Restoring vessel integrity without lowering general ferulic acid levels overcomes dwarfism in ccr1 mutants and reveals monolignol transport from vessels to xylary fibers.
Abstract
Lignocellulosic biomass is recalcitrant toward deconstruction into simple sugars due to the presence of lignin. To render lignocellulosic biomass a suitable feedstock for the bio-based economy, plants can be engineered to have decreased amounts of lignin. However, engineered plants with the lowest amounts of lignin exhibit collapsed vessels and yield penalties. Previous efforts were not able to fully overcome this phenotype without settling in sugar yield upon saccharification. Here, we reintroduced CINNAMOYL-COENZYME A REDUCTASE1 (CCR1) expression specifically in the protoxylem and metaxylem vessel cells of Arabidopsis (Arabidopsis thaliana) ccr1 mutants. The resulting ccr1 ProSNBE:CCR1 lines had overcome the vascular collapse and had a total stem biomass yield that was increased up to 59% as compared with the wild type. Raman analysis showed that monolignols synthesized in the vessels also contribute to the lignification of neighboring xylary fibers. The cell wall composition and metabolome of ccr1 ProSNBE:CCR1 still exhibited many similarities to those of ccr1 mutants, regardless of their yield increase. In contrast to a recent report, the yield penalty of ccr1 mutants was not caused by ferulic acid accumulation but was (largely) the consequence of collapsed vessels. Finally, ccr1 ProSNBE:CCR1 plants had a 4-fold increase in total sugar yield when compared with wild-type plants.
Lignocellulose, being the most abundant biomass on earth, has great potential as a renewable feedstock for the production of carbon-neutral chemicals and polymers in the bio-based economy (Vanholme et al., 2013a; Isikgor and Becer, 2015). Lignocellulosic biomass is composed mainly of secondary thickened cell walls, which primarily consist of cellulose and hemicellulose polysaccharides, impregnated with lignins (Cosgrove, 2005). The latter are aromatic heteropolymers, composed mainly of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, derived from the monolignols p-coumaryl, coniferyl, and sinapyl alcohol, respectively (Boerjan et al., 2003; Vanholme et al., 2010). Lignin provides the plant with the necessary strength and hydrophobicity in order to stand upright and transport water through the vascular system (Weng and Chapple, 2010). In addition, it acts as a physical barrier against pathogens and herbivores (Weng and Chapple, 2010; Miedes et al., 2014). Unfortunately, lignin also is the major limiting factor in the processing of lignocellulosic biomass for downstream applications (Chen and Dixon, 2007; Van Acker et al., 2013; Vanholme et al., 2013b). This aromatic polymer contributes to the recalcitrance of the plant cell wall toward deconstruction by hindering the enzymatic hydrolysis of cell wall polysaccharides into simple sugars (i.e. saccharification; Weng et al., 2008; Pauly and Keegstra, 2010). Therefore, several efforts to reduce cell wall recalcitrance have focused on modifying the lignin content and/or composition (Goujon et al., 2003; Chen and Dixon, 2007; Leplé et al., 2007; Shadle et al., 2007; Jackson et al., 2008; Day et al., 2009; Voelker et al., 2010; Eudes et al., 2012, 2015, 2016; Mansfield et al., 2012; Wilkerson et al., 2014; Mottiar et al., 2016).
Lignin-modified plants that show the highest improvement in saccharification efficiency typically suffer from undesired phenotypes, including biomass and seed yield penalties, a phenomenon also called lignin modification-induced dwarfism (Chen and Dixon, 2007; Shadle et al., 2007; Bonawitz and Chapple, 2013; Van Acker et al., 2013, 2014; Vanholme et al., 2013b). Although the molecular mechanisms for lignin modification-induced dwarfism are still poorly understood, several hypotheses have been postulated to explain this phenomenon. First, the dwarfed phenotype of lignin-modified plants could be caused by the loss of vessel cell wall integrity, which, in turn, results in the inability of the plant to efficiently transport nutrients and water from the roots to the aerial parts. As a consequence, a collapse of the weakened vessel cells occurs under the negative pressure generated by transpiration, which is called the irregular xylem (irx) phenotype (Bonawitz and Chapple, 2013). For example, irregular vessels have been reported for different plant species (Arabidopsis [Arabidopsis thaliana], poplar [Populus tremula × Populus alba], and tobacco [Nicotiana tabacum]) perturbed in the expression of the lignin biosynthesis genes PHENYLALANINE AMMONIA-LYASE (PAL; Huang et al., 2010), CINNAMATE 4-HYDROXYLASE (C4H; Stout and Chapple, 2004), 4-COUMARATE:COENZYME A LIGASE (4CL; Voelker et al., 2010), HYDROXYCINNAMOYL-COENZYME A SHIKIMATE/QUINATE HYDROXYCINNAMOYL TRANSFERASE (HCT; Besseau et al., 2007), p-COUMARATE 3-HYDROXYLASE (C3H; Franke et al., 2002), CAFFEOYL SHIKIMATE ESTERASE (CSE; Vanholme et al., 2013b), CAFFEOYL-COENZYME A O-METHYLTRANSFERASE (CCoAOMT; Zhong et al., 1998), and CINNAMOYL-COENZYME A REDUCTASE (CCR; Piquemal et al., 1998; Jones et al., 2001). Furthermore, a series of dwarfed cellulose and hemicellulose biosynthesis mutants also exhibit the irx phenotype (Turner and Somerville, 1997; Taylor et al., 1999; Brown et al., 2005; Persson et al., 2007; Li et al., 2012).
A second (or additional) cause for the observed yield penalties could be the accumulation of pathway intermediates (or derivatives thereof) that could be toxic for the plant. For example, ccr1 mutants show strongly increased levels of ferulic acid, which was described to drastically decrease the levels of reactive oxygen species (ROS; Xue et al., 2015). Because high levels of ROS are required for the exit from cell proliferation, the defective cell cycle and dwarfed growth of ccr1 mutants have been ascribed to the high levels of ferulic acid leading to reduced levels of ROS (Xue et al., 2015).
A third hypothesis explaining the yield penalty of lignin-modified plants could be the depletion of other phenylpropanoid-related metabolites that are essential for normal plant development (Bonawitz and Chapple, 2013).
Fourth, the triggering of an active cell wall integrity pathway, which allows plants to sense cell wall abnormalities, could result in transcriptional responses that, in turn, cause growth perturbations (Vanholme et al., 2012; Bonawitz and Chapple, 2013). Such transcriptional control mechanisms of the phenylpropanoid metabolism have been shown to be involved in the response to lignin pathway perturbations (Bonawitz et al., 2014; Anderson et al., 2015). More specifically, mutation of genes encoding subunits of the transcriptional coregulatory complex Mediator (Med5A and Med5B) resulted in a (partial) reversion of the growth penalty, the reduced lignin abundance, and the collapsed vessels of c3h1 mutants (Bonawitz et al., 2014).
Efforts have been made to overcome the dwarfed phenotype of lignin mutants while maintaining the beneficial high sugar yield upon saccharification. Some of these attempts focused on the recovery of vessel cell integrity in lignin mutants. In these studies, VASCULAR-RELATED NAC DOMAIN6 (VND6) and VND7 promoter sequences were used to drive the expression of a lignin biosynthesis gene in the respective lignin mutant, thereby aiming at reintroducing lignin biosynthesis specifically in vessel cells. The expression of VND6 has been shown to be restricted to the metaxylem vessels, whereas VND7 had the highest expression level in protoxylem vessels (Kubo et al., 2005; Zhong et al., 2008; Vargas et al., 2016). An example of this strategy includes the partial restoration of the dwarfed phenotype of c4h knockdown mutants by the reintroduction of the C4H gene under the control of a 2,757-bp VND6 promoter sequence, resulting in plants with normal, open vessels (Yang et al., 2013). However, c4h ProVND6:C4H plants showed a reoccurrence of lignin in the interfascicular fiber region, indicating that the complementation strategy used was not highly specific for vessel cells. Moreover, these lines had lower sugar yields per mg of cell wall when compared with c4h mutants. A similar strategy has been used in Arabidopsis cse-2 mutants (Vargas et al., 2016). Here, introducing CSE under the control of a 1,004-bp VND6 or 1,997-bp VND7 promoter sequence partially restored their growth and vascular integrity. However, the specific reoccurrence of lignin in the xylem, and not in the interfascicular fibers, of cse-2 ProVND6:CSE and cse-2 ProVND7:CSE lines resulted in cellulose-to-glucose conversion efficiencies equal to those of cse-2 mutants. Similar results were obtained for a vessel-specific complementation approach of xylan mutants, where the use of 2,757-bp VND6 and 2,009-bp VND7 promoter sequences only partially recovered the irx and dwarfed phenotype of the respective xylan biosynthesis mutants (Petersen et al., 2012). Taken together, these data hint that the VND6 and VND7 promoters are not strong and/or not specific enough to fully restore the yield penalty and at the same time keep the high cellulose-to-glucose conversion efficiency of cell wall biosynthesis mutants.
Here, we completely overcame the total plant biomass penalty of severely dwarfed ccr1 mutants without lowering general ferulic acid levels but while fully maintaining its high saccharification potential. To achieve this, the artificial SECONDARY WALL NAC BINDING ELEMENT of the XYLEM CYSTEINE PROTEASE1 promoter (ProSNBE) was chosen to drive the expression of the CCR1 gene in a ccr1 mutant background (McCarthy et al., 2011). ProSNBE is bound by both VND6 and VND7 and confers expression in both protoxylem and metaxylem vessels (Zhong et al., 2010). The resulting ccr1 ProSNBE:CCR1 lines showed a full recovery in vascular integrity, a strong increase in total stem biomass as compared with wild-type plants, and provided evidence for monolignol transport from vessel cells to the cell wall of xylary fibers.
RESULTS
ProSNBE Confers Vessel-Specific Expression in Both the Protoxylem and Metaxylem of Arabidopsis
To fully restore the integrity of the vessels and the growth of ccr1 mutants, a vessel-specific promoter was required that conferred higher expression levels and/or a broader expression pattern than the previously tested VND6 and VND7 promoters (Petersen et al., 2012; Yang et al., 2013; Vargas et al., 2016). The artificial ProSNBE has been shown to direct the expression of reporter genes to xylem vessel cells of the Arabidopsis inflorescence stem (McCarthy et al., 2011). Furthermore, ProSNBE should confer expression in both protoxylem and metaxylem vessels, because it is bound by both VND6 and VND7 (Zhong et al., 2010). The ProSNBE used here is composed of three tandem repeats of the cis-regulatory SNBE1 originating from the Arabidopsis XYLEM CYSTEINE PROTEASE1 (XCP1) promoter, fused to the cauliflower mosaic virus (CaMV) 35S minimal promoter (Fig. 1). The XCP1 gene has been shown to be expressed specifically in Arabidopsis vessel cells, where the corresponding protein is involved in vessel autolysis during xylogenesis (Ohashi-Ito et al., 2010; Zhong et al., 2010; McCarthy et al., 2011).
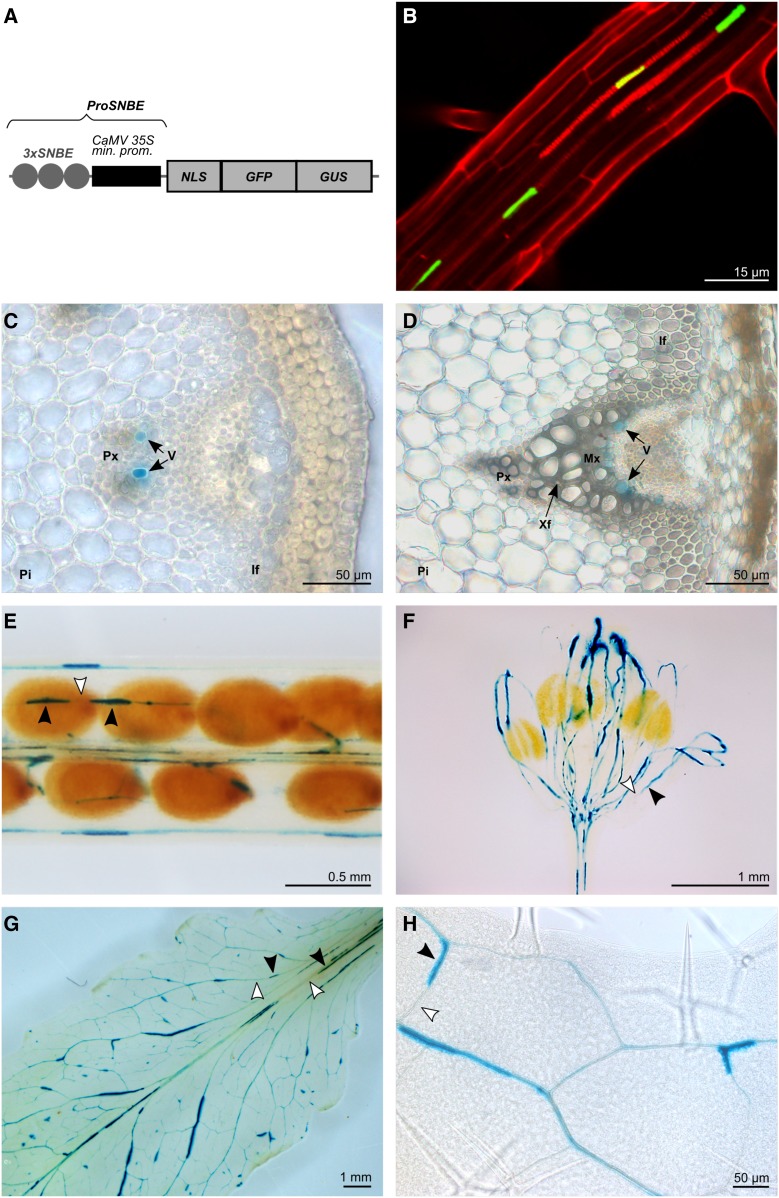
Figure 1.
Expression pattern conferred by ProSNBE. A, Diagram of the ProSNBE:GFP:GUS construct: GFP and GUS reporter genes are driven by three copies of the XCP1-SNBE sequence fused to the CaMV minimal 35S promoter (ProSNBE). NLS, Nuclear localization signal. B, Expression analysis in roots showing GFP in xylary vessels. Propidium iodide was used to counterstain the cell wall. C, Cross section of an elongating internode of the primary inflorescence stem showing GUS staining in developing vessels of the protoxylem. D, Cross section of a nonelongating internode of the primary inflorescence stem showing GUS staining in developing vessels of the metaxylem but not in xylary or interfascicular fibers. E to H, GUS expression analysis in siliques (E), flowers (F), and rosette leaves (G and H), showing reporter gene expression in the vasculature. Black arrowheads indicate cells with GUS staining, and white arrowheads indicate cells lacking GUS staining. For B, transgenic ProSNBE:GFP:GUS seedlings were grown for 20 d in a long-day photoperiod. For C to H, transgenic ProSNBE:GFP:GUS plants were grown for 6 weeks in a short-day photoperiod followed by 5 weeks in a long-day photoperiod. If, Interfascicular fibers; Mx, metaxylem; Pi, pith; Px, protoxylem; V, vessel; Xf, xylary fiber.
