sequence similarity with the Arabidopsis tonoplast monosaccharide transporter gene AtTMT2 (renamed AtTST2; Jung et al., 2015). Therefore, we named this watermelon gene C. lanatus tonoplast sugar transporter 2 (ClTST2). Prediction of transmembrane helices (TMHs) at TMHMM Server v. 2.0 showed that both ClTST2 (Supplemental Fig. S3B) and Arabidopsis TST2 (formerly termed TMT2; Supplemental Fig. S3C) contain 12 predicted TMHs, indicating that they are transmembrane proteins (Fig. 1E).
ClTST2 Is the Most Highly Expressed TST Gene, and Its Expression Is Positively Correlated with Sugar Accumulation in Watermelon Fruit
We identified three ClTST orthologs at the public watermelon genome database (www.icugi.org). A phylogenetic analysis using only functionally analyzed TSTs (Jung et al., 2015; Wingenter et al., 2010) revealed that ClTST1, ClTST2, and ClTST3 are most closely related to the Arabidopsis/Beta vulgaris homologs AtTMT1/BvTST1, AtTMT2/BvTST2, and AtTMT3/BvTST3, respectively (Fig. 1F). To gain insights into the nature of the TST gene family expression in watermelon fruits, we compared the ClTSTs expression in different tissues of sweet-dessert watermelon accession 97103 and the wild low-sugar-content line PI296341-FR. We sampled watermelon fruit flesh (NF) and rind (NR) at critical fruit development stages. Fruit flesh and rind of cultivated watermelon 97103 were sampled at 10, 18, 26, and 34 d after pollination (DAP). In addition, we sampled leaves and roots. The fruit flesh of the unsweet accession PI 296341-FR (PF), a late maturation accession, was sampled at 10, 18, 26, 34, 42, and 50 DAP (Supplemental Figure S4A). These samples were used for both sugar and gene expression analyses. The soluble sugar accumulated in flesh of mature (34 DAP) 97103 watermelon fruit comprises about 50% Suc, 30% Fru, and ∼20% Glc (Supplemental Fig. S4B). As expected, the sweet-dessert watermelon line 97103 contained 42 mg Suc per gram flesh fresh weight at 34 DAP, while the unsweet parental line PI296341-FR contained ∼1.2 mg Suc per gram flesh fresh weight at all analyzed fruit development stages (Supplemental Fig. S4C). The Suc content increased sharply from 10 to 34 DAP in the 97103 fruit (Supplemental Fig. S4C), while the Fru and Glc levels remained constant (Supplemental Fig. S5). These results indicate that Suc is the major contributor to fruit total soluble sugar content.
Using quantitative RT-PCR (qRT-PCR), we measured the relative expression patterns of ClTST1, ClTST2, and ClTST3 in flesh and rind of developing fruits of 97103 versus PI 296341-FR. The expression pattern of these sugar transporter genes was also measured in roots and leaves of 97103. Relative expression of ClTST2 in 97103 fruit flesh (NF) increased 15-fold from 10 to 26 DAP, while it remained at a constant low level in the 97103 rind (NR 10–34 DAP), leaves, roots, and in PI 296341-FR fruits (PF10–50 DAP; Supplemental Fig. S4D). These results indicated that ClTST2 mRNA levels are positively associated with Suc accumulation during maturation of the sweet-dessert watermelon fruit. In contrast, both ClTST1 and ClTST3 have low expression during sugar accumulation in the watermelon fruit (Supplemental Fig. S4D; Supplemental Table S1).
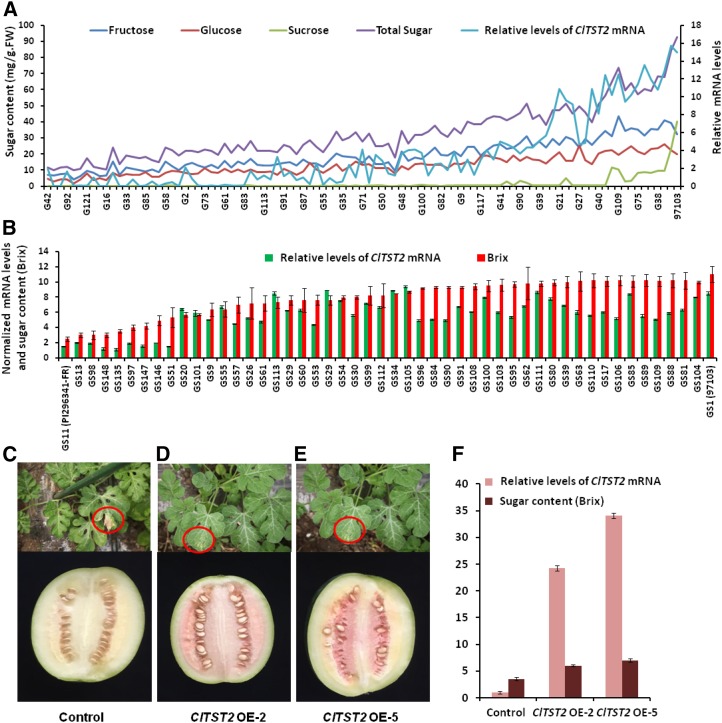
To confirm that relative transcription of ClTST2 is correlated with fruit sugar content, we performed qRT-PCR in mature fruit of all 96 RILs (Fig. 2A) and in a core germplasm collection, comprising 50 accessions (Fig. 2B) selected from 1,197 accessions (Zhang et al., 2016). The expression of ClTST2 in the sweet watermelon lines was ∼5 to 10 times higher than in the unsweet PI 296341-FR, while the expression of ClTST2 in low sugar content lines resembled that of PI 296341-FR (Fig. 2, A and B). In the RIL population and core collection, the ClTST2 mRNA levels were correlated with sugar content with correlation index (r) of 0.79 and 0.68 (P < 0.0001), respectively, showing that transcription of ClTST2 is positively correlated with sugar content in watermelon fruit flesh.
Figure 2.
High expression of ClTST2 increases sugar accumulation. ClTST2-specific mRNA relative levels determined by qRT-PCR in the mature fruit of 96 RILs (A) and 50 core germplasm accessions (B) selected from Supplemental Table S4. Basta resistance in seedling stage (shown in red circles) and fruit phenotype at 30 DAP for transgenic control (C) and representative ClTST2 overexpression T1 lines (D and E). In lines ClTST2 OE-2 and ClTST2 OE-5 with higher levels of ClTST2 mRNA, the fruit sugar contents increased significantly compared with the control (F).
Overexpression of ClTST2 Increases Sugar Accumulation in Watermelon Fruit
To clarify the effects of ClTST2 during sugar accumulation in watermelon fruits, we generated a 35S::ClTST2 construct to transform the white-fleshed semiwild GS24_PI179878 (Supplemental Table S4). Basta-resistance analysis of the overexpression (OE) transgenic population helped to identify 25 independent T0 plants, which were propagated for further analysis. Five transgenic lines were tested with increased ClTST2 mRNA levels, and two of the T1 lines (ClTST2 OE-2 and ClTST2 OE-5) were selected for future sugar accumulation study (Fig. 2, D and E). Transgenic plants, which did not show any Basta resistance in the same T1 segregation population were used as negative controls (Fig. 2C). In these transgenic lines, ClTST2 OE-2 and ClTST2 OE-5 with higher levels of ClTST2 mRNA and Basta resistance (Fig. 2, D–F), the fruit color turned red (Fig. 2, D and E) and sugar contents increased significantly compared with the control (Fig. 2F). Flesh sugar contents in these transgenic lines were consistent with the ClTST2 transcription abundance. Increases of flesh sugar contents in the ClTST2 OE transgenic lines indicated that the higher ClTST2 expression levels are necessary for increased flesh sugar accumulation in watermelon.
ClTST2 Is a Vacuole-Localized Suc and Hexoses Transporter
To corroborate the vacuolar localization of the putative watermelon sugar transporter, we fused the 97103-ClTST2 sequence with the yellow fluorescent protein (YFP) gene and expressed the corresponding fusion protein transiently in protoplasts made from 10 DAP watermelon fruit (white fruit stage without chromoplasts). The yellow fluorescence was well resolved in the fruit cell vacuoles (Supplemental Fig. S4, E–G). The tonoplast localization of the YFP fusion protein observed in transiently transgenic fruit protoplasts led us to investigate the subcellular localization of the 97103-ClTST2 protein in watermelon fruit.
