Prolines in transit peptides are critical for the preprotein translocation through import channels and prevent inappropriate insertion of TMD-containing preproteins into chloroplast envelopes.
Abstract
Chloroplasts import many preproteins that can be classified based on their physicochemical properties. The cleavable N-terminal transit peptide (TP) of chloroplast preproteins contains all the information required for import into chloroplasts through Toc/Tic translocons. The question of whether and how the physicochemical properties of preproteins affect TP-mediated import into chloroplasts has not been elucidated. Here, we present evidence that Pro residues in TP mediate efficient translocation through the chloroplast envelope membranes for proteins containing transmembrane domains (TMDs) or proteins prone to aggregation. By contrast, the translocation of soluble proteins through the chloroplast envelope membranes is less dependent on TP prolines. Proless TPs failed to mediate protein translocation into chloroplasts; instead, these mutant TPs led to protein mistargeting to the chloroplast envelope membranes or nonspecific protein aggregation during import into chloroplasts. The mistargeting of TMD-containing proteins caused by Pro-less TPs in wild-type protoplasts was mimicked by wild-type TPs in hsp93-V protoplasts, in which preprotein translocation is compromised. We propose that the physicochemical properties of chloroplast proteins affect protein translocation through the chloroplast envelope, and prolines in TP have a crucial role in the efficient translocation of TMD-containing proteins.
The plastid is a unique organelle found in land plants and algae. Of the four main types of plastids, chloroplasts are responsible for photosynthesis, amino acid and fatty acid synthesis, and the plant immune response (Padmanabhan and Dinesh-Kumar, 2010). Chloroplasts are thought to contain more than 3,000 different types of proteins; however, the chloroplast genome only encodes approximately 100 proteins. Most proteins in the chloroplast proteome are encoded by the nuclear genome and post-translationally imported into the chloroplast (Lee et al., 2013; Li and Chiu, 2010; Jarvis, 2008).
The import of nuclear-encoded proteins into chloroplasts occurs via a complex process with distinct steps that occur in a sequential manner (Lee et al., 2006; Pilon et al., 1995). First, chloroplast preproteins are translated on cytosolic ribosomes and then navigate through the cytosol to chloroplasts (Lee et al., 2013). During this step, chloroplast preproteins are maintained in an unfolded state by cytosolic chaperones and factors such as 14-3-3 and Hsp70/90, which are essential for efficient targeting to chloroplasts (May and Soll, 2000; Qbadou et al., 2006). The preproteins arrive at the chloroplast surface and are recognized by specific import receptors, such as Translocon at the Outer Envelope of Chloroplasts 159 (Toc159) and Toc33 (Li and Chiu, 2010; aessler and Schnell, 2006, 2009). Subsequently, preproteins are translocated through the import channel (Toc75) at the outer envelope membrane (Hinnah et al., 2002; Paila et al., 2016). Then, preproteins are translocated through the inner envelope membrane by Translocon at the Inner Envelope of Chloroplasts (TIC), in which atTic20 functions as a preprotein channel (Li and Chiu, 2010). Recently, a mega-dalton complex composed of atTic20, atTic21, atTic214, atTic56, and atTic100 was suggested to function as a general Tic translocon (Kikuchi et al., 2009, 2013), although the exact nature of Tic translocon is still elusive (de Vries et al., 2015; Bölter and Soll, 2017; Agne et al., 2017; Jensen and Leister, 2014). Finally, stromal chaperones such as Hsp93, cpHsc70, and Hsp90C, together with atTic110 and atTic40, pull the preproteins into the chloroplast stroma (Flores-Pérez and Jarvis, 2013; Chou et al., 2006; Su and Li, 2010; Kovacheva et al., 2007; Inoue et al., 2013; Shi and Theg, 2010; Liu et al., 2014). Thus, the entire import process contains many distinct steps, each of which involves many proteins and factors. This process is even more complex, as recent studies showed that protein import is regulated according to conditions such as plant age and environmental conditions (Teng et al., 2012; Li and Teng, 2013). Plant cells have another mechanism, the unimported plastid precursor response, which limits the number of preproteins in the chloroplast translocation pathway below a certain threshold to prevent nonspecific aggregate formation (Lee et al., 2009b, 2016).
The precise mechanism that efficiently translocates preproteins through the import channel after binding to the receptor complexes at the chloroplast surface has not been elucidated. Chloroplast preproteins are in a largely unfolded state before reaching their final destination, which may allow efficient translocation across the outer/inner envelopes and increase the chances of interacting with molecular chaperones involved in delivering preproteins to chloroplasts (Flores-Pérez and Jarvis, 2013). The N-terminal transit peptide (TP) of unfolded chloroplast preproteins is inserted into the import channel at the outer envelope membrane and then passed through channels at inner envelope membranes to finally emerge into the stroma. Several complexes are present at the stromal side of the inner envelope membrane that bind to and pull the TP through the envelope (Chou et al., 2006; Li and Chiu, 2010). Several stromal chaperones are thought to be critical for pulling preproteins into chloroplasts (Flores-Pérez and Jarvis, 2013; Su and Li, 2010; Kovacheva et al., 2007; Inoue et al., 2013; Shi and Theg, 2010; Liu et al., 2014). In mitochondria, mtHsp70 functions as the ratchet that pulls substrates into this organelle (Yamano et al., 2008; Esaki et al., 1999). It is not fully understood how the pulling process occurs in chloroplasts, or whether TPs contain any sequence motifs that direct the pulling. Chloroplasts import soluble and membrane proteins that contain transmembrane domains (TMDs; Okawa et al., 2014; Viana et al., 2010; Froehlich and Keegstra, 2011; Lee et al., 2017). The mechanistic function of chloroplast import channels is not fully understood, and it is possible that specific properties of the polypeptides that pass through the import channels may affect their translocation. For example, the translocation rate of nascent polypeptides passing through the Sec61 channel of the endoplasmic reticulum is affected by the type of amino acids present in the polypeptides (Kang et al., 2006; Kim et al., 2002).
In this study, we investigated the mechanisms that mediate efficient translocation of preproteins through chloroplast import channels. We postulated the existence of a pulling activity that efficiently translocates preproteins through import channels. Previous studies reported that the stromal chaperones Hsp93-V and cpHsc70 are crucial for efficient translocation of preproteins (Lee et al., 2015; Su and Li, 2010; Huang et al., 2016; Liu et al., 2014). To identify sequence motifs involved in efficient translocation of preproteins through the channel, we examined the role of Pro residues, which are abundant in most TPs. Pro residues in polypeptides are associated with two main functions: they break the local secondary structure, thereby giving unstructured property to TPs, and they also introduce localized kinks into the secondary structure. Therefore, we predict that Pro residues contribute to the structural flexibility of TPs by terminating secondary structural elements such as α-helices and β-sheets (Guzzo, 1965; Bruce, 2000; Zhang and Glaser, 2002; Zybailov et al., 2008; Bhushan et al., 2006). The structural flexibility of TPs is thought to be important for their function in protein import into chloroplasts, although the importance of Pro residues and Pro-induced TP flexibility in protein import into chloroplasts has not been examined in vivo. Here, we present evidence that Pro residues in TPs have a crucial role in the efficient translocation of preproteins through chloroplast import channels, especially preproteins containing TMDs or those prone to aggregation. We show that both TMD-containing and aggregation-prone preproteins containing TPs that lack Pro localize at the chloroplast outer envelope membrane, and the latter exists as more severely aggregated forms. These combined results indicate that prolines in TPs have crucial roles in translocating preproteins across the chloroplast envelopes.