To investigate whether ProSNBE indeed specifically directs gene expression in the vessels throughout the xylem (including protoxylem and metaxylem), we fused ProSNBE to the GFP and GUS reporter genes and studied the expression pattern in various organs of transformed Arabidopsis plants. In roots, GFP expression was observed in the xylem vessel cells (Fig. 1). In inflorescence stems, GUS staining was detected in the xylem, but it was lacking in interfascicular fibers or pith cells (Fig. 1). Detailed examination of ProSNBE:GFP:GUS lines revealed GUS activity in developing vessels of both protoxylem and metaxylem (Fig. 1; Supplemental Fig. S1). In addition, GUS activity was found in the vasculature of siliques, flowers, and rosette leaves (Fig. 1). As described for ProXCP1:GUS plants, the GUS activity of ProSNBE:GFP:GUS plants appeared to be discontinuous throughout the vasculature, with cells lacking GUS activity alternating with those showing GUS activity (Fig. 1; Funk et al., 2002). This discontinuous staining pattern could be a reflection of the degradation of the GUS protein at the vacuole or protoplast degeneration stage occurring during vessel maturation. As a result, cells that passed this stage will lack the GUS signal. Because ProSNBE was found to restrict expression to both the protoxylem and metaxylem vessel cells, this promoter was used further for the envisioned complementation strategy.
The Reintroduction of CCR1 Expression under the Control of ProSNBE Restores the Dwarfed Phenotype of ccr1 Mutants
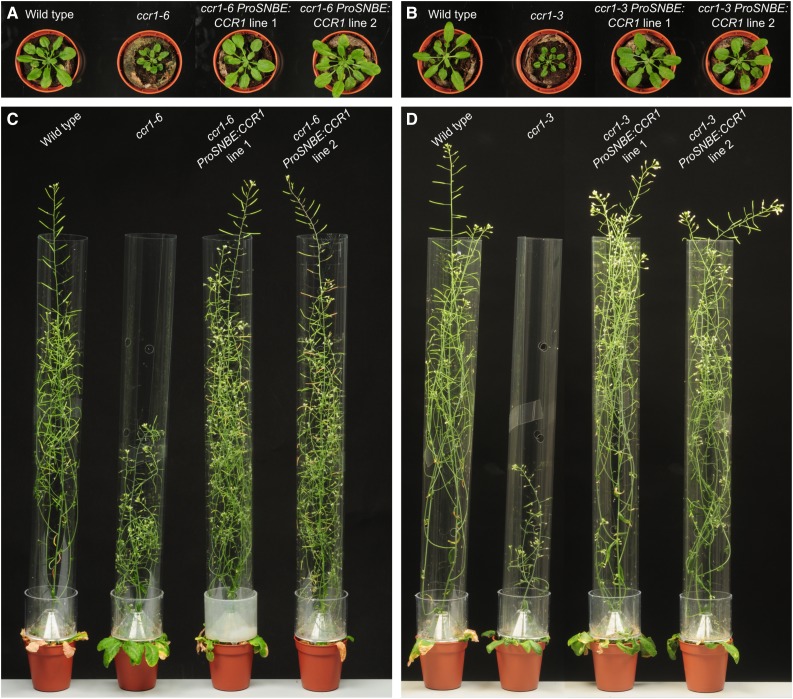
To restrict lignin biosynthesis to the vessel cells, ProSNBE was used to drive expression of the CCR1 gene in both ccr1-3 and ccr1-6 mutant backgrounds. Two independent, homozygous and single-locus ccr1-3 and ccr1-6 lines harboring the ProSNBE:CCR1 construct were selected and used for further analyses. To first evaluate whether the ProSNBE:CCR1 constructs successfully restored plant growth, the ccr1 ProSNBE:CCR1 lines were grown alongside their respective ccr1 background and the wild type under short-day conditions for 6 weeks, after which they were moved to long-day conditions. These growth conditions allowed the development of large rosettes and tall inflorescence stems to maximize secondary cell wall thickening (Vanholme et al., 2012). Whereas the ccr1 rosettes were smaller when compared with the wild type, the rosette size was fully recovered in ccr1 ProSNBE:CCR1 plants (Fig. 2). The final height of the primary inflorescence stem of ccr1 ProSNBE:CCR1 lines was equal to that of the wild type and approximately 2-fold higher than that of their respective ccr1 mutants (Fig. 2; Table I). The final dry weight of the primary (main) inflorescence stem (devoid of siliques and leaves) of both ccr1-6 ProSNBE:CCR1 lines and ccr1-3 ProSNBE:CCR1 line 2 was not significantly different from that of the wild type, whereas all four lines were significantly heavier (78%–124%) than their respective ccr1 background (Table I). The final dry weight of the primary inflorescence stem of ccr1-3 ProSNBE:CCR1 line 1 was even increased (by 19%) when compared with that of the wild type (Table I).
Figure 2.
Phenotype of ccr1 ProSNBE:CCR1 lines. Wild-type, ccr1, and ccr1 ProSNBE:CCR1 plants are shown after cultivation for 6 weeks in a short-day photoperiod followed by 1.5 weeks (A and B) or 5 weeks (C and D) in a long-day photoperiod.
Table I. Biomass measurements of ccr1 ProSNBE:CCR1 lines.
Measurements were performed on fully senesced plants. For the stem measurements, the leaves, siliques, and seeds were removed. The data represent average values ± sd and the number of repeats are indicated by asterisks: *, 20 repeats for the wild type and the ccr1 ProSNBE:CCR1 lines and 40 repeats for the ccr1 mutants; **, 12 repeats for the wild type and the ccr1 ProSNBE:CCR1 lines and 24 repeats for the ccr1 mutants; and ***, six repeats for the wild type and the ccr1 ProSNBE:CCR1 lines and 12 repeats for the ccr1 mutants. Note that the set containing ccr1-3, ccr1-3 ProSNBE:CCR1, and wild-type plants and the set containing ccr1-6, ccr1-6 ProSNBE:CCR1, and wild-type plants were grown in two independent experiments. Different letters represent significant differences at the 0.05 significance level (Dunnett-Hsu adjusted Student’s t test). Total plant biomass = biomass of the aerial part of the plant, just above the rosette.
| Primary Inflorescence Stem |
Secondary Inflorescence Stems |
Seeds |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Line | Height* | Mass* | No. Originating from Rosette** | No. Originating from Primary Inflorescence Stem** | Total** | Mass** | No.*** | Mass per Seed*** | Total Mass* | Total Stem Biomass** | Total Plant Biomass* |
| cm | mg | mg | µg | mg | mg | mg | |||||
| Wild type | 49.9 ± 4.9 a | 43.4 ± 10.5 a | 2.6 ± 1.0 a | 6.2 ± 0.9 a | 8.8 ± 1.2 a | 213 ± 30 a | 9,343 ± 1,002 a | 27.3 ± 1.5 a | 266 ± 48 a | 262 ± 41 a | 765 ± 124 a |
| ccr1-3 | 29.3 ± 3.9 b | 23.0 ± 5.4 b | 3.1 ± 1.2 a | 4.8 ± 1.1 b | 7.9 ± 1.8 a | 108 ± 26 b | 898 ± 431 b | 34.8 ± 1.8 b | 29 ± 15 b | 143 ± 38 b | 288 ± 92 b |
| ccr1-3 ProSNBE:CCR1 line 1 | 51.3 ± 4.6 a | 51.5 ± 6.0 c | 5.7 ± 1.0 b | 6.5 ± 1.1 a | 12.1 ± 1.7 b | 373 ± 67 c | 5,110 ± 944 c | 31.5 ± 1.0 c | 157 ± 58 c | 416 ± 70 c | 844 ± 113 c |
| ccr1-3 ProSNBE:CCR1 line 2 | 51.0 ± 3.8 a | 47.1 ± 7.6 a,c | 5.3 ± 1.8 b | 5.7 ± 0.8 a | 11.0 ± 2.0 b | 318 ± 75 d | 4,409 ± 1,221 c | 30.8 ± 1.7 c | 134 ± 46 c | 373 ± 91 c | 780 ± 79 a,c |
| Wild type | 48.2 ± 3.1 a | 44.0 ± 10.0 a | 1.6 ± 1.1 a | 6.1 ± 0.9 a,b | 7.7 ± 1.4 a | 116 ± 39 a | 8,373 ± 2,390 a | 26.8 ± 2.5 a | 189 ± 71 a | 155 ± 46 a | 522 ± 154 a |
| ccr1-6 | 24.9 ± 2.5 b | 24.8 ± 4.6 b | 4.4 ± 1.1 b | 5.4 ± 0.9 a | 9.8 ± 1.3 b | 96 ± 27a | 1,513 ± 469 b | 33.6 ± 2.2 b | 45 ± 24 b | 120 ± 30 b | 265 ± 73 b |
| ccr1-6 ProSNBE:CCR1 line 1 | 50.2 ± 4.2 a | 48.2 ± 9.6 a | 4.5 ± 1.7 b | 6.5 ± 0.9 b | 11.0 ± 1.4 c | 187 ± 50 b | 4,023 ± 892 c | 32.8 ± 1.3 b | 141 ± 32 c | 229 ± 46 c | 639 ± 113 c |
| ccr1-6 ProSNBE:CCR1 line 2 | 48.7 ± 3.3 a | 44.1 ± 9.8 a | 4.1 ± 1.9 b | 6.6 ± 1.2 b | 10.6 ± 1.6 b,c | 176 ± 38 b | 3,606 ± 1,062 c | 32.2 ± 3.1 b | 123 ± 38 c | 214 ± 40 c | 590 ± 192 a,c |
Although the ccr1 ProSNBE:CCR1 plants were recovered in rosette and primary inflorescence stem biomass, some phenotypic differences from the wild type were noted. First, it was observed that the ccr1 ProSNBE:CCR1 plants had more secondary inflorescence stems in comparison with the wild type (Fig. 2). Hence, we determined the weight and number of secondary inflorescence stems originating (1) from the rosette and (2) directly from the primary inflorescence stem. The ccr1 ProSNBE:CCR1 plants had more secondary inflorescence stems originating from the rosette and an increase in secondary inflorescence stem biomass of 49% to 75% in comparison with the wild type (Table I). The increase in the number of secondary inflorescences was also observed in ccr1-6 mutants but not in the ccr1-3 mutants (Table I). In comparison with that of the wild type, ccr1-3 and ccr1-6 total (primary and secondary) stem biomass was reduced by 45% and 22%, respectively, whereas the ccr1 ProSNBE:CCR1 plants had an increase of 38% to 59% in total stem biomass (Table I).
A second phenotypic difference between ccr1 ProSNBE:CCR1 lines and the wild type was a perturbation in seed development in ccr1 ProSNBE:CCR1 lines. Confirming previous observations (Mir Derikvand et al., 2008), ccr1-3 and ccr1-6 mutants had a lower number of seeds when compared with the wild type (−90% and −82%, respectively; Table I). The total seed mass was reduced by 89% and 76%, respectively, whereas the individual seeds were heavier (+27% and +25%, respectively) when compared with the wild type (Table I). Although less severe than their respective ccr1 backgrounds, the ccr1-3 ProSNBE:CCR1 and ccr1-6 ProSNBE:CCR1 lines still had a lower number of seeds (−49% and −54%, respectively) when compared with the wild type (Table I). The total seed biomass in these lines was decreased by 45% and 30%, respectively, and the seeds were slightly heavier (+14% and +21%, respectively) when compared with the wild type (Table I). The total plant biomass (aerial part of the plant, without the rosette) of the ccr1 ProSNBE:CCR1 lines was equal or higher when compared with the wild type, whereas that of ccr1 was reduced significantly to more than half that of the wild type (Table I). Because the ccr1-3 ProSNBE:CCR1 lines and ccr1-6 ProSNBE:CCR1 lines were phenotypically similar (Table I; Fig. 2), we decided to focus further efforts mainly on the analysis of the ccr1-6 ProSNBE:CCR1 lines.
We hypothesized that the increase in the number of secondary inflorescence stems, and hence the total stem biomass, could have been a secondary consequence of the impaired seed development in the ccr1 ProSNBE:CCR1 lines. To test this hypothesis, immature siliques were systematically removed throughout development, with a frequency of three times per week. As has been described before, the removal of siliques resulted in delayed senescence and the outgrowth of more secondary inflorescences for all lines examined compared with the control (in which all lines were grown without the removal of siliques; Supplemental Table S1; Hensel et al., 1994; Wuest et al., 2016). The ccr1-6 ProSNBE:CCR1 lines were now equal to the wild type in both the number of secondary inflorescences (originating from both the rosette and the main stem) and the total stem biomass (Table II). These results suggest a role for seed development signals in the increase of lignocellulosic biomass in the ccr1 ProSNBE:CCR1 lines.
Table II. Stem biomass measurements of ccr1-6 ProSNBE:CCR1 lines of which developing siliques were repeatedly removed during plant growth.
