To adjust to iron limitation, Arabidopsis rosettes coordinately down-regulate expression of abundant Fe proteins and key components of the iron-sulfur cluster assembly system in the chloroplasts.
Abstract
Iron (Fe) is an essential element for plants, utilized in nearly every cellular process. Because the adjustment of uptake under Fe limitation cannot satisfy all demands, plants need to acclimate their physiology and biochemistry, especially in their chloroplasts, which have a high demand for Fe. To investigate if a program exists for the utilization of Fe under deficiency, we analyzed how hydroponically grown Arabidopsis (Arabidopsis thaliana) adjusts its physiology and Fe protein composition in vegetative photosynthetic tissue during Fe deficiency. Fe deficiency first affected photosynthetic electron transport with concomitant reductions in carbon assimilation and biomass production when effects on respiration were not yet significant. Photosynthetic electron transport function and protein levels of Fe-dependent enzymes were fully recovered upon Fe resupply, indicating that the Fe depletion stress did not cause irreversible secondary damage. At the protein level, ferredoxin, the cytochrome-b6f complex, and Fe-containing enzymes of the plastid sulfur assimilation pathway were major targets of Fe deficiency, whereas other Fe-dependent functions were relatively less affected. In coordination, SufA and SufB, two proteins of the plastid Fe-sulfur cofactor assembly pathway, were also diminished early by Fe depletion. Iron depletion reduced mRNA levels for the majority of the affected proteins, indicating that loss of enzyme was not just due to lack of Fe cofactors. SufB and ferredoxin were early targets of transcript down-regulation. The data reveal a hierarchy for Fe utilization in photosynthetic tissue and indicate that a program is in place to acclimate to impending Fe deficiency.
Iron (Fe) is a pivotal micronutrient for plants because of its role as a cofactor in proteins involved in electron transport, redox reactions, and catalysis (Balk and Schaedler, 2014). Low bio-availability of Fe in arable soils can decrease agricultural yields and affect the nutritional value of edible plant parts (reviewed in Briat et al., 2015). Fe can be utilized in three different types of cofactors: heme, Fe-sulfur (Fe-S) clusters, or nonheme Fe (Balk and Schaedler, 2014). In plants, the plastid has its own Fe-S cluster biosynthesis machinery (Balk and Pilon, 2011), and it is also the site for the synthesis of heme precursors (Moulin and Smith, 2005). In Arabidopsis (Arabidopsis thaliana) the majority of the Fe in the vegetative shoot (68%) was found to be in the chloroplasts (Shikanai et al., 2003), which indicates that these organelles are a major sink for Fe utilization in green leaves. Therefore, plastids should play a central role in maintaining Fe homeostasis in green tissues, especially when Fe supply becomes limiting.
Research on Fe homeostasis in plants has mainly focused on Fe uptake mechanisms in the roots and Fe distribution within the plants, resulting in the identification of several molecular factors involved in whole-plant Fe homeostasis (for recent reviews, see Kobayashi and Nishizawa, 2012; Brumbarova et al., 2015, Krohling et al., 2016). In addition to Fe uptake and redistribution, plants can also acclimate their metabolism when Fe supply cannot meet all needs (López-Millán et al., 2013). A still-understudied aspect of the biology of Fe is its utilization within plant cells. We hypothesize that plants implement a mechanism for Fe economy that allows a preferential allocation of Fe to the most important functions, when the cellular demand for Fe in photosynthetic tissue exceeds its potential supply by the roots. Are certain enzymes maintained while others are lost, and is there a regulatory Fe economy system in place to mediate such an acclimation? These crucial questions need to be addressed to get a comprehensive view of how plants respond to Fe deficiency.
Nutrient “economy” strategies tend to prioritize, recycle, and remobilize limiting nutrients (Blaby-Haas and Merchant, 2013). Studies on nonphotosynthetic unicellular organisms such as bacteria and yeast have suggested that programs exist to acclimate metabolism to Fe deficiency (Oglesby-Sherrousse and Murphy, 2013; Philpott et al., 2012). Compared to heterotrophs, photosynthetic organisms have to cope with a substantially higher demand for Fe because of the added Fe requirement of the chloroplast (for review, see Blaby-Haas and Merchant, 2013). The response to Fe deficiency in Chlamydomonas rheinhardtii, a unicellular alga and a facultative photo-autotroph, depends on whether the cells grow photosynthetically or heterotrophically (Terauchi et al., 2010; Glaesener et al., 2013). Fe economy in Chlamydomonas gives a central role to the chloroplast and relies on reduced de novo Fe-protein synthesis, turnover of Fe proteins, together with up-regulation of specific enzymes such as Fe superoxide dismutase (Moseley et al., 2002; Page et al., 2012). However, unlike Chlamydomonas, plants absolutely require photosynthesis for growth. In the context of copper (Cu) nutrition, another pivotal trace element for photosynthesis, a molecular remodeling has been proposed both in higher plants and in Chlamydomonas, but the targets of regulation are different in the two kinds of organisms (Burkhead et al., 2009).
In this study, a hydroponic system was used to control available Fe to analyze physiological and molecular level changes upon Fe depletion with an emphasis on chloroplastic Fe-dependent proteins in Arabidopsis vegetative shoots. The results suggest that a defined hierarchy is implemented for Fe utilization in chloroplasts when Fe is scarce in the environment.
RESULTS
Experimental Design and Characterization of Fe Deficiency Symptoms in Arabidopsis Rosettes
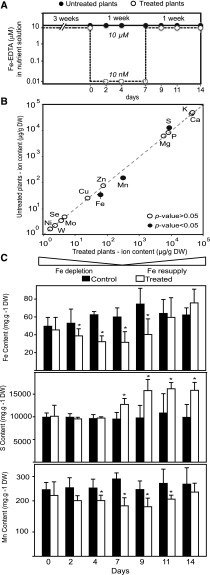
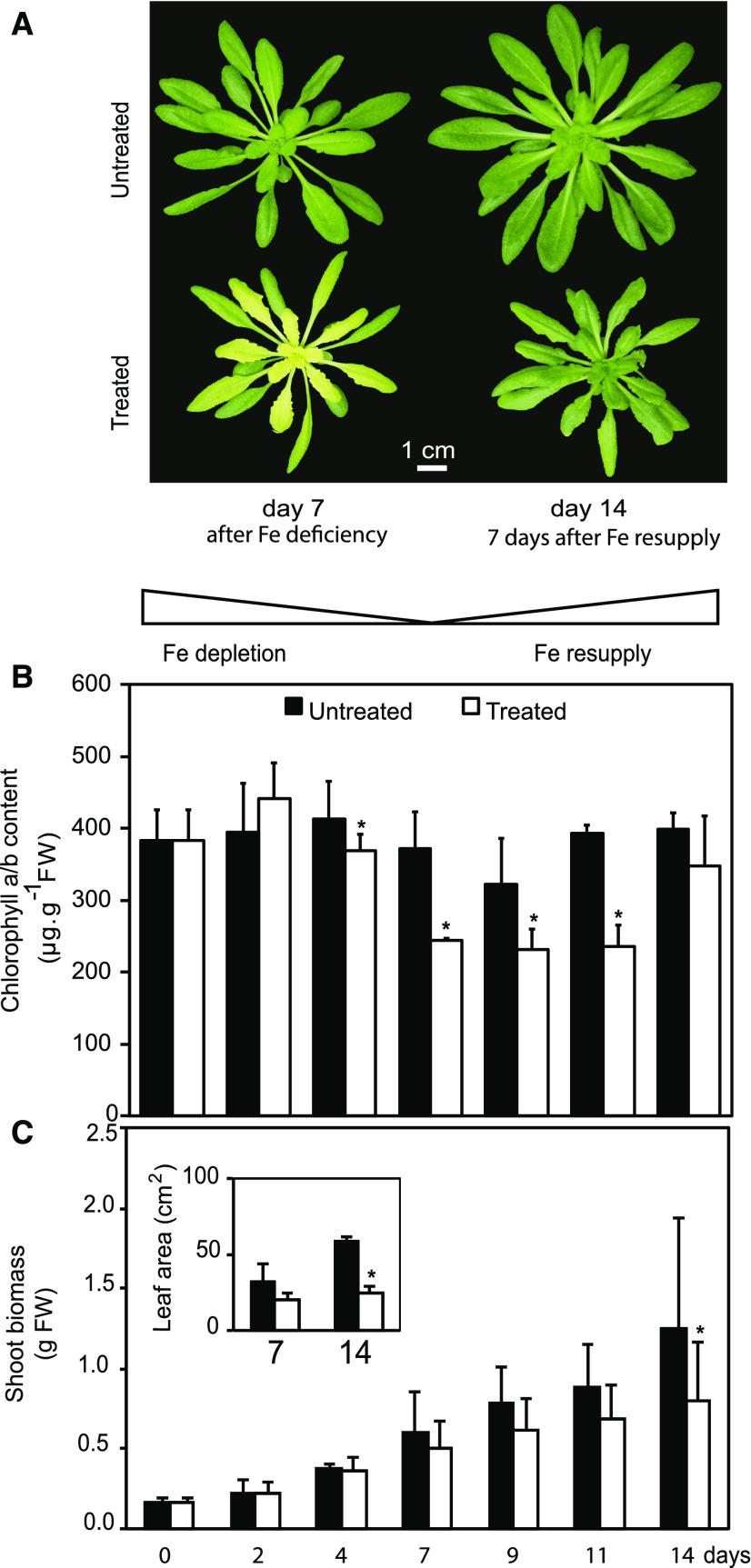
We established hydroponic growth conditions to analyze progressive changes in the vegetative shoot in response to 1 week of low Fe availability followed by resupply (Fig. 1A). As expected, lowering Fe from 10 µm Fe-EDTA in the nutrient solution to 10 nm Fe-EDTA resulted in a strong decrease in Fe content in plant rosettes measured after 7 d (Fig. 1B). A significant decrease in shoot Fe content was measured as soon as 2 d after Fe limitation and became more pronounced after 4 and 7 d (Fig. 1C). For the Fe resupply treatment, Fe in the medium was restored to 10 µm Fe-EDTA 7 d after the initiation of the deficiency treatment (Fig. 1A). This resupply led to a rise of Fe levels in the rosette, which reached control levels after only 4 d of Fe resupply (Fig. 1C). Among the other elements analyzed, only the levels of sulfur (S) and manganese (Mn) showed significant alterations (Fig. 1B). The reduction in the Mn content in shoot tissues correlated with the evolution of Fe levels, albeit the decrease of Mn was not yet significant after 2 d of Fe depletion (Fig. 1C). An increase in S content in shoots was observed after 7 d of Fe deficiency, and the S levels remained high after plants had recovered from the Fe depletion, while Fe and Mn were back to control levels at that time (Fig. 1C). Expression analysis for the Fe status markers ferritin (Petit et al., 2001; Ravet et al., 2009) and IRT1 (Vert et al., 2002; Séguela et al., 2008) indicated that the Fe deficiency response was induced upon depletion and that plants returned to normal Fe status with resupply within a week (Supplemental Fig. S1). As expected, the loss of Fe content during depletion was accompanied by the appearance of chlorosis due to a drop in chlorophyll content, mainly in the young developing leaves of treated plants (Fig. 2, A and B). Importantly, upon Fe resupply, plants were able to fully recover their chlorophyll content (Fig. 2, A and B). Furthermore, leaf morphology was normal, indicating that the visible symptoms of deficiency were fully reversible with resupply (Fig. 2A). Typical of Fe depletion, treated plants had decreased total rosette biomass (Fig. 2C) and lagged in primary root growth (Supplemental Fig. S2). Although Fe-deficient plants quickly recovered after resupply, the Fe deficiency treatment caused a delay in growth and development (Fig. 2, A and C).