We verified the transport function of ClTST2 using the patch-clamp technique in Human Embryonic Kidney cell-line 293T (HEK293T) expressing the 97103-ClTST2 gene (Fig. 3, A–C). This technique has already allowed the identification of proton-coupled sugar antiporters belonging to the TST family in mesophyll cell’s vacuoles of Arabidopsis (Schulz et al., 2011) and sugar beet (Jung et al., 2015). To localize ClTST2 on the cellular level in HEK293T cells, we fused the 97103-ClTST2 with the GFP gene and transiently overexpressed the corresponding fusion gene in HEK293T cells. The green fluorescence of the fusion protein was observed in plasma membranes of HEK293T cells (Supplemental Figure S6, C and D). GFP was also located in the endoplasmic reticulum, nuclear envelope, and Golgi complex, which is common when heterogeneous membrane proteins are overexpressed (Snapp and Lajoie, 2011).
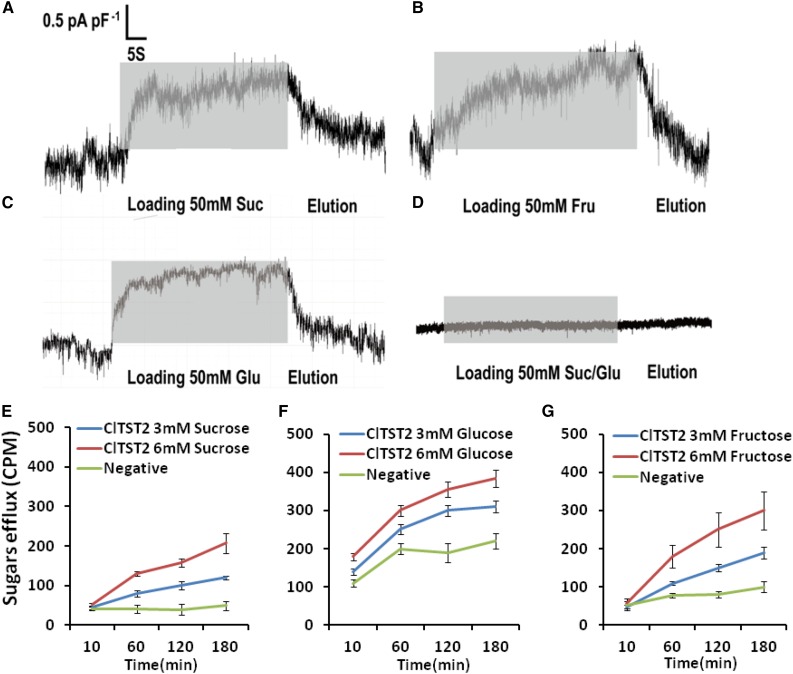
Figure 3.
Transport function of ClTST2. Incubation with 50 mm Suc (A), Fru (B), and Glc (C) solutions induced inward current responses (gray bars) in HEK293T cells expressing the ClTST2. HEK293T cells only did not exhibit a significant change in current in response to incubation of 50-mm Suc/Glc solution (D). Tracer efflux assay in Xenopus oocytes at pH 5.5. [14C] Suc (E), [14C] Glc (F), and [14C] Fru (G) efflux by ClTST2 in Xenopus oocytes in 3- and 6-mm [14C] sugars. As negative controls, we used oocytes injected with water to efflux [14C] sugars (n = 9).
When 50 mm Suc were applied to the bath medium of the HEK293T cell membrane, cells harboring ClTST2 responded with a strong current trace with respect to the background current level (Fig. 3A). These Suc-activated outward currents indicate that Suc moved into the cells at pH 7.5. The same results were observed when 50 mm Fru and Glc were applied to the outside of the HEK293T cell membrane (Fig. 3, B and C, respectively), while in the absence of ClTST2, the HEK293T cells did not exhibit significant sugar transport activity (Fig. 3D). To confirm this result, we carried out radio-labeled sugars efflux assay in oocytes of Xenopus. In this experiment, we transiently overexpressed the ClTST2-GFP in the Xenopus oocytes to reconfirm the plasma membrane localization of ClTST2 (Supplemental Figure S6, A and B). Subsequently, the ClTST2 protein demonstrated Suc, Fru, and Glc transport activity at pH 6.5 by time-dependent efflux of sugars labeled with [14C], injected into Xenopus oocytes cells (Fig. 3, E–G). Here, the ClTST2 demonstrated proton-coupled antiporter activity involved with sugar transport, similar to the Arabidopsis TST family (Schulz et al., 2011) and the sugar beet transporters (Jung et al., 2015).
ClTST2 Expression Increases Sugar Accumulation in Fruits
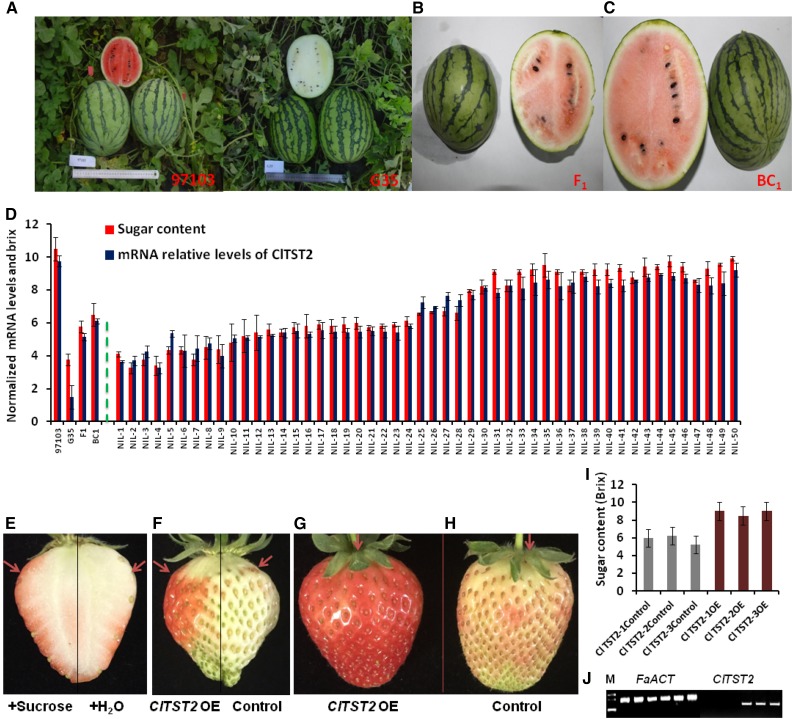
To reduce the background variation, which is contributed by wild accession PI 296341-FR but is unassociated with sugar accumulation, to the 97103 × PI 296341-FR RILs, we generated near-isogenic lines (NILs) by crossing the sweet-dessert elite Chinese watermelon line 97103 with the low sugar content and white hard flesh G35 RIL line, which has high genetic identity and phenotypic similarities (fruit shape, size, and rind stripes) with line 97103 (Fig. 4, A–C). The F1 plant (97103 × G35) was backcrossed to line 97103, which was also used as the recurrent parent to have a BC2 generation. A BC2F1 plant was self-pollinated to produce a segregating BC2F2 population, which enables the analysis of ClTST2 allelic variation effect on fruit sugar accumulation in the same watermelon fruit type and with less genetic background noise. Mature fruit of parental lines and progeny were analyzed for sugar (Brix) and for ClTST2 mRNA levels by qRT-PCR. We observed high levels of ClTST2 mRNA in the high sugar 91703, low levels in the unsweet G35, and medial levels in both F1 and BC1 plants. Similar analyses of 50 selected BC2F2 population, we observed low ClTST2 mRNA levels in the low-sugar-content lines (NILs 1–24) and high ClTST2 mRNA levels in the high-sugar-content lines (NILs 25–50; Fig. 4D). These results suggest positive association of ClTST2 expression with sugar accumulation in watermelon fruit flesh.
Figure 4.
ClTST2 increases sugar accumulation in fruits. A to C, The parental lines 97103, G35, F1, and BC1 fruits for NIL population construction. D, Sugar content and relative expression levels of ClTST2 in (from left to right) 97103 and G35 parental lines, F1, BC1 plants, and 50 segregating (BC2F2) NILs. E, Suc injection induces early maturity in strawberry fruits; red arrows point at the injection site. F to H, ClTST2 overexpressing strawberry fruit sections (OE), coloration compared to the empty vector control. I, Accumulation of sugars in ClTST2 overexpressing strawberry fruit sections (OE) and in the empty vector control. J, Semiquantitative RT-PCR expression analysis of FaACT and ClTST2 in the empty vector controls 1 to 3 and in ClTST2 overexpressing strawberry fruit sections (ClTST2-1-3 OE). The same sample order as in I for FaACT and ClTST2 expression analyses, respectively.
