KinG is a plant-specific calponin-homology domain kinesin that forms a complex with SHR through binding to SIEL, promotes its intercellular trafficking, and facilitates pausing of SHR-associated endosomes on microtubules.
Abstract
Both endogenous plant proteins and viral movement proteins associate with microtubules to promote their movement through plasmodesmata. The association of viral movement proteins with microtubules facilitates the formation of virus-associated replication complexes, which are required for the amplification and subsequent spread of the virus. However, the role of microtubules in the intercellular movement of plant proteins is less clear. Here we show that the SHORT-ROOT (SHR) protein, which moves between cells in the root to regulate root radial patterning, interacts with a type-14 kinesin, KINESIN G (KinG). KinG is a calponin homology domain kinesin that directly interacts with the SHR-binding protein SIEL (SHR-INTERACING EMBRYONIC LETHAL) and localizes to both microtubules and actin. Since SIEL and SHR associate with endosomes, we suggest that KinG serves as a linker between SIEL, SHR, and the plant cytoskeleton. Loss of KinG function results in a decrease in the intercellular movement of SHR and an increase in the sensitivity of SHR movement to treatment with oryzalin. Examination of SHR and KinG localization and dynamics in live cells suggests that KinG is a nonmotile kinesin that promotes the pausing of SHR-associated endosomes. We suggest a model in which interaction of KinG with SHR allows for the formation of stable movement complexes that facilitate the cell-to-cell transport of SHR.
Cell-to-cell movement of transcription factors is a common form of intercellular communication in plants (Lucas et al., 1995; Kurata et al., 2005; Pi et al., 2015; Gallagher et al., 2014). Many of these mobile transcription factors function as positional signals that regulate various aspects of plant development, including embryonic development (Schlereth et al., 2010), shoot apical meristem maintenance (Lucas et al., 1995; Kim et al., 2005; Yadav et al., 2011), floral initiation (Sessions et al., 2000; Wu et al., 2003), root hair formation (Kurata et al., 2005; Savage et al., 2008), stomata differentiation (Raissig et al., 2017), and root patterning (Nakajima et al., 2001; Pi et al., 2015). Intercellular movement of proteins in plants occurs via plasmodesmata, highly specialized channels that form cytoplasmic continuity and allow for the exchange of molecules between two adjacent cells (Oparka, 2004). Blocking of plasmodesmata in specific tissues results in the restriction of protein movement and often leads to cellular patterning defects (Vatén et al., 2011; Benitez-Alfonso et al., 2009; Daum et al., 2014; Wu et al., 2016; Liu et al., 2017).
A well-documented case of transcription factor movement involves the movement of SHORT-ROOT (SHR) between tissues in the root meristem. SHR is made in the stele and moves into the endodermis, quiescent center, and cortical endodermal initial cells (Nakajima et al., 2001). Movement of SHR is required for the asymmetric divisions of the cortical endodermal daughter cells that generate the separate layers of cortex and endodermis (Helariutta et al., 2000). Later in development of the root, a reduction in SHR movement triggers the asymmetric divisions in the endodermis that lead to the formation of a middle cortex (Koizumi et al., 2012). While it is known that SHR moves between cells via plasmodesmata, it is not known how SHR movement is regulated nor how SHR accesses plasmodesmata. Here, we report the interaction between SHR and a type 14 kinesin-like motor protein, KINESIN G (KinG; At1g63640), which directly binds to an essential protein, SHR INTERACTING EMBRYONIC LETHAL (SIEL), and supports the cell-to-cell movement of SHR.
Mechanisms Regulating SHR Cell-to-Cell Movement
The SHR protein moves through plasmodesmata. Semidominant mutations in CALLOSE SYNTHASE3 decrease the size exclusion limit of plasmodesmata and inhibit movement of SHR (Vatén et al., 2011). Structure-function analysis of SHR has shown that the movement of SHR via plasmodesmata is both targeted and regulated; there are factors that promote and factors that restrict movement of SHR. Among the factors that support the cell-to-cell movement of SHR are endosomes, microtubules, and SIEL (Koizumi et al., 2011; Wu and Gallagher, 2013, 2014). SIEL is a Huntingtin, EF3, PP2A, TOR1 (HEAT)-domain-containing protein that directly interacts with SHR. Null alleles of SIEL are embryonic lethal; hypomorphs have reduced movement of SHR. Via interaction with SIEL, SHR localizes to endosomes. In turn, the localization of SIEL to endosomes is tied to microtubules (Wu and Gallagher, 2013). When microtubules are disrupted, SIEL no longer localizes to microtubules, and SHR movement is reduced. Likewise, inhibition of endocytosis or interference with early or late endosomes hinders movement of SHR (Wu and Gallagher, 2014). These results suggest important roles for microtubules and endosomes in promoting the intercellular movement of SHR. However, the mechanism by which endosomes, microtubules, and SIEL support the movement of SHR is not known.
Evidence for the Function of Microtubules and Kinesins in Intercellular Protein Trafficking
In addition to endogenous plant proteins, viruses exploit plasmodesmata for movement between cells (Niehl and Heinlein, 2011; Harries and Ding, 2011). Many plant viruses interact with microtubules via virally encoded movement proteins (MPs) that facilitate transport via plasmodesmata (Heinlein et al., 1995, 1998; Padgett et al., 1996; Serazev et al., 2003; Wright et al., 2010). For example, Tobacco Mosaic Virus (TMV) encodes a MP (TMV-MP) that binds viral RNA and microtubules forming a viral ribonucleoprotein complex (vRNP; Citovsky et al., 1990; Heinlein et al., 1995, 1998). Since the association of MP with microtubules is correlated with the ability of the virus to spread between cells (Boyko et al., 2000, 2007), microtubules were thought to target the vRNP to plasmodesmata (similar to the role that microtubules play in the transport of membrane-bound cargo proteins in animals). In this model, microtubules serve as tracks for the movement of vRNPs to plasmodesmata. However, there is very little evidence for the directional transport of vRNP via microtubules. Instead, most data suggest that microtubules serve in the anchorage and release of viral replication complexes (VRCs; Niehl et al., 2013). Early in the process of infection, microtubules support the formation of VRC – microtubule anchored, endoplasmic reticulum (ER)-derived hubs of viral replication (Boyko et al., 2007; Sambade et al., 2008). Later in the process of infection, microtubules promote the release of VRCs from the ER for movement between cells (Sambade et al., 2008; Sambade and Heinlein 2009). Thus, the primary role of microtubules in the spread of vRNPs between cells is the anchoring of VRCs to the ER (to allow replication) and in later stages of infection, the release of the vRNP from the ER to the cytoplasm for transport to plasmodesmata, which likely occurs via interactions with the actin (Wright et al., 2007; Niehl and Heinlein, 2011). It is less clear what role microtubules play in the intercellular movement of endogenous plant proteins. However, since non-cell-autonomous proteins like SHR associate with the endomembrane (Wu and Gallagher, 2014), microtubules may serve as points of anchorage for the assembly of movement complexes. Insight into the roles that microtubules play in intercellular trafficking of proteins via plasmodesmata may come from studies on MP-BINDING PROTEIN 2C (MPB2C). MPB2C is a microtubule-binding protein with structural similarities to myosins and kinesins (Kragler et al., 2003). This protein interacts with both TMV-MP and the KNOTTED1/SHOOTMERISTEMLESS homeodomain transcription factors (Kragler et al., 2003; Winter et al., 2007). Transient overexpression of MPB2C in Nicotiana benthamiana epidermal cells interferes with the cell-to-cell movement of TMV. Overexpression of MPB2C in A. thaliana or N. benthamiana results in reorganization of cortical microtubules and a loss of KNOTTED1 movement (Winter et al., 2007). In both tobacco and A. thaliana, overexpression of MPB2C appears to trap TMV-MP and KNOTTED1 on microtubules.
Role of Calponin Homology Domain Kinesins (KCH) in Protein Trafficking
There are 61 annotated kinesins in A. thaliana; of these, 21 are putative minus-end-directed, type-14 kinesins (Endow and Waligora, 1998; Reddy and Day, 2001). Within this group of minus-end-directed kinesins is a plant-specific subgroup of seven kinesins with a KCH (Preuss et al., 2004; Lee and Liu, 2004; Reddy and Day, 2001). In both plants and animals, CH domains are found in many classes of actin-binding proteins. A fully functional actin-binding domain is composed of tandem CH domains; however, single CH domain proteins (e.g. like that in calponin) can also bind actin, albeit with lower affinity than an actin-binding domain (Gimona and Mital, 1998; Gimona et al., 2002; Korenbaum and Rivero, 2002). Consistent with this trend, plant KCHs interact with both microtubules and actin filaments. However, there are conflicting data as to the relative affinities of KCH proteins for actin filaments and microtubules (Preuss et al., 2004; Frey et al., 2009; Buschmann et al., 2011; Klotz and Nick, 2012; Schneider and Persson, 2015). The ability of KCH kinesins to dynamically interact with microtubules and actin in interphase cells suggests roles for KCH kinesins in microtubule-microfilament cross linking and the stabilization of cellular structures (Dixit, 2012, 2015). Recently, Dixit (2015) suggested that KCHs function in the microtubule-dependent rearrangement and movement of actin filaments. Thus, KCH proteins likely play regulatory roles in the coordination of the microtubule and actin cytoskeleton.