Figure 1.
Experimental set up and elemental composition of Arabidopsis rosettes over the experimental time course. A, Arabidopsis plants were grown hydroponically on 10 μm FeIIIEDTA for 3 weeks and then a subset of plants was transferred to 10 nM FeIIIEDTA. After 1 week, the deficient plants were resupplied with 10 μm FeIIIEDTA. Circles represent time points at which data were collected for untreated (black) and treated (white) plants. All analyses were completed before bolting to avoid compounding effects of nutrient reallocation during flowering and seed set. B, Ionome of untreated versus treated plants at day 7. The elemental composition was compared in treated and untreated plants at day 7 (n = 5–7). Concentration, as µg· x g−1 dry weight (DW), of a given element in untreated plants was plotted against its concentration in treated plants. Black circles represent elements that differ significantly (P value < 0.05) in treated plants compared to control. C, Changes in elemental content of Fe, S, and Mn in Arabidopsis rosettes with time. Control (black) and treated (white) plants are compared at each time point in the study with statistical significance denoted by an asterisk (indicating that that treated plants differed from untreated plants on a given time point, P value < 0.05; n = 5).
Figure 2.
Symptoms of Fe-deficient Arabidopsis and impacts on chlorophyll content and growth. A, Appearance of untreated (top) and treated (bottom) Arabidopsis plants. Representative plants were photographed at day 7 (left) and day 14 (right). B, Total chlorophyll a/b content in Arabidopsis rosettes. Values are given as averages ± sd (n = 10). FW, Fresh weight. C, Shoot biomass of Arabidopsis during Fe deficiency and resupply. Growth of the shoot was monitored by measurement of rosette fresh weight. Values at the indicated days of treatment are given as averages ± sd (n = 15). Insert, total leaf area per plant for Fe-deficient plants (day 7) and Fe-resupplied (day 14) plants. Values are given as averages ± sd (n = 15). Black and white bars represent untreated and treated plants, respectively. Stars above bars represent significant differences (P value < 0.05) between untreated and treated plants for a given time point.
Fe Deficiency Has Strong Impact on Light-Dependent Carbon Assimilation
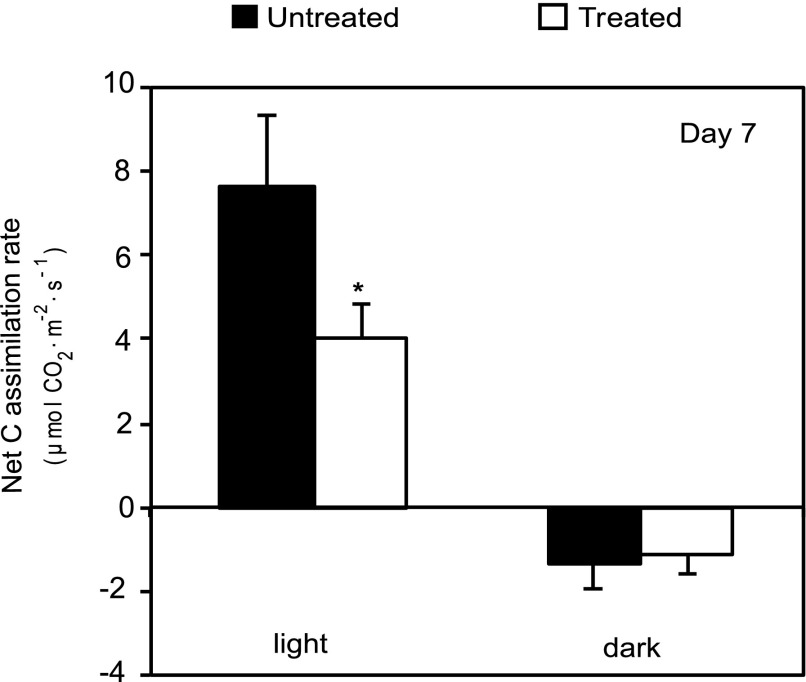
Carbon assimilation and release were measured to get estimates for net photosynthetic capacity in the light and for dark respiration. The net CO2 assimilation rate per leaf area in the light was reduced by about 50% in Fe-deficient leaves after 7 d of treatment, while CO2 release was only slightly, but not significantly, affected in the dark (Fig. 3). These observations suggest that maintenance of mitochondrial respiration activity may be prioritized over photosynthesis when Fe becomes limiting. Seven days after Fe resupply, the net CO2 assimilation rates were no longer different between treated and control plants (not shown).
Figure 3.
Fe deficiency strongly affects photosynthetic CO2 assimilation. Net C assimilation was determined in the light (350 µmol photons x m−2 x s−1) and in darkness at the end of the Fe deficiency treatment. Values are given as averages ± sd (n = 10). Black and white bars represent treated and control plants, respectively. Stars above bars represent significant differences (P value < 0.05) between untreated and treated plants for a given time point.
Fe Deficiency Affects Photosynthetic Electron Transport Primarily in the Younger Leaves
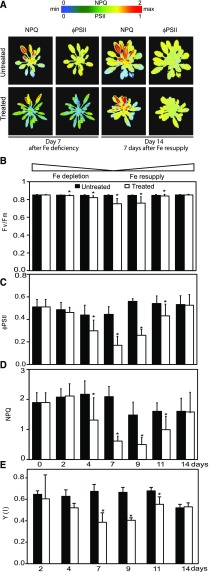
The strong effect of Fe depletion on carbon assimilation led us to investigate the light reactions of photosynthesis. To analyze the spatial distribution of photosynthesis-related symptoms caused by Fe depletion within the rosettes, we used a Fluorcam imaging system. (Fig. 4A). The parameter ΦPSII (PSII efficiency) gives an indication for how much of the light energy absorbed by chlorophyll molecules is used to drive electron transport. The parameter nonphotochemical quenching (NPQ) estimates nonphotochemical quenching in PSII antennae, which depends on acidification of the lumen and thus electron transport activity (Maxwell and Johnson, 2000). Compared to the controls, the Fe-deficient plants had lower values for ΦPSII and NPQ throughout their rosettes, with the strongest effects in the younger leaves (Fig. 4A, left side). Importantly, a week after Fe was resupplied to the plants, both these chlorophyll fluorescence parameters were again identical in control and treated plants, which indicates that the plants had fully recovered their photosynthetic electron transport (Fig. 4A, right).
Figure 4.
Fe deficiency decreases electron transport though the photosynthetic apparatus, primarily in the younger leaves. A, The NPQ and ΦPSII of Arabidopsis untreated (top) and treated plants (bottom). Representative false color chlorophyll fluorescence images are shown for day 7 (Fe deficient) and day 14 (Fe-resupplied) plants. B to D, Chlorophyll fluorescence parameters obtained using a FMS Hansatech system. B, Fv/Fm of dark-adapted plants. C, ΦPSII measured at 250 µmol photons m−2·s−1. D, NPQ measured at 600 µmol photons m−2·s−1. E, Quantum yield of photochemical energy conversion Y(I). Photooxidation/reduction of P700 was monitored as the light-induced absorbance change at 820 nm using a Dual-PAM-100 P700 fluorometer. All PSI parameters were measured over the time course of the treatment with the exception of day 0, because the intermediate leaves of the rosette at this time point were too small to fit in the Dual-PAM-100 leafclip used for the measurements. B to E, Values are given as averages ± sd (n = 6). Black and white bars represent untreated and treated plants, respectively. Stars above bars represent significant differences (P value < 0.05) between untreated and treated plants for a given time point.