RESULTS
Pro Residues in TPs Are Crucial for Translocation through the Chloroplast Envelope Membrane
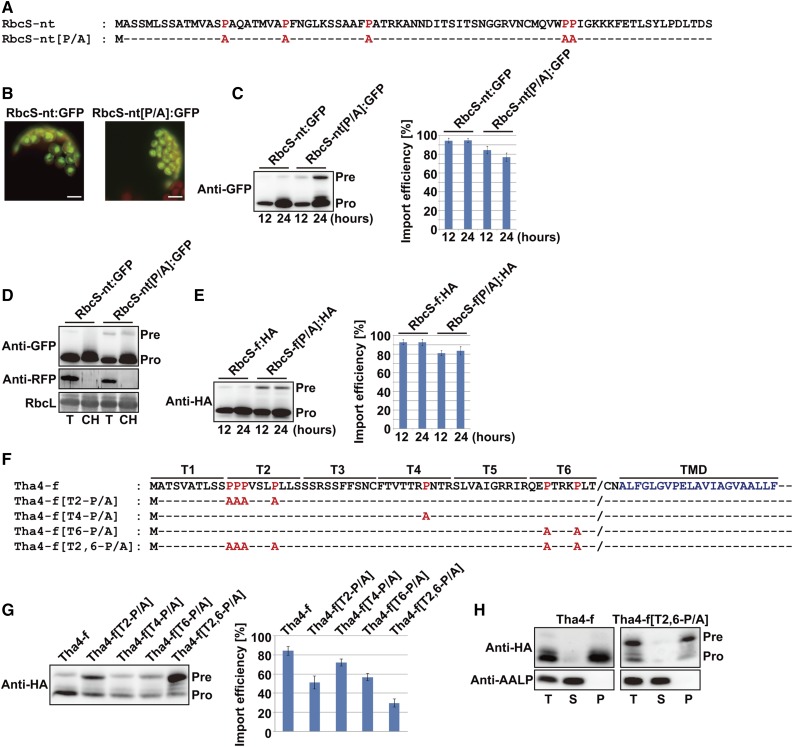
To examine the role of the abundant Pro residues in TPs during protein import into chloroplasts, we substituted five Pro residues in the TP of Rubisco small subunit (RbcS-TP) with alanines to generate RbcS-nt[P/A], which was then fused to GFP (Fig. 1A). The control RbcS-nt:GFP and mutant RbcS-nt[P/A]:GFP constructs were transformed into Arabidopsis protoplasts, and their import into chloroplasts was investigated using a fluorescence microscope (Jin et al., 2001). At 12 to 24 hours after transformation, GFP fluorescence was predominantly observed within chloroplasts for both constructs (Fig. 1B). To confirm these results at the biochemical level, total protein extracts from protoplasts were analyzed by western blotting using anti-GFP antibody. Compared with the wild-type RbcS-nt:GFP, RbcS-nt[P/A]:GFP showed lower import efficiency (Fig. 1C). However, these results were not consistent with microscopy images, which showed that RbcS-nt[P/A]:GFP was not in the cytosol (Fig. 1B). Therefore, we examined whether RbcS-nt[P/A]:GFP precursors were associated with, but not imported into, chloroplasts. Chloroplasts were purified from gently lysed protoplasts using a Percoll gradient and then analyzed by western blotting using anti-GFP antibody (Lee et al., 2008). Unimported RbcS-nt[P/A]:GFP precursors copurified with the chloroplast fraction, suggesting that Pro residues in the RbcS TP have a role in translocation through the envelope membranes but not in navigation through the cytosol or binding to receptors at the chloroplast surface (Fig. 1D). To assess the contribution of Pro residues to preprotein import into chloroplasts in a more physiologically relevant context, we tagged full-length RbcS and RbcS-f[P/A] with hemagglutinin (HA) at the C terminus to generate RbcS-f:HA (control) and RbcS-f[P/A]:HA. RbcS-f[P/A]:HA also displayed lower chloroplast import efficiency than the control (Fig. 1E), in agreement with the reduced chloroplast import efficiency observed for RbcS-nt[P/A]:GFP.
Figure 1.
Pro residues in TPs affect preprotein translocation into chloroplasts. A and F, Sequences of wild-type TPs and their P/A substitution mutants. P/A substitutions are indicated in red. B, Localization of reporter proteins. Protoplasts from Arabidopsis leaf tissues were transformed with the indicated constructs, and GFP patterns were observed 12 to 24 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. C, E, and G, Western-blot analysis of reporter proteins. Protein extracts from protoplasts were analyzed by western blotting using anti-GFP and anti-HA antibodies. Import efficiency is defined as the percentage of the processed form divided by the total amount of expressed protein. The protein band intensity was measured using software installed on the LAS3000 imager (FUJI FILM). Data represent means (n = 3) with sd. Pre, precursor form; Pro, processed form. D, Copurification of RbcS-nt[P/A]:GFP precursors with chloroplasts. Protoplasts were gently lysed 12 h after transformation, and chloroplasts were isolated using a Percoll gradient. Total and chloroplast fractions were analyzed by western blotting with anti-GFP and anti-RFP antibodies. RFP, cotransformed control for cytosolic proteins. Rubisco complex large subunit (RbcL) stained with Coomassie Brilliant Blue (CBB) was used as a loading control. T, total protein; CH, chloroplast fraction; Pre, precursor form; Pro, processed form. H, Subcellular fractionation by ultracentrifugation. Fractions were analyzed by western blotting with anti-GFP antibody. Endogenous AALP was used as a control for soluble proteins. T, total protein; S, soluble fraction; P, pellet fraction; Pre, precursor form; Pro, processed form.
Next, we investigated whether Pro residues in other TPs have the same functions as those in RbcS TP. We previously showed that TPs can be grouped by the sequence motifs in the TP region (Lee et al., 2008). We examined the role of Pro residues in the chlorophyll a/b-binding protein (Cab) TP by generating P/A substitutions (Supplemental Fig. S1A). Cab is a thylakoid membrane protein that contains three hydrophobic TMDs. We substituted six Pro residues in Cab-TP with alanines to generate Cab-nt[P/A], which was then fused to GFP. We also introduced the same mutations in the full-length Cab (Cab-f) tagged with HA to generate Cab-f[P/A]:HA. Cab-nt[P/A]:GFP fluorescence was not observed in the protoplast cytoplasm, consistent with the localization of RbcS-nt[P/A]:GFP fluorescence (Supplemental Fig. S1B). However, both Cab-nt[P/A]:GFP and Cab-f[P/A]:HA displayed more severely defective import into chloroplasts compared with RbcS constructs (Supplemental Fig. S1, C and D). We examined the localization of precursor proteins by separating total protein extracts into soluble and insoluble (pelleted) fractions using ultracentrifugation and then performing western-blot analysis. Unimported precursors of Cab-f[P/A]:HA were largely detected in the pellet, suggesting that they were trapped at the import channel (Supplemental Fig. S1E).
Next, we tested whether other membrane proteins also display more severe defects in chloroplast import than stromal proteins when TP Pro residues are substituted with Ala by examining the import of thylakoid assembly 4 (Tha4; Fig. 1F; Dabney-Smith and Cline, 2009). Tha4 contains a TMD adjacent to the TP (Fig. 1F). The Tha4 TP contains seven Pro residues: four in the second 10 aa (T2), one in T4, and two in T6 (Fig. 1F). We generated HA-tagged full-length Tha4 (Tha4-f:HA) and four P/A substitution mutants, Tha4-f[T2-P/A]:HA, Tha4-f[T4-P/A]:HA, Tha4-f[T6-P/A]:HA, and Tha4-f[T2,6-P/A]:HA, in which all Pro residues in T2, T4, and T6 were replaced with Ala, in addition to both T2 and T6. These Tha4 constructs were transformed into Arabidopsis protoplasts, and their import into chloroplasts was examined. The P/A substitution in single T segments resulted in varying degrees of import defects (Fig. 1G). Simultaneous P/A substitutions in both T2 and T6 segments caused more severe import defects (Fig. 1G), indicating an additive effect of the mutations. Unimported precursors of Tha4-f[T2,6-P/A]:HA were detected in the pellet fraction, similar to Cab-f[P/A]:HA, suggesting that Pro residues in the Tha4 TP region are crucial for efficient translocation through the chloroplast envelope membranes (Fig. 1H).
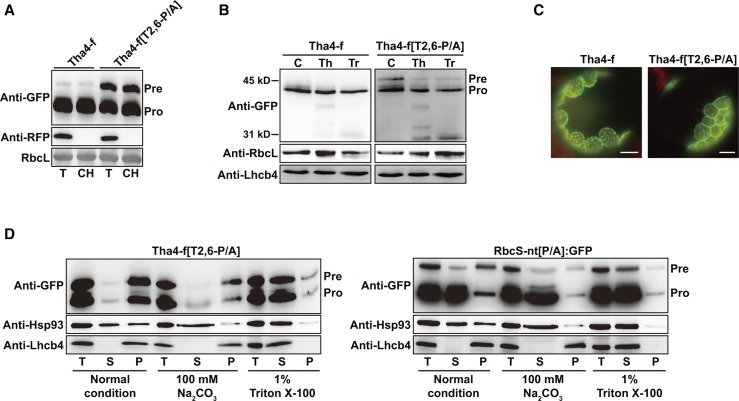
To examine the exact localization of protein precursors detected in the pellet fractions, we generated GFP-tagged Tha4-f[T2,6-P/A] and Tha4-f control constructs. Western-blot analysis revealed that Tha4-f:GFP was largely processed and detected primarily in the chloroplast fraction, confirming its import into chloroplasts (Fig. 2A). By contrast, the Tha4-f[T2,6-P/A]:GFP pool contained a significant amount of unprocessed precursors together with the major processed forms (Fig. 2A). However, there were significantly more processed forms from Tha4-f[T2,6-P/A]:GFP than from Tha4-f[T2,6-P/A]:HA (Fig. 1G), suggesting that the C-terminal GFP moiety somehow facilitates chloroplast import. Despite this difference in import efficiency, these results confirmed that Pro residues have a crucial role in chloroplast import of preproteins with TMDs. The precursor was copurified with chloroplasts (Fig. 2A), indicating that it was successfully targeted to chloroplasts but not efficiently imported. Precursors, but not processed forms, were sensitive to both thermolysin and trypsin (Fig. 2B). The control Tha4-f:GFP also was resistant to both proteinases, confirming that it was successfully imported into chloroplasts (Fig. 2B). These results indicate that the GFP moiety of Tha4-f[T2,6-P/A]:GFP precursors is exposed on the surface of chloroplasts. We examined GFP localization in live protoplasts, and observed that Tha4-f:GFP produced a punctate staining pattern within chloroplasts (Fig. 2C). This localization pattern differed from that of RbcS-nt:GFP in the chloroplast stroma, suggesting that Tha4-f:GFP was inserted into thylakoid membranes. By contrast, Tha4-f[T2,6-P/A]:GFP produced a ring pattern surrounding the chloroplasts, together with only a few punctate structures with weaker fluorescence than those observed with Tha4-f:GFP (Fig. 2C). To examine whether Tha4-f[T2,6-P/A]:GFP was integrated into or peripherally associated with the outer membrane, we performed subcellular fractionation in the presence of 100 mm Na2CO3 or 1% Triton X-100 (Fig. 2D). The precursor of Tha4-f[T2,6-P/A]:GFP was still associated with the membrane fraction under 100 mm Na2CO3 treatment. At this condition, Hsp93 that is peripherally associated with inner membranes was dissociated (Chu and Li, 2011, 2012). By contrast, RbcS-nt[P/A]:GFP copurified with the membrane fraction was significantly washed off at the same condition (Fig. 2D). These combined results suggest that TP prolines have a crucial role in the translocation of TMD-containing cargo proteins through the chloroplast envelope membranes via prevention of inappropriate insertion of TMD into envelope membranes.