Here, we show that the KCH protein KinG supports the intercellular trafficking of SHR. KinG was previously characterized in BY-2 cells, where it was shown to localize to both microtubules and actin. Similarly, we find that KinG associates with both microtubules and actin. When SHR and KinG are coexpressed in tobacco leaf epidermal cells, there is significant overlap between the two proteins. Strikingly, when SHR is associated with KinG, it is transiently immobilized within the cell. Over a 3-min time frame, SHR can be seen moving between regions of KinG localization within the cell, each time pausing for 1 min before dissociating from KinG. Based upon the subcellular localization of KinG and SHR and the dynamics of their interaction in live cells, we propose a model in which KinG serves as a stable platform that facilitates posttranslational processes that promote the trafficking of SHR.
RESULTS
Identification of KinG as a Protein Interacting with the Mobile Form of SHR
To identify proteins involved in SHR movement, coimmunoprecipitation followed by mass spectrometry (coIP/MS) was used on A. thaliana roots expressing the SHR-GFP translational fusion under the control of the SHR promoter (SHR:SHR-GFP; Supplemental Fig. S1, A and D). Two different nonmobile forms of SHR-GFP, unable to move from the stele to the endodermis, were also used to filter the results for specific interaction with the mobile SHR protein. The first was a substitution allele of SHR in which Thr 289 is replaced with an Ile (SHR:SHRT289I-GFP). SHRT289I is a nonmobile and nonfunctional protein (Gallagher et al., 2004; Supplemental Fig. S1, B and E). For the second allele of SHR, the LNELDV motif (residues 342–347) was replaced with three Ala resides (SHR:SHRΔLNELDV-GFP). Mutation of the LNELDV motif results in a loss of movement, but if the mutant protein is ectopically expressed, it is a functional SHR protein (Gallagher and Benfey, 2009) and therefore should maintain interactions associated with the function of SHR as a transcription factor (Supplemental Fig. S1, C and F). From this screen, we found a single protein, KinG, which coprecipitated with the mobile SHR-GFP, but not with SHRT289I-GFP or SHRΔLNELDV-GFP.
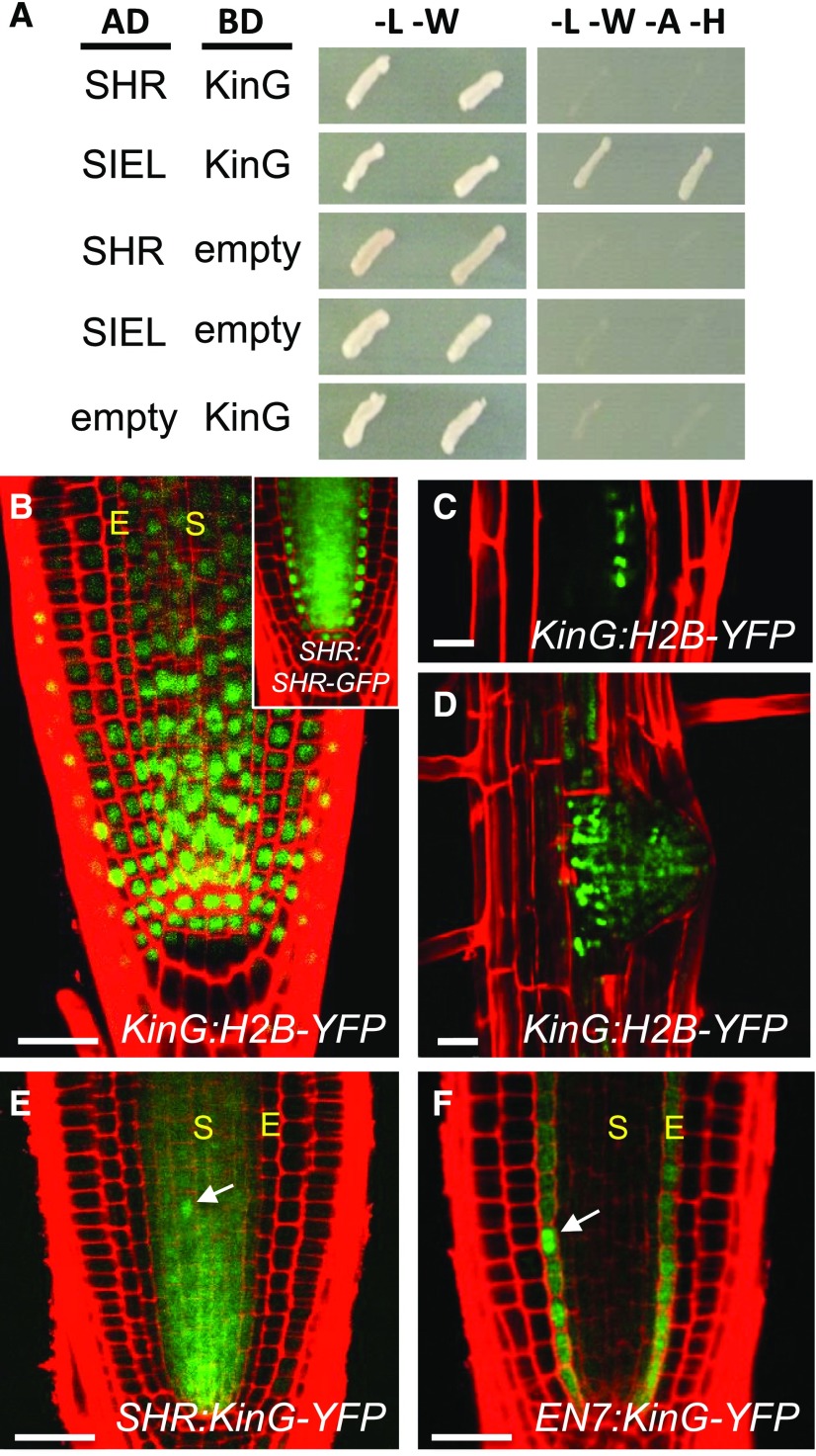
To validate that KinG directly interacts with SHR, targeted yeast two-hybrid assays were performed using a modified SHR protein that lacks autoactivation as bait (in the pDEST22 vector; Wu et al., 2014) and KinG as prey (in the pDEST32 vector). In these assays, all tests for interaction between SHR and KinG were negative. However, in assays where the SHR-interacting protein SIEL was used as bait and KinG as prey (Fig. 1A), we consistently saw interaction. These results were further corroborated using bimolecular fluorescence complementation (BiFC) assays in A. thaliana leaf protoplasts. When KinG, fused to the c-Venus and SIEL, fused to the n-Venus were cotransfected into leaf protoplast, distinct fluorescent punctate were present in the cytoplasm (Supplemental Fig. S2). These results show that KinG directly interacts with SIEL. Since SIEL directly interacts with SHR (Koizumi et al., 2011), SIEL likely serves as a linker between KinG and SHR.
Figure 1.
The kinesin KinG interacts with SIEL and is expressed in the root meristem. A, Diploid yeast expressing SHR or SIEL as bait with the KinG prey protein (as labeled) grown on selective medium. Medium lacking adenine and His was used to select for interaction between the bait and prey proteins. AD, Activating domain vector (bait); BD, binding domain vector (prey). B, Expression of KinG:H2B-YFP in wild-type root meristem. Inset, Expression of SHR:SHR-GFP in wild-type root meristem. C and D, Expression of KinG:H2B-YFP in lateral root primordia. E, Expression of SHR:KinG-YFP in wild-type root meristem. F, Expression of EN7:KinG-YFP in wild-type root meristem. White arrows in D and E mark changes in KinG-YFP localization in dividing cells. Scale bars, 25 μm.
The Expression Domain of KinG Overlaps with SHR in the Root Meristem
To determine whether expression of KinG overlaps with SHR, we cloned the putative KinG promoter (the 2-kb genomic fragment upstream of the ATG) and used it to drive expression of HISTONE 2B-YFP (KinG:H2B-YFP). In three independent lines, KinG:H2B-YFP was expressed throughout the root meristem, including the stele, endodermis, and quiescent center—the known domains of SHR expression or activity—and in regions of the elongation zone above the meristem (Fig. 1B). The KinG promoter was also active in the lateral root primordia (Fig. 1, C and D), both at a time before and during which SHR is expressed (Lucas et al., 2011). These results show that KinG expression overlaps with SHR and that KinG is expressed broadly in the meristem in both mitotic (e.g. the root initials) and nonmitotic cells (e.g. the quiescent center cells and cells of the elongation zone).
KinG Is a Nonmobile Protein in A. thaliana Roots That Localizes to Microtubules and Actin in N. benthamiana Leaf Epidermal Cells
To examine the subcellular localization of KinG and its potential for cell-to-cell movement, the KinG cDNA was fused in-frame to YFP and expressed from the KinG promoter. We examined 20 independently transformed lines and failed to detect YFP fluorescence in any of the KinG:KinG-YFP lines. Likewise, we detected no fluorescence in 50 independent 35S:KinG-YFP and 30 independent Ub10:KinG-YFP lines. Therefore, as an alternative approach to examine KinG localization, promoters with restricted domains of expression were used. KinG-YFP was expressed under the control of the stele-specific SHR promoter (SHR:KinG-YFP; Fig. 1E) or the endodermis-specific ENDODERMIS7 (EN7) promoter (EN7:KinG-YFP; Fig. 1F). In both tissues (stele and endodermis), KinG-YFP was detected only within the domain of promoter activity, indicating that KinG is a cell-autonomous protein. In cells expressing KinG-YFP, the protein was present throughout the cytoplasm and the nucleus. In dividing cells (based upon the appearance of the H2B-mCherry marker), the subcellular localization of KinG-YFP suggested an association with cytoskeletal/mitotic arrays (Supplemental Fig. S3A). The localization pattern of KinG in interphase and dividing cells matched well with the localization of the microtubule marker, mCherry-TUA5 when expressed in the root meristem (Supplemental Fig. S3B). Our inability to recover KinG-YFP-tagged lines when using the KinG, Ub10, or 35S promoters suggests that up-regulation of KinG is either embryo or gametophyte lethal.
