Efficient ER membrane fusion requires dimerization of RHD3 in different ER membranes and an interaction between membrane lipids and the C-terminal amphipathic helix of RHD3.
Abstract
The endoplasmic reticulum (ER) is a network of tubules and sheets stretching throughout the eukaryotic cells. The formation of the ER requires homotypic membrane fusion, which is mediated by a family of Dynamin-like Atlastin GTPase proteins. The Arabidopsis (Arabidopsis thaliana) member ROOT HAIR DEFECTIVE3 (RHD3) has been demonstrated to mediate ER membrane fusion, but how exactly RHD3 is involved in the process is still unknown. Here we conducted systemic structure-function analyses of roles of different RHD3 domains in mediating ER fusion. We showed that efficient ER membrane fusion mediated by RHD3 requires a proper dimerization of RHD3 through the GTPase domain (GD) and the first and second three helix bundles (3HBs) in the middle domain. RHD3 has a 3HB-enriched middle domain longer than that of Atlastins, and we revealed that the third and fourth 3HBs are required for the stability of RHD3. The transmembrane segments of RHD3 are essential for targeting and retention of RHD3 in the ER and can also facilitate an oligomerization of RHD3. Furthermore, we showed that an amphipathic helix in the C-terminal cytosolic tail of RHD3 has a membrane anchoring ability that is required for efficient ER membrane fusion mediated by RHD3. This work contributes to a better understanding of a coordinated action of RHD3 in the fusion of ER membranes.
The endoplasmic reticulum (ER) is a highly conserved, continuous, and interconnected membrane network composed of smooth tubules and sheet-like cisternae that extend throughout the cytoplasm. The ER plays vital roles in protein synthesis, folding, and transport; mitochondrial division; calcium regulation; and lipid synthesis and transport in eukaryotic cells including plant cells. Live-cell imaging revealed that the ER is a highly dynamic organelle, showing continuous growth, retraction, sliding, and fusion of tubules to form strands, cisternae, and polygons (Sparkes et al., 2011). This dynamic is important for the ER to perform its diverse functions. It is known that the ER shaping proteins, reticulons, and DP1/Yop1p play key roles in generating and maintaining the unique reticular morphology of the ER in plant, yeast, and mammalian cells (Hu et al., 2008; Sparkes et al., 2010; Voeltz et al., 2006). It is assumed that these proteins can induce and stabilize a membrane curvature of the tubules and edges of the sheet through their two sets of closely spaced transmembranes (TMs) and a C-terminal amphipathic helix (Brady et al., 2015; Breeze et al., 2016; Voeltz et al., 2006). Connecting ER tubules into the polygonal network requires membrane fusion, which is mediated by a family of Dynamin-like Atlastin GTPase proteins, including Atlastins in metazoans, Sey1p in yeast, and ROOT HAIR DEFECTIVE3 (RHD3) in plants (Anwar et al., 2012; Chen et al., 2011; Hu et al., 2009; Orso et al., 2009; Zhang et al., 2013). The deletion of Atlastin leads to long and nonbranched ER tubules in mammalian cells and ER fragmentation in Drosophila (Hu et al., 2009; Orso et al., 2009). In plants, the loss of RHD3 leads to thick and bundled ER tubules (Zheng et al., 2004). These ER fusogens also play critical physiological roles. In humans, mutations in Atlastin-1 cause a neurodegenerative disease called hereditary spastic paraplegia (Zhao et al., 2001). In yeast Candida albicans, deletions of Sey1p lead to attenuated virulence in mice as well as increased susceptibility to cycloheximide (Yamada-Okabe and Yamada-Okabe, 2002). In plants, mutations in RHD3 result in short and wavy root hair growth (Schiefelbein and Somerville, 1990).
Atlastins, Sey1p, and RHD3 are all ER membrane-bound GTPases and share a conserved protein structure. They all have a cytosolic N-terminal GTPase domain (GD), a three helix bundles (3HB)-enriched middle domain and two TM segments, and a cytosolic C-terminal tail (CT; Liu et al., 2012; Stefano and Brandizzi, 2014). The mechanistic basis of the action of Atlastins and Sey1p in the ER membrane fusion has been studied (Hu and Rapoport, 2016). Based on the revealed homotypic interaction and the crystal structure of the cytosolic N terminus of Atlastin-1 (Bian et al., 2011), it was proposed that two Atlastin-1 molecules in different membranes bind GTP and form a dimer. The GTP hydrolysis and phosphate release trigger a conformational change of the dimer, leading to the membrane fusion (Bian et al., 2011). Because the two TMs are also involved in homotypic interaction of Atlastin-1, it is believed that several Atlastin-1 molecules in the same membrane could also associate with each other through their TMs, and multiple cycles of GTP hydrolysis occur before a successful fusion event (Liu et al., 2015; Liu et al., 2012). It is believed that Sey1p mediates the homotypic ER membrane fusion by a similar paradigm (Yan et al., 2015).
Despite what is unveiled in mammalian and yeast cells, the mechanistic basis of the action of RHD3 in the ER membrane fusion has not been well studied. It is known that overexpression of GDP-locked RHD3 mutant (S51N) and GTP-locked RHD3 mutant (T75A) could exert a dominant-negative effect on the ER morphology and root hair growth (Chen et al., 2011). Also, in vitro membrane fusion mediated by RHD3 requires GTP hydrolysis (Zhang et al., 2013). Moreover, it has been shown that RHD3 can undergo a GTP-dependent homotypic interaction in yeast 2-hybrid and bimolecular fluorescent complementation systems (Chen et al., 2011; Zheng and Chen, 2011). Previous studies indicated that mutations in the GD (Chen et al., 2011; Wang et al., 2015; Zhang et al., 2013), the middle domain (Zhang et al., 2013), and two TMs (Stefano et al., 2012) all can make RHD3 defective, suggesting that the RHD3-mediated ER fusion is a delicate process that may require a coordinated action from different domains of RHD3. However, how the different domains of RHD3 are involved in the RHD3-mediated ER fusion is still not known.
Here, we studied the roles of different RHD3 domains in mediating ER membrane fusion. Our data suggest that, in addition to its GTPase activity, the GD of RHD3 is involved in a dimerization of RHD3 for an efficient ER membrane fusion. While the first and second 3HBs of the middle domain of RHD3 are also involved in the dimerization of RHD3, the third and fourth 3HBs of RHD3 are required for the stability of RHD3. The TMs of RHD3 are essential for the targeting and retention of RHD3 in the ER and can facilitate an oligomerization of RHD3. Furthermore, we showed that an amphipathic helix in the CT of RHD3 has a membrane anchoring ability that is required for efficient ER membrane fusion mediated by RHD3.
RESULTS
Dimerization of RHD3 through GTPase Interface Is Required for an Efficient ER Fusion Mediated by RHD3
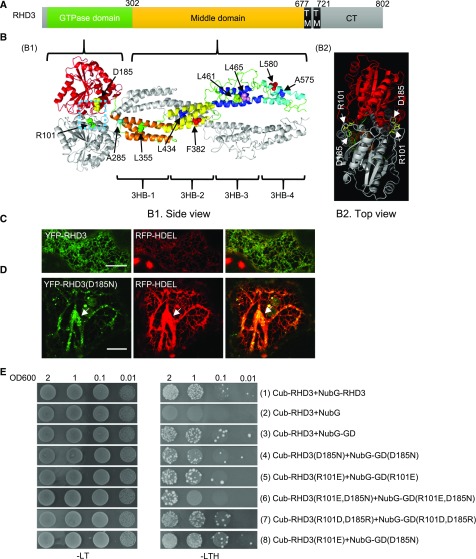
Although the overall structure of the Dynamin-like Atlastin GTPase proteins is conserved, the predicted 3HB-enriched middle domains of RHD3 and Sey1p are much longer than that of Atlastin-1 (Stefano and Brandizzi, 2014; Supplemental Fig. S1A; Fig. 1A). To understand the mechanistic basis of RHD3, we simulated the structure of the cytosolic N terminus of RHD3 based on the crystal structure of the cytosolic N terminus of C. albicans Sey1p (Yan et al., 2015). The side view of the simulated structure suggested that, like Sey1p, the cytosolic N terminus of RHD3 formed a dimer (Fig. 1B1, one RHD3 monomer is colored, another is in gray). The GDs of two RHD3 molecules were facing each other and the middle domains, composed of four 3HBs, form a twisted arm (Fig. 1B1). In the interface of the two GDs, there were three potential polar contacts between Asp-185 (D185) of one RHD3 molecule and Arg-101 (R101) of another (Supplemental Fig. S1B, arrowheads). When the simulated structure of RHD3 was viewed from the top (Fig. 1B2), there were two potential D185-R101 salt bridge sites in the interface of RHD3 (Fig. 1B2). We noted that rhd3-2, an RHD3 mutant allele with a point mutation (D185N), grows short and wavy root hairs (Wang et al., 1997). We first wondered if this point mutation in RHD3 could affect the function of RHD3 in the formation of the ER. To this end, we transiently expressed YFP-RHD3(D185N) together with an ER marker RFP-HDEL in tobacco (Nicotiana tabacum) leaves. Transient expression of wild-type YFP-RHD3 did not change the interconnected polygonal ER network (Fig. 1C), while expressing YFP-RHD3(D185N) made thick and unbranched ER bundles (Fig. 1D, arrowheads). This result suggests that RHD3(D185N) has a negative effect on the formation of the polygonal ER network in vivo.
