HAWAIIAN SKIRT is a cofactor for Arabidopsis thaliana microRNA function and acts in an F-box-dependent manner.
Abstract
As regulators of gene expression in multicellular organisms, microRNAs (miRNAs) are crucial for growth and development. Although a plethora of factors involved in their biogenesis and action in Arabidopsis (Arabidopsis thaliana) has been described, these processes and their fine-tuning are not fully understood. Here, we used plants expressing an artificial miRNA target mimic (MIM) to screen for negative regulators of miR156. We identified a new mutant allele of the F-box gene HAWAIIAN SKIRT (HWS; At3G61590), hws-5, as a suppressor of the MIM156-induced developmental and molecular phenotypes. In hws plants, levels of some endogenous miRNAs are increased and their mRNA targets decreased. Plants constitutively expressing full-length HWS—but not a truncated version lacking the F-box domain—display morphological and molecular phenotypes resembling those of mutants defective in miRNA biogenesis and activity. In combination with such mutants, hws loses its delayed floral organ abscission (“skirt”) phenotype, suggesting epistasis. Also, the hws transcriptome profile partially resembles those of well-known miRNA mutants hyl1-2, se-3, and ago1-27, pointing to a role in a common pathway. We thus propose HWS as a novel, F-box dependent factor involved in miRNA function.
MicroRNAs (miRNAs) are 21-nucleotide-long to 24-nucleotide-long single-stranded RNA molecules that are crucial for regulating and fine-tuning gene expression in multicellular organisms (Bartel, 2009). In plants, transcription of MIRNA genes generates longer precursor RNAs with characteristic stem-loop folding, and is followed by two major nuclear processing (“dicing”) steps mediated by the endoribonuclease DICER-LIKE1 (DCL1), and cofactors including SERRATE (SE) and HYPONASTIC LEAVES1 (HYL1; Bernstein et al., 2001; Kurihara and Watanabe, 2004; Vazquez et al., 2004; Laubinger et al., 2008). The first dicing step, usually near the base of the precursor stem, excises the primary miRNA stem-loop, which is further cut to remove the loop and form the duplex of miRNA and miRNA*. After 3′-O methylation and translocation to the cytoplasm (Yu et al., 2005), the mature miRNA, or guide, associates with an ARGONAUTE (AGO) protein and together they form an active miRNA INDUCED SILENCING COMPLEX (miRISC). By sequence complementarity of the miRNA to its mRNA targets, miRISC selectively inhibits translation or triggers transcript cleavage (Brodersen et al., 2012; Rogers and Chen, 2013). Balancing the abundance of miRNAs, the degradation of both free and AGO1-bound miRNAs involves their 3′-truncation through members of the SMALL RNA DEGRADING NUCLEASE family of exonucleases and consecutive uridylation by HEN1 SUPPRESSOR1 and UTP:RNA URIDYLYLTRANSFERASE1 (Tu et al., 2015; Yu et al., 2017).
Mechanisms that control the levels of miRNAs and thereby adjust target silencing have been described: transcriptional regulation ensures the correct spatio-temporal expression of MIR genes, whereas posttranscriptional steps further fine-tune miRNA accumulation (Achkar et al., 2016). Additionally, mature miRISC activity can be attenuated by miRNA target mimicry (Franco-Zorrilla et al., 2007). In that mechanism, a noncoding RNA containing a sequence motif that is complementary to a miRNA sequesters the respective miRISC and thus renders it unavailable for inhibition of regular mRNA targets (Franco-Zorrilla et al., 2007). In animals, competing endogenous RNAs play a similar role, although their biological significance remains controversial (Bak and Mikkelsen, 2014; Wang et al., 2015; Denzler et al., 2016). Only a single case of natural miRNA target mimicry, that of INDUCED BY PHOSPHATE STARVATION1 (IPS1), which interferes with the activity of miR399, has been studied in detail in plants (Franco-Zorrilla et al., 2007). However, positive posttranscriptional regulation by mismatched miRNAs associating with their target and thereby protecting it, has been proposed as a mimicry-related mechanism (Couzigou et al., 2016).
The miRNA target mimicry principle has been exploited to investigate the biological function of miRNAs (Franco-Zorrilla et al., 2007; Sha et al., 2014). Artificial miRNA target mimicry lines (MIMs) based on the endogenous IPS1 transcript, or on entirely artificial sequences, have been used to interfere with dozens of Arabidopsis (Arabidopsis thaliana) miRNAs (Franco-Zorrilla et al., 2007; Todesco et al., 2010; Yan et al., 2012; Reichel et al., 2015). Many conserved plant miRNAs are encoded by medium-size gene families (Li and Mao, 2007). In Arabidopsis, one such family is the deeply conserved miR156 family with eight MIR genes, encoding miR156a to miR156h (Griffiths‐Jones, 2004; Kozomara and Griffiths-Jones, 2014). Mature miR156 accumulates in the shoots of juvenile plants and gradually decreases in abundance as the plant ages (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009; Wang, 2014). It regulates at least 10 out of 16 members of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factor family, which control a range of biological processes, most of them relating to developmental progression during the vegetative phase of plant growth (Xu et al., 2016).
Plants constitutively expressing the IPS1-based MIM156 transgene have reduced miR156 levels and activity. Accordingly, they have an accelerated juvenile-to-adult phase transition as well as exaggerated adult growth traits such as serrated leaf margins (Franco-Zorrilla et al., 2007). Similar phenotypes have been observed in plants expressing miR156-insensitive versions of SPL targets (Wu and Poethig, 2006; Franco-Zorrilla et al., 2007; Wang et al., 2008). In such plants, the rate at which rosette leaves are initiated during vegetative growth is also greatly reduced, and as in plants overexpressing a MIM156 transgene, cotyledons are bent and spoon-shaped (Todesco et al., 2010). In contrast, ectopic overexpression of miR156, which reduces SPL levels, prolongs the juvenile phase and accelerates plastochron of rosette leaves (Wu and Poethig, 2006).
Fine-tuning of miR156 effects on its targets could be mediated by additional factors. We sought to identify such negative regulators of miR156 activity by screening for suppressors of MIM156-induced developmental alterations, i.e. factors that normally prevent compensation of the effects of MIM-induced lack of miR156. Here, we describe the F-box protein encoded by HAWAIIAN SKIRT (HWS; At3G61590) as a suppressor not only of miR156, but generally of miRNA activity in Arabidopsis. As part of an Skp-Cullin-F-box (SCF) complex, F-box proteins provide E3 ubiquitin ligases with target specificity via recognition of substrates for ubiquitination (Risseeuw et al., 2003). HWS was previously shown to interact with the classical components of an SCF complex, i.e. ARABIDOPSIS SKP1-like 20A and ARABIDOPSIS SKP1-like 20B, which function as a bridge between CULLIN1 and HWS (Kuroda et al., 2002; Ogura et al., 2008). To date, no substrates for HWS, marked either for proteasome-mediated degradation or other molecular processes, have been identified. Mutations in HWS affect root growth via regulation of quiescent-center independent meristem activity as well as stomatal guard cell development (Yu et al., 2015; Kim et al., 2016). Due to delayed abscission, hws mutants also fail to shed sepals, petals, and anthers (González-Carranza et al., 2007). Loss of HWS furthermore results in increased organ growth, whereas overexpression yields smaller plants with elongated, serrated, and hyponastic leaves (González-Carranza et al., 2007).
Our study demonstrates that HWS broadly affects plant miRNA function. Mutations in HWS increase the steady-state levels of several miRNAs, resulting in corresponding decreases in the levels of their respective targets. Overexpression of full-length HWS, on the other hand, reduces the levels of miRNAs and increases their targets, but only when the F-box domain is intact. The characteristic delayed floral organ abscission, or skirt phenotype, of hws mutants is lost when combined with mutants defective in miRNA biogenesis. This indicates that HWS and miRNA factors like SE, HYL1, and AGO1 are epistatic to each other and active in a common pathway, a notion supported by significant overlaps within hws and miRNA mutant transcript profiles. We propose that HWS is a new factor involved in the biogenesis or function of miRNAs, and exerts its role through an F-box-dependent process.
