Abstract
The functional organization of the plant nuclear pore, nuclear envelope, and nucleoplasm marks dynamically changing environmental cues and developmental programs.
In both plants and animals, the nucleus acts as an organizing center for the changes that occur during an organism’s life cycle, whether as part of a developmental program or in response to environmental factors. Here, we cover recent research that explores roles of the nucleus other than gene expression in light response, fertilization, plant-pathogen interactions, bacterial symbiotic events, and hormone signaling. New strides have been made in understanding subnuclear organization, how nuclear organization responds to different environments and developmental stages, and how the cytoplasm-nucleoplasm connections are made that allow these responses. We further highlight new tools that have been developed to study dynamic changes in nuclear organization.
The nucleus is compartmentalized by a double membrane called the nuclear envelope (NE), which consists of the outer nuclear membrane (ONM), the inner nuclear membrane (INM), and the embedded nuclear pore complexes (NPCs). The NPC is the primary pathway of macromolecular transport between the nucleus and the cytoplasm and is conserved throughout eukaryotes. The ONM and INM are populated by a variety of membrane-associated proteins, and in particular the inner membrane hosts a specific subset of proteins (Starr and Fridolfsson, 2010; Meier et al., 2017). In addition to functioning in nuclear morphology, chromatin attachment, and likely signal transduction, plant NE-associated proteins are involved in nuclear positioning and movement within the larger cellular context (Griffis et al., 2014). While plant nuclear ultrastructure reveals an inner nuclear membrane-associated meshwork similar to the animal lamina, plant genomes do not encode obvious lamin homologs. Instead, plant-specific long coiled-coil proteins with structural similarity to animal lamins might contribute to this meshwork. Plants also lack centrosomes, and the NE plays a role as the microtubule-organizing center at the onset of mitosis. NE proteins functioning as calcium channels are involved in perinuclear calcium oscillations, which are an important step in the establishment of plant-symbiont interactions (recently reviewed in Meier et al., 2017).
While the nuclear periphery is emerging as an essential organizing platform for the nucleus, intranuclear functional and structural organization might also originate from independent self-assembly processes, with the most conspicuous manifestation of these processes revealed as the nucleolus. New methods are now robustly adapted for plants to reveal greater details of functional nuclear and nucleolar organization. Due to the increasing realization that spatial organization is a crucial part of biological function at the subcellular level, many of these aspects have been recently reviewed (Meier, 2016; Perrella and Kaiserli, 2016; Grob and Grossniklaus, 2017; Meier et al., 2017; Thorpe and Charpentier, 2017; Yang et al., 2017). Here, we focus on the developments of the past two years that have revealed exciting new connections between nuclear organization and dynamic plant adaptations during development and environmental responses.
THE NUCLEAR PORE AND NUCLEAR TRANSPORT
The eukaryotic nucleus is perforated by NPCs, each consisting of multiple complements of about 30 proteins called nucleoporins (Nups). Different classes of Nups function in anchoring the NPC to the NE, forming the scaffold structure of the NPC and contributing to the inner NPC channel active in transport (Schmidt and Görlich, 2016). The NPC allows for the selective transport of macromolecules between the nucleus and the cytoplasm. This transport is assisted by karyopherins and other transport receptors and energized by the compartmentalized cyclic GTP hydrolysis and nucleotide exchange of the small GTPase Ran (reviewed in Wente and Rout, 2010; Tamura and Hara-Nishimura, 2014; Meier et al., 2017).
Most of the early identification of plant Nups came from mutant screens for phenotypes in auxin response, plant-microbe interactions, or abiotic stress responses (Zhang and Li, 2005; Dong et al., 2006; Parry et al., 2006). Defects in nuclear protein import, mRNA export, and miRNA abundance/compartmentalization were all found in these Nup mutants and were proposed to be causal for the phenotypes (Parry et al., 2006; Jacob et al., 2007; Xu et al., 2007b; Muthuswamy and Meier, 2011). A core question was whether any Nups were pathway specific and could thus claim a “regulatory role.” In a few cases where the same Nups were identified in different screens, there was apparently no such specificity (Zhang and Li, 2005; Parry et al., 2006). In other cases, only some members of a subcomplex affected a pathway, suggesting a more specific role for these Nups (Parry, 2014).
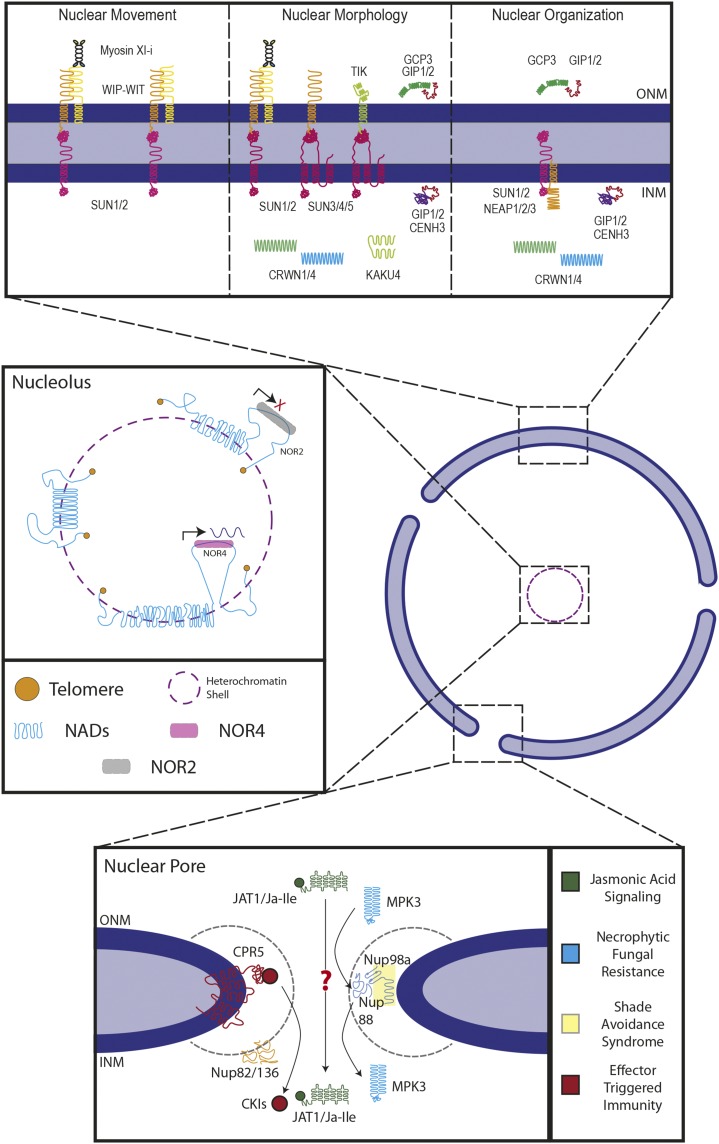
Constitutive expresser of pathogenesis-related genes5 (CPR5), a known inhibitor of programmed cell death and effector-triggered immunity (ETI), was recently described as a Nup by Gu et al. (2016). The authors show that CPR5 is associated with cytoplasmic speckles, the NE, and NPCs. CPR5 is a multispan transmembrane protein that interacts either genetically or physically with a number of other Nups. Overexpression of the CPR5 C terminus—competing with endogenous CPR5 for NPC binding sites—up-regulates a number of different genes, including those responding to light, cold, dehydration, and auxin. Overexpression of full-length CPR5 in protoplasts leads to cytoplasmic retention of NPR1, an important regulator of ETI. Interestingly, CPR5 undergoes homomeric interaction at the NE, disrupted by the initiation of the immune response. The model now is that CPR5 gates the nuclear pore in the absence of ETI, preventing ETI-related molecules from entering the nucleus, and that ETI disrupts this function. This is consistent with the reported phenotypes of cpr5 mutants, which include spontaneous hypersensitive cell death and resistance in the absence of receptor activation (Fig. 1; Wang et al., 2014; Gu et al., 2016).
