Neither PINs nor stable anticlinal microtubules define the lobing patterns of pavement cells; broader analyses of microtubules and cell wall strain are needed.
Abstract
The leaf epidermis is a biomechanical shell that influences the size and shape of the organ. Its morphogenesis is a multiscale process in which nanometer-scale cytoskeletal protein complexes, individual cells, and groups of cells pattern growth and define macroscopic leaf traits. Interdigitated growth of neighboring cells is an evolutionarily conserved developmental strategy. Understanding how signaling pathways and cytoskeletal proteins pattern cell walls during this form of tissue morphogenesis is an important research challenge. The cellular and molecular control of a lobed cell morphology is currently thought to involve PIN-FORMED (PIN)-type plasma membrane efflux carriers that generate subcellular auxin gradients. Auxin gradients were proposed to function across cell boundaries to encode stable offset patterns of cortical microtubules and actin filaments between adjacent cells. Many models suggest that long-lived microtubules along the anticlinal cell wall generate local cell wall heterogeneities that restrict local growth and specify the timing and location of lobe formation. Here, we used Arabidopsis (Arabidopsis thaliana) reverse genetics and multivariate long-term time-lapse imaging to test current cell shape control models. We found that neither PIN proteins nor long-lived microtubules along the anticlinal wall predict the patterns of lobe formation. In fields of lobing cells, anticlinal microtubules are not correlated with cell shape and are unstable at the time scales of cell expansion. Our analyses indicate that anticlinal microtubules have multiple functions in pavement cells and that lobe initiation is likely controlled by complex interactions among cell geometry, cell wall stress patterns, and transient microtubule networks that span the anticlinal and periclinal walls.
Plant leaves are thin, mechanically durable organs. Their size and shape influence the efficiency of photosynthetic light capture and strongly influence crop yield (Zhu et al., 2010). The growth properties of the leaf epidermis can influence the morphology of the organ (Savaldi-Goldstein et al., 2007); therefore, there is a strong desire to understand how the division and growth of epidermal cells contribute to polarized growth at the level of tissues and organs. In the plant kingdom, an undulating cell shape is commonly generated in the leaf epidermis as polyhedral cells exit the cell division cycle and undergo an extended phase of polarized expansion (Panteris et al., 1993; Panteris and Galatis, 2005; Andriankaja et al., 2012). Interdigitated growth may give the leaf mechanical stability (Onoda et al., 2015; Sahaf and Sharon, 2016) and/or influence polarized growth at spatial scales that extend beyond that of individual cells (Elsner et al., 2012; Remmler and Rolland-Lagan, 2012).
The biomechanics of the lobing process is complex. As in all other plant cell types, turgor pressure provides the driving force for pavement cell morphogenesis, and the physical connectivity of adjacent cells strongly influences the resulting mechanical stresses in the cell wall (Szymanski, 2014). For example, mechanical simulations of the pavement cell wall predict that tension forces are maximal on the unpaired outer periclinal (parallel to the leaf surface) wall and at the interface of the periclinal and anticlinal (perpendicular to the leaf surface) walls (Sampathkumar et al., 2014). These forces and the geometry of the cell likely contribute to the patterns of growth in lobed cells.
The molecular and biomechanical mechanisms that control lobe initiation are poorly understood. The directions and rates of plant cell expansion are determined by the mechanical properties of the cell wall (Baskin, 2005; Szymanski and Cosgrove, 2009; Cosgrove, 2016), and local modulation of the cellulose microfibril network is a central feature of all pavement cell growth models (Fu et al., 2005; Panteris and Galatis, 2005; Szymanski, 2014). Cellulose microfibrils are highly anisotropic with considerable tensile strength; as a result, bundles of aligned fibers in the cell wall can resist strain most effectively parallel to the fiber network and promote orthogonal strain (Baskin, 2005). Clear thresholds for microfibril alignment can explain the highly anisotropic strain of leaf trichomes (Yanagisawa et al., 2015).
In leaf epidermal pavement cells that already possess a lobed morphology, the correlation between the presence of microtubule bundles and a local convex shape is widespread in the plant kingdom (Panteris and Galatis, 2005). Localization analyses of lobed cells in many plant species detected clusters of anticlinal microtubules and adjacent regions of increased cellulose content at or near convex regions of the cell (Wernicke and Jung, 1992; Panteris et al., 1993; Kotzer and Wasteneys, 2006). Given that microtubules can pattern cellulose microfibril deposition (Paradez et al., 2006) and the correlation of anticlinal microtubules with convex regions of the lobed cells, a common idea is that anticlinal microtubules persist locally earlier in pavement cell morphogenesis and define lobe initiation sites.
Local regions of perpendicularly oriented microfibrils in the anticlinal wall have been proposed to locally resist strain and generate protrusions at adjacent cortical domains (Fu et al., 2005). However, longitudinal alignment of microtubules along the anticlinal wall is a permissive arrangement for transverse elongation of the anticlinal wall in the plane of the epidermis. For example, longitudinally aligned microtubules are observed not only at the convex region of lobes but also along the growing flanks and tips of pavement cell protrusions (Qiu et al., 2002; Zhang et al., 2011). An alternative model proposes that this arrangement of microtubules and microfibrils could enable increased local strain to accommodate the polarized expansion of both cells at the lobing interface (Szymanski, 2014). Stable anticlinal microtubules also are proposed to generate cell wall thickenings that locally resist strain during lobe formation (Panteris and Galatis, 2005). Thickened cell wall pads are present at or near the interface of the anticlinal and periclinal walls in lobed epidermal cells of many species (Panteris et al., 1994); however, to our knowledge, cell wall thickness gradients have not been detected throughout the entire height of the anticlinal wall, and similar cell wall pads or anticlinal thickenings have not been reported in Arabidopsis (Arabidopsis thaliana).
Periclinal microtubules frequently splay out from anticlinal microtubule bundles (Panteris et al., 1993, 1994). These splayed microtubules may be part of a transfacial network of microtubules and cellulose fibers that are proposed to generate local anisotropic strain and drive lobe initiation (Panteris and Galatis, 2005; Szymanski, 2014). Because stable anticlinal microtubules figure prominently in all current morphogenesis models, this article focuses primarily on their behaviors as a function of lobe formation.
Arabidopsis has been used as a genetic system to analyze the molecular mechanisms of lobe formation and leaf epidermal morphogenesis. Dozens of mutants that affect the microtubule, actin, and cell wall systems cause pavement cell shape defects (Wasteneys and Galway, 2003; Buschmann and Lloyd, 2008; Fujita et al., 2013; Jacques et al., 2014). In the cotyledon, pavement cell morphogenesis is more synchronous compared with leaves (Tsukaya et al., 1994). Using time-lapse imaging, it was shown that lobe initiation is not a continuous iterative process during cell expansion. Instead, it occurs predominantly during a permissive phase of seedling development up to ∼2 d after germination (DAG; Zhang et al., 2011; Armour et al., 2015; Wu et al., 2016). Subsequently, the pavement cells expand for days and increase in area largely independent of lobe initiation (Zhang et al., 2011; Armour et al., 2015; Wu et al., 2016). In highly lobed pavement cells, anticlinal microtubules often are concentrated at the convex region of the cell (Wasteneys et al., 1997; Zhang et al., 2011), and this microtubule arrangement may reflect the ability of the microtubule network to maintain polarized lobe outgrowth and rearrange in response to local cell geometry and the predicted patterns of cell wall stress (Sampathkumar et al., 2014). Transverse microtubules within expanding lobes (concave cell domain) also may pattern microfibrils to reinforce highly polarized lobe outgrowth.
A Rho of Plants (ROP)- (Fu et al., 2005) and auxin- (Xu et al., 2010) centric cell shape control model proposes that polarized subcellular distributions of the PIN-FORMED1 (PIN1) auxin efflux carrier and extracellular AUXIN-BINDING PROTEIN1 (ABP1) generate staggered cortical domains of actin filaments and microtubule bundles that pattern lobe initiation. This model has been challenged by a recently generated null allele of ABP1 that had no pavement cell phenotype (Gao et al., 2015). In this article, we describe a broader genetic analysis of the proposed auxin signaling network and demonstrate that there is no clear evidence for PIN-based control of lobe initiation.
A recent study analyzed microtubule localization as a function of lobe initiation and concluded that microtubules are stable features that mark lobe initiation sites (Armour et al., 2015). However, this analysis relied on imaging microtubules at a single time point before lobe initiation and lacked a plasma membrane marker to closely monitor the cell boundary. This latter technical issue made it difficult to detect subtle cell wall deformations that reflect the earliest events during lobe formation. To more carefully analyze the ability of anticlinal microtubules to predict lobe initiation sites, we conducted long-term quantitative analyses of anticlinal microtubules and cell shape using two-color 3D imaging. We found that, in fields of cells that are competent to form lobes, microtubules are neither long-lived structures that predict sites of lobe initiation nor do they define specific sites of localized anticlinal cell wall thickening. Our data indicate that the anticlinal microtubules have multiple functions in lobing pavement cells, and the subset that controls lobe initiation remains unknown. We did detect cortical locations with persistent anticlinal microtubules, and our data suggest that tissue geometry and cell wall stress patterns may play important roles in patterning the microtubule cytoskeleton.
RESULTS
Genetic Analysis of the Plasma Membrane-Localized PINs
PIN1 is a central player in a current model of pavement cell shape control (Xu et al., 2010). To confirm this result, we analyzed the pavement cell phenotype of the pin1-1 null mutant (Sawchuk et al., 2013). The proposed PIN signaling pathway has reported phenotypes in both cotyledons and true leaves (Fu et al., 2009; Xu et al., 2010). We primarily used the cotyledon system here because the cell types, shape change, and genetic control mechanisms are indistinguishable from those of leaves, and the confounding effect of patchy cell division is minimized. We analyzed pin1-1 plants for pavement cell shape defects at 2, 5, and 10 DAG (Fig. 1); 10 DAG is the terminal phenotype at which cotyledon expansion ceases (Qiu et al., 2002). We used the recently described LobeFinder algorithm to measure cell shape and count lobes because it eliminates the unavoidable variability in lobe number scoring among individuals (Wu et al., 2016).
Figure 1.
Pavement cells from pin1-1 null mutants are indistinguishable from wild-type (WT) cells. Representative images of wild-type (top) and pin1-1 (bottom) cotyledon pavement cells are shown. The time points at which the seedlings were imaged are labeled at top. Bars = 50 μm.