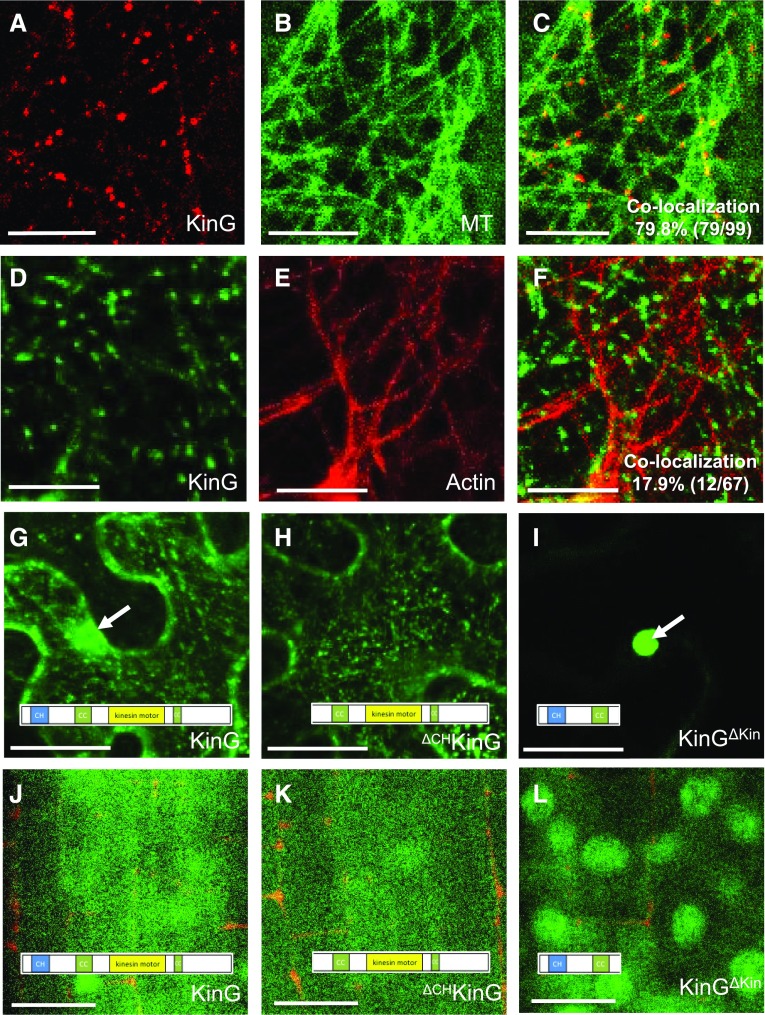
Using BY-2 cells Buschmann et al. (2011) previously showed colocalization of KinG with markers of the microtubule and actin cytoskeleton. To examine KinG localization within an intact tissue, we transiently expressed TagRFP-KinG or YFP-KinG in N. benthamiana leaf epidermal cells. Both the TagRFP and the YFP-labeled KinG showed nuclear and cytoplasmic localization. Within the cytoplasm, TagRFP-KinG showed extensive overlap (79.8%) with the microtubule marker GFP-TUA6 (Fig. 2, A–C; Supplemental Fig. S4A). In contrast, there was moderate but significant (17.9%) overlap between YFP-KinG and the actin marker, TagRFP-UtrCH (Fig. 2, D–F; Supplemental Fig. S4B). Preferential localization of KCHs with perinuclear actin fibers has been reported (Klotz and Nick, 2012); however, we saw no differences in the overlap of YFP-KinG with perinuclear or cortical actin arrays (Supplemental Figure S4, C–E). This suggests that KinG predominantly localizes to microtubules but also maintains a degree of overlap with actin.
Figure 2.
Subcellular localization of KinG. A to C, Coexpression of TagRFP-KinG (A) and the microtubule marker GFP-TUA6 (B) in N. benthamiana epidermal cells. C, Overlay of TagRFP-KinG and GFP-TUA6. D to F, Z-stack maximal projection of a N. benthamiana leaf epidermal cell coexpressing of YFP-KinG (D) and the actin marker TagRFP-UtrCH (E). F, Overlay of YFP-KinG and TagRFP-UtrCH. G to I, Z-stack maximal projection of YFP-KinG (G), YFP-ΔCHKinG (H), or YFP-KinGΔKin (I) in N. benthamiana leaf epidermal cells. J to L, Expression of YFP-KinG (J), YFP-ΔCHKinG (K), or YFP-KinGΔKin (L) in A. thaliana root meristem stele. Scale bars, A to F and J to L, 10 μm; G to I, 50 μm.
The kinesin domain of KCH proteins generally mediates interactions with microtubules, while the CH domain is thought to facilitate association with actin. To test the contribution of the two domains to KinG localization, two truncated versions of KinG were cloned: ΔCHKinG (an N-terminal truncated version of KinG that lacks the CH domain) and KinGΔKin (a C-terminal truncated version of KinG that lacks the kinesin motor domain). Expression of the full version of YFP-KinG in N. benthamiana resulted in punctate localization throughout the cell, with diffuse nuclear localization (Fig. 2G). Expression of YFP-ΔCHKinG resulted in similar punctate localization in the cytoplasm; however, nuclear localization was largely abolished (Fig. 2H). In contrast, YFP-KinGΔKin showed strong exclusive nuclear localization (Fig. 2I). Similar patterns of localization were observed when the ∆CHKinG and KinG∆Kin proteins were stably expressed in the stele of A. thaliana roots using the SHR promoter (Fig. 2, J–L). It should be noted that expression of these constructs did not affect root patterning. To determine if the CH or kinesin domain truncations affected the ability of KinG to interact with SIEL, both truncated versions were expressed as prey in yeast two-hybrid assays with SIEL (Supplemental Fig. S5). SIEL interacted with ΔCHKinG, but not with KinGΔKin. Collectively, these results suggest that the KinG CH domain is not required for association with the cytoskeleton or for interaction with SIEL but may facilitate nuclear localization.
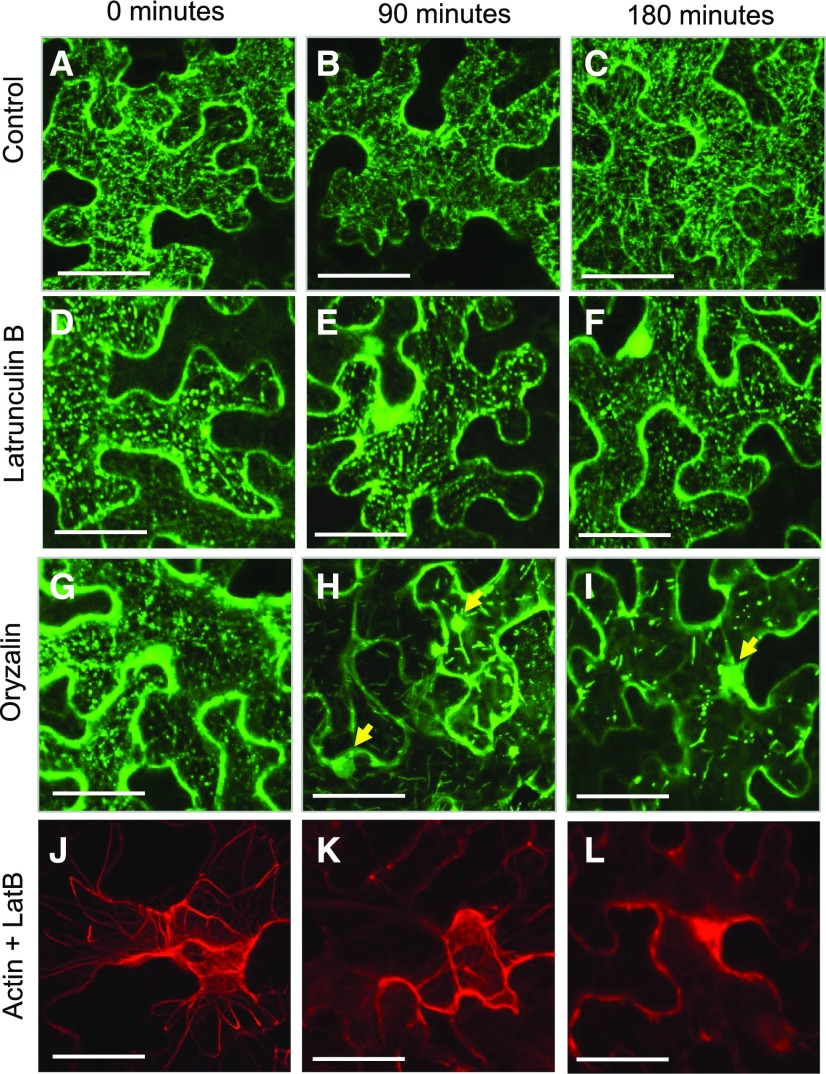
The distinct nuclear localization of KinG∆Kin in both tobacco leaf epidermal cells and A. thaliana roots suggests that interaction of KinG with microtubules prevents its accumulation in the nucleus. However, since the truncated version of KinGΔKin is 600 amino acids shorter than the full-length KinG, nuclear localization could be the result of passive diffusion. To distinguish between these two possibilities, we treated N. benthamiana leaves expressing full-length YFP-KinG with 20 μm latrunculin B, 20 µm cytochalasin D, or 2 μm oryzalin to disrupt actin and microtubules, respectively. Treatment with latrunculin B or cytochalasin D did not impair the localization of KinG (Fig. 3, A–C and D–F; Supplemental Fig. S6); however, it did disrupt most F-actin (Fig. 3, J–L). Treatment with oryzalin dramatically disturbed KinG localization and led to an increase in the accumulation of KinG in the nucleus (Fig. 3, G–I). Collectively, these results suggest the kinesin domain of KinG is essential for its ability to maintain cytoplasmic localization and, based upon the yeast 2- hybrid assays, to interact with SIEL.