Figure 1.
Mutations in the interface of the dimer influence the homotypic interaction of RHD3. A, The overall structure of full-length RHD3. The numbers indicate the corresponding amino acid positions. B, The predicted cytosolic dimer structure of RHD3. B1, the side view of cytosolic dimer structure of RHD3, B2, the top view of cytosolic dimer structure of RHD3. One RHD3 monomer is shown in gray and the other is shown in different colors. RHD3 forms a dimer through its GD (red) and middle domain. The middle domain contains four different 3HBs: 3HB-1 (orange), 3HB-2 (yellow), 3HB-3 (blue), and 3HB-4 (cyan). The mutated amino acids in this paper are indicated as the highlighted spheres. The enlarged structure of the blue dashed box is shown in Supplemental Figure S1B. C, Coexpressing YFP-RHD3 and RFP-HDEL in tobacco leaves. Scale bar = 10 μm. D, Coexpressing YFP-RHD3(D185N) and RFP-HDEL in tobacco leaves. The ER bundles are indicated by the arrow. Scale bar = 10 μm. E, Yeast mating-based SUS for the homotypic interaction of RHD3 and various mutated versions of RHD3 as indicated. Cub fused full-length RHD3 interacts with NubG fused full-length RHD3 (1), but not with NubG alone (2). Cub fused RHD3 interacts with NubG fused GD (3), but Cub fused mutant RHD3(D185N) (4), RHD3(R101E) (5), and RHD3(D185N, R101E) (6) have the reduced interaction with NubG fused mutant GD(D185N), GD(R101E), and GD(D185N, R101E), respectively. The interaction between RHD3 and its GD domain with reciprocal mutagenesis (R101D, D185R) (7) was restored. On the other hand, the interaction between RHD3(R101E) and GD(D185N) (8) was not significantly reduced compared to RHD3 and its GD (3). Mated diploid yeast cells were diluted into DO600 = 2, 1, 0.1, and 0.01.
According to the simulated structure, the D185N mutation could potentially reduce the polar contacts between Asp-185 (D185) of one RHD3 molecule and Arg-101 (R101) of another in the dimerization of RHD3 (Supplemental Fig. S1C, arrowhead). As previously reported, RHD3 undergoes a homotypic interaction in both yeast 2-hybrid and bimolecular fluorescent complementation assays (Chen et al., 2011; Zheng and Chen, 2011), and RHD3 is functional in yeast cells (Zhang, et al., 2013). We thus tested if there is a reduced interaction when D185 or R101 is mutated using yeast mating based split-ubiquitin system (SUS; Grefen et al., 2007). A full-length RHD3 could interact with a full-length RHD3 (Fig. 1E, row 1). More importantly, we found that a full-length RHD3 could also interact with the GD of RHD3 (Fig. 1E, row 3), indicating that the simulated dimerization of RHD3 via the GDs is likely. When D185 was mutated to N (Fig. 1E, row 4) or R101 was mutated to E (Fig. 1E, row 5), either mutation reduced the interaction between the full-length RHD3 and its GD. Strikingly, the double mutant (R101E, D185N) had a much weaker homotypic interaction than either D185N or R101E mutation (Fig. 1E, row 6). In addition, when the interaction between RHD3 and its GD domain with the reciprocal mutation (R101D, D185R) was tested, the interaction between RHD3 and its GD was restored (Fig. 1E, row 7). These results confirmed the interaction linkage between D185 and R101 in the simulated RHD3 dimerization. While there was a reduced interaction when either of the amino acids was mutated, interestingly, no significant reduction was found in the interaction between RHD3(R101E) and GD(D185N) compared with the interaction between RHD3 and its GD (Fig. 1E, compare row 8 with row 3). As mentioned, there were two potential D185-R101 salt bridge sites in the interface of RHD3 dimer (Fig. 1B2). Likely, between RHD3(R101E) and GD(D185N), only one of two salt bridge sites will be affected. This could be a reason we did not detect a significant reduction to the level that was visible in SUS.
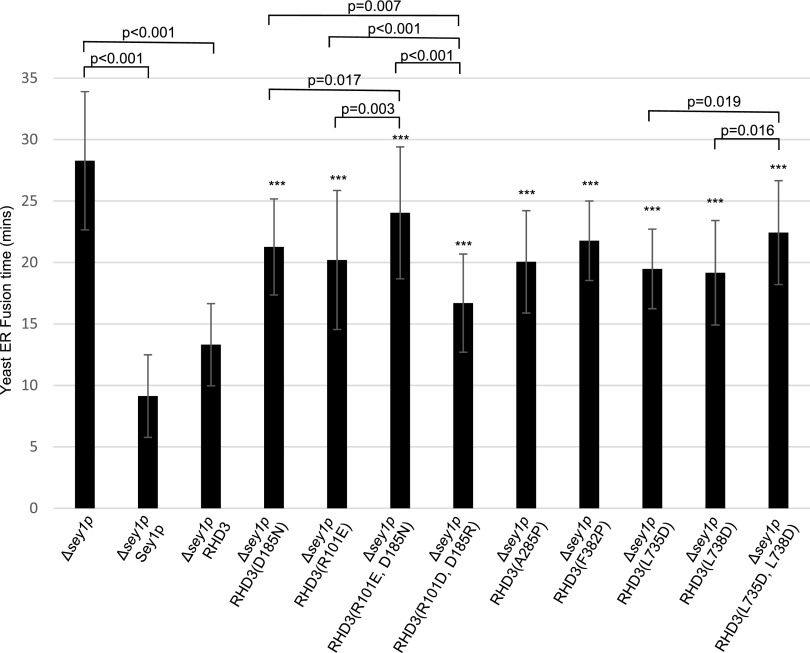
The dimerization of Atlastin-1 or Sey1p plays an important role in the efficient ER membrane fusion. We thus wondered if RHD3(D185N) or RHD3(R101E) is defective in mediating the ER fusion. Because RHD3 is able to rescue the ER fusion deficiency of Δsey1p yeast cells (Zhang et al., 2013), we therefore used a Δsey1p-based yeast ER fusion assay to quantify the ER fusion efficiency (Anwar et al., 2012). Consistent with the previous report, the Δsey1p mutant cells took approximately 28 min to finish the ER fusion (Fig. 2, column 1), while expressing Sey1p and RHD3 in the Δsey1p mutant cells could significantly reduce the fusion time to approximately 9 and 13 min, respectively (Fig. 2, column 2 and 3). The ER fusion time was approximately 21 and 20 min with the D185N and R101E mutations, respectively (Fig. 2, column 4 and column 5). The double mutant RHD3(R101E, D185N) had an enhanced ER fusion deficiency compared to RHD3(R101E) or RHD3(D185N) in rescuing the ER fusion defects of Δsey1p mutant (Fig. 2, column 6). Interestingly, the reciprocal mutant, RHD3(R101D, D185R), has much more efficient ER fusion (Fig. 2, column 7) than double mutant RHD3 (R101R, D185N) as well as single mutants RHD3(R101E) and RHD3(D185N). This was likely due to the restored interaction in the reciprocal mutant RHD3(R101D, D185R) revealed in Figure 1E. Taken together, the dimerization through D185 and R101 in the GD is required for the efficient ER fusion mediated by RHD3.