Figure 2.
Pro residues in Tha4 TP have a crucial role in translocation through import channels. A, Copurification of Tha4-f[T2,6-P/A]:GFP precursors with chloroplasts. Chloroplasts were isolated and analyzed as described in Figure 1D. RFP, cotransformed control for cytosolic proteins. RbcL (stained with CBB) was used as a loading control. T, total protein; CH, chloroplast fraction; Pre, precursor form; Pro, processed form. B, Thermolysin and trypsin sensitivity of Tha4-f[T2,6-P/A]:GFP precursors. Protoplasts transformed with the indicated constructs were gently lysed and treated with thermolysin or trypsin. Subsequently, protein extracts were analyzed by western blotting using anti-GFP, anti-RbcL, and anti-Lhcb4 antibodies. Pre, precur sor form; Pro, processed form. C, Localization of reporter proteins. Transformed protoplasts were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. D, Membrane integration of Tha4-f[T2,6-P/A]:GFP precursors. Protein extracts were treated with 100 mm Na2CO3 or 1% Triton X-100 followed by ultracentrifugation. Untreated extracts were included as control. Soluble and pellet fractions were analyzed by western blotting with anti-GFP antibody. Hsp93 and LHCB4 were used as controls for peripheral and integral membrane proteins, respectively. T, total protein; S, soluble fraction; P, pellet fraction; Pre, precursor form; Pro, processed form.
In hsp93-V Protoplasts, Translocation of Tha4-f:GFP Is Affected in a Similar Way as Tha4-f[T2,6-P/A]:GFP in Wild-Type Protoplasts
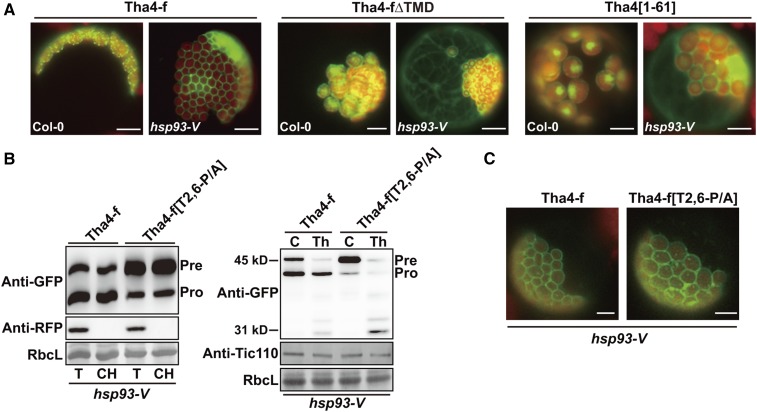
Based on the observed defects in the translocation of TMD-containing preproteins with TPs lacking Pro, we hypothesized that a pulling activity may be required for efficient translocation of preproteins through chloroplast import channels, similarly as for mitochondrial protein import (Lim et al., 2001). A molecular ratchet is involved in pulling incoming mitochondrial proteins into the matrix (Yamano et al., 2008; Esaki et al., 1999), and TPs lacking prolines would not be efficient targets for this pulling activity. Hsp93-V is considered as a component of the stromal import motor that interacts with TPs released from the Tic complex to pull preproteins into the stroma (Huang et al., 2016; Chou et al., 2006; Lee et al., 2015). Plants with a T-DNA insertion in Hsp93-V display severe defects in chloroplast development and exhibit a pale-green phenotype. The efficiency of preprotein import into hsp93-V mutant protoplasts is significantly diminished, and unimported RbcS-nt:GFP precursors are present in the cytosol (Lee et al., 2015, 2016). We used hsp93-V mutant protoplasts to examine chloroplast import of three Tha4 constructs: Tha4-f:GFP, Tha4-fΔTMD:GFP lacking the TMD, and Tha4[1-61]:GFP containing only the TP of Tha4 (Fig. 3). We found that Tha4-f:GFP produced a ring pattern surrounding chloroplasts in hsp93-V protoplasts, similar to Tha4-f[T2,6-P/A]:GFP in wild-type protoplasts, whereas Tha4-fΔTMD:GFP and Tha4[1-61]:GFP localized to the cytosol and chloroplasts, but not to the chloroplast envelope membranes, similar to RbcS-nt:GFP and Cab-nt:GFP (Figs 2C and 3A; Lee et al., 2015). Thus, for TMD-containing protein precursors, the mistargeting pattern observed with TP P/A substitution mutants (lacking Pro) in wild-type protoplasts was identical to that observed with wild-type TPs in hsp93-V protoplasts. These results further support the conclusion that Pro residues in TPs are crucial for efficient translocation of chloroplast preproteins through the envelope membranes.
Figure 3.
Pattern of Tha4-f[T2,6-P/A]:GFP localization in wild-type protoplasts is identical to that of Tha4-f:GFP in hsp93-V protoplasts. A, Localization of reporter proteins. Transformed protoplasts were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. B, Localization of Tha4-f[T2,6-P/A]:GFP in hsp93-V protoplasts. Chloroplasts were isolated and analyzed as described in Figure 1D. Protoplasts were gently lysed and treated with thermolysin. Subsequently, protein extracts were analyzed by western blotting using anti-GFP and anti-Tic110 antibodies. Pre, precursor form; Pro, processed form; T, total protein; CH, chloroplast fraction; RbcL, loading control. C, Localization of Tha4-f[T2,6-P/A]:GFP at chloroplast envelope membranes in hsp93-V protoplasts transformed with the indicated constructs and observed 12 h after transformation.
Next, we examined Tha4-f[T2,6-P/A]:GFP import in hsp93-V protoplasts. The majority of Tha4-f[T2,6-P/A]:GFP was isolated as the protein precursor, and only a minor fraction was isolated as the processed protein, indicating that the Tha4-f[T2,6-P/A]:GFP import efficiency was substantially lower than that of Tha4-f:GFP in hsp93-V protoplasts (Fig. 3B). Tha4-f[T2,6-P/A]:GFP localization was examined in hsp93-V protoplasts. Tha4-f[T2,6-P/A]:GFP formed a ring pattern surrounding the chloroplast, similar to that of Tha4-f:GFP in hsp93-V protoplasts, indicating that Tha4-f[T2,6-P/A]:GFP was inappropriately inserted into the chloroplast envelope membranes in hsp93-V protoplasts (Fig. 3C). We biochemically tested the exact location of GFP in protoplasts and found that precursors copurified with chloroplasts and were sensitive to thermolysin treatment, indicating that the GFP moiety was exposed to the cytosol (Fig. 3B). By contrast, coexpressed cytosolic RFP was not detected in the chloroplast fraction, confirming proper fractionation. These results suggest that the defect caused by the TP P/A mutation is additive to the defect caused by the loss-of-function Hsp93-V mutation.
Inappropriate Insertion of TMD-Containing Preproteins into the Chloroplast Outer Envelope Occurs during Translocation but Not by Direct Targeting from the Cytosol
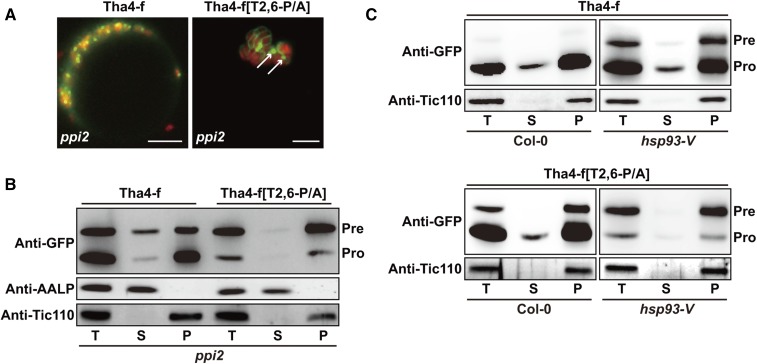
We examined how Tha4-f[T2,6-P/A]:GFP and Tha4-f:GFP localize to the chloroplast envelope membranes in wild-type and hsp93-V protoplasts, respectively. This could occur either during translocation through envelope membranes or by direct insertion into the outer membrane from the cytosol. To test these possibilities, we used ppi2 plants, which have a T-DNA insertion at AtToc159 and display an albino phenotype (Bauer et al., 2000). In ppi2 protoplasts, the initial binding of TP-containing preproteins to the Toc complex is critically disrupted, resulting in cytosolic localization of unimported RbcS-nt:GFP preproteins (Lee et al., 2009a, 2015). However, direct insertion of TMD-containing proteins into the outer envelope membrane does not require AtToc159 (Tu and Li, 2000). First, we examined whether chloroplast import of Tha4-f:GFP is dependent on AtToc159. Tha4-f:GFP was transformed into protoplasts of wild-type and ppi2 plants (Fig. 4). Tha4-f:GFP gave a punctate-staining pattern at chloroplasts in ppi2 protoplasts as in wild-type protoplasts (Figs. 2C and 4A), indicating that Tha4-f:GFP is imported into chloroplasts. In the absence of AtToc159, AtToc159-dependent cargoes can be imported into chloroplasts via a AtToc132/120-dependent manner (Bischof et al., 2011). However, in contrast to wild-type protoplasts, ppi2 protoplasts also showed diffuse signals in the cytosol (Fig. 4A), indicating that a significant portion of Tha4-f:GFP remains in the cytosol. Moreover, the presence of GFP signals in the cytosol confirmed that ppi2 plants have a defect at an early cytosolic step during Tha4-f:GFP import into chloroplasts as reported earlier with RbcS-nt:GFP (Lee et al., 2009a, 2015). Western-blot analysis showed that a significant portion of Tha4-f:GFP was detected as precursor forms in ppi2 protoplasts, confirming the image results (Fig. 4B). Fractionation followed by western-blot analysis showed that Tha4-f:GFP precursors were detected at both the soluble and pellet fractions of ppi2 protoplasts, whereas Tha4-f:GFP precursors were largely detected at the pellet fraction of hsp93-V protoplasts (Fig. 4, B and C). The presence of Tha4-f:GFP precursors at the soluble fraction is consistent with the image of Tha4-f:GFP. In addition, the presence of Tha4-f:GFP precursors at the pellet fraction suggests its association with chloroplasts. However, Tha4-f:GFP did not produce the ring pattern in ppi2 protoplasts as observed in hsp93-V (Fig. 3A), raising the possibility that the location of Tha4-f:GFP precursors at ppi2 chloroplasts is different from that at hsp93-V chloroplasts. These results suggest that insertion of Tha4-f:GFP into chloroplast outer membrane in hsp93-V protoplasts occurs during translocation, but not by direct insertion from the cytosol.