Measurements were performed on fully senesced plants grown without (control) or with the removal of siliques. The data represent average values ± sd of 12 repeats for the wild type and the ccr1-6 ProSNBE:CCR1 lines and 18 repeats for the ccr1-6 mutants. Statistical analysis focused on the differences between the lines within a treatment (control or with siliques removed). Different letters represent significant differences within a treatment at the 0.05 significance level (Dunnett-Hsu adjusted Student’s t test).
| Control |
With Siliques Removed |
|||||||
|---|---|---|---|---|---|---|---|---|
| Secondary Inflorescence Stems |
Secondary Inflorescence Stems |
|||||||
| Line | No. Originating from Rosette | No. Originating from Primary Inflorescence Stem | Total | Total Stem Biomass | No. Originating from Rosette | No. Originating from Primary Inflorescence Stem | Total | Total Stem Biomass |
| mg | mg | |||||||
| Wild type | 2.3 ± 1.0 a | 6.1 ± 1.2 a,b | 8.4 ± 0.5 a | 192.1 ± 47.7 a | 5.4 ± 1.5 a,b | 7.8 ± 1.1 a | 13.3 ± 1.4 a,b | 480.9 ± 111.7 a |
| ccr1-6 | 3.7 ± 0.6 b | 5.1 ± 0.8 a | 8.8 ± 1.1 a | 122.3 ± 45.3 a | 4.5 ± 1.6 a | 7.2 ± 1.6 a | 11.7 ± 1.9 a | 190.3 ± 62.0 b |
| ccr1-6 ProSNBE:CCR1 line 1 | 4.8 ± 1.4 b | 6.6 ± 1.1 b | 11.4 ± 2.1 b | 335.6 ± 102.8 b | 6.0 ± 1.7 b | 7.9 ± 2.0 a | 13.9 ± 2.6 b | 535.1 ± 103.3 a |
| ccr1-6 ProSNBE:CCR1 line 2 | 4.5 ± 1.3 b | 6.4 ± 0.7 b | 10.9 ± 1.6 b | 350.4 ± 74.5 b | 5.8 ± 1.3 a,b | 8.4 ± 1.0 a | 14.2 ± 1.5 b | 479.6 ± 58.7 a |
ProSNBE:CCR1 Reinforces the Vascular System and Partially Restores Lignin Deposition in the Xylem of ccr1 Mutants
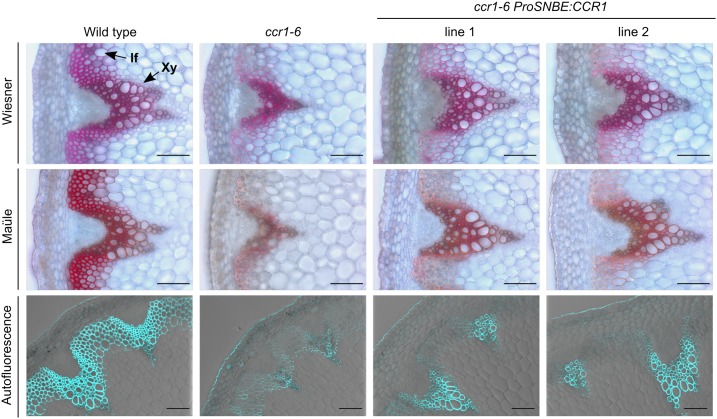
To examine whether the vessel-specific expression of CCR1 in ccr1 leads to vessel-specific lignification and restoration of vessel integrity, the lignin of fully grown wild-type, ccr1, and ccr1 ProSNBE:CCR1 plants was visualized with Wiesner and Mäule staining and via autofluorescence (Fig. 3; Supplemental Fig. S2). In the wild type, the xylem tissue, which contains large, open vessels, and the interfascicular fibers were heavily lignified. In accordance with previous reports (Jones et al., 2001; Goujon et al., 2003; Mir Derikvand et al., 2008), ccr1 mutants showed an overall reduction in lignin deposition and developed irregularly shaped and collapsed vessels. The xylem tissue of the ccr1 ProSNBE:CCR1 lines showed a strong coloration and contained large open vessels, similar to those of the wild type. Remarkably, both vessels and xylary fibers of ccr1 ProSNBE:CCR1 lines appeared to be lignified. On the other hand, the interfascicular fibers of the ccr1 ProSNBE:CCR1 lines showed reduced lignin deposition similar to ccr1 mutants.
Figure 3.
Lignin deposition in inflorescence stems of ccr1-6 ProSNBE:CCR1 lines. Transverse stem sections are shown for the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 lines. Wiesner and Mäule staining and lignin autofluorescence are shown. If, Interfascicular fibers; Xy, xylem. Bars = 100 µm.
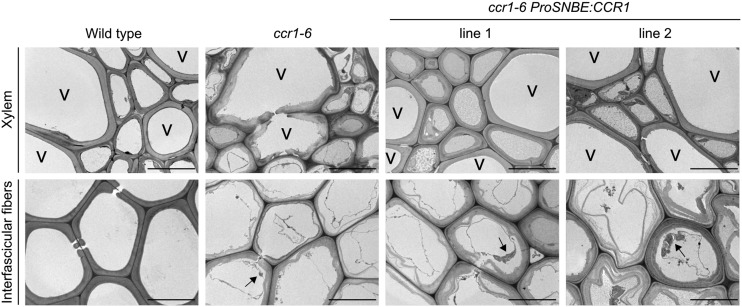
Because the previously described lignin visualization methods do not allow visualization of the anatomical features of the secondary cell walls, transmission electron microscopy was performed on the different lines (Fig. 4). The secondary cell walls of the vessels and (xylary and interfascicular) fibers of wild-type stems were organized, compact, and displayed good cohesion. By contrast, the ccr1-6 mutant exhibited dramatic disorganization and loosening of the secondary walls in both vessels and fibers. The xylem tissue of ccr1-6 ProSNBE:CCR1 lines appeared similar to that of the wild type, indicated by its proper organization and internal cohesion of the walls in both the vessels and xylary fibers. However, the phenotype of the interfascicular fibers of ccr1-6 ProSNBE:CCR1 appeared to be similar to that of ccr1-6, indicated by the loosening of the secondary cell wall. Additionally, the interfascicular fiber cells of the wild type were devoid of cellular content, whereas those of the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines still contained cellular contents. These results indicated that ccr1-6 and ccr1-6 ProSNBE:CCR1 did not complete programmed cell death at the time of harvest, despite the fact that all the lines were harvested at the same age.
Figure 4.
Morphology of cell walls of wild-type, ccr1-6, and ccr1-6 ProSNBE:CCR1 stems. Transmission electron microscopy demonstrates the ultrastructure of the interfascicular fibers and xylem regions. Arrows indicate residual cellular content. V, Xylary vessel. Bars = 10 µm.
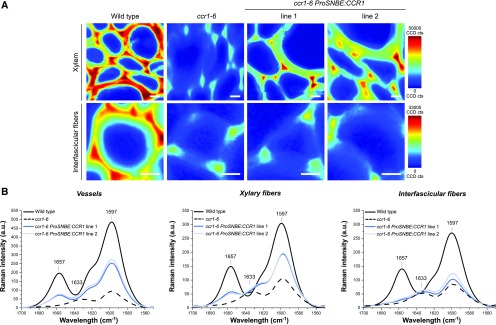
Based on the lignin and cell wall visualization methods, the xylem of the wild type and ccr1 ProSNBE:CCR1 lines appeared to be similar (Figs. 3 and 4; Supplemental Fig. S2). Additionally, both the vessels and xylary fibers of ccr1 ProSNBE:CCR1 seemed to be lignified (Fig. 3; Supplemental Fig. S2). To investigate lignin amount and composition in the different cell types, Raman microscopy analysis was performed (Fig. 5; Supplemental Fig. S3). First, the distribution of components having aromatic ring structures (i.e. building blocks of the lignin polymer) was visualized by integrating the intensity of each spectrum in the range of 1,650 to 1,550 cm−1 into 2D mappings. In the xylem, the intensity of the aromatic ring stretching was high in the wild type, intermediate in the ccr1-6 ProSNBE:CCR1 lines, and low in ccr1-6. In the interfascicular region, the mappings of ccr1-6 and ccr1-6 ProSNBE:CCR1 lines showed a similar intensity, which was considerably lower compared with that of the wild type. In the next step, a more detailed analysis was performed on the aromatic region of lignin (1,700–1,550 cm−1) obtained from vessels, xylary fibers, and interfascicular fibers. In vessels and xylary fibers, ccr1-6 mutants showed a drastic decrease in the aromatic stretching vibration of lignin at 1,597 cm−1 (Agarwal et al., 1997) when compared with the wild type, whereas the ccr1-6 ProSNBE:CCR1 lines had band intensities between those of the wild type and ccr1-6. First, these results indicated that the lignin levels in the vessels of ccr1-6 SNBE:CCR1 were not recovered to wild-type levels but, rather, were intermediate between the high levels found in the wild type and the drastically reduced levels in ccr1-6. Second, we could conclude that the lignin content in the xylary fibers of ccr1-6 ProSNBE:CCR1 lines also was partially recovered to levels between those of the wild type and ccr1-6. In accordance with the lignin staining data, the lignin content in interfascicular fibers (based on peak intensities for the lignin aromatic stretching at 1,597 cm−1) of ccr1-6 and ccr1-6 ProSNBE:CCR1 lines was reduced drastically when compared with the wild type. Another difference was observed for the peak at 1,657 cm−1, which is assigned mainly to coniferyl alcohol (C=C stretching of coniferyl alcohol and C=O stretching of coniferaldehyde; Agarwal et al., 2011). Whereas the wild type showed a high intensity for this peak in vessels, xylary fibers, and interfascicular fibers, it was absent in all regions of ccr1-6. In the vessels and xylary fibers of ccr1-6 ProSNBE:CCR1 lines, the intensity of the peak at 1,657 cm−1 was intermediate between that of the wild type and ccr1-6, whereas in ccr1-6 ProSNBE:CCR1 interfascicular fibers, the peak at 1,657 cm−1 was absent. Interestingly, in vessels and fibers of ccr1-6, a new band appeared at 1,633 cm−1. This band was also present in the interfascicular region of ccr1-6 ProSNBE:CCR1 lines and is known as C=C stretching of ferulic acid (Agarwal and Atalla, 1990; Prinsloo et al., 2004; Meyer et al., 2011; Mateu et al., 2016).
Figure 5.
Raman microscopy analysis of cell walls of wild-type, ccr1-6, and ccr1-6 ProSNBE:CCR1 plants. A, Examples of Raman mapping images taken from the xylem and interfascicular fiber region of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 lines by integrating the aromatic stretching vibration from 1,650 to 1,550 cm−1. CCD cts, Charged Coupled Device counts. Bars = 10 µm. B, Extracted average spectra in the lignin aromatic region between 1,700 and 1,550 cm−1 obtained by a region-of-interest study in vessels, xylary fibers, and interfascicular fibers (n = 18: 3 biological replicates × 2 mappings × 3 regions of interest). The marked bands represent 1,657 cm−1 (C=C stretching of coniferyl alcohol plus C=O stretching of coniferaldehyde), 1,633 cm−1 (C=C stretching from the propenoic acid side chain of ferulic acid), and 1,597 cm−1 (aromatic ring stretching of lignin). a.u., arbitrary units.
Finally, the stiffness of the stems was determined via a two-point bending test (Supplemental Table S2). Whereas wild-type stems had a bending modulus of 112.5 kPa, ccr1-6 ProSNBE:CCR1 lines 1 and 2 had significantly reduced bending moduli of 49.4 and 60.2 kPa, respectively. Similarly, the bending modulus of ccr1-6 also was reduced significantly when compared with the wild type, to a value of 25.7 kPa. Based on this, we could conclude that the stiffness of the stems of ccr1-6 ProSNBE:CCR1 lines was partially restored when compared with that of ccr1-6 but was still decreased when compared with that of the wild type.
The Metabolism of ccr1-6 ProSNBE:CCR1 Lines Shows Characteristics of ccr1-6 Mutants and of Wild-Type Plants
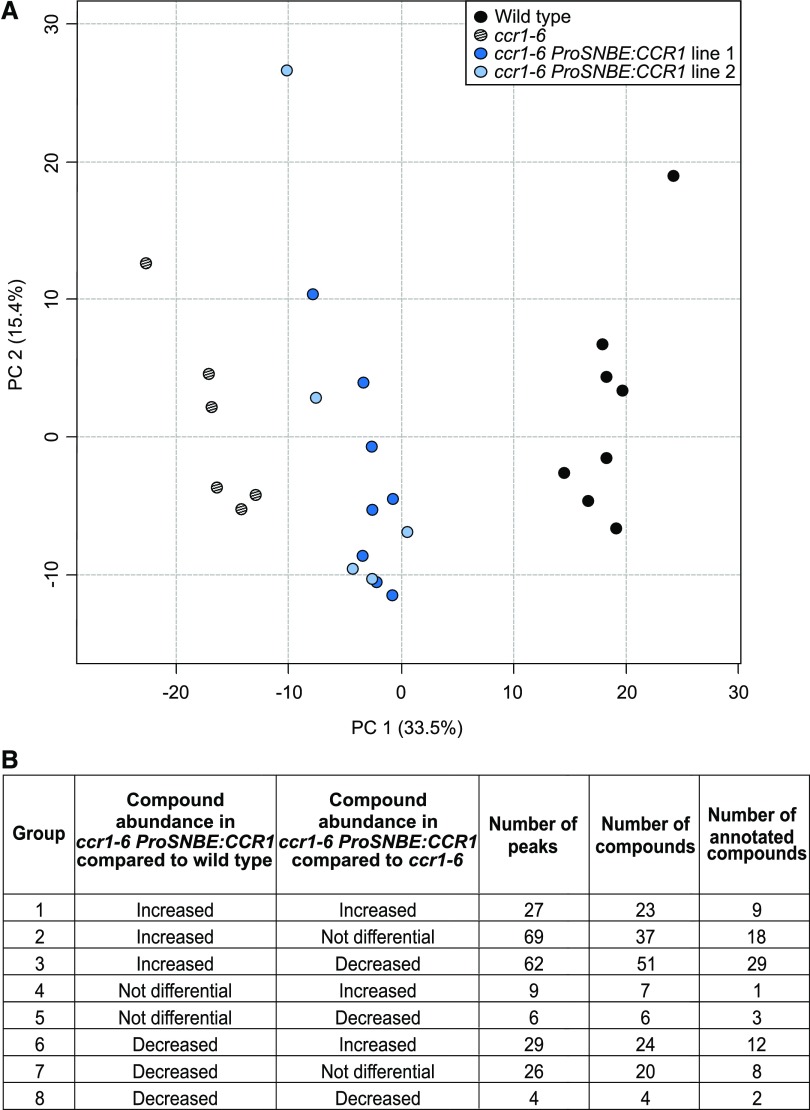
The biomass penalty and vessel collapse provoked by the ccr1 mutation were recovered in ccr1 ProSNBE:CCR1 lines (Table I; Fig. 3; Supplemental Fig. S2). To examine whether the molecular phenotype of ccr1 mutants was also recovered for ccr1 ProSNBE:CCR1 lines, metabolic profiling of their inflorescence stems was performed via ultra-high-pressure liquid chromatography-mass spectrometry (UHPLC-MS). This procedure allows the detection of several classes of phenolic compounds and glucosinolates (Vanholme et al., 2012, 2013b; Sundin et al., 2014). A total of 9,746 peaks (mass-to-charge ratio [m/z] features) were integrated in the chromatograms of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 lines 1 and 2 (Supplemental Data Set S1). After applying stringent filters, 554 peaks were retained for statistical analysis (see “Materials and Methods”). Principal component analysis (PCA) of these 554 peaks showed that the metabolic profiles of ccr1-6 ProSNBE:CCR1 plants were situated between those of ccr1-6 mutants and wild-type plants according to the first principal component, which explains 33.5% of the variation (Fig. 6). The second principal component, which explains 15.4% of the variation, reflects variation within the genotypes (and not between the genotypes) and can be attributed to biological and/or technical variation. One-way ANOVA followed by posthoc Student’s t tests resulted in a list of 232 peaks with significantly different intensities between the ccr1-6 ProSNBE:CCR1 lines and the wild type and/or the ccr1-6 ProSNBE:CCR1 lines and ccr1-6. Based on this, the peaks were classified into eight different groups (Fig. 6; Supplemental Data Set S1). Because no significant differences in peak intensity were found between ccr1-6 ProSNBE:CCR1 lines 1 and 2 for these 232 peaks, the two ccr1-6 ProSNBE:CCR1 lines were treated further as one line. In-depth analysis of the 232 peaks showed that these could be assigned to 172 compounds, of which 82 could be putatively structurally characterized based on their m/z, retention times, and tandem mass spectrometry (MS/MS) fragmentation spectra (Fig. 7; Supplemental Tables S3 and S4; Supplemental Fig. S4). The latter were biochemically classified and situated onto a metabolic map of the phenolic and glucosinolate metabolism in Arabidopsis stems (Fig. 8).