Figure 3.
Response of KinG to latrunculin B and oryzalin N. benthamiana leaf epidermal cells. A to I, Z-stack maximal projection of cells expressing YFP-KinG were incubated with a control solution (A–C), 20 μm latrunculin B (D–F), or 2 μm oryzalin (G–I) for 0 min (A, D, and G), 90 min (B, E, and H) or 180 min (C, F, and I). J to L, Z-stack maximal projection of cells expressing TagRFP-UtrCH incubated with a latrunculin B (LatB) for 0 min (J), 90 min (K), or 180 min (L). Yellow arrows in H and I point to accumulation of KinG in the nucleus. Scale bars, 50 μm.
Cell-to-Cell Movement of SHR Is Reduced in kinG Mutants
Since intact microtubules are required for both SHR movement and KinG localization, we tested whether KinG plays a role in SHR movement. While there are several T-DNA insertion lines annotated as having an insertion within KinG available from the Arabidopsis Biological Resource Center (ABRC), none are within the coding sequences (note that SAIL_754_A01 is annotated as having an insertion 100 bp upstream of the ATG; however, the levels of KinG mRNA are normal in these lines). Therefore, a kinG null was generated using CRISPR/Cas9. Guide RNAs were designed to target the seventh exon, between the CH and kinesin motor domain. We screened 60 lines and found one with a 155-bp insertion in the seventh exon that introduces a premature stop codon (Supplemental Fig. S7, A and B). As a result of this insertion, KinG mRNA was significantly decreased (likely due to nonsense-mediated mRNA decay) in the kinG mutant (Supplemental Fig. S7C). We observed no obvious defects in the growth or overall appearance of the kinG lines or changes in the cellular organization of the root meristem (Supplemental Fig. S7, D–H), indicating that KinG is not essential for plant growth or cellular patterning of the root.
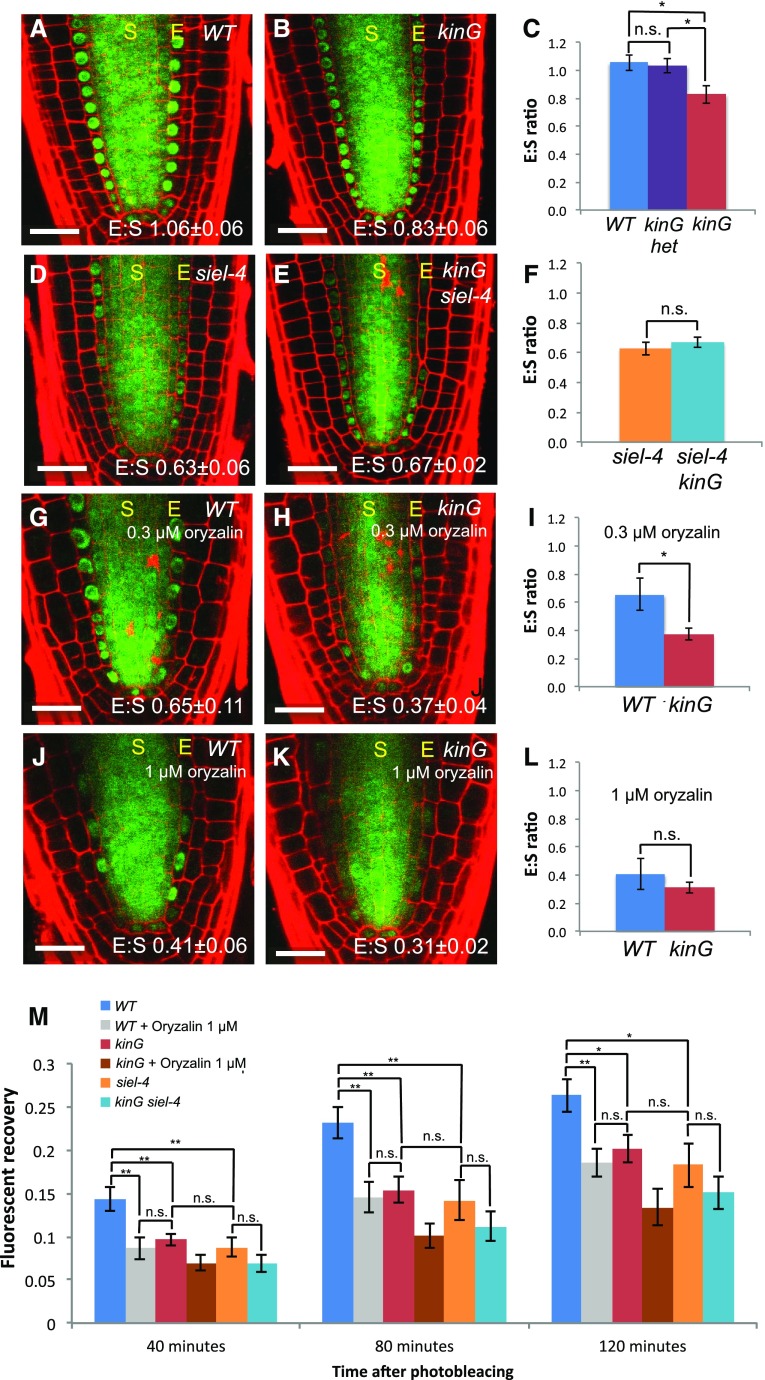
When SHR movement is blocked early in the development of the root, no endodermis is made. However, moderate decreases in SHR movement (in the 20%–30% range) have no effect on root patterning (Koizumi et al., 2011, 2012). To test whether KinG functions in SHR movement, kinG mutants were crossed to SHR:SHR-GFP marker lines. As in previous studies, the ratio of fluorescence intensity between the endodermis and the stele (E:S ratio) was used as a measure of SHR-GFP movement. In 5-d-old wild-type roots, the E:S ratio of SHR-GFP fluorescence was 1.06 (±0.057); similar values were measured in roots heterozygous for kinG (1.03 ± 0.051). In contrast, in kinG homozygotes, the E:S ratio was 0.83 (±0.062), which is significantly lower than in wild type or heterozygote (P < 0.05; Student’s t test, n = 10; Fig. 4, A–C). These results indicate a (21.7%) decrease in SHR movement in the kinG line. Consistent with reduced movement of SHR-GFP, we saw a significant reduction in the recovery of SHR-GFP fluorescence in the endodermis of kinG mutants after photobleaching as compared to wild type (FRAP of SHR-GFP; Fig. 4M; Supplemental Fig. S8). For comparison, mutations in SIEL (siel-4) or short treatment of wild-type seedlings with 1 µm oryzalin also reduced FRAP of SHR-GFP to levels similar to kinG (Fig. 4M). The siel-4;kinG double mutants were indistinguishable from the siel-4 single mutant (Fig. 4, D–F), indicating that the siel-4 phenotype is not significantly enhanced by kinG mutation. Likewise, we examined kinG;kinH (At5g41310.11) double mutants. Of the seven KCH proteins in A. thaliana KinH (named here) is most similar to KinG (66.5% amino acid identity and 76.2% similarity). The kinG/kinH roots were identical to kinG single mutants with respect to both SHR movement and radial patterning of the root (Supplemental Fig. S9), suggesting additional levels of functional redundancy.
Figure 4.
The kinG null mutation reduces the cell-to-cell movement of SHR. A and B, Expression of SHR:SHR-GFP in primary root meristems of (A) wild-type (WT) seedlings and (B) kinG mutants. C, Quantification of SHR-GFP E:S ratio in WT seedlings, kinG heterozygotes, and kinG homozygote mutants. D and E, Expression of SHR:SHR-GFP in D WT and E kinG roots treated with 0.3 μm oryzalin for 12 h. F, Quantification of SHR-GFP E:S ratio in D and E. G to I, Expression of SHR:SHR-GFP in G WT and H kinG roots treated with 1 μm oryzalin for 12 h. I, Quantification of SHR-GFP E:S ratio in G and H. J to L, Expression of SHR:SHR-GFP in (J) siel-4 mutants and (K) kinG/siel-4 double mutants. L, Quantification of SHR-GFP E:S ratio in J and K. M, FRAP of endodermis from the different plant lines and treatments. Fluorescent recovery was measured 40, 80, and 120 min after photobleaching. Scale bars, 25 μm. *P < 0.05, **P < 0.01, by Student’s t test (n > 5).
Chemical inhibitors can be used to overcome functional redundancy by inhibiting multiple members of a protein family at once. Since KinG requires microtubules for localization, we tested the sensitivity of the kinG roots to treatment with oryzalin (Fig. 4, G–L) with the expectation that other members of the protein family would have similar requirements for microtubules for proper localization. As shown in Figure 4, G–I, treatment of roots with 0.3 µm oryzalin for 12 h enhanced the effect of the kinG mutation on the movement of SHR, decreasing the E:S ratio of SHR-GFP by 42.9% as compared to the wild-type. This difference between kinG and wild-type roots in 0.3 µm oryzalin is statistically significant (P < 0.05), suggesting that kinG is more sensitive than wild-type roots to moderate levels of oryzalin. The kinG mutants were similarly more sensitive than wild-type to treatment of with 1 µm oryzalin when assayed at 3 h posttreatment. At the 3 h time point, there is a 21.5% decrease in the E:S ratio of SHR-GFP in oryzalin-treated kinG roots compared to nontreated but no change in wild-type roots (Supplemental Fig. S10). In contrast, when assayed at 12 h after treatment with 1.0 µm oryzalin, the E:S ratio of SHR:GFP in both wild-type and kinG roots is very similar (Fig. 4, J–L). These results suggest that kinG mutants are more sensitive than wild-type roots to short-term treatment with moderate levels of oryzalin, which have little secondary effects on root growth. Note that the sensitivity of kinG to oryzalin appears specific to the movement of SHR-GFP. Other aspects of microtubule-mediated root growth (e.g. cell division, cell elongation) in the kinG mutants are not more sensitive to oryzalin than wild type (Supplemental Fig. S7H). These results indicate that loss of KinG makes SHR movement more sensitive to destabilization of microtubules.