Figure 4.
TPs lacking Pro residues cause mistargeting of TMD-containing preproteins to outer envelope membranes. A, Localization of reporter proteins. The ppi2 protoplasts were transformed with the indicated constructs, and GFP patterns were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. B and C, Subcellular fractionation. Protein extracts were separated into soluble and pellet fractions by ultracentrifugation, and fractions were analyzed by western blotting with anti-GFP antibody. AALP and Tic110 were used as controls for soluble and membrane proteins, respectively. T, total protein; S, soluble fraction; P, pellet fraction; Pre, precursor form; Pro, processed form.
Next, we examined import of Tha4-f[T2,6-P/A]:GFP in ppi2 protoplasts. Tha4-f[T2,6-P/A]:GFP was transformed into ppi2 protoplasts and its localization was examined. In contrast to the localization in wild-type protoplasts, Tha4-f[T2,6-P/A]:GFP formed large aggregates near chloroplasts (indicated by arrows), together with a few rings surrounding the poorly developed chloroplasts in ppi2 protoplasts (Fig. 4A). Western-blot analysis showed that the import efficiency of Tha4-f[T2,6-P/A]:GFP was more severely affected in ppi2 protoplasts than in wild-type protoplasts (Fig. 4, A and B). Fractionation followed by western-blot analysis showed that most Tha4-f[T2,6-P/A]:GFP proteins were detected at the pellet fraction, with only a small proportion at the soluble fraction (Fig. 4B). These results suggest that combination of P/A mutation in TP and the mutation at AtToc159 results in severe inhibition of Tha4-f[T2,6-P/A]:GFP import into chloroplasts. Cytosolically localized precursors of Tha4-f[T2,6-P/A]:GFP may form nonspecific aggregates in ppi2 protoplasts, which might be due to prolonged duration of TMD-containing preproteins with Pro-less TP having reduced flexibility (Bruce, 2000). It is possible that a certain portion of both Tha4-f:GFP and Tha4-f[T2,6-P/A]:GFP was still imported into poorly developed chloroplasts through a AtToc159-independent, but AtToc132/120-dependent manner in ppi2 protoplasts (Bischof et al., 2011). Even if Tha4-f[T2,6-P/A]:GFP was delivered to the import channel via an AtToc132/120-dependent manner, the lack of prolines in TPs may lead to insertion of Tha4-f[T2,6-P/A]:GFP into the envelope membranes, thereby displaying a ring pattern around poorly developed chloroplasts in ppi2 protoplasts (Fig. 4A). These results suggest that membrane insertion of Tha4-f:GFP (in hsp93-V protoplasts) and Tha4-f[2,6-P/A]:GFP precursors (in wild-type and hsp93-V protoplasts) occurs after the step requiring AtToc159, possibly during translocation through the Toc75 channel, but not by direct insertion from the cytosol.
The Role of Pro in Translocation through Chloroplast Import Channels Is a General Feature of TPs
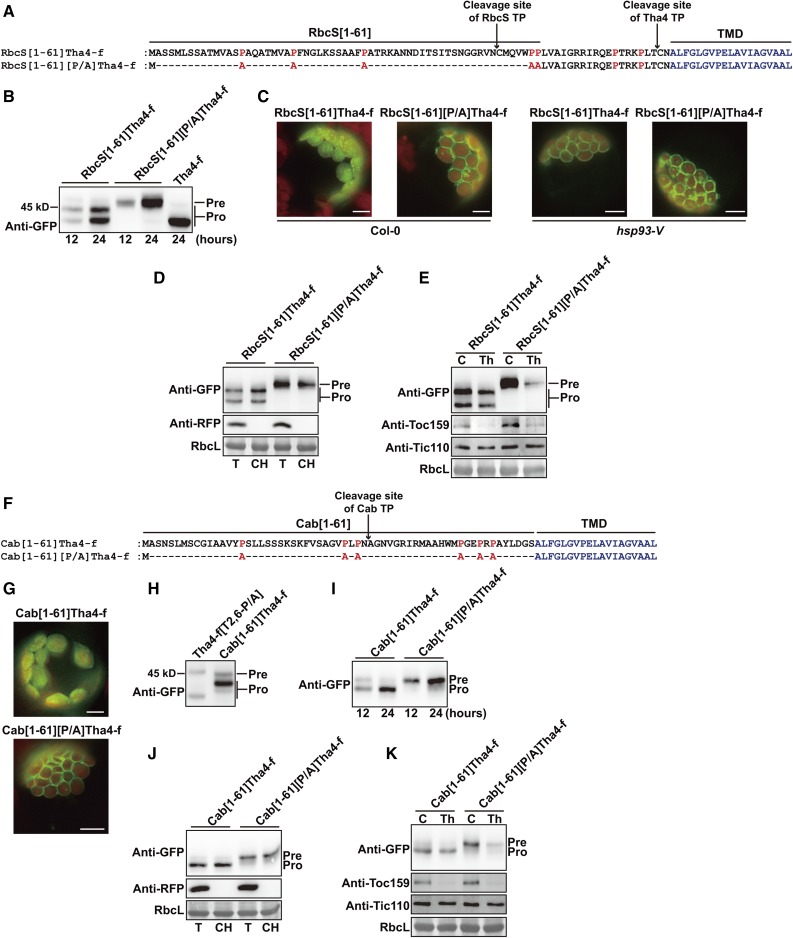
P/A substitution in the TP of Tha4 caused a significant defect in precursor protein import, whereas the equivalent substitution in the RbcS TP caused only a marginal defect (Fig. 1, E and G). To test whether the defect was TP-specific, we generated the chimeric construct RbcS[1-61]Tha4-f:GFP by replacing Tha4[1-41] with RbcS[1-61] in such a way that the chimeric construct contained 81 residues in front of the TMD (Fig. 5A), which was almost the same length as RbcS-nt. Previous studies showed that the length of TP is critical for the efficient import of preproteins into chloroplasts (Lee et al., 2008; Bionda et al., 2010). RbcS[1-61]Tha4-f:GFP was efficiently imported into chloroplasts similarly as Tha4-f:GFP (Fig. 5, B and C), indicating that RbcS[1-61] was functional in the chimeric construct. However, RbcS[1-61]Tha4-f:GFP produced two processed forms (Fig. 5B) and both of them were copurified with chloroplasts, indicating that they are imported into the chloroplast (Fig. 5D). It is possible that RbcS[1-61]Tha4-f:GFP has two processing sites, each of them from RbcS[1-61] and Tha4[41-61]. In fact, RbcS[1-61] has a processing site at the amino acid position 54 and Tha4 TP has a processing site at the amino acid position 59 (Fig. 5A). Next, we introduced the same P/A mutations in RbcS[1-61] that were introduced into RbcS-nt[P/A] to generate RbcS[1-61][P/A]Tha4-f:GFP. RbcS[1-61][P/A]Tha4-f:GFP was not delivered into chloroplasts, thereby showing more severe import defects than Tha4-f[2,6-P/A]:GFP (Fig. 5B). RbcS[1-61][P/A]Tha4-f:GFP exclusively localized to the chloroplast envelope membrane, indicating a conserved role for Pro residues in preprotein translocation through import channels (Fig. 5C). To further test this idea, we examined RbcS[1-61][P/A]Tha4-f:GFP import in hsp93-V protoplasts. Both RbcS[1-61]Tha4-f:GFP and RbcS[1-61][P/A]Tha4-f:GFP localized to the chloroplast envelope membranes in hsp93-V protoplasts (Fig. 5C). The unimported precursor of RbcS[1-61][P/A]Tha4-f:GFP was copurified with chloroplasts and thermolysin-sensitive, indicating that the C-terminal region of this construct is exposed to the cytosol (Fig. 5D and 5E).
Figure 5.