At 2 DAG, the number of lobes per cell was slightly higher in pin1-1 compared with the wild type (Table I). However, this difference was not statistically significant later in development, as the lobe number of pin1-1 and the wild type were indistinguishable at 5 and 10 DAG. Circularity is a dimensionless shape descriptor based on normalized cell perimeter-to-area ratios, with a circle having a circularity of 1. Circularity values decrease for wild-type cells as they adopt a more convoluted shape, and there were no differences between pin1-1 and the wild type at any developmental stage (Table I). The pavement cells in the midblade of pin1-1 and wild-type leaves were very similar in size and shape (Supplemental Fig. S1), indicating that PIN1 had little or no effect on pavement cell lobing. This unexpected result prompted us to analyze the expression pattern of PIN1 in pavement cells that were undergoing lobe initiation using a validated live cell probe.
Table I. Population-level analyses of cell area and shape in pin1-1 and wild-type pavement cells at 2, 5, and 10 DAG.
Values are means ± sd. Asterisks indicate significant statistical differences from the wild type at the same DAG by Wilcoxon-Mann-Whitney test (P < 0.05). n ≥ 44.
| Time | Genotype | Area | Circularity | Lobes |
|---|---|---|---|---|
| µm2 | ||||
| 2 DAG | pin1-1 | 1,428 ± 389 | 0.46 ± 0.09* | 11 ± 2* |
| Wild type | 1,447 ± 481 | 0.49 ± 0.07 | 10 ± 2* | |
| 5 DAG | pin1-1 | 4,389 ± 2,615* | 0.29 ± 0.08 | 16 ± 3 |
| Wild type | 5,290 ± 2,452* | 0.32 ± 0.09 | 16 ± 3 | |
| 10 DAG | pin1-1 | 11,956 ± 5,045 | 0.22 ± 0.07 | 17 ± 4 |
| Wild type | 10,111 ± 4,641 | 0.24 ± 0.07 | 16 ± 4 |
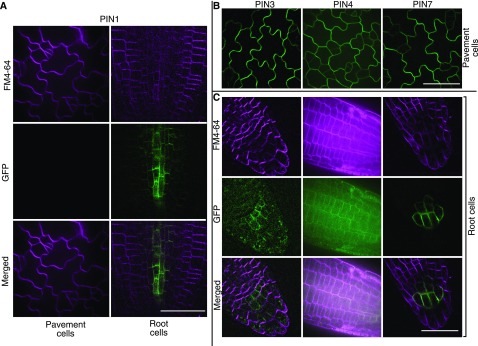
PIN1 was reported to have a polarized localization in lobes following microprojectile bombardment and transient overexpression (Xu et al., 2010; Guo et al., 2015). To analyze the expression pattern and localization of PIN1 in cotyledon pavement cells, we imaged PIN1::PIN1-GFP (Benková et al., 2003) using spinning disk confocal microscopy. In living cells, PIN1 signal was not detected in 2-DAG pavement cells (Fig. 2A), but it was highly expressed in root cortex as described previously (Yan et al., 2013). In young leaves that contain polyhedral pavement cells that were transitioning to polarized growth, PIN1 was expressed along the leaf margin, but not in other fields of pavement cells that were presumably in the process of lobe formation (Supplemental Fig. S2). These genetic and gene expression results indicated that PIN1 does not affect cell morphogenesis in the cotyledon and leaf epidermis.
Figure 2.
PIN3, PIN4, and PIN7, but not PIN1, are expressed in expanding cotyledon pavement cells. A, PIN1 is expressed in root cells but not in 2-DAG cotyledon pavement cells. B, PIN3, PIN4, and PIN7 are expressed in 2-DAG cotyledon pavement cells. C, Expression of PIN3, PIN4, and PIN7 in the root cortex. FM4-64 was used to visualize the plasma membrane when necessary. Bars = 50 µm.
We wanted to test the possibility that other plasma membrane-localized PIN proteins were involved in the process of lobe formation. The PIN family consists of eight proteins that can be subdivided into two major groups. The distinguishing characteristic of the group termed long PINs is a large hydrophobic loop (PIN1–PIN4, PIN6, and PIN7). The second group lacks this hydrophobic loop and is termed short PINs (PIN5 and PIN8). Short PINs appear to localize to the endoplasmic reticulum (Zazímalová et al., 2010). Of the long PINs, PIN6 localizes to the endoplasmic reticulum membrane as well (Sawchuk et al., 2013). PIN2 is implicated in the gravitropism response and is expressed primarily in the root and the hypocotyl (Wiśniewska et al., 2006). PIN4 is expressed very weakly in the seedling epidermis and is a candidate pavement cell shape regulator (Zádníková et al., 2010). The expression patterns of PIN3 (Zádníková et al., 2010), PIN4, and PIN7 in the epidermis were tested directly. We found that all three PINs were expressed and plasma membrane localized in cotyledon pavement cells (Fig. 2B) and the root (Fig. 2C). We tested selected combinations of pin mutants for cell shape defects in 10-DAG pavement cells (Supplemental Fig. S3). The number of lobes per cell were not significantly different for the single, double, and triple pin mutants (Table II). The pin quadruple mutant pin1;3;4;7 had a slightly reduced lobe number that was statistically significant; however, cell area was reduced nearly by a factor of 2 in this dwarfed quadruple mutant, so the lobe per cell ratio was much higher than that of the wild type. The circularity of the pin quadruple pavement cells was higher than in the wild type, indicating that the degree of lobe outgrowth was affected, but not lobe initiation. In addition, the leaf pavement cells of the pin quadruple mutant were highly lobed (Supplemental Fig. S1). Our analyses indicated that plasma membrane PINs promote cell expansion, but they are not necessary for lobe formation in cotyledon and leaf pavement cells. We also failed to detect a phenotype for ric4 (Table II; Supplemental Fig. S3), a proposed effector of ROP signaling in the PIN signaling network (Fu et al., 2005) that was placed in an actin-dependent pathway (Table II). To validate the use of LobeFinder as a useful tool to detect lobe number phenotypes, we conducted a similar analysis on the ARP2/3 complex mutant distorted2 (dis2)/arpc2, which has a reported small reduction in lobe number compared with the wild type (El-Din El-Assal et al., 2004; Le et al., 2006). LobeFinder could reveal a significantly reduced lobe number in dis2 (Table II; Supplemental Fig. S3). The results we present here are consistent with the negative results on ABP1 (Gao et al., 2015) and indicate that auxin efflux carriers do not control lobe initiation.
Table II. Cell area and shape quantification of pin1, pin3, pin4, and pin7 mutant combinations and ric4-2.
Values are means ± sd. Asterisks indicate significant statistical differences between the null mutant line and the wild type by Wilcoxon-Mann-Whitney test (P < 0.05). n ≥ 44; n = 16 for ric4-2.
| Genotype | Area | Circularity | Lobes |
|---|---|---|---|
| µm2 | |||
| Wild type | 10,111 ± 4,641 | 0.24 ± 0.07 | 16 ± 4 |
| pin3 | 6,839 ± 3,177* | 0.26 ± 0.08 | 16 ± 4 |
| pin3;7 | 8,671 ± 5,543 | 0.28 ± 0.10 | 15 ± 4 |
| pin3;4;7 | 7,834 ± 4,109* | 0.22 ± 0.07 | 17 ± 4 |
| pin1;3;4;7 | 4,797 ± 1,718* | 0.33 ± 0.08* | 15 ± 3* |
| ric4-2 | 5,169 ± 1,441* | 0.26 ± 0.05 | 15 ± 2 |
| dis2 | 9,098 ± 3,143 | 0.43 ± 0.12* | 11 ± 4* |
Development of an Image-Analysis Pipeline to Test for Correlations between Anticlinal Microtubule Behaviors and Lobe Initiation
We shifted our analysis to microtubule localization along the anticlinal wall as a function of lobe formation. Existing genetic and pharmacological data indicate that microtubules are involved in lobe formation (Burk et al., 2001; Whittington et al., 2001; Qiu et al., 2002; El-Din El-Assal et al., 2004; Ambrose et al., 2007; Armour et al., 2015); however, the details of how microtubules pattern the cell are under debate (Panteris and Galatis, 2005; Xu et al., 2010; Szymanski, 2014). A growth restriction model for lobe initiation includes a component in which anticlinal microtubule bundles persist for extended periods of time prior to a symmetry-breaking event generating local cell wall thickening, and this local thickening resists strain and leads to adjacent cell outgrowth and lobe initiation (Panteris and Galatis, 2005). In Arabidopsis, an initial growth restriction model was driven largely by the detection of anticlinal microtubule bundles at the convex regions of lobed cell types (Fu et al., 2002). To our knowledge, the article of Armour et al. (2015) is the only one to attempt to image microtubules in the same cell prior to and after lobe formation. They concluded that stable anticlinal microtubules predict lobe initiation sites. However, as mentioned above, this study had limited sampling and statistical analyses of microtubules prior to lobe formation. In addition, the cell boundary was inferred based on the cytosolic signal of the microtubule marker, raising doubts about the accuracy with which lobe initiation was detected. In our time-lapse imaging studies, we immediately noticed that anticlinal microtubules were quite transient. Therefore, we initiated a new approach in which long-term time-lapse imaging of anticlinal microtubules and the plasma membrane-localized protein was quantitatively analyzed to test for correlations between microtubule localization in the anticlinal cortex and cell shape change.
The first step was to develop methods to enable two-channel 3D imaging over the time scales of 6 to 16 h to capture lobe initiation events. The 1.5-DAG seedlings expressing both PIP2:mCherry (Nelson et al., 2007) and GFP:TUB6 (Abe et al., 2004; Abe and Hashimoto, 2005) were mounted in a chambered slide and imaged hourly, which allowed us to capture the ∼300-nm cell boundary deformations without bleaching the microtubules (Fig. 3; Supplemental Movies S1 and S2). Our time-lapse imaging protocol did not adversely affect cell expansion, because the areal strain rates of individual pavement cells ranged from 5.2% to 9.6% per hour. These values were comparable to previously reported rates for pavement cells at the early growth stages (Zhang et al., 2011; Elsner et al., 2012; Armour et al., 2015). Segments are defined here as the shared cell wall between two three-way cell wall junctions, and their shape was traced manually at the junction of outer periclinal walls and the anticlinal wall.