KinG and SHR Localize to Stable Structures in Tobacco Leaf Epidermal Cells
In both A. thaliana and in N. benthamiana leaf epidermal cells, SHR localizes to endosomes (Wu and Gallagher, 2014; Supplemental Fig. S11, A and D). In N. benthamiana leaf epidermal cells, endosome-associated SHR-GFP shows punctate localization (shown here with YFP-SHR and Rab2Fa-mCherry; inset in Supplemental Fig. S11D). In marked contrast, the immobile variants of SHR, YFP-SHRT289I, and YFP-SHRΔLNELDV show no association with endosomes and diffuse localization throughout the cytoplasm (Supplemental Fig. S11, B C, E, and F). Disruption of endosomes in intact roots inhibits the cell-to-cell movement of SHR (Wu and Gallagher, 2014). Likewise, disruption of microtubules in A. thaliana disrupts the association of SIEL with endosomes and the movement of SHR between cells (Wu and Gallagher, 2013). To explain how KinG fits into this scenario and facilitates the intercellular movement of SHR, we carefully examined interactions among SHR, endosomes, KinG, and actin using tobacco leaf epidermal cells.
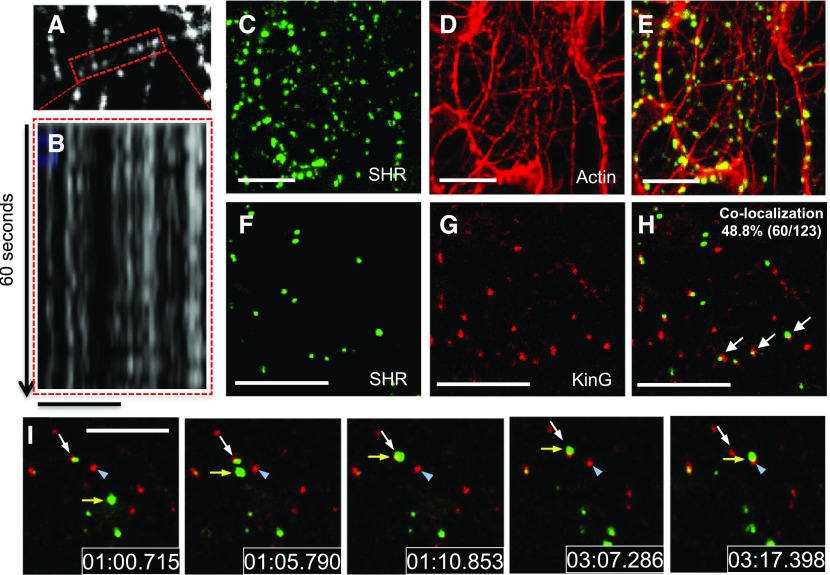
In tobacco leaf cells, YFP-SHR shows punctate localization that correlates with an association with endosomes. YFP-SHR foci are highly dynamic moving both within the cell cortex (Supplemental Movie 1) and in the cytoplasm, often clustering around the nucleus (Supplemental Movie 2). To test whether KinG might drive the intracellular movement of SHR foci (SHR-associated endosomes), we examined YFP-KinG motility. Using confocal microscopy, epidermal cells expressing YFP-KinG were imaged every 3 s for 60 s. In these assays, the YFP-KinG signal was remarkably stable, showing almost no movement (kymograph analysis, Fig. 5, A and B). These results indicate that the high degree of mobility observed for SHR-GFP is not a result of KinG-directed mobility.
Figure 5.
Interaction between SHR-associated endosomes, actin strands, and KinG foci. A and B, Kymograph of KinG along a microtubule (highlighted section in A) showing lack of motility (B). C to E, Z-stack maximal projection of a N. benthamiana leaf epidermal cell coexpressing YFP-SHR (C) and the actin marker TagRFP-UtrCH (D). E, Overlay of YFP-SHR and TagRFP-UtrCH. F to H, Coexpression of YFP-SHR (F) and TagRFP-KinG (G) in N. benthamiana epidermal cells. H, Overlay of YFP-SHR and TagRFP-KinG. White arrows point to colocalization events. I, Time course of YFP-SHR associated vesicles pausing on a TagRFP-KinG sites. The yellow arrow points to mobile YFP-SHR; the white arrow in frame 1:00.715 indicates TagRFP-KinG and YFP-SHR. Between time 1:00.715 and 1:10.853, YFP-SHR (yellow arrow) moves to colocalize with TagRFP-KinG and YFP-SHR (white arrow). After pausing there for approximately 2 min, YFP-SHR then moves to colocalize with TagRFP-KinG in a different region of the cell (blue arrowhead). Scale bars, B, 5 μm; C to E, 25 μm; F to I, 10 μm.
In plants, organelles generally move along actin filaments and pause at microtubules (Hamada et al., 2012). To test whether SHR associates with actin, TagRFP-UtrCH was expressed along with SHR-GFP in tobacco leaf epidermal cells. As shown in Figure 5, C–E, there is significant overlap between SHR-GFP and the TagRFP-UtrCH, suggesting that while in association with endosomes, SHR moves along actin. The dynamic movement of YFP-SHR, the immotility of KinG, and the preferential localization of KinG with microtubules suggest that there are different populations of YFP-SHR and TagRFP-KinG in the cell. To test for colocalization between SHR and KinG, YFP-SHR was expressed together with TagRFP-KinG in tobacco (Fig. 5, F–H). In these experiments, we saw between 33.3% and 48.8% overlap in signal depending upon whether TagRFP-KinG or YFP-SHR was used (respectively) to determine the region of interest. In instances were YFP-SHR foci colocalized with KinG, YFP-SHR was immobile with an average velocity of 0.01 (±0.002) μm/s. In contrast, when SHR-GFP was not associated with KinG, SHR was mobile, with an average velocity of 0.36 (±0.048) μm/s (Supplemental Movie 3; Fig. 5G). This difference between the two populations of SHR was statistically significant (P < 0.001; n = 40). Remarkably, in instances where mobile YFP-SHR encountered TagRFP-KinG, SHR movement paused, often for several seconds to minutes, before resuming movement (Fig. 5I; Supplemental Movies 4 and 5). As can be seen in Figure 5I (images time stamped and extracted from Supplemental Movie 4), YFP-SHR moves (yellow arrow) to colocalize with TagRFP-KinG (white arrow). YFP-SHR is stable in this location (for approximately 2 min) before moving to colocalize again with TagRFP-KinG in a different position in the cell (Fig. 5I, blue arrowhead). These data suggest that KinG may serve as a linker between actin and microtubules and thus promote pausing of SHR-associated endosomes in regions where actin and microtubules overlap.
DISCUSSION
The plant cytoskeleton is a highly dynamic network of proteins that interact to promote nearly all aspects of plant growth. Here, we characterized the localization and function of KinG, a type 14 KCH. KinG showed both cytoplasmic and nuclear localization in various cell types in the A. thaliana root and N. benthamiana leaf epidermal cells. In N. benthamiana, coexpression of fluorescently tagged KinG with markers of the microtubule or actin cytoskeleton revealed extensive overlap between KinG and TUA6 and moderate but significant colocalization with UtrCH. Limited structure-function analysis suggests that the aminoterminal kinesin domain of KinG is required for localization of the full-length protein to microtubules. Truncated alleles of KinG lacking the kinesin domain showed near-exclusive nuclear localization in N. benthamiana leaf epidermal cells and nuclear enrichment in A. thaliana roots, suggesting that interactions with microtubules may prevent its accumulation in the nucleus. Similar results were seen when YFP-KinG cells were treated with oryzalin. These results suggest that KinG is a bona fide microtubule and actin binding protein.
In the context of this study, KinG was identified as a protein that coimmunoprecipitated with mobile versions of SHR (SHR-YFP or SHR-GFP) but not with immobile variants (SHRΔLNELDV-GFP or SHRT289I-GFP), suggesting a role for KinG in the intercellular movement of SHR. Indeed, the movement of SHR-GFP from the stele into the endodermis is significantly reduced in kinG loss-of-function lines. Since previous analysis of SHR movement identified SIEL, endosomes, and microtubules as elements that facilitate SHR movement, we analyzed KinG activity with respect to these other factors. We found that KinG directly interacts with SIEL and that the SHR movement phenotype of kinG is largely epistatic to siel. Loss of KinG sensitized roots to the effects of oryzalin, but only with respect to SHR movement (other microtubule-dependent processes like cell elongation were not more sensitive to oryzalin), suggesting that KinG promotes the movement of SHR in a microtubule-dependent manner. Examination of the intracellular dynamics of KinG and SHR suggest that interaction of KinG promotes the pausing of SHR-associated vesicles perhaps in regions of the cell where microtubules and actin overlap. It is unclear how pausing might promote the intercellular movement of SHR; however, one possibility is that it allows for the posttranslational modification of SHR or the transfer of SHR to complexes that are destined for plasmodesmata.
