The ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 ethylene receptors signal via a noncanonical pathway to control sensitivity to the hormone abscisic acid during seed germination.
Abstract
Ethylene is a gaseous plant hormone perceived by a family of receptors in Arabidopsis (Arabidopsis thaliana) including ETHYLENE RESPONSE1 (ETR1) and ETR2. Previously we showed that etr1-6 loss-of-function plants germinate better and etr2-3 loss-of-function plants germinate worse than wild-type under NaCl stress and in response to abscisic acid (ABA). In this study, we expanded these results by showing that ETR1 and ETR2 have contrasting roles in the control of germination under a variety of inhibitory conditions for seed germination such as treatment with KCl, CuSO4, ZnSO4, and ethanol. Pharmacological and molecular biology results support a model where ETR1 and ETR2 are indirectly affecting the expression of genes encoding ABA signaling proteins to affect ABA sensitivity. The receiver domain of ETR1 is involved in this function in germination under these conditions and controlling the expression of genes encoding ABA signaling proteins. Epistasis analysis demonstrated that these contrasting roles of ETR1 and ETR2 do not require the canonical ethylene signaling pathway. To explore the importance of receptor-protein interactions, we conducted yeast two-hybrid screens using the cytosolic domains of ETR1 and ETR2 as bait. Unique interacting partners with either ETR1 or ETR2 were identified. We focused on three of these proteins and confirmed the interactions with receptors. Loss of these proteins led to faster germination in response to ABA, showing that they are involved in ABA responses. Thus, ETR1 and ETR2 have both ethylene-dependent and -independent roles in plant cells that affect responses to ABA.
Ethylene is a phytohormone that affects the growth and development of plants and mediates plant stress responses (Mattoo and Suttle, 1991; Abeles et al., 1992). Many studies over the last several decades have identified constituents of the ethylene signaling pathway. This model proposes that activity of the receptors is reduced upon binding ethylene leading to reduced activity of a protein kinase, CONSTITUTIVE TRIPLE RESPONSE1 (CTR1; Kieber et al., 1993). Lower activity of CTR1 leads to a reduction in the phosphorylation of ETHYLENE INSENSITIVE2 (EIN2), causing a decrease in the ubiquitination of EIN2 and a rise in EIN2 protein levels; this allows for the proteolytic release of the C-terminal portion of the protein via an unidentified protease (Qiao et al., 2009, 2012; Chen et al., 2011; Ju et al., 2012; Wen et al., 2012). The EIN2 C-terminal portion modulates two transcription factors, EIN3 and EIN3-LIKE1, leading to the majority of ethylene responses (Chao et al., 1997; Solano et al., 1998; Alonso et al., 1999; Guo and Ecker, 2003; Yanagisawa et al., 2003; Binder et al., 2004a, 2004b; Gagne et al., 2004; Qiao et al., 2012).
Ethylene receptors are localized to the membrane of the endoplasmic reticulum (Chen et al., 2002; Ma et al., 2006; Grefen et al., 2008; Bisson et al., 2009). In Arabidopsis (Arabidopsis thaliana), there are five ethylene receptor isoforms known as ETHYLENE RESPONSE1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR1 (ERS1), ERS2, and EIN4 (Chang et al., 1993; Hua and Meyerowitz, 1998; Hua et al., 1998; Sakai et al., 1998; Gao et al., 2003). Based on sequence comparisons, the ethylene receptors fall into two subfamilies with ETR1 and ERS1 in subfamily 1 and ETR2, ERS2, and EIN4 in subfamily 2. All of the receptor isoforms contain an ethylene binding domain at the N terminus that is composed of three membrane-spanning α-helices (Schaller and Bleecker, 1995; Schaller et al., 1995; Rodríguez et al., 1999). They all also contain a cytosolic portion with a GAF domain and a kinase domain. Three of the Arabidopsis receptors (ETR1, ETR2, and EIN4) contain a receiver domain at the C terminus. All five of the receptors have kinase activity with ETR1 containing His kinase activity, ETR2, ERS2, and EIN4 containing Ser/Thr kinase activity, and ERS1 displaying both depending on the assay conditions (Gamble et al., 2002; Moussatche and Klee, 2004; Bisson and Groth, 2010).
Although the receptors overlap in the control of many traits, they also have nonredundant roles in the regulation of various traits (Shakeel et al., 2013). For instance, in Arabidopsis, the subfamily-1 receptors have a larger role in the control of growth than the subfamily-2 receptors and this is not simply a matter of different levels of the receptors (O’Malley and Bleecker, 2003; Wang et al., 2003; Qu et al., 2007). It is possible that these differences are in part due to different interactors. For instance, ETR1, and to a lesser extent ERS1, are affected by REVERSION TO ETHYLENE SENSITIVITY1, whereas, the other three receptors are not (Resnick et al., 2006; Deslauriers et al., 2015). A second example is that ETR1, ETR2, and EIN4 (receptors containing a receiver domain) are important for growth recovery after ethylene removal, but ERS1 and ERS2 are not (Binder et al., 2004a, 2004b). Additionally, in Arabidopsis, silver ions, which are well known to inhibit the ethylene receptors (Beyer, 1976), act predominantly via the ETR1 receptor (McDaniel and Binder, 2012). Surprisingly, there are also some phenotypes where the different ethylene receptor isoforms have contrasting roles. For example, ethylene-stimulated nutational bending of Arabidopsis hypocotyls requires the ETR1 receptor, whereas, the other four receptor isoforms inhibit nutations (Binder et al., 2006; Kim et al., 2011). Another example is that ERS1 and ETR1 have opposite roles in the control of growth (Liu et al., 2010). Recently, we found that ETR1 and ETR2 have contrasting roles in the control of Arabidopsis seed germination in darkness and during NaCl stress in the light where ETR1 hinders seed germination and ETR2 increases seed germination (Wilson et al., 2014a, 2014b; Bakshi et al., 2015). EIN4 has a smaller role similar to ETR1 in the control of seed germination during these stresses, whereas ERS1 and ERS2 have no measurable roles in regulating this trait (Wilson et al., 2014a, 2014b). These contrasting roles seem to be independent of changes in ethylene biosynthesis or sensitivity, and likely involves a change in abscisic acid (ABA) responsiveness (Wilson et al., 2014a, 2014b). These data are not explained by existing models of ethylene signal transduction, and point to unique roles for the ETR1 and ETR2 receptors in the control of ABA signal transduction.
A large body of research has led to models for ABA signaling (Cutler et al., 2010; Umezawa et al., 2010; Yang et al., 2017). In these models, ABA binds to PYRABACTIN RESISTANCE1 (PYR1) and PYR1-like (PYL) receptors leading to inactivation of some type 2C protein phosphatases from group A (PP2CA; Fujii and Zhu, 2009; Fujita et al., 2009; Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Park et al., 2009). Most of the PP2CAs involved in ABA signaling act as negative regulators of the pathway so that when inactivated by the receptors, ABA signaling proceeds. This occurs by the activation of SUC NONFERMENTING1-RELATED PROTEIN KINASE2s (SnRK2s), which in turn activate the ABA-RESPONSIVE ELEMENTS BINDING FACTOR (ABF) transcription factors causing an alteration in the transcription of ABA-responsive genes (Mustilli et al., 2002; Yoshida et al., 2006a, 2006b; Boudsocq et al., 2007; Nakashima et al., 2009; Umezawa et al., 2009; Vlad et al., 2009; Soon et al., 2012; Waadt et al., 2015). Several PP2CAs are positive regulators of the pathway. ABA also leads to changes in anion channels (Fujii et al., 2009; Geiger et al., 2009; Xue et al., 2011; Brandt et al., 2012). The gene transcripts encoding for many of these proteins in the ABA signaling pathway are up-regulated by ABA and various stresses (Chan, 2012).
Prior research suggests models where the ethylene receptors have roles independent of the canonical ethylene signaling pathway (Gamble et al., 1998; Beaudoin et al., 2000; Desikan et al., 2005; Binder et al., 2006; Resnick et al., 2006; Dong et al., 2008, 2010; Qiu et al., 2012; Kim et al., 2013; Wilson et al., 2014a, 2014b; Bakshi et al., 2015), which may underlie these observations. Establishing the mechanisms for noncanonical receptor signaling is of interest across kingdoms because noncanonical signal transduction has been observed in other signal transduction pathways of animals and bacteria (Federle and Bassler, 2003; Jenkins, 2009; Miller and McCrea, 2010; Chen et al., 2015). To understand the mechanism for the contrasting roles of ETR1 and ETR2 in germination, we determined that these receptors are predominantly affecting ABA signaling to alter germination in response to many stresses. Unexpectedly, we found that the role of ETR1 and ETR2 in seed germination occurs independently of the canonical ethylene signal transduction pathway. Because of this, we used yeast two-hybrid to screen for interacting partners with ETR1 and ETR2 and identified many putative nonoverlapping interaction proteins that may underlie their contrasting roles in the control of seed germination. We confirmed three of these interactions, and showed that loss of any of these three proteins led to altered germination in response to ABA. Thus, ETR1 and ETR2 signal via a noncanonical pathway to affect ABA signal transduction.
RESULTS
ETR1 and ETR2 Have Opposite Roles in the Control of Seed Germination under Various Inhibitory Conditions
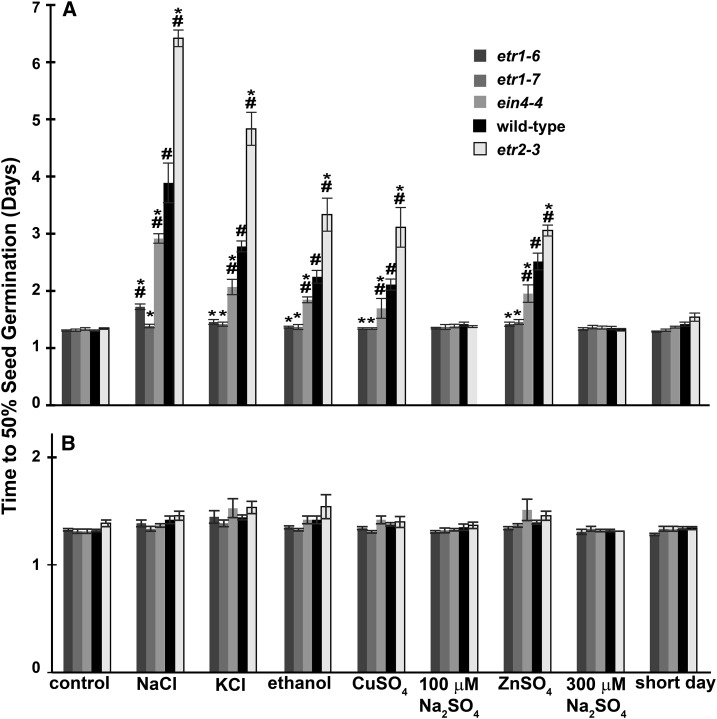
We have previously shown that ETR1, and to a lesser extent EIN4, inhibits and ETR2 promotes seed germination under NaCl stress and in darkness (Wilson et al., 2014a, 2014b). We aimed to know if these opposite roles extended to other conditions that delay Arabidopsis seed germination such as stress caused by other salts, heavy metals, ethanol, and short-day conditions (Shinomura et al., 1994; Hirayama et al., 2004; Li et al., 2005; Wang et al., 2007, 2008). To examine this, we measured the germination time-courses for wild-type, etr1-6, etr1-7, ein4-4, and etr2-3 seeds under control conditions and conditions that introduced one of the following variables: 150 mm NaCl, 150 mm KCl, 100 µM CuSO4, 300 µM ZnSO4, 100 mm ethanol, and short-day conditions (8 h light/16 h dark). Seed germination was considered to be complete upon rupture of the testa. As a control for sulfate ions in the CuSO4 and ZnSO4 conditions, we examined the effect of 100 μM and 300 μM Na2SO4 on the time-course of germination. We calculated the time for 50% germination under each of these conditions. Consistent with our prior results (Wilson et al., 2014a, 2014b), NaCl delayed seed germination of all three seed lines and etr1-6, etr1-7, and ein4-4 mutant plants germinated faster than wild-type and etr2-3 mutant plants germinated slower under this condition (Fig. 1A; Supplemental Fig. S1). Germination was delayed in wild-type seeds to a lesser extent by KCl, CuSO4, ZnSO4, and ethanol compared to NaCl at the concentrations used. In all these conditions, the etr1-6, etr1-7, and ein4-4 seeds germinated faster and the etr2-3 seed germinated slower than wild-type. Consistent with our prior study using NaCl (Wilson et al., 2014a, 2014b), there was a smaller effect of the ein4-4 mutation compared to loss of ETR1. Treating seeds with either 100 μM or 300 μM NaSO4 had no measurable effect on seed germination, indicating that the delay in germination caused by CuSO4 and ZnSO4 is due to the heavy metals, not the sulfate ions. In short-day conditions, the etr1-6, etr1-7, and ein4-4 mutant plants germinated slightly faster than wild-type seeds and the etr2-3 mutant plants slightly slower than wild-type seeds. But these differences were not statistically significant (P > 0.05).