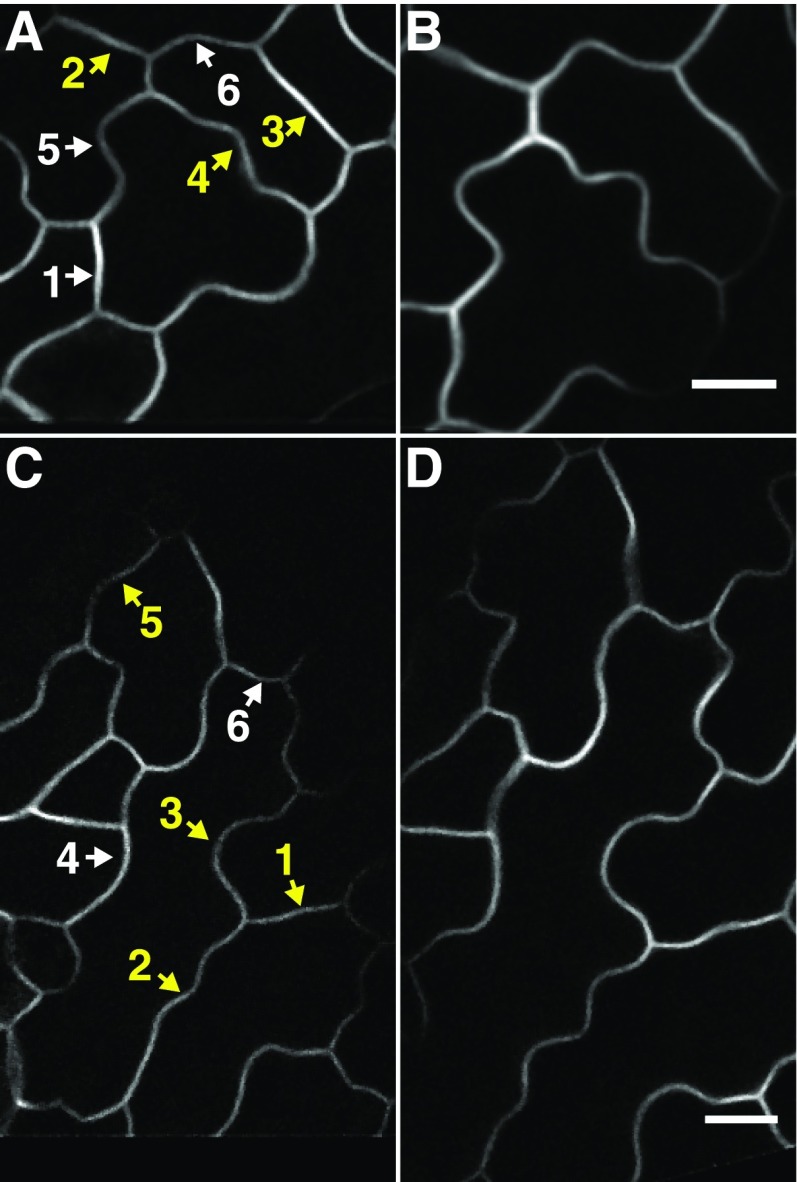
Figure 3.
Pavement cell shape change at high temporal and spatial resolution. A and B, Example image field at time point 1 (A) and time point 11 (B) ∼9 h later. For full time lapse, see Supplemental Movie S1. C and D, Example image field at time point 1 (C) and time point 11 (D) ∼10 h later. For full time lapse, see Supplemental Movie S2. Yellow labels are defined as segments that produce a new lobe(s). White labels are defined as lobed segments that grow but do not have a symmetry-breaking event. Bars = 10 µm.
The next challenge was to create a method to quantify the timing and location of a lobe initiation event within a cell segment. The LobeFinder algorithm serves this purpose (Wu et al., 2016); however, it operates on closed geometric shapes. Using the findpeaks algorithm in Matlab on these segments, we found that lobes began as very small but persistent local deflections in the cell boundary that became progressively larger (Supplemental Fig. S4). Three example segments at the beginning and end of the time lapse are shown within the cell field (Fig. 4, A and B), and the corresponding segmented images of the cell-cell boundary are displayed in Figure 4C. Lobe initiation events were defined as local prominence peaks of at least 286 nm or one pixel that persisted in subsequent images. Using such a small deviation to call an event was valid, because spurious lobes were not identified and all the features detected using this method expanded into easily recognizable lobes at later time points (Fig. 4C, segments II and III). With this algorithm, 86% (n = 29) of the lobes were detected continuously after their first appearance. For the other identified lobes, severe local warping within an adjacent region of the segment distorted the cell boundary in a way that the new lobe was still present but fell below our detection threshold momentarily (Supplemental Fig. S5; Supplemental Movie S3). This severe localized warping must include uneven lateral expansion rates along the anticlinal wall. This growth behavior invalidates a major assumption of uniform anticlinal wall expansion that was used to create a strain model to explain lobe initiation (Armour et al., 2015).
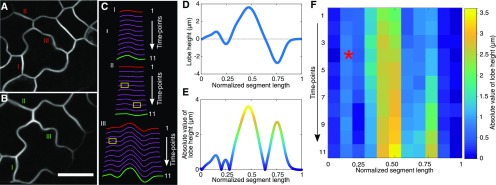
Figure 4.
Development of a method to quantify the timing and location of lobe initiation in pavement cell segments that span three-way cell junctions. A and B, Live-cell images of the cell boundaries at time points 1 and 11, respectively. The total elapsed time between these two images is 9.8 h. Bar = 20 µm. C, Segmented cell boundaries illustrating different types of wall reshaping that occur in developing cells. Yellow boxes label the time points and locations of new lobe formation. Red and green segments correspond to the segments shown in A and B, respectively. D, A plot of lobe height defined as the orthogonal distance from the cell boundary to a reference line that connects the two three-way junctions. E, Absolute value of the lobe height in D color coded with a heat map to reflect differing lobe height along the segment length. F, Heat map of lobe height of segment III as a function of normalized segment length (x axis) over time (y axis). The red star shows new lobe formation.
Based on this lobe detection method, we scored segments as either those that formed new lobes (lobing) or as those that were already lobed and failed to initiate any additional features (lobed; Fig. 3, A and C). In our time-lapse images, we observed segments in which an existing lobe(s) increased in height and width (Fig. 4C, segment I), relatively straight segments that produced one or more new lobes (Fig. 4C, segment II), and already lobed segments that produced additional lobes (Fig. 4C, segment III). In agreement with previous reports (Zhang et al., 2011; Elsner et al., 2012; Armour et al., 2015), we found that different segments of the same cells undergo very different types of shape change behaviors (Supplemental Movie S1, segments 3, 4, and 6). The geometric features of a cell segment that influence the probability that a new lobe will form are not known. There is evidence for a minimal lobe-spacing rule (Staff et al., 2012) and a weak correlation between the initial segment length and the probability of lobe formation (Wu et al., 2016). As expected, we found that lobes could form on segments with varying initial geometries; however, new lobes did not form within recently formed lobes. Instead, initiation tended to occur between an existing lobe and a three-way cell wall junction. This does not necessarily reflect direct inhibition within existing lobes. For example, the cytoskeletal polarizations, cell wall modifications, and geometric transformations that occur as the lobe expands may make the structure insensitive to the forces and/or signaling activities that promote lobe initiation.
In order to graph segment shape from the same cell over time, their Euclidian distances were normalized from 0 to 1 and the cell boundary features, termed lobe height, were plotted as a function of vertical distance from the cell edge to the line that connected the two three-way junctions (Fig. 4D). The absolute value of lobe height was plotted as a function of position along the segment, so that convex features of both cells at the segment interface could be captured and eventually compared with microtubule distributions (Fig. 4E). We restricted our cell shape-microtubule cross-correlation analyses to protrusions that crossed the baseline and allowed us to detect the convex regions of both cells. The full time course of shape change of segment III includes the expansion of existing lobes and the appearance of new features over time (Fig. 4F).
Quantification of Anticlinal Microtubule Positions over Time
Because anticlinal microtubules are a central component to all existing models of lobe initiation (Fu et al., 2005; Panteris and Galatis, 2005; Szymanski, 2014), including the auxin-based patterning model (Xu et al., 2010), we focused primarily on the localization and persistence of microtubules in the anticlinal cell wall. Current models predict that anticlinal microtubules are expected to accumulate at defined locations prior to lobe formation and persist at the apex of convex surfaces along the cell segment interface. Therefore, we set out to test for long-lived anticlinal microtubules at cortical locations that would give rise to future lobes. We developed methods to quantify the locations and persistence of anticlinal microtubules over extended periods of time for which high-resolution data also were available for cell shape. We were able to obtain high-quality images of anticlinal microtubules along the anticlinal wall using the validated GFP:TUB6 (Abe et al., 2004; Abe and Hashimoto, 2005) live cell microtubule marker (Fig. 5, A–C). Quantification of microtubule signals from a face-down view is possible (Armour et al., 2015); however, from this perspective, weak microtubule signals are missed and information about their orientation is lost. Therefore, we resliced the image stack through the x-y planes in the region of interest and quantified signals along the x-z plane, using face-on views of the anticlinal microtubules (Fig. 5, B and C). The microtubule signals along the anticlinal walls were summed as a function of position along the segment perimeter (Fig. 5C) at each time point, and their signal intensity was normalized from 0 to 1 (Fig. 5D). An example time-lapse movie of anticlinal microtubules in Figure 5 is provided as Supplemental Movie S4. Summing microtubule signals along the length of the anticlinal wall perimeter did not cause excessive peak broadening because they were oriented orthogonally to the leaf surface (Supplemental Fig. S6). This allowed us to plot microtubule signals in the cortex of the anticlinal surface as a function of location and time (Fig. 5E). At each time point, the heat map reflects the relative height, shape, and position of the microtubule peaks.
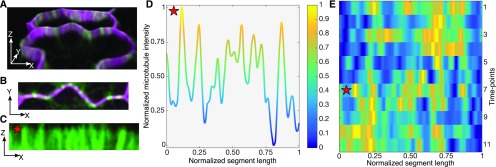
Figure 5.
Quantification of microtubule signals and their positions along the segment. A, Live-cell image of a lobing segment: plasma membrane marker in magenta and microtubule marker in green. Cell volume is tilted to highlight the anticlinal wall. B, Projected face-on view of microtubules along the segment. C, The segment is digitally straightened and resliced to provide a face-on view of the microtubules along the anticlinal wall. D, The signal from C is summed as a function of segment location and normalized from 0 to 1. E, Heat map of the normalized microtubule signals as a function of normalized segment length (x axis) over time (y axis). The red star labels the time point and location of microtubule data that are displayed in C and D.
Anticlinal Microtubules Are Not Stable Structures That Predict the Sites of Lobe Initiation
It was immediately apparent that anticlinal microtubules were not strictly linked with the patterns of cell shape change. Microtubules were broadly distributed along the segment length at a given time point (Fig. 5E). The anticlinal microtubules were not stable features in the cortex, as the same peak was rarely if ever present in the subsequent image (Fig. 5E; Supplemental Movie S4). This broad localization and temporal instability of anticlinal microtubules were consistent among a population of lobing and lobed segments (Supplemental Figs. S7 and S8, respectively). Example images of the anticlinal microtubules are shown for each segment. These results strongly suggested that anticlinal microtubules or microtubule bundles are not stable features that predict lobe initiation and that they are not restricted to most convex regions of the cell cortex. The observed broad distribution of microtubules along the anticlinal wall suggests that microtubules are polymerized and depolymerized along extended convex regions of the cell and also is consistent with previous reports that showed that anticlinal microtubules also reside within the concave region of protruding lobes (Qiu et al., 2002; Zhang et al., 2011).