To validate sugar transport activity of ClTST2 in vivo, we transiently expressed this gene in cultivated strawberry (F. × ananassa) at the early, white flesh stage. Three days after transformation, the transiently transformed sides of the strawberry fruit overexpressing 97103-ClTST2 accumulated 1.5-fold more sugars (Fig. 4I) and induced ripening, reflected by the fruit’s red coloration, compared to the control empty-vector-injected transgenic fruit sides (Fig. 4, F–H). We confirmed ClTST2 expression in transgenic strawberry by semiquantitative RT-PCR analysis of RNA extracted at the third day after transformation. The transiently transgenic strawberry fruit section showed high ClTST2 mRNA levels, versus undetectable levels in the controls (Fig. 4J). In a separate experiment, injection of 50 mm Suc to young strawberry fruits at the early white color stage induced ripening and higher sugar accumulation than the control fruits 3 days after injection (Fig. 4E). Exogenous treatment with sugar has been reported to induce rapid fruit maturation and anthocyanin accumulation in strawberry (Jia et al., 2013) and grapes (Lecourieux et al., 2014). In the watermelon RIL population, the fruit ripening time (DAP) was correlated negatively with the sugar content with correlation index (r) of −0.768 (P < 0.0001), showing that higher sugar concentrations can accelerate fruit ripening transition (Supplemental Table S3) in watermelon.
Identification of Functional Fragments in the ClTST2 Promoter
We sequenced 1,825 bp in the ClTST2 upstream (5′) promoter region of the sweet 97103 and unsweet PI 296341-FR. Two small deletions of 4 bp and 29 bp were detected at nucleotides −1,237 and −1,741, respectively in 97103 (Supplemental Fig. S7). The 29-bp deletion contains one ARR1 recognition site, while the 4-bp deletion resulted in gaining GTGANTG10, a transcription factor binding site (Supplemental Fig. S8). Two SNPs in the 1,825-bp promoter region led to changes in two predicted transcription factor recognition sites. The SNP at −1,568 (T/G; T in PI296341-FR/G in 97103) resulted in loss of a MYB motif (ACCATCC) while the SNP at −1368 (A/C; A in PI 296341-FR/C in 97103) changed one out of five W boxes (TGACT), known to serve as WRKY transcription factor binding site, which resides within a SURE (sugar responsive element; Supplemental Figs. S7 and S8).
To date, there has not been any report describing regulation of a TST gene by a particular transcription factor in plants. To functionally determine the major expression-regulating regions, DNA fragments of different lengths from the 5′ end of the ClTST2 promoter region were fused to the luciferase (LUC) gene and expressed transiently in strawberry fruits and Nicotiana benthamiana leaves. In total, a 5′ full-length (−1,825 bp) and six 5′-truncated fragments were fused to the LUC reporter gene (reporter 1–7 in Supplemental Fig. S7B). The results showed that there were no significant changes in LUC activity when the constructs missing one or both of the 4-bp and 29-bp deletions were expressed in leaves, as compared to reporter 1 (−1,825 bp; Supplemental Fig. S7, C and D). This suggests that there are probably no major regulatory regions controlling ClTST2 expression near the 4-bp and 29-bp deletions. LUC activities declined sharply by a factor of five (Supplemental Fig. S7, E and F) when reporters 3 (−1,000 bp) and 4 (−500 bp; Supplemental Fig. S7B). In addition, there was no difference in LUC activity between reporter 1 (−1,825 bp) and reporter 2 (−1,500 bp). Therefore, the DNA sequence elements regulating expression of ClTST2 are likely to be located between −1,000 and −1,500 bp. These results indicate that the A/C transversion SNP (–1,368) changed W box3 (Supplemental Fig. S8), which may serve as the binding site of a WRKY transcription factor that regulates ClTST2 expression and plays a critical role in sugar accumulation in the sweet-dessert watermelon (Supplemental Fig. S7).
Effects of Domestication on the ClTST2 Promoter Region
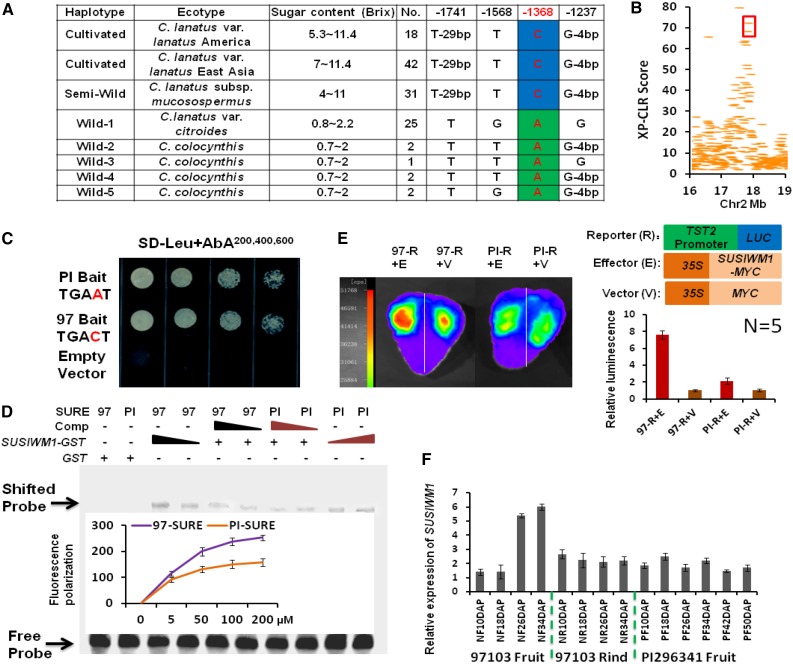
We show here that the relative expression of ClTST2 is higher in sweet (97103) than in unsweet watermelon (PI 296341-FR) and the association of this difference with a single SNP (–1,368, A/C) in the ClTST2 promoter region (Supplemental Fig. S7A). To characterize the general meaning of these findings at the population level, we analyzed the ∼2 kb upstream promoter-sequence of 124 watermelon accessions; including 33 unsweet watermelon accessions of C. colocynthis and C. lanatus var citroides, 31 egusi type watermelons (also known as C. lanatus subsp. mucosospermus), and 60 sweet-desert cultivars (C. lanatus var lanatus). The geographic distributions of these accessions included East Asia (mainly China, Japan, and Korea), the United States, and Africa (Supplemental Table S4). The ClTST2 promoter sequences of the sweet-dessert and egusi type watermelons shared the same haplotype, while the wild watermelons had five haplotypes (Fig. 5A). The –1,368 SNP (A/C) in W box3 of the ClTST2 promoter region explains most of the sugar content variation among the 124 germplasm accessions (r = 0.827; Supplemental Table S4). This result indicates that ClTST2 activity is required for fruit sugar accumulation in the sweet-dessert watermelons. Moreover, this result revealed a possible selection for higher expression of ClTST2 during domestication, leading to increased sugar accumulation in watermelon.
Figure 5.
SUSIWM1 regulates expression of ClTST2. A, Association of sugar content with ClTST2 promoter haplotype in cultivated, semiwild, and wild watermelons. B, XP-CLR estimation of the ClTST2 genome region. ClTST2 is localized in the 40-kb regions surrounded by red frame, which resides in the genome region that went through selective sweep during watermelon domestication. C, Y1H assays showing that SUSIWM1 activated AUR1-C expression when both the 97103 (97) and PI296341-FR (PI) sequence motifs were used in the presence of 200 to 600 ng/mL AbA. No activation was detected for the empty vector. D, EMSA of GST-SUSIWM1 fusion protein binding to the SURE cis-elements in ClTST2 from 97103 (97) and PI296341-FR (PI). FP assay showing probed SUSIWM1 dose-dependent binding activity to SURE cis-elements in ClTST2 from 97 and PI. E, LUC assay of strawberry fruits cotransformed with p35S::SUSIWM1::tNOS as the effector or with p35S::MYC::tNOS as the control, with the −1,500 bp promoter region of the sweet and unsweet lines fused to LUC. Relative fluorescence results are presented as histograms, representing the means of five replicates. F, Digital expression (in RPKM) of SUSIWM1 in high-sugar-content fruit line 97103, in low-sugar fruit (PI296341-FR) and low-sugar tissue (97103 rind).
To evaluate the selection for ClTST2 during domestication, we used the cross-population composite likelihood ratio test (XP-CLR; Sabeti et al., 2007), which can approximate the effect of a selective sweep on SNPs in the vicinity of ClTST2 promoter region. Results of this analysis indicated that the ClTST2 was involved in the 40-kb domesticated region of this QTL interval (Fig. 5B). Gene ontology analysis indicated that the fruit development related cellular metabolic genes, and vesicle-mediated and intracellular transporters were enriched significantly during selection. This result indicates a selection sweep for ClTST2 during domestication that aimed for increasing sugar accumulation in the watermelon fruit.