Several KCH proteins colocalize with actin (Preuss et al., 2004; Frey et al., 2009; Buschmann et al., 2011; Klotz and Nick, 2012). This finding was attributed to the actin-binding properties of the CH domain. However, no functional analyses were ever performed to determine if the CH domain alone has the capacity to bind actin in vivo. Our results (Fig. 2) show that fusion of the KinG CH domain to YFP (YFP-KinGΔKin) is insufficient to localize the protein to actin. This finding may indicate that the binding of the CH domain to actin is weak/transient or that binding of kinesin motor domain to microtubules is required for the CH domain to function as an actin-binding motif. Gimona et al. (2002) suggested that single CH domains could serve as scaffolds, rather than actin cross linkers. The findings that KinG forms a complex with SIEL and SHR, and that endosomes pause on KinG support either hypothesis.
Hamada et al. (2012) suggested that pausing on microtubules could accommodate the interaction and exchange of molecules between different cellular organelles. In addition, the authors suggested that this pausing could be facilitated by specific kinesins that cross link organelles and microtubules. KinG may serve as one such kinesin. Further experiments may determine if KinG is associated with pausing of other organelles such as peroxisomes, Golgi vesicles, or p bodies and shed additional light on the relatively unexplored phenomenon of organelle pausing and the role KCHs play in that process. Roles for KinG in the nucleus remain to be elucidated. Insights into the nuclear function of SIEL were recently provided by Liu et al. (2016), who showed that SIEL (referred to as DSP3) functions as a scaffold in the assembly and function of the small nuclear RNA (snRNA) processing complex. KinG may interact with SIEL in the nucleus to promote the assembly of this complex as well.
Various plant organelles traffic on actin tracks and pause when encountering microtubules. Interestingly, in assays performed by Hamada et al. (2012), organelle pausing persisted in presence of oryzalin, suggesting that the plant cell provides alternate mechanisms to sustain this process. This indicates the existence of redundancy not only at the genetic level but also at the cellular level. We observe a similar phenomenon of “cellular redundancy” with respect to the cell-to-cell trafficking of SHR. While microtubules, actin, and the endomembrane promote movement of SHR, blocking any of them using chemical inhibitors does not completely eliminate trafficking of SHR (Wu and Gallagher, 2013, 2014). Null alleles of siel are embryonic lethal, and siel partial loss of function still allow for the trafficking of SHR, making it difficult to conclude that SIEL is indispensable for the movement of SHR (Koizumi et al., 2011). Similarly, the kinG null mutant only partially inhibits the transport of SHR and does not enhance the siel-4 mutant phenotype. The partial role of KinG in promoting the movement of SHR can be explained by additional KCH proteins that may serve similar functions. However, the fact that treatment of kinG with oryzalin only decreases movement by 68.9% argues against an essential role. Given that plasmodesmata-mediated signaling is essential for plant development and survival, it is possible that multiple cellular pathways have evolved to maintain trafficking. This may explain the limited ability of genetics to elucidate plasmodesmata-mediated protein trafficking and our consistent observations that inhibition of no one specific pathway is sufficient to completely block the movement of SHR.
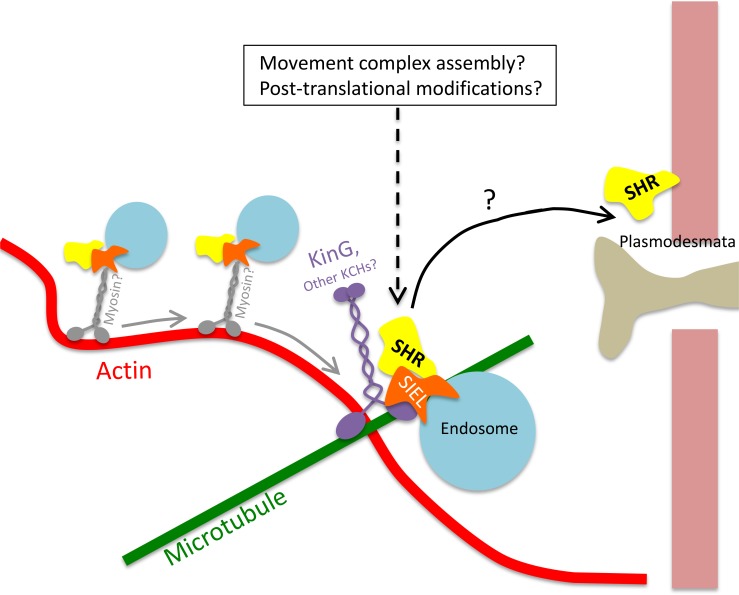
Oparka (2004) proposed the “grab-a-Rab” hypothesis to explain how proteins are targeted to plasmodesmata. In this model, non-cell-autonomous proteins associate with endosomes through interaction with Rab proteins. The whole protein-Rab-endosome complex is shuttled to plasmodesmata. This concept is mostly based on the high enrichment of specific cargo proteins in and around plasmodesmata and the localization of the N terminus fragment of Rab11 to plasmodesmata (Escobar et al., 2003). Our results, however, suggest that the cytoskeleton and endomembrane systems do not play a dynamic part in delivering SHR to plasmodesmata, but rather serve as a stable platform that likely facilitates the assembly or modification of a movement-competent SHR complex. This is mainly supported by the findings that microtubule stabilization or misorientation do not hinder the trafficking of SHR (Wu and Gallagher, 2013) and that KinG serves as a microtubule pausing site for SHR-associated endosomes. According to our model (Fig. 6), SHR associates to endomembrane vesicles via interaction with SIEL. These vesicles move along actin strands, likely via myosins, and pause on microtubules. This pausing is facilitated by interaction of SIEL with KinG. Pausing events may facilitate processes that promote trafficking such as assembly of movement-competent complex or posttranslational modifications that enable directing SHR to plasmodesmata. The exact manner by which SHR is targeted to plasmodesmata, however, is not known.
Figure 6.
A hypothetical model for the involvement of KinG in the intercellular movement of SHR. SIEL binds to SHR and localizes it to endosomes. These endosomes then move along actin strands, likely via myosin motor proteins. Subsequently, the endosomes reach an actin-microtubule junction in which they pause via interaction of SIEL and KinG. Pausing on KinG may then facilitate the assembly of a movement-competent protein complex and/or posttranslational modifications that promote the cell-to-cell movement of SHR through plasmodesmata.
MATERIALS AND METHODS
CRISPR/Cas9 Mutagenesis
The kinG null mutant was generated using the CRISPR/Cas9 mutagenesis method described previously (Mao et al., 2013; Feng et al., 2014) with several modifications. A CRISPR/Cas9 target site within the KinG ORF, between the CH and kinesin motor domain, was chosen using the CRISPR-PLANT platform (http://www.genome.arizona.edu/crispr/). The guide RNA was constructed using the DNA oligos kinG_CRISPR_F and kinG_CRISPR_R (Supplemental Table S1). These oligos were phosphorylated using PNK (New England Biolabs, https://www.neb.com/), annealed, and cloned into the psgR-Cas9-At vector using the BbsI restriction enzyme (Thermo Fisher Scientific, https://www.thermofisher.com/us/en/home.html). The generated cassette was then cloned into the pCAM-NAP:eGFP binary vector, in which seed-coat-expressed eGFP serves as selection marker (Wu et al., 2015). T1 seeds containing the CRISPR/Cas9 cassette were screened based upon seed coat eGFP fluorescence. T1 plants containing large insertions were screened using the kinG_fla_F and kinG_fla_R primers, flanking a 302-bp fragment surrounding the gRNA target site. The CRISPR/Cas9 cassette was segregated out, and homozygous T3 kinG mutants were backcrossed twice with Col-0 wild-type plants to eliminate possible nonspecific mutations.
Plasmid Construction and Transformation
The plasmids used in this research were cloned using the Gateway cloning system (Thermo Fisher Scientific, https://www.thermofisher.com/us/en/home.html). A pDONR207-KinG plasmid containing the KinG ORF with a stop codon (Buschmann et al., 2011) was a kind gift from Dr. Henrik Buschmann (University of Osnabruck, Germany). To clone p35S:YFP-KinG and p35S:TagRFP-KinG, pDONR207-KinG was recombined to the pEarleyGate104 (Earley et al., 2006) and pSiteII-6C1 (Martin et al., 2009) respectively. ΔCHKinG is an N terminus-truncated version of KinG starting 420 bp from the original start codon. This version was cloned by amplifying the KinG CDS with the KinGΔCH_F and KinGΔCH_R primers (Supplemental Table S1). KinGΔKin is a C terminus-truncated version of KinG that ends 1,806 bp upstream to the original stop codon. This version was cloned by amplifying the KinG CDS with the KinGΔKin_F and KinGΔKin_R primers (Supplemental Table S1). These versions were further cloned into a pENTR/d-topo entry plasmid (Thermo Fisher Scientific, https://www.thermofisher.com/us/en/home.html). p35S:YFP-KinGΔCH and p35S:YFP-KinGΔKin were cloned by recombining pENTR-KinGΔCH and pENTR-KinGΔKin with the pEarleyGate104 plasmid. To clone C-terminal YFP fusion constructs, the stop codon was deleted from pDONR207-KinG using the QuickChange II site-directed mutagenesis kit (Agilent Technologies, http://www.agilent.com/). This plasmid was then recombined with modified versions of pGreenBarT containing SHR:attR1/R2-YFP or pEN7:attR1/R2-YFP (Wu et al., 2014) to create SHR:KinG-YFP and pEN7:KinG-YFP. The cloned KinG promoter is a 2 kb fragment upstream to the KinG start codon. This fragment was amplified from genomic Col-0 DNA using the KpnI_pKING_F and XhoI_pKinG_R primers (Supplemental Table S1). The fragment was then used to replace the SHR promoter from the SHR:H2B-YFP using KpnI and XhoI restriction sites (Wu et al., 2014), forming the KinG:H2B-YFP vector. p35S:YFP-SHR, p35S:YFP-SHRT289I, and p35S:YFP-SHRΔLNELDV were cloned by the recombination of pDONR221 containing either the SHR, SHRT289I, or SHRΔLNELDV ORF to pEarleyGate104. All plasmids were transformed into the Agrobacterium strain GV3101-pSouppMP. A. thaliana (Arabidopsis thaliana; Col-0) transformation was done using the floral-dip method (Clough and Bent, 1998). Transgenic plants were screened by resistance to glufosinate-ammonium (Basta) in soil.