To more quantitatively analyze microtubule signals as a function of cell shape, we tested for cross correlations between cell shape (Fig. 4F) and microtubule signals (Fig. 5E) that were plotted on a common coordinate system. As mentioned above, we conducted this analysis without knowing in which cell the microtubule signals originated. This uncertainty stems from the fact that the thickness of the anticlinal cell wall is below the resolution of the light microscope, and even with the plasma membrane markers, we cannot clearly assign a microtubule to a particular cell. This technical issue exists for any study that analyzes anticlinal microtubules using live cell imaging. Bombardment of a microtubule marker into individual cotyledon pavement cells is possible, but by the time expression levels are detectable, the cells rarely initiate new lobes (Zhang et al., 2011). Another way to assign anticlinal microtubules to a particular cell is to trace the anticlinal microtubules from the anticlinal to the periclinal cell face of the occupied cell. However, our imaging setup was optimized to collect data as deeply as possible without saturating the signals from the anticlinal microtubules. We could not reliably trace microtubules back to an origin at the anticlinal cell face. This is not completely due to artifacts of poor image quality, as close inspection of the image stacks made it clear that many strong microtubule signals along the anticlinal cell face (Fig. 5C; Supplemental Movie S4) clearly terminated at the interface with the periclinal wall. It has been shown previously that only subsets of anticlinal microtubules in later stage cotyledon pavement cells are associated with a periclinal array (Zhang et al., 2011).
Nonetheless, the technical limitation of not knowing the cell of origin for the microtubule does not preclude a statistical test of the growth control model in which anticlinal microtubules are stable cortical landmarks that predict and mark lobe initiation sites (Fu et al., 2005; Panteris and Galatis, 2005; Armour et al., 2015). In this scenario, anticlinal microtubules should always localize to cortical regions that become convex regions of the cell and persist at the extreme convex regions of the cell after lobe formation. The extreme convex regions of the cell were accurately quantified in our lobe height heat maps (Fig. 4E) and allowed us to test for this expected correlation. We conducted Pearson cross-correlation analyses of microtubule signals and cell shape in seven lobing (Supplemental Fig. S7) and five lobed (Supplemental Fig. S8) segments. As expected, the overall Pearson correlation coefficient (PCC) values were very low as a function of time or space. The PCC values fluctuated due to microtubule peak instability over time, but overall, the correlations were low for both classes of cell segments.
A similar analysis was conducted on 5-DAG pavement cells. These cells are highly lobed and their shape change is symmetrical, as the cells increase in area but maintain their overall shape (Zhang et al., 2011). Subsets of convex regions of pavement cells at this stage have periclinal microtubules that splay from anticlinal microtubule bundles (Zhang et al., 2011; Sampathkumar et al., 2014). In cells at this stage, cell shape was quite stable over the time course (Supplemental Fig. S9, A–D). In each highly lobed segment, the anticlinal microtubules were broadly distributed along the cortical surface. In three of four segments, the signals were unstable over time. In one case, the microtubules were more stable but their locations were inversely correlated, with a convex cell shape residing at the distal flanks of a lobe (Supplemental Fig. S9C). Clearly, the geometry of the cell segment alone is not sufficient to predict anticlinal microtubule distributions in pavement cells at several developmental stages.
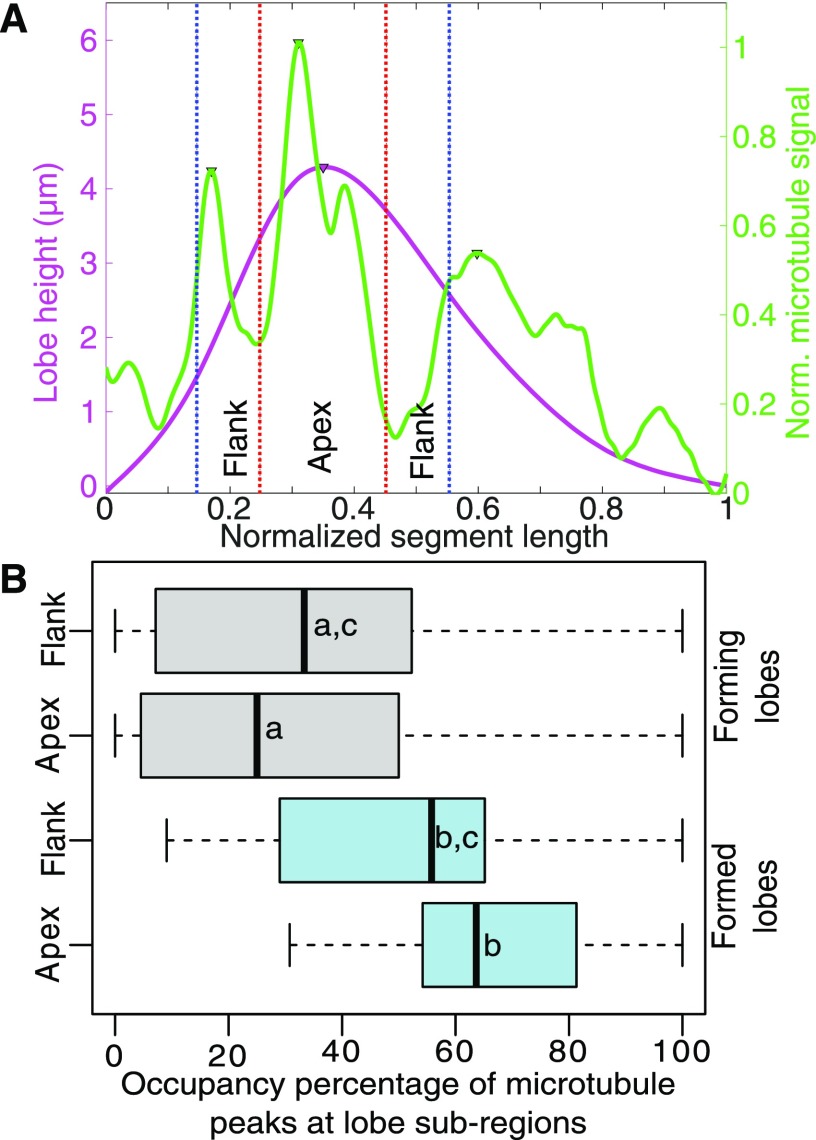
Although it was clear that there was no global correlation between cell segment shape and anticlinal microtubules, we wanted to test for an enrichment of microtubules at lobe apices compared with flanking regions in recently formed lobes. Therefore, we analyzed anticlinal microtubule distributions in a more directed manner as a function of the local lobe geometry. To do this, individual lobes were subdivided into apex and flanking regions of similar areas based on the full width at half-maximal lobe height (Fig. 6A). The flanking region was defined so that it was clearly separated from the convex regions of the adjacent cell. Microtubule occupancy was calculated simply as the percentage of time points at which a microtubule peak was detected within either the apex or the flanking region. This analysis indicated that, for both lobing and lobed segments, the apex and the flank had similar microtubule peak occupancies (Fig. 6B). Our data suggest that anticlinal microtubules are not restricted to subregions within lobes. However, it is likely that there is some bias for microtubule localization within convex regions along the cell-cell interface, and in some cell segments, the microtubule signals may arise predominantly from the convex region of cells. As mentioned above, we cannot rule this out because we cannot define the cell of origin for most of the microtubules. However, our analyses indicate that, if this is occurring, it is occurring very broadly across the convex surface and the microtubules are unstable at the time scale of hours.
Figure 6.
The microtubule peak occupancy at the lobe apex is not significantly higher than that of the adjacent flank regions. A, Example of lobe subdivision based on the lobe width (W) at half-maximal lobe height. The apex is 1/2 W centered at the lobe apex. The flank region total width is 1/2 W surrounding the apex region. B, Population-level analysis of microtubule signal peak occupancy for recently formed lobes (forming) and lobes present at the start of the time series (formed). Letters represent statistically equivalent groups.
Although the anticlinal microtubules were not stable features of developing pavement cells, we wanted to evaluate if there were cortical domains along the anticlinal wall in which microtubules were more likely to reside. We plotted the microtubule signals as a type of persistence plot, summing the values of the microtubule signals along segment length over time (Fig. 7, A and C). Interestingly, there were peaks of accumulated signal for all lobing (Supplemental Fig. S10) and lobed (Supplemental Fig. S11) segments, suggesting that subregions of the cortex were more likely to polymerize and/or stabilize microtubules. In most cases, there were more microtubule persistence peaks than there were lobes, and their locations did not globally correlate with a particular geometric feature of the cell. However, there were examples in both lobing (Supplemental Fig. S10, B–E) and lobed segments (Supplemental Fig. S11, A–D), in which a subset of microtubule peaks were located near lobe apices. We next tested for microtubule persistence in cortical regions that would develop into lobes at a future time point. To do this, we identified the cortical domain that would generate a lobe, projected that cortical domain onto an equivalent region of the same segment at previous time points, and measured the accumulated microtubule signal in the cell segment up to the point of lobe initiation (Supplemental Fig. S12). Most of the lobing segments contained a subset of microtubule peaks at or near cortical regions that would subsequently become part of a lobe (Supplemental Fig. S12, A, B, D, F, and G) and other instances in which there was no obvious relationship between microtubule signals and lobe formation (Supplemental Fig. S12, C, E, and H). This does not rule out a role for persistent anticlinal microtubules during lobe formation: it simply shows that they are not restricted solely to convex or future convex regions of the cell. It is also likely that microtubules that influence lobe initiation were likely missed in hourly image acquisitions.
Figure 7.
Persistence plots of microtubule signals show unequal accumulation of microtubule signals along a segment. A, Microtubule persistence plot of a segment that forms two new lobes. B, Absolute value of lobe height along the segment analyzed in A. C, Microtubule persistence plot for a lobed segment. D, Absolute value of lobe height along the segment analyzed in C. PCC values in B and C correspond to the correlation analysis between the microtubule persistence signal and the final segment shape.