Figure 6.
Summary of metabolite profiling of primary inflorescence stems of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1. A total of 9,746 peaks were detected over the different samples (wild type, n = 8; ccr1-6, n = 6; and ccr1-6 ProSNBE:CCR1, n = 13). After applying stringent filters, 554 peaks were selected for PCA and statistical analysis. A, PCA of the selected peaks revealed that the first principal component (PC 1; 33.5% of variation) explained mainly the difference between genotypes. The second principal component (PC 2; 15.4% of the variation) reflects the variation within the genotypes. B, Further statistical analysis revealed that the peak intensities of 232 compounds were significantly different in ccr1-6 ProSNBE:CCR1 when compared with the wild type and when compared with ccr1-6. The differentially accumulating metabolites were classified into eight groups. Per group, the number of peaks, corresponding compounds, and annotated compounds are given.
Figure 7.
List of putatively structurally characterized metabolites with a different abundance in inflorescence stems of ccr1-6 ProSNBE:CCR1 plants as compared with the wild type or ccr1-6 mutants. Eighty-two compounds were characterized, of which the abundance was significantly different between ccr1-6 ProSNBE:CCR1 and the wild type, on the one hand, and/or ccr1-6 ProSNBE:CCR1 and ccr1-6, on the other hand. The differentially accumulating metabolites were classified into eight different groups (Fig. 6B). Each putatively structurally characterized metabolite has a unique number that represents (1) the group that the metabolite belongs to and (2) the ranking within this group. The metabolites are listed per metabolic class, and the dashed lines separate the different groups present in a specific metabolic class. The average peak intensities are represented by a heat map, ranging from low values represented in blue to high values represented in red (wild type, n = 8; ccr1-6, n = 6; and ccr1-6 ProSNBE:CCR1, n = 13).
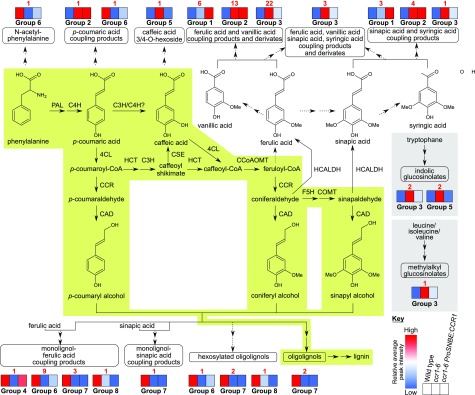
Figure 8.
Metabolic map of phenolic and glucosinolate metabolism in the primary inflorescence stems of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 lines. The green highlighted part of the pathway is considered to be the major route for lignin biosynthesis. The gray highlighted part depicts glucosinolate biosynthesis. Dashed arrows represent suggested pathways. Metabolites were classified into one of eight specific groups, based on their abundance in the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 (Figs. 6B and 7; Supplemental Table S3). In each group, different metabolite classes are represented (Fig. 7). In the pathway, these different metabolite classes are indicated by round-angled boxes. Per metabolite class, the respective group(s) containing this specific class is (are) indicated, accompanied by heat maps. These heat maps represent the average peak intensities of all characterized metabolites belonging to a specific group for the wild type (first block), ccr1-6 (second block), and ccr1-6 ProSNBE:CCR1 (third block), indicated by a range from blue (lowest values) to red (highest values; see key at bottom). The red number indicates the number of metabolites belonging to this group in a specific metabolic class. Terms not defined in the text are as follows: F5H, FERULATE 5-HYDROXYLASE; COMT, CAFFEIC ACID O-METHYLTRANSFERASE; CAD, CINNAMYL ALCOHOL DEHYDROGENASE; HCALH, HYDROXYCINNAMALDEHYDE DEHYDROGENASE (Vanholme et al., 2012; Vanholme et al., 2013b). C3H/C4H?: Interaction between C3H and C4H has been shown for both Arabidopsis and poplar, but activity of the C3H/C4H heterodimer for the conversion of p-coumaric acid to caffeic acid has only been shown for poplar (Bassard et al., 2012; Chen et al., 2011).
Of the 82 putatively structurally characterized metabolites, 52 belonged to the classes of ferulic acid, vanillic acid, sinapic acid, and syringic acid coupling products and derivatives. Previously, ccr1 mutants have been described to accumulate members of these metabolic classes (Vanholme et al., 2012). The abundances of these metabolites also were higher in ccr1-6 ProSNBE:CCR1 when compared with the wild type, of which nine compounds had levels that were even higher than those in ccr1-6 (group 1), 17 compounds had levels not significantly different from those in ccr1-6 (group 2), and 26 compounds had levels lower than those in ccr1-6 (group 3). Furthermore, six compounds were classified as oligolignols and hexosylated oligolignols. The abundance of these metabolic classes was severely reduced in ccr1-6 mutants when compared with the wild type (Vanholme et al., 2012). In the ccr1-6 ProSNBE:CCR1 lines, the peak intensities of these metabolites were also reduced when compared with the wild type, to levels higher than in ccr1-6 (one hexosylated oligolignol in group 6), not significantly different from in ccr1-6 (two hexosylated oligolignols and two oligolignols in group 7), or lower than in ccr1-6 (one hexosylated oligolignol in group 8). Another class of phenylpropanoic acid-derived metabolites in Arabidopsis comprises coupling products of monolignols and ferulic acid or sinapic acid. Similar to (hexosylated) oligolignols, the abundance of monolignol-ferulic acid coupling products was reduced in ccr1-6 mutants (Vanholme et al., 2012). In ccr1-6 ProSNBE:CCR1 lines, the abundance of one monolignol-ferulic acid coupling product was not significantly different when compared with the wild type and increased significantly as compared with ccr1-6 (group 4). However, the abundance of 14 monolignol-ferulic acid and -sinapic acid coupling products was still reduced in the ccr1-6 ProSNBE:CCR1 lines as compared with their abundance in the wild type, of which nine had abundances intermediate between those of the wild type and ccr1-6 (group 6), four had abundances that were not significantly different from those of ccr1-6 (group 7), and one had an abundance lower than that of ccr1-6 (group 8). In addition, five glucosinolates that accumulated to high levels in ccr1-6 were (partially) restored to wild-type levels in ccr1-6 ProSNBE:CCR1 (groups 3 and 5). Furthermore, the abundances of N-acetyl-phenylalanine and p-coumaroyl glutamate were decreased in the ccr1-6 mutant when compared with the wild type, whereas the ccr1-6 ProSNBE:CCR1 lines had levels intermediate between those of the wild type and ccr1-6 (group 6). The abundance of p-coumaroyl hexose was increased to a similar level in ccr1-6 and the ccr1-6 ProSNBE:CCR1 lines when compared with that of the wild type (group 2). Finally, caffeic acid 3/4-O-hexoside accumulated to high levels in ccr1-6 mutants when compared with the wild type but was reduced again to wild-type levels in the ccr1-6 ProSNBE:CCR1 lines (group 5). Despite the notable exceptions, we found that the majority of metabolic shifts present in dwarfed ccr1-6 mutants were still largely present in the phenotypically fully recovered ccr1 ProSNBE:CCR1 lines.
The Effect of Ferulic Acid Content on Cell Proliferation and Growth
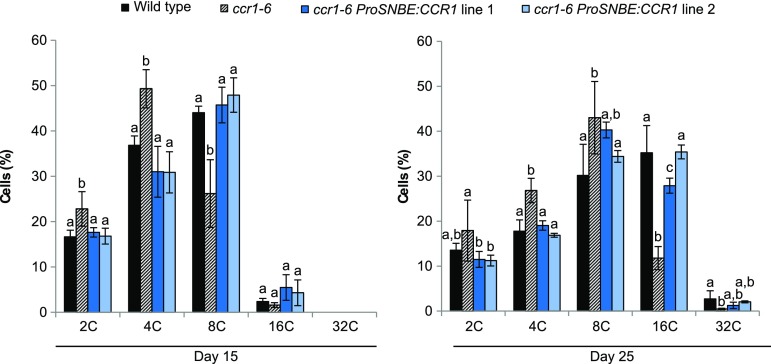
It has been proposed that the dwarfed phenotype of ccr1 mutants is caused by the dramatically increased level of ferulic acid (Xue et al., 2015). These high levels of ferulic acid were reported to delay the exit from cell proliferation, thereby reducing the average nuclear ploidy level and causing the observed growth defects of ccr1 mutants (Xue et al., 2015). Elaborating on this reasoning, the ferulic acid levels of the growth-restored ccr1-6 ProSNBE:CCR1 lines should be reduced when compared with ccr1-6, leading to restoration of the cell cycle and growth. To test this hypothesis, we determined the ferulic acid levels and nuclear ploidy level of ccr1-6 ProSNBE:CCR1 lines using the wild type and ccr1-6 as a control. In analogy with Xue et al. (2015), all experiments were performed on the first pair of leaves of 15- and 25-d-old plants. At both time points, the rosette sizes of ccr1-6 ProSNBE:CCR1 plants were similar to those of the wild type, whereas ccr1-6 rosettes were significantly smaller (Supplemental Fig. S5). Additionally, the morphology of the vasculature of the first leaves was investigated via microscopy. Notably, wild-type and ccr1-6 ProSNBE:CCR1 plants had large and open vessels in the xylem, whereas the vessels of ccr1-6 mutants were collapsed (Supplemental Fig. S6).
In contrast to the findings of Xue et al. (2015), the levels of ferulic acid remained below the detection limit in all samples in our analysis. Therefore, a series of ferulic acid coupling products was used as a measure for the total ferulic acid content (Table III). On day 15, the levels of all ferulic acid coupling products were increased in ccr1-6 ProSNBE:CCR1 lines when compared with the wild type, to levels equal to those in the ccr1-6 mutant (Table III). The same trend was observed for seedlings on day 25. At this time point, the levels of ferulic acid coupling products were increased in the ccr1-6 ProSNBE:CCR1 lines when compared with the wild type, to levels equal to or lower than in the ccr1-6 mutant (Table III). In accordance with the literature, the average nuclear ploidy level of cells from ccr1-6 mutants was lower as compared with that of the wild type at both time points (Fig. 9; Xue et al., 2015). However, the average ploidy level of cells in both ccr1-6 ProSNBE:CCR1 lines was similar to that of the wild type and significantly higher than that of the ccr1-6 mutant at both time points (Fig. 9). Taken together, these results show that the overall level of ferulic acid was similar in both ccr1-6 and ccr1-6 ProSNBE:CCR1 seedlings, whereas cells of the ccr1-6 mutant, but not those of ccr1-6 ProSNBE:CCR1, retained their mitotic state for a prolonged time.
Table III. Analysis of ferulic acid content in leaves of 15- and 25-d-old seedlings of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 lines.
The identified ferulic acid coupling products and their average peak area ± se are shown (n = 5). Statistics are per compound and per time point (see “Materials and Methods”). Different letters represent significant differences at the 0.01 significance level (Dunnett-Hsu adjusted Student’s t test). b.d., below detection limit. The asterisk indicates a compound detected as a formic acid adduct.
| No. | Retention Time | m/z | Compound Name | Day 15 |
Day 25 |
||||
|---|---|---|---|---|---|---|---|---|---|
| Wild Type | ccr1-6 | ccr1-6 ProSNBE:CCR1 | Wild Type | ccr1-6 | ccr1-6 ProSNBE:CCR1 | ||||
| min | |||||||||
| 1 | 3.76 | 355.103 | ferulic acid 4-O-hexoside | 3 ± 3 a | 282 ± 11 b | 272 ± 25 b | 5 ± 2 a | 258 ± 21 b | 169 ± 12 c |
| 2 | 5.41 | 355.103 | feruloyl hexose | 30 ± 27 a | 1,178 ± 123 b | 1,416 ± 298 b | 22 ± 13 a | 595 ± 119 b | 179 ± 38 c |
| 3 | 5.52 | 355.103 | ferulic acid + hexose | 34 ± 24 a | 1,453 ± 98 b | 1,553 ± 131 b | 85 ± 6 a | 1,624 ± 117 b | 1,220 ± 75 b |
| 4 | 8.99 | 309.060 | feruloyl malate | 0 ± 0 a | 140 ± 10 b | 150 ± 14 b | b.d. | b.d. | b.d. |
| 5 | 6.39 | 322.093 | feruloyl glutamate | 11 ± 9 a | 1,375 ± 181 b | 1,194 ± 172 b | 16 ± 3 a | 1,007 ± 100 b | 321 ± 37 c |
| 6 | 5.77 | 322.093 | ferulic acid + glutamate | 15 ± 11 a | 3,119 ± 433 b | 3,043 ± 420 b | 4 ± 4 a | 2,829 ± 272 b | 916 ± 132 c |
| 7 | 2.62 | 563.161 | feruloyl hexose + hexose* | 5 ± 5 a | 170 ± 22 b | 141 ± 25 b | 2 ± 2 a | 520 ± 65 b | 221 ± 44 c |
| 8 | 5.86 | 471.114 | ferulic acid + malate + hexose | 4 ± 3 a | 1,335 ± 166 b | 795 ± 114 b | 7 ± 1 a | 1,783 ± 184 b | 1,011 ± 109 c |
| 9 | 8.76 | 455.118 | ferulic acid + malate + deoxyhexose | 0 ± 0 a | 114 ± 10 b | 149 ± 26 b | 0 ± 0 a | 74 ± 15 b | 115 ± 13 b |
Figure 9.
Nuclear ploidy levels of wild-type, ccr1-6, and ccr1-6 ProSNBE:CCR1 cells. Flow cytometry analysis is shown for the first leaves at 15 and 25 d post stratification (15 d, n = 4; 25 d, n = 5). Error bars indicate sd. Different letters represent significant differences at the 0.05 significance level (Dunnett-Hsu adjusted Student’s t test).