The role of Pro residues in efficient translocation through import channels is independent of the type of transit peptides. A and F, Sequences of the chimeric TP and its P/A substitution mutant. P/A substitutions are indicated in red. B, H, and I, Western-blot analysis. Protein extracts from protoplasts transformed with the indicated constructs were analyzed by western blotting using anti-GFP antibody. Pre, precursor form; Pro, processed form. C and G, Localization of reporter proteins. Protoplasts were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. D and J, Chloroplast isolation. Chloroplasts were isolated and analyzed as described in Figure 1D. E and K, Thermolysin sensitivity of precursors. Protoplasts transformed with the indicated constructs were gently lysed and treated with thermolysin. Subsequently, protein extracts were analyzed by western blotting using anti-GFP, anti-Toc159, and anti-Tic110 antibodies. Pre, precursor form; Pro, processed form.
Next, to test whether the TP among different TMD-containing preproteins is functionally equivalent in protein import into chloroplasts, we generated Cab[1-61]Tha4-f:GFP in which Cab[1-61] substituted Tha4[1-61] in Tha4-f:GFP (Fig. 5F). This construct was efficiently imported into chloroplasts and mostly present as a processed form (Fig. 5, G–K), indicating that Cab[1-61] is functionally equivalent with Tha4[1-61] in protein import into chloroplasts. However, in the case of its P/A substitution mutant, Cab[1-61][P/A]Tha4-f:GFP, the majority of proteins was not imported into chloroplasts, but remained at the outer envelope membrane (Fig. 5, G–K), indicating that again Pro residues in Cab TP also play a critical role in translocation of a nonnative TMD-containing cargo.
Pro Residues in TP of Inner Envelope Membrane Protein AtTic110 Also Are Critical for Translocation across Chloroplast Envelope Membranes
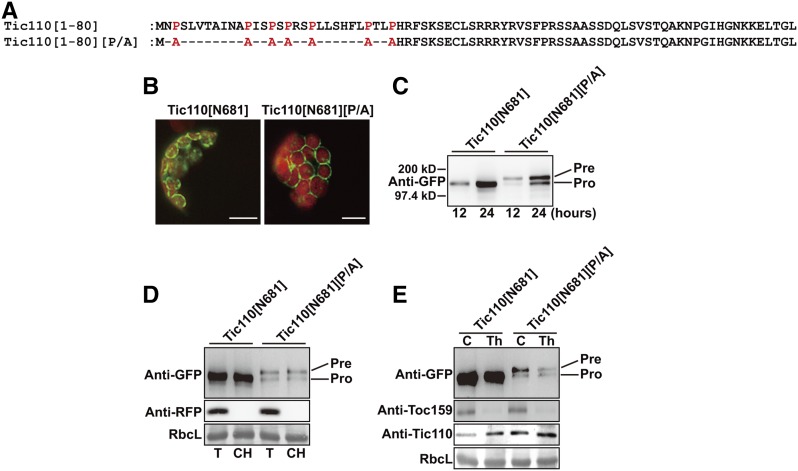
Most inner envelope membrane proteins are imported through Toc/Tic translocons (Lee et al., 2017). Among them, certain proteins such as Tic110 and Tic40 are inserted into inner envelope membranes via a soluble intermediate in the stroma (Li and Schnell, 2006; Lee et al., 2017). We examined whether Pro residues of Tic110 TP are important for translocation. Tic110[N681]:GFP was generated by fusing the N-terminal fragment containing 681 amino acid residues of AtTic110 to the N terminus of GFP (Fig. 6A). This construct displayed a ring-like pattern around chloroplasts (Fig. 6B). In addition, western-blot analysis showed that Tic110[N681]:GFP was present predominantly as the processed form and resistant to thermolysin, indicating that Tic110[N681]:GFP localizes at the inner envelope membrane (Fig. 6, B–E). To examine the role of Pro residues in TP of Tic110, we generated Tic110[N681][P/A]:GFP in which 7 Pro residues in the TP were substituted with alanines and examined for localization in protoplasts. Tic110[N681][P/A]:GFP also localized to chloroplasts and gave a ring pattern (Fig. 6, B and C). However, Tic110[N681][P/A]:GFP produced largely precursor forms. Unimported precursor of Tic110[N681][P/A]:GFP was copurified with chloroplasts but sensitive to thermolysin, indicating that Tic110[N681][P/A]:GFP localizes to outer envelope membranes (Fig. 6, D and E). These results suggest that Pro residues in TPs are crucial for the efficient translocation of membrane proteins destined to both inner envelope membranes and thylakoid membranes.
Figure 6.
Pro residues in the TP are crucial for efficient import of an inner envelope membrane protein AtTic110 into chloroplasts. A, Sequences of AtTic110[1-80] and its P/A substitution mutant. P/A substitutions are indicated in red. B, Localization of reporter proteins. Protoplasts were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and overlap between green and red signals, respectively. Scale bar = 20 μm. C, Western-blot analysis. Protein extracts from protoplasts transformed with the indicated constructs were analyzed by western blotting using anti-GFP antibody. Pre, precursor form; Pro, processed form. D, Chloroplast isolation. Chloroplasts were isolated and analyzed as described in Figure 1D. E, Thermolysin sensitivity of precursors. Protoplasts transformed with the indicated constructs were gently lysed and treated with thermolysin. Subsequently, protein extracts were analyzed by western blotting using anti-GFP, anti-Toc159, and anti-Tic110 antibodies. Pre, precursor form; Pro, processed form.
Pro Residues in TPs Are More Important in TMD-Containing Proteins Than Soluble Stromal Proteins
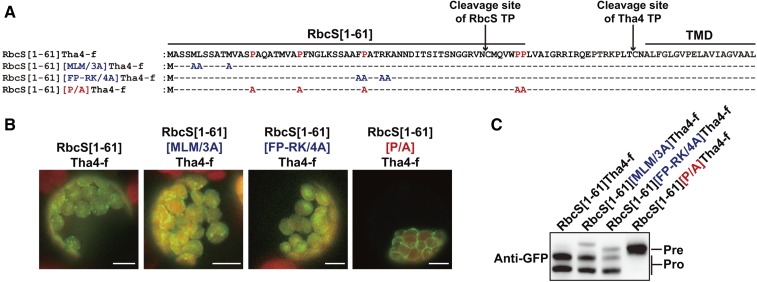
Previously, multiple sequence motifs were identified in RbcS TP (Lee et al., 2006, 2009a). In addition, we showed that the functionality of sequence motifs is dependent on the overall sequence contexts of TPs (Lee et al., 2015). We tested functionality of sequence motifs in RbcS-nt in the context of RbcS[1-61]Tha4-f:GFP. Among many sequence motifs, the N-terminal MLM and internal FP-RK are the most crucial motifs in protein import into chloroplasts in the context of RbcS-nt:GFP (Lee et al., 2006, 2009a, 2015; Razzak et al., 2017). We substituted MLM and FP-RK motifs with alanines in RbcS[1-61]Tha4-f:GFP to give RbcS[1-61][MLM/3A]Tha4-f:GFP and RbcS[1-61][FP-RK/4A]Tha4-f:GFP, respectively (Fig. 7A) and examined for their import into chloroplasts. Both of these proteins were modestly affected whereas RbcS[1-61][P/A]Tha4-f:GFP showed a severe defect in protein import into chloroplasts (Fig. 7, B and C). By contrast, RbcS-nt[P/A]:GFP showed a mild defect in protein import into chloroplasts (Fig. 1, B and C). These results indicate that MLM and FP-RK, the two critical sequence motifs in RbcS TP, are less crucial in import of the hybrid construct, RbcS[1-61]Tha4-f:GFP. By contrast, the Pro residues in RbcS TP that were not crucial for import of RbcS-nt:GFP became crucial in import of RbcS[1-61]Tha4-f:GFP, suggesting that Pro residues are more critical for TMD-containing proteins. Previous studies showed that multiple sequence motifs in TPs function together for efficient protein import into chloroplasts (Lee et al., 2006, 2008, 2009a, 2015; Razzak et al., 2017). Thus, although we did not pursue in this study, it is likely that sequence motifs other than prolines also are important for translocation of RbcS[1-61][P/A]Tha4-f:GFP.
Figure 7.
Pro residues in TP are more important for import of TMD-containing membrane proteins than soluble proteins. A, Sequences of the chimeric TP and its Ala substitution mutants. B, Localization of reporter proteins. Protoplasts were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and overlap between green and red signals, respectively. Scale bar = 20 μm. C, Western-blot analysis. Protein extracts from protoplasts transformed with the indicated constructs were analyzed by western blotting using anti-GFP antibody. Pre, precursor form; Pro, processed form.