Although our time-lapse imaging was not optimized for individual periclinal microtubules, we tested for an enrichment of cortical microtubules along the periclinal wall prior to lobe formation. We were able to analyze the density of periclinal microtubules in two adjacent cells prior to lobe formation. It was reported previously that periclinal microtubules were enriched in the cell that would adopt a convex shape prior to lobe formation (Armour et al., 2015). We used our time-lapse image stacks to quantify the cortical occupancy of microtubules at opposing periclinal surfaces as a function of lobe initiation (Fig. 8, A–D). To do this, we focused on cell segments for which we had images of periclinal microtubules for at least seven time points prior to lobe initiation. The mean cortical occupancies (Higaki et al., 2010) of periclinal microtubules over time were calculated within opposing regions of interest (ROIs) in the two adjacent cells at the lobing interface. The ROIs were termed V, to denote the microtubule signals in the cell that would adopt a convex shape, and C, to denote the microtubule signals in the adjacent cell that would adopt a concave shape (Fig. 8D). In five of six lobing segments, there was no significant difference in the cortical occupancy of microtubules between the two cells, and in one instance, microtubules were more dense in the future concave-shaped cell (Fig. 8E). This latter observation is the opposite of what would be predicted from current models, and collectively, the above data indicate that simple differences in periclinal microtubule density in opposing cells are not sufficient to explain the location and timing of lobe initiation. However, it is likely that differences in microtubule alignment in opposing cells and the polymerization of microtubules that span the anticlinal and periclinal cell faces are important parameters that control lobe initiation.
Figure 8.
The cortical density of periclinal microtubules does not differ in opposing cells prior to lobe formation. A, Image of periclinal microtubules at the interface of a segment that will form a lobe. B, Processed microtubule image using the cortical occupancy method of Higaki et al. (2010). C, Skeletonized version of the processed image showing segmented microtubules. D, Overlay of the GFP:TUB6 image from B and the microtubule skeleton in C. The ROIs of the future convex (V) and future concave (C) cells are shown at the lobing interface. E, Box plots of the mean cortical occupancy of microtubules in paired convex (V) and concave (C) cells at the lobing interface. ***, Significant difference between the paired cell regions by Student’s t test (P < 0.05).
Anticlinal Microtubule Signals Are Depleted Near Three-Way Cell Wall Junctions
The above analyses did not reveal a particular microtubule feature that could easily explain the patterns of lobe initiation. However, we did notice examples in which anticlinal microtubule signals and cell shape appeared to be correlated in early-stage pavement cells. These tended to be cases in which the initial segment shape was simple and a single broad lobe was present (Supplemental Fig. S11, A and C). This correlation appeared to be driven by the local depletion of anticlinal microtubules adjacent to three-way cell wall junctions. Previously, microtubules were reported to be depleted at three-way junctions at a single time point in lobed pavement cells (Higaki et al., 2016). Our time-lapse data showed that microtubule signals tended to be lower near three-way junctions in cell segments of varying geometries over time (Supplemental Figs. S10 and S11). To test for a robust pattern of localized microtubule depletion, the microtubule signals adjacent to the three-way junctions were compared with an equivalent proportion of the cell cortex in the center of the segment (Fig. 9, A and B). For both lobing and lobed segments, microtubule signals were reduced significantly near three-way junctions compared with the central region (Fig. 9C). The pattern correlates with the reduced magnitude of cell wall stress near three-way junctions that has been predicted by finite element computational models of pavement cell clusters (Sampathkumar et al., 2014).
Figure 9.
Microtubule signals are significantly lower near three-way junctions compared with a central region of the segment. A, Live-cell image of a segment with microtubule marker in green and plasma membrane marker in magenta. B, Microtubule signal intensity plot of the segment in A. Orange is the area under the microtubule signal curve 1 µm from the three-way junctions. Light blue is the area under the microtubule signal curve 2 µm from the cortex centered at the segment midpoint. C, Box plot of the microtubule signal intensity from three-way junctions and center of segments for both lobing and lobed segments. Letters represent statistically equivalent groups.
Anticlinal Microtubule Bundles Are Not Correlated with Regions of Increased Cell Wall Thickness
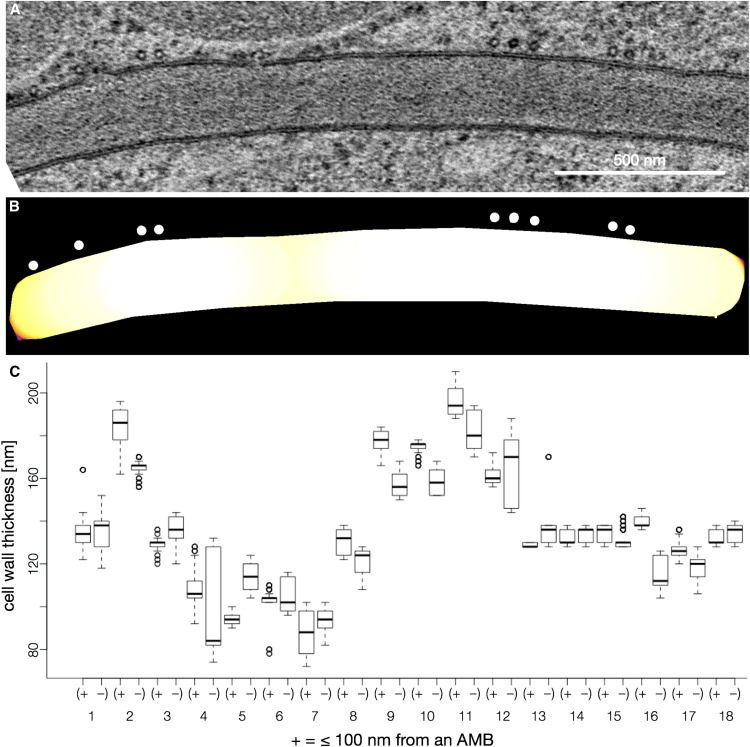
Our time-lapse data indicated that anticlinal microtubules did not selectively persist at the lobe initiation sites and, therefore, were unlikely to define stable locations to direct the synthesis of a thick cellulose-rich cell wall. We wanted to conduct an independent test of the hypothesis that anticlinal microtubule bundles, defined as two or more microtubules in very close proximity, generate local regions of cell wall thickening in lobing pavement cells, using high-pressure freeze fixation and transmission electron microscopy (TEM) of paradermal sections of early-stage cotyledon pavement cells at 1.5 to 2 DAG. Thin sections were then analyzed for cell wall thickness in the regions that overlay an anticlinal microtubule bundle. Properly oriented paradermal sections were identified based on the appearance of the cell at low magnification and the gap in electron density in the plasma membrane lipid bilayer at high magnification. The railroad track-like appearance of the bilayer is only apparent in sections that are exactly parallel to lipid head groups and fatty acid chains. Microtubules in anticlinal microtubule bundles were clearly detected in cross sections and segmented from the images based on their appearance as hollow circles with a diameter of 20 to 22 nm adjacent to the plasma membrane (Fig. 10A). The cell wall also could be segmented from the images, and its thickness was measured as a function of the proximity to anticlinal microtubule bundles. The analysis was conducted on relatively long and straight sections of cell wall from six independent cells that were in a stage of permissive lobe initiation. Among the 36 regions of anticlinal wall that we detected and imaged by TEM, the cell wall thickness varied from approximately 80 to 200 nm. However, the heterogeneity could not be explained by the presence or absence of microtubules. For example, Figure 10C displays box plots of the cell wall thickness values at cell wall domains that overlay anticlinal microtubule bundles and a box plot of wall thickness at an adjacent segment of equal length free of anticlinal microtubule bundles. Among the 18 cell wall regions from six different cells, there was no clear correlation between the presence of anticlinal microtubule bundles and cell wall thickness. In general, microtubules were closely associated with the plasma membrane and tended to reside within one of the two cells. If the cell had a local convex geometry, the microtubules were often, but not always, localized along the convex cell surface. A gallery of TEM images that were analyzed as part of Figure 10 is provided as Supplemental Figure S13. In bundles 4 and 6, there were five or more microtubules (some with visible cross-links to the plasma membrane) within a bundle, and the local wall thickness in these two regions was among the thinnest in the entire data set (Fig. 10C). These data indicate that microtubule bundles do not necessarily mark regions of the wall with an increased cell wall thickness. We could not distinguish the thickness of the two individual anticlinal cell walls in these TEM images because the middle lamella was not clearly detected, so we simply divided the total cell wall width by 2. The geometries of the adjacent cells hardly varied at the spatial scale of our cell wall thickness analyses, and alternating patterns of microtubules in adjacent cells were not detected. Therefore, there was no indication that either microtubules or complementary cell shapes could generate a constant total wall thickness but alternating patterns of varying wall thickness in the opposing cells. However, we are not saying that the material properties in the wall do not vary as a function of anticlinal microtubule bundle localization. For example, an anticlinal microtubule bundle could increase the local proportion of aligned cellulose fibers in the wall, which could influence the local strain behaviors of the cell.
Figure 10.
The domains of the anticlinal wall that underlie anticlinal microtubule bundles (AMBs) are not thicker than adjacent cell wall domains lacking microtubules. A, TEM image of a paradermal section that was perpendicular to the anticlinal wall. B, Thresholded image of A in which the cell wall is color coded from thinnest (yellow) to thickest (white). Microtubules are indicated with white circles. C, Box plot of cell wall thickness values in cell wall domains that overlaid the AMBs (+) and in adjacent regions of the wall not associated with microtubules (−).
DISCUSSION
Leaf epidermal morphogenesis is a multiscale process in which interdigitated growth of expanding polyhedral pavement cells drives epidermal growth (Panteris et al., 1993, 1994; Frank et al., 2003; Zhang et al., 2011). This evolutionarily conserved form of tissue morphogenesis may influence the morphology or mechanical integrity of the leaf. Existing models describe a feedback control mechanism in which the output of signaling is a stable pattern of actin filaments and microtubules in opposing cells that pattern the wall and specify the sites for lobe initiation (Fu et al., 2005; Xu et al., 2010). Such alternating patterns are widespread in lobed cells and may reflect a valid cell shape control mechanism in many cell types such as the crenulated epidermal cells in maize (Zea mays) leaves (Panteris and Galatis, 2005). Our data indicate that there are no such stable cytoskeletal arrays in the anticlinal cortex of Arabidopsis pavement cells, and their localization does not predict or reflect the changing geometry of the cell boundary in a simple way.