RESULTS
The hws-1 Mutant Suppresses miRNA156 Target Mimicry-induced Developmental Alterations
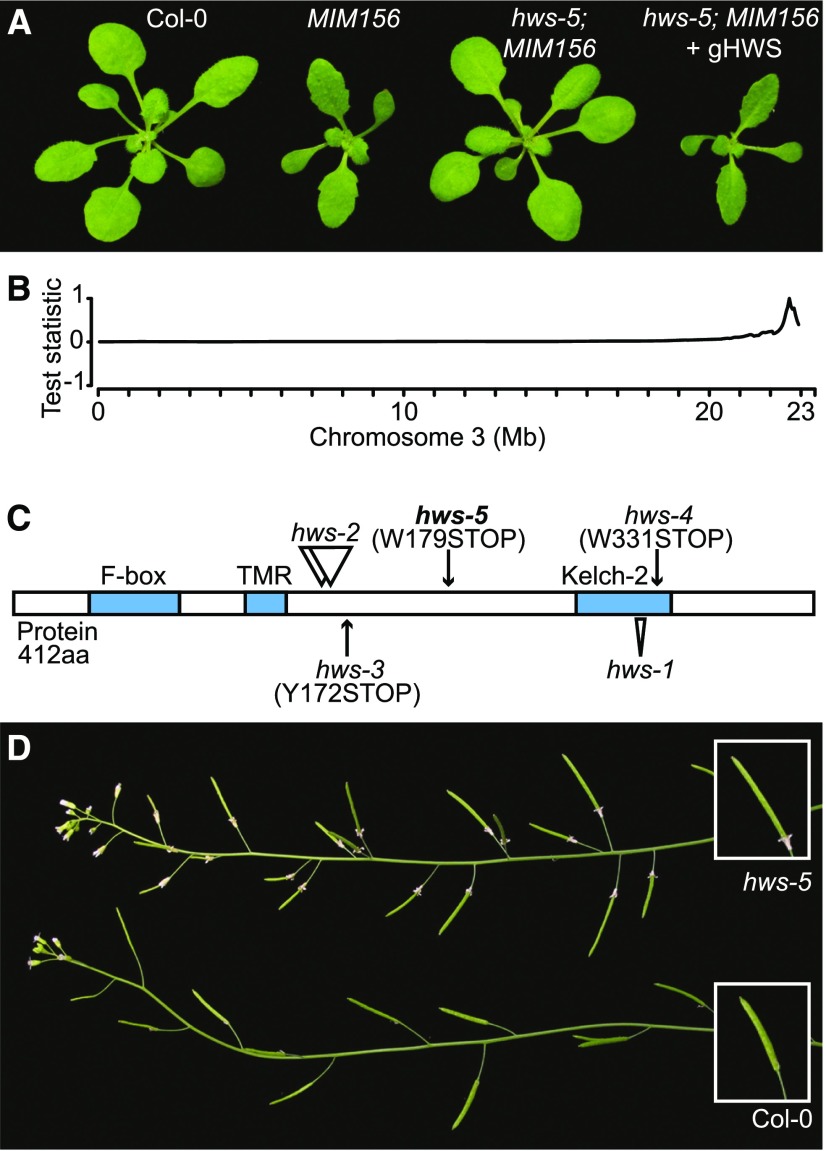
To identify genetic modifiers of plants expressing an IPS1-based MIM156 transgene, we focused on three easily monitored developmental abnormalities characteristic of lines ectopically expressing this transgene: spoon-shaped cotyledons, premature rosette leaf serration, and a reduced leaf initiation rate during vegetative growth. One line that we found after ethyl methanesulfonate (EMS) mutagenesis displayed suppression of all investigated MIM156-induced developmental abnormalities (Fig. 1). Using mapping by sequencing, we localized the causal mutation to a region on the right arm of chromosome 3 (Fig. 1; see “Materials and Methods” for details).
Figure 1.
Characterization of the hws-5 mutant. A, Phenotype of 21-d-old Col-0, MIM156, hws-5; MIM156 and hws-5; and MIM156 + gHWS plants. B, Chromosome 3 SHOREmap results for hws-5. C, Location and effects of the mutations on the HWS protein. Annotated or predicted domains are marked in blue. D, Sepal-fusion skirt phenotype and phyllotactic distortion in hws-5 compared to Col-0.
Within this region, a G-to-A substitution (G537A, Chr3:22793585, TAIR10) was identified that caused a premature termination codon (W179STOP) in HAWAIIAN SKIRT (HWS). HWS is a 412-amino acid protein with an N-terminal F-box domain, a predicted transmembrane domain, and a C-terminal Kelch-2 domain (Fig. 1; González-Carranza et al., 2007). The typical MIM156 phenotype was restored in mutants transformed with a genomic construct of HWS, confirming that HWS was indeed the causal locus (Fig. 1). We henceforth refer to the mutant allele as “hws-5”. Levels of HWS transcript detected by reverse transcription quantitative PCR (RT-qPCR) are decreased in hws-5 compared to wild type (Supplemental Fig. S1; see below for description of mutant).
Other hws mutant alleles have previously been isolated and shown to affect root meristem activity (hws-3 and hws-4; Kim et al., 2016), or to be impaired in the abscission of floral organs (hws-1 and hws-2; González-Carranza et al., 2007). Incomplete separation of sepals imposes a structural barrier that prevents the shedding of sepals, petals, and stamens throughout completion of the plant’s lifecycle (González-Carranza et al., 2007). Both with and without the MIM156 transgene, we observed similarly impaired abscission in hws-5 plants (Fig. 1, Supplemental Fig. S1). Partial fusion of cauline leaves to the inflorescence stem was evident in both hws-1 and hws-5 mutants, and like sepal abscission persisted in the presence of the MIM156 transgene (Supplemental Fig. S1).
Mutations in HAWAIIAN SKIRT Affect MIM-induced Phenotypes
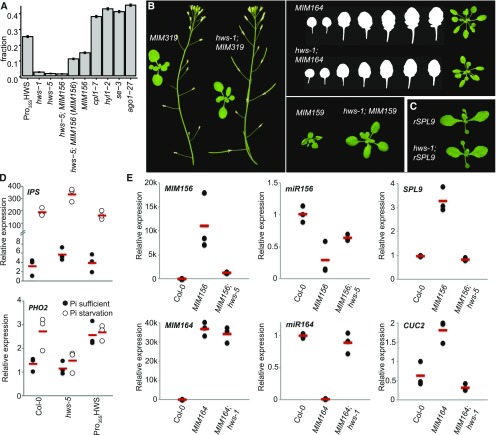
To determine whether HWS’s activity was specific to miR156, we tested if other mimicry lines were equally affected. We combined hws-1 with three additional, ubiquitously expressed mimicry transgenes: MIM159, MIM164, and MIM319. Each of these transgenes causes distinct developmental alterations, including hyponastic leaves in MIM159, increased leaf serrations in MIM164, and reduced fertility in MIM319 lines (Todesco et al., 2010). We found that the hws-1 mutation could suppress the characteristic phenotypes of all three mimicry lines (Fig. 2), implying HWS action upstream of MIM156-specific factors like the miR156-targeted SPL transcripts. As expected, presence of the MIM319 and MIM159 transgenes did not affect the hws abscission phenotype (Supplemental Fig. S2 and see below).
Figure 2.
Effects of hws mutation on MIM transgene-dependent and -independent phenotypes. A, Fraction of differentially expressed genes in 35S::HWS, hws-1, hws-5, hws-5; MIM156 (compared to Col-0 and to MIM156), MIM156 (n = 2), miRNA-mutants cpl1-7, hyl1-2, and se-3 (n = 3; data from Manavella et al., 2012) and ago1-27 (n = 3, data from SRA PRJNA309714) with 95% confidence intervals. For P values, see Supplemental Table S4. B, Suppression of MIM159, MIM319, and MIM164 phenotypes in hws-1 background. C, miR156-resistant rSPL9 and rSPL9; hws-5 plants. D, Relative expression of IPS and PHO2 in Col-0, hws-5, and hws-1 plants harboring 35S::HWS. E, Relative expression of MIM156, miR156, SPL9 in Col-0, MIM156, and hws-5; MIM156 and of MIM164, miR164 and CUC2 in Col-0, MIM164, and hws-1; MIM164. Dots represent biological replicates as means of technical replicates (n = 3); bars indicate means of biological replicates.