The Lignocellulosic Composition of ccr1 ProSNBE:CCR1 Is Highly Similar to That of the ccr1 Mutant
Lignocellulosic biomass is recalcitrant toward deconstruction mainly because of the presence of lignin. Because ccr1 ProSNBE:CCR1 lines do not suffer from a yield penalty but have a reduced amount of lignin in both the xylem and interfascicular fibers (Fig. 5), translation of this strategy in a bioenergy crop could be interesting for the biorefinery. To study the lignocellulosic biomass composition of ccr1-6 ProSNBE:CCR1, the lignin content and composition and cellulose content of senesced inflorescence stems were determined (Table IV). First, soluble compounds were removed from the stems by applying a sequential extraction to produce CWR (Van Acker et al., 2013). In compliance with the literature, ccr1-6 mutants had 12% less CWR, and thus relatively more soluble compounds, than the wild type (Van Acker et al., 2013). On average, the ccr1-6 ProSNBE:CCR1 lines had 6% less CWR than the wild type. Second, the fraction of lignin in these prepared CWRs was determined via the Klason method. The lignin amount of the ccr1-6 ProSNBE:CCR1 lines did not differ significantly from that of ccr1-6 but was approximately half that of the wild type. Third, the lignin composition was analyzed via thioacidolysis, which allows quantification of the H, G, S, and other minor units that are linked by β-O-4 interunit bonds in the lignin polymer. Lignins from both the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines released substantially fewer monomers (H + G + S) than the lignin from wild-type samples. This indicates that the lignins of the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines have fewer β-O-4 interunit bonds and, thus, are enriched in carbon-carbon (mainly β-5 and β-β) interunit bonds. The H monomers were barely detectable in the wild type and constituted only 1.8% of the total identified thioacidolysis-released units. By contrast, the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines showed a relative increase in thioacidolysis-released H units by approximately 3-fold. Furthermore, the S/G ratio was decreased for the ccr1-6 mutant when compared with that of the wild type. Interestingly, this decrease was even more strikingly pronounced for the ccr1-6 ProSNBE:CCR1 lines. Incorporation of ferulic acid (FA), which is a known minor constituent of lignin, results in the release of three different units after thioacidolysis: two are linked via conventional β-O-4 structures (the β-O-4-FA-I and β-O-4-FA-II units), while the third, derived from the bis-β-O-4 coupling of ferulic acid, results in a truncated side chain (Ralph et al., 2008). In agreement with previously reported results for plants deficient in CCR, the relative abundance of all three ferulic acid units was increased in the ccr1-6 mutant when compared with the levels in the wild type (Goujon et al., 2003; Leplé et al., 2007; Mir Derikvand et al., 2008; Ralph et al., 2008; Van Acker et al., 2014). Interestingly, the ccr1-6 ProSNBE:CCR1 lines also showed a relative increase in all three thioacidolysis-released ferulic acid units when compared with the wild type, to levels not significantly different from those in the ccr1-6 mutant. Fourth, crystalline cellulose content was analyzed via the spectrophotometric phenol-sulfuric acid assay. In accordance with previously published results (Van Acker et al., 2013), ccr1-6 mutants had less crystalline cellulose than the wild type (with an average relative decrease of about 17%). The crystalline cellulose content in the ccr1-6 ProSNBE:CCR1 lines did not differ significantly from that of the ccr1-6 mutants but was reduced as compared with that of the wild type.
Table IV. Cell wall characteristics.
The cell wall residue (CWR) expressed as a percentage of dry weight was determined gravimetrically after a sequential extraction (n = 6). Lignin content was determined with the Klason method and expressed as percentage of CWR (n = 3). Lignin composition was determined with thioacidolysis (n = 6). The sum of H, G, and S is expressed in μmol g−1 Klason lignin. The relative proportions of the different lignin units were calculated based on the total thioacidolysis yield (including the minor nonconventional lignin units). S/G ratio was calculated based on the absolute values for S and G. β-O-4-FA-I, G-CH=CH-COOH; β-O-4-FA-II, G-CHR-CH2-COOH; bis-β-O-4-FA, G-CHR-CHR2 (R = thioethyl). Cellulose was expressed as a percentage of CWR (n = 6). Values given are averages ± sd. Different letters represent significant differences at the 0.05 significance level (Dunnett-Hsu adjusted Student’s t test).
| Line | CWR | Lignin Content and Composition |
Crystalline Cellulose | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Klason Lignin | H + G + S | H | G | S | S/G Ratio | β-O-4-FA-I | β-O-4-FA-II | bis-β-O-4-FA | |||
| % | % CWR | μmol g−1 | % | % | % | % | % | % | % CWR | ||
| Wild type | 78.5± 1.3 a | 18.0± 0.9 a | 23.6± 3.5 a | 1.8± 0.6 a | 71.5± 1.2 a | 26.6± 0.9 a | 0.37± 0.02 a | 0.03± 0.03 a | 0.02± 0.00 a | 0.09± 0.01 a | 44.7± 2.2 a |
| ccr1-6 | 69.4± 2.0 b | 9.5± 1.0 b | 8.9± 1.9 b | 5.5± 2.9 b | 70.5± 1.5 a | 21.9± 3.3 b | 0.31± 0.05 b | 0.35± 0.25 b | 0.40± 0.13 b | 1.34± 0.76 b | 34.4± 3.2 b |
| ccr1-6 ProSNBE:CCR1 line 1 | 74.2± 1.7 c | 9.7± 0.5 b | 6.9± 1.7 b | 5.5± 2.7 b | 77.8± 2.6 b | 14.2± 1.5 c | 0.18± 0.02 c | 0.41± 0.26 b | 0.46± 0.09 b | 1.56± 0.92 b | 38.6± 2.5 b |
| ccr1-6 ProSNBE:CCR1 line 2 | 73.6± 1.2 c | 9.9± 0.3 b | 7.4± 1.5 b | 5.5± 1.7 b | 78.1± 2.7 b | 14.0± 2.3 c | 0.18± 0.04 c | 0.38± 0.17 b | 0.54± 0.12 b | 1.45± 0.58 b | 35.5± 2.3 b |
The ccr1 ProSNBE:CCR1 Lines Have a 4-Fold Increase in Total Plant Saccharification Yield When Compared with the Wild Type
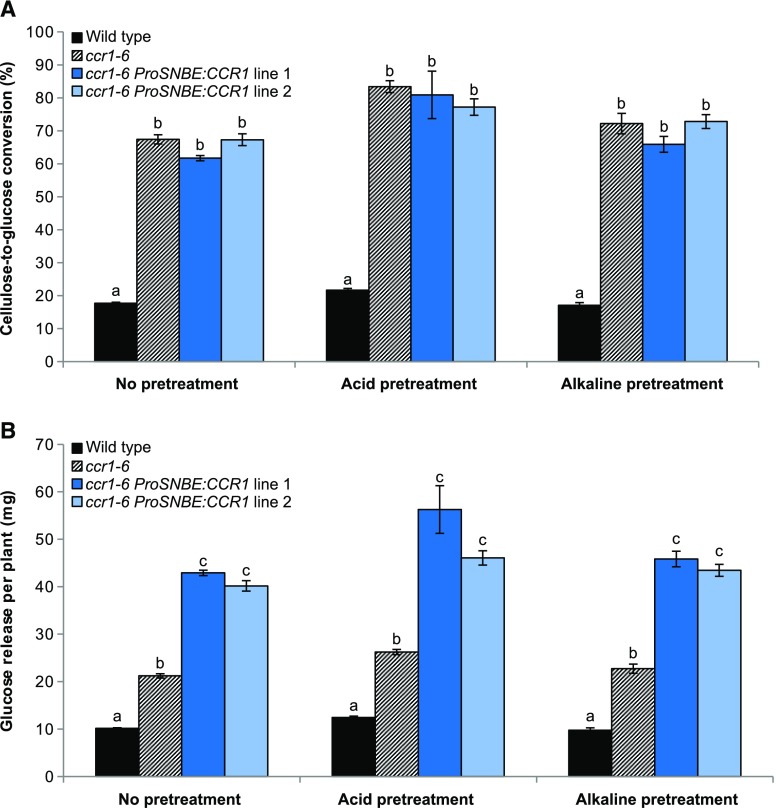
Cell wall analysis revealed that the lignin content of the ccr1-6 ProSNBE:CCR1 lines was reduced when compared with that of the wild type, to levels similar to those of the ccr1-6 mutant (Table IV). Because the lignin amount has a negative effect on the saccharification efficiency, the saccharification potential of ccr1-6 ProSNBE:CCR1 biomass after acid, alkaline, or no pretreatment was investigated further. Pretreatment (with acid or alkali) allowed the samples to reach the plateau much sooner (i.e. after 48 h instead of 96 h compared with no pretreatment; Supplemental Fig. S7). Cellulose-to-glucose conversion for the ccr1-6 ProSNBE:CCR1 lines was similar to that of the ccr1-6 lines and much higher than that of the wild type, independent of the pretreatment (Fig. 10; Supplemental Fig. S7; Supplemental Table S5). More specifically, the cellulose-to-glucose conversion of the unpretreated samples had increased from 18% in the wild type to, on average, 65% in the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines (i.e. a relative increase of 261%). In the case of acid pretreatment, the conversion increased from 22% in the wild type to, on average, 81% in the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines (i.e. a relative increase of 268%). Finally, in the case of alkaline pretreatment, the conversion increased from 17% in the wild type to, on average, 70% in the ccr1-6 and ccr1-6 ProSNBE:CCR1 lines (i.e. a relative increase of 312%).
Figure 10.
Saccharification efficiency of ccr1-6 ProSNBE:CCR1 plants. A, Cellulose-to-glucose conversions after 192 h of saccharification of senesced inflorescence stems of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1. B, Glucose release after 192 h of saccharification per total stem biomass. Samples were saccharified using no pretreatment, acid pretreatment (1 m HCl), or alkaline pretreatment (62.5 mm NaOH). Error bars indicate se; n = 6. Different letters represent significant differences at the 0.01 significance level (Dunnett-Hsu adjusted Student’s t test) per pretreatment.
The glucose yield after saccharification was also expressed per plant (i.e. total stem biomass; Fig. 10). In the case of the dwarfed ccr1-6 mutants, the total glucose release per plant was increased by about 2-fold in comparison with the wild type for each of the tested pretreatments. Due to the combined effect of the increase in total inflorescence stem biomass and the increase in saccharification efficiency, the ccr1-6 ProSNBE:CCR1 plants showed more than a 4-fold increase in glucose yield per plant when compared with the wild type in each of the tested pretreatments.
DISCUSSION
Vessels Act as Good Neighbors to Xylary Fibers
Complementing ccr1 mutants with ProSNBE:CCR1 resulted in plants with restored vessel integrity and growth. Using light, fluorescence, Raman, and electron microscopy, we analyzed the organization of the cell wall and lignin deposition in vessels and (xylary and interfascicular) fibers. Interestingly, lignification in ccr1 ProSNBE:CCR1 lines was not only (partially) restored in the vessels, but the cell walls of the xylary fibers also showed increased lignification and restored cell wall integrity. The latter is in apparent contrast to the vessel-specific expression pattern conferred by ProSNBE (McCarthy et al., 2011; this study). This observation demonstrates that monolignols synthesized in the vessel cells not only lignify the vessel cell wall but also contribute to the lignification of the cell walls of neighboring cells. Previous research in Arabidopsis has revealed that nonlignifying parenchyma cells provide monolignols to vessels and xylary fibers during the lignification process, a property that was termed good neighbors (Smith et al., 2013). In addition, xylary fibers also play a role in the lignification of neighboring vessel elements during xylem development (Smith et al., 2017b). These combined observations show that monolignols can be transported between the different xylem cell types.
Raman microscopy analysis showed that the lignin deposition in the xylem cells of ccr1 ProSNBE:CCR1 lines is increased by approximately 2-fold when compared with that in the ccr1 background but is not restored to wild-type levels. This observation could be explained by the fact (1) that ProSNBE and the native CCR1 promoter differ in timing and/or strength of expression in the vessel cells or (2) that the expression of CCR1 in the vessels alone is insufficient to compensate for the lack of monolignols that otherwise originate from the vessels, xylary fibers, and parenchyma cells (Smith et al., 2013, 2017b). (3) Furthermore, vessels have been shown to continue lignification following programmed cell death by the donation of monolignols from neighboring parenchyma cells and xylary fibers (Pesquet et al., 2013; Serk et al., 2015; Smith et al., 2017b). Because CCR1 expression is restored only in the vessels of ccr1 ProSNBE:CCR1 lines, postmortem lignification is unlikely. Nevertheless, the partial restoration of lignin deposition in the xylem of ccr1 ProSNBE:CCR1 appeared to be sufficient for the complete recovery of the stem biomass yield of ccr1 ProSNBE:CCR1 lines.
Despite Their Phenotypic Recovery, ccr1-6 ProSNBE:CCR1 Lines Share Many Characteristics with ccr1-6
Although being restored in growth, the lignocellulosic composition and metabolome of ccr1-6 ProSNBE:CCR1 lines were very similar to those of ccr1-6. Interestingly, ccr1-6 and ccr1-6 ProSNBE:CCR1 also share a similar delay in cell development: whereas the interfascicular fibers of wild-type plants have completed cell death, the interfascicular fibers of ccr1-6 and ccr1-6 ProSNBE:CCR1 plants still contain their cellular contents, indicating a delay in programmed cell death. Furthermore, ccr1-6 mutants had a lower S/G ratio, which also is characteristic of their delay in lignification and development (Jones et al., 2001; Laskar et al., 2006; Leplé et al., 2007; Ruel et al., 2009). A reduced S/G ratio also was observed for ccr1-6 ProSNBE:CCR1, to a level that was even lower than that of ccr1-6. The latter also is a consequence of the lignin deposition pattern in ccr1-6 ProSNBE:CCR1: because the xylem region is more enriched in G units (while the fibers contain a higher level of S lignin; Pradhan Mitra and Loqué, 2014), (partial) restoration of lignification in the xylem will lead to a further decrease in the S/G ratio in the ccr1-6 ProSNBE:CCR1 lines.