Figure 1.
Key elements involved in nuclear dynamics. Top box, Nuclear envelope proteins involved in nuclear dynamics. SUN-WIP-WIT complexes have been implicated in nuclear movement in pollen tubes. The SUN-WIP-WIT-Myosin XI-i LINC complex has been shown to be required for nuclear movement and nuclear positioning in root hairs. The SUN-WIP-WIT2-MyosinXI-i complex has also been shown to be required for nuclear shape determination in root hairs, as have the Mid-SUN domain proteins (SUN3/4/5). Mid-SUNs have been shown to interact with the KASH proteins WIPs and TIK. The plant lamin-like proteins CRWN1 and CRWN4 are required for nuclear shape determination and play a role in nuclear organization as well. KAKU4 interacts with CRWN1/4 and is additionally required for nuclear shape determination. The NE-associated proteins NEAP1–3 physically interact with SUN1/2 and are in some cell types required for nuclear shape determination. The NE-associated proteins GIP1/2 play a role in nuclear morphology, as well as nuclear organization, in concert with their interactors GCP3, in the cytoplasm, and CENH3, in the nucleoplasm. Middle box, The nucleolus is surrounded by a heterochromatic shell (purple dashes), which includes dense networks of chromatin known as nucleolus-associated chromatin domains (NADs, blue lines). Nucleolus organizer regions (NORs) contain rRNA genes, where NOR4 associates with the nucleolus and is actively expressed (pink shaded box; arrow depicting high transcriptional activity), while NOR2 is not associated with the nucleolus and has little transcriptional activity (gray shaded box; red X depicts low transcriptional activity). Telomeres (orange circles) are generally located within the nucleolus. Bottom box, Several nucleoporins are implicated in plant biotic and abiotic responses. Colors indicate association with a particular pathway, as marked on the right. Nups are drawn at predicted positions within the nuclear pore. Close association between Nups indicates confirmed protein-protein interaction. For details, see text.
Nups that were reported in the past for one phenotype or pathway are often later associated with another pathway, either related or unrelated, as is the case for Nup88 and Nup98 in Genenncher et al. (2016) and Gallemí et al. (2016), respectively. Nup88—also called MOS7—was known as a contributor to the autoimmunity of the suppressor of npr1-1, constitutive1 mutant (Cheng et al., 2009). It was also found that MOS7 was required for basal resistance, ETI, and systemic acquired resistance against biotropic and hemibiotropic pathogens (Cheng et al., 2009). Genenncher et al. (2016) expanded on this work by demonstrating that MOS7 is also involved in resistance to a necrotrophic fungus. The mos7-1 mutation causes a 4-amino acid in-frame deletion within the MOS7 protein and affects protein abundance and nuclear accumulation of certain MAP kinases involved in resistance signaling, such as MPK3, without affecting their mRNA abundance. Furthermore, the amino acid exchange in mos7-1 weakens the interaction between MOS7 and Nup98a/b, onsistent with the deletion being located at the start of a loop that, in yeast, contributes to the interface for NUP98. These data led the authors to propose a model in which the weakened interaction between MOS7 and Nup98a/b limits the accumulation of MPK3 in the nucleus (Fig. 1; Genenncher et al., 2016). Indeed, a nuclear-excluded version of MPK3 cannot rescue the mpk3 mutant, and although this effect is more drastic than what was found in the mos7-1 mutant, the result supports the model. A nup98a mutant also recapitulates mos7-1 in weakening fungal resistance (Genenncher et al., 2016). It would be interesting to know if nup98a also recapitulated the molecular phenotypes of mos7-1, and this would further strengthen the authors’ model.
Nup98a is also known as DRACULA2, based on a mutant screen for genes involved in shade avoidance syndrome (SAS). In nup98a mutant alleles, the SAS response is diminished (Gallemí et al., 2016). In addition to nup98a, several mutant alleles of NUP160 and NUP96 (scaffold Nups) as well as NUP58, NUP54, and NUP62 (like Nup98a, predicted to be peripheral Nups) were also tested. Specific mutant alleles of NUP160, NUP96, NUP62, and NUP58 indeed showed defects in SAS, again suggesting an overall pleiotrophic effect of disturbing NPC composition and thus likely transport functions. Interestingly, Nup98a down-regulation leads to a sharp up-regulation of Nup98b, suggesting a compensatory response, similar to those seen after down-regulation of Nup62, Nup160 (Parry, 2014), and HOS1 (MacGregor et al., 2013). It would thus be interesting to also investigate SAS—as well as the disease phenotypes described above—in a nup98a nup98b double mutant to test if a more pronounced loss-of-function phenotype could be revealed.
While new roles for known Nups shed light on the complexity of NPC disturbance, new plant Nups are still being discovered. Tamura et al. (2017) found an ortholog of Arabidopsis (Arabidopsis thaliana) Nup136 that is similar to the N-terminal half of Nup136 and that appears to be limited to angiosperms. This new protein, Nup82 (named after its Mr), is redundantly involved in salicylic-acid-dependent plant defense with Nup136.
While protein and RNA transport through the nuclear pore are tightly regulated, small molecule trafficking from the cytoplasm to the nucleoplasm is typically considered diffusion controlled (Ghavami et al., 2016, and references therein). Li et al. (2017) now describe evidence in plants that the active form of jasmonic acid (JA-Ile) is imported into the nucleus by a newly discovered JA transporter and that this is biologically relevant for JA signaling. The transporter is a member of the ATP-binding cassette (ABC) transporter family and is also involved in exporting JA-Ile from the cell via transport through the plasma membrane. The similarity of JA-Ile nuclear import to that of calcium (Charpentier et al., 2016) comes to mind, with the difference that JA-Ile is synthesized in the cytoplasm and not stored for release in the endoplasmic reticulum (ER) lumen. Given the topology of the nuclear envelope as a double membrane system contiguous with the ER, the role of membrane-spanning transporters in this process is somewhat enigmatic. The model proposes that the hormone-transporter complex enters the nucleus through the aqueous phase of the nuclear pore instead (Fig. 1). Clearly, more future work is needed to dissect this interesting process and to distinguish transmembrane transport of JA-Ile from karyopherin-mediated nuclear import.