To confirm that HWS acts upstream of miRNA-regulated transcription factors, we introduced a miR156-resistant SPL9 (rSPL9) transgene into hws mutants. This transgene expresses a version of SPL9 that avoids regulation by miR156 due to the presence of five base substitutions in its miR156 target site (Wang et al., 2008). Like MIM156, rSPL9 plants accumulate higher levels of SPL9 and display MIM156-like phenotypes, including spoon-shaped cotyledons and a slower leaf initiation rate (Fig. 2). We found that the rSPL9 phenotype was similar in wild-type and hws-5 backgrounds (Fig. 2), indicating that HWS plays a role upstream of miRNA target stability and/or activity.
We speculated that a more general role of HWS could be to impede MIM action at the level of the IPS1-based MIM transcript. In this case, hws mutation would affect transgene and endogenous IPS1 transcripts similarly. We thus monitored IPS1 steady-state levels, both under normal conditions and phosphorus (Pi) starvation, inducing the otherwise lowly expressed endogenous IPS1 to facilitate detection (Martín et al., 2000). Independent of Pi supply, IPS1 accumulation was increased in hws-5 compared to the wild type (Fig. 2). Overabundance of mimicry transcripts (i.e. IPS1) should increase miRNA sequestration, and consequently release target suppression. Levels of PHO2, the endogenous target of IPS1-bound miR399, however, remained unaffected (Fig. 2). This may indicate a defect in miRNA-mediated target regulation or result from feedback regulation within the IPS1-miR399-PHO2 module buffering fluctuations of PHO2 accumulation (Fujii et al., 2005; Bari et al., 2006; Chiou et al., 2006; Franco-Zorrilla et al., 2007). We therefore turned to analysis of plants harboring the engineered MIM transgenes, which are supposedly uncoupled from endogenous, IPS1-promoter-based feedback loops.
The effects of the hws-5 mutation on engineered IPS1-based transcripts were variable, but miR156 and miR164 overaccumulated. Expression of their targets was reduced in hws-5; MIM156 and hws-5; MIM164 plants, respectively, indicating restoration of miRNA-mediated target regulation (Fig. 2).
HWS Reduces miRNA Accumulation
Leaf serrations are reduced in hws mutants, whereas they are typically enhanced in miRNA biogenesis mutants such as alleles of se or ago1 (Supplemental Fig. S3; Morel et al., 2002; Laubinger et al., 2008). Moreover, both the skirt and cauline leaf fusions to the stem have been described as results of miR164 overexpression (Mallory et al., 2004b; Schwab et al., 2005). Further, when we overexpressed the HWS coding sequence from the constitutive CaMV 35S promoter, plants developed severe abnormalities, including upwards-pointing, highly serrated and hyponastic leaves, similar to what has been described before (Fig. 3; González-Carranza et al., 2007). This was also reminiscent of mutants with impaired miRNA activity, for example hyl1-2, ago1-25, and ago1-27 or hasty-3 (hst-3; Supplemental Fig. S3; Morel et al., 2002; Bollman et al., 2003; Vazquez et al., 2004). We therefore decided to test whether miRNAs were directly affected in HWS-overexpressing plants, as well as in hws mutants in a nontransgenic wild-type background. Using RT-qPCR and small RNA northern blots, we observed that miRNA levels tended to be higher in hws mutant tissue and lower in 35S::HWS plants compared to the wild-type control (Fig. 3). We also saw a decrease in the levels of miRNA-targeted transcripts of SPL3 (miR156), AGO1 (miR168), TEOSINTE BRANCHED1, CYCLOIDEA, and PCF4 (miR319), LEAF CURLING RESPONSIVENESS (miR394), and TARGET OF EARLY ACTIVATION TAGGED2 (miR172) in hws mutants, and a matching increase in 35S::HWS plants (Fig. 3). In addition, IPS1 accumulation in 35S::HWS was similar to what was seen in wild type, whereas PHO2, the IPS1/miR399 target, was upregulated, independent of Pi supply (Fig. 2).
Figure 3.
Effects of hws and HWS overexpression on miRNA and miRNA target steady-state levels. A, Phenotype of 28-d-old T1 35S::HWS plant in the hws-1 background. Note hyponastic, serrated leaves. B and C, Levels of mature miRNAs and miRNA targets in Col-0, hws-1, and 35S::HWS as measured by RT-qPCR. Dots represent biological replicates (n = 3 to 4); bars indicate mean of biological replicates as means of technical replicates. D, Mature miRNA levels in Col-0, hws-1, hws-5, MIM156, hws-5; MIM156 and 35S::HWS as determined by RNA blotting.
The particularly pronounced effects on miR164 and miR156 might be attributed to HWS being specifically active in regions where those miRNAs are expressed. Both miR156 and miR164 accumulate to high levels in emerging leaves (Nikovics et al., 2006; Wu and Poethig, 2006; Wang et al., 2008, 2009; Bazzini et al., 2009), with miR164 also showing a more restricted expression pattern around the veins and the points of leaf serration (Supplemental Fig. S3; Nikovics et al., 2006). A similar expression pattern of the HWS reporter was seen in ProHWS::GUS plants (Supplemental Fig. S3). This is in agreement with the reduction of serration found in hws plants; as in the emerging leaves of wild-type plants, miR164 acts as a suppressor of leaf serration by targeting members of the CUP-SHAPED COTYLEDON (CUC) family of transcription factor genes (Supplemental Fig. S3; Nikovics et al., 2006).
Genetic Interactions with Other miRNA Factors
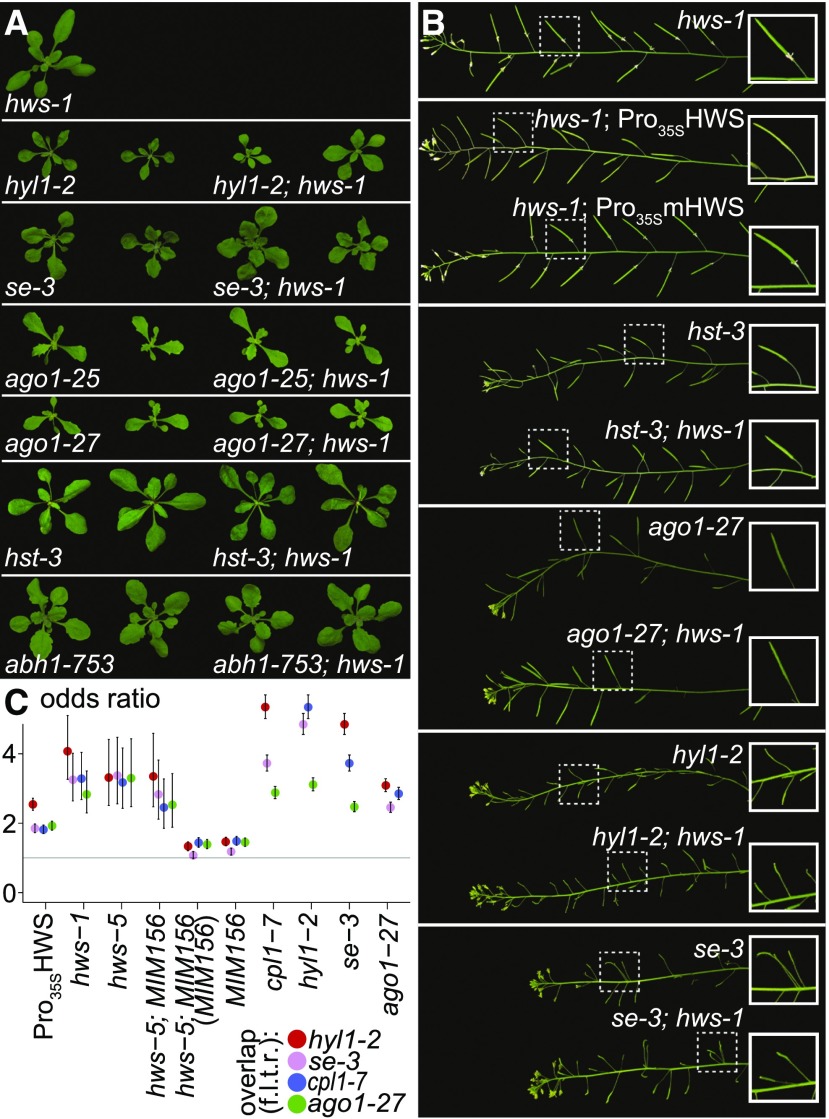
To further substantiate a connection between HWS and miRNA pathway components, we tested for genetic interactions. For this purpose, we combined the hws-1 allele with different mutant alleles of AGO1 (ago1-25, ago1-27; Morel et al., 2002), ABA HYPERSENSITIVE1 (ABH1, abh1-753; Laubinger et al., 2008), HST (hst-3; Bollman et al., 2003), HYL1 (hyl1-2; Vazquez et al., 2004), and SE (se-3; Fig. 4; Grigg et al., 2005). In all cases, hws-1 did not modify the phenotype of these mutants; this indicates that all these miRNA factors, even though they affect different aspects of miRNA biogenesis or activity, are epistatic to HWS (Fig. 4).