Figure 2.
Mutations in RHD3 compromise the ER fusion efficiency of RHD3. Sey1p, RHD3, and different RHD3 mutants as indicated were expressed in the Δsey1p mutant, and their average ER fusion time was quantified from 20 to 50 cells per sample. Each error bar represents mean ± sd. Student tests were used to check the significant difference between two groups of data and the corresponding P values are shown. *** Indicates significant difference from wild-type RHD3 (P value < 0.001, t test).
First and Second 3HBs Are Also Involved in RHD3 Dimerization for Efficient ER Fusion While Third and Fourth 3HBs Are Required for Protein Stability of RHD3
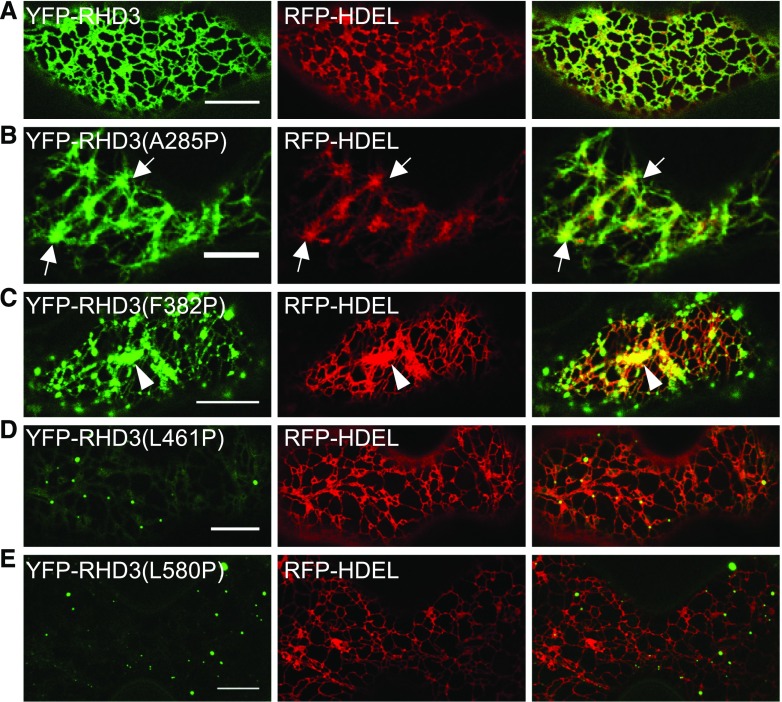
The simulated structure for the cytosolic N terminus of RHD3 suggests that there are four different 3HBs in the middle domain (Fig. 1B1). To gain the insight into the roles of the four 3HBs in the middle domain may play, two different conserved hydrophobic residues in each 3HB were replaced by Pro to disrupt the α-helix structure. The mutated versions of RHD3 were fused with YFP and then transiently expressed in tobacco leaves with an ER marker RFP-HDEL to assess their actions in the formation of the ER. The expression of 3HB-1 mutants RHD3(A285P) and RHD3(L355P) created thick and unbranched ER bundles (Fig. 3B; Supplemental Fig. S2B, arrows), while the expression of 3HB-2 mutants RHD3(F382P) and RHD3(L434P) produced aggregations of ER tubules (Fig. 3C; Supplemental Fig. S2C, arrowheads). On the other hand, transient expression of 3HB-3 mutants RHD3(L461P) and RHD3(L465P) and 3HB-4 mutants RHD3(A575P) and RHD3(L580P), including RHD3-1 with a point mutation (A575V) in 3HB-4, had no obvious negative effect on the formation of the polygonal tubular ER network (Fig. 3, D and E; Supplemental Fig. S2, D–F). Interestingly, while 3HB-1 mutants RHD3(A285P) and RHD3(L355P) and 3HB-2 mutants RHD3(F382P) and RHD3(L434P) were still targeted to the ER (Fig. 3, B and C; Supplemental Fig. S2, B and C), all 3HB-3 mutants, RHD3(L461P) and RHD3(L465P), and 3HB-4 mutants, RHD3(A575P), RHD3(L580P), and RHD3-1, had only a faint YFP signal in the form of small punctates (Fig. 3, D and E; Supplemental Fig. S2D–F).
Figure 3.
Transient expression of RHD3 with mutations in the middle domain in tobacco leaves. A, Coexpressing YFP-RHD3 and RFP-HDEL. Scale bar = 10 μm. B, Expression of 3HB-1 mutant RHD3(A285P) generates thick ER bundles (arrows). Scale bar = 10 μm. C, Expression of 3HB-2 mutant RHD3(F382P) generates aggregations of ER tubules (arrowhead). Scale bar = 10 μm. D and E, Expression of 3HB-3 mutant RHD3(L461P) and 3HB-4 mutant RHD3(L580P) only marks weak punctates on the ER and has no influence on the formation of the ER indicated by RFP-HDEL. Scale bar = 10 μm.
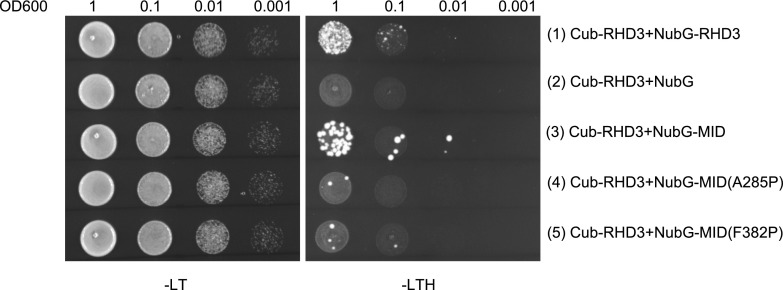
According to the simulated dimerization of RHD3, the first two 3HBs of different RHD3 monomers may hold each other tightly in the dimerization of RHD3. Consistent with this hypothesis, our SUS assay showed that there was an interaction between a full length of RHD3 and its middle domain (Fig. 4, row 3). More importantly, mutations in 3HB-1 (A285P) and 3HB-2 (F382P) significantly reduced the interaction between RHD3 and its middle domain (MID; Fig. 4, compare the interaction in row 3 with row 4 and 5). The yeast-based ER fusion assay also showed that RHD3(A285P) and RHD3(F382P) mutants had a reduced ER fusion efficiency (Fig. 2, column 8 and 9, respectively), suggesting that this dimerization through 3HB-1 and 3HB-2 is also required for efficient ER fusion mediated by RHD3 in vivo.
Figure 4.
3HB-1 and 3HB-2 of RHD3 are important for the homotypic interaction of RHD3. (Rows 1–5) Yeast mating-based SUS for interaction of RHD3 with its middle domain (MID) and mutated middle domains as indicated. Cub fused full-length RHD3 interacts with NubG-RHD3 (1) and NubG-MID (3), while the A285P or F382P mutations significantly compromised the interaction between RHD3 and its middle domain (4, 5, respectively). Mated diploid yeast cells were diluted into DO600 = 1, 0.1, 0.01, and 0.001.