Additionally, ccr1-6 mutants have been shown to have a lot of differences in their metabolism when compared with the wild type (Vanholme et al., 2012). These differences could be attributed to their developmental delay and/or the pathway perturbation itself. Despite their recovery in growth and partial restoration in CCR1 expression, ccr1-6 ProSNBE:CCR1 lines still share most of the shifts in their metabolism with ccr1-6. Of the 232 selected peaks, 15 (belonging to groups 4 and 5) were not differential between the wild type and ccr1-6 ProSNBE:CCR1, whereas 95 (belonging to groups 2 and 7) were not differential between ccr1-6 and ccr1-6 ProSNBE:CCR1. In addition, both ccr1-6 and ccr1-6 ProSNBE:CCR1 accumulate ferulic acid, vanillic acid, sinapic acid, and syringic acid coupling products. Moreover, both lines show a reduction in (hexosylated) oligolignols and monolignol-ferulic acid and -sinapic acid coupling products when compared with the wild type. As a notable exception, glucosinolate levels in ccr1-6 ProSNBE:CCR1 plants were (partially) restored to wild-type levels. Glucosinolates are involved in plant defense, and their biosynthesis is up-regulated in the case of tissue or cell damage (Del Carmen Martínez-Ballesta et al., 2013). Transcripts of the glucosinolate biosynthesis genes are more abundant in ccr1 mutants when compared with the wild type (Vanholme et al., 2012). It seems plausible that the accumulation of glucosinolates in ccr1-6 lines is a consequence of the general stress status of these mutants. Therefore, the lower levels of glucosinolates in ccr1-6 ProSNBE:CCR1 lines as compared with those in ccr1-6 could be a reflection of their reduced stress levels caused by the reversal of the irx phenotype.
Ferulic Acid Accumulation Is Not the Cause for the Yield Penalty of ccr1 Mutants
According to Xue et al. (2015), CCR1 plays a dual role in plants: next to being important for lignin biosynthesis, it is required for the termination of cell division. For the latter, CCR1, ferulic acid, and ROS coordinate the cell proliferation exit for leaf development. From day 11 onward, CCR1 expression covers the entire leaf, resulting into decreased ferulic acid levels in all leaf cells. As ferulic acid antagonizes the effect of ROS, the lower levels of ferulic acid upon CCR1 expression will lead to an accumulation of ROS throughout the leaf, which results into the exit from cell proliferation and the initiation of endoreduplication. The increased endoreduplication rate will result in a higher nuclear ploidy level. Based on this reasoning, Xue et al. (2015) conclude that the ccr1 dwarfed phenotype is not the result of the reduced lignin content but, instead, is the consequence of the dramatically increased levels of ferulic acid that keep the cellular oxidation state low, resulting in a prolonged phase of cell proliferation, a lower average nuclear ploidy level, and dwarfism (Xue et al., 2015). In the ccr1-6 ProSNBE:CCR1 lines, CCR1 expression is only partially restored in vessels. In these lines, all other cells, including leaf mesophyll cells, are still deficient in CCR1. As a consequence, ccr1-6 ProSNBE:CCR1 lines accumulate ferulic acid to levels similar to those in ccr1-6, but only the latter has a prolonged cell proliferation stage and stunted growth. Thus, our results suggest that high levels of ferulic acid are not the reason for the cell cycle defects and stunted growth of ccr1 mutants. However, irrespective of its cause, the fact that endoreduplication is restored in ccr1-6 ProSNBE:CCR1 plants could still indicate that the biomass yield penalty observed in ccr1-6 mutants is the consequence of the disturbed endoreduplication itself. Interestingly, high endopolyploidy was shown to be associated with increased leaf size in mapping populations (Gegas et al., 2014). Furthermore, the link between endoreduplication and dwarfism of cell wall mutants was made before: the stunted growth of an esk1-5 mutant, which is impaired in the acetylation of hemicellulose, could be largely suppressed by mutating the KAKTUS gene (Bensussan et al., 2015). The latter is described as an endoreduplication repressor and could control cell enlargement in different tissues.
The Yield Penalty of Low-Lignin Mutants Can Be Fully Overcome without Settling in Sugar Yield by Sufficiently Reinforcing the Vessels
Previous vessel-complementation attempts using VND6 and VND7 promoter sequences did not achieve a full restoration of vessel integrity and growth while maintaining the high sugar yield for Arabidopsis plants mutated in genes involved in lignin and hemicellulose biosynthesis (Petersen et al., 2012; Yang et al., 2013; Vargas et al., 2016). This could be the consequence of (1) the targeted cell wall biosynthesis gene or (2) the expression level and pattern conferred by the chosen promoter. VND6 expression is restricted to the metaxylem vessels, whereas VND7 has the highest expression level in protoxylem vessels (Kubo et al., 2005; Zhong et al., 2008). ProSNBE is a target of both VND6 and VND7 and confers expression in both the metaxylem and protoxylem (Zhong et al., 2010; McCarthy et al., 2011). The latter was sufficient to fully restore vessel integrity in ccr1 ProSNBE:CCR1 stems and leaves, as judged here from the open vessels in the xylem and the appearance of the cell wall observed by light microscopy and transmission electron microscopy. Despite having many characteristics that were similar to ccr1, restoration of vascular integrity was sufficient to restore the total plant biomass in ccr1 ProSNBE:CCR1 lines. These data suggest that the irx phenotype of ccr1 mutants is responsible for their dwarfed growth and possibly at the origin of the observed cell cycle defect. Additional proof for the collapsed vessel hypothesis as the reason for the dwarfism of cell wall mutants was given by the analysis of other cell wall-modified plants. Arabidopsis cell wall mutants that displayed collapsed vessels (e.g. ccr1, cse, c3h1, c4h, hct, and pal1/2/3/4 mutated in lignin biosynthesis; irx1, irx2, and irx3 mutated in cellulose biosynthesis; and irx7, irx8, and irx9 mutated in xylan biosynthesis) all suffer from growth perturbations (Jones et al., 2001; Franke et al., 2002; Besseau et al., 2007; Huang et al., 2010; Vanholme et al., 2013b; Yang et al., 2013). Moreover, vessel integrity seems to be correlated directly with plant growth. For example, ccr1-6 mutants and cad-c cad-d double mutants have been shown to have similarly decreased lignin contents, but only ccr1-6 suffered from collapsed vessels and prominent developmental defects (Thévenin et al., 2011). These observations are probably the consequence of the differences in lignin composition providing strength to the vessels: whereas lignin of ccr1-6 mutants had an S/G ratio of 0.48, cad-c cad-d double mutants had an S/G ratio of 0.081. Furthermore, despite the fact that the lignin amount of med5a med5b c3h triple mutants was restored to wild-type levels, they still suffered from collapsed vessels and were only partially restored in growth (Bonawitz et al., 2014). In the latter case, it is conceivable that the lignin of med5a med5b c3h, which consists mainly of H units and shorter lignin chains, does not provide sufficient strength to the vessels in order to avoid their collapse. In conclusion, we hypothesize that the dwarfism of cell wall mutants can be fully overcome by sufficiently reinforcing the vessels, thereby avoiding their collapse, by optimizing the cell wall composition.
When compared with the wild type, the ccr1 ProSNBE:CCR1 plants were fully recovered in height, but they also showed an increased number of secondary inflorescences and stem biomass. The increase in secondary inflorescences was also observed for ccr1-6 mutants and other CCR1-deficient Arabidopsis plants (Goujon et al., 2003; Van Acker et al., 2013). Here, experiments with ccr1-6 ProSNBE:CCR1, ccr1-6, and the wild type that were not allowed to set seeds showed that the increase in stem biomass in ccr1-6 ProSNBE:CCR1 lines resulted (largely) from their reduction in seed yield. The latter is supported by the fact that other plants perturbed in silique development (e.g. ms1-1 and ap3-1) also produce more axillary branches (Hensel et al., 1994; Wuest et al., 2016). Moreover, field-grown ccr1 maize (Zea mays) mutants, which also suffered from reduced seed weights, produced significantly more stem biomass when compared with wild-type maize plants (Smith et al., 2017a). The underlying signal leading to an increased biomass in seed-compromised plants remains largely unknown. Elucidation of this signal could open an interesting avenue to increase lignocellulosic biomass and, thus, fermentable sugar yield.
Cellulose-to-glucose conversions of ccr1-6 and ccr1-6 ProSNBE:CCR1 lines were 4-fold higher than those of the wild type, independent of the pretreatment. Given the negative correlation between saccharification and lignin amount, the increase in saccharification yield observed in these lines could be largely attributed to the reduced amount of lignin but possibly also to the increased levels of ferulic acid and H units (Ziebell et al., 2010; Van Acker et al., 2013). On a plant basis and compared with the wild type, the total plant sugar yield from the dwarfed ccr1-6 mutants was increased by 2-fold, whereas the ccr1-6 ProSNBE:CCR1 lines exhibited a 4-fold increase. The latter is the consequence of the full recovery in height and the increased stem biomass in ccr1-6 ProSNBE:CCR1 lines when compared with the wild type. To our knowledge, the ccr1-6 ProSNBE:CCR1 lines are the highest sugar-yielding Arabidopsis lines reported so far.
Potential of the Vessel-Specific Lignin Biosynthesis Strategy To Be Translated into Crops
In addition to their high sugar yield, the ccr1-6 ProSNBE:CCR1 lines do not suffer from a yield penalty, which makes the translation of this research into biomass crops attractive. Hybrid poplar shows great potential as a woody energy crop (Carroll and Somerville, 2009), and wood of CCR down-regulated poplar had up to 161% increased ethanol yield per unit of biomass (Van Acker et al., 2014). However, this strategy resulted in unstable down-regulation of the CCR gene and significant yield penalties in the respective trees. Since we have proven that the yield penalty of low-lignin ccr1 mutants can be fully overcome by allowing sufficient lignification to occur in the cell walls of vessels, a similar approach could be used for the yield recovery of poplar. For example, the stability problem of the down-regulation can be overcome by making stable ccr mutants using the efficient CRISPR/Cas9 genome-engineering technique (Tsai and Xue, 2015; Zhou et al., 2015). Recovery of the yield penalty could be achieved by reintroducing CCR expression specifically in the vessels. However, because we have proven that, in Arabidopsis, vessels provide monolignols to (xylary) fibers in order to lignify their cell wall, also in poplar the vessels may act as good neighbors to the surrounding fiber cells, as was hypothesized before (Gorzsás et al., 2011). Nevertheless, in Arabidopsis, the vessels were only able to recover the lignin amount in the xylem cells to approximately half that of the wild type. For this reason, a stable reduction in the lignin amount of poplars can potentially be achieved without the corresponding yield penalty. Potential alternative approaches to achieve vessel-specific lignification in poplar is via perturbing the lignification specifically in fibers. Because poplars do not (normally) flower when grown in short rotation for biomass, it is unlikely that they will also show an increase in lignocellulosic biomass, as was observed for the ccr1 ProSNBE:CCR1 lines. However, translation of this strategy into biomass crops that set seeds but of which the seeds are not the prime target (such as switchgrass [Panicum virgatum]) could potentially result in an increased lignocellulosic biomass yield.
MATERIALS AND METHODS
Plant Material and Vector Construction
Arabidopsis (Arabidopsis thaliana; ecotype Columbia-0) wild-type, ccr1-3 (SALK_123689), and ccr1-6 (GABI_622C01) mutant plants were used as controls and for plant transformation (Mir Derikvand et al., 2008; Ruel et al., 2009; Vanholme et al., 2012; Van Acker et al., 2013). The artificial SNBE promoter contained three copies of XCP1-SNBE1 fused to the CaMV 35S minimal promoter (from −46 to −1), as described by McCarthy et al. (2011; Supplemental Table S6). To clone this synthetic promoter, the 103-bp construct was first synthesized by Invitrogen (Life Technologies). Next, the construct was PCR amplified using primers containing the restriction sites for BamHI and XhoI (Supplemental Table S6). Subsequently, the PCR product was cloned into the Gateway pEN-L4-R1 vector using T4 DNA Ligase (Invitrogen) to generate the ProSNBE entry vector pEN-L4-ProSNBE-R1, whose identity was confirmed by sequencing. For the GFP and GUS reporter line, the ProSNBE building block was introduced into the destination vector pMK7S*NFm14GW by using LR Clonase (Invitrogen), which fused ProSNBE to a nuclear localization signal:GFP:GUS reporter construct, resulting in the ProSNBE:GFP:GUS expression clone. By using a nuclear localization signal, the resulting fusion protein would accumulate in the nucleus to higher concentrations than when it would remain in the cytoplasm, making the GFP fluorescence signal more easy to observe using fluorescence microscopy (Chytilova et al., 1999). For complementation, the ccr1-3 and ccr1-6 mutants were transformed with the ProSNBE:CCR1 construct. To this end, the coding sequence of CCR1 was PCR amplified and cloned into the pDONR221 vector using BP Clonase (Invitrogen; Supplemental Table S6). The sequence identity was confirmed by sequencing. Subsequently, the two building blocks pEN-L4-ProSNBE-R1 and pDONR221-L1-CCR1-L2 were introduced into the destination vector pK7m24GW-FAST via Multisite LR Clonase Plus (Invitrogen), which resulted in the ProSNBE:CCR1 expression clone. All the recombinant plasmids were introduced into Agrobacterium tumefaciens strain C58C1 PMP90 by electroporation. After plant transformation using the floral dip method, the identification of transformed seeds was based on kanamycin resistance (ProSNBE:GFP:GUS reporter lines) or seed fluorescence (ccr1 ProSNBE:CCR1 lines; Shimada et al., 2010). For the reporter lines, 30 independent T1 plants were analyzed. For the ccr1 ProSNBE:CCR1 lines, two independent, single-locus, homozygous T3 lines per ccr1 background were selected for further analysis.
Reporter Gene Analysis
For the reporter line analysis of the aerial parts, 20 independent, single-locus, homozygous T3 plants were cultivated in soil under short-day conditions (8-h-light/16-h-dark photoperiod, 21°C, and 55% humidity) during 6 weeks, after which they were transferred to long-day conditions (16-h-light/8-h-dark photoperiod, 21°C, and 55% humidity). After 5 and/or 8.5 weeks in long-day conditions, primary inflorescence stems and other plant organs were harvested for GUS analysis. The bottom of the inflorescence stem represents nonelongating internodes, while the top of the inflorescence stem represents elongating internodes.
For inflorescence stem cross sections, the bottom 1 cm of the main stem was removed and the above 3 cm was embedded in 7% (w/v) agarose. Sections of 100 μm thick were made using a vibratome (Campden Instruments) and subsequently stained for the presence of GUS by incubating at 37°C (in the dark) in a staining buffer containing 1 mm 5-bromo-4-chloro-3-indolyl β-d-glucopyranoside sodium salt, 0.5% Triton X-100, 1 mm EDTA, pH 8, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 500 mm sodium phosphate buffer, pH 7. The staining was performed for 2 h (Fig. 1; Supplemental Fig. S1) or overnight (Supplemental Fig. S1) and subsequently stopped by replacing the staining buffer with 70% (v/v) ethanol (overnight). Next, the sections were transferred to tap water and imaged using a Zeiss Axioskop 2 microscope with an EC Plan-Neofluar 20× (0.5 dry) objective.