THE NUCLEAR PERIPHERY
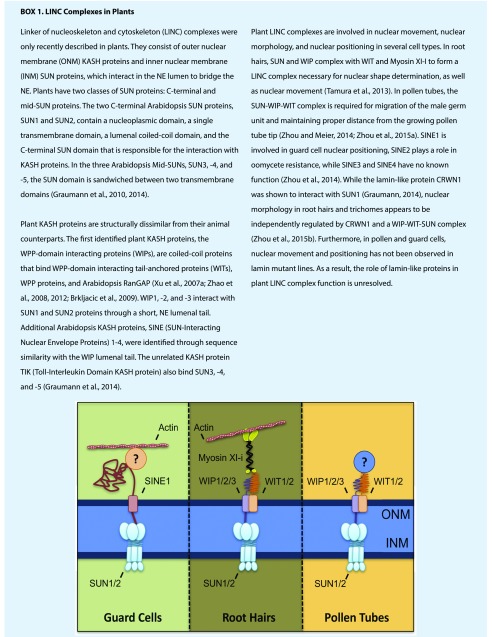
Linker of nucleoskeleton and cytoskeleton (LINC) complexes (Box 1; Brkljacic et al., 2009; Graumann, 2014; Graumann et al., 2010, 2014; Tamura et al., 2013; Xu et al., 2007a; Zhao et al., 2008; Zhou et al., 2012, 2015a, 2015b) bridge the nuclear envelope and are involved in nuclear positioning and migration as well as chromatin organization (reviewed in animals in Starr and Fridolfsson, 2010; Chang et al., 2015; Meier, 2016). Plant LINC complexes have only recently been identified and function in nuclear movement in pollen tubes and male fertility, as well as in nuclear positioning in the leaf epidermis, guard cells, and root hairs (Fig. 1; Box 1; Meier et al., 2017, and therein). A recent study by Poulet et al. (2017b) examined phylogenetic relationships of LINC complex components in plants. While some Klarsicht/ANC-1/Syne-1 Homology (KASH) proteins are broadly conserved among land plants, suggesting they are evolutionarily old, others are limited to angiosperms or even a single angiosperm species. C-terminal Sad-1/UNC-84 (SUN) proteins are conserved in land plants, while mid-SUNs are conserved throughout the plant kingdom (Poulet et al., 2017b). This distribution of broadly conserved and highly specialized plant LINC complexes resembles the situation in animals and might help to shed light on the specific functions of the individual complexes (Zhou et al., 2014).
One currently understudied aspect of plant LINC complex biology is assessing whether they have an influence on chromatin association with the nuclear periphery and subsequent regulation of gene expression. Toward that goal, Poulet et al. (2017a) have developed a 3D imaging protocol to query a number of aspects of nuclear and chromatin organization in Arabidopsis mutants of LINC complex components and related proteins. They found that chromocenters were preferentially associated with the nuclear periphery, but that this association and chromocenter compaction were changed in some mutants. Interestingly, transcriptional repression of heterochromatic repeat sequences was alleviated in some LINC mutants, suggesting that the Arabidopsis nuclear periphery is indeed involved in regulatory chromatin compaction (Poulet et al., 2017a).
Gene redundancy occurs in Arabidopsis LINC complexes like in many other gene families. Zhou et al. (2015c) probed into the potential redundancy within WPP domain-interacting proteins (WIPs), WPP domain-interacting tail-anchored proteins (WITs), and SUN protein gene families in relationship to nuclear movement in root hairs and the nuclear morphology of leaf epidermal cells. They found that WIT2, but not WIT1, is essential for nuclear shape determination and that SUN1 is more important for this process than SUN2. The nuclear morphology changes are independent of the plant lamin-like protein Crowded Nuclei1 (CRWN1; Fig. 1; Box 1), suggesting that nuclear morphology in plants can be affected both by cytoplasmic forces transferred to the NE and nucleoplasmic filaments formed under the NE.
In comparison to the ONM, significantly less is known about the plant INM proteome. In a search for novel plant INM proteins, a bioinformatic screen for a combination of a coiled-coil domain, a nuclear localization signal, and C-terminal hydrophobic domain returned the four proteins nuclear envelope associated protein1–4 (NEAP1–4), with NEAP4 being a truncated form of NEAP3 (Pawar et al., 2016). NEAP1–3 are located at the NE and interact preferentially with SUN2 (Fig. 1). NEAP1 and NEAP2 display protein mobility comparable to NE-embedded proteins such as the SUNs, while NEAP3 is more mobile (Pawar et al., 2016). NEAPs have not yet been thoroughly investigated in terms of biological roles, but first hints come from the study of a neap1 neap3 double mutant, which displays reduced primary root growth, altered nuclear organization and nuclear morphology, and reduced heterochromatin formation. A novel putative interactor of NEAP1 was identified as the transcription factor bZIP18.
CRWN proteins are long coiled-coil proteins that were first identified as Daucus carota (carrot) nuclear matrix constituent proteins (NMCPs; Masuda et al., 1997). They may act as the plant equivalent of animal nuclear lamins (Ciska and Moreno Díaz de la Espina, 2013) and are present throughout land plants. Arabidopsis CRWN1 and CRWN4 are located at the nuclear periphery, while CRWN2 and CRWN3 are nucleoplasmic (Fig. 1; Wang et al., 2013). CRWNs have been implicated to be determinants of nuclear shape, as crwn mutants have small, spherical nuclei in several cell types, including leaf epidermal cells and trichomes (Dittmer et al., 2007; Sakamoto and Takagi, 2013; Wang et al., 2013). CRWNs also play a role in nuclear organization, as crwn4 mutants display aberrant chromocenter distribution (Wang et al., 2013). Because animal nuclear lamins interact with both SUN proteins and nucleoplasmic factors (Gruenbaum and Foisner, 2015), a yeast-two-hybrid screen was performed with carrot NMCP1. Putative interactors include actin-related protein7, Seven in absentia family protein 1 (SINAT1), MYB-type transcription factor 3 (MYB3), and BES-interacting MYC-like1 (Mochizuki et al., 2017). All four NMCP1 interactors are localized to the nucleus, with SINAT1 localized to the cytoplasm as well. It will now be interesting to identify the biological role of these proteins in connection with known CRWN functions.