SUSIWM1 Regulates the Expression of ClTST2
The A/C transversion (SNP –1,368), which had the most significant value in association study (Fig. 1D), changed W box3 that resides in a SURE cis-element (Supplemental Fig. S8). To determine which protein can bind to regulatory elements of the ClTST2 gene promoter and whether SNP –1,368 affects the binding activity, we performed a yeast one-hybrid (Y1H) experiment that measures protein-DNA interactions. For the Y1H assay, five watermelon fruit-expressed WRKY transcription factors were fused with the AUR1-C transcriptional activation domain as prey, respectively. The SURE cis-elements that include the W box3, in which SNP –1,368 (A/C) resides from 97103 and PI 296341-FR watermelon accessions were cloned into pAbAi-bait. Remarkably, after cotransformation of yeast with the prey and individual baits, only the Cla009557 protein activated AUR1-C expression when both the 97103 motif (TGACT) and PI 296341-FR (TGAAT) sequences were used in the presence of 200 to 600 ng/mL Aureobasidin A (AbA). At the same time, no activation was detected for the empty vector (Fig. 5C). Cla009557 is the most SUSIBA2-like gene in watermelon genome (62% homology at the protein level). SUSIBA2 (sugar signaling in barley) is a barley (Hordeum vulgare) WRKY transcription factor, whose expression response to Suc signal (Su et al., 2015). Additionally, we produced a glutathione-S-transferase (GST)-Cla009557 recombinant protein in E. coli that was used as a probe in an electrophoretic mobility shift assay (EMSA). Bands with retarded migrations were observed when both 97103 and PI296341-FR SURE cis-elements were incubated with the GST-Cla009557 protein. The bands became significantly weaker in the presence of competitive probes, and no shifted probe band was detected when only GST tag protein was present (Fig. 5D). As showed by EMSA, the bands binding by PI 296341-FR SURE cis-elements were weaker than that of 97103. To quantify the binding rate difference, we performed an fluorescence polarization (FP) assay, which is more sensitive and samples required are reduced by at least 100-fold from the amount needed for EMSA (Wan and Le, 2000). Two-fold FP value was detected in the presence of GST-Cla009557 when fluorescein-labeled 97103 SURE cis-elements than the PI296341-FR did (Fig. 5D). These results together suggested that Cla009557 is capable to bind the SURE cis-elements in the ClTST2 gene promoter and shows differences in binding activity in sweet versus unsweet watermelons; thus, we called this gene SUSIWM1 (sugar signaling in watermelon).
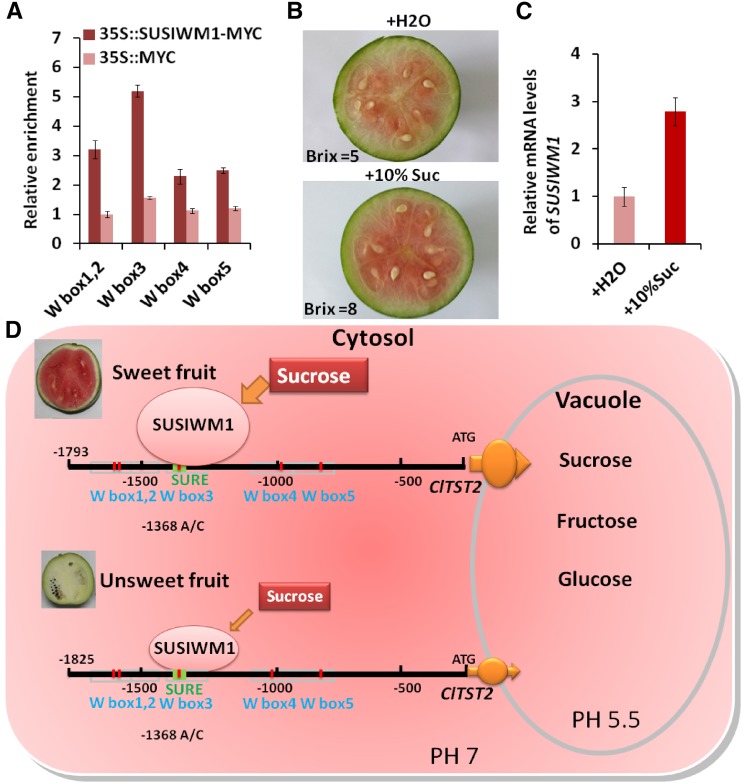
Next, we asked whether SUSIWM1 is able to activate expression of ClTST2 in plants. We performed an in vivo LUC assay by cotransformation of strawberry fruits with p35S::SUSIWM1::tNOS and with a –1,500-bp promoter region of the sweet and the unsweet lines (97103-p ClTST2::LUC and PI 296341-FR-p ClTST2::LUC). The 97103 ClTST2 promoter induced significantly higher LUC activity (∼7-fold) than the PI 296341-FR ClTST2 promoter did in the presence of SUSIWM1 (Fig. 5E). These results indicated that 97103 ClTST2 promoter is more active than its PI 296341-FR counterpart and that SUSIWM1 transcription factor activates ClTST2 expression. To identify the exact binding site of SUSIWM1 in ClTST2 expression in watermelon, we performed chromatin immunoprecipitation (ChIP) coupled to detection by qRT-PCR using watermelon protoplasts that were transiently expressing a SUSIWM1-Myc fusion protein. The qRT-PCR analysis revealed that SUSIWM1 was selectively recruited to the ClTST2 promoter region containing W box elements and that SUSIWM1 interacts directly with the W box3, which contains the –1,368 SNP (A/C), in the SURE region of ClTST2 promoter (Fig. 6A).
Figure 6.
Regulation of ClTST2 and SUSIWM1. A, ChIP analysis of SUSIWM1 recruitment to the ClTST2 promoter W box3, which contains the −1,368 SNP (A/C) in a SURE element. The results are expressed as means ± sd. B, Changes in fruit sugar content (Brix) and color 3 d after injection of 1 mL 10% Suc into 10 DAP immature watermelon fruit. C, Relative level of SUSIWM1 mRNA after treatment with Suc. D, Proposed model for regulation of ClTST2 by SUSIWM1 based on RNA-seq transcriptome data. Suc induces the expression of SUSIWM1, which up-regulates ClTST2 in cultivar sweet 97103 fruit flesh, while transcription of ClTST2 is repressed during all stages of fruit maturation in low-sugar-content tissues such as PI296341-FR fruit and 97103 fruit rind.
ClTST2 had increased expression during sweet watermelon fruit maturation (Supplemental Fig. S4D). To explain how this high expression is regulated by SUSIWM1, we analyzed the RNA-seq transcriptome data (Guo et al., 2015) and found that SUSIWM1 was highly expressed in the high-sugar-content 97103 fruit flesh during the rapid sugar increasing stage (26 and 34 DAP), while it was significantly lower in the low-sugar fruit (PI 296341-FR) and tissue (97103 rind; Fig. 5F). Moreover, the SUSIWM1 expression has increased 6-fold in 97103 fruit flesh during fruit maturation (Fig. 5F).
To investigate if Suc signal can regulate SUSIWM1 expression, we injected 1 mL of 10% Suc into 97103 watermelon fruits at 10 DAP. Three days after Suc injection, fruit sugar content (Brix) increased by 60% compared with the water-injected control. This increase was also associated with darker fruit coloration (Fig. 6B). Correspondingly, relative mRNA level of SUSIWM1 increased 3-fold at 3 days after Suc injection (Fig. 6C). These results strongly suggest that sugar is an important signal for SUSIWM1 expression regulation leading to a positive feedback in increasing sugar accumulation in watermelon cultivar. Our model shows that Suc induce expression of SUSIWM1, which up-regulates expression of ClTST2 in the sweet line 97103 fruit flesh (but not in the rind), while transcription of ClTST2 is suppressed during all stages of the low-sugar PI 296341-FR fruit maturation (Fig. 6D).
Soluble sugars translocation to the developing fruit is likely regulated by additional QTL that control uploading from source (leaves), translocation, and downloading to sink (fruit). We showed here that the sink is powered, at least in part, by the activity of ClTST2 regulated by SUSIWM1.