Figure 4.
Epistasis analysis of hws-1 and mutations in major miRNA biogenesis factors. A, Rosette phenotype of single and double homozygous F3 plants between hws-1, hyl1-2, se-3, ago1-25, ago1-27, hst-3, and abh1-753 at approximately 21 DAS. B, Abscission phenotype of double mutants of hws-1, hws-5, hyl1-2, hst-3, se-3, and ago1-27 as well as T1 of 35S::HWS and 35S::mHWS in hws-1 background. C, Odds ratio of enrichment of differentially expressed genes also found in the miRNA-mutants hyl1-2, se-3, cpl1-7, and ago1-27 as calculated by two-tailed Fisher’s exact test in 35S::HWS, hws-1, hws-5, hws-5; MIM156 (compared to Col-0 and to MIM156; n = 2), MIM156 and miRNA-mutants cpl1-7, hyl1-2, and se-3 (n = 3; data from Manavella et al., 2012) and ago1-27 (n = 3, data from SRA; PRJNA309714), with 95% confidence intervals. For P values, see Supplemental Table S2.
We reasoned that functional relatedness should be reflected in an overlap of differentially expressed (DE) genes, and thus compared those observed in hws alleles and the suppressor line hws-5; MIM156 to DE genes in the known miRNA mutants hyl1-2, se-3, and cpl1-7 (Manavella et al., 2012) and ago1-27 (SRA, PRJNA309714). Shared differential expression was twice as likely as expected by chance, not too dissimilar from pairwise comparisons among the previously described miRNA mutants themselves (two- to four-fold likelihood of shared DE, see Fig. 4; for P values of two-tailed Fisher’s exact test and odds ratios, see Supplemental Table S2). Identification of this bias, irrespective of the hws lines displaying the lowest fraction of total DE genes within our samples, emphasizes the relevance of these similarities with miRNA mutants (Fig. 2). Neither DE genes in MIM156 nor between hws-5; MIM156 and MIM156 showed a similar tendency, supporting a postulated role of HWS in miRNA function also independently of the MIM156 transgene.
Owing to its F-box domain, HWS’s mode of action could involve an SCF-complex, as HWS has been shown to interact with the common SCF component Arabidopsis-Skp protein ASK1 (Kuroda et al., 2002; Ogura et al., 2008). There is precedence for involvement of F-box proteins in miRNA function, examples being the viral silencing suppressor P0 (Pazhouhandeh et al., 2006; Baumberger et al., 2007; Bortolamiol et al., 2007) and F-BOX WITH WD-40 2 (FBW2; Earley et al., 2010). Full-length 35S::HWS complemented the hws-1 phenotype, whereas transformants expressing a version that lacks the F-box domain (35S:mHWS transgene; Supplemental Fig. S5) retained the hws-characteristic skirt (Supplemental Fig. S1). Furthermore, 35S::mHWS did not induce the hyl1-like and ago1-like leaf phenotypes observed in 35S::HWS plants (Fig. 3; Supplemental Fig. S3). Consistently, steady-state levels of miRNAs and their targets in 35S::mHWS were more similar to wild type (Supplemental Fig. S3).
To detect HWS interactors in planta, we harvested rosette leaves from 35S::Citrine-HWS plants, immunoprecipitated the fusion protein with a GFP-antibody, and performed mass spectrometry. As controls, we included leaves from both 35S::Citrine-mHWS and 35S::GFP. Enrichment of ASK1 and two other SCF-complex proteins, ARABIDOPSIS SKP-LIKE2 (At5g42190) and CULLIN1 (At4g02570), in the 35S::Citrine-HWS fraction, but in neither of the two controls supported HWS’s function as a classical F-box protein (Supplemental Table S3). Another protein detected via mass spectroscopy—albeit at lower levels also in the controls—was the miRNA-factor AGO1, known to associate with F-box proteins (Pazhouhandeh et al., 2006; Baumberger et al., 2007; Bortolamiol et al., 2007; Earley et al., 2010; Csorba et al., 2015). Similarities between the HWS overexpression phenotype and ago1 supported a potential functional connection, and AGO1’s role in miRISC assembly fits with presumed HWS action upstream of miRNA targets. However, we could not confirm an interaction with HWS using yeast-2-hybrid (Y2H) or coimmunoprecipitation assays (Supplemental Figs. S4 and S5; Fields and Song, 1989; Manavella et al., 2012)). Although presence of the F-box had a positive influence on HWS’s stability itself, we did not observe effects of HWS overexpression on AGO1 accumulation, or on AGO1 ubiquitination mediated by a HWS-associated SCF-complex (Supplemental Figs. S5 and S6). We further did not detect substantial effects of HWS overexpression on the abundance of SE, HYL1, and DCL1—three additional core miRNA function factors (Supplemental Fig. S7).
Beyond this, we did not detect strong associations of HWS with known miRNA-related proteins. Hence, it is possible that HWS contributes to the miRNA pathway through a protein not yet described in this context—that the interaction with an already known factor is weak or transient, as described for other F-box proteins (Earley et al., 2010; Coyaud et al., 2015)—or that the concentration of the interactor in entire leaves is low and therefore not detectable by our approach. Although we are confident that the F-box domain is important for HWS’s function in the miRNA context, its mode of action and interaction(s) with miRNA factors remain to be determined.
Transcriptome Profiles Support a Role of HWS as miRNA Factor
Analyzing RNA profiles by sequencing (RNA-seq), we identified DE genes in the suppressor line hws-5; MIM156 compared to the isogenic MIM156 parent (false discovery rate adjusted P value < 0.05). Up-regulation of IPS1 (At3G09922) transcripts in both lines, although more strongly in MIM156, confirmed that suppression was not simply due to loss of expression of the IPS1-based MIM156 transgene. DE genes in Col-0 and hws-5; MIM156 compared to MIM156 plants are largely overlapping and similarly up- or down-regulated (99% of genes in the overlap). This indicates that the suppression of the MIM156 morphological phenotypes in hws-5; MIM156 seedlings is broadly reflected in a normalization of the transcriptional profile (Supplemental Fig. S1; Supplemental Table S1).