Figure 1.
ETR1 and ETR2 have contrasting roles on seed germination under a range of inhibitory conditions. Germination time-courses for wild-type, etr1-6, etr1-7, ein4-4, and etr2-3 seeds were conducted as described in the “Materials and Methods” and the times for 50% of seeds to germinate were calculated. Germination experiments were conducted in the (A) absence or (B) presence of 100 μM NF to inhibit ABA biosynthesis. Conditions used were: control (using standard conditions described in the “Materials and Methods”), 150 mm NaCl, 150 mm KCl, 100 mm ethanol, 100 μM CuSO4, 100 μM NaSO4, 300 μM ZnSO4, 300 NaSO4, and short days (8-h light:16-h dark). Data represents the average ± sd. Data were analyzed using two-way ANOVA and Tukey’s multiple comparisons test. (*) Statistically different from wild type in that condition. (#) Statistically different from untreated seeds of that seed line (P < 0.05).
We have previously concluded that ETR1 and ETR2 probably affects sensitivity of seeds to ABA to alter seed germination (Wilson et al., 2014a, 2014b). Abiotic stress conditions are known to cause an increase in ABA biosynthesis leading to inhibition of germination (Vishwakarma et al., 2017). We therefore treated germinating seeds with 100 μM norflurazon (NF) to inhibit the biosynthesis of ABA in the germinating seeds. Treatment with NF drastically reduced or eliminated the differences in seed germination seen among wild-type, etr1-6, etr1-7, ein4-4, and etr2-3 in NaCl, KCl, CuSO4, ZnSO4, and ethanol (Fig. 1B; Supplemental Fig. S2). Application of NF had no significant effect on seed germination under control conditions. This indicates that ETR1, EIN4, and ETR2 are likely to also be affecting ABA under these various conditions that inhibit seed germination.
The Receiver Domain of ETR1 Is Required for its Role in Seed Germination under Various Inhibitory Conditions
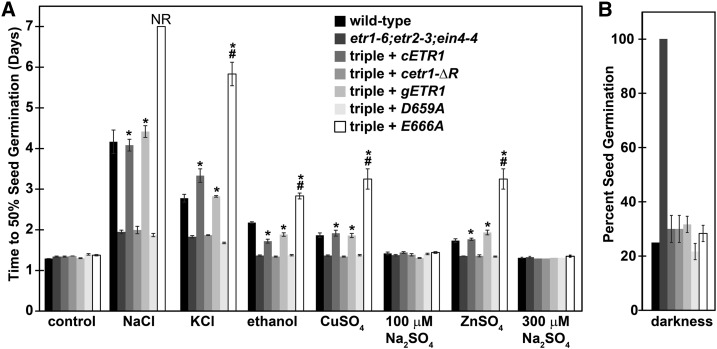
We have previously shown that the receiver domain of ETR1 is required for its inhibitory role on seed germination during NaCl stress, but not in darkness (Wilson et al., 2014a, 2014b). Additionally, several amino acids in the receiver domain are critical for normal ETR1 function related to germination on NaCl, but not in the control of other traits such as growth inhibition and ethylene-stimulated nutations (Bakshi et al., 2015). To determine whether the receiver domain is required for the control of seed germination by ETR1 under other inhibitory conditions, we examined germination time-courses of etr1-6;etr2-3;ein4-4 triple-mutant plants lacking ethylene receptor isoforms with a receiver domain, and this triple mutant transformed with cDNA for the full-length ETR1 transgene (cETR1) or a truncated ETR1 transgene lacking the receiver domain (cetr1-ΔR). Consistent with our previous results (Wilson et al., 2014a, 2014b), the etr1-6;etr2-3;ein4-4 triple mutant plants germinated faster than wild-type seeds under NaCl stress; the cETR1 transgene caused germination to be slower and comparable to wild type, whereas the cetr1-ΔR transgene failed to delay germination (Fig. 2A; Supplemental Fig. S3). We observed a similar pattern with KCl, CuSO4, ZnSO4, and ethanol, suggesting that ETR1 functions similarly in all of these conditions. Thus, the receiver domain is required for the function of ETR1 in regulating seed germination under these inhibitory conditions.
Figure 2.
The rapid germination of etr1;etr2;ein4 triple mutant plants under various inhibitory conditions is differentially rescued by truncated and etr1 receiver domain mutants. A, Germination time-courses were determined for etr1-6;etr2-3;ein4-4 triple mutant plants and these triple mutants were transformed with a cDNA for full-length ETR1 (cETR1) or a truncated ETR1 lacking the receiver domain (cetr1-ΔR) or genomic DNA for full-length wild-type ETR1 (gETR1), a D659A mutant, or an E666A mutant. The times for 50% germination were then calculated. Conditions used were control (using standard conditions described in the “Materials and Methods”), 150 mm NaCl, 150 mm KCl, 100 mm ethanol, 100 μM CuSO4, 100 μM NaSO4, 300 μM ZnSO4, and 300 μM NaSO4. B, Percent seed germination of seeds kept in darkness 7 d after planting. For both panels, data are the average ± sd. Data were analyzed using two-way ANOVA and Tukey’s multiple comparisons test. NR, never reached 50% germination in the time-course of the experiment. (*) Denotes the etr1-6;etr2-3;ein4-4 triple mutant transformed with the indicated transgene is statistically different from the triple mutant. (#) The transformant is statistically slower than triple mutant transformed with full-length ETR1 transgene (P < 0.05).
In a prior study (Bakshi et al., 2015), we found that transforming the etr1-6;etr2-3;ein4-4 triple-mutant plants with genomic DNA encoding for an ETR1 transgene containing a D659A mutation (D659A) in the receiver domain resulted in ETR1 having a reduced function in the control of germination in seeds treated with NaCl, because it poorly rescued the time-course of germination to wild-type level. By contrast, an ETR1 transgene with an E666A mutation (E666A) causes ETR1 to be hyperfunctional for this trait so that germination on NaCl is slower than the triple mutant transformed with the wild-type genomic ETR1 (gETR1) transgene. Both of these mutant transgenes have normal function in controlling other traits (Bakshi et al., 2015). We conducted seed germination time-course experiments with these transformants, and observed a similar pattern for all of these inhibitory conditions where D659A was not functional and E666A was hyperfunctional in the conditions tested (Fig. 2A; Supplemental Fig. S3). This further supports the idea that ETR1 has a similar role in the control of germination under these various conditions that inhibit germination.
Unlike the above results, the receiver domain of ETR1 is not needed for ETR1 to inhibit germination in darkness (Wilson et al., 2014a, 2014b). We were therefore curious to know if the D659A or E666A mutations affected germination in darkness. Germination in darkness is very poor and wild-type seeds failed to reach 50% germination 7 d after planting (Supplemental Fig. S4), consistent with our prior study (Wilson et al., 2014a, 2014b). Therefore, we examined the extent of germination of etr1-6;etr2-3;ein4-4 triple-mutant plants and transformants 7 d after planting and maintained in darkness. We found, like our prior study, that wild-type seeds germinated very poorly, that etr1-6;etr2-3;ein4-4 triple-mutant plants germinated to 100% and that both cETR1 and cetr1-ΔR transgenes reversed this, causing the seeds to germinate poorly (Fig. 2B). Similarly, the D659A or E666A mutant transgenes caused the triple-mutant plants to germinate poorly. All seed lines reached 100% germination within 2.5 d of being transferred from darkness to long-day conditions as previously reported (Wilson et al., 2014a, 2014b), showing that all seeds were viable. Thus, unlike the above stress conditions, the ETR1 receiver domain is not needed nor influences germination in darkness.
ETR1, EIN4, and ETR2 Affect Responsiveness to ABA
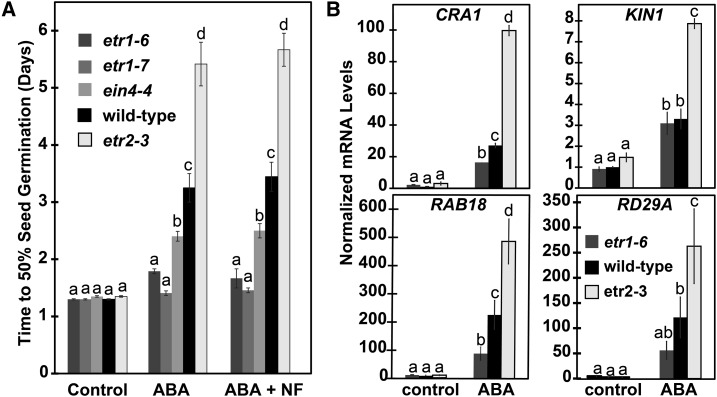
The above results, as well as our previous findings (Wilson et al., 2014a, 2014b), suggest that ETR1, EIN4, and ETR2 are affecting ABA sensitivity or levels or both. In the control of seed germination, we previously showed that etr1-6 seeds are less sensitive to ABA, and etr2-3 seeds are more sensitive to ABA compared to wild-type (Wilson et al., 2014a, 2014b). However, it is unclear if ABA synthesis may also be affected in these mutants. To distinguish between these possibilities, we examined the effect of 1 μM ABA in the presence and absence of 100 μM NF on seed germination. We posited that if the ETR1, EIN4, and ETR2 receptors are only affecting ABA sensitivity, then NF should have no effect on germination time-courses under these conditions. However, if these receptors are affecting biosynthesis of ABA, then NF should eliminate or reduce germination differences. Consistent with Wilson et al. (2014a, 2014b), in the presence of ABA, the etr1-6 mutant plants germinated faster than wild-type seeds and the etr2-3 mutant plants germinated slower than wild-type seeds (Fig. 3A). Similar to the results examining germination under stress, both the etr1-7 and ein4-4 seeds also germinated faster than wild-type in the presence of ABA. The application of NF had no measurable effect on this, supporting the idea that ETR1, EIN4, and ETR2 mainly affect sensitivity to ABA.
Figure 3.
ETR1 and EIN4 affect responses to ABA oppositely from ETR2. A, Germination time-courses of wild-type, etr1-6, etr1-7, ein4-4, and etr2-3 seeds in response to 1 μM ABA and 1 μM ABA plus 100 μM NF to block ABA biosynthesis were conducted and the times for 50% seed germination were determined. Data represents the average ± sd. B, The transcript abundance of RAB18, CRA1, KIN1, and RD29A were measured in wild-type, etr1-6, and etr2-3 seeds using real-time qRT-PCR. Seeds were germinated for 2 d in the presence or absence of 1 μM ABA and mRNA extracted. Data were normalized to the levels of At3g12210 in each seed line to calculate the relative transcript level for each gene. These were then normalized to levels of the transcript in untreated wild-type seeds. The average ± se for two biological replicates with three technical replicates each is shown. For both panels, 0.01% (v/v) ethanol was used as a solvent control. Data were analyzed using a two-way ANOVA and Tukey’s multiple comparisons test. In each panel, the different letters indicate significant difference (P < 0.05).
To confirm that ABA responsiveness is altered, we used real-time qRT-PCR to examine the transcript levels of several ABA-responsive genes including CRUCIFERIN1 (CRA1) encoding for a seed storage protein, RESPONSIVE TO ABA18 (RAB18) encoding for a dehydrin protein, a stress-responsive protein called KIN1, and RESPONSIVE TO DESICATION 29A (RD29A) encoding for a Leu zipper transcription factor (Pang et al., 1988; Lång and Palva, 1992; Chan, 2012; Gliwicka et al., 2012). We compared transcript levels of these genes in seeds germinated for 2 d in the presence or absence of 1 μM ABA. Because ETR1 and ETR2 are having the largest effects on germination, we compared the transcript levels in etr1-6 and etr2-3 mutant seeds. Consistent with Bakshi et al. (2015), application of ABA caused CRA1 and RAB18 transcript levels to rise in wild-type seeds (Fig. 3B). Similarly, ABA also increased the transcript abundance of KIN1 and RD29A in wild-type seeds. In the absence of exogenous ABA, loss of either ETR1 or ETR2 had no statistically significant effect on the transcript levels of these four genes. However, when ABA was applied, the induction for these gene transcripts was larger in etr2-3 seeds and generally reduced in etr1-6 seeds compared to wild type. Interestingly, we previously showed that ABA has little or no effect on the transcript levels of CRA1 and RAB18 in etr1-6;etr2-3;ein4-4 triple-mutant plants (Bakshi et al., 2015). Transformation of the triple-mutant plants with gETR1 rescued this response; by contrast, transformation with D659A failed to rescue this induction, and transformation with E666A led to an enhanced induction of these genes that correlates with the effects of these point mutants on germination (Bakshi et al., 2015). Together, these data show that ETR1 and ETR2 are affecting ABA signal transduction oppositely and this is influenced by the ETR1 receiver domain.