DISCUSSION
Fruit sweetness is a major quality component of many fruits. Fruit sugar accumulation is controlled by environmental and genetic factors that affect sugar translocation to the fruit, metabolism, and storage in the developing fruits. Sugar accumulation starts with the loading from photosynthetic source tissues (leaves) into the phloem that translocates sugars to the fruits, which represent nonphotosynthetic sink tissues. In the fruit, sugar is downloaded and can be further metabolized, and the final step involves uploading of the sugars into the vacuoles where it is stored. Thus, fruit sugar accumulation is a multifactorial trait, genetically regulated by quantitative trait loci.
The tetrasaccharide stachyose and the trisaccharide raffinose are the main phloem translocated sugars in cucurbit plants while mainly Suc, but also Glc and Fru, accumulate in the fruit vacuole of sweet cucurbit cultivars, including in watermelon (Zhang et al., 2010a). Once stachyose and raffinose arrive to the fruit, they are metabolized to the accumulating sugars, which are then finally uploaded form the cytosol to the vacuoles of the sugar accumulating cells, maintaining the fruit sink strength. Various approaches have been made to uncover the mechanisms that underlie sugar accumulation in watermelon fruit, including cloning and characterization of sugar metabolism genes and enzymes (Zhang et al., 2010a), comparative transcriptome analysis of sweet and unsweet watermelons (Guo et al., 2015), and intense mapping efforts that led to the identification of several major QTLs (Sandlin et al., 2013; Ren et al., 2014). These studies contributed to our understanding of the events associated with sugar accumulation in watermelon fruit. However, none of these approaches identified a causal gene that underlies any of the QTLs responsible for sugar accumulation in watermelon fruits. In this study, QBRX2-1, previously identified as a major watermelon sugar QTL (Sandlin et al., 2012; Ren et al., 2014), was genetically dissected to identify this QTL causal gene and to gain insight into the mechanisms controlling sugar accumulation in watermelon fruit. As one of four major QTLs, QBRX2-1 showed a significant contribution (R2 = 23.4%) to the variation in total soluble sugars (Brix) in the 97103 × PI 296341-FR RIL population (Ren et al., 2014; Zhang et al., 2004).
Higher Expression of ClTST2 Was Selected during Sweet-Dessert Watermelon Domestication
Sweetness has been the main impetus in domestication and further breeding of many fruits and vegetables. However, there is no sufficient knowledge about the molecular mechanisms associated with increased sugar accumulation in edible crop organs, especially since sweetness has been evolved independently in a polyphyletic manner in different crops.
Until recently, our knowledge and understanding of TST genes was mainly limited to the TST gene family of the model species Arabidopsis, and our understanding of the molecular mechanism that drives Suc import capacity across the tonoplast of high-Suc-accumulating plant organs had been limited (Hedrich et al., 2015). Here, we show that the watermelon genome contains three TST orthologs (Fig. 1F), the BvTST2 homolog, ClTST2 is the main Suc and hexoses uploader (Fig. 3) in sweet-dessert watermelon fruit flesh. The ClTST2 expression level, regulated by SUSIWM1 (Fig. 5), makes the difference in sweetness between sweet and unsweet watermelon fruits and tissues. Nondessert type watermelons have been cultivated and consumed by native people in Africa for many years, mainly for seed consumption. Human desire for sweetness has played a major role in the domestication of many fruit and vegetable crops. Thus, it is likely that the occurrence of a sweet type watermelon commenced its domestication. Subsequent selection for horticultural and fruit qualities led to today’s high-quality sweet-dessert watermelon.
The pronounced phenotypic differences and at the same time the limited variation in coding regions of genes of unsweet versus sweet-red flesh watermelons (Guo et al., 2013) suggest that changes in promoters or transcriptional regulators played a major role in evolution and domestication of watermelon plants. Our population study strongly suggests that this has been the case in gaining sweetness during sweet-dessert watermelon domestication (Fig. 5). Our previous QTL study identified at least six additional QTLs associated with fruit sweetness. Finding the causal genes that underlie these QTLs could elucidate the events involved in the evolution and domestication of the sweet-dessert watermelon. The haplotype uniformity in ClTST2 promoter among sweet watermelons implies that a sequence change in this promoter occurred in an early stage of domestication.
Suc-Induced SUSIWM1 Positively Regulates ClTST2 Expression
Our results suggest a positive regulation of ClTST2 expression by SUSIWM1, a SUSIBA2-like transcription factor, which binds to a W box in a SURE element located in the ClTST2 promoter region (Fig. 6D). SURE element was first described as a binding site of a putative transcription factor that positively modulates the expression of the patatin genes, a multigene family that encodes a major potato (Solanum tuberosum) tuber protein, in a Suc-dependent manner (Grierson et al., 1994). SURE elements were present in the promoters of Suc-regulated expression genes (Grierson et al., 1994). Later, a fundamental study in barley showed that SUSIBA2, a member of the plant transcription factor WRKY superfamily, binds to a SURE element in the promoter region of isoamylase1 to enhance the endosperm-specific expression of isoamylase1 and that the expression of SUSIBA2 is induced by Suc (Sun et al., 2003). More recently, ectopic expression of the barley SUSIBA2 transcription factor in rice seeds and stems favored the allocation of photosynthesis products to above-ground biomass over allocation to roots by increasing biomass and starch content in the seeds and stems (Su et al., 2015). We show here that SUSIWM1 (Cla009557), the watermelon SUSIBA2-like transcription factor, induces the expression of ClTST2 that creates a sugar sink in the fruit by tonoplast loading. The promoter region of ClTST2 includes additional putative transcription factor binding sites (Supplemental Fig. S8). However, our data indicate that binding of SUSWM1 to the W box3, which includes the A/C transversion (SNP −1,368), is the major expression regulator of ClTST2.
Sugar content in watermelon fruit is determined by QTLs, indicating that this trait is regulated by several genes. The injection of sugar into early-stage developing fruit of strawberry (Fig. 4E), which accumulates anthocyanin pigments, and watermelon (Fig. 6B), which accumulates carotenoid pigments, triggered red fruit pigmentation, most probably due to increased fruit ripening. This may suggest that sugar regulates or induces, at least in part, additional processes associated with sweet-dessert watermelon domestication, such as pigmentation, earliness, and softening of the fruit flesh.
Our results indicate that ClTST2, a sugar transporter gene localized within a watermelon sugar content QTL region (Fig. 1), encodes a vacuole-localized transporter of Suc, Fru, and Glc (Fig. 3). Transgenic overexpression of ClTST2 increased sugar contents significantly compared with the control line (Figs. 2C-2F). ClTST2 mRNA levels, which are upregulated by SUSIWM1 (Fig. 5) are significantly positively correlated with sugar accumulation during fruit ripening (Fig. 2A–2B Supplemental Fig. S4C). These results led us to conclude that ClTST2 is the key transporter protein responsible for accumulating Suc, Fru, and Glc in the vacuole of watermelon fruit cells. Our genetic analysis of a population that included sweet and unsweet watermelon accessions indicates the sweetness trait was selected during early stages of domestication of sweet-dessert watermelon.
MATERIALS AND METHODS
Plant Materials and Sugar Content Phenotypic Data
An F8 population consisting of 96 RILs derived from a cross between the elite Chinese sweet-dessert watermelon line 97103 (Citrullus lanatus var lanatus) and the unsweet accession PI 296341-FR (C. lanatus var citroides) was used to develop the SNP bin linkage map. PI 296341-FR is a watermelon collected in southern Africa with unsweet, thick, hard flesh, and round, medium-sized light-green striped fruit that matures at ∼50 DAP. The 97103 × PI 296341-FR RIL population and parents were grown in Beijing (39.48°N, 116.28°E) over 2 years in a completely randomized block design with two replications. To ensure that the fruits were ripe, we divided the RIL population into early-maturing, midmaturing, and late-maturing subgroups based on previous maturity data for each RIL line. The early-maturing and midmaturing fruits were harvested at 30 and 35 DAP, while the late-maturing citron-type fruits were harvested at 40 to 45 DAP. Each fruit was cut lengthwise, and only the flesh from the center of the fruit was used for sugar content measurements. Degrees Brix were measured using a pocket refractometer (model pal-1; ATAGO) from a sample of juice collected from the center of each watermelon. Subsequently, the same center juice from each watermelon was used for Fru, Suc, and Glc content measurements by HPLC (LC-10A VP; Shimadzu). A Shimadzu low-temperature evaporative light scattering detector (Elsd-LT) was used for HPLC sugars detection. The Elsd-LT was operated at 40°C, 250 kPa, and air was used as the nebulizing gas (Agblevor et al., 2004). The samples were quantified using Shimadzu CLASS-VP7 chromatographic software.