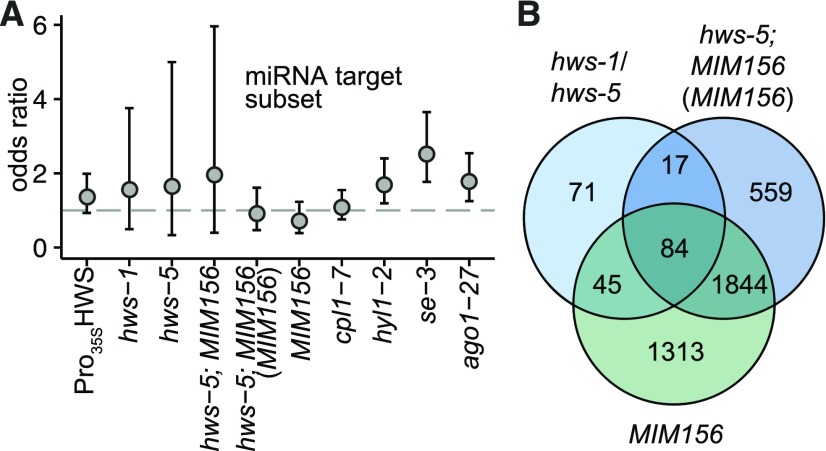
For further analysis of expression changes in hws, we compiled a “silencing” list, containing genes involved in miRNA biogenesis, predicted and validated miRNA targets and miRNA encoding genes (Supplemental Table S6). We used this list to determine whether genes regulated in a HWS-dependent manner would support a role of HWS as a miRNA factor and furthermore whether they might point to the place of action of HWS within the pathway. Silencing genes, and particularly miRNA targets alone, were overrepresented among the genes differentially expressed in hyl1-2, se-3, and ago1-27 (P values of two-tailed Fisher’s exact test odds ratios P = 0.001358, P = 3.65*10^-5, P = 0.036262 and P = 0.002467, P = 8.10*10^-8, P = 0.000811, see Supplemental Table S2; and SRA PRJNA309714; Manavella et al., 2012). In hws lines, we did not detect such a general silencing gene overrepresentation. Still, 35S::HWS displayed miRNA target enrichment (P = 0.092868)—more pronounced even than the miRNA mutant cpl1-7 (Fig. 5). Therefore, our annotation seems to reliably determine the miRNA-relatedness of main pathway components such as HYL1, SE, and AGO1, whereas weaker and functionally diverse mutants like cpl1-7 (and hws) are more difficult to assign. Further, RT-qPCR in our case appears to have higher sensitivity than RNA-seq for detecting miRNA target level changes (Fig. 3). Analysis of the target-subset nevertheless supports the observed similarities between 35S::HWS and miRNA-mutants, suggesting HWS activity after miRNA transcription and biogenesis.
Figure 5.
Transcriptome effects of hws. A, Odds ratio of enrichment of differentially expressed genes in the validated miRNA target gene subset as calculated by two-tailed Fisher’s exact test in 35S::HWS, hws-1, hws-5, hws-5; MIM156 (compared to Col-0 and to MIM156; n = 2), MIM156 and miRNA-mutants cpl1-7, hyl1-2, and se-3 (n = 3; data from Manavella et al., 2012) and ago1-27 (n = 3, data from SRA PRJNA309714), with 95% confidence intervals. For P values, see Supplemental Table S2; for gene list, see Supplemental Table S6. B, Venn diagram of differentially expressed genes in the hws mutants combined compared to MIM156 and hws-5; MIM156.
Looking more closely at additional transcript changes, we found that approximately half of the DE genes in each of the two hws mutants were the same and changed in the same direction relative to wild type (Fig. 5; Supplemental Fig. S8; Supplemental Table S5). Among those 217 broadly HWS-dependent genes, three were part of our silencing gene list, and even found amid the overlapping DE genes in hws-5; MIM156: the miRNA-biogenesis-related GLY-RICH RNA-BINDING PROTEIN7 (GRP7, At2G21660, also detected in mass spectroscopy), and the two miRNA-targets COPPER/ZINC SUPEROXIDE DISMUTASE 1 (CSD1, AT1G08830) and OLIGOPEPTIDE TRANSPORTER1 (OPT1, AT5G55930; van Nocker and Vierstra, 1993; Sunkar et al., 2006; Ahmed et al., 2014; Köster et al., 2014). Two of those, GRP7 and CSD1, are absent in the comparison of MIM156 to Col-0. Consequently, they are part of the fraction of DE genes unrelated to the phenotypic reversion of MIM156-related developmental alterations, and thus potentially indicative of general HWS downstream effects independent of alterations in miR156 activity, maybe involving other miRNAs (Supplemental Table S8).
To find genes regulated by HWS, irrespective of the presence or absence of miR156-related transgenes (including those affected solely in the MIM156 background; see Supplemental Table S7), we intersected three groups of genes: those changed in hws-5; MIM156 with respect to its MIM156 parent, those changed in both hws mutants, and those changed in the MIM156 line (the latter three relative to the Col-0 wild type). The resulting list of genes affected in all three comparisons again includes GRP7, CSD1, and OPT1, as well as 98 additional genes, of which around half are annotated as enzymes (Supplemental Table S9; TAIR; Berardini et al., 2015). Of the whole list, 33 (33%) were also differentially regulated in at least three out of the four analyzed miRNA mutants. Assuming that HYL1, SE, and AGO1 primarily function in (mi)RNA metabolism, the overlap of numerous DE genes, many encoding regulatory proteins, in hws and the miRNA mutants could suggest a joint merging point upstream of those.
DISCUSSION
MiRNA production and function involve a multitude of both general and more specialized factors (Rogers and Chen, 2013; Reis et al., 2015; Achkar et al., 2016). Using a MIM156-based genetic screen, we have identified the F-box protein HWS as a new factor involved in plant miRNA biology. Both genetic and molecular evidence support a role of HWS in miRNA-dependent processes that goes beyond miR156.
Our observation that MIM156 levels are reduced in hws mutants (Fig. 2; Supplemental Fig. S1) points to a disruption of the equilibrium between MIM and miRNA accumulation. This raises the question where HWS interferes with this balance, and if it is acting down- or upstream of the miR156/SPL regulon. Suppression of additional, miR156-unrelated MIM-phenotypes by hws hints at a more general effect on miRNAs or their targets (Fig. 2; Supplemental Fig. S2).
Substantial overlap of transcriptome changes among hws and se-3, hyl1-2, cpl1-7, and ago1-27—mutants with known defects in miRNA biogenesis (Fig. 4)—further points toward involvement of HWS in the same pathway as the other four genes, possibly upstream of the miRNA-target interaction level. Epistasis of several miRNA related mutations over hws-1 supports this notion: combination of hws-1 with such mutants does not change their respective phenotypes, but leads to suppression of the characteristic hws skirt phenotype (Fig. 4). (Mallory et al., 2004a; Hibara et al., 2006; Nikovics et al., 2006). The skirt phenotype is likely caused by overaccumulation of miR164 in floral organs, as it is lost in the presence of MIM164 (Supplemental Figs. S2 and S3). This is consistent with earlier observations that continuous overexpression of miR164b, as well as simultaneous CUC1 and CUC2 inactivation, induces fused sepals and stamens and hence floral skirts (Mallory et al., 2004a; Hibara et al., 2006; Nikovics et al., 2006), which has recently also been suggested by others (González-Carranza et al., 2017). It also correlates with the observed positive effect of hws on miRNA abundance (Fig. 3).
Involvement of HWS in miRNA biogenesis or action and analogous shifts in the MIM/miRNA balance in hws could explain the observed suppression of various MIM phenotypes. MIMs largely function as miRNA “sponges”, which have also been described for animal miRNAs (Ebert et al., 2007), specifically sequestering the miRNAs they can bind to. Because these “sponges” can sequester only a limited amount of miRNAs, overexpression of miRNAs can counter their effects, a scenario potentially reflected in the miRNA-overaccumulating (Fig. 3) and MIM-suppressing (Fig. 2) hws mutant. The effects of MIM constructs on the cognate miRNAs differ substantially: some are greatly reduced, whereas others are only mildly affected (Todesco et al., 2010). Whether this reflects differential efficacies of MIM constructs, or how essential a miRNA is, is not known. Thus, even a small change in MIM transcript levels as in hws-1; MIM164 (Fig. 2) could already be sufficient to tip the equilibrium between MIM and miRNA necessary for MIM efficacy, and cause suppression of the conspicuous phenotype.