Pro Residues in TPs Are Crucial for Translocation of Aggregation-Prone Preproteins
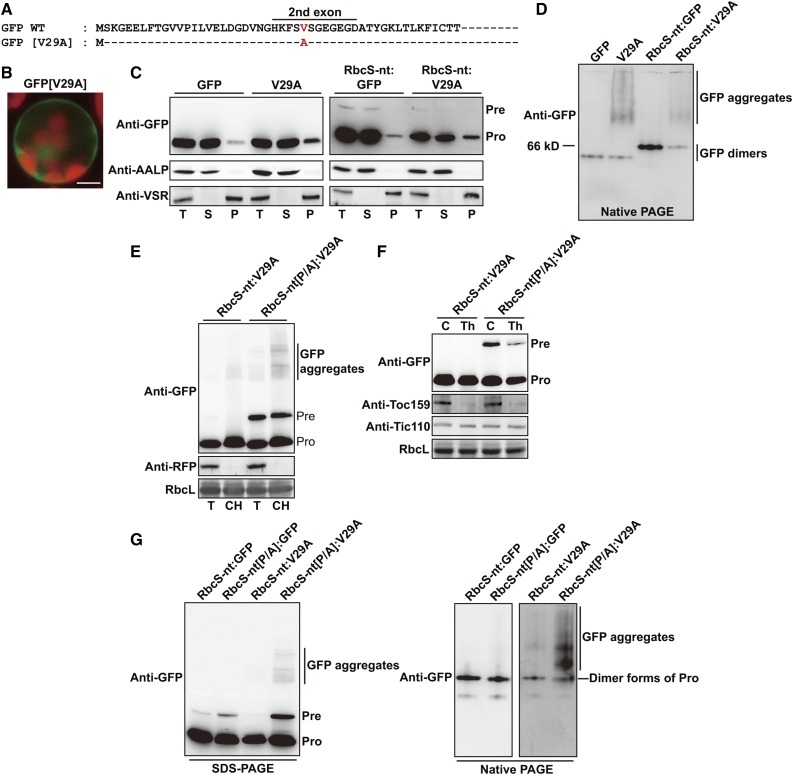
The results shown in Figures 5 to 7 strongly suggest that the degree of defective import may be dependent on the cargo type; the presence of a hydrophobic TMD in a mature protein renders the translocation step highly dependent on TP Pro residues, whereas soluble proteins are less dependent on the Pro residues. To further test this idea, we examined the import of a protein which does not have any TMD, but is prone to form nonspecific aggregates due to a defect in folding. We generated a mutant form of GFP by substituting V29 with Ala to generate GFP[V29A] (Fig. 8A). V29 is located in the second exon, which forms a β-barrel structure that is critical for proper folding and stability of GFP (Prasad et al., 2010; Lee et al., 2016). IUPred analysis predicted that the V29A substitution would disrupt the protein secondary structure (Supplemental Dataset S1; Dosztányi et al., 2005). First, we tested the localization of GFP[V29A] in protoplasts. When GFP[V29A] alone was expressed in protoplasts, the GFP fluorescence was still visible, suggesting that GFP[V29A] underwent proper folding (Fig. 8B). However, a significant portion of GFP[V29A] was present in the pellet fraction after ultracentrifugation, and was present as nonspecific aggregates under nondenaturing conditions, consistent with the proposal that the V29A mutation causes a certain degree of defective folding (Fig. 8, C and D). Next, we examined the localization of GFP[V29A] during import into chloroplasts. RbcS-nt:GFP[V29A] was imported into chloroplasts with an efficiency comparable to that of RbcS-nt:GFP (Fig. 8C). However, a significant portion of processed (imported) forms were present in the pellet fraction (Fig. 8C), indicating that a fraction of imported GFP[V29A] was not properly folded in chloroplasts, consistent with the idea that it exhibits a defect in folding.
Figure 8.
Pro residues in TPs are crucial for efficient translocation of aggregation-prone preproteins through import channels. A, Partial sequences of wild-type GFP and GFP[V29A]. B, Localization of reporter proteins. Protoplasts were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. C, Subcellular fractionation. Protein extracts were separated into soluble and pellet fractions by ultracentrifugation, and fractions were analyzed by western blotting with anti-GFP antibody. AALP and VSR were used as controls for the soluble and pellet fractions, respectively. T, total protein; S, soluble fraction; P, pellet fraction; Pre, precursor form; Pro, processed form. D and G, Analysis of reporter proteins using SDS-PAGE and nondenaturing native PAGE. Total protein extracts were analyzed by SDS-PAGE and nondenaturing native PAGE, followed by western blotting using anti-GFP antibody. Pre, precursor form; Pro, processed form. E, Association of RbcS-nt[P/A]:GFP[V29A] preproteins with chloroplasts. Protoplasts were transformed with RbcS-nt[P/A]:GFP[V29A] together with RFP. Chloroplasts were isolated from protoplasts 12 h after transformation using a Percoll gradient. Total and chloroplast fractions were analyzed by western blotting using anti-GFP and anti-RFP antibodies. RbcL was used as a loading control. T, total protein; CH, chloroplast fraction; Pre, precursor form; Pro, processed form. F, Sensitivity of RbcS-nt[P/A]:GFP[V29A] preproteins to thermolysin. Gently lysed protoplast extracts were treated with thermolysin and then analyzed by western blotting using anti-GFP, anti-Toc159, and anti-Tic110 antibodies. Pre, precursor form; Pro, processed form; T, total protein; CH, chloroplast fraction; RbcL, loading control.
To test the effect of the P/A mutation on GFP[V29A] import, we generated RbcS-nt[P/A]:GFP[V29A] and examined its import into chloroplasts. The amount of precursors was significantly higher than that with RbcS-nt:GFP[V29A] and RbcS-nt[P/A]:GFP (Fig. 1C and 8E), indicating that Pro residues were crucial for the efficient import of GFP[V29A]. We determined the localization of precursors by purifying chloroplasts and performing western blotting using anti-GFP antibody. Precursors and processed forms were both primarily localized in chloroplasts, confirming that precursors of RbcS-nt[P/A]:GFP[V29A] were targeted to chloroplasts but not fully imported (Fig. 8E). Precursors were sensitive to thermolysin, indicating that the GFP moiety was exposed to the cytosol (Fig. 8F). A significant portion of RbcS-nt[P/A]:GFP[V29A] was detected as high Mr aggregates in both SDS-PAGE and nondenaturing native conditions (Fig. 8G).
To examine whether the effect of P/A substitution on GFP[V29A] also occurred with other TPs, we generated Cab-nt:GFP, Cab-nt:GFP[V29A], Cab-nt[P/A]:GFP, and Cab-nt[P/A]:GFP[V29A], and transformed them into protoplasts (Supplemental Fig. S2). Cab-nt:GFP[V29A] was imported into chloroplasts as efficiently as Cab-nt:GFP. However, the import efficiency of Cab-nt[P/A]:GFP[V29A] was greatly affected like RbcS-nt[P/A]:GFP[V29A] (Fig. 8E), confirming that prolines in TPs have a crucial role in the import of preproteins that are prone to forming aggregates (Supplemental Fig. S2, A and B).
DISCUSSION
In this study, we present evidence that Pro residues in TPs have a crucial role in the translocation (import) of chloroplast proteins, particularly for TMD-containing or aggregation-prone proteins. Previously, Pro residues were thought to be crucial for the unstructured nature of the TP (Pilon et al., 1995; Bruce, 2000), as they prevent the formation of secondary structural elements in polypeptides (Guzzo, 1965). However, our results showed that all precursors of RbcS, Tha4, Cab, and Tic110 with TPs containing P/A substitutions largely localized to chloroplast envelope membranes in live cells, and were detected in membrane fractions after centrifugal separation into soluble and membrane fractions. These results suggest that prolines in TPs are not involved at early stages of protein import such as binding to the import receptors. The import of proteins supported by mutant TPs with P/A substitutions was still dependent on AtToc159, which was observed previously for RbcS-nt (Lee et al., 2009a, 2015). The amount of Tha4-f:GFP precursors at the envelope membrane was significantly reduced in ppi2 protoplasts, suggesting that the insertion of both Tha4-f:GFP (in hsp93-V protoplasts) and Tha4-f[T2,6-P/A]:GFP (in wild-type and hsp93-V protoplasts) into the chloroplast envelope membrane occurs after the Toc159-dependent step. This is consistent with a previous study showing that the central Pro-rich region (P15−P26) of ferredoxin TP serves as a motif for a later step during chloroplast protein import (Pilon et al., 1995; Bruce, 2000).
What is the role of Pro residues during late steps of protein import into chloroplasts? We reasoned that the localization of preproteins with TPs lacking prolines may provide insight into the role of prolines in protein import into chloroplasts. The localization pattern of Tha4-f[T2,6-P/A]:GFP was phenocopied by Tha4-f:GFP in hsp93-V plants, and import of Tha4-f[T2,6-P/A]:GFP was more severely affected in hsp93-V plants. These results may provide an important clue to the role of Pro residues in TPs. Hsp93-V is a stromal chaperone that is physically associated with the Tic complex and proposed to function in pulling preproteins released from the Tic complex (Chou et al., 2006; Huang et al., 2016; Chu and Li, 2012). The dependency of import on Pro residues became more pronounced with proteins containing TMDs in their mature regions, but not with the type of TPs. Consistent with this result, an identically mutated P/A TP showed more severe import defects with the aggregation-prone GFP[V29A] than with the wild-type GFP. Thus, proteins with a hydrophobic domain or an unstructured region are more difficult to pass through the channels than soluble polypeptides. These results raise an intriguing possibility that the nature of the mature regions has a large influence on the translocation efficiency. Indeed, previous studies showed that mature regions of chloroplast proteins can affect the import efficiency (Ko and Ko, 1992). Currently, it is not known why TMD-containing proteins or aggregation-prone polypeptides have greater difficulty in translocation through import channels.