Further evidence for the connection of nuclear periphery and chromatin organization come from the study of the NE-associated GCP3-interacting proteins GIP1 and GIP2. They are required for γ-tubulin complex protein localization, spindle integrity, and chromosomal stability (Batzenschlager et al., 2013). gip1 gip2 null mutants display centromere cohesion defects, including increased distance between homologous chromosomes (Batzenschlager et al., 2015). Additionally, gip1 gip2 mutants have an increase in the number of chromocenters, also observed in mutants of the epigenetic regulator mgo3, as well as the double mutant gip1 mgo3 (Batzenschlager et al., 2017).
While nuclear-associated calcium oscillations are involved in the establishment of both nodulation and arbuscular mycorrhizal symbiosis (Charpentier and Oldroyd, 2013) and the potassium antiporter in this process has been known for some time (Charpentier et al., 2008; Capoen et al., 2011), the identity of the calcium channels was unknown until recently. Working in the model legume Medicago truncatula, Charpentier et al. (2016) determined that cyclic-nucleotide gated channels (CNGCs), specifically MtCNGC15a, b, and c, are required for these calcium oscillations. CNGC15s are associated with the NE and interact with the INM-located potassium-permeable channel that modulates nuclear calcium release (DMI1; Capoen et al., 2011). Further, cngc15 mutants display deficiencies in both nodulation and arbuscular mycorrhizal symbiosis (Charpentier et al., 2016). It will now be very interesting to see if MtCNGC15a, b, and c, or their homologs in other plant species, are also involved in other processes regulated by nuclear calcium release.
DYNAMIC ORGANIZATION OF THE NUCLEOPLASM
General Nuclear Organization
The organization of the nucleus, as well as the folding and packaging of the genome into higher-order structures, likely contributes to the regulation of eukaryotic gene expression, but the process by which this occurs is unclear (Even-Faitelson et al., 2016). Through the study of tandem repeat sequences and chromocenters in mammalian and plant cells, important insights into the relationship between chromatin movement and gene regulation have been gained (Dittmer et al., 2007; Wang et al., 2013; Bonev and Cavalli, 2016; Nikumbh and Pfeifer, 2017). In Drosophila melanogaster, chromatin mobility is important for interacting with distinct nuclear compartments during differentiation (Thakar and Csink, 2005). Furthermore, altering nuclear morphology changes chromatin organization and disrupts genome function (Lanctôt et al., 2007; Ramdas and Shivashankar, 2015; Smith et al., 2015).
Association of chromatin with lamin proteins and NE transmembrane proteins may serve to tether genes to the nuclear periphery and act as a determinant of genome organization (Lanctôt et al., 2007; Ramdas and Shivashankar, 2015). In mammals, the association of genes with NE proteins causes repression, while those genes found in the interior of the nucleus are actively transcribed. However, there are examples in S. cerevisiae where this trend is reversed, with gene activation being associated with localization to the nuclear periphery and gene silencing with a location in the interior of the nucleus (Lanctôt et al., 2007; Dultz et al., 2016). The relationship between gene positioning and transcriptional activity in plants is largely unclear.
Alterations in plant nuclear architecture and how this may pertain to gene expression control, adaptability, and response to extracellular cues is also gaining attention. An example is how light-mediated development (photomorphogenesis) can influence changes in chromatin organization and nuclear organization (recently reviewed in Perrella and Kaiserli, 2016; Rodriguez-Granados et al., 2016).
Recently, Bourbousse et al. (2015) investigated the relationship between developmental stages and light responsiveness. During germination, nuclear size and chromatin organization are independent of light conditions. However, later developmental stages undergo a light-dependent increase in both nuclear size and chromatin condensation. Additionally, a transgene containing a transposable element fused to a GUS reporter was transcriptionally silenced in a light-dependent manner. Thus, light-triggered changes in nuclear organization are correlated with gene expression changes that occur at the interface of different stages of plant development.
Gene expression changes are also associated with nuclear positioning. Feng et al. (2014) visualized light-mediated chlorophyll a/b-binding protein (CAB) loci repositioning from the nuclear interior to the periphery, which was associated with increased CAB gene expression. Smith et al. (2015) showed that artificially tethering a locus to a nucleoporin enhances its expression. Another group performed an in vivo study that identified and characterized Arabidopsis chromatin regions that preferentially associated with the nuclear periphery. Pericentromeric chromatin was enriched at the periphery, particularly highly methylated transposable element genes and pseudogenes, while distal chromosome arms were depleted at the nuclear periphery (Bi et al., 2017).
It would be interesting to further investigate if photomorphogenesis-triggered nuclear reorganization is required for the transcriptional regulation of all light-inducible genes, how this movement occurs, and why this repositioning is needed. These studies begin to elucidate nuclear architectural dynamics and their role in gene expression but the mechanism by which the nucleus is sensing and responding to changes in light conditions remains to be elucidated.
Nucleolus Organizer Regions
The nucleolus is well known for its role in rRNA gene expression and ribosome biogenesis (McStay and Grummt, 2008). Nucleoli are formed around specific regions within distinct chromosomes that contain multiple copies of rRNA genes known as nucleolus-organizer regions (NORs). In mammals, active rRNA genes are located within fibrillar centers and inactive genes are outside these areas. The mammalian nucleolar shell consists of a dense chromatin network of nucleolus-associated chromatin domains (NADs), which contain inactive chromosomal regions composed of specific satellite repeats, in addition to the NORs (Németh et al., 2010).
Pontvianne et al. (2016) recently identified and characterized plant NADs in Arabidopsis. By visualizing the overlap between rDNA-FISH signals and DAPI staining, the authors saw a substantial number of uncorrelated signals. Sequencing of nuclear versus nucleolar DNA revealed an enrichment of nucleolar genomic regions, which they identified as NADs. Similar to mammals, plant NADs consist of both transcriptionally active (rRNA genes and the adjacent short arm of chromosome 4 or NOR4) and silent genes (transposable elements, pseudogenes, and tRNA genes). Plants contain two NORs: NOR4 associates with the nucleolus and is actively transcribed, whereas its counterpart on the short arm of chromosome 2, NOR2, is inactive and is excluded from the nucleolus (Fig. 1).
In null mutants for the nucleolar protein Nucleolin1 (nuc1), NOR2 becomes associated with the nucleolus. Subsequently, there is a severe decrease in expression of NOR4 NAD genes and an increase in NOR2 NAD-gene expression. nuc1 mutants also display increased heterochromatin foci within the nucleoli. These data suggest a link between nucleolar association and gene activity as well as a role for NUC1 in heterochromatin distribution. Interestingly, this NUC1 role appears to oppose the action of the Drosophila Nucleolin homolog, Modullo, which is required for centromere sequestration at the nucleolus periphery (Padeken et al., 2013). NUC1 also plays a role in the localization of FIB2, a nucleolar protein located in fibrillary centers within the nucleolus and involved in methylation of rRNA precursors (Picart and Pontvianne, 2017). This suggests a correlation between ribosome biogenesis and genome organization within the nucleolus.