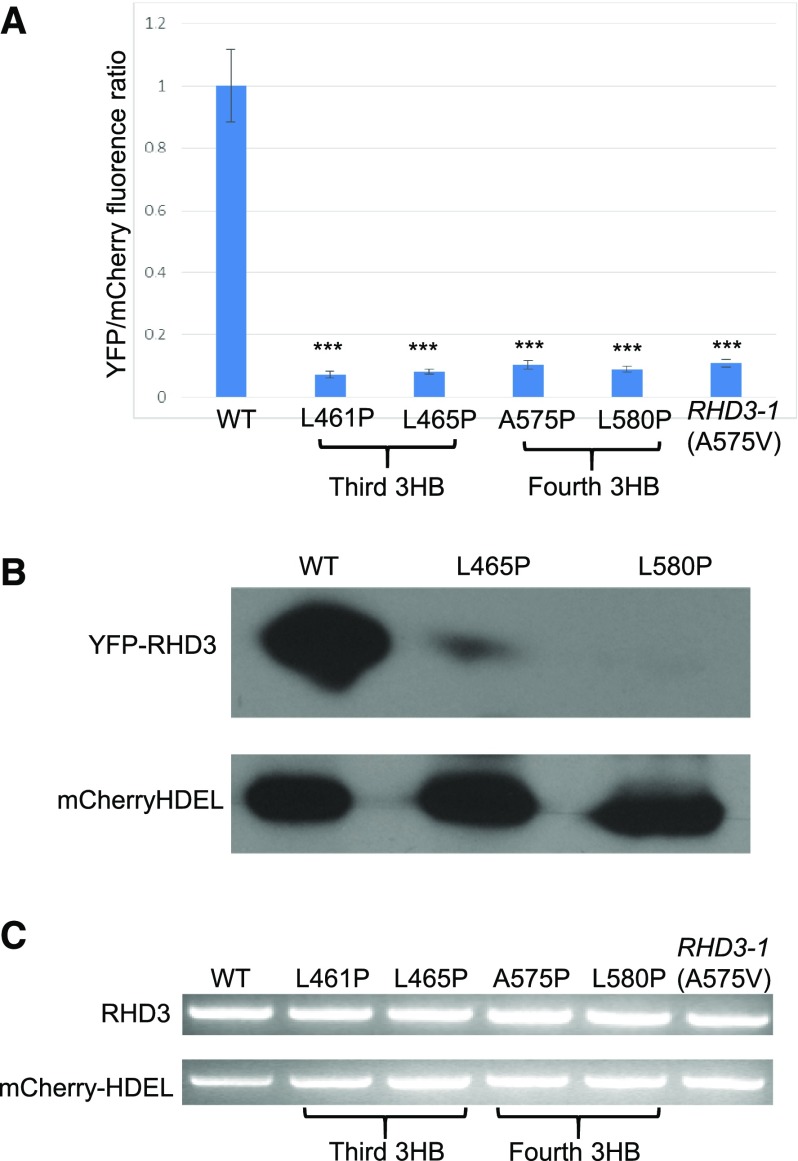
RHD3-1(A575V), located in the fourth 3HB, is showed to have reduced protein stability (Zhang et al., 2013). Because of the faint YFP signal produced by mutations in 3HB-3 and 3HB-4 of RHD3, we developed an RY (red and yellow) vector to quantify the expression level of the 3HB-3 and 3HB-4 RHD3 mutants. The RY vector can express mCherry-HDEL and YFP-RHD3 simultaneously (Supplemental Fig. S3A) so that mCherry-HDEL can be used as a reference for the expression of RHD3 (Zhang et al., 2015). With this system, we found that the 3HB-3 mutants RHD3(L461P) and RHD3(L465P), and 3HB-4 mutants RHD3(A575P), RHD3(L580P) and RHD3-1(A575V), again showed a faint YFP signal in the form of small punctates (Supplemental Fig. S3B). The fluorescence quantification showed that the YFP signal of these RHD3 mutants was dramatically lower than that of wild-type RHD3 (Fig. 5A). Western blot confirmed that the 3HB-3 and 3HB-4 RHD3 mutants had a reduced protein level (Fig. 5B), while at the transcription level, different 3HB-3 and 3HB-4 RHD3 mutants did not have significant difference compared to wild-type RHD3 (Fig. 5C). These results indicate that both 3HB-3 and 3HB-4 of RHD3 play a role in the protein stability of RHD3.
Figure 5.
3HB-3 and 3HB-4 of RHD3 are essential for the RHD3 protein stability. A, The ratio of YFP fluorescence of different YFP-fused RHD3 mutants as indicated compared to mCherry-HDEL fluorescence intensity. The YFP/mCherry values of different RHD3 mutants were quantified relatively to the wild-type value which was set as 1. The fluorescent intensity from 20 cells was quantified for each mutant. Each bar represents mean ± sd. *** Indicates significant difference from YFP-RHD3 (P value < 0.001, t test). B, Western blot for the protein expression level of YFP fused 3HB-3 (RHD3(L465P)) and 3HB-4 (RHD3(L580P)) mutants. Total proteins were extracted 48 hours after the infiltration of respective constructs. Anti-GFP was used to detect YFP fused proteins and anti-RFP was used to detect mCherry-HDEL as the loading control. C, RT-PCR for the transcription of YFP fused 3HB-3 and 3HB-4 mutants. Compared to the transcription of the reference gene mCherry-HDEL, different RHD3 mutants as indicated have no significant difference at the transcription level.
Transmembrane Domains Play an Important Role in Targeting and Retention of RHD3 in ER
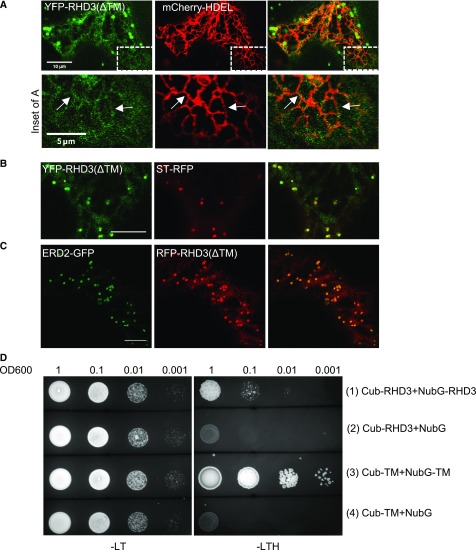
RHD3 is an ER localized membrane protein (Chen et al., 2011). RHD3 with a point mutation (P701S) between the two TMs was present in some large structures, which were different from the ER network distribution of wild-type RHD3 (Stefano et al., 2012). We therefore wondered what determines the ER localization of the RHD3 protein. To this end, the two putative TMs (aa677-721) were deleted to create RHD3(ΔTM) and then RHD3(ΔTM) was fused to either YFP or RFP. We found that RHD3(ΔTM) was present largely in the cytosol (Fig. 6, A–C). In addition, occasional reticulum-like structures (Fig. 6A, arrows) and many punctates (Fig. 6, B and C) were also visible. Coexpression with RFP-HDEL indicated that those reticulum-like structures were ER tubules (Fig. 6A, arrows), while coexpression with ERD2-GFP or ST-RFP indicated that those punctates were cis-Golgi (Fig. 6, B and C). The results suggested that TMs of RHD3 play an important role in targeting and retention of RHD3 in the ER.
Figure 6.
Transmembrane segments of RHD3 determine the ER localization of RHD3 and can self-interact. A, Although largely cytosolic, YFP-RHD3(ΔTM) is occasionally colocalized with RFP-HDEL (inset for A). The arrows in the inset of A point at ER tubules colocalized with mCherry-HDEL. B, Part of YFP-RHD3(ΔTM) is partially overlapped with ST-RFP (trans-Golgi). C, Part of RFP-RHD3(ΔTM) is fully overlapped with donut-shaped Cis-Golgi (ERD2-GFP). Scale bars = 10μm except the scale bar in the inset of A = 5 μm. D, Yeast mating based split-ubiquitin system (SUS) for self-interaction of the TMs of RHD3. Note the Cub fused TMs of RHD3 can efficiently interact with the NubG fused TMs of RHD3 (3), but not with NubG alone (4).
Atlastin-1 undergoes oligomerization in the same membrane through its TMs (Liu et al., 2012), so we wondered if the TMs of RHD3 have a similar mechanism. SUS test showed that the TMs of RHD3 could efficiently interact with each other (Fig. 6D, row 3), which suggested that RHD3 could also form oligomerization through TMs. These results indicate that in addition to determining the ER localization of RHD3, the TMs of RHD3 can also facilitate homotypic interaction of RHD3.