Whereas the imaging system provides a spatial analysis of chlorophyll fluorescence, this setup could only reach an actinic light intensity of about 120 µmol photons x m−2 x s−1, which is less than one-half the light intensity used to grow the plants and below the level where saturation of the light reactions typically occurs (Maxwell and Johnson, 2000). Therefore, a FMS fluorometer was used for a more quantitative analysis of chlorophyll fluorescence (Fig. 4, B–D). Because the FMS clamp does not allow measurements on the smallest leaves, we conducted the measurements on intermediate leaves considering that these reflect a behavior representative of the entire rosette (Fig. 4A). The maximum photochemical efficiency of PSII in the dark-adapted state (FV/FM) was only slightly decreased by Fe deficiency (Fig. 4B). This indicates that the majority of PSII centers are still functional after 1 week and that photoinhibition was minor. This observation was confirmed by Fluorcam measurements (not shown). Importantly, the FV/FM recovered with Fe resupply, indicating that long-term damage to PSII was avoided (Fig. 4B). The ΦPSII was measured over a range of light intensities, which showed a consistent trend where the Fe-depleted plants had reduced activity compared to the control plants. For simplicity, only the data obtained at an actinic light intensity close to what was used to grow plants are presented for ΦPSII (Fig. 4C). ΦPSII is strongly affected by Fe deficiency but is also recovered rapidly upon Fe resupply (Fig. 4C). Overall, the time course analysis of the ΦPSII over the depletion and resupply period (Fig. 4C) mirrors the evolution of shoot Fe levels (Fig. 1C). Because NPQ is induced by high light, we chose to analyze the evolution of NPQ over the time course of depletion and resupply at a relatively high light intensity. (Fig. 4D). The NPQ values strongly diminished after Fe levels dropped, indicative of a lack of electron transport and proton pumping. NPQ recovered with Fe resupply (Fig. 4D).
To gain insight into how PSI was affected, changes in the redox state of the PSI were analyzed. The Y(I) parameter indicates the quantum yield of photochemical energy conversion in PSI (Klughammer and Schreiber, 2008; Fig. 4E). Y(I) was significantly diminished by Fe depletion (Fig. 4E). In untreated plants, the PSI quantum yield was already limited by electron donors upstream of PSI, and this limitation was exacerbated further by Fe depletion (Supplemental Fig. S3).
In conclusion, the analyses of the light reactions suggest that PSII is only mildly affected by Fe deficiency (small changes in FV/FM) but that strong defects are seen in electron transport downstream of PSII. On the other hand, PSI function is strongly compromised by Fe deficiency, and this may be in large part because of upstream limitations. The observations thus suggest that Fe deficiency causes a backup in electron transport at the Fe-containing cytochrome-b6f complex that functions downstream of PSII and donates electrons to PSI via the copper protein plastocyanin.
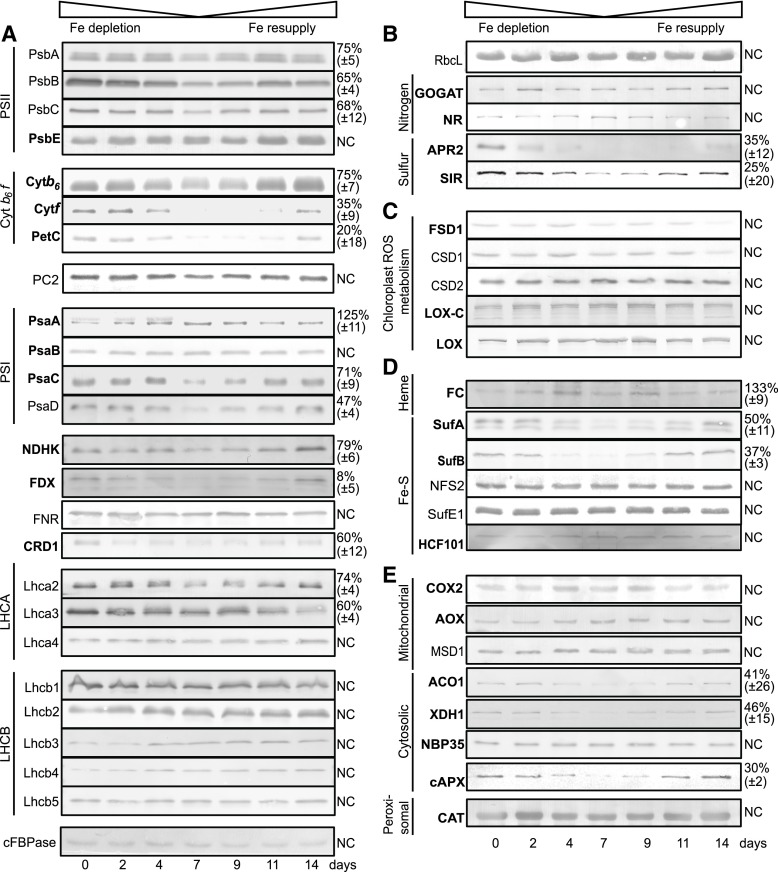
Fe Deficiency Triggers Specific Changes in the Abundance of Fe-Dependent Chloroplast Proteins and Their Transcripts
To study changes in the abundance of selected Fe- and photosynthesis-related chloroplast proteins over the time course of Fe depletion and Fe resupply, we used quantitative immunoblotting (Fig. 5). Fe deficiency caused a decrease in the abundance of several Fe binding proteins and affected all complexes of the electron transport chain (Fig. 5A). However, only mild effects were observed for the subunits of PSII. The three core subunits, PSBA (D1 protein), PSBB, and PSBC, which harbor nonheme Fe (Barber, 2016), were reduced to 65% to 75% of control levels at 7 d of Fe depletion, whereas no major change was noted before this time. No change was seen for the heme-binding protein PSBE (Fig. 5A). PSI contains 12 Fe atoms in three 4Fe-4S clusters bound by the PSAA, B, and C subunits (Merchant and Sawaya, 2005). Surprisingly, the PSAA subunit of PSI, which carries the FeSx cluster, appeared to be slightly increased in abundance after Fe depletion (Fig. 5A). PSAB and PSAC carry two 4Fe-4S clusters between them. These Fe-binding PSI core proteins were only mildly affected by Fe depletion in our conditions (Fig. 5A). However, a stronger effect on protein levels was observed for PSAD, which mediates interactions between the PSI core and ferredoxin but does not bind Fe itself (Sétif et al., 2002), and which was reduced to one-half of the control levels at day 7 of depletion. In comparison to the two photosystems, the effect of Fe depletion was stronger and observed earlier for subunits of the cytochrome-b6/f complex. The strong decrease in cytochrome-f and the [2Fe-2S]-Rieske proteins became evident at 2 d of depletion. NDHK, which is an Fe-S protein of the NDH complex that mediates cyclic electron flow, was also affected. By far the strongest effect of Fe depletion was seen for ferredoxin (FDX). The major FDX2 isoform (Hanke et al., 2004) accumulated to only 8% of control levels and showed already very clear changes at day 2. All proteins that changed abundance in response to Fe returned to control levels with 7 d of Fe resupply. The two Fe-independent electron carriers, plastocyanin (PC) and FNR, were not affected by Fe treatment, indicating that Fe deficiency does not affect all electron transport chain components.
Figure 5.
Impact of Fe deficiency on the abundance of Fe proteins in the Arabidopsis rosette. Total protein extracts (20 µg) from treated Arabidopsis rosettes were fractionated by SDS-PAGE and blotted onto nitrocellulose membranes. The cytosolic Fru-1,6-bisphosphatase was used as a loading control. Immunoblots are presented for: A, proteins related to the photosynthetic light reactions; B, chloroplast metabolism; C, plastid ROS related proteins; D, plastid Fe cofactor assembly, and E, cytosolic, mitochondrial, and peroxisomal Fe related proteins. Protein name is denoted to the left of the immunoblot. Fe-binding proteins are in bold face. Shown are representative blots of at least four independent biological replicates. Immunoblots were quantified on day 7 comparing treated (low Fe) and untreated (control medium) plant samples. Numbers (right) represent the remaining amount of protein (as %) in Fe-deficient plants (treated plants, day7) relative to controls (untreated, day 7) for significantly (P value < 0.05) affected proteins. For proteins that did not show a significant change, NC for “no change” is written to the right of the blot.
The di-Fe enzyme chlorophyll cyclase (CRD1/CHl27) that is required for chlorophyll synthesis (Tottey et al., 2003; Bang et al., 2008) was affected by Fe depletion with about 60% of the protein left after 7 d of depletion (Fig. 5A), which matches chlorophyll levels (Fig. 2B). Similar down-regulation was observed for 7-Hydroxymethyl Chlorophyll a Reductase, a Fe-S cluster enzyme required for chlorophyll a formation (not shown). However, most of the chlorophyll binding light-harvesting complex (LHC) proteins that we could detect were unaffected by Fe depletion with the exception of LHCA2 and LHCA3, which showed significant lower abundance at day 7 (Fig. 5A).
In summary, the analysis of proteins involved in the light reactions indicate that Fe deficiency triggers a large drop in ferredoxin abundance followed by a progressive loss of cytochrome-b6/f complex components. These changes can help explain the loss of photosynthetic electron transport. In comparison, PSI and PSII and associated LHCs are less affected.
No changes were observed for the abundance of Rubisco (C fixation) and the chloroplastic Glu 2 oxoglutarate aminotransferase, a 3Fe-4S enzyme (GOGAT, N assimilation). By contrast, the abundance of two key proteins for S assimilation, adenosine 5′-phosphosulfate reductase (APR), a Fe-S protein, and sulfite reductase (SIR), a heme and Fe-S cluster protein, was strongly decreased. Fe depletion had minor effects on the major chloroplast Fe- superoxide dismutase FSD1 and Cu/zinc (Zn)-superoxide dismutases CSD1 (cytosol) and CSD2 (chloroplast). Two plastid lipopoxygenases that are nonheme Fe proteins were also unaffected (Fig. 5C).