Plant Material and Growth Conditions
The A. thaliana Col-0 ecotype was used as the wild type in all experiments. Seeds were sterilized in 70% commercial bleach, rinsed with sterile Milli-Q water three times, and imbibed at 4°C for 2 d prior to plating. Plants were germinated vertically on 1× Murashige and Skoog medium (Caisson, www.caissonlabs.com) containing 0.05% w/v MES (pH 5.7), 1.0% w/v Suc, and 1.0% granulated agar (DIFCO, www.bd.com) in a growth chamber at 19°C, 16-h light/8-h dark cycle. Root imaging was conducted 4 to 5 d after plating in all experiments. Transgenic plants expressing SHR:SHR-GFP were crossed to the homozygous kinG mutant. Detection of homozygous kinG mutants in the F2 population was done using PCR using the kinG_fla_F and kinG_fla_R primers (Supplemental Table S1). Detection of homozygous mutants expressing SHR:SHR-GFP was done on F3 plants based upon fluorescence. To generate kinG siel-4 double mutants, the kinG homozygous mutants containing the SHR:SHR-GFP marker was crossed to siel-4 mutants (Koizumi et al., 2011). Selection for siel-4 T-DNA lines was done using the siel-4_LP, siel-4_RP, and the LBb1 primers (Supplemental Table S1). T-DNA lines with an insertion in the third exon of KinH (SALK_117796.49.20.x) were acquired from the ABRC. Homozygous mutants were identified using kinH_LP and kinH _RP primers (Supplemental Table S1) and crossed to the kinG homozygous mutants containing the SHR:SHR-GFP construct.
CoIP/MS/MS Analysis
All seedlings were germinated and grown for 5 d on standard Murashige and Skoog medium (Caisson, www.caissonlabs.com) containing 1% Suc. After 5 d of growth, roots were excised from several hundred seedlings and processed as previously described (Michniewicz et al., 2007). The two mobile SHR lines used were SHR:SHR-GFP (Sena et al., 2004) and SHR:SHR-YFP (provided prior to publication by Dr. Ben Scheres; Long et al., 2015). The two nonmobile SHR proteins used were SHR:SHRΔLNELDV-GFP and SHR:SHRT289I-GFP (Gallagher et al., 2004; Gallagher and Benfey, 2009). To identify potential mediators of SHR movement, the MS results were filtered to identity proteins (with a minimum of two unique peptides) that are expressed in the stele of the root meristem (Birnbaum et al., 2003; Brady et al., 2007) and coimmunoprecipitated with both of the mobile SHR, but not the immobile variants
Transient Expression in Nicotiana benthamiana
Well-expanded leaves of 3- to 4-week-old N. benthamiana plants were infiltrated according to the procedure previously described (Goodin et al., 2002). For colocalization of KinG with microtubules, Agrobacterium culture containing the 35S:TagRFP-KinG construct was infiltrated into transgenic N. benthamiana expressing p35S:GFP-TUA6 (Gillespie et al., 2002), a kind gift from Dr. Karl Oparlka (University of Edinboro, UK). For colocalization of KinG and Actin, Agrobacterium culture containing 35S:YFP-KinG was coexpressed with the actin marker 35S:TagRFP-UtrCH (Levy et al., 2015), a kind gift from Dr. Amit Levy (University of Florida). Both cultured were mixed in a 1:1 OD ratio prior to infiltration.
Chemical Inhibitor Treatments
Stock solutions of 20 mm latrunculin B, 20 mm cytochalasin D (Sigma, www.sigmaaldrich.com), or 2 mm Oryzalin (Sigma) were prepared in dimethyl sulfoxide and stored at −20°C. For the treatment of A. thaliana roots, 4- to 5-d-old seedlings were grown on regular Murashige and Skoog agar plates and transferred to the Murashige and Skoog plates containing the indicated concentration of oryzalin for the specified extent of time. Treatment of N. benthamiana leaves was 48 h after agroinfiltration with 35S:YFP-KinG. An MES 10 mm solution was supplemented with either oryzalin 2 μm, latrunculin B 20 μm, cytochalasin D 20 μm, or no inhibitor as control was infiltrated to the leaf. Response to the different inhibitors was monitored over time using confocal microscopy.
Yeast Two-Hybrid Assay
The coding sequences of SHR and SIEL from the Col-0 ecotype were cloned into pDEST22 as bait and transformed into the yeast strain Y187. The coding sequences of KinG, ΔCHKinG, and KinGΔKin was cloned into pDEST32 as prey and transformed into the yeast strain AH109. Protein−protein interactions were tested in diploid yeast cells by mating the two yeast strains as described by the Matchmaker protocol (Clontech).
BiFC Analysis
The BiFC plasmids are the modified versions of pDEST-VYCE(R) and pDEST-VYNE(R) (Wu et al., 2014; Gehl et al., 2009), to which the ORF of SIEL and KinG were cloned, respectively. Protoplasts were isolated from well-expanded source leaves of 3-week-old plants grown under normal light conditions. The enzyme solution consisted of 1.5% (w/t) Cellulase R-10 and 0.5% Macerozyme R-10 (Yakult Pharmaceutical). In brief, 10 μg of plasmid DNA was mixed to a solution containing an equal volume of 40% (v/v) polyethylene glycol (MW 4000; Fluka) with 0.1 m CaCl2 and 0.2 m mannitol. The mix was incubated at room temperature for 13 min and then washed in W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm Glc, and 2 mm MES, pH 5.7). After 24 h incubation in low-light conditions, protoplasts were imaged on a Leica TCS SL microscope using a 20× water-immersion lens.
Confocal Microscopy and Image Analysis
For imaging, roots were counterstained with 0.01 μg/mL propidium iodide in water. All confocal images were obtained using a 20× water-immersion lens on a Leica TCS SL microscope equipped with an argon-krypton ion laser. For colocalization analysis, dual channel observation was conducted as sequential scan, to prevent the detection of nonspecific signals. Colocalization analysis was done using ImageJ (https://imagej.nih.gov/ij/). For KinG colocalization with actin or microtubules, a segmented line was drawn through several KinG foci. For SHR colocalization with KinG, a segmented line was drawn through several SHR puncta. Relative fluorescent intensities were then quantified using the plot profile tool for the red and green channel separately. An overlap between a red peak and a green peak was considered one colocalization event. For each experiment, at least four independent images were analyzed. Kymograph construction and analysis was done on a time series imaging of YFP-KinG expressed in N. benthamiana epidermal cells as previously described (Martínez de Alba et al., 2015) using the Multi Kymograph tool in ImageJ (https://imagej.nih.gov/ij/). Vesicle velocity for YFP-SHR was measured using confocal time series. Distance was measured by tracking a given vesicle from the first frame to the last frame it appeared in the focal plane. The ratio between this value and the number of seconds the vesicle remained in frame was defined as vesicle velocity.
FRAP
Fluorescence recovery after photobleaching (FRAP) analysis was done according to (Wu and Gallagher, 2015) with the following modifications: In brief, photobleaching of GFP in the endodermis was done using 20 iterations of the 488-nm laser at 45% power on a Leica TCS SL microscope equipped with an argon-krypton ion laser. The microscope slides holding the seedlings were then placed in a moist petri dish during the increments. The endodermis-to-stele ratio of SHR-GFP in the different time points was then determined using ImageJ. Fluorescent recovery was determined by subtracting the SHR-GFP endodermis-to-stele ratio measured immediately after photobleaching from the endodermis-to-stele ratio at a given time point (E:S[tx] − E:S[t0]).
Accession Numbers
KinG, At1g63640. KinH, At5g41310. SHR, At4g37650. SIEL, At3g08800.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GFP marker lines used in coIP experiment to detect proteins potentially involved in SHR movement.
Supplemental Figure S2. Interaction of KinG with SIEL.
Supplemental Figure S3. KinG MT-binding activity in dividing root meristem cells.
Supplemental Figure S4. Subcellular localization of KinG in N. benthamiana leaf epidermal cells.
Supplemental Figure S5. The KinG kinesin motor domain is required for interaction with SIEL.
Supplemental Figure S6. Response of KinG to cytochalasin D in N. benthamiana leaf epidermal cells.
Supplemental Figure S7. The kinG null mutant displays normal root patterning and growth.
Supplemental Figure S8. FRAP analysis of SHR movement.
Supplemental Figure S9. The kinG × kinH double mutant.
Supplemental Figure S10. Movement of SHR-GFP in kinG mutants treated with oryzalin.
Supplemental Figure S11. Cell-to-cell mobility of SHR in A. thaliana is associated with its endosomal localization in N. benthamiana leaf epidermal cells.
Supplemental Table S1. DNA oligos used in this study.