For the other aerial organs, plant material was placed in GUS staining solution (as described above), vacuum infiltrated for 1 min, and incubated subsequently overnight at 37°C (in the dark). To terminate the reaction, the staining solution was removed, and plants were incubated overnight in 70% ethanol. Next, the organs were incubated for 1 week in GUS destaining solution (50% glycerol, 25% lactic acid, and 25% Milli-Q water). Images were obtained using a Nikon AZ100M microscope with AZ Plan Apo 0.5× (0.05) objective.
For reporter line analysis of the root, 10 independent, single-locus, homozygous T3 plants were grown for 20 d in long-day conditions on one-half-strength Murashige and Skoog plates. Then, root cell walls were stained with 30 mm propidium iodide at the onset of the experiment. The excitation energy of 488 nm was from an argon laser. The propidium iodide fluorescence emission was collected between 550 and 650 nm, and GFP fluorescence emission was collected between 500 and 550 nm. All images were captured with an inverted LSM 710 META confocal microscope equipped with 20×-Air objectives (Zeiss).
Plant Growth and Harvest
Unless mentioned otherwise, plants were grown as follows. The ccr1 ProSNBE:CCR1 lines and their respective controls (the wild type and ccr1-3 or ccr1-6) were cultivated in soil under short-day conditions during 6 weeks, after which they were transferred to long-day conditions. After 4 weeks in long-day conditions, main stems were harvested for lignin microscopy and bending tests, whereas after 5 weeks in long-day conditions, main stems were harvested for metabolic profiling (with plants having a height of approximately 26 cm for ccr1 and 50 cm for the other lines). For all other analyses, fully senesced plants/stems were used.
For the analysis of the first leaves, two ccr1-6 ProSNBE:CCR1 lines, the wild type, and ccr1-6 mutants were grown on soil in long-day conditions. Leaf 1 and leaf 2 were harvested 15 and/or 25 d post stratification.
Biomass Measurements
Plants were fully senesced, on average, after 6 weeks in short-day conditions followed by 10 weeks in long-day conditions. A first set of plants with ccr1-3, ccr1-3 ProSNBE:CCR1, and their wild-type control and a second set with ccr1-6, ccr1-6 ProSNBE:CCR1 lines, and their wild-type control were grown in two independent experiments. The inflorescence of completely senesced plants was harvested in full. First, the primary inflorescence stem (the main stem) was obtained by stripping off the leaves, axillary inflorescences, and siliques, after which the weight and height were determined and the number of secondary (axillary) inflorescence stems originating from the rosette and directly from the main stem were counted. Second, the secondary inflorescence weight was determined by stripping off the leaves and siliques. Third, seeds of the full plant were harvested for number and weight determinations. The total stem biomass is defined here as the weight of the primary and secondary inflorescences, without seeds, siliques, and leaves. The total plant biomass is defined here as the weight of the harvested aerial part of the plant, including the seeds, siliques, and cauline leaves, but without the rosette leaves.
For the biomass measurements on plants of which the developing siliques were repeatedly removed, the plants were grown as described above. With a frequency of three times per week, all siliques were removed from all plants. When the plants were fully senesced (after, on average, 6 weeks in short-day conditions and 10 weeks [for the control] or 14 weeks [for the plants of which the siliques were removed] in long-day conditions), the number of secondary inflorescence stems originating from the rosette and main stem was determined, after which the total stem biomass was determined.
Two-Point Bending Tests
Bending tests were carried out on 7-cm-long basal segments of primary inflorescence stems. To reduce the effect of turgor loss, stems were tested within 5 min of being harvested. The average cross-sectional area of the stem piece (A) was estimated by considering the cross section as a perfect circle using the formula A = π.(D/2)2. Here, D is the average of the diameter measured with a caliper at the basal and at the apical sides of the 7-cm piece. Approximately 2 cm of the basal side of the stem was taped to a support, to keep the stem in a horizontal position. Then, a weight (here, 0.001 kg) was attached to the apical side, after which the vertical deformation of the stem was measured. The bending modulus (Pa) was calculated as (F.L)/(A.Δx), where F is the force exerted by the weight on the stem segments (here, F = m.g = 0.0099 n), L is the distance between the support and the position of the weight (here, L = 0.05 m), A (m2) is the cross-sectional area through which the force is applied, and Δx (m) is the vertical displacement of the stem as a consequence of the applied force.
Light and Fluorescence Microscopy
After removing the bottom 1 cm of the main stem, the above 3 cm was embedded in 7% (w/v) agarose, and slices of 100 μm thick were made using a vibratome (Campden Instruments). Lignin staining with Wiesner and Mäule reagents was performed as described by Sundin et al. (2014). Images were acquired using a Zeiss Axioskop 2 microscope with an EC Plan- Neofluar 20× (0.5 dry) objective. Lignin autofluorescence was imaged using the Zeiss LSM 780 microscope with a Plan-Apochromat 10× (0.45 M27) objective. The fluorescence signal for lignin was obtained using 350 nm for excitation and the emission wavelength ranging from 407 to 479 nm.
For microscopy on 25-d-old first leaves, the harvested leaves were cut into small pieces and immersed in a fixative solution of 2.5% glutaraldehyde and 4% formaldehyde in 0.1 m sodium cacodylate buffer, placed in a vacuum oven for 30 min, and then left rotating for 3 h at room temperature. This solution was later replaced with fresh fixative, and samples were left rotating overnight at 4°C. After washing, samples were postfixed in 1% OsO4 with potassium ferricyanide in 0.1 m sodium cacodylate buffer, pH 7.2. Samples were dehydrated through a graded ethanol series, including a bulk staining with 2% uranyl acetate at the 50% ethanol step, followed by embedding in Spurr’s resin. Semithin sections were cut at 0.5 µm using the Leica EM UC6 ultramicrotome (Leica Microsystems) and stained with Toluidine Blue. Images were acquired using a Zeiss Axioskop 2 microscope with a Plan-Neofluar 100× Ph 3 (1.25 oil)objective.
Transmission Electron Microscopy
After removing the bottom 1 cm of the main stem, the above 3 cm was fixated, dehydrated, and embedded in Spurr’s resin as described above for the first leaves. Ultrathin sections of a gold interference color were cut using the Leica EM UC6 ultramicrotome, followed by poststaining with uranyl acetate and lead citrate in the Leica EM AC20, and collected on formvar-coated copper slot grids. They were viewed with the JEM-1400plus transmission electron microscope (JEOL) operating at 60 kV.
Raman Microscopy
After removing the bottom 1 cm of the main stem, the above 3 cm was embedded in polyethylene glycol matrix as described previously (n = 3; Gierlinger et al., 2012). Next, 18-µm-thick cross sections were cut using a rotary microtome (Leica RM2255). After polyethylene glycol was removed by rinsing with water, the samples were placed with a drop of water on a glass slide and sealed with a coverslip and nail polish. For each biological replicate, one cross section was analyzed using a confocal Raman microscope (InVia) equipped with a linearly polarized red laser (λ = 633 nm). A 100× oil-immersion lens was used for high spatial resolution. On each cross section, four Raman mappings were conducted: two in the xylem region and two in the interfascicular region. For the mapping, full spectra were obtained with a 0.3-µm step size. The integration time of one spectrum was set to 3 s using 1,800 I mm−1 grating and 18-mW laser power. After data acquisition, a cosmic ray removal was applied using Wire software (Renishaw; version 3.7). The Raman spectra were further analyzed using Cytospec software (Cytospec; version 2.00.01). By integrating the intensity of the band in the range of 1,550 to 1,650 cm−1 (which is assigned to the aromatic skeletal vibrations), cell corners, middle lamella, and secondary cell walls could be distinguished. Then, a more detailed region-of-interest study was performed on the secondary cell wall of vessels, xylary fibers, and interfascicular fibers. Three regions of interest were chosen for each anatomical region on each mapping, and an average spectrum was calculated for each region of interest. Consequently, a total number of 18 average spectra was obtained for each anatomical region (vessels, xylary fibers, and interfascicular fibers) of each genotype. The average spectra were then baseline corrected and analyzed further using the software Opus (Bruker; version 7).
Metabolic Profiling
After removing the bottom 1 cm of the stem, the next 11 cm from the bottom of the stem was frozen and ground. Subsequently, the stem tissue was extracted in 2-mL tubes at 70°C by shaking for 15 min with 1 mL of methanol. After centrifugation, the supernatant was transferred to new 1.5-mL tubes and lyophilized. Subsequently, the pellet was resuspended in equal volumes of cyclohexane and ultrapure water (100 µL each). After vortexing, samples were centrifuged, and the aqueous phase was subjected to UHPLC-MS on a Waters Acquity UHPLC device connected to a Synapt HDMS Q-TOF mass spectrometer (Waters). In the chromatograms of the wild type, ccr1-6, and the ccr1-6 ProSNBE:CCR1 lines, a total of 9,746 peaks were processed with Progenesis QI software version 2.1 (Waters), as described by Vanholme et al. (2013b). For peak selection, the following filters were applied: (1) retention time greater than 2 min, (2) value greater than 0 for at least all replicates in one line, and (3) average peak intensity greater than 5,000 for at least one line. After this, 554 peaks were retained for further PCA and statistical analysis. PCA (autoscaling) was performed in MetaboAnalyst (MetaboAnalyst software version 3.0; Xia and Wishart, 2016). For statistics, a one-way ANOVA was performed on arcsinh-transformed peak areas, which resulted in a list of 274 peaks that were significantly different between at least two lines (P < 0.001). Additional posthoc Student’s t tests were used to find peaks with significantly different peak areas in the ccr1-6 ProSNBE:CCR1 lines as compared with those in the wild type and in the ccr1-6 mutants. Peaks that had P < 0.01 and a ratio of at least 2-fold (greater than 2 or less than 0.5) when compared with the wild type and/or ccr1-6 were considered as statistically significant. Another posthoc Student’s t test showed that there were no significant differences (P < 0.001) in peak intensity between ccr1-6 ProSNBE:CCR1 line 1 and line 2 for these peaks. Therefore, the two ccr1-6 ProSNBE:CCR1 lines were treated as one line for subsequent analyses. In total, 232 peaks were selected and classified into eight different groups (Fig. 6). The peaks that were significantly different as compared with both the wild type and ccr1-6 were classified accordingly in groups 1, 3, 6, and 8. Those that were only significantly different from the wild type were reclassified at milder cutoff values when compared with ccr1-6: P of 0.05 irrespective of the fold change. As such, additional compounds were classified in groups 1, 3, 6, and 8. Similarly, peaks that were only significantly different as compared with ccr1-6 were reclassified at milder cutoff values when compared with the wild type: P of 0.05 irrespective of the fold change. Again, additional compounds were classified in groups 1, 3, 6, and 8. The remaining peaks were classified accordingly into groups 2, 4, 5, and 7. Structural characterization was initially performed by matching m/z, retention times, and MS/MS fragmentation against our in-house library, created by Instant Jchem for Excel software (Chemaxon). Structural characterization of the remaining unknown compounds was done manually by comparing m/z and MS/MS fragmentation with those of compounds described in the literature and online databases.
For the analysis of the first leaves, two different sets of plants were grown to be harvested 15 and 25 d post stratification. During harvesting, the first leaves (leaves 1 and 2) originating from the same plant were pooled together and used further as one sample for metabolite extraction. Hereby, the frozen ground tissue was extracted in 2-mL tubes at 70°C by shaking for 15 min with 500 µL of methanol. After centrifugation, the supernatant was transferred to new 1.5-mL tubes and the methanol was evaporated. Subsequently, the pellet was resuspended in equal volumes of cyclohexane and ultrapure water (75 µL each). After vortexing, samples were centrifuged, and the aqueous phase was analyzed using UHPLC-MS. UHPLC was performed on an ACQUITY UPLC I-Class system (Waters) consisting of a binary pump, a vacuum degasser, an autosampler, and a column oven. Chromatographic separation was carried out on an ACQUITY UPLC BEH C18 (150 × 2.1 mm, 1.7 μm) column from Waters, and temperature was maintained at 40°C. A gradient of two buffers was used: buffer A (99:1:0.1 water:acetonitrile:formic acid, pH 3) and buffer B (99:1:0.1 acetonitrile:water:formic acid, pH 3), as follows: 95% A for 0.1 min decreased to 50% A in 30 min. The flow rate was set to 0.35 mL min−1, and the injection volume was 10 μL. The UHPLC system was coupled to a Vion IMS QTOF hybrid mass spectrometer (Waters). The LockSpray ion source was operated in negative electrospray ionization mode under the following specific conditions: capillary voltage, 2.5 kV; reference capillary voltage, 2.5 kV; cone voltage, 30 V; source offset, 50 V; source temperature, 120°C; desolvation gas temperature, 400°C; desolvation gas flow, 600 L h−1; and cone gas flow, 50 L h−1. The collision energy for full MS was set at 6 eV; for MS/MS (data-dependent analysis) purposes, the collision energy was ramped from 10 to 15 eV for low mass and from 20 to 50 eV for high mass. Nitrogen (greater than 99.5%) was employed as desolvation and cone gas. Leucin-enkephalin (250 pg μL−1 solubilized in water:acetonitrile 1:1 [v/v], with 0.1% formic acid) was used for the lock mass calibration, with scanning every 2 min at a scan time of 0.1 s. Profile data were recorded through a UNIFI Scientific Information System (Waters). Data processing was performed with Progenesis QI software version 2.1 (Waters) as described by Vanholme et al. (2013b). After targeted selection of the ferulic acid coupling products, the following filters were applied: (1) value greater than 0 for at least all replicates in one line, (2) average peak intensity greater than 2 for at least one line, and (3) average peak intensity greater than 2 for at least one line. After performing a one-way ANOVA on arcsinh-transformed peak areas (P < 0.001), a posthoc Student’s t test showed that there were no significant differences (P < 0.001) in peak intensity between ccr1-6 ProSNBE:CCR1 line 1 and line 2 for the selected peaks. Therefore, the two ccr1-6 ProSNBE:CCR1 lines were treated as one line for subsequent statistical analysis. Additional posthoc Student’s t tests were used to find significant differences between the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 (P < 0.01). Structural characterization was done as described above for stem samples.
Flow Cytometry
Leaves 1 and 2 were harvested and snap frozen in liquid nitrogen 15 and 25 d post stratification for a first and second set of plants, respectively. Subsequently, 15-d-old leaves were pooled in pairs, whereas 25-d-old leaves were analyzed individually. First, the leaves were chopped with a razor blade. Second, the nuclei were isolated by adding 200 μL of Cystain UV Precise P nuclei extraction buffer and stained using 800 μL of Cystain UV Precise P staining buffer (Sysmex-Partec) before filtering using a 30-μm mesh. Flow cytometry was performed using a Cyflow flow cytometer (Sysmex-Partec), and the results were analyzed using Cyflogic software version 1.2.1 (Cyflogic).