Moreover, consistent with a broader role of HWS, transcriptome changes in hws indicate that its effects go beyond merely correcting the aberrant gene expression in MIM156 plants (Fig. 4, Fig. 5). Because HWS does not encode a transcription factor, the observed changes in the transcriptome are likely cascading effects of its actual, direct effect on one or several proteins. Attempts to pinpoint specific HWS-regulated processes through approaches like GO-term enrichment have, however, been inconclusive.
Although HWS’s targets and its precise function in miRNA function remain somewhat enigmatic, it seems highly likely that HWS regulates the stability of a protein important for miRNA biogenesis, action, or degradation. This conclusion is based on the finding that miRNA-like phenotypes and mutant rescue in HWS complementation lines depend on the presence of the HWS F-box domain (Supplemental Figs. S1, S3, S5; Fig. 3). HWS was previously shown to have F-box activity (Kuroda et al., 2002; Takahashi et al., 2004) and confirmed to interact with SCF-complex proteins (this work, Supplemental Table S3; Arabidopsis Interactome Mapping Consortium, 2011). Other F-box proteins have been implicated in miRNA function before: AGO1, the core protein of miRISC, is targeted for degradation by the viral suppressor and F-box protein P0, and its levels also decrease upon overexpression of the F-box protein FBW2 (Pazhouhandeh et al., 2006; Baumberger et al., 2007; Bortolamiol et al., 2007; Csorba et al., 2010; Earley et al., 2010).
However, F-box proteins often interact with their targets only transiently, complicating detection of these interactions, especially with heterologous approaches like Y2H (Bortolamiol et al., 2007; Earley et al., 2010). HWS interaction with miRNA biogenesis factors, if present, is thus likely much weaker and more transient than the HWS F-box connection to SCF complexes. Accordingly, although we could detect enrichment of both SKPs and CUL in the mass spectrometry analysis, indications of AGO1 enrichment in potential 35S::Citrine-HWS complexes could so far not be confirmed in additional experiments. Although we might not yet have found a direct HWS target, HWS action may also require additional, Arabidopsis-specific factors, precluding its activity in transient assays. Possibly, a bridging factor is necessary for HWS to indirectly affect miRNA biology, or HWS-mediated targeting depends on prior modifications, as for example phosphorylation-dependent F-box protein recruitment (Skaar et al., 2013). As HWS overaccumulated to higher levels without the F-box (Supplemental Figs. S5 and S6), and because 35S::HWS, but not 35S::mHWS plants, display a strong miRNA-related phenotype, it is also possible that HWS is part of a feedback-loop involving miRNAs. Interaction with or recognition of its target(s) might destabilize HWS, explaining the observed stabilized protein levels in plants lacking the F-box. Alternatively, HWS might to a certain amount self-ubiquitinate, or be degraded together with its target.
The hws mutant phenotypes are, apart from the characteristic skirts, rather subtle. Strong defects are visible only in the overexpressor line or within a sensitized MIM context, whereas the weak, but pleiotropic effects of HWS deficiency are hardly detectable in an otherwise wild-type background. Effects of hws beyond MIM156 suppression hint at a broader impact, possibly on general metabolic pathways. Genes that are differentially expressed in hws mutants and in at least three miRNA mutants tend to be related to cell wall, anisotropic growth, and wounding, which is in agreement with HWS affecting cell division activity in the root meristem as well as guard cell growth (Fig. 4; Yu et al., 2015; Kim et al., 2016). In addition to this function, HWS has also been reported to affect general plant and organ growth (González-Carranza et al., 2007; Yu et al., 2015). Further, it is reminiscent of relatively weak, pleiotropic defects in other miRNA factors like CPL1 (Manavella et al., 2012; Jeong et al., 2013; Cui et al., 2016) and suggests that HWS may affect growth processes via the miRNA pathway, either through direct involvement in miRNA function, stability or even degradation, or upstream of miRNA-related processes.
CONCLUSIONS
Using an artificial miRNA target mimic, we identified the F-box protein HAWAIIAN SKIRT as a negative regulator of miR156. Molecular and genetic analyses reveal phenotypic similarities between hws and well-known miRNA mutants hyl1-2, se-3, and ago1-27, indicating an F-box-dependent involvement of HWS in miRNA function. Effects at the transcriptome level place HWS action between miRNA biogenesis and miRNA target inhibition. Direct miRNA-related target(s) of ubiquitination mediated by a HWS-containing SCF-complex remain enigmatic and are a promising subject for further investigation.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) seeds of the Col-0 accession were surface sterilized with 10% (v/v) bleach, 0.5% (v/v) SDS and stratified for 2 to 3 d at 4°C. Plants were grown at 23°C either on Murashige & Skoog (MS) plates (1/2 MS, 0.8% [w/v] agar, pH 5.7) or in soil in either short day (8-h light/16-h dark) or long day conditions (16-h light/8-h dark) in growth chambers with 65% humidity. A mixture of Cool-White and Gro-Lux Wide Spectrum fluorescent lights (Sylvania) with a fluence rate of 125 μmol to 175 μmol m−2 s−1 was used.
For Pi starvation, plants were germinated on MS plates for 7 d, then shifted to plates with full media lacking Pi (0.8% [w/v] agar, pH 5.7; Conn et al., 2013) and grown for four more days. Nicotiana benthamiana seeds were surface sterilized and vernalized as described above and grown on soil in long day conditions. Mutant alleles hyl1-2 (N564863, SALK_064863), abh1-753 (N516753, SALK_016753), se-3 (N583196, SALK_083196), ago1-25, ago1-27, hst-3 (N24278), miR164a-4 and hws-1 as well as miRNA mimicry lines MIM156 (N783223), MIM159 (N783226), MIM319 (N783243), MIM164 (N783232), the ProMIR156c::GUS, ProMIR164a::GUS and the miR156 overexpressor-line 35S::MIR156B have been described and were obtained either from the Nottingham Arabidopsis Stock Center or from colleagues (Morel et al., 2002; Bollman et al., 2003; Vazquez et al., 2004; Grigg et al., 2005; Schwab et al., 2005; Nikovics et al., 2006; Franco-Zorrilla et al., 2007; González-Carranza et al., 2007; Todesco et al., 2010; Rubio-Somoza et al., 2014).
For phenotypic analysis of double mutants between hws-1 and mutant alleles of AGO1, HYL1, ABH1, HST, and SE, F2 or F3 plants were selected phenotypically, then genotyped to confirm homozygosity of both mutations. The oligonucleotides used for genotyping can be found in Supplemental Table S10.
Photographs of plants were taken with a PowerShot G12 (Canon; Figs. 1 to 5; Supplemental Figs. S1 and S3). Microscopy was done using an MZ FLIII Dissecting Scope (Leica) with an attached AxioCam HRc (Carl Zeiss; Supplemental Fig. S1; Fig. 3) and the AxioVision software (Rel. 4.8.2; Carl Zeiss).
Transgenes
The HWS promoter (2460 bp), genomic (4190 bp), and coding sequences (1236 bp) were PCR-amplified from genomic DNA and cDNA, respectively. They were cloned into pCR8GWTOPO and recombined with a ProQuest Two-Hybrid System (Life Technologies) and pGREEN vectors (Hellens et al., 2000). A detailed list of constructs used in this work can be found in Supplemental Table S11. All oligonucleotides used to amplify the HWS fragments are listed in Supplemental Table S10.
Transient expression in N. benthamiana after Agrobacterium-mediated plant transformation has been described (Yang et al., 2000).