Further work detailing the function of NORs in Arabidopsis utilized the atxr5 atxr6 (trithorax-related proteins 5 and 6) double mutant, renamed here altered subtype content1 (ASC1) because it caused the replacement of NOR4 with NOR2, resulting in two NOR2 regions (Mohannath et al., 2016). Interestingly, this resulted in activation of the usually silenced NOR2-derived rRNA genes. These results indicate that rRNA gene silencing is a result of gene position, not sequence variations. Although it has been shown that NOR4 associates with the nucleolus and NOR2 does not, the subnuclear location of the altered NORs in ASC1 were not determined. It would be worthwhile to examine the position of the ASC1 NORs within the nucleus to examine any correlation between the change in gene expression with a possible relocation to the nucleolus. This would add clarity to our understanding of how gene regulation occurs, whether expression is effected more by the chromatin location within the nucleus (i.e. spatial location) or the role of individual gene positioning within a given chromatin region (i.e. nearby elements within the chromatin region itself).
Together, these studies imply that the position of NORs relative to the nucleolus might directly regulate gene expression, with nucleolus-association enhancing gene activity. Adding to the findings on chromatin association with the nuclear periphery, these data point at another important subnuclear location that may act toward regulating gene expression.
Advancement of Cytogenetic Techniques
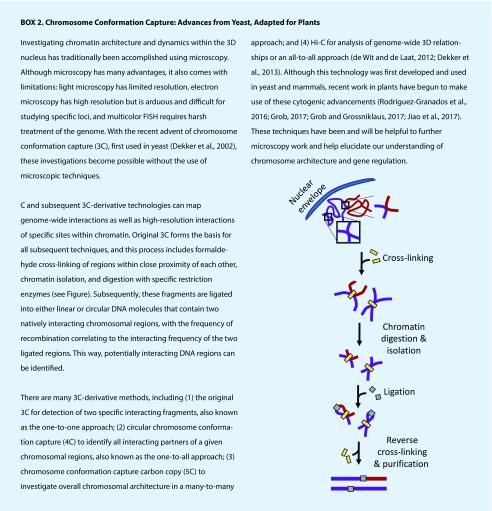
Visualization of 3D chromatin interactions has recently become accessible through chromosome conformation capture (3C; Box 2; de Wit and de Laat, 2012; Dekker et al., 2002, 2013; Grob, 2017; Jiao et al., 2017). Wang et al. (2017) utilized previous data from Hi-C technology to categorize Arabidopsis chromosomal interactions into seven groups based on gene densities, gene types, histone modifications, and interacting frequencies. Consistent with similar studies in humans (Lan et al., 2012), intrachromosomal interactions appear to predominate in Arabidopsis, compared to interchromosomal interactions, which occur at sites of transposable elements and typically result as a response to stimuli. Grob and Grossniklaus (2017) developed a new Hi-C protocol that is optimized for Arabidopsis seedling tissue and shows promise to provide further specificity and to verify these findings.
A modified chromatin immunoprecipitation protocol utilizes restriction enzyme digestion instead of high-energy sonication. The use of restriction enzymes allows for milder sonication and minimizes disruptions to chromatin-interacting loci, increasing the identification of chromatin-interacting partners (Bi et al., 2017). The authors used this to identify the enrichment of transcriptionally inactive chromatin at the nuclear periphery in Arabidopsis. Considering chromatin patterns during DNA replication, in vivo 5-ethynyl-20-deoxyuridine (EdU) pulse labeling combined with flow cytometry of fixed nuclei allowed for isolation of distinct stages of S phase in maize root-tip cells. EdU pulse labeling allowed for the discovery of a novel middle S phase chromatin pattern unique to plants (Bass et al., 2015).
Chromatin architecture studies are also reaching less-well-characterized plant model species. In Cowpea (Vigna unguiculata [L.] Walp), 11 distinct chromosomal structures were identified using chromosome-specific bacterial artificial chromosome clones for FISH, allowing for the analysis of heterochromatin distribution, centromere position, chromosome lengths, and positions of various rDNA regions (Iwata-Otsubo et al., 2016). In Brachypodium distachyon root-tip cells, chromosomal aberrations were analyzed using multicolor FISH during chemical mutagen-induced micronuclei formation. The authors found two unique rRNA genes on distinct chromosomes, allowing for identification of these specific chromosomes and adding to general chromosomal markers (i.e. telomeric, centromeric, and interstitial sites). The authors were able to use these markers to analyze chromosome and chromosome fragment involvement in micronuclei formation for the first time in Brachypodium, and similarly to other species, chromosomal aberrations are not random (Kus et al., 2017).
In summary, advances in both techniques and molecular characterization further enhance our knowledge of chromatin architecture, function, and gene expression in plants and will help to lay the foundation to study how these dynamics are altered during development and plant stress response. The cytogenetic characterization of plant crop species allows for direct comparison to traditional model organisms and can be used to determine whether these principles are conserved across plant species.
NUCLEI ON THE MOVE: NUCLEAR POSITIONING AND MIGRATION
While nucleocytoplasmic transport, the composition and interactions of nuclear envelope protein complexes, and the organization of chromatin all add to the complex dynamics of nuclear functional composition, the nucleus itself is also a highly dynamic organelle. Nuclear movement occurs in multiple plant cell types and developmental processes, as well as in response to both biological and mechanical stimuli (Griffis et al., 2014, and references therein). Several recent papers have focused on two nuclear movement events: nuclear photorelocation (Kawashima and Berger, 2015; Suetsugu et al., 2015; Iwabuchi et al., 2016; Suetsugu et al., 2016a, 2016b) and male germ unit migration in pollen tubes (Zhou and Meier, 2014; Zhou et al., 2015c). In addition, nuclei have been traced during root-hair cell death, in collet root hairs, and during gamete fusion (Kawashima and Berger, 2015; Sliwinska et al., 2015; Tan et al., 2016).
Blue Light-Dependent Nuclear Movement
In Arabidopsis leaf epidermal and mesophyll cells, nuclei move from central “dark” positions to peripheral “light” positions in response to blue light, termed photorelocation (Iwabuchi et al., 2007). When irradiated on their sides with blue light, epidermal cell nuclei have two modes of movement: parallel to actin bundles (“parallel movement”) and away from the point of irradiation (“avoidance movement”); similarly applied red light induces only parallel movement (Higa et al., 2014). Avoidance movement is dependent upon both nuclear-adjacent chloroplasts and chloroplast-adjacent actin filaments (cp-actin; Higa et al., 2014). C2-domain proteins PMI1 and PMIR1 have also been implicated in cp-actin-dependent nuclear photorelocation in pavement cells (Suetsugu et al., 2015).