Amphipathic Helix of C-Terminal RHD3 Has Ability for Membrane Targeting
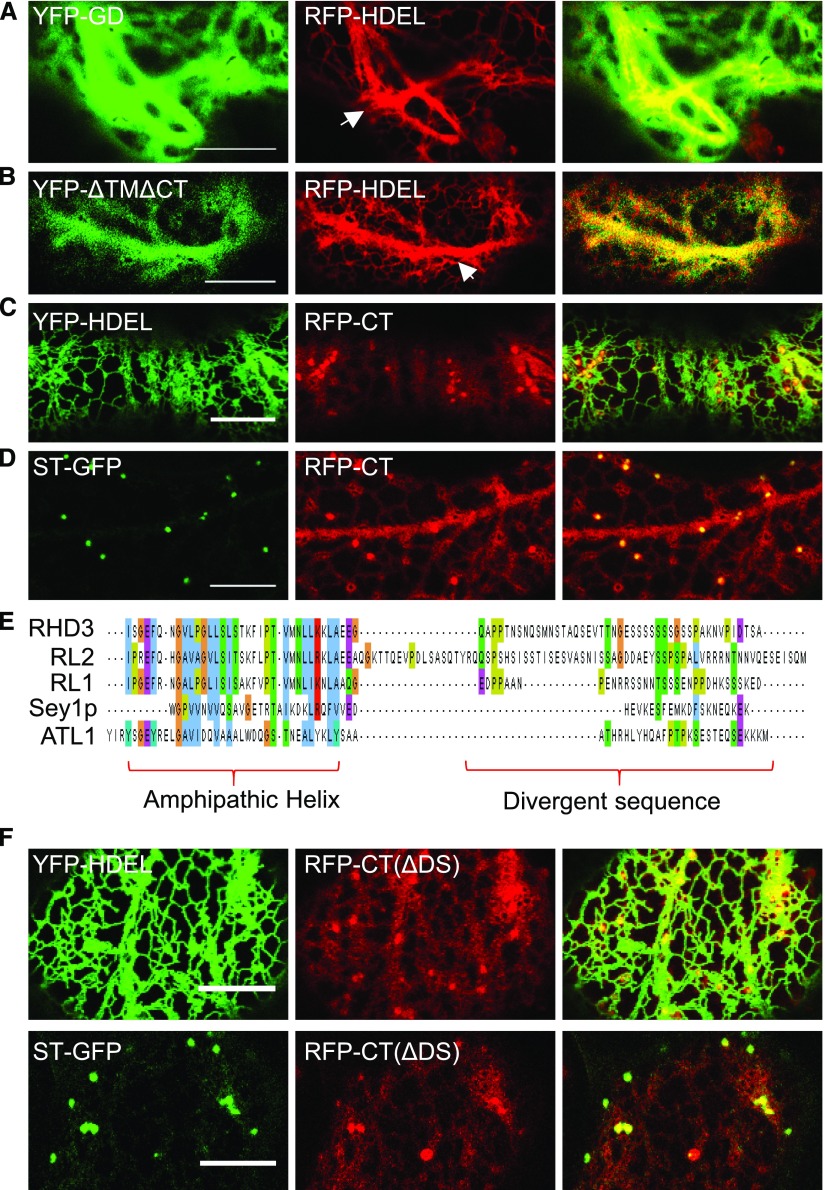
Although the TMs of RHD3 play an important role in targeting and retention of RHD3 in the ER, the unexpected ER and Golgi membrane association of RHD3(ΔTM) also suggests that there are membrane targeting sequences outside of the TMs of RHD3. To determine this, the GD of RHD3, the N-terminal cytosolic part of RHD3 (RHD3(ΔTMΔCT)), and the CT of RHD3 were fused with YFP or RFP and transiently expressed in tobacco leaves. The GD of RHD3 or RHD3(ΔTMΔCT) was seen only in the cytosol (Fig. 7, A and B). Interesting, although the GD of RHD3 or RHD3(ΔTMΔCT) was seen only in the cytosol, the expression of both parts of RHD3 has a negative effect on the formation of the polygonal ER network as indicated by RFP-HDEL (Fig. 7, A and B, arrows). On the other hand, the CT of RHD3, like RHD3(ΔTM), was localized to the ER membrane and Golgi, in addition to the cytoplasm (Fig. 7, C and D). This indicates that the CT of RHD3 contains a membrane targeting sequence(s). To gain deeper insight into what determines the membrane targeting in the CT of RHD3, a multiple sequence alignment was performed within the CT of proteins in the Dynamin-like Atlastin GTPase protein family (Fig. 7E). We revealed that there were a conserved amphipathic helix and a divergent sequence (DS) in the end. The DS is involved in the phosphorylation of RHD3 (Ueda et al., 2015). When the DS was deleted, the membrane targeting of the CT of RHD3 was not abolished (Fig. 7F). This indicated that the amphipathic helix in the C-terminus of RHD3 has an ability for membrane targeting.
Figure 7.
The amphipathic helix in the CT of RHD3 has a membrane targeting ability. A, The YFP-GD is cytosolic and has a negative effect on the formation of the polygonal ER network as indicated by RFP-HDEL. The arrow indicates the thick and bundled ER tubules. Scale bar = 10 μm. B, the cytosolic N-terminal domain of RHD3(ΔTMΔCT) (AA1-676) is also in the cytosol and has a negative effect on the formation of the polygonal ER network. The arrow indicates the thick and bundled ER tubules. Scale bar = 10 μm. C, Coexpressing the RFP fused CT of RHD3 (RFP-CT) (AA722-802) and YFP-HDEL. Although largely cytosolic, the occasional tubular structure of RFP-CT is colocalized with YFP-HDEL. Scale bar = 10 μm. D, Coexpressing RFP-CT and ST-GFP. The punctate structure of RFP-CT is colocalized with ST-GFP. Scale bar = 10 μm. E, Multiple alignment of the cytosolic C-terminal sequences of the selected atlastin proteins. The alignment was analyzed by MUSCLE. Sequences used were from RHD3(AA722-802), RHD3-LIKE2(RL2)(AA729-834), RHD3-LIKE1(RL1)(AA728-795), Sey1p(AA727-776), and ATL1(AA493-558). The amphipathic helix region and divergent sequence are highlighted. F, Coexpression of the RFP fused CT(ΔDS) of RHD3 with YFP-HDEL and ST-GFP. Divergent sequence deleted RFP-CT(ΔDS) exhibits the same localization as full C terminus of RHD3. Scale bar = 10 μm.