Two major Fe cofactor types are Fe-S clusters and heme. Fe-S clusters for plastidial proteins are synthesized from Fe and sulfide within the chloroplast by the SUF system (Balk and Pilon, 2011). Strikingly, two of the Fe-S assembly system components, SUFA and SUFB, were strongly reduced in abundance (−50%; Fig. 5D). Sulfide of Fe-S clusters originates from desulfurization of Cys. This is achieved in plant plastids by the Cys desulfurase NFS2, which is activated by SUFE1 (Pilon-Smits et al., 2002; Ye et al., 2006). Both NFS2 and SUFE1 remained unaffected by Fe depletion. Similarly, the putative Fe-S scaffold HCF101 was also not affected. Heme synthesis requires ferrochelatase (FC) to incorporate Fe into the porphyrin structure (Cornah et al., 2003). Strikingly, FC protein levels were maintained during the Fe depletion (Fig. 5D).
COX2, which is a core subunit of mitochondrial cytochrome-c oxidase, a heme- and Cu-containing complex, was not affected in abundance by Fe depletion. Similarly, the mitochondrial di-Fe protein alternative oxidase (AOX) was also not affected (Fig. 5E). However, strongly affected by Fe depletion were the cytosolic proteins Aconitase1 (ACO1, 4Fe-4S cofactor), xanthine dehydrogenase (2Fe-2S cofactor) as well as heme-containing cAPX (Fig. 5E). However, the cytosolic nitrate reductase, which also carries heme, did not change in abundance. We tested NBP35, a component of the cytosolic Fe-S biosynthesis machinery (Bych et al., 2008), but we did not observe an effect of the Fe treatment on its abundance (Fig. 5E). Catalase, a major peroxisomal Fe protein, was maintained during Fe deficiency (Fig. 5E).
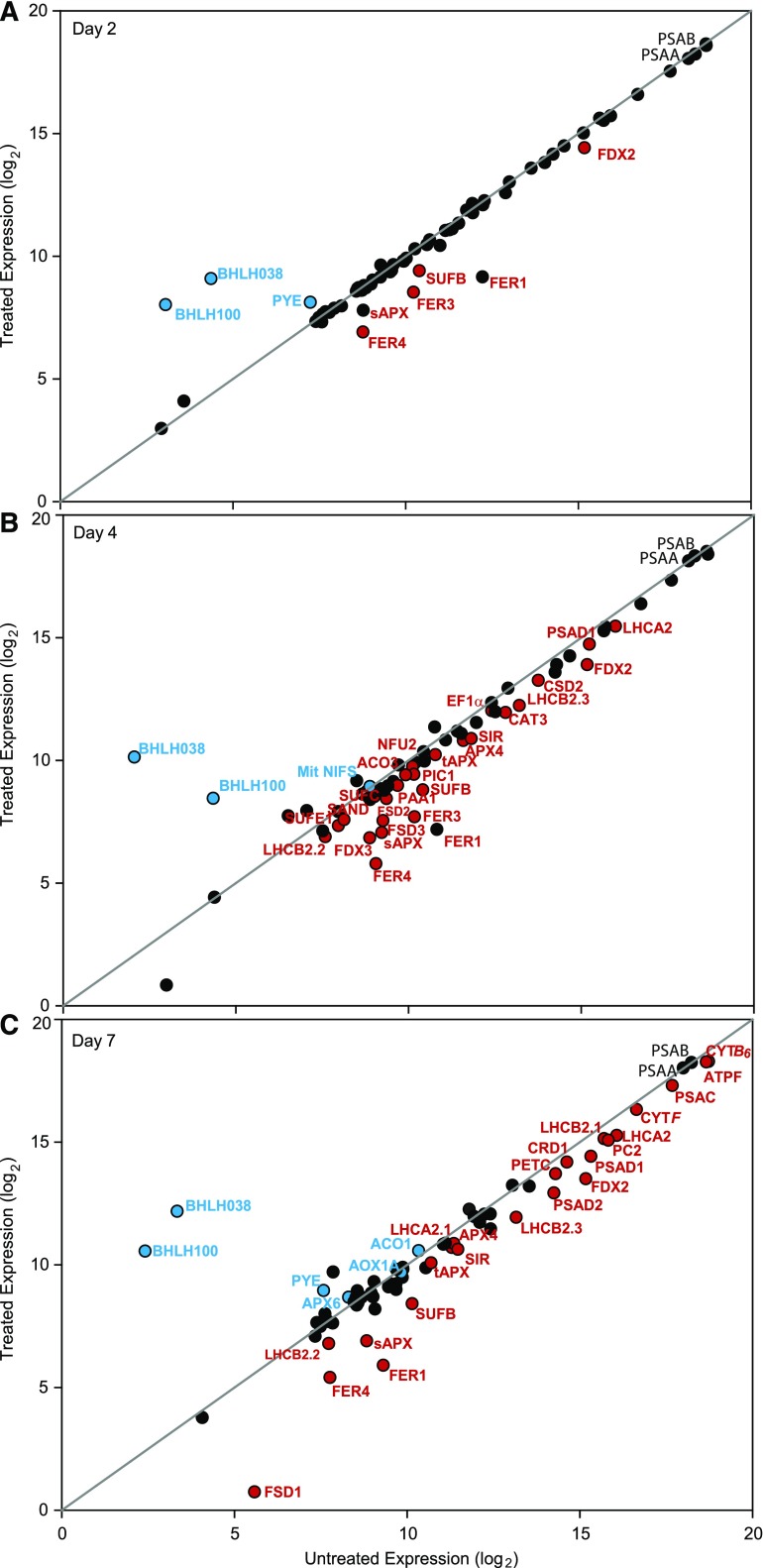
We used NanoString Technology for transcript analysis (Fig. 6), because it can measure transcripts of both nuclear and plastid encoded genes; it is suitable for measurements with multiple time points with low variability between biological replicates and good dynamic range (Malkov et al., 2009; Veldman-Jones et al., 2015). In our set of genes for interrogation, we included the mRNA for all proteins that had shown a change in response to Fe deficiency (Fig. 5). Multiple isoforms were tested when appropriate. We complemented our list with genes encoding major Fe binding protein subunits and known Fe homeostasis factors for which antibodies are lacking. A complete list of target genes is presented in Supplemental Table S1.
Figure 6.
Impact of Fe deficiency on the abundance of Fe-related transcripts in the Arabidopsis rosette. Total leaf RNA was analyzed for transcript abundance with NanoString Technology. Normalized mRNA expression for treated and control plants is compared using a log2 scale. UBIQUITIN11 and ACTIN2 were used for normalization. A, Day 2 after Fe deficiency (n = 4). B, Day 4 after Fe deficiency (n = 3). C, Day 7 after Fe deficiency (n = 4). Black circles represent unchanged transcripts, red circles represent significantly decreased transcripts, and blue circles represent significantly increased transcripts (P value < 0.05).
The transcript abundance of each gene after 2, 4, or 7 d of Fe deficiency was plotted using a log2 scale against its control value (Fig. 6, A–C). Unaffected transcripts remained on the diagonal, transcripts with lower expression after deficiency fell below the diagonal, whereas up-regulated transcripts were above. Abundance under control conditions increases to the right in the plots. As expected, after 2 d of Fe deficiency, FER1 expression was decreased, while PYE, BHLH38, and BHLH100, markers for plant Fe deficiency (Long et al., 2010; Sivitz et al., 2012), were up-regulated. Generally, a good correlation was observed between protein and mRNA abundance. For proteins that did not change in abundance such as chloroplast encoded PSAA and PSAB, the corresponding transcript levels were also stable. In some cases (AOX1, CSD1), we noted a slight increase in transcript abundance late in the deficiency treatment. Conversely, a decrease in protein level was generally accompanied by a lower transcript level also. There were, however, noticeable exceptions. Whereas SUFA, APR2 (the major APR isoform), and ACO1 proteins showed much lower accumulation at the protein levels under Fe depletion, the transcripts for these genes did not diminish over the course of 7 d. It is also noteworthy that transcripts encoding proteins proposed to be involved in Fe transport over the plastid envelope (PIC1, FRO6, FRO7, YSL4, YSL6) showed small, statistically not significant changes after Fe depletion.
Strikingly, among the Fe-related components tested at the transcript level, only SUFB and FDX2 were decreased significantly in Fe-starved plants compared to control at 2 d after Fe depletion (Fig. 6A; Supplemental Fig. S4). For sAPX, a trend toward reduced transcript accumulation was already seen at day 2, but it was not yet significantly different from the control (Supplemental Fig. S4). Transcripts for SUFB, FDX2, and sAPX remained low over the 1-week Fe depletion. After 4 and 7 d, many additional genes showed reductions in transcript levels in treated versus control plants, including both chloroplastic and cytosolic transcripts encoding abundant subunits of the photosynthetic machinery (Fig. 6C). PSAA and PSAB transcripts, however, were unchanged (Fig. 6C), consistent with protein levels. SIR expression was reduced by Fe depletion, but in contrast, the transcript for nitrite reductase (NIR), which carries the same Fe-S siroheme cofactor, was not affected (Fig. 6). Some of the transcripts such as CSD1, AOX1, and ACO1 appeared to become up-regulated toward the end of the deficiency treatment.
DISCUSSION
Responses to Low Fe Target Specific Chloroplast Functions Progressively
The main effects on Fe-related chloroplast proteins are summarized in Supplemental Figure S5. It can be envisioned that Fe deficiency causes a loss of Fe proteins due to lack of cofactor availability, but in addition, the Fe proteome can be affected by the regulation of gene expression. The data presented in Figures 5 and 6 indicate that during Fe deficiency, specific abundant Fe proteins of the chloroplast such as ferredoxin and the cytochrome-b6f complex are targeted for down-regulation at the transcript level, which has consequences for photosynthesis and other chloroplast metabolism. There are therefore priorities in the regulation of protein and transcript levels in response to low Fe. The implication is that novel, thus-far unrecognized regulatory circuits to regulate Fe economy must exist and that Fe protein abundance is not just controlled passively by cofactor availability. This regulation of Fe utilization in green tissue has not been studied in detail in plants before. We propose that Fe economy is an important mechanism to cope with impending Fe deficiency and functions in concert with the up-regulation of root Fe uptake, and the adjustment of metabolism (López-Millán et al., 2013).