Statistical Analysis of Phenotypic Data
PROC GLM in SAS8.0 was used for ANOVA. The broad-sense heritability (h2) for each trait was calculated on a plot basis as h2 = σG2/σG2 + σGE2/n + σe2/nr), where σG2, σGE2, and σe2 are the variance estimates for genotype, genotype-by-environment interaction, and experimental error, respectively; n is the number of environments and r is the number of replications. The h2 confidence intervals were calculated according to reported method (Knapp et al., 1989). The Pearson’s phenotypic correlation coefficients among traits in all environments were calculated on a mean basis using SAS PROC CORR (Littell et al., 1998).
Sugar Content QTL Detection and Association Study
In our study, we carried out two mapping analyses as follows: (1) analysis for each single year (environment) and (2) joint analysis across all environments. The single-environment and multienvironment joint analyses were performed using QTL Network software based on a mixed-model-based composite interval mapping. Mixed-model-based composite interval mapping was performed by using forward-backward stepwise regression with a threshold of P = 0.05 to select cofactors. The threshold for declaring the presence of a significant QTL was defined by 1,000 permutations at a significance level of P = 0.05. The confidence interval calculated by the odds ratio reduced by a factor of 10 was averaged for each QTL according to previous publication (Yang and Zhu, 2005). The final genetic model incorporated significant additive and epistatic effects, as well as their interactions with the environments. QTLs detected in different environments for the same trait were considered the same if their confidence intervals overlapped. Association study was performed with the compressed Mixed Liner model implemented in the software TASSEL (Zhang et al., 2010b); a total of 21,249 SNPs were used for this analysis. The 400 representative watermelon accessions used for association study were collected worldwide, including 44 wild accessions, 30 semiwild (lanatus subsp. mucosospermus), and 326 cultivars (C. lanatus ssp. vulgaris) representing the major of cultivated varieties bred by breeding institutions and worldwide seed companies in recent decades.
RNA Extraction and qRT-PCR
Total RNA was extracted using the Quick RNA isolation kit (Huayueyang Biotechnologies). cDNA was synthesized from 0.2 to 1 μg of total RNA using SuperScript III transcriptase (Invitrogen). For qRT-PCR, SYBR Green I was added to the reaction system, and the experiments were run on a Roche LightCycler 480 instrument according to the manufacturer’s instructions using the following thermal profile: 60 s hot start at 95°C followed by 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 30 s. Three replicates were carried out for each gene, and primer efficiency was calculated by the following equation: Efficiency = 10(−1/slope)−1. The watermelon ClYLS8 and ClACTIN genes (Kong et al., 2014) were used as the internal control in the analysis. The strawberry FaACTIN was used as reference gene (Amil-Ruiz et al., 2013). The relative expression levels of ClTST2, ClTST1, ClTST3, and SUSIWM1 were normalized to that of ACTIN gene transcription and averaged from the three biological replicates.
RACE and DNA Constructs
cDNA was generated by reverse transcription of RNA isolated from watermelon fruit and leaf tissues, respectively. The 5′-and 3′-RACE of the transcripts was performed using the SMARTer RACE 5′/3′ KiT (Clontech Laboratories, Takara Biomedical Technology Beijing). All PCR products were amplified with Phusion-HF DNA Polymerase (New England Biolabs). The p35S:ClTST2 eYFP fusion construct was generated using the Gateway-specific destination vector pX-YFP_GW (Chen et al., 2010b). For this construct, the ClTST2 cDNA was amplified and the stop codon removed by PCR using ClTST2-specific primers harboring the attB1 and attB2 sites (Supplemental Table S5) then cloned via the BP reaction into pDONR221F1 (Invitrogen), followed by an LR reaction to transfer the ClTST2 sequence into pX-YFP_GW. The ClTST2 gene was inserted between the EcoRI and BamHI sites in the HEK293T cell expression vector pCMV6. To generate the reporter construct pClTST2:LUC, the 1,825-, 1,500-, 1,000-, and 500-bp upstream promoter sequences of ClTST2 from lines 97103 and PI296341-FR were fused with the luciferase reporter gene and cloned into the BamHI and EcoRI sites of the vector pCAMBIA1381Z (CAMBIA).
Agrobacterium-Mediated Transformation in Watermelon
The watermelon explants were transformed according to a modified method of Yu et al. (2011). In brief, surface-sterilized watermelon seeds were sown on basic Murashige and Skoog solid medium supplemented with 3% Suc for 3 d. Then cotyledons without embryo were cut into 2 × 2 mm pieces. Agrobacterium tumefaciens strain EHA105 that harbors the binary vector was used for transformation. The cotyledon explants were cocultivated in the dark for 4 d and then transferred onto selective induction medium containing 1.5 mg/L 6 BA, 2 mg/L Basta. The regenerated adventitious buds were excised and transferred onto selective elongation medium, containing 0.1 mg/L 6 BA, 0.01 mg/L NAA, 2 mg/L Basta.
Watermelon Fruit Protoplast Isolation, Subcellular Localization, and Radioactive Sugar Accumulation Assays
To examine the subcellular localization of ClTST2 in watermelon, fruit protoplasts were isolated from cells of 5 to 10 DAP-old fruits according to the method of Yoo et al. (2007). The p35S:ClTST2 eYFP fusion construct was purified and transformed into watermelon fruit protoplasts according to a previously reported method (Malnoy et al., 2016). Protoplast volume 200 μL (2 × 105 cells) and 20 μL plasmid (20 mg) was used for transformation. Prior to the transformation, protoplast and plasmid were premixed and incubated at room temperature for 10 min. The protoplast and plasmid mix was mixed with an equal volume (220 μL) of PEG 4000, gently and immediately mixed the tube before aggregation occurred and incubating it for 10 min at 25°C. Four hundred forty microliters of W5 solution was added, mixed, and incubated at room temperature for further 10 min. Additional 1 mL W5 solution was added, mixed, and incubated at room temperature for a further 10 min. The tube was centrifuged at 100g for 5 min and discarded the supernatant, and 1 mL of W5 solution was added, followed by incubation at 25°C in dark for 16 h. Yellow fluorescence was analyzed using a confocal microscope (Zeiss LSM700) at excitation and emission wavelengths of 488 nm and 530 nm, respectively. The observed ClTST2-YFP overexpressed protoplasts were incubated in W5 solution containing 50 mm [14C] Glc or 50 mm [14C] Fru at room temperature for a further 10, 30, and 60 min. Protoplasts were collected at 100×g for 5 min for radioactivity determination by scintillation counting.
Patch-Clamp and Sugar Efflux Analyses of Transformed HEK293T and Xenopus laevis Oocyte Cells
HEK293T cells were grown in high-Glc DMEM medium (Invitrogen) with 10% fetal calf serum, 50 μg/mL penicillin, and 50 μg/mL streptomycin (Invitrogen). The cells were transiently transfected at 50% to 70% confluence using Lipofectamine 2000 Reagent (Invitrogen) in Opti-MEM I reduced-serum medium (Invitrogen). After transfection, the cells were incubated for 48 h before electrophysiological measurement. The electrophysiological recordings were obtained under visual control with a microscope (Olympus IX71). An amplifier (HEKA EPC10) was used to record the electrophysiological signal. Offset potentials were nulled directly before formation of a seal. HEK293T cell (in pF) was made from whole-cell capacitance compensation. The data were stored and analyzed with Patchmaster software. All of the experiments were performed at room temperature. A fast perfusion system was used to apply sugars directly to the cell in seconds. Each concentration was perfused over 5 min or until the current reached a steady-state level. Bath and pipette solutions were symmetrical in Hanks’ buffer with the bath and pipette solutions adjusted to pH 7.5 (HEPES/Tris) and pH 6.5 (MES/Tris), respectively. To monitor sugar-induced currents, Glc or Suc (50 mm) was applied to the bath medium of the HEK293T cell membrane. For efflux assays, oocytes were injected on day 3 after injection of the cRNA with 30 nL sugar solution containing 50-mm or 100-mm sugars (0.1 μCi μl−1 [14C] sugars) as described by Chen and coworkers (2012). The oocyte water space was estimated to be 480 nL (450 nL and 30 nL injected radiolabeled sugar) as reported by Chen et al. (2012), the corresponding estimate of the sugar concentration in oocyte was 3 mm or 6 mm.
Deciphering Positive Selection
Cross-population composite likelihood ratio test (XP-CLR) was performed by using XP-CLR software with 20-kb nonoverlapping windows. The XP-CLR calculated allele frequency differentiation between two populations by two models: the Brownian motion to model neutralized genetic drift and the deterministic model to approximate the selective sweep effect on nearby SNPs (Chen et al., 2010a). We designated windows with an XP-CLR value in the genome-wide top 1% of the empirical distribution.