Amphipathic Helix of C-Terminal RHD3 Is Required for Efficient ER Membrane Fusion
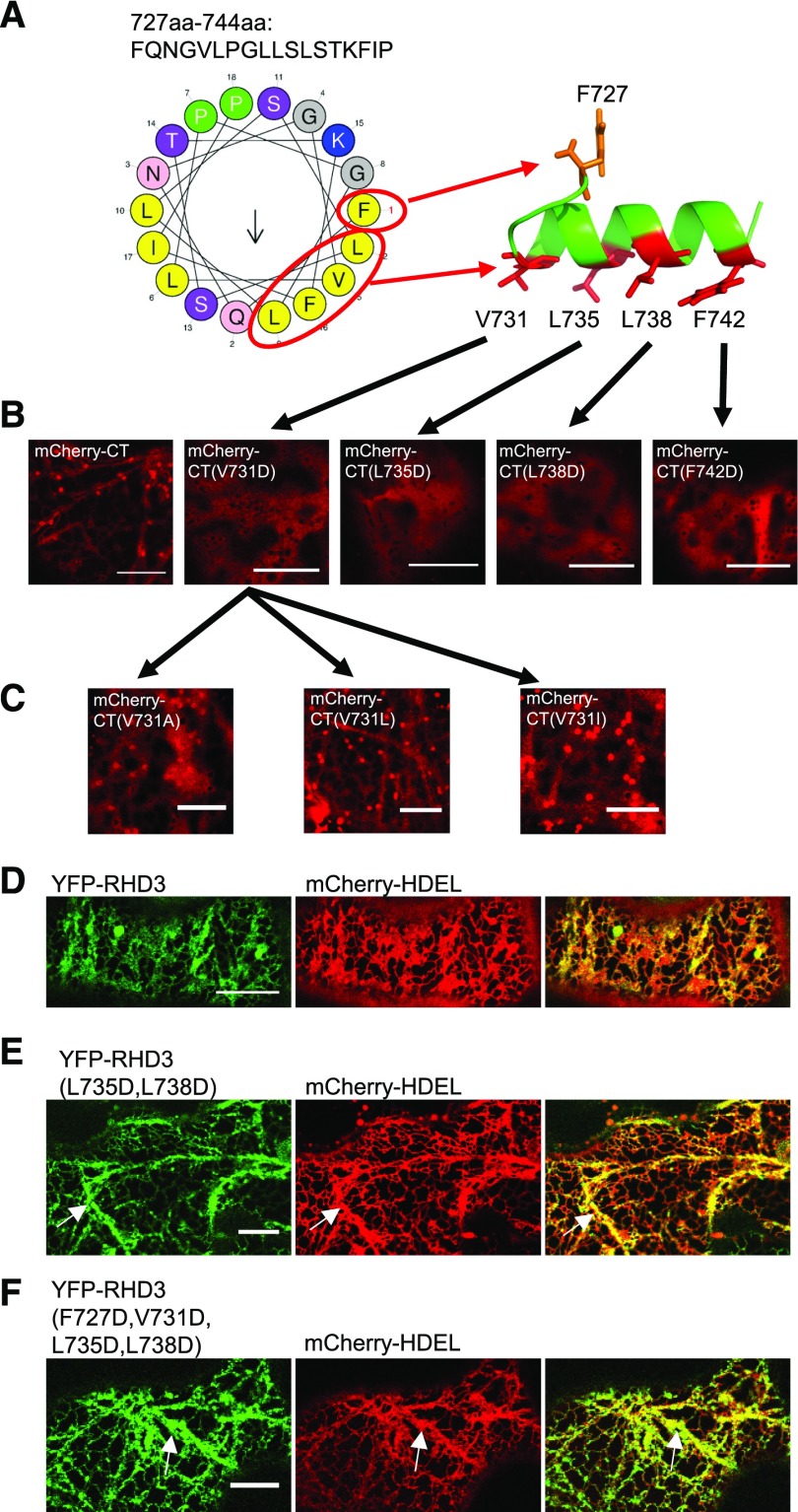
To understand how this amphipathic helix of RHD3 could direct a membrane targeting of a portion of RHD3(ΔTM) or the CT of RHD3 as well as the biological function of the amphipathic helix of RHD3, the possible arrangement of the helix (AA727-744) was first predicted using HeliQuest and PEP-FOLD (Fig. 8A). There was an obvious hydrophobic face in the helix and four of them (V731, L735, L738, and F742) are facing to the same surface, while F727 was not (Fig. 8A). When the hydrophobic residues were replaced by the acidic residue, Asp(D), all the mutations on the hydrophobic face, V731D, L735D, L738D, and F742D, abolished the membrane anchoring ability of the CT of RHD3 (Fig. 8B), as indicated by the lack of punctate Golgi labeling. On the other hand, the F727D mutation did not change the membrane localization of the CT of RHD3 (Supplemental Fig. S4A). This suggests that the hydrophobic residues in the hydrophobic face are important for the membrane anchoring of the CT of RHD3. Moreover, changing V731 into the other hydrophobic residues (V731A, V731L, and V731I) did not significantly diminish the membrane association of the CT of RHD3 (Fig. 8C), suggesting that the hydrophobic property of the hydrophobic face in the amphipathic helix is more important than the amino acid sequence specificity.
Figure 8.
The hydrophobic face in the amphipathic helix of RHD3 is essential for membrane anchoring. A, Amphipathic helix model. Left: The possible arrangement of the amphipathic helix of RHD3, predicted by HeliQuest. The hydrophobic, positively charged and polar uncharged residues are shown in yellow, blue, and magenta, respectively. Right, The 3D model of the amphipathic helix of RHD3 was generated by PEP-FOLD. The hydrophobic residues are shown in red and their corresponding positions in the HeliQuest prediction are circled and pointed out. B, Acidic mutations in the hydrophobic face abolish the membrane association of the CT of RHD3. mCherry fused wild type CT of RHD3, and CT mutants (CT(V731D), CT(L735D), CT(L738D), and CT(F742D)) were transiently expressed in the tobacco leaves. All mutated CTs of RHD3 were localized in the cytoplasm. Scale bar = 10 μm. C, Different hydrophobic substations of V731 do not change the membrane association of the CT of RHD3. V731 was changed into Ala (V731A), Leu (V731L), or Ile (V731I), and all mutants show a similar membrane association as the wild type CT of RHD3, as indicated by punctate Golgi localization. Scale bar = 10 μm. D, Transient expression of wild-type RHD3 and mCherry-HDEL in tobacco leaves. Scale bar = 10 μm. E, Transient expression of double CT mutant RHD3(L735D, L738D) and mCherry-HDEL in tobacco leaves. Thick and bundled ER tubules are indicated by arrows. Scale bar = 10 μm. F, Transient expression of quadruple CT mutant RHD3(F727D, V731D, L735D, L738D) and mCherry-HDEL in tobacco leaves. Thick and bundled ER tubules are indicated by arrows. Scale bar = 10 μm.
The potential biological function of the amphipathic helix was then tested in the yeast ER fusion assay. We found that RHD3 with single mutation RHD3(L735D) and RHD3(L738D) in the amphipathic helix had a reduction in the efficiency of rescuing ΔSey1p (Fig. 2, column 10 and 11, respectively), and RHD3 with double mutations RHD3(L735D, L738D) in the amphipathic helix had severe reduction in the efficiency of rescuing ΔSey1p [Fig. 2, column 12; the P value between RHD3(L735D) and RHD3(L735D, L738D) is 0.019 < 0.05 and between RHD3(L738D) and RHD3(L735D, L738D) is 0.016 < 0.05]. When transiently expressed in the tobacco leaves, the double mutant RHD3(L735D, L738D) and quadruple mutant RHD3(F727D, V731D, L735D, L738D) created thick and bundled ER tubules (Fig. 8, E and F, arrows). Furthermore, the CT deleted mutant RHD3(ΔCT), and the quadruple mutant RHD3(F727D, V731D, L735D, L738D) could not rescue the ER defects revealed in the rhd3-8 mutant (Supplemental Fig. S4B). Therefore, the membrane anchoring mediated by the amphipathic helix of RHD3 is required for the efficient ER fusion mediated by RHD3.
DISCUSSION
As a member of the Atlastin GTPase family functioning in mediating the fusion of different ER tubules in plant cells, the mechanistic basis of the action of RHD3 in the ER membrane fusion is still not well studied. Previous studies showed that RHD3 undergoes a GTP-dependent homotypic interaction, which can be enhanced by the phosphorylation on the divergent sequences of the CT of RHD3 (Chen et al., 2011; Ueda et al., 2015). Although the structure of RHD3 has not been resolved, based on the high structure similarity between RHD3 and Sey1p, a yeast member of the Atlastin GTPases, we simulated the structure of the cytosolic N terminus of RHD3. The simulated cytosolic structure of RHD3 suggests that the GDs and the first two 3HBs of two RHD3 monomers could twist together to form a dimer. In consistence with this simulation, we find that a full-length RHD3 can form homotypic interactions with its GD as well as its middle domain. When the potential polar linkage between D185 of one RHD3 monomer and R101 of another monomer in the interface of the dimer is disrupted, the interaction between a full-length RHD3 and the GD of RHD3 is weakened. Similarly, mutations in the first two 3HBs in the middle domain could also reduce the interaction between a full-length RHD3 and its middle domain. We suggest that the dimerization of RHD3 mediated by its GD and 3HBs is highly likely. Interestingly, when we expressed the GD or cytosolic N terminus of RHD3 in plant cells, the expression has a negative effect on the formation of the polygonal ER network, suggesting that the function of endogenous RHD3 is affected. This influence is possibly through the interaction between the GD or cytosolic N terminus of RHD3 and full-length, endogenous RHD3. In animal cells, it is believed that, to facilitate different ER membrane fusion, the cytosolic N terminus of Atlastin-1 in two different ER membranes first undergo a dimerization between GDs to tether different ER membrane together. This dimerization is then stabilized by 3HBs in the middle domain, after which a GTP-hydrolysis dependent conformational change occurs to fuse the ER membranes. Because the interaction between a full-length RHD3 and its GD or first two 3HBs was required for the efficient ER fusion, we think that a similar mechanism may exist for RHD3.