The import process can be divided into multiple distinct steps that occur immediately after translation on ribosomes. Nascent preproteins must navigate through the cytosol to bind to chloroplast-localized import receptors or chloroplast lipids. This cytosolic step may be facilitated by the guidance complex involving Hsp70/14-3-3 and Hsp90, depending on the type of TPs (May and Soll, 2000; Qbadou et al., 2006). The hydrophobicity of the N-terminal region may contribute to binding of preproteins to chloroplasts. For example, the MLM motif in the RbcS TP is important at the cytosolic step (Lee et al., 2006). During the early steps of protein import into chloroplasts, preproteins are recognized at the chloroplast surface by receptors such as Toc159, Toc132/120, Toc90, Toc33/34, and Toc64 (Li and Chiu, 2010). Specific sequence motifs such as FNGLK, Ser residues, or the T5 segment of the RbcS TP are thought to be involved in this step (Chotewutmontri et al., 2012; Lee et al., 2009a). The next step is translocation through import channels at both outer and inner envelope membranes, which should be mediated by the N-terminal TPs. At the outer envelope membrane, Toc75 constitutes the import channel. At the inner envelope membrane, Tic20 is thought to form the import channel (Kikuchi et al., 2013). Currently, it is not fully understood how the TPs mediate translocation through the import channels. Many proteins are present at the stromal side of inner envelope membranes, including atTic40 and atTic110, and chaperones such as Hsp93, cpHsc70, and Hsp90C. They may function in pulling chloroplast proteins through the import channel into the stroma (Su and Li, 2010; Chu and Li, 2012; Inoue et al., 2013). However, the exact sequence motifs involved in these steps are not well defined, and it is not fully understood how this occurs. One prevailing hypothesis is that chaperone complexes are involved in the pulling process, which resembles the molecular ratchet involved in protein import into mitochondria (Yamano et al., 2008; Esaki et al., 1999). Multiple chaperones such as Hsp93-V/III, cpHsc70-1/2, and Hsp90C are essential for pulling preproteins and ensuring complete translocation. Our results suggest that TPs lacking prolines can deliver proteins to chloroplasts, but cannot support their efficient translocation through import channels. These results raise an intriguing possibility that these chaperones may not be able to pull preproteins with TPs containing few prolines into the stroma. However, at this point we cannot rule out the possibility that the Pro residues in TPs also have a role during the TP translocation through the import channel before they bind to the pulling machinery.
Another important feature resulting from the import defect is that TMD-containing proteins are inserted into the outer envelope membranes. This conclusion is based on the results showing that preproteins formed a ring pattern similar to that of outer envelope membrane proteins such as GFP-tagged OEP7 (Lee et al., 2001). The exact mechanism of how preproteins are inserted into the outer envelope membrane is not clearly understood. Preproteins that engaged import channels may have four options to proceed during import. One is forward movement, which leads to complete translocation. However, if the pulling activity is low, precursors stay longer in the import channel, which may result in (1) trapping within the import channel, (2) retrograde translocation back into the cytosol, or (3) lateral insertion into the envelope membranes, depending on the type of protein. Soluble proteins may undergo retrograde trafficking to the cytosol (Chou et al., 2003), where they are subject to degradation via the ubiquitin-26S proteosomal pathway (Lee et al., 2009b, 2016). However, retrograde trafficking to the cytosol for preproteins with TMDs may be more problematic because they are highly prone to form cytotoxic nonspecific aggregates in the aqueous cytosolic environment. Instead, they may be subject to lateral insertion into the envelope membranes. Specific mitochondrial and chloroplast proteins are proposed to be inserted into the outer or inner envelope membrane by lateral translocation from the import channel (Viana et al., 2010; Okawa et al., 2014; Harner et al., 2011), and endoplasmic reticulum membrane proteins are laterally translocated from import channels to membranes (Ismail et al., 2012). The lateral translocation of inefficiently translocating preproteins with TMDs to outer envelope membranes may constitute a mechanism by which the import channel is preserved for efficient translocation of cargoes. Clogging of the import channels would be detrimental to biogenesis of the chloroplast proteome. Consistent with this hypothesis, loss-of-function mutants lacking chaperones involved in translocation exhibit embryonic lethal phenotypes (Inoue et al., 2013; Kovacheva et al., 2007; Su and Li, 2010). Further elucidation of the mechanisms involved in lateral insertion from the Toc complex will provide important clues in understanding protein import into chloroplasts.
In summary, we present evidence that Pro residues in TPs have a crucial role in efficient translocation through import channels during protein import into chloroplasts. The dependency on TP Pro residues was much more pronounced with preproteins that contain hydrophobic or aggregation-prone domains in their mature regions. Moreover, prolines in TPs prevented the inappropriate insertion of unimported precursors into envelope membranes of chloroplasts. In the future, elucidating the mechanisms of how TP prolines play roles in preprotein translocation will be necessary to fully understand the mechanism of protein import into chloroplasts.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) (Col-0) was grown on Gamborg B5 plates in a growth chamber at 22°C to 23°C with a 16-h-light/8-h-dark cycle. Leaf tissues were harvested from 2- to 3-week-old plants and used immediately for protoplast transformation. The hsp93-V (Col-0) plants used in this study were described previously (Su and Li, 2010).
Construction of Reporter Plasmids
P/A substitution constructs were generated using PCR-based mutagenesis (Lee et al., 2006). Primer sequences used to generate the reporter constructs are shown in Supplemental Table S1. To construct Cab-f:HA, the full-length Cab (Cab-f) cDNA was PCR amplified from an Arabidopsis cDNA library using primers XbaI-Cab-f_forward and BglII-Cab-f_reverse. PCR products were digested with XbaI and BglII, and digested products were ligated to HA-containing expression vectors also digested with XbaI and BamHI restriction endonucleases. To generate Cab-f[P/A]:HA, the primers used to generate the Cab-nt[P/A]:GFP were used for PCR-based mutagenesis. To generate Tha4-f:HA and Tha4-f:GFP, the full-length Tha4 cDNA was PCR-amplified from an Arabidopsis cDNA library using primers XbaI-Tha4-f_forward and BamHI-Tha4-f_reverse. PCR products were digested with XbaI and BamHI and ligated to HA-containing expression vectors also digested with XbaI and BamHI. To generate Tic110[N681]:GFP, the construct containing the full-length cDNA of AtTic110 was digested with XbaI and BamHI restriction endonucleases. The digested DNA fragment was ligated to GFP-containing expression vector-digested XbaI and BamHI. To generate Tic110[N681][P/A]:GFP construct, the primers Tic110[P/A]-T and sGFP-s5 were used for PCR-amplification. The PCR product was digested with XbaI and BamHI restriction endonucleases, and the digested DNA fragment was ligated to GFP-containing expression vector digested XbaI and BamHI.
PEG-Mediated Transformation and Import of Reporter Proteins into Chloroplasts
The reporter plasmid DNA used for PEG-mediated transformation was purified using Qiagen columns according to the manufacturer’s instructions. Plasmid DNA was introduced into protoplasts by PEG-mediated transformation (Jin et al., 2001; Kim et al., 2002; Lee and Hwang, 2011). Images of transformed protoplasts were captured using a fluorescence microscope (Zeiss Axio Plan 2; Jin et al., 2001; Lee et al., 2008). More than 50 GFP-positive protoplasts were observed in each transformation, and GFP images shown in each figure are representative of those observed in more than 95% of protoplasts.
Subcellular Fractionation of Protoplast Extracts and Chloroplast Isolation
Protoplasts were collected and resuspended in 300 μL buffer (20 mm Tris-HCl, pH 7.5, 2.5 mm MgCl2, 1 mm EDTA, pH 8.0, 160 mm NaCl, and protease inhibitor cocktail), followed by brief sonication. Samples were centrifuged at 3,000g for 10 min to remove debris. Soluble fractions were further incubated on ice or treated with Na2CO3 (pH 11.5) or Triton X-100 at final concentrations of 100 mm or 1%, respectively, on ice for 30 min, followed by ultracentrifugation at 100,000g. Supernatant (soluble) and pellet fractions were collected separately for further analysis.
Chloroplasts were isolated from protoplasts using a Percoll gradient (Lee et al., 2008). Protein extracts from total and chloroplast fractions were prepared as follows. Protoplasts and isolated chloroplasts were resuspended in 300 μL buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and protease inhibitor cocktail), followed by brief sonication. Sonicated samples were subject to centrifugation at 3,000g for 10 min to remove debris. Supernatants were mixed with protein loading buffer and then boiled for 5 min. After separation by SDS-PAGE, protein samples were analyzed by western blotting.
Antibodies
Western-blot analysis was performed using mouse anti-GFP (Clontech, 1:5,000 dilution), rabbit anti-RFP (1:2,000 dilution; Lee et al., 2006, 2008, 2009a), rabbit anti- Arabidopsis aleurain-like protein (AALP; 1:5,000 dilution; Lee et al., 2006), rabbit anti-VSR (1:2,000 dilution; Kang et al., 2012), rabbit anti-Tic110 (1:2,000 dilution; Lee et al., 2001), rabbit anti-Hsp93 (Agrisera, AS01 001, 1:2,500 dilution), and anti-Lhcb4 (Agrisera, AS04 045, 1:2,500 dilution) antibodies.
Thermolysin and Trypsin Treatments
Protoplasts were gently lysed in 300 μL ice-cold HMS buffer (50 mm HEPES, pH 8.0, 3 mm MgSO4, and 300 mm sorbitol) by repeated pipetting. Subsequently, lysed protoplasts were treated with thermolysin or trypsin on ice, followed by quenching with 10 mm EDTA and soybean (Glycine max) trypsin inhibitor (Froehlich, 2011). After that, protein extracts were mixed with protein loading buffer and boiled for 5 min. Protein samples were analyzed by western blotting using anti-GFP, anti-RbcL, and anti-Lhcb4 antibodies.