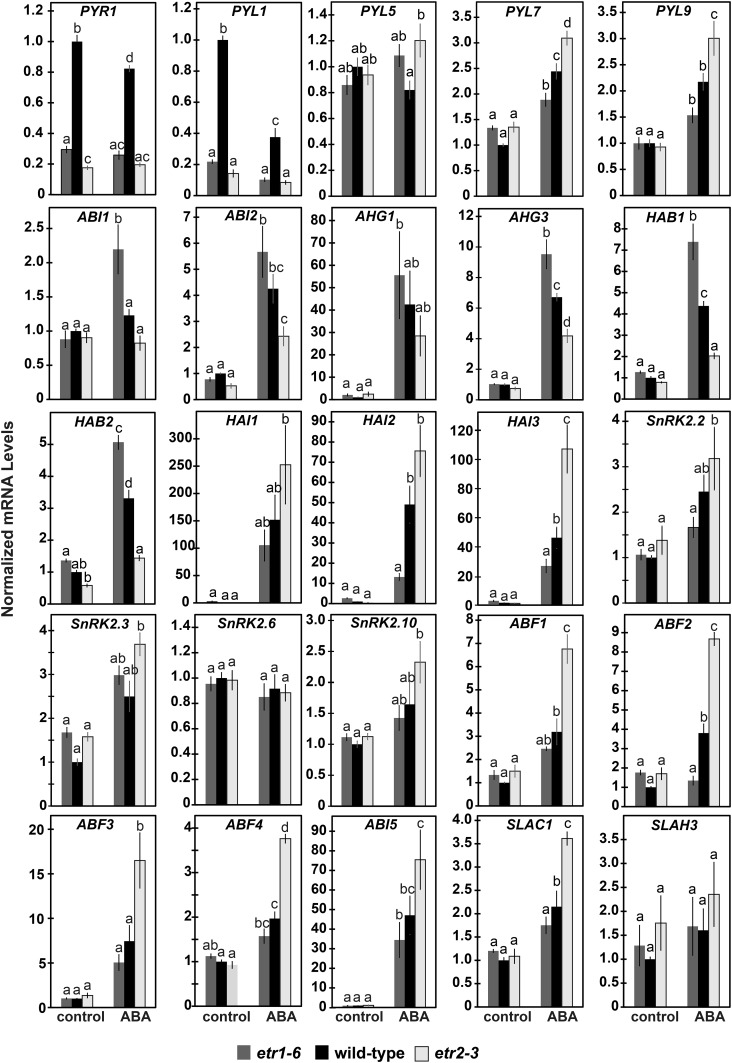
We wished to determine what aspects of ABA signaling are altered in these mutants. The gene transcripts encoding for many of ABA signaling proteins are up-regulated by ABA and various stresses (Chan, 2012). We therefore used real-time qRT-PCR to study the transcript abundance of 25 genes that encode for proteins involved in ABA signaling (Fig. 4). This included genes that encode for the ABA receptors PYR1, PYL1, PYL5, PYL7, and PYL9, the PP2CAs ABA INSENSTIVE1 (ABI1), ABI2, ABA-HYPERSENSITIVE GERMINATION1 (AHG1), AHG3, HOMOLOGY TO ABI1 (HAB1), HAB2, HIGHLY ABA-INDUCED1 (HAI1), HAI2, and HAI3, the SnRK2s SnRK2.2, SnRK2.3, SnRK2.6, and SnRK2.10, the ABFs ABF1, ABF2, ABF3, ABF3, and ABI5, and the SLOW ANION CHANNEL-ASSOCIATED1 (SLAC1) and SLAC1-HOMOLOG3 (SLAH3) ion channels.
Figure 4.
ETR1 and ETR2 oppositely affect the transcript abundance of many genes encoding for ABA signaling proteins. The transcript levels of genes in germinating seeds encoding for proteins in the ABA signal transduction pathway were analyzed with real-time qRT-PCR as described in Figure 3. The average ± se for two to three biological replicates with three technical replicates each is shown. In all panels, 0.01% (v/v) ethanol was used as a solvent control. Data were analyzed using a two-way ANOVA and Tukey’s multiple comparisons test. In each panel, different letters indicate significant difference (P < 0.05).
In the absence of exogenous ABA, loss of either ETR1 or ETR2 generally had no effect on the transcript abundance of most of these genes (Fig. 4). The exceptions were PYR1 and PYL1, where loss of either ETR1 or ETR2 led to a 3-to 5-fold decrease in the transcript abundance of these genes that encode for receptors. There was also a slight effect on the transcript levels of HAB2, where etr1-6 seeds had slightly higher levels than etr2-3 seeds and wild-type seeds had intermediate levels in the absence of added ABA.
In wild-type seeds, application of ABA caused an increase in the transcript abundance of PYL7, PYL9, ABI2, AHG3, HAB1, HAB2, HAI2, HAI3, ABF1, ABF2, ABF4, ABI5, and SLAC1 and a decrease in PYR1 and PYL1 transcript levels (P < 0.05; Fig. 4). Application of ABA to wild-type seeds caused little or no change in the transcript levels of ABI1, AHG1, HAI1, SnRK20.2, SnRK2.3, SnRK2.6, SnRK2.10, ABF3, and SLAH3, or else the changes observed were below the statistical cutoff we used (P > 0.05).
In most cases, ABA caused the smallest increase in etr1-6 mutant plants and the largest increase in etr2-3 mutant plants with wild-type seeds having intermediate responses (Fig. 4). This pattern was seen with PYL7, PYL9, HAI1, HAI2, HAI3, SnRK2.2, SnRK2.10, ABF1, ABF2, ABF3, ABF4, ABI5, and SLAC1. By contrast, application of ABA caused the largest change in the transcript levels of six gene transcripts encoding the PP2CAs ABI1, ABI2, AHG1, AHG3, HAB1, and HAB2 in etr1-6 seeds and smallest change in etr2-3 seeds. Contrary to the genes mentioned above, these PP2CAs inhibit ABA signaling (Gosti et al., 1999; Merlot et al., 2001; Leonhardt et al., 2004; Saez et al., 2004; Kuhn et al., 2006; Yoshida et al., 2006a, 2006b; Nishimura et al., 2007; Rubio et al., 2009). Application of ABA had little or no effect on the transcript levels of PYR1, PYL1, PYL5, SnRK2.3, SnRK2.6, and SLAH3 in the etr1-6 and etr2-3 mutant seeds.
Together, these data support a model where, in the presence of ABA, ETR1 and ETR2 lead to opposite changes in the transcript abundance of many genes that encode for proteins involved in ABA signal transduction leading to changes in ABA sensitivity.
The Receiver Domain of ETR1 Is Involved in the Induction of ABA Signaling Genes by ABA
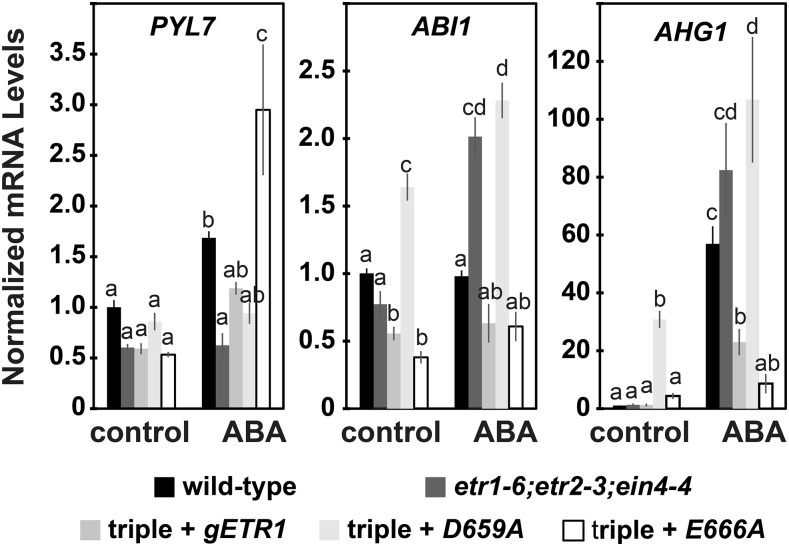
We were also interested to know if the ETR1 receiver domain was important for the induction of these gene transcripts. We therefore examined the transcript abundance of three of the genes that encode for ABA signaling components in the in etr1-6;etr2-3;ein4-4 triple-mutant plants and the triple mutants transformed with the gETR1, D659A, or E666A transgenes. We chose one positive (PYL7) and two negative (ABI1, AHG1) regulators of ABA signaling that showed a clear difference in the etr1-6 versus etr2-3 mutants in the presence of ABA (Fig. 4).
In the absence of exogenous ABA, there were no statistically significant differences (P > 0.05) in the transcript levels of the three genes in wild-type versus the triple-mutant seeds (Fig. 5). However, under this condition, transformation of the triple mutant with D659A led to an increase in transcript levels of ABI1 and AHG1 compared to the other seed lines.
Figure 5.
The D659A and E666A mutant transgenes oppositely affect the transcript abundance of genes encoding for ABA signaling proteins. The transcript levels of selected genes encoding for ABA signaling proteins were determined for etr1-6;etr2-3;ein4-4 triple mutant plants and these triple mutants transformed with genomic DNA for full-length wild-type ETR1 (gETR1), a D659A mutant, or an E666A mutant. For comparison, data for wild-type seeds are shown. The transcript levels of genes in germinating seeds encoding for proteins in the ABA signal transduction pathway were analyzed with real-time qRT-PCR as described in Figure 3. The average ± se for two biological replicates with three technical replicates each is shown. Data were analyzed using a two-way ANOVA and Tukey’s multiple comparisons test. In each panel, different letters indicate significant difference (P < 0.05).
In the presence of exogenous ABA, transformation of the triple mutant with the D659A transgene led to comparable levels of PYL7, ABI1, and AHG1 compared to the triple mutants, indicating this transgene was minimally functional. By contrast, transformation of the triple mutant with the E666A transgene led to an increase of PYL7 transcript abundance compared to the gETR1 transformant and lower levels of AHG1 transcript abundance compared to the gETR1 transformant. Additionally, the levels of PYL7 gene transcript was higher in the E666A seed line compared to all the other seed lines including wild-type. By contrast, the levels of ABI1 and AHG1 transcripts were lower than wild type in the E666A seed line. Thus, in the presence of ABA, D659A is causing lower levels of the positive ABA regulator and higher levels of the negative regulators correlating with less ABA signaling and faster germination of this transformant. By contrast, E666A is having the opposite effect, which correlates with higher levels of ABA signaling and slower germination of this transformant. These results are consistent with the ETR1 receiver domain being involved in the changes in the levels of genes encoding for ABA signaling genes.