Supplemental Movie 1. Expression of YFP-SHR in a N. benthamiana leaf epidermal cell.
Supplemental Movie 2. The nuclear region of a N. benthamiana leaf epidermal cell expressing YFP-SHR.
Supplemental Movie 3. Co-expression of YFP-SHR and TagRFP-KinG in a N. benthamiana leaf epidermal cell.
Supplemental Movie 4. Pausing of YFP-SHR on TagRFP-KinG foci.
Supplemental Movie 5. Pausing of YFP-SHR on TagRFP-KinG foci.
Acknowledgments
The coIP/MS assays were done in the laboratory of Dr. Dolf Weijers (Wageningen University, the Netherlands) with the assistance of Dr. Siobhan Brady. We thank Dr. Henrik Buschmann, Dr. Karl Oparka, Dr. Amit Levy, and Dr. Jian-Kang Zhu for sharing material used in this manuscript. We also thank the members of the Gallagher lab, Jason Diaz, and Ruthsabel O’Lexy for their critical reading of the manuscript and valuable inputs.
Footnotes
Z.S. and C.-M.L. were partially supported by National Science Foundation grant 1243945 awarded to K.L.G. Z.S. was partially supported by BARD, the United States - Israel Binational Agricultural Research and Development Fund, Vaadia-BARD Postdoctoral Fellowship Award FI-525-2015.
[CC-BY]: Articles can be viewed without a subscription.
References
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Heinlein M (2000) Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J 22: 315–325 [DOI] [PubMed] [Google Scholar]
- Boyko V, Hu Q, Seemanpillai M, Ashby J, Heinlein M (2007) Validation of microtubule-associated Tobacco mosaic virus RNA movement and involvement of microtubule-aligned particle trafficking. Plant J 51: 589–603 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Buschmann H, Green P, Sambade A, Doonan JH, Lloyd CW (2011) Cytoskeletal dynamics in interphase, mitosis and cytokinesis analysed through Agrobacterium-mediated transient transformation of tobacco BY-2 cells. New Phytol 190: 258–267 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Knorr D, Schuster G, Zambryski P (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637–647 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R. (2012) Putting a bifunctional motor to work: Insights into the role of plant KCH kinesins. New Phytol 193: 543–545 [DOI] [PubMed] [Google Scholar]
- Dixit R. (2015) Kinesin motors: Teamsters’ union. Nat Plants 1: 15126. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Endow SA, Waligora KW (1998) Determinants of kinesin motor polarity. Science 281: 1200–1202 [DOI] [PubMed] [Google Scholar]
- Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K (2003) High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15: 1507–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey N, Klotz J, Nick P (2009) Dynamic bridges—a calponin-domain kinesin from rice links actin filaments and microtubules in both cycling and non-cycling cells. Plant Cell Physiol 50: 1493–1506 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN (2009) Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J 57: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN (2004) Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Sozzani R, Lee CM (2014) Intercellular protein movement: Deciphering the language of development. Annu Rev Cell Dev Biol 30: 207–233 [DOI] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ (2002) Functional plasticity of CH domains. FEBS Lett 513: 98–106 [DOI] [PubMed] [Google Scholar]
- Gimona M, Mital R (1998) The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J Cell Sci 111: 1813–1821 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Hamada T, Tominaga M, Fukaya T, Nakamura M, Nakano A, Watanabe Y, Hashimoto T, Baskin TI (2012) RNA processing bodies, peroxisomes, Golgi bodies, mitochondria, and endoplasmic reticulum tubule junctions frequently pause at cortical microtubules. Plant Cell Physiol 53: 699–708 [DOI] [PubMed] [Google Scholar]
- Harries P, Ding B (2011) Cellular factors in plant virus movement: At the leading edge of macromolecular trafficking in plants. Virology 411: 237–243 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Kim JY, Rim Y, Wang J, Jackson D (2005) A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev 19: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz J, Nick P (2012) A novel actin-microtubule cross-linking kinesin, NtKCH, functions in cell expansion and division. New Phytol 193: 576–589 [DOI] [PubMed] [Google Scholar]
- Koizumi K, Hayashi T, Wu S, Gallagher KL (2012) The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc Natl Acad Sci USA 109: 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Wu S, MacRae-Crerar A, Gallagher KL (2011) An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr Biol 21: 1559–1564 [DOI] [PubMed] [Google Scholar]
- Korenbaum E, Rivero F (2002) Calponin homology domains at a glance. J Cell Sci 115: 3543–3545 [DOI] [PubMed] [Google Scholar]
- Kragler F, Curin M, Trutnyeva K, Gansch A, Waigmann E (2003) MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol 132: 1870–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. (2005) Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Lee YR, Liu B (2004) Cytoskeletal motors in Arabidopsis. Sixty-one kinesins and seventeen myosins. Plant Physiol 136: 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Zheng JY, Lazarowitz SG (2015) Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 25: 2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li S, Chen Y, Kimberlin AN, Cahoon EB, Yu B (2016) snRNA 3′ end processing by a CPSF73-containing complex essential for development in Arabidopsis. PLoS Biol 14: e1002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu M, Liang N, Zheng Y, Yu Q, Wu S (2017) Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis. Proc Natl Acad Sci USA 114: 4005–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Smet W, Cruz-Ramírez A, Castelijns B, de Jonge W, Mähönen AP, Bouchet BP, Perez GS, Akhmanova A, Scheres B, et al. (2015) Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27: 1185–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6: 2008–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Martínez de Alba AE, Moreno AB, Gabriel M, Mallory AC, Christ A, Bounon R, Balzergue S, Aubourg S, Gautheret D, Crespi MD, et al. (2015) In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res 43: 2902–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Niehl A, Heinlein M (2011) Cellular pathways for viral transport through plasmodesmata. Protoplasma 248: 75–99 [DOI] [PubMed] [Google Scholar]
- Niehl A, Peña EJ, Amari K, Heinlein M (2013) Microtubules in viral replication and transport. Plant J 75: 290–308 [DOI] [PubMed] [Google Scholar]
- Oparka KJ. (2004) Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci 9: 33–41 [DOI] [PubMed] [Google Scholar]
- Padgett HS, Epel BL, Kahn TW, Heinlein M, Watanabe Y, Beachy RN (1996) Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J 10: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Kovar DR, Lee YR, Staiger CJ, Delmer DP, Liu B (2004) A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiol 136: 3945–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Matos JL, Anleu Gil MX, Kornfeld A, Bettadapur A, Abrash E, Allison HR, Badgley G, Vogel JP, Berry JA, et al. (2017) Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355: 1215–1218 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Day IS (2001) Kinesins in the Arabidopsis genome: A comparative analysis among eukaryotes. BMC Genomics 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M (2008) Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 9: 2073–2088 [DOI] [PubMed] [Google Scholar]
- Sambade A, Heinlein M (2009) Approaching the cellular mechanism that supports the intercellular spread of Tobacco mosaic virus. Plant Signal Behav 4: 35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage NS, Walker T, Wieckowski Y, Schiefelbein J, Dolan L, Monk NA (2008) A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol 6: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schneider R, Persson S (2015) Connecting two arrays: The emerging role of actin-microtubule cross-linking motor proteins. Front Plant Sci 6: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN (2004) A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Serazev TV, Kalinina NO, Nadezhdina ES, Shanina NA, Morozov SY (2003) Potato virus X coat protein interacts with microtubules in vitro. Cell Biol Int 27: 271–272 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D (2000) Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–782 [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Wright KM, Wood NT, Roberts AG, Chapman S, Boevink P, MacKenzie KM, Oparka KJ (2007) Targeting of TMV movement protein to plasmodesmata requires the actin/ER network; evidence from FRAP. Traffic 8: 21–31 [DOI] [PubMed] [Google Scholar]
- Winter N, Kollwig G, Zhang S, Kragler F (2007) MPB2C, a microtubule-associated protein, regulates non-cell-autonomy of the homeodomain protein KNOTTED1. Plant Cell 19: 3001–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KM, Cowan GH, Lukhovitskaya NI, Tilsner J, Roberts AG, Savenkov EI, Torrance L (2010) The N-terminal domain of PMTV TGB1 movement protein is required for nucleolar localization, microtubule association, and long-distance movement. Mol Plant Microbe Interact 23: 1486–1497 [DOI] [PubMed] [Google Scholar]
- Wu X, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, Weigel D (2003) Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Wu S, Gallagher KL (2013) Intact microtubules are required for the intercellular movement of the SHORT-ROOT transcription factor. Plant J 74: 148–159 [DOI] [PubMed] [Google Scholar]
- Wu S, Gallagher KL (2014) The movement of the non-cell-autonomous transcription factor, SHORT-ROOT relies on the endomembrane system. Plant J 80: 396–409 [DOI] [PubMed] [Google Scholar]
- Wu S, Gallagher KL (2015) Techniques for assessing the effects of pharmacological inhibitors on intercellular protein movement. Methods Mol Biol 1217: 245–258 [DOI] [PubMed] [Google Scholar]
- Wu S, Lee CM, Hayashi T, Price S, Divol F, Henry S, Pauluzzi G, Perin C, Gallagher KL (2014) A plausible mechanism, based upon Short-Root movement, for regulating the number of cortex cell layers in roots. Proc Natl Acad Sci USA 111: 16184–16189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, O’Lexy R, Xu M, Sang Y, Chen X, Yu Q, Gallagher KL (2016) Symplastic signaling instructs cell division, cell expansion, and cell polarity in the ground tissue of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 113: 11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Rossidivito G, Hu T, Berlyand Y, Poethig RS (2015) Traffic lines: new tools for genetic analysis in Arabidopsis thaliana. Genetics 200: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]