Nondenaturing Native PAGE
Nondenaturing native PAGE analysis was performed as described previously (Bollag and Edelstein, 1991).
Accession Numbers
Sequence data generated during this work can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RbcS, At1g67090; Cab, At3g54890; Hsp93-V, At5g50920; Tha4, At5g28750; and AtTic110, At1g06950.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Pro residues in the Cab TP affect preprotein translocation into chloroplasts.
Supplemental Figure S2. Pro residues in the Cab TP are crucial for efficient translocation of an aggregation-prone preprotein.
Supplemental Table S1. Primers used in this study.
Supplemental Dataset S1. IUPred analysis of GFP[V29A] predicts a partially unstructured conformation.
Acknowledgments
We thank Dr. Hsou-min Li (Academia Sinica, Taipei, Taiwan) for providing the hsp93-V mutant seeds.
Footnotes
This research was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning (2016R1E1A1A02922014). This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ010953012017)” Rural Development Administration, Republic of Korea. D.W.L. was supported by grant NRF-2017R1C1B1006784 from the National Research Foundation, Ministry of Science, Technology and Future Planning, Korea.
References
- Agne B, Köhler D, Baginsky S (2017) Protein import-independent functions of Tic56, a component of the 1-MDa translocase at the inner chloroplast envelope membrane. Plant Signal Behav 12: e1284726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 [DOI] [PubMed] [Google Scholar]
- Bhushan S, Kuhn C, Berglund AK, Roth C, Glaser E (2006) The role of the N-terminal domain of chloroplast targeting peptides in organellar protein import and miss-sorting. FEBS Lett 580: 3966–3972 [DOI] [PubMed] [Google Scholar]
- Bionda T, Tillmann B, Simm S, Beilstein K, Ruprecht M, Schleiff E (2010) Chloroplast import signals: the length requirement for translocation in vitro and in vivo. J Mol Biol 402: 510–523 [DOI] [PubMed] [Google Scholar]
- Bischof S, Baerenfaller K, Wildhaber T, Troesch R, Vidi PA, Roschitzki B, Hirsch-Hoffmann M, Hennig L, Kessler F, Gruissem W, et al. (2011) Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag DM, Edelstein SJ (1991) Protein Methods, Wiley-Liss, John Wiley & Sons. Inc. [Google Scholar]
- Bölter B, Soll J (2017) Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Mol Plant 10: 219–221 [DOI] [PubMed] [Google Scholar]
- Bruce BD. (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Chotewutmontri P, Reddick LE, McWilliams DR, Campbell IM, Bruce BD (2012) Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell 24: 3040–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Chu CC, Chen LJ, Akita M, Li HM (2006) Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J Cell Biol 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Li HM (2011) Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol Biol 774: 339–350 [DOI] [PubMed] [Google Scholar]
- Chu CC, Li HM (2012) The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo. Plant Physiol 158: 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney-Smith C, Cline K (2009) Clustering of C-terminal stromal domains of Tha4 homo-oligomers during translocation by the Tat protein transport system. Mol Biol Cell 20: 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Sousa FL, Bölter B, Soll J, Gould SB (2015) YCF1: a green TIC? Plant Cell 27: 1827–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmok V, Tompa P, Simon I (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21: 3433–3434 [DOI] [PubMed] [Google Scholar]
- Esaki M, Kanamori T, Nishikawa Si, Endo T (1999) Two distinct mechanisms drive protein translocation across the mitochondrial outer membrane in the late step of the cytochrome b(2) import pathway. Proc Natl Acad Sci USA 96: 11770–11775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Jarvis P (2013) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833: 332–340 [DOI] [PubMed] [Google Scholar]
- Froehlich J. (2011) Studying Arabidopsis envelope protein localization and topology using thermolysin and trypsin proteases. Methods Mol Biol 774: 351–367 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Keegstra K (2011) The role of the transmembrane domain in determining the targeting of membrane proteins to either the inner envelope or thylakoid membrane. Plant J 68: 844–856 [DOI] [PubMed] [Google Scholar]
- Guzzo AV. (1965) The influence of amino-acid sequence on protein structure. Biophys J 5: 809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Neupert W, Deponte M (2011) Lateral release of proteins from the TOM complex into the outer membrane of mitochondria. EMBO J 30: 3232–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah SC, Wagner R, Sveshnikova N, Harrer R, Soll J (2002) The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys J 83: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PK, Chan PT, Su PH, Chen LJ, Li HM (2016) Chloroplast Hsp93 directly binds to transit peptides at an early stage of the preprotein import process. Plant Physiol 170: 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Li M, Schnell DJ (2013) An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc Natl Acad Sci USA 110: 3173–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N, Hedman R, Schiller N, von Heijne G (2012) A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol 19: 1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jensen PE, Leister D (2014) Chloroplast evolution, structure and functions. F1000Prime Rep 6: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Kim SY, Song K, Sohn EJ, Lee Y, Lee DW, Hara-Nishimura I, Hwang I (2012) Trafficking of vacuolar proteins: the crucial role of Arabidopsis vacuolar protein sorting 29 in recycling vacuolar sorting receptor. Plant Cell 24: 5058–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS (2006) Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127: 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Schnell D (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr Opin Cell Biol 21: 494–500 [DOI] [PubMed] [Google Scholar]
- Kessler F, Schnell DJ (2006) The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic 7: 248–257 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Oishi M, Hirabayashi Y, Lee DW, Hwang I, Nakai M (2009) A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 21: 1781–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS (2002) Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell 2: 207–217 [DOI] [PubMed] [Google Scholar]
- Ko K, Ko ZW (1992) Carboxyl-terminal sequences can influence the in vitro import and intraorganellar targeting of chloroplast protein precursors. J Biol Chem 267: 13910–13916 [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P (2007) Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Lee DW, Hwang I (2011) Transient expression and analysis of chloroplast proteins in Arabidopsis protoplasts. Methods Mol Biol 774: 59–71 [DOI] [PubMed] [Google Scholar]
- Lee DW, Jung C, Hwang I (2013) Cytosolic events involved in chloroplast protein targeting. Biochim Biophys Acta 1833: 245–252 [DOI] [PubMed] [Google Scholar]
- Lee DW, Kim JK, Lee S, Choi S, Kim S, Hwang I (2008) Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20: 1603–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kim SJ, Oh YJ, Choi B, Lee J, Hwang I (2016) Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol Plant 9: 1428–1431 [DOI] [PubMed] [Google Scholar]
- Lee DW, Lee J, Hwang I (2017) Sorting of nuclear-encoded chloroplast membrane proteins. Curr Opin Plant Biol 40: 1–7 [DOI] [PubMed] [Google Scholar]
- Lee DW, Lee S, Lee GJ, Lee KH, Kim S, Cheong GW, Hwang I (2006) Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol 140: 466–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Lee S, Oh YJ, Hwang I (2009a) Multiple sequence motifs in the rubisco small subunit transit peptide independently contribute to Toc159-dependent import of proteins into chloroplasts. Plant Physiol 151: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Woo S, Geem KR, Hwang I (2015) Sequence motifs in transit peptides act as independent functional units and can be transferred to new sequence contexts. Plant Physiol 169: 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jürgens G, Hwang I (2009b) Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chiu CC (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Li HM, Teng YS (2013) Transit peptide design and plastid import regulation. Trends Plant Sci 18: 360–366 [DOI] [PubMed] [Google Scholar]
- Li M, Schnell DJ (2006) Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J Cell Biol 175: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Martin F, Guiard B, Pfanner N, Voos W (2001) The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J 20: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, McNeilage RT, Shi LX, Theg SM (2014) ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa K, Inoue H, Adachi F, Nakayama K, Ito-Inaba Y, Schnell DJ, Uehara S, Inaba T (2014) Targeting of a polytopic membrane protein to the inner envelope membrane of chloroplasts in vivo involves multiple transmembrane segments. J Exp Bot 65: 5257–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Dinesh-Kumar SP (2010) All hands on deck—the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol Plant Microbe Interact 23: 1368–1380 [DOI] [PubMed] [Google Scholar]
- Paila YD, Richardson LG, Inoue H, Parks ES, McMahon J, Inoue K, Schnell DJ (2016) Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. eLife 5: e12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Wienk H, Sips W, de Swaaf M, Talboom I, van ’t Hof R, de Korte-Kool G, Demel R, Weisbeek P, de Kruijff B (1995) Functional domains of the ferredoxin transit sequence involved in chloroplast import. J Biol Chem 270: 3882–3893 [DOI] [PubMed] [Google Scholar]
- Prasad R, Kawaguchi S, Ng DT (2010) A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell 21: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Mirus O, Tews I, Soll J, Schleiff E (2006) The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J 25: 1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzak MA, Lee DW, Yoo YJ, Hwang I (2017) Evolution of rubisco complex small subunit transit peptides from algae to plants. Sci Rep 7: 9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Chan PT, Li HM (2012) Differential age-dependent import regulation by signal peptides. PLoS Biol 10: e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Li HM (2000) Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell 12: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana AA, Li M, Schnell DJ (2010) Determinants for stop-transfer and post-import pathways for protein targeting to the chloroplast inner envelope membrane. J Biol Chem 285: 12948–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Kuroyanagi-Hasegawa M, Esaki M, Yokota M, Endo T (2008) Step-size analyses of the mitochondrial Hsp70 import motor reveal the Brownian ratchet in operation. J Biol Chem 283: 27325–27332 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Glaser E (2002) Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7: 14–21 [DOI] [PubMed] [Google Scholar]