ABA Affects the Transcript Levels of ETR1, EIN4, and ETR2, But Not ERS1 and ERS2
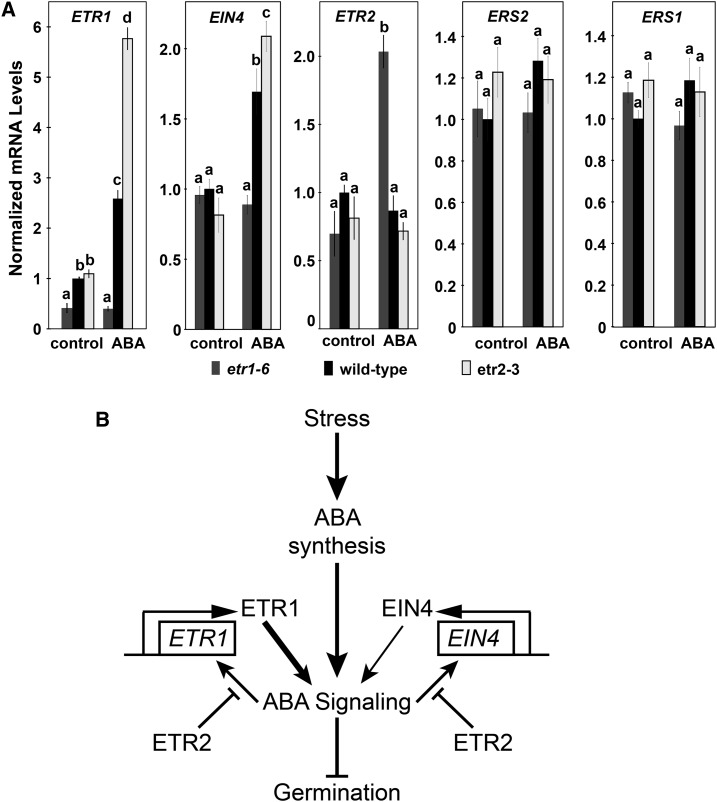
Using epistasis analysis, we have previously posited that ETR2 signals via ETR1 and EIN4, to control germination in response to stress or application of ABA (Wilson et al., 2014a, 2014b). In this model, the ETR1 and EIN4 receptors function in parallel to stimulate ABA signal transduction, with ETR1 having the larger role in this. The function of ETR2 is to inhibit both receptors. It is unclear how ETR2 is having this effect. It is known that stress and application of ABA can affect the transcript levels of the ethylene receptors in various plant species (Zhao and Schaller, 2004; Martín-Rodríguez et al., 2011; Mou et al., 2016). We were therefore curious to know if application of ABA altered the transcript levels of the five ethylene receptors, and whether loss of either ETR1 or ETR2 affected ABA-induced changes in the other receptors.
In the absence of exogenous ABA, loss of ETR1 or ETR2 had no measurable effect on transcript levels of the remaining four receptor isoforms (Fig. 6A). Application of 1 μM ABA caused an increase in the transcript levels of ETR1 and EIN4 in wild-type seeds. Interestingly, ABA had no effect on EIN4 transcript abundance in the etr1-6 seeds, suggesting that ABA signaling is reduced to a level where it does not affect EIN4 transcription. By contrast, the ABA-stimulated increase in both ETR1 and EIN4 transcript abundance was larger in etr2-3 mutant seeds. Application of ABA caused no measurable change in the levels of ETR2, ERS1, or ERS2 transcripts in wild-type seeds. However, loss of ETR1 resulted in ABA treatment causing an increase in ETR2 transcript abundance. Application of ABA had no measurable effect on the transcript abundance of either ERS1 or ERS2 in the etr1-6 and etr2-3 seeds. These data suggest that output from the ABA signal transduction pathway is affecting the levels of the ETR1, EIN4, and ETR2 transcripts. In the case of ETR1 and EIN4 transcript levels, ETR2 is diminishing, but not completely blocking the ABA-stimulated increase in transcript abundance. By contrast, ETR1 is preventing the ABA-stimulated increase in ETR2 transcript abundance so that the increase is only seen when ETR1 is removed.
Figure 6.
ETR1 and ETR2 affect ABA-induced changes in ETR1, EIN4, and ETR2, but not ERS1 and ERS2 transcripts. A, The transcript levels of the five ethylene receptor isoforms in germinating seeds were analyzed with real-time qRT-PCR as described in Figure 3. The average ± se for two to three biological replicates with three technical replicates each is shown. In all panels, 0.01% (v/v) ethanol was used as a solvent control. Data were analyzed using the two-way ANOVA and Tukey’s multiple comparisons test, and in each panel the different letters indicate significant difference (P < 0.05). B, Model of roles of ETR1, EIN4, and ETR2 in the control of ABA signaling during seed germination. In this model, it is proposed that various stresses increase ABA levels. Previous epistasis indicates that in the presence of ABA, ETR2 inhibits ETR1 and EIN4, which act in parallel to enhance ABA signaling (Wilson et al., 2014a, 2014b). Results here indicate that ETR2 inhibits ETR1 and EIN4 by reducing, but not eliminating, ABA-induced increases in ETR1 and EIN4 transcription. Not shown here, we also found that ETR1 suppresses ABA-induced changes in ETR2 transcript abundance. It is possible that in the presence of ABA, EIN4 is also affecting transcript abundance of ETR2, but this has not yet been studied. The width of the arrows from ETR1 and EIN4 denote the relative signaling strength affecting ABA signaling. It is unknown whether these are direct effects on ABA signaling or happening via intermediaries.
These results lead to a model for how ETR2 inhibits ETR1 and EIN4 by preventing accumulation of ETR1 and EIN4 transcripts (Fig. 6B). These data also suggest that ETR1 is negatively affecting the accumulation of ETR2 transcript in the presence of ABA, leading to feedback on this circuit to cause more signaling from ETR1 due to higher ETR1 transcript abundance. The ERS1 and ERS2 receptors have no apparent role in regulating ABA to affect seed germination because loss of either ERS1 or ERS2 has no effect on seed germination in response to NaCl stress (Wilson et al., 2014a, 2014b) and ABA fails to cause changes in ERS1 and ERS2 transcript levels.
ETR1 and ETR2 Signal Independently of CTR1 to Control Seed Germination
Ethylene is known to stimulate Arabidopsis seed germination (Bleecker et al., 1988). However, we previously showed that the effects of the etr1-6 and etr2-3 mutations on germination are likely to not involve ethylene signaling because etr2-3 seeds are more sensitive to application of ethylene than wild-type and etr1-6 seeds (Wilson et al., 2014a, 2014b). This is opposite to what is predicted if ethylene is involved. Additionally, ethylene biosynthesis is minimally affected in the mutants compared to wild-type during germination under NaCl stress conditions. Current models of ethylene signaling posit that all receptor signaling occurs via CTR1 and EIN2. However, various studies have provided evidence that the receptors may signal via alternate pathways in addition to the canonical pathway (Gamble et al., 1998; Beaudoin et al., 2000; Desikan et al., 2005; Binder et al., 2006; Qiu et al., 2012; Kim et al., 2013; Wilson et al., 2014a, 2014b; Bakshi et al., 2015).
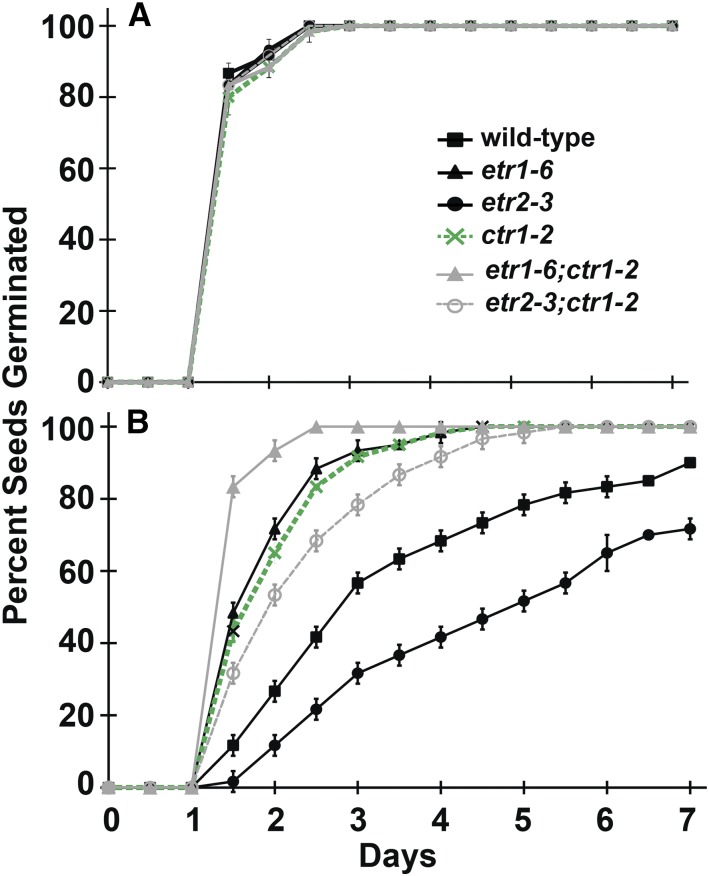
To more thoroughly test whether ETR1 and ETR2 are affecting seed germination independently of the canonical ethylene signaling pathway, we generated etr1-6;ctr1-2 and etr2-3;ctr1-2 double mutant plants. It has previously been demonstrated that ctr1 mutant plants germinate better in the presence of salt than wild-type seeds (Lin et al., 2012). Therefore, we predicted that if ETR1 and ETR2 are signaling via the canonical signaling pathway to control ABA sensitivity, in the presence of ABA, the double mutant plants containing ctr1-2 should germinate similarly to ctr1-2 single mutant plants. Double mutants with phenotypes that diverge from these predictions would support a model invoking signaling via a noncanonical pathway.
We generated crosses between either etr1-6 or etr2-3 and ctr1-2. We only obtained one cross of these because these crosses were very stunted and generated few seeds. Each double mutant was grown to the F4 generation. We used PCR to confirm that the double-mutant plants were homozygous for either etr1-6 or etr2-3 (Supplemental Fig. S5, A and B). When grown in the dark, Arabidopsis seedlings have long hypocotyls and treatment with ethylene results in short hypocotyls (Bleecker et al., 1988; Guzmán and Ecker, 1990). Using this assay, we confirmed that double mutant plants containing ctr1-2 had a constitutive growth inhibition response when grown in ethylene-free air (Supplemental Fig. S5C) similar to the findings of Kieber et al. (1993). Consistent with prior studies (Hua and Meyerowitz, 1998; Cancel and Larsen, 2002), the etr1-6 mutant plants were slightly shorter than wild type in air and had a slightly stronger growth inhibition response upon addition of ethylene (Supplemental Fig. S5C).
In the absence of exogenous ABA, all seed lines had similar germination time-courses and times for 50% germination (Fig. 7A; Supplemental Fig. S6). In the presence of 1 µM ABA, the ctr1-2 seeds germinated faster than wild-type seeds with a germination time-course slightly slower than etr1-6 seeds (Fig. 7B; Supplemental Fig. S6). Interestingly, the etr1-6;ctr1-2 mutant plants germinated faster than either the etr1-6 or ctr1-2 single mutant plants and the etr2-3;ctr1-2 mutant plants gave a germination time-course intermediate between the single mutant parents. This additive behavior between the receptors and CTR1 suggest that the receptors are signaling, at least in part, independently of CTR1 to affect seed germination.
Figure 7.
ETR1 and ETR2 function independently of the canonical ethylene signaling pathway to alter response to ABA. Double etr1-6;ctr1-2 and etr2-3;ctr1-2 mutant plants were generated and physiologically evaluated. For comparison, wild-type and etr1-6, etr2-3, and ctr1-2 single mutant plants were included. Seed germination time-courses were conducted in the (A) absence and (B) presence of 1 μM ABA.