Our live cell imaging pipeline allowed us to analyze lobe formation at high spatial resolutions over the time scale of hours during which lobe formation occurs. The first detectable landmark of lobe initiation is a shallow deflection of ∼300 nm at the interface of the anticlinal and periclinal walls (Supplemental Fig. S4). Therefore, the challenge is to discover the cytoskeletal control and cell wall patterning mechanisms that drive a subtle but stable cell wall feature that sets the polarized growth process into motion. Our data are not consistent with a model in which a plasma membrane-localized PIN auxin efflux carrier is a key molecule for pavement cell polarization. We found that a pin1-1 mutant had no noticeable effect on lobe formation at several developmental stages (Table I), and the functional PIN1::PIN1:GFP reporter (Benková et al., 2003) was not expressed in fields of leaf and cotyledon pavement cells that are making new lobes. PIN3, PIN4, and PIN7 are expressed in fields of lobing pavement cells, but with no clear clustering within protrusions as described previously (Xu et al., 2010). A broader genetic analysis of plasma membrane-localized PINs also failed to detect a lobing phenotype (Table II). The pin3, the pin3;7 double, and the pin3;4;7 triple mutants had wild-type lobe numbers. The severely dwarfed pin1;3;4;7 quadruple mutant had much smaller cells but only a slight reduction in lobe number. We also failed to detect a lobing defect in ric4 (Table II), which is proposed to be a target of PIN1 signaling that controls cortical actin polymerization and functions in a negative feedback loop that is needed to generate alternating patterns of cortical actin and microtubules in developing pavement cells (Fu et al., 2005; Xu et al., 2010). These negative results do not reflect our inability to detect a subtle lobing initiation phenotype using LobeFinder, because the dis2/arpc2 phenotypes were clear (Table II). The negative data with ric4, and the lack of stable patterns of microtubules along the anticlinal wall, also suggest that there are no stable cortical actin domains that are proposed to alternate with microtubules via a PIN-based mechanism (Fu et al., 2005; Nagawa et al., 2012). We are not claiming that auxin has no role in lobe formation, as long-term treatments with exogenous auxin can increase cell lobe number (Xu et al., 2010; Gao et al., 2015). Exogenous auxin may extend the developmental window during which pavement cells are competent to form lobes, or auxin may have a general effect on the cell wall that makes lobing more permissive. Our results suggest that the details of the ROP-based signaling pathways that influence lobe formation (Fu et al., 2002; Qiu et al., 2002) remain to be discovered.
Our results on PINs do not agree with those from previous publications (Fu et al., 2005; Xu et al., 2010; Guo et al., 2015). Conflicting data can arise because pavement cell morphogenesis is far more complicated and variable than previously thought. Their shapes and sizes depend on their developmental stages and are strongly affected by their positions on the organ. Cell population-based phenotyping is unreliable, and the recent movement toward time-lapse analysis of lobing is a step in the right direction (Zhang et al., 2011; Armour et al., 2015). The prior lack of an objective lobe detection tool like LobeFinder (Wu et al., 2016), and the peak prominence feature (Supplemental Fig. S4) used here, made it more likely that subjective phenotyping could cause variability among different groups working on the same mutants with weak phenotypes. We attempted to minimize these errors by (1) analyzing pin1-1 at several developmental stages at specific leaf positions; (2) using LobeFinder as a validated lobe-counting tool; and (3) using the circularity parameter to objectively analyze cell shape (Tables I and II). A polarized localization of PIN proteins has been reported in lobe tips following transient ectopic overexpression (Xu et al., 2010; Li et al., 2011; Guo et al., 2015). However, the interpretation of localization data in pavement cells also is complicated, even when tagged proteins are expressed with native promoters. First, the brightness of tagged protein signals along the cell perimeter is highly sensitive to small tilt angles of the sample and the highly variable cytoplasmic density and membrane geometry along the cell perimeter. More importantly, given the unpredictable and heterogenous growth behaviors of lobing pavement cells, it is impossible to relate the localization of a candidate regulatory factor to lobe initiation based on a single image. At present, we are not aware of any protein that localizes to and predicts the sites of lobe initiation.
Based on a previous report (Armour et al., 2015) and current models in which long-lived anticlinal microtubule bundles generate local wall thickenings that influence lobe formation (Fu et al., 2005; Panteris and Galatis, 2005), we expected to detect populations of stable anticlinal microtubules that predict lobe initiation sites. Our quantitative long-term time-lapse imaging analysis of microtubules and cell geometry showed that this clearly was not the case. The same microtubule signal peak was rarely, if ever, present at the same location 1 h later in regions undergoing lobe formation (Fig. 5E). In our time-lapse experiments, it was possible to backtrack to the time points that immediately preceded lobe initiation and test for anticlinal microtubules in the cortical regions that were expected to give rise to lobes at later time points (Supplemental Fig. S12). Anticlinal microtubules were not continuously present at these locations, conclusively showing that these microtubules are not stable structures that predict the site of lobe initiation.
Microtubule turnover also was observed in highly lobed cells (Supplemental Fig. S8), and by graphing microtubule and cell geometry dynamics on a common coordinate system, it was clear that microtubule signals were not strictly correlated with a specific cell shape in either lobing or lobed segments (Supplemental Figs. S7 and S8). The limitation of not knowing from which cell the microtubules originate does not weaken our ability to test the hypothesis that stable anticlinal microtubules are enriched in highly curved regions of the cell compared with the relatively straight flanking region of the lobe. If anticlinal microtubules were stable markers of lobe initiation and lobe outgrowth, the microtubule signals would strongly correlate with regions of high local cell curvature, and they do not. We examined the persistence of microtubule peak signals within existing lobes by dividing the cortex into an extreme apical domain and an adjacent flanking domain that was spatially isolated from the convex region of both cells at the cell-cell interface. Even in lobes that were present through the entire time lapse, anticlinal microtubules were equally likely to be observed at the apex and flanking regions (Fig. 6B). Our data clearly show that cell geometry is not sufficient to predict the patterns of anticlinal microtubules.
However, we are not stating that there is never a correlation between cell geometry and microtubule localization. Microtubule localization to the convex regions of cells has been reported for lobed pavement cells for many species (Panteris and Galatis, 2005) and has been reported repeatedly in Arabidopsis pavement cells (Wasteneys et al., 1997; Fu et al., 2002, 2005; Qiu et al., 2002; Zhang et al., 2011; Sampathkumar et al., 2014). Our TEM analyses here detected a similar bias for microtubule localization to the convex regions of cells (Fig. 10; Supplemental Fig. S13). Microtubules may be more likely to reside in convex regions of one cell, and if there is a transcellular mechanism to inhibit microtubule polymerization in the adjacent cortical region of a neighboring cell, this would generate an offset pattern of microtubules in curved regions of opposing cells. This offset pattern is not always observed (Supplemental Fig. S9C), but when it is present, it cannot be resolved in our analysis because we cannot distinguish the cell of origin for the microtubules. In any case, microtubule localization to convex regions of the cell does not explain how a convex shape is generated in the first place.
It was claimed previously that cortical periclinal microtubules were enriched near the convex region of the cell compared with the concave region of the adjacent cell prior to lobe formation (Armour et al., 2015). We found no clear differences in the occupancy of cortical microtubules in the periclinal cortex of the two adjacent cells prior to lobe formation (Fig. 8). We do not believe that we successfully detected and segmented every microtubule in the periclinal cortex, because the image acquisitions were optimized to measure the brighter anticlinal microtubule bundles. However, assuming that all regions of the cell were equally affected, our data suggest that the cortical density of microtubules does not differ greatly among subcellular domains of adjacent cells during lobe formation. We believe that our conclusions differ from those of Armour et al. (2015) because we more accurately detected the timing and location of lobe formation and we sampled microtubule and cell shape dynamics repeatedly prior to lobe initiation. Further analysis of local microtubule order, origin of nucleation, and persistence as a function of lobe initiation will provide greater insight (see below).
Anticlinal and periclinal microtubules clearly have a role in lobe formation based on convincing genetic and pharmacological data (Qiu et al., 2002; Kotzer and Wasteneys, 2006; Armour et al., 2015). However, our results indicate that there are diverse populations of anticlinal and periclinal microtubules that likely have multiple functions in the cell. Lobe initiation is superimposed on diffuse growth and planar increases in cell area (Supplemental Movies S1 and S2), and the most obvious global function of microtubules is to modulate the strain patterns of the anticlinal and periclinal cell faces. Anticlinal microtubule localization within lobes has been reported previously and discussed by our group (Qiu et al., 2002; Zhang et al., 2011), and the results here further indicate that anticlinal microtubules have a general role to promote anisotropic growth of the anticlinal wall. Anticlinal microtubules may serve as transient cortical landmarks to pattern the local synthesis of aligned cellulose fibers that limit the height of the anticlinal wall and promote the expansion of the anticlinal wall in the plane of the epidermis. The anticlinal microtubules do not appear to generate local regions of increased cell wall thickness. A direct TEM analysis of cell wall thickness as a function of the location of anticlinal microtubule bundles also failed to detect an increased cell wall thickness where it overlaid anticlinal microtubule bundles (Fig. 10). An anticlinal microtubule position and their persistence may have more to do with the tension and shear forces in the wall that vary depending on the geometry of cell/tissue and the local growth behaviors of adjacent cells. Periclinal microtubules with variable orientations may pattern a relatively isotropic but thicker outer cell wall that would resist bulging out of the plane of the leaf surface and enable broad regions of the outer cell wall to expand symmetrically. Therefore, the observation that average microtubule density does not vary greatly along the periclinal cell cortex between two cells is easily explained (Fig. 8).
The distribution of microtubules in the anticlinal cortex is not completely random. They are highly aligned parallel to the leaf surface (Supplemental Fig. S6). The microtubule array in cotyledon pavement cells can align parallel to predicted patterns of cell wall stress, and a static finite element model of a pavement cell predicts that tension forces are maximal at the interface of the outer periclinal and anticlinal walls (Sampathkumar et al., 2014). Tension forces in the anticlinal wall would be expected to be oriented toward the outer periclinal wall, and these forces may dictate the observed alignment of microtubules perpendicular to the outer periclinal wall (Fig. 5C; Supplemental Fig. S6). If microtubules can sense and align parallel to cell wall stress tensors, this may be a plausible mechanism to bias microtubules in the anticlinal wall of any elongated epidermal cell toward a transverse alignment. Time-lapse imaging detected many instances in which a subset of microtubule signal persistence peaks correlated weakly with future sites of lobe initiation (Supplemental Fig. S12). However, their locations were not correlated with cell curvature. Instead, the spatial distributions of persistent microtubule signals were most strongly correlated with tissue geometry: microtubule signals increased away from three-way cell wall junctions in lobing pavement cells (Fig. 9). This pattern also is consistent with predicted cell wall stress patterns, as the magnitude of cell wall stress was predicted to be reduced near three-way junctions, where the cell wall is mechanically reinforced by adjacent cells (analogous to a supported beam; Sampathkumar et al., 2014). Cell wall stress also depends on cell wall thickness (Yanagisawa et al., 2015). Perhaps the localized clustering of microtubules in the thin sections of the anticlinal wall (Supplemental Fig. S13) reflects the localized response of the microtubule system to local regions of increased growth, transient wall thinning, and elevated cell wall stress. Collectively, the above results suggest that stress patterns in the cell wall, due in part to tissue geometry, may pattern the microtubule system and explain why simple parameters like local cell curvature do not explain microtubule patterns. Finite element computational modeling combined with multivariate live-cell imaging is a powerful method with which to analyze how cytoplasmic components, cell wall mechanical properties, and wall stress interact during polarized diffuse growth (Yanagisawa et al., 2015). The development of realistic finite element growth models of pavement cell clusters has the potential to guide experiments that unravel the interactions and feedback controls among cell geometry, cell signaling, and cell wall patterning during epidermal morphogenesis.