Transient Gene Expression Assays in Strawberry Fruits and N. benthamiana Leaves
Agrobacterium-mediated infiltration of strawberry fruits was performed as previously described (Chen et al., 2011). For protein interaction detection, the indicated cell suspensions were mixed in equal volumes and coinfiltrated into fully expanded N. benthamiana leaves on plants ∼5 to 7 weeks old using a needleless syringe. Forty hours after infiltration, N. benthamiana leaves were sprayed with luciferin solution (100 μm luciferin, 0.1% Triton X-100, 1 mm NaOH) and kept in the dark for 6 min to quench the fluorescence. Images were captured with a low-light cooled CCD imaging apparatus (NightSHADE LB985 with Indigo software). Five to ten independent determinations were made, and experiments were repeated at least three times.
Y1H Assay
Primers used for cloning and promoter information are in Supplemental Table S5. Yeast cells were cotransformed with the pAbAi bait vector harboring the W-box of ClTST2 and the pGADT7-Rec prey vector harboring the open reading frame of SUSIWM1. As negative controls, yeast cells were transformed with the empty pGADT7 and pAbAi vectors harboring the corresponding promoter. Transformed yeast cells were grown in SD-Leu-Ura medium and were selected on SD-Leu-Ura medium plates supplemented with AbA (Sigma Aldrich). The plates were incubated for 5 d at 30°C, and positive clones were confirmed and sequenced.
EMSA and FP Assays
The full-length SUSIWM1 protein gene was fused in-frame with GST and expressed in Escherichia coli. The recombinant protein was purified by GST-agarose affinity chromatography. EMSA was performed using the Light Shift Chemiluminescent EMSA Kit (Thermo Fisher) according to the manufacturer’s instructions. The biotin-labeled DNA fragments listed in Supplemental Table S5 were synthesized and used as probes, and the same sequences with 2- or 4-fold the amount of unlabeled fragments were used as competitors in this assay. The GST protein was used as the negative control. Binding reactions were carried out using 20 and 40 ng of fusion protein, and 10 ng of each biotin-labeled promoter fragment at 25°C for 30 min. Samples were separated on 5% polyacrylamide gels (19:1 acrylamide: bisacrylamide) in 0.5× Tris-borate-ethylenediaminetetraacetic acid (EDTA) (45 mm Tris-borate, 1 mm EDTA) at 4°C. The DNA fragments were transferred to a nylon filter that was exposed to ultraviolet light to cross link the samples. The membrane was then soaked in a blocking buffer for 20 min and washed three times (each time for 5 min) with a standard buffer (provided in the kit). The pictures were captured with Azure Biosystems c300 imaging system. For FP assays, the FAM-labeled DNA fragments listed in Supplemental Table S5 were synthesized and used as fluorescence probes. Assays were performed in 96-well black plates in an EMSA binding kit. Fluorescence was measured in plate reader (M1000, TECAN) 15 to 30 min after GST-SUSIWM1 protein, and SURE DNA elements were added (Wan and Le, 2000).
Suc Treatment of Developing Cultivar Fruits
The 10% Suc solution and the control distilled water were gently injected into 10 DAP 97103 fruits. At the third day after injection, the treated fruits were sampled for sugar content and transcriptional analysis.
ChIP Assays
ChIP assays were performed as previously described (Shang et al., 2014). Protoplasts of transiently expressing 6× Myc-tagged SUSIWM1 were used in ChIP assays. Harvested protoplasts were subjected to nuclear protein extraction. The crude nuclear suspension was then sonicated for five cycles, each with a 20-s pulse with 60% of the maximal power, followed by a 40-s cooling period on ice. After sonication, the sample was incubated on ice for 20 min. Then sample was centrifuged twice at 16,000g for 10 min at 4°C. The 200-µL supernatant was used as the control. About 5 mg of chromatin was incubated with 50 µL of protein A/G agarose beads (Santa Cruz Biotechnology) for 1.5 h at 4°C with gentle rotation to preclear the sample. The precleared sample was then incubated with 30 µL antic-Myc Agarose Affinity Gel antibody (Sigma-Aldrich A7470) overnight at 4°C (sample treated with 30 µL of protein A/G agarose was used as negative control). The beads-antibody-TF-DNA complex was collected at 1,000g for 5 min at 4°C and washed once with WB1 (50 mm HEPES-KOH, pH 7.5, 150 mm NaCl, 1 mm EDTA pH 8.0, 1% [w/v] sodium deoxycholate, 1% [v/v] Triton X-100, 0.1% [w/v] sodium dodecyl sulfate [SDS], protease inhibitor), once with WB2 (50 mm HEPES-KOH, pH 7.5, 500 mm NaCl, 1 mm EDTA, pH 8.0, 0.1% [w/v] sodium deoxycholate, 1% [v/v] Triton X-100, 0.1% [w/v] SDS, protease inhibitor), once with WB3 (50 mm Tris-Cl, pH 8.0, 0.25 mm LiCl, 1 mm EDTA, 0.5% [v/v] NP-40, and 0.5% [w/v] sodium deoxycholate, protease inhibitor), and twice with TE (10 mm Tris-Cl, pH 8.0, 1 mm EDTA, protease inhibitor) at 4°C. The beads-antibody-TF-DNA complex was collected and resuspended in 400 µL proteinase K buffer (50 mm Tris-Cl, pH 8.0, 25 mm EDTA, 1.25% [w/v] SDS) and incubated at 45°C for 2 h with addition of 10 µL proteinase K (20 mg/mL). In parallel, the control sample was diluted to 400 µL and incubated with 10 µL proteinase K at 55°C for 2 h. Cross links were reversed by incubation at 65°C for at least 6 h or overnight. DNA was then extracted and precipitated using standard procedures. The ChIP DNA samples were suspended with 100 µL TE. The primers used for qPCR amplification for different promoters are listed in Supplemental Table S5.
Accession Numbers
Sequence data from this article can be found in the watermelon genome database (www.icugi.org) and GeneBank under the following accession numbers: the watermelon ACTIN gene (Cla007792, KY367356), ClYLS8 (Cla020175), ClTST2 (Cla000264, KY367352), ClTST1 (Cla021919, KY367354), ClTST3 (Cla014359, KY367355), and SUSIWM1 (Cla009557, KY367353).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Representative fruit of the RILs that were derived from a cross between the red fruit elite Chinese line 97103 and the white fruit PI296341-FR.
Supplemental Figure S2. Watermelon SNP-based bin map of chromosome 2 in the RIL population.
Supplemental Figure S3. The primary structure of ClTST2 shows two amino acid variations between the sweet cultivar watermelon 97103 and the wild-type low-sugar-content watermelon accession PI296341-FR.
Supplemental Figure S4. Transcriptome profiles and the vacuole-localized ClTST2 transporter.
Supplemental Figure S5. Sugar contents in high-sugar-content watermelon line 97103 fruit, rind, leaf, root, and fruit of line PI296341-FR.
Supplemental Figure S6. Localization of the ClTST2 protein in the Xenopus oocytes and HEK293T cells.
Supplemental Figure S7. Functional analysis of the ClTST2 promoter in N. benthamiana leaves and strawberry (Fragaria × ananassa) fruits.
Supplemental Figure S8. Deletion endpoints and predicted transcription factor recognition sites in the 1,793- and 1,825-bp promoter regions of ClTST2 from the 97103 and PI296341-FR genomes.
Supplemental Table S1. Mean value, variance components, and heritability for sugar content traits of the 97103 × PI 296341-FR watermelon RIL population in 3 years.
Supplemental Table S2. Gene ID, location, and expressed candidate genes in different tissues in the QTL mapping region during watermelon fruit maturation.
Supplemental Table S3. Watermelon ripening time (DAP) had negative correlation (r = –0.768) with sugar content in the 97 RILs.
Supplemental Table S4. SNP variation in the ClTST2 promoter region and sugar content in 124 watermelon germplasm accessions.
Supplemental Table S5. Name, sequence, amplification efficiency, and quantfication cycle (Cq) value of oligonucleotide primers used in this study.
Footnotes
This research was supported by grants from the National Key R&D Program of China (2016YFD0100506), National Natural Science Foundation of China (31772328, 31772329), Beijing Scholar Program (BSP026), Beijing Agriculture Innovation Consortium (BAIC10-2017), the Beijing Nova Program (Z171100001117032, Z161100004916081), the Beijing Natural Science Foundation (6141001), the Beijing Excellent Talents Program (2014000021223TD03), and the Ministry of Agriculture of China (CARS-26).