A major difference between RHD3 and Atlastin-1 is that the predicted helical bundle enriched middle domain of RHD3 is much longer than that of Atlastin-1 (Stefano and Brandizzi, 2014). Like RHD3, Sey1p also has a middle domain longer than Atlastin-1. It has been shown that the deletion of 3HB-3 and 3HB-4 inactivates Sey1p (Yan et al., 2015). Here we find that in RHD3, 3HB-3 and 3HB-4 play a vital role in the protein stability of RHD3. Although both 3HB-1 and 3HB-2 are required for possible dimerization of RHD3, it is interesting to note that 3HB-1 mutants RHD3(A285P) and RHD3(L355P) localized to thick bundled ER tubules, while 3HB-2 mutants RHD3(L379P) and RHD3(A434P) form punctates and aggregations on the ER tubules. This suggests that the mutations in 3HB-2 (L379P and A434P) may also influence the distribution of RHD3 on ER tubules.
RHD3 is an ER localized protein. Our results indicate that the TMs of RHD3 are important for the targeting and retention of RHD3 in the ER. The TMs not only serve as an ER membrane anchor, they are also able to interact with each other. Atlastin-1 in animal cells undergo a GTP-independent association through its TMs. This TM-mediated association is proposed to increase the density of Atlastin-1 at the site of membrane tethering (Liu et al., 2012; Yan et al., 2015), so multiple cycles of dimerization of Atlastin-1 in the different membranes can occur for the efficient ER membrane fusion. Likely, RHD3 may also undergo a TM-mediated association for the efficient ER membrane fusion in plant cells.
Although the TMs are important for the targeting and retention of RHD3 in the ER, we also found the amphipathic helix located in the CT of RHD3 has an ability for the ER and Golgi targeting. Given the fact that a large portion of RHD3(ΔTM) or the CT of RHD3 is in the cytosol, we think a portion of RHD3(ΔTM) or the CT of RHD3 is anchored to the membrane of the ER and Golgi. When the hydrophobic residues on the same hydrophobic face in the amphipathic helix are replaced by the acidic residues, all the mutations abolished the membrane attachment ability of the CT of RHD3; however, different hydrophobic residue replacement did not compromise the membrane association of the CT of RHD3. Thus, it is highly likely that the membrane anchoring of RHD3(ΔTM) or the CT of RHD3 is due to a hydrophobic interaction between the amphipathic helix and membrane lipids. The amphipathic helix of Atlastin-1 has been proposed to facilitate the ER membrane fusion by providing the driving force for outer leaflet mixing of two different membranes through interacting with and destabilizing the lipid bilayer (Liu et al., 2012). The mutations in the hydrophobic residues on the same hydrophobic face in the helix not only abolish the membrane attachment ability of the CT of RHD3, the full-length RHD3 with such mutations in the amphipathic helix also has reduced ability to rescue ΔSey1p and rhd3-8 mutants. This indicates that membrane anchoring mediated by the amphipathic helix of RHD3 also play a role in the efficient ER membrane fusion, possibly through its interaction with lipid bilayers like the amphipathic helix of Atlastin-1.
It is interesting to note that a portion of RHD3(ΔTM) or the CT of RHD3 is not only anchored to the ER membrane, but also the Golgi membrane. This is probably a reflection of the specific organization of the ER-Golgi interface in plant cells that Golgi is physically linked to the ER (Sparkes et al., 2009). However, when the CT of yeast Sey1p was expressed in plant cells, no Golgi targeting is revealed (Supplemental Fig. S5A), but replacing the amphipathic helix in the CT of yeast Sey1p with the amphipathic helix of RHD3 will target YFP to the Golgi or ER membrane (Supplemental Fig. S5B), so the Golgi targeting of RHD3(ΔTM) or the CT of RHD3 is RHD3 specific and is determined by the amphipathic helix of the CT of RHD3. However, the full-length RHD3 is ER localized (Chen et al., 2011), so the physiological relevance of this Golgi membrane preference of the CT of RHD3 is not clear.
CONCLUSIONS
Based on our systemic structure-function analysis of RHD3, we hypothesize that, to mediate different ER membrane fusion, different RHD3 molecules in different ER membranes need to undergo homotypic interaction through their GDs and middle domains. Likely, RHD3 may also undergo a TM-mediated association to increase the density of RHD3 in the same ER membrane for efficient ER membrane fusion. Meanwhile, the amphipathic helix of the C terminus of RHD3 would attach to the ER membrane to disturb the lipid bilayer to facilitate the membrane fusion.
MATERIALS AND METHODS
Molecular Cloning
RHD3 was first cloned into pCR8/GW/TOPO entry vector (Invitrogen). RHD3(ΔTM) was generated by overlap PCR. RHD3(ΔTM), GD of RHD3, RHD3(ΔTMΔCT), CT of RHD3, and CT of Sey1p were cloned into pCR8/GW/TOPO. All the point mutations were performed in pCR8/GW/TOPO by Quick Change Site-Directed Mutagenesis kit (Agilent Technologies). The RHD3 mutants were gateway cloned into pEarleyGate104 to produce YFP tagged RHD3 mutants. RY vectors were created based on pEarleyGate104 YFP-RHD3 with AQUA Cloning assay (Beyer et al., 2015). The original Basta resistance gene in pEarleyGate104 was replaced by mCherry-HDEL. To express in plants, all the constructs were transformed into Agrobacterium tumefaciens GV3101. For SUS, all the mutated RHD3 or different domains were gateway cloned into pNCW-GWRFC to generate N-terminal Cub fusion or pNX32-DEST to generate N-terminal NubG fusion. For yeast ER fusion assay, endogenous Sey1p promoter and terminator were amplified from yeast genome and cloned into a p416GPD vector (URA3) with KpnI-SacI sites. All the RHD3 mutants were cloned between Sey1p promoter and terminator with BamHI-XbaI sites. All the ER and Golgi markers were described by Zheng et al. (Zheng et al., 2005).
Structure Simulation
The N-terminal sequence (AA1-668) of RHD3 or D185N sequence was used in Swiss-model and Sey1p homo-dimer (5CA9, binding with GDPAlF4-) was used as the template for modeling. Multiple sequence alignment was analyzed by MUSCLE. Sequences were from RHD3: AA722-802, RHD3-LIKE2(RL2): AA729-834, RHD3-LIKE1(RL1): AA728-795, Sey1p: AA727-776, ATL1: AA493-558. The possible arrangement of the amphipathic helix (AA727-744) was predicted by HeliQuest. 3D model of the short peptide was generated by PEP-FOLD. All the predicted structures were analyzed by PyMOL.
Mating Based SUS
The Cub tagged RHD3 mutants and NubG tagged RHD3 mutants were transformed into haploid yeast strains THY.AP4 and THY.AP5, respectively (Obrdlik et al., 2004). After transformation, colonies were picked and inoculated in SC selection medium (-leu for AP4 and -trp for AP5) for overnight at 28°C. The THY.AP4 (Cub) and THY.AP5 (NubG) suspensions were mixed and plated on YPD plates. After 6 to 8 h at 28°C, mated cells were streaked on -LT(-Leu-Trp) selection plates and incubated at 28°C for 2 d. Diploid cells were collected and inoculated in -LT liquid medium and incubate at 28°C overnight. Yeast cells were resuspended in water and measured the OD600 value. Suspensions were diluted into OD600 = 2, 1, 0.1, and 0.01 or OD600 = 1, 0.1, 0.01, and 0.001. Then 15 μL per spot was dropped on -LT plates for mating control or -LTH (-Leu-Trp-His) plates for interaction test. Plates were incubated at 28°C for 2 to 3 d. The interaction efficiency was evaluated by counting the number of yeast colonies on -LTH plates.