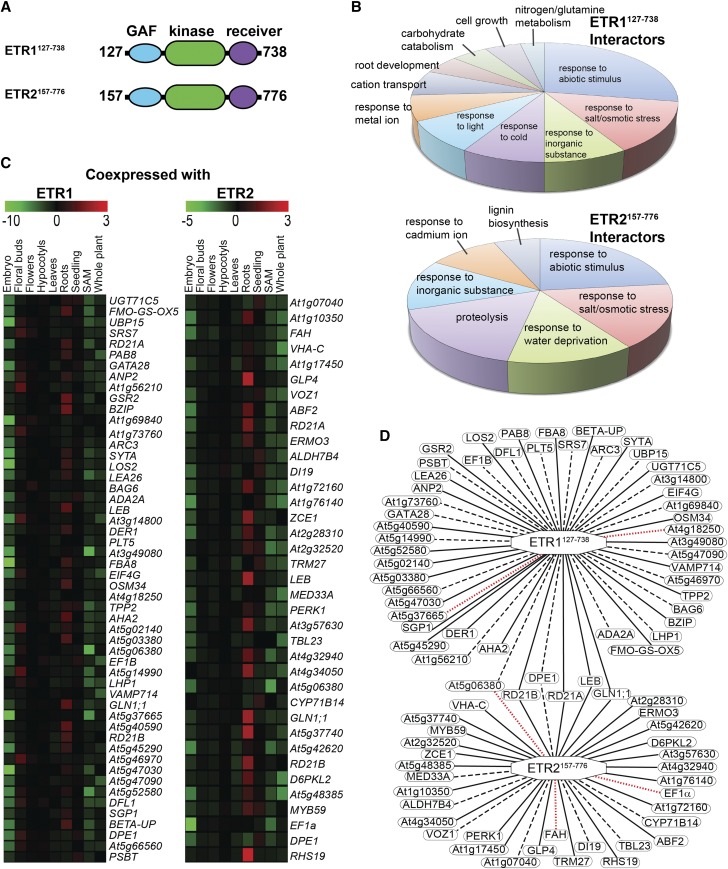
ETR1 and ETR2 Have Both Overlapping and Nonoverlapping Interaction Partners
The above results raise the possibility that ETR1 and ETR2 are involved in unidentified protein interactions to affect ABA signaling. To study this, we used yeast two-hybrid analysis to screen for interacting partners where we made bait constructs encoding for the cytosolic portion of each receptor. For ETR1, this included expressing amino acids 127 through 738 (ETR1127-738) and for ETR2, amino acids 157 through 776 (ETR2157-776), which included the GAF, kinase, and receiver domain of each but eliminated the N-terminal portion containing the transmembrane α-helices (Fig. 8A). Each construct was fused to the GAL4 DNA binding domain of pGBKT7 vector and then introduced into the yeast (Saccharomyces cerevisiae) strain Y187 to generate the bait strain. These were then used to screen cDNA prey libraries generated in yeast strain AH109 as a fusion to the GAL4 activation domain of the pGADT7 vector (Hewezi et al., 2008). A total of 52 possible interaction partners for ETR1127-738 and 37 for ETR2157-776 was identified; there were six that overlapped for both (Supplemental Table S1). A gene ontogeny (GO) classification analysis showed that more than half were related to stress responses including abiotic stresses, water stress, cold stress, and salt and osmotic stress (Fig. 8B). To map possible tissue coexpression of these with ETR1 or ETR2, we used the Multiple Experiment Viewer (http://mev.tm4.org/) to construct a heatmap of gene coexpression patterns in various tissues such as embryos, floral buds, flowers, hypocotyls, leaves, roots, shoot apical meristem, seedlings, and whole plants (Fig. 8C). From this information, we generated a gene coexpression map where we show all the interactions uncovered, as well as those that are predicted to occur in root tissue (Fig. 8D). It was interesting to note that the majority of interacting pairs are coexpressed in only one organ/tissue with most coexpressed in the roots. In a few cases, the putative interacting partners coexpress in two or, to a much lesser extent, in three tissues. The network contained 83 nodes and 87 edges representing the interacting combinations. However, two that were identified as potentially interacting with ETR1 (At4g18250, At5g37665) and two that were identified as possibly interacting with ETR2 (EF1α, FAH) appear to not be coexpressed in any tissue with their respective receptor partner. One putative interacting partner (At5g06380) that was identified as potentially interacting with both ETR1 and ETR2 appears to only be coexpressed with ETR1.
Figure 8.
Yeast two-hybrid screen using the cytosolic domains of ETR1 and ETR2 reveals putative overlapping and nonoverlapping interaction partners. A yeast two-hybrid screen was carried out using the cytosolic portion of ETR1 (ETR1127-738) or ETR2 (ETR2157-776) as described in the “Materials and Methods”. A, Diagram of cytosolic portions of ETR1 and ETR2 used for yeast two-hybrid screen. Numbers represent the amino acids included in these constructs. B, GO categorization of proteins identified in this screen as interacting with ETR1127-738 or ETR2157-776. GO enrichment analysis was carried out using DAVID (https://david.ncifcrf.gov/). Only GO categories with a false discovery rate value < 0.05 were included. C, Heatmap showing coexpression patterns for ETR1 and ETR2 interacting proteins. Red indicates the protein has highly correlated gene coexpression with the receptor and green represents highly anticorrelated coexpression. D, Gene coexpression map of the ETR1 and ETR2 interacting partners. Solid edges represent proteins with significantly correlated expression profiles with the receptor in roots. Dashed edges represent proteins with significantly correlated coexpression profiles in at least one tissue, but not roots. Dotted red edges represent no significant correlation in the coexpression profile between the receptor and the gene in question.
Analysis of Selected Protein Interactors
We next selected some of the potentially interesting interacting protein candidates for further analysis. For this, we focused on the DROUGHT-INDUCED19 (DI19) transcription factor, a β-glucosidase called LONG ER BODY (LEB), and the RESPONSIVE TO DEHYDRATION21A (RD21A) protease identified in the yeast two-hybrid screen. All three of these proteins are documented to be involved in stress responses or response to ABA and are predicted to be expressed in either the cytosol or endoplasmic reticulum. It is interesting to note that out of these three proteins, DI19 interacts with CPK11, which encodes for a Ca2+-dependent, calmodulin-independent protein kinase that functions as a positive regulator of ABA signaling pathway by phosphorylating ABA-responsive element binding protein factors ABF1 and ABF4 (Hayashi et al., 2001; Milla et al., 2006; Ma and Bohnert, 2007; Sherameti et al., 2008; Liu et al., 2013; Qin et al., 2014; Song et al., 2016).
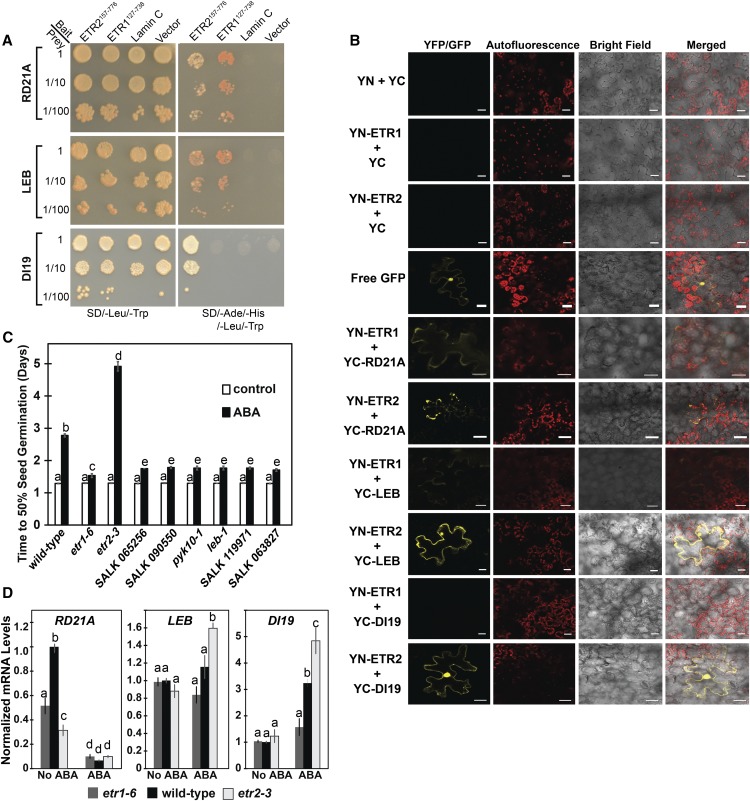
To further test whether the identified proteins interacted with ETR1 and ETR2, we used yeast cotransformation assays where we cotransformed yeast (strain AH109) with a full-length cDNA encoding one of the identified targets and either ETR1127-738 or ETR2157-776. The full-length sequence encoding the individual interacting partners was cloned in a prey vector (pGADT7), whereas the C-terminal coding sequences of ETR1 or ETR2 were cloned into a bait vector (pGBKT7). Subsequently, yeast AH109 cells were cotransformed with the bait and prey vectors and potential interactions were visualized by differential growth on the nonselective synthetic dropout (SD) medium (SD/-Leu/-Trp) and on the selective medium (SD/-Leu/-Trp/-His/-Ade; Fig. 9A). Growth on the selective medium confirmed the yeast two-hybrid results that showed that RD21A and LEB interact with both ETR1 and ETR2, whereas DI19 only interacts with ETR2. Serial dilutions of yeast cotransformed cells were used to measure the strength of the interaction and we observed interactions with 1:100 dilutions.
Figure 9.
Three interaction partners affect seed germination in response to ABA. A, Cotransformation analysis was carried out as described in the “Materials and Methods”. Yeast strain AH109 was cotransformed with full-length coding sequences of the selected interacting partners cloned in the prey vector (pGADT7) and C-terminal coding sequences of ETR1 (ETR1127-738) or ETR2 (ETR2157-776) cloned in the bait vector (pGBKT7). Protein-protein interactions were visualized by differential growth on the nonselective synthetic drop-out medium (SD/-Leu/-Trp; left) and on the selective medium (SD/- Leu/-Trp/-His/-Ade; right). B, BiFC in N. benthamiana was carried out with the full-length clones of ETR1 or ETR2 fused to YN and the full-length clones of the indicated proteins fused to the YC as described in the “Materials and Methods”. RD21A, LEB, and DI19 were labeled at their N terminus with YC. ETR1 and ETR2 were labeled at their N terminus (YN-ETR1 and YN-ETR2, respectively) for interaction studies with LEB and RD21A and at their C terminus (ETR1-YN, ETR2-YN) for experiments with DI19. YN+YC, YN-ETR1+YC, and YN-ETR2+YC were included as negative controls and free GFP as a positive control. YFP or GFP fluorescence, chlorophyll autofluorescence, and bright field images were acquired and merged. Scale bar is 25 μm. C, Time for 50% germination of mutants for RD21A (SALK 065256, SALK 090550), LEB (pyk10-1, leb-1), and DI19 (SALK 119971, SALK 063827). Seeds were germinated in the absence or presence of 1 μM ABA as indicated, and the percent germination was determined every 12 h for 7 d and the time for 50% seed germination was calculated. For comparison, the germination of wild-type, etr1-6, and etr2-3 seeds is shown. Data represents the average ± sd. D, The transcript levels of the RD21A, LEB, and DI19 in germinating seeds were analyzed with real-time qRT-PCR as described in Figure 3. The average ± se for two to three biological replicates with three technical replicates each is shown. In (C) and (D), days were analyzed using a two-way ANOVA and Tukey’s multiple comparisons test. Different letters indicate significant difference (P < 0.05).
To confirm the protein-protein interactions in planta, bimolecular fluorescence complementation (BiFC) assays were conducted, where we used the full-length coding sequences of the receptors fused to the N-terminal half of a yellow fluorescent protein gene (nEYFP), whereas the full-length clones of DI19, RD21A, or LEB were fused to the C-terminal half of the yellow fluorescent protein gene (cEYFP; Fig. 9B). The different combinations between nEYFP and cEYFP fusions were coexpressed in Nicotiana benthamiana cells via biolistic bombardment. As expected, negative controls gave no YFP signal. Consistent with the yeast two-hybrid screen and yeast cotransformation analysis, we found that DI19 only interacted with ETR2, whereas LEB and RD21A interacted with both receptors by reconstituting the fluorescent YFP in the transformed N. benthamiana cells. The DI19 and ETR1 interacting combination yielded no YFP fluorescence. The interactions of LEB and RD21A with ETR1 gave a weaker signal than what was observed with ETR2. Together, these data indicate that LEB and RD21A interact with both receptors and DI19 interacts with ETR2 both in yeast and in planta.
We wanted to know if these proteins are involved in ABA-induced changes in germination. We therefore measured the seed germination time-courses of two loss-of-function alleles each for DI19, LEB, and RD21A. These included the pyk10-1 and leb-1 mutants for LEB that have previously been characterized (Nagano et al., 2008, 2009), the SALK 065256 and SALK 090550 lines with mutations in RD21A, and the SALK 119971 and SALK 063827 lines with mutations in DI19. The SALK lines were confirmed to be homozygous using PCR as described in the “Materials and Methods”. In these mutants, germination time-courses and time for 50% germination were similar to wild-type seeds in the absence of exogenous ABA (Fig. 9C; Supplemental Fig. S7). By contrast, loss of any one of these proteins led to faster germination than wild type in response to ABA. The time for 50% germination of these mutant plants in the presence of ABA was slightly slower than what was observed for the etr1-6 mutant plants. These data indicate these proteins are involved in the control of seed germination in response to ABA.
We were curious to know if loss of either ETR1 or ETR2 affected the transcript abundance of DI10, RD21A, or LEB. In the absence of ABA, loss of these receptors had no measurable effect on the transcript abundance of DI19 or LEB (Fig. 9D). By contrast, loss of either receptor caused a reduction in the transcript levels of RD21A in the absence of ABA. Application of ABA to wild-type seeds caused an increase of DI19 transcript, a decrease in RD21A transcript, and no change in LEB transcript. The increase in DI19 transcript was lost in etr1-6 seeds and enhanced in etr2-3 seeds. Similarly, loss of ETR2 caused a larger increase in LEB transcript abundance upon application of ABA; however, loss of ETR1 had no effect on LEB transcript levels. Loss of either receptor had no effect on transcript abundance of RD21A in the presence of ABA. Thus, under certain conditions the ETR1 and ETR2 receptors are affecting the transcript abundance of these genes that encode for interacting partners.