We are still left with unanswered questions about which subset of microtubules influences lobe formation. Our data indicate that the microtubules that pattern lobes are transient features that cannot be resolved in an approximately hourly sampling regime. It is also apparent from this and previous work that future experiments should thoroughly analyze the presence and order of periclinal microtubules as a function of lobe initiation (Panteris and Galatis, 2005; Szymanski, 2014). Anticlinal microtubules can exist either as bundles that are restricted to the anticlinal wall or as a component of a transfacial microtubule array that spans the anticlinal and periclinal walls (Zhang et al., 2011). We propose that this later class of aligned transfacial microtubules can pattern cellulose fibers that persist in the primary cell wall to generate a localized patch of anisotropic cell wall that expands asymmetrically at both the anticlinal and periclinal faces (Szymanski, 2014). Locally aligned periclinal microtubules and cellulose microfibrils adjacent to the cell-cell interface could generate an anisotropic strain that could be sufficient to break symmetry and initiate a lobe. Given that a very small deformation of the anticlinal wall is needed to initiate a lobe, it may be that relatively few, short-lived microtubules are sufficient to pattern the cell wall and symmetry-breaking events. Perhaps subsets of specialized transfacial microtubules are decorated with microtubule-associated proteins that efficiently recruit cellulose synthase complexes and orient their motility in the plasma membrane. Solving the puzzle of microtubule-dependent lobing will require genetic and cellular analyses of microtubule order, cell wall strain, and lobe initiation at high spatial and temporal resolutions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seedlings were grown at 22°C on one-half-strength Murashige and Skoog medium with 1% Suc (w/v) and 0.8% (w/v) Bacto agar under continuous illumination. Arabidopsis ecotype Columbia-0 was used as the wild type. PIN null mutants were described previously: pin1-1 (Sawchuk et al., 2013), pin3-3 (Friml et al., 2002), pin4-2 (Friml et al., 2002), and pin7EN (Blilou et al., 2005). The pin double, triple, and quadruple mutants were obtained by crossing the above lines. PIN3::PIN3:GFP was described previously (Zádníková et al., 2010). PIN7::PIN7:GFP is a translational fusion of PIN7 (AT1G23080; −1,537 to +2,830; primers: PIN7 prom SalI forw and PIN7 UTR KpnI rev) to EGFP (Clontech; insertion at +921 of PIN7; primers: EGFP SacI forw and EGFP SacI rev). Primer sequences are as follows: PIN7 prom SalI forw, 5′-TAAGTCGACAAAAATAATATTTTTATTTAAGATAATTATG-3′; PIN7 UTR KpnI rev, 5′-TATGGTACCCTTTCTCAAATAATCTC-3′; EGFP SacI forw, 5′-TAAGAGCTCAGGTGAGCAAGGGCGAGGAG-3′; and EGFP SacI rev, 5′-TATGAGCTCCCTTGTACAGCTCGTCCATGC-3′. PIN4::PIN4:GFP is a translational fusion of PIN4 (AT2G01420; −4,598 to +3,095; primers: PIN4 prom PstI forw and PIN4 1032 SalI rev, PIN4 1033 SalI forw and PIN4 UTR EcoRI rev) to EGFP (Clontech; insertion at +1,032 of PIN4; primers: EGFP SalI Forw and EGFP SalI Rev). Primer sequences are as follows: PIN4 prom PstI forw, 5′-TCTCTGCAGTTTGTGTATCTTAATTATTTGAGTATG-3′; PIN4 1032 SalI rev, 5′-TATGTCGACGTCATGGCTCGCTTTGCTATC-3′; PIN4 1033 SalI forw, 5′-TATGTCGACGCTAAGGAGCTTCACATG-3′; PIN4 UTR EcoRI rev, 5′-TACGAATTCCAGTATAAACCACTTAACTAGAAAC-3′; EGFP SalI Forw, 5′-TATGTCGACGTGAGCAAGGGCGAGGAG-3′; and EGFP SalI Rev, 5′-TATGTCGACCTTGTACAGCTCGTCCATGC-3′. ric4-2 is a T-DNA insertional line obtained from the Arabidopsis Biological Resource Center (SALK_015799), and its genotype was confirmed by PCR analysis. The plasma membrane marker plasmid, pm-RB (CD3-1008; Nelson et al., 2007), was obtained from the Arabidopsis Biological Resource Center and introduced using Agrobacterium tumefaciens-mediated transformation to a GFP:TuB6-expressing line in the Columbia-0 background. GFP-tagged PINs and GFP:TUB6 were imaged as described below for confocal microscopy and time-lapse imaging.
Imaging and Analysis of Cotyledon Pavement Cell Shape: Population-Level Studies
Whole seedlings were stained using FM4-64 (1 µm) for 15 to 30 min. Cotyledons were dissected and imaged using a chambered slide and imaged using a Bio-Rad 2100 laser-scanning confocal microscope mounted on a Nikon Eclipse E800 stand. Samples were excited with a 488-nm laser, and the fluorescence signal was collected using a 490-nm long-pass dichroic and a 500- to 550-nm band-pass emission filter. A 40× oil-immersion, 1.3 numerical aperture (NA) objective was used for 2- and 5-DAG cotyledons, and a 20× 0.5 NA objective was used for 10-DAG cotyledons. Image fields from the apical half of the cotyledon were obtained from 2 DAG, and the apical third of the cotyledon was used for 5 and 10 DAG. Complete pavement cells that were not part of the stoma cell linage were traced manually using the polygon tool in FIJI 4.0 and ROIs were splined using the line tool option in FIJI. ROIs were analyzed using LobeFinder (Wu et al., 2016), and statistical analyses were done in R.
Confocal Microscopy and Time-Lapse Imaging of Lobe Initiation
The 1.5-DAG whole seedlings were mounted in our homemade long-term chamber system as described previously (Yanagisawa et al., 2015). Confocal fluorescence microscopy was performed using a 60× C-Apo 1.2 NA water-immersion objective. Images were acquired using a spinning disk CSU-10 confocal head (Yokogawa Electric) mounted on a Zeiss Oberver.Z1 inverted microscope controlled using Slidebook software (Intelligent Imaging Innovations). mCherry and GFP were excited by 561- and 488-nm laser lines, respectively. Approximately hourly sequential image acquisition was done using an Evolve 512 camera (Photometrics) through band-pass filters (617/73 and 482/35; Semrock).
Analysis of Microtubules in Segments
Using the plasma membrane channel, cell boundaries spanning from three-way junctions were segmented manually using the segmented line tool with the spline function activated in FIJI and saved as an ROI. After all time points were obtained for a segment, the ROIs were saved as a zip file from the ROI Manager window in FIJI. To obtain information about anticlinal microtubules and segment geometry, the macro AMB.IJM was used, which asks for an input folder containing microtubule-tagged image stacks, an output folder, and the ROI.zip file for the segment. The output will produce, for each time point, a .csv file containing information about the summed resliced anticlinal microtubules and a .txt file containing the xy coordinates of the segment. For details, including input requirements and additional options this macro, refer to the AMB_README file included as part of the documentation provided with the GitHub submission (see below). In Matlab, the code InputFiji.m will do the following processes to the xy coordinates: move one end to the xy origin, rotate the segment so that both ends are in the x axis line, interpolate the segment to 500 points and normalize its length from 0 to 1, identify lobes using the prominence method in the findpeaks algorithm, returning for each lobe in a segment its peak position and midheight width, which, in turn, are used to calculate the lobe apex and flanking regions. To the AMB file (.csv), it will do the following: calculate a line scan of anticlinal microtubules in the segment by summing the column values of the image text file and normalize the signal intensity from 0 to 1, interpolate the line to 500 points and normalize the segment length from 0 to 1, identify anticlinal microtubule peaks using the prominence method in the findpeaks algorithm, obtaining the peak position for each identified peak. The code will produce an overlapping plot containing both the normalized anticlinal microtubule signal and the normalized segment with identified lobes subdivided into apex and flank for each time point. It will also calculate a Pearson correlation coefficient for both temporal and location base analysis between the anticlinal microtubule and the absolute lobe height value. For details including additional analysis and options about this code, refer to InputFiji_README. For sublobe microtubule localization, overlapping plots of anticlinal microtubule signals and segment shape produced from Matlab were combined as a stack in FIJI and counted for each subregion. The sum of occupancy was divided by the number of time points and multiplied by 100 to obtain a percentage. Persistence plots were done by plotting the anticlinal microtubule signals using the area tool plot in Matlab. PCC analysis of the final shape with anticlinal microtubule persistence was conducted using the sum of anticlinal microtubule signal with the final segment shape acquired. For microtubule analyses at three-way junctions, the areas of anticlinal microtubules on the first and last five pixels (1.06 µm) of the segment were calculated using the trapz function in Matlab. For the middle section, 10 pixels (2.12 µm) were used. PCC analyses were performed in Matlab. Other statistical analyses were done in R. Code is available at https://github.com/yamsissamy/PlantPhys2017.
Analysis of Microtubules along the Periclinal Cortex
Using the plasma membrane channel, ROIs of equal area dimensions at the convex and concave sides of a lobe were created using the polygon selection in FIJI. The ROIs were analyzed from the beginning of the time lapse to the time of lobe detection. The images were processed in the following manner: smooth, background reduction using a 15-pixel ball, sharpening, despeckle, Z-projection using the max intensity option, band-pass filtering from one to five pixels, binarize, skeletonized, and saved as an eight-bit image. The total integrated intensity was measured for each ROI and divided by 255 to obtain the number of occupied pixels (OP). The total number of pixels (TP) in the ROI was determined. The occupancy percentage was calculated as follows: occupancy = 100 × (OP/OT).