Cell Wall Characterization and Saccharification
For the wild type and ccr1 ProSNBE:CCR1 lines, the bottom 36 cm of the main stem was chopped into pieces of 2 mm, and samples were pooled per three individuals. For the ccr1-6 mutants, the bottom 18 cm of the main stem was chopped into pieces of 2 mm, and samples were pooled per six individuals. Preparation of cell wall residue, cellulose quantification, and thioacidolysis were performed as described previously by Van Acker et al. (2013). Saccharification assays were performed as described by Van Acker et al. (2016). For the latter, measurements were performed at different time points after adding the saccharification enzymes: 3, 7, 24, 48, and 197 h. In case of no pretreatment, an extra time point at 97 h was added to obtain more resolution. To calculate the total plant sugar yield, the saccharification efficiency of the primary inflorescence stem was used as representative for all inflorescences. Lignin content was measured using a modified Klason method (Van den Bosch et al., 2015), where the 4-h incubation with a soxhlet extractor was replaced by a 1-h incubation at 121°C in an autoclave.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GUS expression analysis in growing and senescing ProSNBE:GFP:GUS inflorescence stems.
Supplemental Figure S2. Lignin deposition in inflorescence stems of ccr1-3 ProSNBE:CCR1 lines.
Supplemental Figure S3. Raman microscopy analysis of vessels, xylary fibers, and interfascicular fibers.
Supplemental Figure S4. MS/MS spectra of the 82 putatively structurally characterized metabolites with differential abundance between ccr1-6 ProSNBE:CCR1 and the wild type or ccr1-6 ProSNBE:CCR1 and ccr1-6.
Supplemental Figure S5. Phenotype of ccr1-6 ProSNBE:CCR1 seedlings.
Supplemental Figure S6. Impact of ProSNBE:CCR1 expression in ccr1-6 mutants on vascular integrity in leaves.
Supplemental Figure S7. Cellulose-to-glucose conversion during saccharification of the senesced inflorescence stems of the wild type, ccr1-6, and ccr1-6 ProSNBE:CCR1 lines.
Supplemental Table S1. Removal of siliques during development results in plants with increased number of secondary inflorescences and higher stem biomass.
Supplemental Table S2. Bending modulus determined by two-point bending tests.
Supplemental Table S3. List of putatively structurally characterized metabolites with different abundance in the inflorescence stems of ccr1-6 ProSNBE:CCR1 plants as compared with the wild type and/or as compared with ccr1-6 mutants.
Supplemental Table S4. Mass accuracy of the 82 putatively structurally characterized metabolites.
Supplemental Table S5. Statistical analysis of cellulose-to-glucose conversion efficiencies using different pretreatments.
Supplemental Table S6. Sequence information.
Supplemental Data Set S1. Metabolic profiling data showing the 9,746 peaks detected in wild-type plants, ccr1-6, and the ccr1-6 ProSNBE:CCR1 lines.
Acknowledgments
We thank Annick Bleys for help in preparing the article.
Footnotes
This work was supported by grants from Ghent University (Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” Grant 01MR0510W), from the Agency for Innovation by Science and Technology (IWT) through the IWT-SBO project BIOLEUM (grant no. 130039) and the IWT-FISCH-SBO project ARBOREF (grant no. 140894). B.D.M., L.d.V, and S.C. are indebted to the IWT for a predoctoral fellowship, and R.V. is indebted to the Research Foundation Flanders for a postdoctoral fellowship.
Articles can be viewed without a subscription.
References
- Agarwal UP, Atalla RH (1990) Formation and identification of cis/trans ferulic acid in photoyellowed white spruce mechanical pulp. J Wood Chem Technol 10: 169–190 [Google Scholar]
- Agarwal UP, McSweeny JD, Ralph SA (2011) FT-Raman investigation of milled-wood lignins: softwood, hardwood, and chemically modified black spruce lignins. J Wood Chem Technol 31: 324–344 [Google Scholar]
- Agarwal UP, Ralph SA, Atalla RH (1997) FT Raman spectroscopic study of softwood lignin. In 9th International Symposium on Wood and Pulping Chemistry: Proceedings, Montréal, Québec, 1997. [Google Scholar]
- Anderson NA, Bonawitz ND, Nyffeler K, Chapple C (2015) Loss of FERULATE 5-HYDROXYLASE leads to mediator-dependent inhibition of soluble phenylpropanoid biosynthesis in Arabidopsis. Plant Physiol 169: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard JE, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, De Jaeger G, Mely Y, Goossens A, Werck-Reichhart D (2012) Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensussan M, Lefebvre V, Ducamp A, Trouverie J, Gineau E, Fortabat MN, Guillebaux A, Baldy A, Naquin D, Herbette S, et al. (2015) Suppression of dwarf and irregular xylem phenotypes generates low-acetylated biomass lines in Arabidopsis. Plant Physiol 168: 452–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60: 165–182 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen H-C, Li Q, Shuford CM, Liu J, Muddiman DC, Sederoff RR, Chiang VL (2011) Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytilova E, Macas J, Galbraith DW (1999) Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann Bot (Lond) 83: 645–654 [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Day A, Neutelings G, Nolin F, Grec S, Habrant A, Crônier D, Maher B, Rolando C, David H, Chabbert B, et al. (2009) Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem 47: 9–19 [DOI] [PubMed] [Google Scholar]
- Del Carmen Martínez-Ballesta M, Moreno DA, Carvajal M (2013) The physiological importance of glucosinolates on plant response to abiotic stress in Brassica. Int J Mol Sci 14: 11607–11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A, George A, Mukerjee P, Kim JS, Pollet B, Benke PI, Yang F, Mitra P, Sun L, Çetinkol ÖP, et al. (2012) Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol J 10: 609–620 [DOI] [PubMed] [Google Scholar]
- Eudes A, Sathitsuksanoh N, Baidoo EE, George A, Liang Y, Yang F, Singh S, Keasling JD, Simmons BA, Loqué D (2015) Expression of a bacterial 3-dehydroshikimate dehydratase reduces lignin content and improves biomass saccharification efficiency. Plant Biotechnol J 13: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A, Zhao N, Sathitsuksanoh N, Baidoo EE, Lao J, Wang G, Yogiswara S, Lee TS, Singh S, Mortimer JC, et al. (2016) Expression of S-adenosylmethionine hydrolase in tissues synthesizing secondary cell walls alters specific methylated cell wall fractions and improves biomass digestibility. Front Bioeng Biotechnol 4: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Funk V, Kositsup B, Zhao C, Beers EP (2002) The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol 128: 84–94 [PMC free article] [PubMed] [Google Scholar]
- Gegas VC, Wargent JJ, Pesquet E, Granqvist E, Paul ND, Doonan JH (2014) Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J Exp Bot 65: 2757–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierlinger N, Keplinger T, Harrington M (2012) Imaging of plant cell walls by confocal Raman microscopy. Nat Protoc 7: 1694–1708 [DOI] [PubMed] [Google Scholar]
- Gorzsás A, Stenlund H, Persson P, Trygg J, Sundberg B (2011) Cell-specific chemotyping and multivariate imaging by combined FT-IR microspectroscopy and orthogonal projections to latent structures (OPLS) analysis reveals the chemical landscape of secondary xylem. Plant J 66: 903–914 [DOI] [PubMed] [Google Scholar]
- Goujon T, Ferret V, Mila I, Pollet B, Ruel K, Burlat V, Joseleau JP, Barrière Y, Lapierre C, Jouanin L (2003) Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: effects on phenotype, lignins and cell wall degradability. Planta 217: 218–228 [DOI] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB (1994) The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol 106: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isikgor FH, Becer CR (2015) Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym Chem 6: 4497–4559 [Google Scholar]
- Jackson LA, Shadle GL, Zhou R, Nakashima J, Chen F, Dixon RA (2008) Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. BioEnergy Res 1: 180–192 [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskar DD, Jourdes M, Patten AM, Helms GL, Davin LB, Lewis NG (2006) The Arabidopsis cinnamoyl CoA reductase irx4 mutant has a delayed but coherent (normal) program of lignification. Plant J 48: 674–686 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bhargava A, Qiang W, Friedmann MC, Forneris N, Savidge RA, Johnson LA, Mansfield SD, Ellis BE, Douglas CJ (2012) The class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 194: 102–115 [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kang KY, Chapple C (2012) Designed for deconstruction: poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol 194: 91–101 [DOI] [PubMed] [Google Scholar]
- Mateu BP, Hauser MT, Heredia A, Gierlinger N (2016) Waterproofing in Arabidopsis: following phenolics and lipids in situ by confocal Raman microscopy. Front Chem 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH (2011) Secondary wall NAC binding element (SNBE), a key cis-acting element required for target gene activation by secondary wall NAC master switches. Plant Signal Behav 6: 1282–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MW, Lupoi JS, Smith EA (2011) 1064 nm dispersive multichannel Raman spectroscopy for the analysis of plant lignin. Anal Chim Acta 706: 164–170 [DOI] [PubMed] [Google Scholar]
- Miedes E, Vanholme R, Boerjan W, Molina A (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir Derikvand M, Sierra JB, Ruel K, Pollet B, Do CT, Thévenin J, Buffard D, Jouanin L, Lapierre C (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD (2016) Designer lignins: harnessing the plasticity of lignification. Curr Opin Biotechnol 37: 190–200 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2010) Plant cell wall polymers as precursors for biofuels. Curr Opin Plant Biol 13: 305–312 [DOI] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PD, Lau J, Ebert B, Yang F, Verhertbruggen Y, Kim JS, Varanasi P, Suttangkakul A, Auer M, Loqué D, et al. (2012) Engineering of plants with improved properties as biofuels feedstocks by vessel-specific complementation of xylan biosynthesis mutants. Biotechnol Biofuels 5: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J, Lapierre C, Myton K, O’Connell A, Schuch W, Grima-Pettenati J, Boudet AM (1998) Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J 13: 71–83 [Google Scholar]
- Pradhan Mitra P, Loqué D (2014) Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J Vis Exp 2014: 51381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinsloo LC, du Plooy W, van der Merwe C (2004) Raman spectroscopic study of the epicuticular wax layer of mature mango (Mangifera indica) fruit. J Raman Spectrosc 35: 561–567 [Google Scholar]
- Ralph J, Kim H, Lu F, Grabber JH, Leplé JC, Berrio-Sierra J, Derikvand MM, Jouanin L, Boerjan W, Lapierre C (2008) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ruel K, Berrio-Sierra J, Derikvand MM, Pollet B, Thévenin J, Lapierre C, Jouanin L, Joseleau JP (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Serk H, Gorzsás A, Tuominen H, Pesquet E (2015) Cooperative lignification of xylem tracheary elements. Plant Signal Behav 10: e1003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadle G, Chen F, Srinivasa Reddy MS, Jackson L, Nakashima J, Dixon RA (2007) Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68: 1521–1529 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Smith RA, Cass CL, Mazaheri M, Sekhon RS, Heckwolf M, Kaeppler H, de Leon N, Mansfield SD, Kaeppler SM, Sedbrook JC, et al. (2017a) Suppression of CINNAMOYL-CoA REDUCTASE increases the level of monolignol ferulates incorporated into maize lignins. Biotechnol Biofuels 10: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Karlen SD, Bird D, Tokunaga N, Sato Y, Mansfield SD, Ralph J, Samuels AL (2017b) Defining the diverse cell populations contributing to lignification in Arabidopsis stems. Plant Physiol 174: 1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L (2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25: 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J, Chapple C (2004) The phenylpropanoid pathway in Arabidopsis: lessons learned from mutants in sinapate ester biosynthesis. Recent Adv Phytochem 38: 39–67 [Google Scholar]
- Sundin L, Vanholme R, Geerinck J, Goeminne G, Höfer R, Kim H, Ralph J, Boerjan W (2014) Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol 166: 1956–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin J, Pollet B, Letarnec B, Saulnier L, Gissot L, Maia-Grondard A, Lapierre C, Jouanin L (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4: 70–82 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Xue LJ (2015) CRISPRing into the woods. GM Crops Food 6: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Leplé JC, Aerts D, Storme V, Goeminne G, Ivens B, Légée F, Lapierre C, Piens K, Van Montagu MCE, et al. (2014) Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc Natl Acad Sci USA 111: 845–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Piens K, Boerjan W (2016) Saccharification protocol for small-scale lignocellulosic biomass samples to test processing of cellulose into glucose. Bio Protoc 6: e1701 [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch S, Schutyser W, Vanholme R, Driessen T, Koelewijn SF, Renders T, De Meester B, Huijgen WJJ, Dehaen W, Courtin C, et al. (2015) Reductive lignocellulose fractionation into soluble lignin-derived phenolic monomers and dimers and processable carbohydrate pulps. Energy Environ Sci 8: 1748–1763 [Google Scholar]
- Vanholme B, Desmet T, Ronsse F, Rabaey K, Van Breusegem F, De Mey M, Soetaert W, Boerjan W (2013a) Towards a carbon-negative sustainable bio-based economy. Front Plant Sci 4: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013b) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas L, Cesarino I, Vanholme R, Voorend W, de Lyra Soriano Saleme M, Morreel K, Boerjan W (2016) Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotechnol Biofuels 9: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, et al. (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Weng JK, Li X, Bonawitz ND, Chapple C (2008) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol 19: 166–172 [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, Mansfield SD, Lu F, Withers S, Park JY, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, et al. (2014) Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344: 90–93 [DOI] [PubMed] [Google Scholar]
- Wuest SE, Philipp MA, Guthörl D, Schmid B, Grossniklaus U (2016) Seed production affects maternal growth and senescence in Arabidopsis. Plant Physiol 171: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS (2016) Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 55: 14.10.11–14.10.91 [DOI] [PubMed] [Google Scholar]
- Xue J, Luo D, Xu D, Zeng M, Cui X, Li L, Huang H (2015) CCR1, an enzyme required for lignin biosynthesis in Arabidopsis, mediates cell proliferation exit for leaf development. Plant J 83: 375–387 [DOI] [PubMed] [Google Scholar]
- Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS, Sun L, Zheng K, Tang K, Auer M, et al. (2013) Engineering secondary cell wall deposition in plants. Plant Biotechnol J 11: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Morrison WH III, Negrel J, Ye ZH (1998) Dual methylation pathways in lignin biosynthesis. Plant Cell 10: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol 208: 298–301 [DOI] [PubMed] [Google Scholar]
- Ziebell A, Gracom K, Katahira R, Chen F, Pu Y, Ragauskas A, Dixon RA, Davis M (2010) Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. J Biol Chem 285: 38961–38968 [DOI] [PMC free article] [PubMed] [Google Scholar]