References
- Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thévenot P, Lemoine R, Atanassova R, Laloi M (2010) The Vitis vinifera sugar transporter gene family: Phylogenetic overview and macroarray expression profiling. BMC Plant Biol 10: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agblevor FA, Murden A, Hames BR (2004) Improved method of analysis of biomass sugars using high-performance liquid chromatography. Biotechnol Lett 26: 1207–1211 [DOI] [PubMed] [Google Scholar]
- Amil-Ruiz F, Garrido-Gala J, Blanco-Portales R, Folta KM, Muñoz-Blanco J, Caballero JL (2013) Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS One 8: e70603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D (2010. a) Population differentiation as a test for selective sweeps. Genome Res 20: 393–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010. b) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J-I, Burla B, Lee D-W, Ryoo N, Hong S-K, Kim H-B, Eom J-S, Choi S-B, Cho M-H, Bhoo SH, et al. (2010) Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytol 186: 657–668 [DOI] [PubMed] [Google Scholar]
- Chomicki G, Renner SS (2015) Watermelon origin solved with molecular phylogenetics including Linnaean material: Another example of museomics. New Phytol 205: 526–532 [DOI] [PubMed] [Google Scholar]
- Dai N, Cohen S, Portnoy V, Tzuri G, Harel-Beja R, Pompan-Lotan M, Carmi N, Zhang G, Diber A, Pollock S, et al. (2011) Metabolism of soluble sugars in developing melon fruit: A global transcriptional view of the metabolic transition to sucrose accumulation. Plant Mol Biol 76: 1–18 [DOI] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, et al. (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549 [DOI] [PubMed] [Google Scholar]
- Giaquinta RT. (1979) Sucrose translocation and storage in the sugar beet. Plant Physiol 63: 828–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt EE, Huber SC (1992) Regulation of photosynthesis by end-product accumulation in leaves of plants storing starch, sucrose, and hexose sugars. Plant Physiol 99: 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Du JS, de Torres Zabala M, Beggs K, Smith C, Holdsworth M, Bevan M (1994) Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J 5: 815–826 [DOI] [PubMed] [Google Scholar]
- Guo S, Sun H, Zhang H, Liu J, Ren Y, Gong G, Jiao C, Zheng Y, Yang W, Fei Z, Xu Y (2015) Comparative transcriptome analysis of cultivated and wild watermelon during fruit development. PLoS One 10: e0130267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, Min J, Guo X, et al. (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45: 51–58 [DOI] [PubMed] [Google Scholar]
- Hawker JS, Hatch MD (1965) Mechanisms of sugar storage by mature stem tissue of sugarcane. Physiol Plant 18: 444–453 [Google Scholar]
- Hedrich R, Sauer N, Neuhaus HE (2015) Sugar transport across the plant vacuolar membrane: Nature and regulation of carrier proteins. Curr Opin Plant Biol 25: 63–70 [DOI] [PubMed] [Google Scholar]
- Jia H, Wang Y, Sun M, Li B, Han Y, Zhao Y, Li X, Ding N, Li C, Ji W, Jia W (2013) Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol 198: 453–465 [DOI] [PubMed] [Google Scholar]
- Jung B, Ludewig F, Schulz A, Meißner G, Wöstefeld N, Flügge U-I, Pommerrenig B, Wirsching P, Sauer N, Koch W, et al. (2015) Identification of the transporter responsible for sucrose accumulation in sugar beet taproots. Nat Plants 1: 14001. [DOI] [PubMed] [Google Scholar]
- Knapp SJ, Bridges-Jr WC, Yang MH (1989) Nonparametric confidence interval estimators for heritability and expected selection response. Genetics 121: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Yuan J, Gao L, Zhao S, Jiang W, Huang Y, Bie Z (2014) Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS One 9: e90612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux F, Kappel C, Lecourieux D, Serrano A, Torres E, Arce-Johnson P, Delrot S (2014) An update on sugar transport and signalling in grapevine. J Exp Bot 65: 821–832 [DOI] [PubMed] [Google Scholar]
- Li M, Feng F, Cheng L (2012) Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One 7: e33055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell RC, Henry PR, Ammerman CB (1998) Statistical analysis of repeated measures data using SAS procedures. J Anim Sci 76: 1216–1231 [DOI] [PubMed] [Google Scholar]
- Liu J, Guo S, He H, Zhang H, Gong G, Ren Y, Xu Y (2013) Dynamic characteristics of sugar accumulation and related enzyme activities in sweet and non-sweet watermelon fruits. Acta Physiol Plant 35: 3213–3222 [Google Scholar]
- Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Kanchiswamy CN (2016) DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Ren Y, McGregor C, Zhang Y, Gong G, Zhang H, Guo S, Sun H, Cai W, Zhang J, Xu Y (2014) An integrated genetic map based on four mapping populations and quantitative trait loci associated with economically important traits in watermelon (Citrullus lanatus). BMC Plant Biol 14: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. ; International HapMap Consortium (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandlin K, Prothro J, Heesacker A, Khalilian N, Okashah R, Xiang W, Bachlava E, Caldwell DG, Taylor CA, Seymour DK, et al. (2012) Comparative mapping in watermelon [Citrullus lanatus (Thunb.) Matsum. et Nakai]. Theor Appl Genet 125: 1603–1618 [DOI] [PubMed] [Google Scholar]
- Schulz A, Beyhl D, Marten I, Wormit A, Neuhaus E, Poschet G, Büttner M, Schneider S, Sauer N, Hedrich R (2011) Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J 68: 129–136 [DOI] [PubMed] [Google Scholar]
- Shang Y, Ma Y, Zhou Y, Zhang H, Duan L, Chen H, Zeng J, Zhou Q, Wang S, Gu W, et al. (2014) Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346: 1084–1088 [DOI] [PubMed] [Google Scholar]
- Slewinski TL. (2011) Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol Plant 4: 641–662 [DOI] [PubMed] [Google Scholar]
- Snapp EL, Lajoie P (2011) Imaging of membrane systems and membrane traffic in living cells. Cold Spring Harb Protoc 2011: 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Hu C, Yan X, Jin Y, Chen Z, Guan Q, Wang Y, Zhong D, Jansson C, Wang F, et al. (2015) Expression of barley SUSIBA2 transcription factor yields high-starch low-methane rice. Nature 523: 602–606 [DOI] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15: 2076–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan QH, Le XC (2000) Studies of protein-DNA interactions by capillary electrophoresis/laser-induced fluorescence polarization. Anal Chem 72: 5583–5589 [DOI] [PubMed] [Google Scholar]
- Wang TD, Zhang HF, Wu ZC, Li JG, Huang XM, Wang HC (2015) Sugar uptake in the Aril of litchi fruit depends on the apoplasmic post-phloem transport and the activity of proton pumps and the putative transporter LcSUT4. Plant Cell Physiol 56: 377–387 [DOI] [PubMed] [Google Scholar]
- Wingenter K, Schulz A, Wormit A, Wic S, Trentmann O, Hoermiller II, Heyer AG, Marten I, Hedrich R, Neuhaus HE (2010) Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol 154: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ejiri Y, Tanaka K (2002) Glycation by ascorbic acid causes loss of activity of ribulose-1,5-bisphosphate carboxylase/oxygenase and its increased susceptibility to proteases. Plant Cell Physiol 43: 1334–1341 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhu J (2005) Methods for predicting superior genotypes under multiple environments based on QTL effects. Theor Appl Genet 110: 1268–1274 [DOI] [PubMed] [Google Scholar]
- Yativ M, Harary I, Wolf S (2010) Sucrose accumulation in watermelon fruits: genetic variation and biochemical analysis. J Plant Physiol 167: 589–596 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu TA, Chiang CH, Wu HW, Li CM, Yang CF, Chen JH, Chen YW, Yeh SD (2011) Generation of transgenic watermelon resistant to Zucchini yellow mosaic virus and Papaya ringspot virus type W. Plant Cell Rep 30: 359–371 [DOI] [PubMed] [Google Scholar]
- Zhang B, Tolstikov V, Turnbull C, Hicks LM, Fiehn O (2010. a) Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proc Natl Acad Sci USA 107: 13532–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fna J, Guo S, Ren Y, Gong G, Zhang J, Weng Y, Davis A, Xu Y (2016) Genetic diversity, population structure, and formation of a core collection of 1197 citrullus accessions. HortScience 51: 23–29 [Google Scholar]
- Zhang R, Xu Y, Yi K, Zhang H, Liu L, Gong G (2004) A genetic linkage map for watermelon derived from recombination inbred lines. J Amer Soc Hort Sci 129: 237–243 [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES (2010. b) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]