The role of cp-actin filaments in nuclear photorelocation was further investigated by Suetsugu et al. (2016a, 2016b). CHLOROPLAST UNUSUAL POSITIONING1 (chup1) and KINESIN-LIKE PROTEIN FOR ACTIN-BASED CHLOROPLAST MOVEMENT1 and 2 (kac1 kac2) mutants are impaired in cp-actin filament formation and avoidance movement. Dark-adapted kac1 kac2 and chup1 kac1 kac2 mutant plants were defective in “dark” nuclear positioning. When the leaf was irradiated with a high dose of blue light, nuclei of chup1 mutants retained only a minimal avoidance response, kac1 kac2 mutants had a moderate avoidance response, while chup1 kac1 kac2 mutants had no avoidance response. These data suggest that CHUP1, KAC1, and KAC2 act redundantly to position nuclei in response to high-intensity blue light, while KAC1 and KAC2 are important for dark-induced nuclear positioning (Suetsugu et al., 2016a, 2016b).
If chloroplast movement is the primary cause of nuclear movement during blue light irradiation (Higa et al., 2014), is nuclear movement a side effect or biologically relevant? Iwabuchi et al. (2016) address one theoretical function of nuclear photorelocation: UV damage protection. They show that cells containing peripherally positioned nuclei are less likely to undergo UVB-induced DNA damage and cell death. Positioning of nuclei to the cell periphery was both actin- and high-light-dependent. These data suggest that plants may use blue light as a proxy for UV irradiation in order to avoid DNA damage (Iwabuchi et al., 2016).
Nuclear Movement and Plant Fertilization
Recent research has shown the importance of LINC complexes (Box 1) in nuclear movement in pollen tubes (Zhou and Meier, 2014; Zhou et al., 2015c). Disruption of either WIP or WIT genes perturbs pollen vegetative nuclear movement and decouples it from pollen tube tip growth (Zhou and Meier, 2014). Zhou et al. (2015c) addressed whether WIP and WIT act in this context as part of a LINC complex. To work around the lethality of the sun1 sun2 double null mutant (Varas et al., 2015), they retargeted remaining SUN proteins in a viable, leaky sun1 sun2 mutant background to the ER and saw a significant effect on both pollen vegetative nuclear movement and fertilization. Taken together, these papers suggest that the entire LINC complex is involved in moving the vegetative nucleus during pollen tube growth.
In the absence of centrosomes in plants, migration of the gamete pronuclei is controlled not by microtubules but actin (Kawashima et al., 2014). Interestingly, while F-actin is required for the migration of the male nucleus toward the female nucleus in both egg cell and central cell, the central cell nuclear position is also dependent on F-actin, but this position is dispensable for double fertilization (Kawashima and Berger, 2015).
Nuclear Movement in Root Hairs
Two recent papers investigated new aspects of nuclear movement in root hairs. Nuclei maintain a set distance from the tip of growing root hairs, moving to a random location once tip growth has ceased (Ketelaar et al., 2002). By contrast, in naturally dying root hairs, Tan et al. (2016) observed basally located nuclei. In root hairs undergoing programmed cell death, nuclei move basipetally, dependent on a nuclear-associated actin network that remains around the nucleus after programmed cell death. Cell death also impacted nuclear shape: While nuclei in growing root hairs are spindle shaped, those in dead root hairs are spherical (Tan et al., 2016).
In the rhd3 mutant, which develops wavy collet hairs, the nucleus is decoupled from tip growth. Additional evidence indicating that RHD3 may be involved in vesicle trafficking (Schiefelbein and Somerville, 1990; Galway et al., 1997; Wang et al., 1997) suggests that this latter process may be involved in maintaining nuclear tip distance in collet hairs. Furthermore, nuclear shape and endoreduplication level also varied widely in rhd3, with a mixture of round nuclei with fewer genome copies and elongated nuclei with more genome copies (Sliwinska et al., 2015).
CONCLUSIONS AND PERSPECTIVES
The connection between the dynamic three-dimensional organization of the nuclear interior and the regulation of gene expression is a rapidly expanding field in many eukaryotic model systems. Silencing, activation, and epigenetic changes are associated with locus position and proximity to nuclear landmarks such as nuclear pores, the nuclear envelope, or the nucleolus. An increasing number of mutants that change 3D chromatin organization, nuclear morphology, and gene expression as well as refined methods to reveal such changes are emerging. Plant research has already revealed interesting novelties, such as plant-specific regulatory nucleoporins, a unique set of nuclear envelope protein complexes, and—importantly—connections between spatial nuclear functionalities and plants’ reactions to the environment. Many dots still need to be connected, but the field promises not only to lead to novel explanations for known phenomena from the cell biologist’s point of view but also to create unique new tools for the targeted manipulation of the underlying pathways.
Acknowledgments
Work on this topic in I.M.’s lab is supported by grants from the National Science Foundation
Glossary
- NE
nuclear envelope
- ONM
outer nuclear membrane
- INM
inner nuclear membrane
- NPC
nuclear pore complex
- ETI
effector-triggered immunity
- SAS
shade avoidance syndrome
- LINC
linker of nucleoskeleton and cytoskeleton
Footnotes
Articles can be viewed without a subscription.