The symptoms observed in Arabidopsis within the first 7 d of Fe depletion treatment were almost all fully reversible with resupply of Fe (Figs. 1, 2, 4, and 5). It is thus likely that secondary and irreversible damage was avoided in the depletion protocol, especially in the first days. Therefore, the changes at the molecular level after the onset of Fe-depletion (Figs. 5 and 6), especially the earlier events, are likely to reflect programmed responses that could help the plant to acclimate to a temporary lack of Fe.
The Fe depletion treatment affected photosynthesis, especially in the younger leaves, much more than respiration, albeit that photosynthesis was not completely blocked even after 7 d (Fig. 4). Chloroplast Fe proteins and their transcripts were among the most affected, while generally milder effects were seen for the mitochondrial Fe proteins (Figs. 5 and 6). Several cytosolic proteins were also affected (Fig. 5E), but transcripts for these were mostly stable relative to transcripts for chloroplast proteins, which dominated the pool of differentially regulated genes (Fig. 6). These observations may be taken as an indication that photosynthesis is more dispensable under Fe deficiency compared to respiration, which also has a strong demand for Fe (Balk and Schaedler, 2014). However, the effect of Fe depletion on the abundance of Fe proteins of the chloroplast was not uniform. For instance, plastid enzymes involved in S assimilation (APR and SIR) were strongly affected, whereas expression of the N assimilation enzymes plastid GOGAT (plastid), NIR (plastid), and nitrate reductase (NR, cytosol) seemed to be less affected (Figs. 5 and 6).
For practical reasons, protein and mRNA levels were analyzed in the entire rosette. It is possible that protein and mRNA were more affected in the youngest leaves, which showed the strongest physiological response. However, the timing of effects also varied for different genes, and the temporal analysis of gene expression was especially useful in discerning early and later responses (Supplemental Fig. S4). Lowering Fe in the media resulted in a measurable reduction in shoot Fe levels after 2 d, when other elements and physiological parameters were not significantly changed yet. Nevertheless, already at day 2 on the molecular level, a clear and notable reduction in the accumulation of the SUFB and leaf ferredoxin (FDX) proteins was observed, which was also accompanied by a reduction in transcript levels. These observations suggest that down-regulation of FDX, a 2Fe-2S protein, and SUFB, a factor required for all plastid FeS synthesis, is a priority under Fe deficiency in the vegetative shoot.
Chlorophyll levels and photosynthetic electron transport parameters became significantly affected at 4 d of depletion, when symptoms had become more severe and remained low until after Fe resupply. Most light harvesting complex proteins were mildly affected by Fe depletion, which agrees with previously reported effects of Fe depletion on thylakoid proteomes (Laganowsky et al., 2009). Exceptions were LHCA2 and LHCA3, which also showed transcriptional regulation. The latter may reflect a remodeling of the PSI antennae to adjust low Fe (Laganowsky et al., 2009; Rodríguez-Celma et al., 2013), as has also been suggested for Chlamydomonas cells depleted of Fe (Moseley et al., 2002). Of the Fe proteins in the major complexes of the photosynthetic electron transport chain, the subunits of the cytochrome-b6f complex were most affected, more than the Fe binding subunits of PSI, while minor damage and no transcriptional down-regulation was observed in PSII as a result of Fe depletion.
Fe Sparing in Chloroplasts Targets Abundant Fe Proteins
By far the largest loss in protein abundance was observed for ferredoxin, especially for FDX2 (Supplemental Fig. S5). Does the down-regulation of FDX2 make sense in the context of Fe economy? In regard to this question, it is of interest to see if photosynthesis-related symptoms of Fe deficiency resemble published phenotypes of ferredoxin mutants (Hanke and Hase, 2008; Voss et al., 2011; Liu et al., 2013). There are several FDX isoforms in Arabidopsis (Hanke et al., 2004). Due to the possible partial redundancy of ferredoxin isoforms, mutants in FDX2 show mild phenotypes (Hanke and Hase, 2008; Voss et al., 2011). This may also explain why in Fe deficiency we see FDX2 protein decrease early (day 2), but we do not see effects on photosynthesis until later (day 4) when also the cytochrome-b6f complex and to a lesser extent PSI and the NDHK subunit of the NDH complex become affected. Even at 7 d of deficiency when FDX protein levels had dropped by approximately 90%, photosynthetic electron transport was still at about 50% of capacity compared to control Fe-replete conditions. FDX2 comprises roughly 90% of the ferredoxin protein in green tissue (Hanke et al., 2004), and the transcript analysis indicates that FDX2 is the most abundant nuclear encoded mRNA for an Fe protein (Fig. 6). FDX is a 2Fe-2S protein and its down-regulation thus will have a relatively large effect on Fe quota. Because FDX proteins seem to accumulate in excess in replete conditions, down-regulation of FDX2 seems to be an efficient way to economize Fe utilization.
How do the symptoms of Fe deficiency compare to electron transport phenotypes of FDX mutants? Genetic loss of FDX2 is reported to result in mild phenotypes and affected mainly photoreduction of NADP, while cyclic electron flow via the less abundant FDX1 isoform or alternative Fe-S proteins may help maintain NPQ in FDX2 mutants (Voss et al., 2011; Liu et al., 2013). Chlorophyll fluorescence measurements indicated that both ΦPSII, which is indicative of linear electron flow, and NPQ, which depends on both linear and cyclic electron flow (Maxwell and Johnson, 2000), are affected by Fe deficiency. The observed lower NPQ in Fe deficiency is expected in view of the observed loss in abundance of not only ferredoxin but also cytochrome-b6f complex subunits and a subunit of the NDH complex, which are all required for both linear and cyclic electron flow and, therefore, induction of NPQ.
The second most abundant nucleus-encoded transcript for an Fe protein is PETC (Fig. 6), which encodes for the Rieske 2Fe-2S-binding subunit of the cytochrome-b6f complex. Rieske protein was also strongly down-regulated, which involved transcript abundance changes, albeit this occurred later than for FDX. It is interesting that the two heme-containing and chloroplast-encoded subunits of the cytochrome-b6f complex, Cytf and Cytb6, also undergo transcriptional down-regulation. The dimeric cytochrome-b6f complex is reported to contain 12 Fe atoms, the same number as is present in PSI (Merchant and Sawaya, 2005). Apparently, under impending Fe deficiency, down-regulation of the cytochrome-b6f complex is preferred over removal of PSI, for which the core subunits are chloroplast encoded and the transcripts have an abundance comparable to the cytochrome-b6f complex subunits. However, the nucleus encoded PSAD subunit, which does not itself bind Fe, was strongly down-regulated in coordination with FDX, for which it contributes to the docking site on PSI.
It is of interest to place our findings in the context of studies that have reported on the general effects of Fe deficiency on chlorophyll synthesis and photosynthesis that have been well documented in the literature (Spiller and Terry, 1980; Andaluz et al., 2006; Nishio and Terry, 1983; Nishio et al., 1985; Sharma, 2007; Timperio et al., 2007; Laganowski et al., 2009; Msilini et al., 2011; Ciaffi et al., 2013; Paolacci et al., 2014; Rodríguez-Celma et al., 2013). Some studies on Fe depletion in plants reported that PSI was the major target of Fe deficiency (Nishio et al., 1985; Timperio et al., 2007), whereas we saw a major effect on the cytochrome-b6f complex and less on PSI. The severity of the applied Fe deficiency, differences in light conditions, and the use of different species may all help determine if PSI or the cytochrome-b6f complex is more affected. Our data are consistent, however, with a study on the effects of mild Fe deficiency in hydroponics on the thylakoid proteome of Arabidopsis (Laganowsky et al., 2009). In Chlamydomonas, Fe deficiency activates a genetic program in which remodeling of pigment binding proteins associated with PSI occurs (Moseley et al., 2002). Furthermore, in Chlamydomonas maintenance of respiration is favored over photosynthesis in the chloroplast (Terauchi et al., 2010). Within the Chlamydomonas chloroplast abundant Fe proteins such as ferredoxin and cytochrome-f were strongly diminished by Fe deficiency (Page et al., 2012), similar to what we found in Arabidopsis. However, in Chlamydomonas, FeSOD was maintained and even up-regulated under deficiency (Page et al., 2012) but this is not the case in Arabidopsis (Fig. 6), where the major control over FSD1 expression depends on Cu levels and is mediated via the SPL7 transcription factor (Yamasaki et al., 2009).
To start to uncover potential mechanisms of Fe economy, we have focused our studies on major Fe proteins of the chloroplast. Untargeted proteomics approaches, as first pioneered with wild-type tomatoes and cloronerva mutans that lack the metal-chelating nicotianamine molecule (Herbik et al., 1996), have the potential to uncover other mechanisms of acclimation to low Fe such as the adjustment of metabolism. A number of studies in various species have investigated the proteome changes after Fe deficiency, the majority focusing on the root proteomic response (for review, see López-Millán et al., 2013). What can be found in proteomic studies will depend heavily on the experimental protocol and the methods used to fractionate samples and detect proteins even if the same plant species is used. For example, it is not surprising that studies in Arabidopsis aimed to detect changes upon low Fe treatment in either the thylakoid proteome (Laganowsky et al., 2009) or the root phosphoproteome (Lan et al., 2012) upon low Fe treatment show no overlap. Proteomic studies can however help reveal specific responses that might help plants to acclimate to low Fe by adjustments of metabolism (López-Millán et al., 2013). In general, proteomic studies have shown changes in protein abundance of oxidative stress response-related proteins such as increases in CuZnSOD, MnSOD, and peroxidase and a decrease in catalase (López-Millán et al., 2013). However, proteomic studies on tomato leaves showed a decrease in CuZnSOD and an increase in cytosolic ascorbate peroxidase with Fe depletion (Herbik et al., 1996). This study shows little effect of Fe deficiency on CuZnSODs or cytosolic ascorbate peroxidase, which might be because the Fe limitation was mild and reversible.