Mutant Screen and Segregation of MIM156 Transgene
Plants from a stable miR156 mimicry (MIM156) line in Col-0 background were subjected to EMS treatment as described (Weigel and Glazebrook, 2002). M2 plants grown in SD conditions were visually inspected for suppression of MIM156 developmental alterations. Candidate plants were crossed to the Ws-0 accession and genomic DNA of 200 to 300 pooled F2 plants was extracted using a CTAB protocol. Sequencing libraries (Illumina TruSeq DNA Sample Preparation Kit) were 10-plexed (Illumina adapters Set A) per flow-cell lane and sequenced on an Illumina HiSEquation 2000 instrument to obtain at least 10-fold genome coverage. The SHOREmap technique was used to identify SNPs and mapping intervals (Schneeberger et al., 2009).
The MIM156 transgene was removed through outcrossing to the Col-0 accession. Presence or absence of the transgene was deduced from BASTA resistance/sensitivity.
RNA Analysis and Sequencing
Total RNA was isolated from pools of approximately 50 plate-grown seedlings 9 d after sowing using TRIZOL reagent (Life Technologies) and DNase A (Life Technologies) treatment according to manufacturer’s instructions. With a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific), reverse transcription was performed on 1 μg to 2 μg of total RNA. Quantitative RT-PCR on HWS, mature miRNAs, and miRNA targets was executed with Maxima SYBR Green 2X Master Mix (Thermo Fisher Scientific) on a CFX384 Real-Time PCR system (Bio-Rad), performing technical triplicates on each sample of biological triplicates using ACTIN2 (At3G18780) as reference gene. Biological replicates are averaged from technical triplicates; horizontal bars show the mean of the biological replicates (Figs. 2 and 3; Supplemental Figs. S1 and S3). All oligonucleotides used for RT-PCR experiments are listed in Supplemental Table S10.
For RNA-seq, total RNA was extracted from pooled flowers, using TRIZOL reagent (Life Technologies) and DNase A (Life Technologies) treatment according to manufacturer’s instructions and a final cleanup using RNeasy Mini spin columns from the RNeasy Plant Mini Kit (Qiagen). Transcriptome libraries were prepared from 1 μg total RNA with a TruSeq RNA Library Prep Kit v2 (Ilumina) and single-end sequenced on a HiSEquation 3000 (Illumina) with 100-bp reads. Reads were mapped to the TAIR10 Arabidopsis genome and gene expression was quantified using the software RSEM (v1.2.30, https://deweylab.github.io/RSEM/; Li and Dewey, 2011; Berardini et al., 2015). All data analysis after initial read cleanup was executed in the software environment R (https://www.r-project.org/). After exclusion of a contaminated sample, differential expression and all subsequent analysis was conducted with two biological replicates, using the software DESeq2 (v1.14.1; Bioconductor). Differential expression was assessed in comparison to the common wild-type Col-0, and for the suppressor line additionally to MIM156. Published transcriptome data of Col-0, cpl1-7, hyl1-2, and se-3 seedlings (Manavella et al., 2012; biological triplicates), and Col-0, ago1-27 flower buds (SRA, PRJNA309714; biological triplicates) were used for further comparisons.
Histochemistry
Seedlings from at least five independent GUS reporter T2 lines were inspected 10 d after sowing (DAS). Activity of the GUS reporter was assessed as described (Weigel and Glazebrook, 2002), using 20 mm potassium-ferro- and 20 mm potassium-ferricyanide.
Protein Analyses
T1 seedlings expressing 35S::Citrine-HWS and 35S::Citrine-mHWS were BASTA-selected on soil and harvested at 21 d for total protein extraction from three to six whole rosettes as tissue pools. Protein was extracted from approximately 300 mg to 1000 mg of ground tissue using equal amounts [w/v] of extraction buffer (50 mm Tris pH 7.5; 150 mm NaCl; 1 mm EDTA; 10% [v/v] glycerol; 1 mm DTT; one tablet of Roche Complete Protease Inhibitor Cocktail per 10 mL buffer). Protein concentration was measured using Bradford solution (Bio-Rad). Expression of the fusion protein was tested by western blot using GFP-trap (ChromoTek), and appropriate pools were chosen for coimmunoprecipitation with GFP-trap or anti-AGO1 (Agrisera) antibodies.
For protein expression analyses, leaves of N. benthamiana transiently cotransformed with 35S::Citrine-HWS or 35S::Citrine-mHWS and 35S::AGO1-HA were harvested 3 d after infiltration. Protein abundance was measured by western blot using anti-AGO1 (AS09 527, 1:10,000; Agrisera), anti-GFP (sc-8334, 1:10,000; Santa Cruz Biotechnology), anti-UBQ (sc-8017, 1:2000; Santa Cruz Biotechnology), anti-DCL1 (AS12 2102, 1:1000; Agrisera), anti-HYL1 (AS06 136, 1:10,000; Agrisera), or anti-SE (AS09 532; 1:5000, Agrisera), and equal loading was confirmed using protein staining with either Ponceau Red or Coomassie Blue.
Subcellular localization was performed on 5-d-old seedlings of two independent stable 35S::Citrine-HWS and 35S::Citrine-mHWS T2 lines each. Seedlings were imaged using an LSM 780 Laser Scanning Microscope (Carl Zeiss) and the software ImageJ (v.2.0.0; https://imagej.nih.gov/ij/) for image processing and fluorescence quantification.
Interaction experiments in yeast were performed using the ProQuest Two-Hybrid System (Life Technologies) and yeast strain AH109.
Mass Spectrometry
Pools of BASTA-selected T1 seedlings expressing 35S::Citrine-HWS and 35S::Citrine-mHWS and GFP-overexpressing control plants were frozen in liquid N and total protein was extracted from up to 1 g finely ground tissue with equal amounts [w/v] of extraction buffer (140 mm NaCl; 8 mm Na2HPO4*7H2O; 2 mm KH2PO4, pH 7.4; 1 mm EDTA; 0.1% [v/v] Triton X-100; 1 tablet of Roche Complete Protease Inhibitor Cocktail per 10 mL buffer). Protein concentration was measured using Bradford solution (Bio-Rad) and fluorescence-marker-expression was verified by western blot.
Total protein extracts were purified using GFP-trap metal beads (ChromoTek). A small fraction was resolved on a PAGE gel for staining with the SilverQuest Silver Stain Kit (Life Technologies). LC-MS/MS analysis (120 min, Top15HCD) was performed after tryptic in-gel digestion, using an Easy-nLC (Proxeon Biosystems) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific; Borchert et al., 2010). Resulting data were analyzed with the software MaxQuant (v.1.2.2.9; http://www.biochem.mpg.de/5111795/maxquant; Cox and Mann, 2008; Cox et al., 2011). Spectra were searched against an Arabidopsis database including the protein sequences of the Citrine::HWS fusion proteins. Raw data were processed with a setting of 1% for the false discovery rate (Supplemental Table S3).
Accession Numbers
Total RNA sequencing data are available via ArrayExpress under the accession E-MTAB-5788.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Differentially expressed genes overlapping in MIM156 (compared to Col-0) and hws-5; MIM156 (compared to MIM156), i.e. HWS-dependent genes.
Supplemental Table S2. Subsets according to overlap with manually curated silencing gene list (see Supplemental Table S6) or differentially expressed genes in miRNA mutants. Counts and results of two-tailed Fisher’s exact test.
Supplemental Table S3. Mass spectrometry results.
Supplemental Table S4. Differentially expressed genes in all lines, data for Figure 2A.
Supplemental Table S5. Differentially expressed genes overlapping in hws-1 and hws-5, and those also differentially expressed in hws-5; MIM156.
Supplemental Table S6. List of genes involved in silencing, i.e. genes related to miRNA biogenesis, predicted and validated miRNA targets, and miRNA encoding genes.
Supplemental Table S7. Differentially expressed genes in hws-5; MIM156 (compared to MIM156) only, i.e. additional hws-induced genes in MIM-background.
Supplemental Table S8. Differentially expressed genes in hws-5; MIM156 (compared to MIM156), hws-1 and hws-5, but not in MIM156 (all compared to Col-0).