References
- Bass HW, Hoffman GG, Lee TJ, Wear EE, Joseph SR, Allen GC, Hanley-Bowdoin L, Thompson WF (2015) Defining multiple, distinct, and shared spatiotemporal patterns of DNA replication and endoreduplication from 3D image analysis of developing maize (Zea mays L.) root tip nuclei. Plant Mol Biol 89: 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzenschlager M, Lermontova I, Schubert V, Fuchs J, Berr A, Koini MA, Houlné G, Herzog E, Rutten T, Alioua A, et al. (2015) Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc Natl Acad Sci USA 112: 8656–8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzenschlager M, Masoud K, Janski N, Houlné G, Herzog E, Evrard JL, Baumberger N, Erhardt M, Nominé Y, Kieffer B, et al. (2013) The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana. Front Plant Sci 4: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzenschlager M, Schmit AC, Herzog E, Fuchs J, Schubert V, Houlné G, Chabouté ME (2017) MGO3 and GIP1 act synergistically for the maintenance of centromeric cohesion. Nucleus 8: 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Cheng YJ, Hu B, Ma X, Wu R, Wang JW, Liu C (2017) Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res 27: 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cavalli G (2016) Organization and function of the 3D genome. Nat Rev Genet 17: 661–678 [DOI] [PubMed] [Google Scholar]
- Bourbousse C, Mestiri I, Zabulon G, Bourge M, Formiggini F, Koini MA, Brown SC, Fransz P, Bowler C, Barneche F (2015) Light signaling controls nuclear architecture reorganization during seedling establishment. Proc Natl Acad Sci USA 112: E2836–E2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic J, Zhao Q, Meier I (2009) WPP-domain proteins mimic the activity of the HSC70-1 chaperone in preventing mistargeting of RanGAP1-anchoring protein WIT1. Plant Physiol 151: 142–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Sun J, Wysham D, Otegui MS, Venkateshwaran M, Hirsch S, Miwa H, Downie JA, Morris RJ, Ané JM, et al. (2011) Nuclear membranes control symbiotic calcium signaling of legumes. Proc Natl Acad Sci USA 108: 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG (2015) Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 208: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, Parniske M (2008) Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Oldroyd GE (2013) Nuclear calcium signaling in plants. Plant Physiol 163: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Sun J, Vaz Martins T, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Véry AA, Sanders D, Morris RJ, et al. (2016) Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciska M, Moreno Díaz de la Espina S (2013) NMCP/LINC proteins: Putative lamin analogs in plants? Plant Signal Behav 8: e26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W (2012) A decade of 3C technologies: Insights into nuclear organization. Genes Dev 26: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA (2013) Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat Rev Genet 14: 390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ (2007) LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19: 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH, Zhu JK (2006) A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 26: 9533–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Tjong H, Weider E, Herzog M, Young B, Brune C, Müllner D, Loewen C, Alber F, Weis K (2016) Global reorganization of budding yeast chromosome conformation in different physiological conditions. J Cell Biol 212: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Faitelson L, Hassan-Zadeh V, Baghestani Z, Bazett-Jones DP (2016) Coming to terms with chromatin structure. Chromosoma 125: 95–110 [DOI] [PubMed] [Google Scholar]
- Feng CM, Qiu Y, Van Buskirk EK, Yang EJ, Chen M (2014) Light-regulated gene repositioning in Arabidopsis. Nat Commun 5: 3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallemí M, Galstyan A, Paulišić S, Then C, Ferrández-Ayela A, Lorenzo-Orts L, Roig-Villanova I, Wang X, Micol JL, Ponce MR, et al. (2016) DRACULA2 is a dynamic nucleoporin with a role in regulating the shade avoidance syndrome in Arabidopsis. Development 143: 1623–1631 [DOI] [PubMed] [Google Scholar]
- Galway ME, Heckman JW Jr., Schiefelbein JW (1997) Growth and ultrastructure of Arabidopsis root hairs: The rhd3 mutation alters vacuole enlargement and tip growth. Planta 201: 209–218 [DOI] [PubMed] [Google Scholar]
- Genenncher B, Wirthmueller L, Roth C, Klenke M, Ma L, Sharon A, Wiermer M (2016) Nucleoporin-regulated MAP kinase signaling in immunity to a necrotrophic fungal pathogen. Plant Physiol 172: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavami A, van der Giessen E, Onck PR (2016) Energetics of transport through the nuclear pore complex. PLoS One 11: e0148876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K. (2014) Evidence for LINC1-SUN associations at the plant nuclear periphery. PLoS One 9: e93406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE (2010) Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 61: 134–144 [DOI] [PubMed] [Google Scholar]
- Graumann K, Vanrobays E, Tutois S, Probst AV, Evans DE, Tatout C (2014) Characterization of two distinct subfamilies of SUN-domain proteins in Arabidopsis and their interactions with the novel KASH-domain protein AtTIK. J Exp Bot 65: 6499–6512 [DOI] [PubMed] [Google Scholar]
- Griffis AH, Groves NR, Zhou X, Meier I (2014) Nuclei in motion: Movement and positioning of plant nuclei in development, signaling, symbiosis, and disease. Front Plant Sci 5: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S. (2017) Circular chromosome conformation capture in plants. Methods Mol Biol 1610: 73–92 [DOI] [PubMed] [Google Scholar]
- Grob S, Grossniklaus U (2017) Chromatin conformation capture-based analysis of nuclear architecture. Methods Mol Biol 1456: 15–32 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R (2015) Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 84: 131–164 [DOI] [PubMed] [Google Scholar]
- Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, Dong X (2016) Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell 166: 1526–1538.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa T, Suetsugu N, Kong S-G, Wada M (2014) Actin-dependent plastid movement is required for motive force generation in directional nuclear movement in plants. Proc Natl Acad Sci USA 111: 4327–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K, Hidema J, Tamura K, Takagi S, Hara-Nishimura I (2016) Plant nuclei move to escape ultraviolet-induced DNA damage and cell death. Plant Physiol 170: 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K, Sakai T, Takagi S (2007) Blue light-dependent nuclear positioning in Arabidopsis thaliana leaf cells. Plant Cell Physiol 48: 1291–1298 [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Lin JY, Gill N, Jackson SA (2016) Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis. Chromosome Res 24: 197–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD (2007) The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol 144: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao WB, Accinelli GG, Hartwig B, Kiefer C, Baker D, Severing E, Willing EM, Piednoel M, Woetzel S, Madrid-Herrero E, et al. (2017) Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res 27: 778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Berger F (2015) The central cell nuclear position at the micropylar end is maintained by the balance of F-actin dynamics, but dispensable for karyogamy in Arabidopsis. Plant Reprod 28: 103–110 [DOI] [PubMed] [Google Scholar]
- Kawashima T, Maruyama D, Shagirov M, Li J, Hamamura Y, Yelagandula R, Toyama Y, Berger F (2014) Dynamic F-actin movement is essential for fertilization in Arabidopsis thaliana. eLife 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, de Ruijter NCA, Grierson CS, Dogterom M, Emons AMC (2002) Positioning of nuclei in Arabidopsis root hairs: An actin-regulated process of tip growth. Plant Cell 14: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus A, Kwasniewska J, Hasterok R (2017) Brachypodium distachyon—a useful model in the qualification of mutagen-induced micronuclei using multicolor FISH. PLoS One 12: e0170618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Witt H, Katsumura K, Ye Z, Wang Q, Bresnick EH, Farnham PJ, Jin VX (2012) Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res 40: 7690–7704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115 [DOI] [PubMed] [Google Scholar]
- Li Q, Zheng J, Li S, Huang G, Skilling SJ, Wang L, Li L, Li M, Yuan L, Liu P (2017) Transporter-mediated nuclear entry of jasmonoyl-isoleucine is essential for jasmonate signaling. Mol Plant 10: 695–708 [DOI] [PubMed] [Google Scholar]