TEM and Cell Wall Thickness Analysis
The 2-DAG cotyledons were high-pressure frozen with a Leica EMPact2 high-pressure freezer in 0.2-mm-deep planchets (Leica Microsystems) using 0.15 m Suc as a cryoprotectant. Next, freeze substitution was carried out in a Leica AFS2 automated freeze substitution unit in 1% OsO4 (EMS). The samples were dehydrated by solution exchange with increasing concentrations of acetone. After dehydration, the samples were infiltrated with Eponate 812 (EMS) by incubating at room temperature for several hours to overnight in increasing concentrations of resin diluted in acetone. The samples were transferred to capsules, and the resin was polymerized in a 60°C oven overnight. Resin-embedded samples were sectioned to ∼60 nm with a Diatome diamond knife on a Leica EM UTC ultramicrotome (Leica Microsystems). Sections were collected on 0.5% formvar-coated slot grids (SPI Supplies) and were poststained for 3 min with 2% aqueous uranyl acetate and for 2 min with Reynolds lead citrate. Images were taken with a four-megapixel Gatan UltraScan 1000 camera on a FEI Tecnai G2 20 Twin 200-kV LaB6 transmission electron microscope. To analyze cell wall thickness in relation to anticlinal microtubule bundles, TEM micrographs were segmented using FIJI’s circle selection tool for microtubules and the polygon selection tool for cell walls. ROIs were selected to include cell wall segments within 100 nm of a point on the cell wall adjacent to microtubules within a bundle. Anticlinal microtubule bundles contained two or more microtubules that were within ∼50 nm. Cell wall thickness was compared with an equivalent length of adjacent cell that did not overlay anticlinal microtubule bundles. The images were then converted to binary and duplicated. One binary image was processed to a Eulerian distance map and the other to an image skeleton. The FIJI image calculator tool was used to identify cell wall thickness values from the distance map along the skeleton points. Those values were saved as xy value coordinates. The middle lamella was not detectable in these samples, so the cell wall thickness values were divided by 2 to account for the presence of two primary cell walls and were analyzed and displayed as box plots using R. Cell wall segments from six independent cells were analyzed.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Neither pin1-1 nor pin1;3;4;7 leaf pavement cells have a detectable lobing phenotype.
Supplemental Figure S2. PIN1 is expressed only in leaf margins of young developing leaves.
Supplemental Figure S3. Lobe formation is not inhibited in plasma membrane-localized PIN mutants or its effector RIC4 but is affected in distorted mutants.
Supplemental Figure S4. A robust method for lobe detection along the cell segment boundary.
Supplemental Figure S5. Expansion in subregions of a segment can warp an adjacent subregion so that it temporarily falls below the identification threshold.
Supplemental Figure S6. Anticlinal microtubules are perpendicular to the leaf surface plane.
Supplemental Figure S7. Pearson correlation analysis between microtubule signals and segment shape for lobing segments.
Supplemental Figure S8. Pearson correlation analysis between microtubule signals and segment shape for lobed segments.
Supplemental Figure S9. Pearson correlation analysis between microtubule signals and segment shape for segments of 5-DAG seedlings.
Supplemental Figure S10. Additional examples of the correlation study between microtubule persistence and final segment shape for lobing segments.
Supplemental Figure S11. Additional examples of the correlation study between microtubule persistence and final segment shape for lobed segments.
Supplemental Figure S12. Microtubule signal persistence before lobe formation occurred but is not highly correlated with new lobe formations.
Supplemental Figure S13. Anticlinal microtubule bundles do not correspond to regions of increased thickening of the anticlinal cell wall.
Supplemental Movie S1. Time-lapse capture of a field of expanding pavement cells.
Supplemental Movie S2. Time-lapse capture of a field of expanding pavement cells.
Supplemental Movie S3. Time lapse of anticlinal microtubules for segment 4 in Supplemental Movie S1.
Supplemental Movie S4. Time-lapse images of anticlinal microtubules that are illustrated in Figure 5.
Acknowledgments
We thank Austin Blackwell and Sarah Mendoza for help with pin genotyping and Peter Ciesielski for assistance with TEM sample preparation. Thanks also to Joe Turner and David Umulis for helpful pavement cell discussions.
Footnotes
This work was supported by NSF MCB Grant No. 1121893 and MCB Grant No. 1715544 to D.B.S. and by Discovery Grants of the Natural Sciences and Engineering Research Council of Canada (NSERC) to E.S. B.S.D. was supported by the Center for Direct Catalytic Conversion of Biomass to Biofuels, an Energy Frontier Research Center funded by DOE, Office of Science, BES, under Award DE-SC0000997. M.G.S. was supported, in part, by an NSERC CGS-M Scholarship and an NSERC CGS-D Scholarship.
Articles can be viewed without a subscription.
References
- Abe T, Hashimoto T (2005) Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J 43: 191–204 [DOI] [PubMed] [Google Scholar]
- Abe T, Thitamadee S, Hashimoto T (2004) Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol 45: 211–220 [DOI] [PubMed] [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO (2007) The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Armour WJ, Barton DA, Law AM, Overall RL (2015) Differential growth in periclinal and anticlinal walls during lobe formation in Arabidopsis cotyledon pavement cells. Plant Cell 27: 2484–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. (2005) Anisotropic expansion of the plant cell wall. Annu Rev Cell Dev Biol 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Buschmann H, Lloyd CW (2008) Arabidopsis mutants and the network of microtubule-associated functions. Mol Plant 1: 888–898 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2016) Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67: 463–476 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Le J, Basu D, Mallery EL, Szymanski DB (2004) DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J 38: 526–538 [DOI] [PubMed] [Google Scholar]
- Elsner J, Michalski M, Kwiatkowska D (2012) Spatiotemporal variation of leaf epidermal cell growth: a quantitative analysis of Arabidopsis thaliana wild-type and triple cyclinD3 mutant plants. Ann Bot 109: 897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Cartwright HN, Smith LG (2003) Three Brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130: 753–762 [DOI] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. (2002) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Himmelspach R, Ward J, Whittington A, Hasenbein N, Liu C, Truong TT, Galway ME, Mansfield SD, Hocart CH, et al. (2013) The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol 162: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA 112: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Qin Q, Yan J, Niu Y, Huang B, Guan L, Li Y, Ren D, Li J, Hou S (2015) TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiol 167: 1058–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Akita K, Takigawa-Imamura H, Yoshimura K, Miura T (2016) A theoretical model of jigsaw-puzzle pattern formation by plant leaf epidermal cells. PLOS Comput Biol 12: e1004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Kondo N, Hasezawa S (2010) Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Jacques E, Verbelen JP, Vissenberg K (2014) Review on shape formation in epidermal pavement cells of the Arabidopsis leaf. Funct Plant Biol 41: 914–921 [DOI] [PubMed] [Google Scholar]
- Kotzer AM, Wasteneys GO (2006) Mechanisms behind the puzzle: microtubule-micofilament cross-talk in pavement cell formation. Can J Bot 84: 594–603 [Google Scholar]
- Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB (2006) Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol 16: 895–901 [DOI] [PubMed] [Google Scholar]
- Li H, Lin D, Dhonukshe P, Nagawa S, Chen D, Friml J, Scheres B, Guo H, Yang Z (2011) Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Res 21: 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Onoda Y, Schieving F, Anten NPR (2015) A novel method of measuring leaf epidermis and mesophyll stiffness shows the ubiquitous nature of the sandwich structure of leaf laminas in broad-leaved angiosperm species. J Exp Bot 66: 2487–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteris E, Apostolakos P, Galatis B (1993) Microtubule organization and cell morphogenesis in two semi-lobed cell types of Adiantum capillus-veneris L. leaflets. New Phytol 125: 509–520 [DOI] [PubMed] [Google Scholar]
- Panteris E, Apostolakos P, Galatis B (1994) Sinuous ordinary epidermal cells: behind several patterns of waviness, a common morphogenetic mechanism. New Phytol 127: 771–780 [DOI] [PubMed] [Google Scholar]
- Panteris E, Galatis B (2005) The morphogenesis of lobed plant cells in the mesophyll and epidermis: organization and distinct roles of cortical microtubules and actin filaments. New Phytol 167: 721–732 [DOI] [PubMed] [Google Scholar]
- Paradez A, Wright A, Ehrhardt DW (2006) Microtubule cortical array organization and plant cell morphogenesis. Curr Opin Plant Biol 9: 571–578 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14: 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmler L, Rolland-Lagan AG (2012) Computational method for quantifying growth patterns at the adaxial leaf surface in three dimensions. Plant Physiol 159: 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahaf M, Sharon E (2016) The rheology of a growing leaf: stress-induced changes in the mechanical properties of leaves. J Exp Bot 67: 5509–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jönsson H, Meyerowitz EM (2014) Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife 3: e01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Sawchuk MG, Edgar A, Scarpella E (2013) Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet 9: e1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staff L, Hurd P, Reale L, Seoighe C, Rockwood A, Gehring C (2012) The hidden geometries of the Arabidopsis thaliana epidermis. PLoS ONE 7: e43546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB. (2014) The kinematics and mechanics of leaf expansion: new pieces to the Arabidopsis puzzle. Curr Opin Plant Biol 22: 141–148 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Cosgrove DJ (2009) Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Tsuge T, Uchimiya H (1994) The cotyledon: a superior system for studies of leaf development. Planta 195: 309–312 [Google Scholar]
- Wasteneys GO, Galway ME (2003) Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54: 691–722 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Willingale-Theune J, Menzel D (1997) Freeze shattering: a simple and effective method for permeabilizing higher plant cell walls. J Microsc 188: 51–61 [DOI] [PubMed] [Google Scholar]
- Wernicke W, Jung G (1992) Role of cytoskeleton in cell shaping of developing mesophyll of wheat (Triticum aestivum L.). Eur J Cell Biol 57: 88–94 [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613 [DOI] [PubMed] [Google Scholar]
- Wiśniewska J, Xu J, Seifertová D, Brewer PB, Růžička K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wu TC, Belteton SA, Pack J, Szymanski DB, Umulis DM (2016) LobeFinder: a convex hull-based method for quantitative boundary analyses of lobed plant cells. Plant Physiol 171: 2331–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan DW, Wang J, Yuan TT, Hong LW, Gao X, Lu YT (2013) Perturbation of auxin homeostasis by overexpression of wild-type IAA15 results in impaired stem cell differentiation and gravitropism in roots. PLoS ONE 8: e58103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Desyatova AS, Belteton SA, Mallery EL, Turner JA, Szymanski DB (2015) Patterning mechanisms of cytoskeletal and cell wall systems during leaf trichome morphogenesis. Nat Plants 1: 15014. [DOI] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, et al. (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617 [DOI] [PubMed] [Google Scholar]
- Zazímalová E, Murphy AS, Yang H, Hoyerová K, Hošek P (2010) Auxin transporters: why so many? Cold Spring Harb Perspect Biol 2: a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Halsey LE, Szymanski DB (2011) The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biol 11: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235– 261 [DOI] [PubMed] [Google Scholar]