Supplemental Table S9. Differentially expressed genes overlapping in hws-5; MIM156 (compared to MIM156), hws-1, hws-5 (all compared to Col-0), i.e. general, core HWS-dependent genes, and those only overlapping in hws-5; MIM156, hws-1, and hws-5, including their overlap with miRNA mutants hyl1-2, se-3, cpl1-7, and ago1-27.
Supplemental Table S10. DNA oligonucleotide primers and probes.
Supplemental Table S11. Plasmids. Constructs for plant transformation are based on pGREEN and confer either Basta or kanamycin resistance in plants.
Supplemental Figure S1. Phenotypic characterization of hws, 35S::HWS and hws; MIM156.
Supplemental Figure S2. Skirt phenotype in MIM159, MIM319, MIM164b, and miR164a ko plants with wild-type and hws-1 background.
Supplemental Figure S3. Phenotypic connection between HWS and miRNA biogenesis.
Supplemental Figure S4. Y2H interaction screen of mHWS with a number of miRNA biogenesis factors.
Supplemental Figure S5. Functional connection among HWS, AGO1, and ubiquitination.
Supplemental Figure S6. Subcellular localization of HWS.
Supplemental Figure S7. Effect of HWS overexpression on AGO1, DCL1, HYL1, and SE.
Supplemental Figure S8. Overlap in hws-1 and hws-5 differentially expressed genes.
Acknowledgments
We thank Diep Tran for support and suggestions with protein work; Jim Carrington and co-workers for ago1-25 and ago1-27; Jeremy A. Roberts for hws-1; Jia-Wei Wang for ProMIR156c::GUS and EMS-mutagenized MIM156 populations; Patrick Laufs for ProMIR164a::GUS seeds; Sascha Laubinger for the DCL1-antibody; and Pablo A. Manavella and Delfina Ré for parts of the silencing gene list. Mass spectrometry and confocal microscopy were conducted at the Proteome Center Tübingen and the MPI for Developmental Biology’s Light Microscopy Facility, respectively. We are grateful to Ignacio Rubio Somoza, Pablo Manavella, and Moisés Exposito Alonso for valuable discussion, input for experiments, analysis, and reviewing the manuscript.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through SFB1101, a DAAD scholarship to E.S.D., an EMBO long-term fellowship and a Swiss National Science Foundation fellowship to E.S., and by the Max Planck Society.
Articles can be viewed without a subscription.
References
- Achkar NP, Cambiagno DA, Manavella PA (2016) miRNA biogenesis: a dynamic pathway. Trends Plant Sci 21: 1034–1044 10.1016/j.tplants.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Ahmed F, Senthil-Kumar M, Lee S, Dai X, Mysore KS, Zhao PX (2014) Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biol 11: 1414–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak RO, Mikkelsen JG (2014) miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA 5: 317–333 [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Tsai C-H, Lie M, Havecker E, Baulcombe DC (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Almasia NI, Manacorda CA, Mongelli VC, Conti G, Maroniche GA, Rodriguez MC, Distéfano AJ, Hopp HE, del Vas M, Asurmendi S (2009) Virus infection elevates transcriptional activity of miR164a promoter in plants. BMC Plant Biol 9: 152 10.1186/1471-2229-9-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park M-Y, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Borchert N, Dieterich C, Krug K, Schütz W, Jung S, Nordheim A, Sommer RJ, Macek B (2010) Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res 20: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol 17: 1615–1621 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Schaller H, Khafif M, Schott G, Bendahmane A, Voinnet O (2012) Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci USA 109: 1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, Su C-L (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, Tyerman SD, Gilliham M (2013) Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 4 10.1186/1746-4811-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou J-M, Lauressergues D, André O, Gutjahr C, Guillotin B, Bécard G, Combier J-P (2016) Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 10.1016/j.chom.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Coyaud E, Mis M, Laurent EMN, Dunham WH, Couzens AL, Robitaille M, Gingras A-C, Angers S, Raught B (2015) BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol Cell Proteomics 14: 1781–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyán J (2015) Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479-480: 85–103 [DOI] [PubMed] [Google Scholar]
- Csorba T, Lózsa R, Hutvágner G, Burgyán J (2010) Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J 62: 463–472 [DOI] [PubMed] [Google Scholar]
- Cui P, Chen T, Qin T, Ding F, Wang Z, Chen H, Xiong L (2016) The RNA polymerase II C-Terminal Domain Phosphatase-Like Protein FIERY2/CPL1 interacts with eIF4AIII and is essential for nonsense-mediated mRNA decay in Arabidopsis. Plant Cell 28: 770–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, Stoffel M (2016) Impact of MicroRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol Cell 64: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Smith M, Weber R, Gregory B, Poethig R (2010) An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou T-J, Lin S-I, Aung K, Zhu J-K (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- González-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA (2007) Hawaiian skirt: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol 144: 1370–1382 10.1104/pp.106.092288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Zhang X, Peters JL, Boltz V, Szecsi J, Bendahmane M, Roberts JA (2017) HAWAIIAN SKIRT controls size and floral organ number by modulating CUC1 and CUC2 expression. PLoS One 12: e0185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. (2004) The microRNA Registry. Nucleic Acids Res 32: D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M (2005) SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong IS, Aksoy E, Fukudome A, Akhter S, Hiraguri A, Fukuhara T, Bahk JD, Koiwa H (2013) Arabidopsis C-terminal domain phosphatase-like 1 functions in miRNA accumulation and DNA methylation. PLoS One 8: e74739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E-S, Choe G, Sebastian J, Ryu KH, Mao L, Fei Z, Lee J-Y (2016) HAWAIIAN SKIRT regulates the quiescent center-independent meristem activity in Arabidopsis roots. Physiol Plant 157: 221–233 10.1111/ppl.12443 [DOI] [PubMed] [Google Scholar]
- Köster T, Meyer K, Weinholdt C, Smith LM, Lummer M, Speth C, Grosse I, Weigel D, Staiger D (2014) Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res 42: 9925–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Mao L (2007) Evolution of plant microRNA gene families. Cell Res 17: 212–218 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B (2004a) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004b) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D (2012) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870 [DOI] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Morel J-B, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Ihara N, Komatsu A, Tokioka Y, Nishioka M, Takase T, Kiyosue T (2008) Gene expression, localization, and protein-protein interaction of Arabidopsis SKP1-like (ASK) 20A and 20B. Plant Sci 174: 485–495 [Google Scholar]
- Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V (2006) F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA 103: 1994–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel M, Li Y, Li J, Millar AA (2015) Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J 13: 915–926 [DOI] [PubMed] [Google Scholar]
- Reis RS, Eamens AL, Waterhouse PM (2015) Missing pieces in the puzzle of plant microRNAs. Trends Plant Sci 20: 721–728 [DOI] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou C-M, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang J-W, Weigel D (2014) Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol 24: 2714–2719 [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen J-E, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Sha A, Zhao J, Yin K, Tang Y, Wang Y, Wei X, Hong Y, Liu Y (2014) Virus-based microRNA silencing in plants. Plant Physiol 164: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu J-K (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kuroda H, Kuromori T, Hirayama T, Seki M, Shinozaki K, Shimada H, Matsui M (2004) Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol 45: 83–91 [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, Zhao Y, Yu Y, Ramachandran V, Ren G, Yu B, Li S, et al. (2015) Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet 11: e1005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Vierstra RD (1993) Two cDNAs from Arabidopsis thaliana encode putative RNA binding proteins containing glycine-rich domains. Plant Mol Biol 21: 695–699 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Wang J-W. (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Wang J-W, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang J-W, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhi H, Zhang Y, Liu Y, Zhang J, Gao Y, Guo M, Ning S, Li X (2015) miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database (Oxford) 2015: bav098 10.1093/database/bav098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park M-Y, Earley KW, Wu G, Yang L, Poethig RS (2016) Developmental functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Murchie EH, González-Carranza ZH, Pyke KA, Roberts JA (2015) Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development. J Exp Bot 66: 1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ji L, Le BH, Zhai J, Chen J, Luscher E, Gao L, Liu C, Cao X, Mo B, Ma J, Meyers BC, et al. (2017) ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol 15: e2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]