Effects of Fe Deficiency on Plastid Fe-S Assembly and S Metabolism
Two members of the chloroplast Fe-S assembly system, SUFA and SUFB, were strongly affected by Fe depletion. SUFB is a key component of the plastid SUF-BCD complex that functions as a scaffold required for the assembly of all plastid Fe-S clusters (Hu et al., 2017b). Down-regulation of SUFB in Fe deficiency had been reported but only at the transcript level, both in Arabidopsis grown on agar media (Xu et al., 2005; Sivitz et al., 2012; Rodríguez-Celma et al., 2013) and in rice (Oryza sativa; Liang et al., 2014). The now observed very early down-regulation of SUFB protein and transcript (Figs. 5 and 6; Supplemental Fig. S4) when other Fe deficiency symptoms are still mild suggests that SUFB may play a special role in the acclimation to low Fe. SUFB is also unique in its regulation in mycobacteria where SUFB protein activity is regulated via an intein, a self-cleaving peptide sequence inserted into the polypeptide (Huet et al., 2005; Topilina et al., 2015). Furthermore, in apicomplexan parasites, SufB is the only suf component of the apicoplast (plastid) that is encoded within the small apicoplast genome (Lim and McFadden, 2010) that may allow for regulation of SufB in response to local conditions within the organelle. SufB and SufC are the evolutionarily most ancient core of the Fe-S system (Boyd et al., 2014), and it is likely that the first association of Fe and S requires SufB (Balk and Pilon, 2011). In the absence of sufficient Fe, the risk of forming incomplete clusters may have to be avoided to prevent oxidative damage via Fenton chemistry.
The protein abundance of the putative SUFA/CpIscA (Abdel-Ghany et al., 2005b) Fe-S carrier was lower after Fe deficiency, whereas the transcript was not affected. It is possible that for this protein a lack of cofactor causes instability of the resulting apoprotein. A similar mechanism may apply to the Fe-S enzyme APR. For SUFA/CpIscA, it had been observed that the protein is also reduced in abundance in mutants for nfu2, a potential scaffold or transfer protein in the plastid Fe-S assembly pathway (Yabe and Nakai, 2006). The chloroplast Cys desulfurase NFS2 may also be required for other S-dependent cofactor synthesis pathways, which are likely important to maintain (Pilon-Smits and Pilon, 2005) and may be why NFS2 was maintained with Fe deficiency together with its SUFE1 and SUFE3 partners. It is possible that plastid heme synthesis remains required for other cellular compartments (Tanaka et al., 2011; Balk and Schaedler, 2014), which could be why ferrochelatase protein was not affected by Fe depletion.
Two key chloroplastic enzymes of the S reduction pathway, APR and SIR, were strongly affected by Fe depletion. The rapid loss of APR may help prevent the accumulation of toxic sulfite in the absence of sufficient SIR activity. Together a lack of APR and SIR should result in lower levels of reduced S compounds including glutathione. Lack of reduced S compounds should cause up-regulation of plant sulfate uptake, which explains the higher S content after Fe deficiency (Koprivova and Kopriva, 2014); indeed, Fe deficiency was shown to increase S uptake capacity in tomato and wheat (Ciaffi et al., 2013; Zuchi et al., 2015)
In plant shoots, micronutrient economy, the idea that certain proteins under deficiency are preferred targets for down-regulation to benefit cofactor delivery to other essential functions, is documented for Cu (Burkhead et al., 2009) and Zn (Li et al., 2013). The down-regulation at the transcript level of abundant Fe proteins, coordinated with the SUFB key factor of the Fe-S assembly system, should help to safely economize and perhaps prioritize Fe during Fe deficiency.
Iron Economy Involves Specific Response Programs
The transcriptional responses are specific. For instance, the transcript for SIR is down-regulated by low Fe, but the transcript for NIR, which carries the same Fe-S siroheme cofactor, is not. Therefore, at least some of the observed effects seem to be caused by Fe deficiency proper and not just by downstream defects due to lack of cofactor assembly. We can also compare consequences of Fe deficiency at the molecular level with effects caused by genetically induced loss of Fe-S assembly factors NFS2 (Van Hoewyk et al., 2007) and SUF-BCD (Hu et al., 2017a, 2017b). Loss via RNAi of NFS2 activity (Van Hoewyk et al., 2007) or SUF-BCD activity (Hu et al., 2017b) caused physiological effects after 1 week that are very similar to the Fe deficiency described here (Figs. 1, 3, and 4). RNAi lines induced for loss of NFS2 or SufBCD complex components had shown a general loss of all Fe-S proteins, including strong effects on PSI subunits PSAA, and PSAB on GOGAT (Van Hoewyk et al., 2007; Hu et al., 2017b). However, we did not see this general loss of plastid Fe-S cluster proteins for Fe deficiency. Conversely, whereas we noted down-regulation of the heme containing subunits of the cytochrome-b6f subunits together with PETC (Rieske protein) in Fe deficiency, the genetically induced loss of SUFB, SUFC, or SUFD strongly affected Rieske protein but had marginal effects on CYTb6 and CYTf protein accumulation (Hu et al., 2017b).
Some of the late effects on transcript levels (Fig. 6C) could be indirect, however. Plastocyanin expression depends on active photosynthesis (Vorst et al., 1993), which may be why we observed slightly lower transcript levels for the major PC2 isoform at day 7 of Fe depletion (Fig. 6). After 7 d of Fe depletion, the major FeSOD, FSD1, was down-regulated at the transcript level and the cytosolic Cu/ZnSOD, CSD1, was up-regulated at the transcript level, albeit protein levels were not notably changed. FSD1 and CSD1 transcripts respond to Cu (Abdel-Ghany et al., 2005a, Yamasaki et al., 2009), which might become more available when Fe is limited. Even if total shoot Cu levels did not change significantly, the lack of biomass formation could have shifted intracellular available Cu pools, affecting FSD1 and CSD1 expression (Pilon, 2017).
An important implication of the specific regulation of chloroplast Fe proteins is that there has to be a mechanism to sense the Fe status and relay this signal. As expected, ferritin transcripts (FER1, 3, 4) were also down-regulated at 2 d after the start of low Fe treatment, similar to SUFB and FDX2 (Fig. 6). At the same time, the transcripts for selected known low Fe markers BHLH38, BHLH100, and PYE were up. FIT and PYE are major regulators of root Fe uptake (for review, see Brumbarova et al., 2015). PYE associates with similar BHLH transcription factors (Selote et al., 2015). Four other BHLH transcription factors, BHLH38, BHLH39, BHLH100, and BHLH101, are Fe responsive (Wang et al., 2007). BHLH100 and BHLH101 act separately from PYE and from BHLH38, BHLH39, and FIT, which interact to mediate regulation of root Fe uptake (Yuan et al., 2008) but all are implied in the regulation of Fe uptake or distribution between tissues (Long et al., 2010, Sivitz et al., 2012, Selote et al., 2015). The bHLH family of transcription factors thus plays an important role in Fe homeostasis. Lack of whole-plant Fe uptake or lack of Fe allocation to the shoot as observed with mutants for PYE, FIT, BHLH100, and BHLH101 caused stronger Fe deficiency for shoots. Indeed, in an experiment on agar media with a BHLH100/101 double-mutant, the down-regulation of low Fe-responsive genes including FDX2 was stronger, not weaker, indicating exacerbated Fe depletion (Sivitz et al., 2012). Therefore, these bHLH transcription factors do not directly mediate the observed local down-regulation of photosynthetic Fe proteins and ferritin, but other bHLH proteins might still be involved. Early changes in FDX2 and SUFB transcript and protein levels due to Fe deficiency suggest transcript abundance regulation. We have analyzed the promoter regions of SUFB and FDX2 for promoter motifs via the online tool, AthaMap (Steffens et al., 2004). The SUFB promoter does not show any bHLH binding motifs, while the FDX2 promoter shows only a very low probability of bHLH binding, with only one putative bHLH binding site. At this point, the mechanism for sensing Fe status in the plant shoot and the relay mechanism is unclear, as it is in Chlamydomonas (Blaby-Haas and Merchant, 2013). Knowing how the chloroplast Fe proteome responds locally gives a starting point for unraveling this important novel regulatory circuit in the future.
CONCLUSION
Fe depletion causes specific effects on the chloroplast Fe proteome, with strong and early down-regulation of SufB and ferredoxin, followed by the cytochrome-b6f complex. These changes affect photosynthesis. Importantly, these observations imply the existence of a hitherto undiscovered program that mediates Fe economy. The observations provide a foundation to investigate further the mechanisms by which Fe is sensed locally in photosynthetically active plant cells.
MATERIALS AND METHODS
Plant Material, Hydroponic Growth Conditions, and Plant Sampling
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were surface sterilized by three consecutive 5-min rinses with 70%, 95%, and 70% ethanol, air dried, and stratified for 3 d at 4°C. Seeds were germinated on vertical half-strength Murashige and Skoog (MS) medium (Sigma-Aldrich) supplemented with 1% (w/v) Suc (Sigma-Aldrich), 1% (w/v) phytoagar (Research Products International), and 10 µm FeIII-EDTA (Caisson Labs) in a growth chamber under controlled conditions (in a 8-h-/16-h-light/-dark cycle, 200 µmol photons x m−2 x s−1, HR 70%, 23°C/20°C day/night). The hydroponics setup was in the same growth chamber. One-week-old seedlings were transferred to hydroponics, in 5-L containers, holding one-fifth strength modified Hoagland’s solution (Ravet et al., 2011), containing 10 µm FeIII-EDTA. Preliminary experiments showed that 10 µm FeIII-EDTA is the lowest amount of Fe allowing Arabidopsis growth without visible signs of Fe deficiency symptoms such as growth retardation or chlorosis. The nutrient solution was replaced once a week. Seedlings were grown for 3 weeks in hydroponics before treatment. Then untreated plants (control) were maintained on 10 µm FeIII-EDTA for 2 additional weeks, while the treated plants were subjected to one week of Fe depletion (Fe limited to 10 nm FeIII-EDTA), followed by 1 week of Fe resupply (restored to 10 µm FeIII-EDTA). Pilot experiments had shown that all the Fe deficiency symptoms were reversible with 1 week of Fe resupply, albeit the plants that had been subjected to depletion lagged in size. To obtain enough biological material, we grew 32 plants per hydroponics container. For each experiment, five containers were started, two for deficiency treatment, two for control, with an extra container to replace plants whose roots had sustained damage during transfer from agar media. Data are reported for plants from eight different hydroponics experiments, which passed quality control by visual inspection and chlorophyll fluorescence imaging that was utilized as a noninvasive method to assess plant health and Fe deficiency.