Yeast ER Fusion Assay
Yeast ER fusion test was performed as previously described (Anwar et al., 2012). Different RHD3 mutants were transformed into two different Δsey1p mutant haploid cells, ACY53(ss-RFP-HDEL) and ACY54 (free GFP). Cells were grown to an OD (600 nm) of 0.1 to 0.4, mixed and concentrated in YPD medium. A 5-μL cell suspension was dropped on a 1-mm-thick SC medium pad and grown at 30°C for 40 to 60 min. Then images were taken at 20-second intervals.
Transient Expression in Nicotiana tabacum and Nicotiana benthamiana
Plants (Nicotiana tabacum or Nicotiana benthamiana) with three or four leaves were used for infiltration. Agrobacterium with different constructs was grown at 28°C overnight and resuspended in infiltration buffer (100 μM acetosyringone and 10 mm MgCl2). The final OD600 of agrobacterium was 0.01. Results were checked 2 to 3 d after infiltration.
Western Blot
Two days after infiltration of RY constructs, total proteins were extracted with 1×SDS loading buffer from Nicotiana benthamiana. Samples were boiled for 5 min. After 2 min of 13,000 rpm centrifuge, the supernatants were loaded on 10% SDS-PAGE gel. SDS-PAGE was performed on a Protean III apparatus (Bio-Rad), and separated proteins were transferred onto a PVDF membrane. Western blotting was carried out with a rabbit anti-GFP antibody (Abcam, ab32146) at 1:5,000 dilution or anti-RFP (Abcam, ab34771) at 1:2,000 dilution. The secondary anti-Rabbit IgG-peroxidase (Sigma-Aldrich) was used at 1:5,000 dilution. Signals were detected using Invitrogen Novex ECL (HRP Chemiluminescent Substrate Reagent Kit) according to the manufacturer’s recommendations.
Confocal Microscopy
The infiltration results were observed with a Leica SP8 point-scanning confocal system on a Leica DMI6000B inverted microscope equipped with spectral fluorescent light detectors (three PMT, one HyD high sensitivity detector). A 63x/1.4 oil objective was used for all the imaging. The 488-nm laser was used to excite GFP or YFP, and the 552-nm laser was used to excite RFP or mCherry. Two channels were excited sequentially. Emission filter was set as 490 to 560 nm for GFP or YFP, and 580 to 660 nm for RFP or mCherry. For post image editing and fluorescence signal quantification, Fiji (Fiji is just Image J) was used (Schindelin et al., 2012).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession number: AtRHD3 (At3g13870).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A, The overall structure of full-length ATL1, caSey1p, and RHD3.
Supplemental Figure S2. Transient expression of RHD3 with mutations in the middle domain in tobacco leaves.
Supplemental Figure S3. Transient expression of different RY constructs in tobacco leaves.
Supplemental Figure S4. A, Transient expression of mutated CT of RHD3 (CT(F727D)) in tobacco leaves.
Supplemental Figure S5. Expression of the CT of Sey1p (Sey1pCT) in plant cells.
Acknowledgments
We thank Sylvie Lalonde (Carnegie Institution, Stanford, CA) for pNX32 and pXN22 mbSUS Gateway vectors; David Bird (University of Manitoba, Winnipeg, Canada) for pNCW-GWRFC.1; William A. Prinz (National Institute of Diabetes and Digestive and Kidney Diseases, NIH, Bethesda, MD) for providing the yeast cells (ACY53 and ACY54 strains) for Yeast ER fusion test; and the Cell Imaging and Analysis Network (CIAN) in McGill Biology department for microscopy imaging support.
Footnotes
This research was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to H.Z. J.S. was supported by a scholarship from the Chinese Scholarship Council.
References
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA (2012) The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol 197: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer HM, Gonschorek P, Samodelov SL, Meier M, Weber W, Zurbriggen MD (2015) AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS One 10: e0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J (2011) Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA 108: 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JP, Claridge JK, Smith PG, Schnell JR (2015) A conserved amphipathic helix is required for membrane tubule formation by Yop1p. Proc Natl Acad Sci USA 112: E639–E648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Dzimitrowicz N, Kriechbaumer V, Brooks R, Botchway SW, Brady JP, Hawes C, Dixon AM, Schnell JR, Fricker MD, et al. (2016) A C-terminal amphipathic helix is necessary for the in vivo tubule-shaping function of a plant reticulon. Proc Natl Acad Sci USA 113: 10902–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Stefano G, Brandizzi F, Zheng H (2011) Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J Cell Sci 124: 2241–2252 [DOI] [PubMed] [Google Scholar]
- Grefen C, Lalonde S, Obrdlik P (2007). Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr Protoc Neurosci 41: 5.27.1–5.27.41 [DOI] [PubMed] [Google Scholar]
- Hu J, Rapoport TA (2016) Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin Cell Dev Biol 60: 105–111 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA (2008) Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C (2009) A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Bian X, Romano FB, Shemesh T, Rapoport TA, Hu J (2015) Cis and trans interactions between atlastin molecules during membrane fusion. Proc Natl Acad Sci USA 112: E1851–E1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J (2012) Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc Natl Acad Sci USA 109: E2146–E2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460: 978–983 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Hawes C, Frigerio L (2011) FrontiERs: movers and shapers of the higher plant cortical endoplasmic reticulum. Curr Opin Plant Biol 14: 658–665 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Tolley N, Aller I, Svozil J, Osterrieder A, Botchway S, Mueller C, Frigerio L, Hawes C (2010) Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Ketelaar T, de Ruijter NC, Hawes C (2009) Grab a Golgi: laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10: 567–571 [DOI] [PubMed] [Google Scholar]
- Stefano G, Brandizzi F (2014) Unique and conserved features of the plant ER-shaping GTPase RHD3. Cell Logist 4: e28217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G, Renna L, Moss T, McNew JA, Brandizzi F (2012) In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J 69: 957–966 [DOI] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kuwata K, Kutsuna N, Mano S, Shimada T, Tamura K, Stefano G, Fukao Y, Brandizzi F, et al. (2015) Phosphorylation of the C-terminus of RHD3 has a critical role in homotypic ER membrane fusion in Arabidopsis. Plant Physiol 170: 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang Y, Yang J, Ma C, Zhang Y, Ge T, Qi Z, Kang Y (2015) Arabidopsis ROOT HAIR DEFECTIVE3 is involved in nitrogen starvation-induced anthocyanin accumulation. J Integr Plant Biol 57: 708–721 [DOI] [PubMed] [Google Scholar]
- Yamada-Okabe T, Yamada-Okabe H (2002) Characterization of the CaNAG3, CaNAG4, and CaNAG6 genes of the pathogenic fungus Candida albicans: possible involvement of these genes in the susceptibilities of cytotoxic agents. FEMS Microbiol Lett 212: 15–21 [DOI] [PubMed] [Google Scholar]
- Yan L, Sun S, Wang W, Shi J, Hu X, Wang S, Su D, Rao Z, Hu J, Lou Z (2015) Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion. J Cell Biol 210: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Karnik R, Wang Y, Wallmeroth N, Blatt MR, Grefen C (2015) The Arabidopsis R-SNARE VAMP721 Interacts with KAT1 and KC1 K+ Channels to Moderate K+ Current at the Plasma Membrane. Plant Cell 27: 1697–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, Hu J (2013) ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol 163: 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L, et al. (2001) Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 29: 326–331 [DOI] [PubMed] [Google Scholar]
- Zheng H, Camacho L, Wee E, Batoko H, Legen J, Leaver CJ, Malhó R, Hussey PJ, Moore I (2005) A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17: 2020–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chen J (2011) Emerging aspects of ER organization in root hair tip growth: lessons from RHD3 and Atlastin. Plant Signal Behav 6: 1710–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I (2004) A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J 37: 398–414 [DOI] [PubMed] [Google Scholar]