DISCUSSION
The signal transduction pathway for ethylene has been well studied. Nonetheless, many questions remain, including understanding the function of the ethylene receptors. This is complicated by a growing body of evidence indicating that the ethylene receptors have both overlapping and nonoverlapping roles (Shakeel et al., 2013). We previously reported that ETR1 and ETR2 have contrasting roles in the control of seed germination under NaCl stress and in response to ABA, and posited that these contrasting roles were likely independent of ethylene (Wilson et al., 2014a, 2014b). Here we demonstrated that ETR1 and ETR2 have contrasting roles under other conditions that inhibit germination. The receiver domain of ETR1 has been implicated as being important for subfunctionalization of this receptor in other traits such as growth recovery after removal of ethylene, ethylene-stimulated nutations, and germination under NaCl stress (Binder et al., 2006; Wilson et al., 2014a, 2014b; Bakshi et al., 2015). The ETR1 receiver domain was also important for the control of germination under most of the additional stress conditions used in this study, suggesting a common mechanism for the control of germination by ETR1 under these conditions. The exception to this was that, consistent with a prior study (Wilson et al., 2014a, 2014b), the receiver domain of ETR1 is not required for ETR1 to affect germination in darkness.
We believe that ABA is involved in mediating the contrasting roles of ETR1 and ETR2 in germination under many stress conditions because ABA is known to be part of abiotic stress responses in plants (Vishwakarma et al., 2017) and NF eliminated germination differences under these stress conditions. Application of ABA caused the largest delay in the seed germination of etr2-3 mutant plants and the smallest delay in etr1-6 mutant plants, and this effect was unaffected by the application of NF to block ABA biosynthesis. This suggests that ETR1 and ETR2 are predominantly affecting ABA sensitivity, rather than ABA biosynthesis. Additional support for this is that ABA caused a smaller increase in the transcript levels of several ABA-responsive genes in the etr1-6 mutant plants compared to etr2-3 mutant plants. Unexpectedly, in the presence of ABA, both ETR1 and ETR2 affected the transcript abundance of many genes that encode for proteins in the ABA signaling pathway. Where such an effect was seen, in most cases the lowest gene transcript levels were seen in the etr1-6 seeds and the highest in the etr2-3 seeds. This included genes for several receptors (PYL7 and PYL9), PP2CAs (HAI1, HAI2, and HAI3), SnRK2s (SnRK2.2 and SnRK2.10), and ABFs (ABF1, ABF2, ABF3, ABF4, and ABI5). Because these genes are positive regulators of ABA signaling (Zhang et al., 2004; Fujii et al., 2007, 2011; Fujita et al., 2009; Bhaskara et al., 2012), the lower levels observed in the etr1-6 seeds correlate with the smaller ABA responses we observe in this mutant during seed germination. Conversely, the higher levels of these genes in the etr2-3 mutant plants correlate with the larger ABA responses we observed with etr2-3. The transcript abundance of six PP2CAs (ABI1, ABI2, AHG1, AHG2, HAB1, and HAB2) showed the opposite pattern with the highest transcript abundance in the etr1-6 seeds and the lowest transcript abundance in the etr2-3 seeds. Because these are negative regulators of ABA signaling (Leonhardt et al., 2004; Saez et al., 2004; Kuhn et al., 2006; Nishimura et al., 2007; Rubio et al., 2009; Antoni et al., 2012), this also correlates with the ABA responses we see in the etr1-6 and etr2-3 mutant plants. In the presence of ABA, the E666A mutation in ETR1 led to a larger induction and the D659A mutation to a smaller induction of ABA-responsive genes (Bakshi et al., 2015). Similarly, these mutations led to alterations in ABA-induced changes of gene transcripts for ABA signaling proteins that correlate with the changes these mutations have on germination during stress. Thus, ETR1 and ETR2 are having a widespread effect on transcript abundance of genes that encode for ABA signaling proteins and this is likely causing changes in ABA responsiveness. In the case of ETR1, this effect involves the receiver domain.
Using epistasis analysis among ETR1, ETR2, and EIN4, we previously generated a model where ETR1 and EIN4 inhibit seed germination by stimulating ABA signaling and ETR2 inhibits both receptors (Wilson et al., 2014a, 2014b). We found that ABA increased the transcript levels of ETR1 and EIN4, but not ETR2, ERS1, or ERS2 in wild-type seeds. The observation that loss of ETR2 leads to higher ETR1 and EIN4 transcript levels in the presence of ABA supports a model where ETR2 inhibits ETR1 and EIN4 via regulation of transcription (Fig. 6B). Interestingly, ABA caused an increase in ETR2 transcript levels in etr1-6 seeds, suggesting that ABA can affect both receptors, but that ETR1 inhibits this effect on ETR2 overcoming the effect of ABA. These data suggest that there is feedback from ABA signaling to alter both receptors, and there is reciprocal regulation between ETR1 and ETR2 where each is negatively affecting the transcript levels of the other under these conditions (Fig. 6B). The role of the interacting proteins identified in this study remains unknown. We demonstrate that loss of LEB, RD21A, or DI19 leads to reduced ABA responsiveness and faster seed germination similar to etr1 loss-of-function mutants. However, the mechanism by which their interaction with ethylene receptors is important to affect germination is unknown. It is also likely that loss of other interactors will have an etr2 loss-of-function phenotype where the mutants are more responsive to ABA and germinate more slowly. A simple genetic working model (Supplemental Fig. S8) illustrates how these proteins might interact to control ABA responsiveness. This opens up the possibility for future research using epistasis analysis between the ethylene receptors and proteins of interest to delineate the pathway(s) involved in affecting the ABA signaling pathway.
The biochemical outputs of ETR1 and ETR2 remain unknown. In the case of ETR1, signaling to inhibit seed germination under inhibitory stress conditions or in response to ABA requires the D659 amino acid residue in the receiver domain. This amino acid residue is thought to be the target of phosphorelay from the ETR1 His kinase (Gamble et al., 1998). However, neither this residue, nor the entire receiver domain, are required for ethylene signaling to control most traits (Gamble et al., 2002; Binder et al., 2004a, 2004b; Kim et al., 2011; Hall et al., 2012; Bakshi et al., 2015). Similarly, ETR1 His kinase activity is not required for ethylene signaling (Wang et al., 2003; Qu et al., 2007). This suggests that ETR1 His kinase activity and phosphotransfer have alternative roles outside of ethylene signal transduction. Because ABA signaling involves changes in the phosphorylation of various proteins (Cutler et al., 2010; Umezawa et al., 2010; Yang et al., 2017), it is possible that one such role for the ETR1 His kinase, and perhaps the ETR2 Ser/Thr kinase, is to modulate ABA signaling. Support for this is the report that Ser/Thr kinase activity in the NTHK1 (for N. tabacum HIS KINASE1) subfamily 2 ethylene receptor affects the sensitivity of tobacco (N. tabacum) to NaCl stress (Chen et al., 2009). It remains to be determined if ABA sensitivity is also altered in NTHK1 mutant plants.
All five ethylene receptors in Arabidopsis have been documented to physically interact with CTR1 (Clark et al., 1998; Cancel and Larsen, 2002; Gao et al., 2003; Bisson et al., 2009; Bisson and Groth, 2010) and most models suggest that all signaling from these receptors requires CTR1. However, ETR1 and ETR2 have opposite roles for seed germination in response to ABA. We previously showed that these contrasting roles do not correlate with biosynthesis of ethylene or sensitivity to ethylene in these mutants (Wilson et al., 2014a, 2014b). Therefore, a likely model is that there is an alternative, noncanonical signaling pathway from these two ethylene receptors that is independent of CTR1. Our epistasis analysis between receptor loss-of-function mutants and CTR1 support this model. Intermediate seed germination phenotypes in response to ABA were observed in the etr1-6;ctr1-2 and etr2-3;ctr1-2 as predicted if signaling is occurring, at least partially, independently of CTR1.
Receptor signaling independent of CTR1 raises the question of what proteins are involved in this alternative pathway. Our yeast two-hybrid analyses show that there are numerous potential candidates to examine because there were many nonoverlapping receptor-protein interactions identified. The fact that many of these potential interacting partners are annotated as stress-related and that mutations in several of these genes affect germination in response to ABA and NaCl strengthens the argument that these interacting proteins could be involved in mediating signaling from these receptors to the ABA signaling pathway. One candidate from the yeast two-hybrid experiments is ABF2, which is part of the ABA signaling pathway. Yeast two-hybrid analysis showed that it potentially physically interacts with ETR2. If true, this provides one pathway by which ETR2 levels might affect ABA signaling. However, this does not explain how ETR1 is affecting ABA signaling. An additional problem with this simple model is that loss of ABF2 alone, or in combination with loss of ABF3 and ABF4, has no measurable effect on seed germination in response to ABA (Fujita et al., 2005; Yoshida et al., 2010), indicating that ETR1 and ETR2 are having a more widespread effect to alter ABA signaling. Our observation that the transcript abundance of genes encoding for many ABA signaling components is affected by ETR1 and ETR2 supports this. If true, it will make it more difficult to determine a specific pathway between these ethylene receptors and ABA signaling. This is especially true because our data also suggest that ETR1 and ETR2 are indirectly affecting ABA signaling.
Cross talk between ethylene signaling and other hormone signaling pathways has been described and usually involves signaling from downstream components such as EIN2 or EIN3 as hub points between the ethylene pathway and other hormone signaling pathways (Muday et al., 2012; van de Poel et al., 2015). Of particular note, in Arabidopsis, CTR1 and EIN2 mutants affect responses to ABA, indicating cross talk from these core components of ethylene signaling to the ABA signaling pathway (Beaudoin et al., 2000; Ghassemian et al., 2000; Wang et al., 2007; Subbiah and Reddy, 2010; Thole et al., 2014). In Physcomitrella patens, CTR1 modulates both ethylene and ABA signal transduction, suggesting an ancestral role of CTR1in both pathways (Yasumura et al., 2015). However, in this study we describe cross talk from the ethylene receptors to affect ABA signaling independently of the canonical ethylene signal transduction pathway. Additionally, we show that ABA affects ethylene receptor transcription and this transcriptional regulation is affected by the ethylene receptors. Even though the exact pathway for these complex interactions still needs to be resolved, the fact that this cross talk originates at receptors for one hormone to affect signaling for another hormone is intriguing, and suggests that there are likely to be other instances where receptors are signaling to other pathways via noncanonical pathways.
MATERIALS AND METHODS
Plant Materials and Chemicals
The Arabidopsis (Arabidopsis thaliana) mutants used are in the Columbia background, which was used as the wild-type control. The etr1-6, etr2-3, and etr1-6;etr2-3;ein4-4 triple mutants are lab stocks originally described by Hua and Meyerowitz (1998). The triple mutant plants transformed with cDNA encoding the full-length ETR1 (cETR1) or truncated ETR1 lacking the receiver domain (cetr1-ΔR), full-length gETR1, a full-length genomic transgene with a D659A or E666A point mutation, have previously been described (Wang et al., 2003; Binder et al., 2004a, 2004b, 2006; Kim et al., 2011; Bakshi et al., 2015). The plant hormone, ABA, was obtained from ACROS Organics and the ABA biosynthesis inhibitor, NF, was from Fluka.
The ctr1-2 mutant plants are lab stocks originally described by Kieber et al. (1993). The etr1-6;ctr1-2, and etr2-3;ctr1-2 double-mutant plants were generated by crossing the appropriate parents. The resultant crosses were allowed to self-pollinate and taken to the F4 generation. In each generation, seedlings were grown in the dark for 3 d in the presence or absence of 1 μL L−1 ethylene to identify ctr1-2-containing plants. The ctr1-2-containing crosses were identified by the constitutive ethylene response phenotype where seedlings grew slowly and with an exaggerated apical hook when grown in ethylene-free air in the dark. The plants were then genotyped for etr1-6 or etr2-3. To distinguish ETR1 from etr1-6, we used the primers and methods of Kim et al. (2011), where etr1-6 gives a larger product than ETR1. For ETR2 and etr2-3, we used the ETR2-1w, ETR2-1m, and ETR2-41 primers and methods described by Hua and Meyerowitz (1998). A product formed by the ETR2-1m and ETR2-4 primer pair indicated the presence of etr2-3, whereas a product formed with the ETR2-1w and ETR2-41 primer pair indicated the presence of the wild-type ETR2.