For shoot material, three whole rosettes were pooled as a unique sample. For one root material sample, the entire root system from three plants was pooled. Material for molecular analyses was immediately frozen in liquid nitrogen, and stored frozen until further analysis. Samples from both treated and untreated plants were collected at day 0 (before treatment), after 2, 4, and 7 d (Fe-depletion period), and after day 9, 11, and 14 (Fe resupply period). All sampling was at 2 h after the start of the light period.
Elemental Analysis
Rosettes were dried for a week at 55°C. Fifty milligrams of homogenized dried tissue was digested in 1 mL of nitric acid and heated for 2 h at 60°C and for 6 h at 130°C. Resulting digests were diluted up to 10 mL with di-deionized water and analyzed using inductively coupled plasma-atomic emission spectrometry as described (Ravet et al., 2011).
Chlorophyll, Leaf Area Determination, and Gas Exchange Measurements
Total chlorophyll was extracted from frozen tissue and chlorophyll a and b concentrations were determined using A662 and 645 nm as previously described (Porra et al., 1989). Leaf area was measured with a conveyor belt system attached to the LI-3050C in RealTime Capture mode (Li-Cor, Inc.). Measurements were made using electronic rectangular approximation with 1-mm2 resolution. Gas exchange measurements were obtained using a LI-6400 portable photosynthesis system (Li-Cor, Inc.) with a custom-built cuvette that enclosed the whole rosette of Arabidopsis (Christman et al., 2008). For daytime measurements, cuvette light intensity was at 350 µmol photons x m−2 x s−1. CO2 was maintained at 400 ppm. Temperature and relative humdity in the gas exchange cuvette were set to ambient growth chamber conditions.
Chlorophyll Fluorescence and Redox State of P700 Analyses
Chlorophyll fluorescence images were captured at room temperature using a FluorCam (PSI) as described (Ravet et al., 2011). Chlorophyll fluorescence numeric measurements were obtained using a FMS2 Fluorometer (Hansatech Instruments). Plants were dark adapted for 30 min prior to analysis. PSII maximum capacity (FV/FM), efficiency of the PSII (ΦPSII), and NPQ were calculated according to Maxwell and Johnson (2000). Photooxidation/reduction of P700 was monitored in dark-adapted leaves at 22°C as the light-induced absorbance change at 820 nm (ΔA820) using a Dual-PAM-100 P700 fluorometer (Walz; Klughammer and Schreiber, 2008). The quantum yield of photochemical energy conversion Y(I), the quantum yield of nonphotochemical energy conversion due to donor side limitation Y(ND), and the quantum yield of nonphotochemical energy conversion due to acceptor side limitation Y(NA), where Y(I) +Y(ND) + Y(NA) = 1, were calculated according to Klughammer and Schreiber (2008). Data shown are results from analyses conducted at a light intensity of 354 µmol photons x m−2 x s−1.
Immunoblotting and Protein Quantitation
Soluble proteins for SDS-polyacrylamide gel analysis were extracted as described (Abdel-Ghany et al., 2005b). Protein concentration was determined according to Bradford (1976) using bovine serum albumin as a standard. For western blotting, 20 μg of total protein was separated by 10 to 15% SDS-PAGE and then transferred onto a nitrocellulose membrane. Each experiment was done at least in biological triplicate with identical results, and representative gels are shown. Quantification of the signals was performed as described (Ravet el al., 2011) using calibration curves based on dilution series (1.5, 1, 0.75, 0.5, 0.25, 0) of control (untreated day 7) samples. Signal intensity was determined using ImageJ software, and regression curves (R2 > 0.9) were determined using SigmaPlot software (version 7.4; Systat Software). Information related to the antibodies used for immunodetection of the various proteins is listed in Supplemental Table S1. In untreated plants, kept on sufficient Fe, all tested proteins were equally expressed over the time course of the experiments except for APR, which increased slightly in abundance in older rosettes in the control plants (not shown).
mRNA Expression Analysis
Total RNA was extracted using the Trizol reagent (Life Technologies) according to the manufacturer’s recommendations. The RNA concentration of samples was determined using a Nanodrop Fluorospectrometer and quality was assessed by agarose electrophoresis of aliquots (Sambrook and Russell, 2001) before shipping RNA to the nanostring core facility. Gene expression was analyzed at the University of AZ Genetics Core in Tucson, AZ by NanoString Technology (Geiss et al., 2008). RNA quality was further assessed by analyzing a 1:10 dilution of each total RNA sample in a Fragment Analyzer, which determines the quality by the ratio and resolution of the large and small rRNA peaks (https://www.aati-us.com/instruments/fragment-analyzer/). In a multiplexed gene expression analysis, 100 ng of total RNA was hybridized to reporter probes and capture probes specific to each gene target (Supplemental Table S1). Following purification and binding of the hybridized probes to the optical cartridge, the cartridge was scanned on the nCounter Digital Analyzer. SAND, EF1a, UBIQUITIN11, and ACTIN2 were used as potential reference genes. The SAND and EF1a mRNA levels varied with treatment at day 4 of treatment, whereas the transcripts for UBIQUITIN11 and ACTIN2 were stable under Fe deficiency and therefore the latter two were used for normalization. Raw counts for each individual gene were imported into the nSolver Analysis Software 3.0 and were normalized against background and the two reference genes.
Statistical Analysis
JMP software (version 9.0.2; SAS Institute) was used for statistical analysis. Figures and data represent average and SD values based on sampling from at least three independent biological replicates. The number of total samples (n) is given when appropriate. Student’s t test was used to calculate significant differences (P value < 0.05). Statistical analysis of gene expression was performed in R (version 3.3.3) using the lsmeans and plyr packages. A one-way ANOVA was used to compare treated and control plants for each day, using pairwise comparisons. P values < 0.05 were considered significant comparisons and are indicated in figures.
Accession Numbers
Accession numbers of genes investigated are given in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Response of Fe status markers to Fe deficiency treatment.
Supplemental Figure S2. Growth of Arabidopsis roots during the Fe deficiency and the Fe recovery treatment.
Supplemental Figure S3. Loss of PSI quantum yield results mainly from limitations upstream of PSI.
Supplemental Figure S4. Transcript changes over the course of Fe deficiency.
Supplemental Figure S5. Proposed model of the effect of Fe deficiency on photosynthetic related proteins.
Supplemental Table S1. Proteins and transcripts tested over the course of Fe deficiency and resupply.
Acknowledgments
We thank Dr Daniel J. Kliebenstein, Dr Janneke Balk, Dr Catherine Curie, Dr Jean-Francois Briat, Dr Stanislas Kopriva, Dr Hisashi Ito, Dr Toshiharu Shikanai, and Dr Atsushi Sakamoto for providing us with antisera. We thank Dr. Ann Hess for advice with transcriptome analysis.
Footnotes
This work was supported by NSF grant numbers MCB 0950726 and MCB 1244142 to M.P. G.K. was supported by an NSF Graduate Research Fellowship (grant no. DGE-1321845). K.R. was supported by a European-FP7-International Outgoing Fellowship (Marie Curie, IntegrRegulFeSPlast; PIOF-GA-2010-273586).
Articles can be viewed without a subscription.