- MacGregor DR, Gould P, Foreman J, Griffiths J, Bird S, Page R, Stewart K, Steel G, Young J, Paszkiewicz K, et al. (2013) HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell 25: 4391–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M (1997) Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res 232: 173–181 [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I (2008) The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157 [DOI] [PubMed] [Google Scholar]
- Meier I. (2016) LINCing the eukaryotic tree of life—towards a broad evolutionary comparison of nucleocytoplasmic bridging complexes. J Cell Sci 129: 3523–3531 [DOI] [PubMed] [Google Scholar]
- Meier I, Richards EJ, Evans DE (2017) Cell biology of the plant nucleus. Annu Rev Plant Biol 68: 139–172 [DOI] [PubMed] [Google Scholar]
- Mochizuki R, Tsugama D, Yamazaki M, Fujino K, Masuda K (2017) Identification of candidates for interacting partners of the tail domain of DcNMCP1, a major component of the Daucus carota nuclear lamina-like structure. Nucleus 8: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohannath G, Pontvianne F, Pikaard CS (2016) Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc Natl Acad Sci USA 113: 13426–13431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy S, Meier I (2011) Genetic and environmental changes in SUMO homeostasis lead to nuclear mRNA retention in plants. Planta 233: 201–208 [DOI] [PubMed] [Google Scholar]
- Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G (2010) Initial genomics of the human nucleolus. PLoS Genet 6: e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikumbh S, Pfeifer N (2017) Genetic sequence-based prediction of long-range chromatin interactions suggests a potential role of short tandem repeat sequences in genome organization. BMC Bioinformatics 18: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeken J, Mendiburo MJ, Chlamydas S, Schwarz HJ, Kremmer E, Heun P (2013) The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell 50: 236–249 [DOI] [PubMed] [Google Scholar]
- Parry G. (2014) Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J Exp Bot 65: 6057–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M (2006) The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar V, Poulet A, Détourné G, Tatout C, Vanrobays E, Evans DE, Graumann K (2016) A novel family of plant nuclear envelope-associated proteins. J Exp Bot 67: 5699–5710 [DOI] [PubMed] [Google Scholar]
- Perrella G, Kaiserli E (2016) Light behind the curtain: Photoregulation of nuclear architecture and chromatin dynamics in plants. New Phytol 212: 908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picart C, Pontvianne F (2017) Plant nucleolar DNA: Green light shed on the role of Nucleolin in genome organization. Nucleus 8: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Carpentier MC, Durut N, Pavlištová V, Jaške K, Schořová Š, Parrinello H, Rohmer M, Pikaard CS, Fojtová M, et al. (2016) Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Reports 16: 1574–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Duc C, Voisin M, Desset S, Tutois S, Vanrobays E, Benoit M, Evans DE, Probst AV, Tatout C (2017a) The LINC complex contributes to heterochromatin organisation and transcriptional gene silencing in plants. J Cell Sci 130: 590–601 [DOI] [PubMed] [Google Scholar]
- Poulet A, Probst AV, Graumann K, Tatout C, Evans D (2017b) Exploring the evolution of the proteins of the plant nuclear envelope. Nucleus 8: 46–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas NM, Shivashankar GV (2015) Cytoskeletal control of nuclear morphology and chromatin organization. J Mol Biol 427: 695–706 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Granados NY, Ramirez-Prado JS, Veluchamy A, Latrasse D, Raynaud C, Crespi M, Ariel F, Benhamed M (2016) Put your 3D glasses on: Plant chromatin is on show. J Exp Bot 67: 3205–3221 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Takagi S (2013) LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol 54: 622–633 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Görlich D (2016) Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci 41: 46–61 [DOI] [PubMed] [Google Scholar]
- Sliwinska E, Mathur J, Bewley JD (2015) On the relationship between endoreduplication and collet hair initiation and tip growth, as determined using six Arabidopsis thaliana root-hair mutants. J Exp Bot 66: 3285–3295 [DOI] [PubMed] [Google Scholar]
- Smith S, Galinha C, Desset S, Tolmie F, Evans D, Tatout C, Graumann K (2015) Marker gene tethering by nucleoporins affects gene expression in plants. Nucleus 6: 471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN (2010) Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26: 421–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Higa T, Gotoh E, Wada M (2016a) Correction: Light-induced movements of chloroplasts and nuclei are regulated in both Cp-actin-filament-dependent and -independent manners in Arabidopsis thaliana. PLoS One 11: e0168318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Higa T, Gotoh E, Wada M (2016b) Light-induced movements of chloroplasts and nuclei are regulated in both Cp-actin-filament-dependent and -independent manners in Arabidopsis thaliana. PLoS One 11: e0157429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Higa T, Kong SG, Wada M (2015) PLASTID MOVEMENT IMPAIRED1 and PLASTID MOVEMENT IMPAIRED1-RELATED1 mediate photorelocation movements of both chloroplasts and nuclei. Plant Physiol 169: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Hatsugai N, Katagiri F, Hara-Nishimura I (2017) Nup82 functions redundantly with Nup136 in a salicylic acid-dependent defense response of Arabidopsis thaliana. Nucleus 8: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Hara-Nishimura I (2014) Functional insights of nucleocytoplasmic transport in plants. Front Plant Sci 5: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 23: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Tan K, Wen C, Feng H, Chao X, Su H (2016) Nuclear dynamics and programmed cell death in Arabidopsis root hairs. Plant Sci 253: 77–85 [DOI] [PubMed] [Google Scholar]
- Thakar R, Csink AK (2005) Changing chromatin dynamics and nuclear organization during differentiation in Drosophila larval tissue. J Cell Sci 118: 951–960 [DOI] [PubMed] [Google Scholar]
- Thorpe SD, Charpentier M (2017) Highlight on the dynamic organization of the nucleus. Nucleus 8: 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas J, Graumann K, Osman K, Pradillo M, Evans DE, Santos JL, Armstrong SJ (2015) Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J 81: 329–346 [DOI] [PubMed] [Google Scholar]
- Wang H, Dittmer TA, Richards EJ (2013) Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol 13: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X (2014) A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 16: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou Y, Li X, Meng X, Fan M, Chen H, Xue J, Chen M (2017) Genome-wide analysis of the distinct types of chromatin interactions in Arabidopsis thaliana. Plant Cell Physiol 58: 57–70 [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP (2010) The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Meulia T, Meier I (2007a) Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol 17: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I (2007b) NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang W, Chu Z, Zhu JK, Zhang H (2017) Roles of nuclear pores and nucleo-cytoplasmic trafficking in plant stress responses. Front Plant Sci 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X (2005) A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Brkljacic J, Meier I (2008) Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Evans DE, Meier I (2012) Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol 196: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Meier I (2015a) The plant nuclear envelope as a multifunctional platform LINCed by SUN and KASH. J Exp Bot 66: 1649–1659 [DOI] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Wirthmueller L, Jones JD, Meier I (2014) Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. J Cell Biol 205: 677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Groves NR, Meier I (2015b) Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 6: 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Groves NR, Meier I (2015c) SUN anchors pollen WIP-WIT complexes at the vegetative nuclear envelope and is necessary for pollen tube targeting and fertility. J Exp Bot 66: 7299–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Meier I (2014) Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc Natl Acad Sci USA 111: 11900–11905 [DOI] [PMC free article] [PubMed] [Google Scholar]