The pyk10-1, leb-1, SALK 065256, SALK 090550, SALK 119971, and SALK 063827 mutants were obtained from the Arabidopsis Biological Resource Center. The pyk10-1 and leb-1 mutants have previously been described (Nagano et al., 2008, 2009). To confirm that the four SALK lines were homozygous T-DNA insertional mutants, we ran PCR reactions with gene-specific primer pairs for each SALK line. The primers used for genotyping were: SALK 065256 5′-CTGAAGAAGAAATGGGGTTCC-3′ (forward), SALK 065256 5′-GTTTATTCCCTCCACTGCTCC-3′ (reverse), SALK 090550 5′-ATACACGAAACCCAACAGCTG-3′ (forward), SALK 090550 5′-GAAAGCAGTTGCTCATCAACC-3′ (reverse), SALK 119971 5′-ATTGGTACTATGTGCGGGTTG-3′ (forward), SALK 119971 5′-GGAAGAGAGGAGGCACAAATC-3′ (reverse), SALK 063827 5′-GTTTCTCACCAGATCGGGATC-3′ (forward), and SALK 063827 5′-GCAATACCAAAAGCAAGATGC-3′ (reverse).
We also confirmed the presence of the insertion by using the reverse primer for each SALK line paired with the LBb1.3 left border primer (5′-ATTTTGCCGATTTCGGAAC-3′) designed by the Salk Institute Genomic Analysis Laboratory for the T-DNA insertion. PCR cycling was performed at 95°C for 4 min for one cycle, followed by 40 cycles consisting of an initial denaturation at 95°C for 40 s, annealing at 52°C for 40 s, and extension at 72°C for 3 min. The final cycle was followed by a 5-min extension phase at 72°C. The lack of a product with the gene-specific primer pair and the presence of a product when using the left border-reverse primer pair indicated the plants were homozygous mutants for the gene in question.
Seed Germination Experiments
Seed germination studies were conducted using the methods of Wilson et al. (2014a, 2014b). To reduce biological variation, plants were grown at the same time under uniform conditions with seeds collected on the same day, then stored in a desiccator. After at least 3 weeks, seeds were mechanically sorted so that we used seeds between 250 and 300 μm in size (Hensel et al., 1993; Elwell et al., 2011). We surface-sterilized the seeds for 30 s with 70% (v/v) ethanol, then allowed the seeds to dry and placed them on 0.8% (w/v) agar plates that contained half-strength Murashige and Skoog salt (Murashige and Skoog, 1962), pH 5.7 fortified with vitamins and no added sugar. At least three plates with 20 seeds each were plated for each seed line and condition as indicated. Plates were wrapped with porous surgical tape to avoid accumulation of ethylene (Buer et al., 2003). Unless otherwise specified, plates were kept at 20°C to 21°C under 12 to 13 µmol m−2 s−1 white light with a long-day photoperiod (16 h light/8 h dark). Seed germination was evaluated every 12 h and considered complete with the rupture of the testa (seed coat). For experiments with ABA and NF, control plates contained 0.01% (v/v) ethanol as a solvent control. For experiments where seed germination was carried out in darkness, seeds were placed on agar plates under dim light, then treated with far-red illumination for 5 min as previously described (Oh et al., 2007; Wilson et al., 2014a, 2014b), and kept in darkness for 7 d at which time the percent of seed germination was determined.
Plasmid Construction
Full-length coding sequences of RD21A, LEB, and DI19 and C-terminal coding sequences of ETR1 and ETR2 were isolated from cDNA of wild-type Columbia seedlings. The coding sequences of these genes were PCR-amplified using forward and reverse primers containing specific restriction enzyme sites (Supplemental Table S2). PCR amplification was performed using New England Taq DNA polymerase following the manufacturer’s instructions. PCR products of the cytosolic domains of ETR1 or ETR2 were restriction-enzyme-digested using EcoRI and BamHI restriction enzymes, purified and fused to the GAL4 DNA binding domain of pGBKT7 vector (Clontech) to generate pGBKT7-ETR1 and pGBKT7-ETR2. Similarly, PCR products of the full-length coding sequences of the interacting proteins were double-restriction-enzyme digested, purified, and fused to the GAL4 DNA activation domain of pGADT7 vector (Clontech) to generate pGADT7-RD21A, pGADT7-LEB, and pGADT7-DI19. All the constructs were verified by double-restriction-enzyme digestion giving the correct size products and by sequencing.
Yeast Two-Hybrid Screens
To conduct yeast two-hybrid screens, bait constructs encoding for the cytosolic portion of ETR1 or ETR2 fused to the GAL4 DNA binding domain of pGBKT7 vector were introduced into yeast (Saccharomyces cerevisiae) strain Y187 to generate the bait strain. These were used to screen cDNA prey libraries generated in yeast strain AH109 as a fusion to the GAL4 activation domain as described in Hewezi et al. (2008). Screening for interacting protein partners and subsequent analyses were performed as described in BD Matchmaker Library Screening Kits (Clontech). Selected interactions identified in the yeast two-hybrid screens were first tested using yeast cotransformation assays. For the cotransformation assay, S. cerevisiae strain AH109 cells were cotransformed with pGBKT7-ETR1 or pGBKT7-ETR2 and pGADT7-RD21A, pGADT7-LEB, and pGADT7-DI19 constructs and interactions were assayed using a stringent selection on synthetic quadruple drop-out media SD/-Ade/-His/-Leu/-Trp selective medium performed in triplicate. Control growth conditions were carried out on double drop-out media (SD/-Leu/-Trp). Serial dilutions of yeast cotransformed cells were used to measure the strength of the interactions.
Gene Coexpression Network Analysis
To determine whether an interaction partner was coexpressed with ETR1 or ETR2, we used methods modified from Piya et al. (2014). Briefly, the coexpression profiles of ETR1, ETR2, and target genes were analyzed from different Arabidopsis tissues and organs including embryos, floral buds, flowers, hypocotyls, leaves, roots, shoot apical meristem, seedlings, and whole plants using 63 different RNAseq datasets from the Sequence Read Archive (Leinonen et al., 2011). The individual gene expression level from all the RNAseq data sets was quantified and represented as values measured in fragments per kilobase of transcript per million mapped reads (FPKMs). FPKM value of 1.0 was set as the threshold for expressed genes and hence, only those genes having FPKM values greater than 1 in at least one tissue were included in the gene coexpression analysis. To identify the tissues where the interacting pairs were coexpressing, we calculated the Z-scores for each of the FPKM values. The Z-score values were averaged across different samples of a given tissue and positive Z-score values with P < 0.05 represent high expression. Next, the sample contribution scores were calculated by multiplying Z-scores of ETR1 or ETR2 with the interacting partners for each tissue as described in Obayashi et al. (2014) and then the values were used to construct the heatmap using the Multiple Experiment Viewer. Red color represented pairs with highly correlated gene coexpression profiles and green represented highly anticorrelated pairs. Positive sample contribution score values obtained from multiplying two negative Z-scores were considered negative. Based on this information, we generated a gene coexpression network using Cytoscape (Shannon et al., 2003). The network contained 83 nodes and 87 edges representing the interacting combinations. Gene coexpression profiles of ETR1, ETR2, and target genes were analyzed in Arabidopsis root tissues using 10 different RNAseq datasets from the SRA. Pairwise coexpression values of genes encoding ETR1, ETR2, and target proteins in root tissues were used to generate the coexpression network of the ETR1, ETR2, and the target interacting proteins. Continuous edges indicated significant coexpression in root, dashed edges represented significant coexpression with the receptor in at least one tissue other than root, and red dotted edges indicated no significant coexpression in any tissue with their respective receptor partner.
BiFC
To determine whether selected proteins interacted with ETR1 or ETR2 in planta, we used BiFC assays as described by Hewezi et al. (2008). The full-length coding sequence of ETR1 or ETR2 were PCR-amplified from cDNAs of wild-type Columbia seedlings using forward and reverse primers containing specific restriction enzyme sites (Supplemental Table S2). After double-restriction digestion of the constructs, the PCR products were cloned into pSAT4-nEYFP-C1 to generate either N terminus-tagged ETR1 and ETR1 (YN-ETR1 and YN-ETR2, respectively) or C terminus-tagged receptors (ETR1-YN and ETR2-YN) fusions using the specific restriction enzymes. Similarly, the full-length coding sequences of RD21A, LEB, and DI19 were cloned into pSAT4-cEYFP-C1B to generate YC-RD21A, YC-LEB, and DI19-YC fusions. All these generated constructs were confirmed by double-restriction-enzyme digestion and sequencing. All combinations of nEYFP and cEYFP fusions, in addition to negative and positive controls, were coexpressed in Nicotiana benthamiana leaves using particle bombardment as previously described by Crawford and Zambryski (2000). All the bombardments were performed in triplicates in two independent experiments. Cotransformed leaf tissues were incubated at 25°C in dark for 16 h to 24 h before being detected for YFP signal. Bright and fluorescent images were captured using a Leica SP8 White Light Laser Confocal System.
RNA Isolation and Real-Time qRT-PCR
To evaluate the transcript abundance of Arabidopsis genes, we used real-time qRT-PCR. To do this, total RNA was isolated from 25 mg (dry weight) of seeds that were germinated in the presence or absence of 1 μM ABA or 150 mm NaCl for 2 d. We used the methods of Meng and Feldman (2010) as modified by Wilson et al. (2014a, 2014b) to isolate RNA. Transcript data were normalized to At3g12210 (Dekkers et al., 2012) using the method of Livak and Schmittgen (2001) for each seed line for each condition to obtain the relative amounts of target gene transcripts between plant backgrounds for each treatment. These levels were then normalized to the levels observed in untreated wild-type seeds. The primer pairs used for qRT-PCR of the receptors were: 5′-AGTGTTAAGACTCGGGAGCTT-3′ (forward) and 5′-GTTTCTTCCTGAGTTCGAATCAAT-3′ (reverse) for ETR1, 5′-ATGGCGTTTACTGTTTTCAAGATG-3′ (forward) and 5′-CAAAATCAAACCAACTTCACGACC-3′ (reverse) for ETR2, 5′-ACTCATTTGCTTAACGCTTGGACGTAT-3′ (forward), and 5′-AAGCTCTCTAACTTTCCATTTCAACAG-3′ (reverse) for EIN4, 5′-ATTGCTAAAGTCTCTTGCGCGGTTGTG-3′ (forward) and 5′-TCTCTATCTAACTCATCAGCTTTCTTC-3′ (reverse) for ERS1, and 5′-TCACTGGCCTTGGGTCATGACAGCTGT-3′ (forward) and 5′-CTCTGGTCTTCTTACTCAACATAAACT-3′ (reverse) for ERS2. All other primers used for this analysis have been described previously (Fujii et al., 2007; Miura et al., 2009; Bhaskara et al., 2012; Chan, 2012; Gliwicka et al., 2012; Singh et al., 2015; Yoshida et al., 2015; Zhou et al., 2015; Zhang et al., 2016).
Accession Numbers
Arabidopsis Genome Initiative accession numbers for genes studied in this article are ABF1, At1g49720; ABF2, At1g45249; ABF3, At4g34000; ABF4, At3g19290; ABI1, At4g26080; ABI2, At5g57050; ABI5, At2g36270; AHG1, At5g51760; AHG3, At3g11410; CRA1, At5g44120; CTR1, At5g03730; DI19, At1g56280; EIN4, At3g04580; ERS1, At2g40940; ERS2, At1g04310; ETR1, At1g66340; ETR2, At3g23150; HAB1, At1g72770; HAB2, At1g17550; HAI1, At5g59220 ; HAI2, At1g07430; HAI3, At2g29380; KIN1, At5g15960; LEB1, At3g09260; PYL1, At5g46790; PYL5, At5g05449; PYL7, At4g01026; PYL9, At1g01360; PYR1, At4g17870; RAB18, At5g66400; RD21A, At1g47128; RD29A, At5g52310; SLAC1, At1g12480; SLAH3, At5g24030; SnRK2.2, At3g50500; SnRK2.3, At5g66880; SnRK2.6, At4g33950; SnRK2.10, At1g60940.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Seed germination time-courses under different inhibitory conditions.
Supplemental Figure S2. The effect of NF on seed germination under different inhibitory conditions.
Supplemental Figure S3. Seed germination time-courses in various inhibitory conditions for ETR1 receiver domain mutants.
Supplemental Figure S4. Seed germination time-courses in the dark for ETR1 receiver domain mutants.
Supplemental Figure S5. Genotyping and phenotyping of double mutant plants containing ctr1-2.
Supplemental Figure S6. Time for 50% seed germination of receptor mutant plants crossed with ctr1-2 in response to ABA.
Supplemental Figure S7. Seed germination time-courses of mutant seed lines in response to ABA.
Supplemental Figure S8. Simple working genetic model for relationships among ETR1, ETR2, and interacting proteins in the control of seed germination.