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T (2005a) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany SE, Ye H, Garifullina GF, Zhang L, Pilon-Smits EA, Pilon M (2005b) Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol 138: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andaluz S, López-Millán A-F, De las Rivas J, Aro EM, Abadía J, Abadía A (2006) Proteomic profiles of thylakoid membranes and changes in response to iron deficiency. Photosynth Res 89: 141–155 [DOI] [PubMed] [Google Scholar]
- Balk J, Pilon M (2011) Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65: 125–153 [DOI] [PubMed] [Google Scholar]
- Bang WY, Jeong IS, Kim DW, Im CH, Ji C, Hwang SM, Kim SW, Son YS, Jeong J, Shiina T, et al. (2008) Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell Physiol 49: 1350–1363 [DOI] [PubMed] [Google Scholar]
- Barber J. (2016) ‘Photosystem II: the water splitting enzyme of photosynthesis and the origin of oxygen in our atmosphere’. Q Rev Biophys 49: e14. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas CE, Merchant SS (2013) Iron sparing and recycling in a compartmentalized cell. Curr Opin Microbiol 16: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Thomas KM, Dai YY, Boyd JM, Outten FW (2014) Interplay between Oxygen and Fe-S Cluster Biogenesis: Insights from the Suf Pathway. Biochemistry 53: 5834–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Briat J-F, Dubos C, Gaymard F (2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci 20: 33–40 [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R (2015) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci 20: 124–133 [DOI] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KAG, Abdel-Ghany SE, Cohu CM, Pilon M (2009) Copper homeostasis. New Phytol 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Bych K, Netz DJA, Vigani G, Bill E, Lill R, Pierik AJ, Balk J (2008) The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J Biol Chem 283: 35797–35804 [DOI] [PubMed] [Google Scholar]
- Christman MA, Richards JH, McKay JK, Stahl EA, Juenger TE, Donovan LA (2008) Genetic variation in Arabidopsis thaliana for night-time leaf conductance. Plant Cell Environ 31: 1170–1178 [DOI] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Celletti S, Catarcione G, Kopriva S, Astolfi S (2013) Transcriptional and physiological changes in the S assimilation pathway due to single or combined S and Fe deprivation in durum wheat (Triticum durum L.) seedlings. J Exp Bot 64: 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornah JE, Terry MJ, Smith AG (2003) Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci 8: 224–230 [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, et al. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Glaesener AG, Merchant SS, Blaby-Haas CE (2013) Iron economy in Chlamydomonas reinhardtii. Front Plant Sci 4: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke GT, Hase T (2008) Variable photosynthetic roles of two leaf-type ferredoxins in Arabidopsis, as revealed by RNA interference. Photochem Photobiol 84: 1302–1309 [DOI] [PubMed] [Google Scholar]
- Hanke GT, Kimata-Ariga Y, Taniguchi I, Hase T (2004) A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol 134: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbik A, Giritch A, Horstmann C, Becker R, Balzer HJ, Bäumlein H, Stephan UW (1996) Iron and copper nutrition-dependent changes in protein expression in a tomato wild type and the nicotianamine-free mutant chloronerva. Plant Physiol 111: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Page MT, Sumida A, Tanaka A, Terry MJ, Tanaka R (2017a) The iron-sulfur cluster biosynthesis protein SUFB is required for chlorophyll synthesis, but not phytochrome signaling. Plant J 89: 1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kato Y, Sumida A, Tanaka A, Tanaka R (2017b) The SUFBC2 D complex is required for the biogenesis of all major classes of plastid Fe-S proteins. Plant J 90: 235–248 [DOI] [PubMed] [Google Scholar]
- Huet G, Daffé M, Saves I (2006) Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen’s survival. J Bacteriol 187: 6137–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C, Schreiber U (2008) Saturation pulse method for assessment of energy conversion in PSI. PAM Appl Notes 1: 11–14 [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S (2014) Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Front Plant Sci 5: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohling CA, Eutropio FJ, Bertolazi AA, Dobbss LB, Campostrini E, Dias T, Ramos AC (2016) Ecophysiology of iron homeostasis in plants. Soil Sci Plant Nutr 62: 39–47 [Google Scholar]
- Laganowsky A, Gómez SM, Whitelegge JP, Nishio JN (2009) Hydroponics on a chip: analysis of the Fe deficient Arabidopsis thylakoid membrane proteome. J Proteomics 72: 397–415 [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Schmidt W (2012) Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiol 159: 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Shi D, Liu X, Qin J, Ge Q, Xu L, Pan X, Li W, Zhu Y, et al. (2013) Spatial-temporal analysis of zinc homeostasis reveals the response mechanisms to acute zinc deficiency in Sorghum bicolor. New Phytol 200: 1102–1115 [DOI] [PubMed] [Google Scholar]
- Liang X, Qin L, Liu P, Wang M, Ye H (2014) Genes for iron-sulphur cluster assembly are targets of abiotic stress in rice, Oryza sativa. Plant Cell Environ 37: 780–794 [DOI] [PubMed] [Google Scholar]
- Lim L, McFadden GI (2010) The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci 365: 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang P, Liu B, Feng D, Zhang J, Su J, Zhang Y, Wang JF, Wang HB (2013) A deficiency in chloroplastic ferredoxin 2 facilitates effective photosynthetic capacity during long-term high light acclimation in Arabidopsis thaliana. Plant J 76: 861–874 [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán AF, Grusak MA, Abadía A, Abadía J (2013) Iron deficiency in plants: an insight from proteomic approaches. Front Plant Sci 4: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, Fare T (2009) Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes 2: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence--a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Merchant S, Sawaya MR (2005) The light reactions: a guide to recent acquisitions for the picture gallery. Plant Cell 17: 648–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JL, Allinger T, Herzog S, Hoerth P, Wehinger E, Merchant S, Hippler M (2002) Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J 21: 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, Smith AG (2005) Regulation of tetrapyrrole biosynthesis in higher plants. Biochem Soc Trans 33: 737–742 [DOI] [PubMed] [Google Scholar]
- Msilini N, Zaghdoudi M, Govindachary S, Lachaâl M, Ouerghi Z, Carpentier R (2011) Inhibition of photosynthetic oxygen evolution and electron transfer from the quinone acceptor QA- to QB by iron deficiency. Photosynth Res 107: 247–256 [DOI] [PubMed] [Google Scholar]
- Nishio JN, Abadía J, Terry N (1985) Chlorophyll-proteins and electron transport during iron-mediated chloroplast development. Plant Physiol 78: 296–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio JN, Terry N (1983) Iron nutrition-mediated chloroplast development. Plant Physiol 71: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby-Sherrouse AG, Murphy ER (2013) Iron-responsive bacterial small RNAs: variations on a theme. Metallomics 5: 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MD, Allen MD, Kropat J, Urzica EI, Karpowicz SJ, Hsieh SI, Loo JA, Merchant SS (2012) Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii. Plant Cell 24: 2649–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Celletti S, Catarcione G, Hawkesford MJ, Astolfi S, Ciaffi M (2014) Iron deprivation results in a rapid but not sustained increase of the expression of genes involved in iron metabolism and sulfate uptake in tomato (Solanum lycopersicum L.) seedlings. J Integr Plant Biol 56: 88–100 [DOI] [PubMed] [Google Scholar]
- Petit J-M, Briat J-F, Lobréaux S (2001) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem J 359: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Leidgens S, Frey AG (2012) Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta 1823: 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M. (2017) The copper microRNAs. New Phytol 213: 1030–1035 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Garifullina GF, Abdel-Ghany S, Kato S, Mihara H, Hale KL, Burkhead JL, Esaki N, Kurihara T, Pilon M (2002) Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol 130: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Pilon M (2005) Sulfur metabolism in plastids. In Wise RR, Hoober JK, eds, Advances in Photosynthesis and Respiration, The Structure and Function of Plastids, Vol 23 Kluwer Academic Publishers, The Netherlands, pp 387–402 [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Ravet K, Danford FL, Dihle A, Pittarello M, Pilon M (2011) Spatiotemporal analysis of copper homeostasis in Populus trichocarpa reveals an integrated molecular remodeling for a preferential allocation of copper to plastocyanin in the chloroplasts of developing leaves. Plant Physiol 157: 1300–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat J-F, Gaymard F, Cellier F (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Pan IC, Li W, Lan P, Buckhout TJ, Schmidt W (2013) The transcriptional response of Arabidopsis leaves to Fe deficiency. Front Plant Sci 4: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Vol 1 Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- Séguéla M, Briat J-F, Vert G, Curie C (2008) Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J 55: 289–300 [DOI] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sétif P, Fischer N, Lagoutte B, Bottin H, Rochaix J-D (2002) The ferredoxin docking site of photosystem I. Biochim Biophys Acta 1555: 204–209 [DOI] [PubMed] [Google Scholar]
- Sharma S. (2007) Adaptation of photosynthesis under iron deficiency in maize. J Plant Physiol 164: 1261–1267 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK, Pilon M (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G, Bennett M (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS One 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S, Terry N (1980) Limiting factors in photosynthesis: II. Iron stress diminishes photochemical capacity by reducing number of photosynthetic units. Plant Physiol 65: 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R (2004) AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res 32: D368–D372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9: e0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi AM, Peers G, Kobayashi MC, Niyogi KK, Merchant SS (2010) Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth Res 105: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timperio AM, D’Amici GM, Barta C, Loreto F, Zolla L (2007) Proteomics, pigment composition, and organization of thylakoid membranes in iron-deficient spinach leaves. J Exp Bot 58: 3695–3710 [DOI] [PubMed] [Google Scholar]
- Topilina NI, Green CM, Jayachandran P, Kelley DS, Stanger MJ, Piazza CL, Nayak S, Belfort M (2015) SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc Natl Acad Sci USA 112: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Abdel-Ghany SE, Cohu CM, Herbert SK, Kugrens P, Pilon M, Pilon-Smits EAH (2007) Chloroplast iron-sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc Natl Acad Sci USA 104: 5686–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, Wappett M, Sharpe A, Dymond M, Barrett JC, et al. (2015) Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res 75: 2587–2593 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat J-F, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorst O, Kock P, Lever A, Weterings B, Weisbeek P, Smeekens S (1993) The promoter of the Arabidopsis thaliana plastocyanin gene contains a far upstream enhancer-like element involved in chloroplast-dependent expression. Plant J 4: 933–945 [DOI] [PubMed] [Google Scholar]
- Voss I, Goss T, Murozuka E, Altmann B, McLean KJ, Rigby SEJ, Munro AW, Scheibe R, Hase T, Hanke GT (2011) FdC1, a novel ferredoxin protein capable of alternative electron partitioning, increases in conditions of acceptor limitation at photosystem I. J Biol Chem 286: 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Y, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Xu XM, Adams S, Chua NH, Møller SG (2005) AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J Biol Chem 280: 6648–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Nakai M (2006) Arabidopsis AtIscA-I is affected by deficiency of Fe-S cluster biosynthetic scaffold AtCnfU-V. Biochem Biophys Res Commun 340: 1047–1052 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) The Interplay between Sulfur and Iron Nutrition in TomatoSQUAMOSA Promoter Binding Protein-Like 7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Abdel-Ghany SE, Anderson TD, Pilon-Smits EAH, Pilon M (2006) CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J Biol Chem 281: 8958–8969 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zuchi S, Watanabe M, Hubberten HM, Bromke M, Osorio S, Fernie AR, Celletti S, Paolacci AR, Catarcione G, Ciaffi M, et al. (2015) The interplay between sulfur and iron nutrition in tomato. Plant Physiol 169: 2624–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]