Supplemental Table S1. List of proteins that interact with the cytosolic portion of ETR1 or ETR2 or both in the yeast two-hybrid screen.
Supplemental Table S2. List of primers used for cotransformation and BiFC analyses.
Acknowledgments
The authors acknowledge the technical help of Andrew Ward, Joanna Nosarzewska, and NaSha Austin, and the helpful advice of Rebecca Wilson, Tessa Burch-Smith, Eric Schaller, and Anna Stepanova.
Footnotes
This project was supported by United States National Science Foundation (NSF) grants IOS-1254423 and MCB-1517032 to B.M.B.
Articles can be viewed without a subscription.
References
- Abeles F, Morgan P, Saltveit MJ (1992) Ethylene in Plant Biology, 2nd Ed. Academic Press, San Diego, CA [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PL (2012) Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol 158: 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi A, Wilson RL, Lacey RF, Kim H, Wuppalapati SK, Binder BM (2015) Identification of regions in the receiver domain of the ETHYLENE RESPONSE1 ethylene receptor of Arabidopsis important for functional divergence. Plant Physiol 169: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer EM. (1976) A potent inhibitor of ethylene action in plants. Plant Physiol 58: 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Nguyen TT, Verslues PE (2012) Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 160: 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Mortimore LA, Stepanova AN, Ecker JR, Bleecker AB (2004a) Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol 136: 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, O’Malley RC, Wang W, Moore JM, Parks BM, Spalding EP, Bleecker AB (2004b) Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol 136: 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, O’Malley RC, Wang W, Zutz TC, Bleecker AB (2006) Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol 142: 1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson MMA, Bleckmann A, Allekotte S, Groth G (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6 [DOI] [PubMed] [Google Scholar]
- Bisson MMA, Groth G (2010) New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol Plant 3: 882–889 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard M-J, Barbier-Brygoo H, Laurière C (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63: 491–503 [DOI] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Wasteneys GO, Masle J (2003) Ethylene modulates root-wave responses in Arabidopsis. Plant Physiol 132: 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Z. (2012) Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 100: 110–115 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen AC, Donovan A, Ned-Sykes R, Andrews NC (2015) Noncanonical role of transferrin receptor 1 is essential for intestinal homeostasis. Proc Natl Acad Sci USA 112: 11714–11719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Binder BM, Garrett WM, Tucker ML, Chang C, Cooper B (2011) Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol Biosyst 7: 2637–2650 [DOI] [PubMed] [Google Scholar]
- Chen T, Liu J, Lei G, Liu Y-F, Li Z-G, Tao J-J, Hao Y-J, Cao Y-R, Lin Q, Zhang W-K, Ma B, Chen S-Y, et al. (2009) Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol 50: 1636–1650 [DOI] [PubMed] [Google Scholar]
- Chen Y-F, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC (2000) Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr Biol 10: 1032–1040 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53: 28–37 [DOI] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Bright J, Harrison J, Weir I, Hooley R, Neill SJ (2005) A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol 137: 831–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslauriers SD, Alvarez AA, Lacey RF, Binder BM, Larsen PB (2015) Dominant gain-of-function mutations in transmembrane domain III of ERS1 and ETR1 suggest a novel role for this domain in regulating the magnitude of ethylene response in Arabidopsis. New Phytol 208: 442–455 [DOI] [PubMed] [Google Scholar]
- Dong C-H, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem 285: 40706–40713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C-H, Rivarola M, Resnick JS, Maggin BD, Chang C (2008) Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J 53: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell AL, Gronwall DS, Miller ND, Spalding EP, Brooks TLD (2011) Separating parental environment from seed size effects on next generation growth and development in Arabidopsis. Plant Cell Environ 34: 291–301 [DOI] [PubMed] [Google Scholar]
- Federle MJ, Bassler BL (2003) Interspecies communication in bacteria. J Clin Invest 112: 1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S-Y, Cutler SR, Sheen J, Rodriguez PL, Zhu J-K (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J-K (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J-K (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu J-K (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA 95: 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE (2002) Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem 278: 34725–34732 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KAS, Romeis T, Hedrich R (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gliwicka M, Nowak K, Cieśla E, Gaj MD (2012) Expression of seed storage product genes (CRA1 and OLEO4) in embryonic cultures of somatic tissues of Arabidopsis. Plant Cell Tissue Organ Cult 109: 235–245 [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Städele K, Růzicka K, Obrdlik P, Harter K, Horák J (2008) Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol Plant 1: 308–320 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, Schaller GE (2012) Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol 159: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yamada K, Shimada T, Matsushima R, Nishizawa NK, Nishimura M, Hara-Nishimura I (2001) A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol 42: 894–899 [DOI] [PubMed] [Google Scholar]
- Hensel LL, Grbić V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, Davis EL, Baum TJ (2008) Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell 20: 3080–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Fujishige N, Kunii T, Nishimura N, Iuchi S, Shinozaki K (2004) A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell Physiol 45: 703–711 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins D. (2009) Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal 21: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, Cooper B, Kieber JJ, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim H, Helmbrecht EE, Stalans MB, Schmitt C, Patel N, Wen C-K, Wang W, Binder BM (2011) Ethylene receptor ETHYLENE RECEPTOR1 domain requirements for ethylene responses in Arabidopsis seedlings. Plant Physiol 156: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Patterson SE, Binder BM (2013) Reducing jasmonic acid levels causes ein2 mutants to become ethylene responsive. FEBS Lett 587: 226–230 [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lång V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M; International Nucleotide Sequence Database Collaboration (2011) The sequence read archive. Nucleic Acids Res 39: D19–D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Khan AM, Yamaguchi S, Kamiya Y (2005) Effects of heavy metals on seed germination and early seedling growth of Arabidopsis thaliana. Plant Growth Regul 46: 45–50 [Google Scholar]
- Lin Y, Wang J, Zu Y, Tang Z (2012) Ethylene antagonizes the inhibition of germination in Arabidopsis induced by salinity by modulating the concentration of hydrogen peroxide. Acta Physiol Plant 34: 1895–1904 [Google Scholar]
- Liu W-X, Zhang F-C, Zhang W-Z, Song L-F, Wu W-H, Chen Y-F (2013) Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol Plant 6: 1487–1502 [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu C, Wen C-K (2010) Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol 10: 60 10.1186/1471-2229-10-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Ma B, Cui M-L, Sun H-J, Takada K, Mori H, Kamada H, Ezura H (2006) Subcellular localization and membrane topology of the melon ethylene receptor CmERS1. Plant Physiol 141: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Martín-Rodríguez JÁ, León-Morcillo R, Vierheilig H, Ocampo JA, Ludwig-Müller J, García-Garrido JM (2011) Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol 190: 193–205 [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Suttle JC (1991) The Plant Hormone Ethylene. CRC Press, Boca Raton, FL [Google Scholar]
- McDaniel BK, Binder BM (2012) ETHYLENE RECEPTOR1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J Biol Chem 287: 26094–26103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng L-M, Zhou XE, Soon F-F, Xu Y, Suino-Powell KM, Park S-Y, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Feldman L (2010) A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J 5: 183–186 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Milla MAR, Townsend J, Chang I-F, Cushman JC (2006) The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol Biol 61: 13–30 [DOI] [PubMed] [Google Scholar]
- Miller RK, McCrea PD (2010) Wnt to build a tube: contributions of Wnt signaling to epithelial tubulogenesis. Dev Dyn 239: 77–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang H-J, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, Yoshida T, Kodaira K-S, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Mou W, Li D, Bu J, Jiang Y, Khan ZU, Luo Z, Mao L, Ying T (2016) Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PLoS One 11: e0154072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P, Klee HJ (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279: 48734–48741 [DOI] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17: 181–195 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol 49: 969–980 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Maekawa A, Nakano RT, Miyahara M, Higaki T, Kutsuna N, Hasezawa S, Hara-Nishimura I (2009) Quantitative analysis of ER body morphology in an Arabidopsis mutant. Plant Cell Physiol 50: 2015–2022 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1, and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Okamura Y, Ito S, Tadaka S, Aoki Y, Shirota M, Kinoshita K (2014) ATTED-II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol 55: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee H-S, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Bleecker AB (2003) Ethylene perception in Arabidopsis by the ETR1 Receptor Family: evaluating a possible role for two-component signaling in plant ethylene responses. In Inouye M, Dutta R, eds, Histidine Kinase in Signal Transduction. Academic Press, Amsterdam, the Netherlands, pp 439-457 [Google Scholar]
- Pang PP, Pruitt RE, Meyerowitz EM (1988) Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol Biol 11: 805–820 [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN Jr., Hewezi T (2014) Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5: 744 10.3389/fpls.2014.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L-X, Li Y, Li D-D, Xu W-L, Zheng Y, Li X-B (2014) Arabidopsis drought-induced protein Di19-3 participates in plant response to drought and high salinity stresses. Plant Mol Biol 86: 609–625 [DOI] [PubMed] [Google Scholar]
- Qiu L, Xie F, Yu J, Wen C-K (2012) Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol 159: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao Z, Schaller GE (2007) A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen C-K, Shockey JA, Chang C (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103: 7917–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim T-H, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB (1995) The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem 270: 12526–12530 [DOI] [PubMed] [Google Scholar]
- Shakeel SN, Wang X, Binder BM, Schaller GE (2013) Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants 5: 10.1093/aobpla/plt01010.1093/aobpla/plt010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I, Varma A, Grundler FM, Oelmüller R (2008) PYK10, a β-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J 54: 428–439 [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by Phytochrome A. Plant Physiol 104: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Jha SK, Bagri J, Pandey GK (2015) ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One 10: e0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yu H, Dong J, Che X, Jiao Y, Liu D (2016) The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet 12: e1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon F-F, Ng L-M, Zhou XE, West GM, Kovach A, Tan MHE, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, Brunzelle JS, Zhang H, et al. (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah V, Reddy KJ (2010) Interactions between ethylene, abscisic acid and cytokinin during germination and seedling establishment in Arabidopsis. J Biosci 35: 451–458 [DOI] [PubMed] [Google Scholar]
- Thole JM, Beisner ER, Liu J, Venkova SV, Strader LC (2014) Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3 (Bethesda) 4: 1259–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Poel B, Smet D, van der Straeten D (2015) Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol 169: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M, Sharma S (2017) Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front Plant Sci 8: 161 10.3389/fpls.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Manalansan B, Rauniyar N, Munemasa S, Booker MA, Brandt B, Waadt C, Nusinow DA, Kay SA, Kunz H-H, Schumacher K, DeLong A, et al. (2015) Identification of open stomata1-interacting proteins reveals interactions with Sucrose Non-Fermenting1-Related Protein Kinases2 and with Type 2A Protein Phosphatases that function in abscisic acid responses. Plant Physiol 169: 760–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu C, Li K, Sun F, Hu H, Li X, Zhao Y, Han C, Zhang W, Duan Y, Liu M, Li X (2007) Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol Biol 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang T, Li K, Li X (2008) Genetic analysis of involvement of ETR1 in plant response to salt and osmotic stress. Plant Growth Regul 54: 261–269 [Google Scholar]
- Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22: 1613–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Bakshi A, Binder BM (2014a) Loss of the ETR1 ethylene receptor reduces the inhibitory effect of far-red light and darkness on seed germination of Arabidopsis thaliana. Front Plant Sci 5: 433 10.3389/fpls.2014.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Kim H, Bakshi A, Binder BM (2014b) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol 165: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Hu H, Ries A, Merilo E, Kollist H, Schroeder JI (2011) Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J 30: 1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yang W, Zhang W, Wang X (2017) Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol J 15: 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Pierik R, Kelly S, Sakuta M, Voesenek LACJ, Harberd NP (2015) An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol 169: 283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38: 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006a) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006b) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Zhang A, Ren H-M, Tan Y-Q, Qi G-N, Yao F-Y, Wu G-L, Yang L-W, Hussain J, Sun S-J, Wang Y-F (2016) S-type anion channels SLAC1 and SLAH3 function as essential negative regulators of inward K+ channels and stomatal opening in Arabidopsis. Plant Cell 28: 949–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase D α 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XC, Schaller GE (2004) Effect of salt and osmotic stress upon expression of the ethylene receptor ETR1 in Arabidopsis thaliana. FEBS Lett 562: 189–192 [DOI] [PubMed] [Google Scholar]
- Zhou S-F, Sun L, Valdés AE, Engström P, Song Z-T, Lu S-J, Liu J-X (2015) Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in Arabidopsis. New Phytol 208: 188–197 [DOI] [PubMed] [Google Scholar]