Abstract
Dynamic changes maintain a multipartite mitochondrial genome meets the changing needs of plant cells.
In plant somatic cells, the number of mitochondria is usually larger than the copy number of their genome. The constant fission and fusion of mitochondria make it possible to share the internal materials between them (Arimura et al., 2004b). Therefore, the chondriome (the collective mitochondria in a cell) is thought to exist as a discontinuous whole (Logan, 2006). In this Update, I describe recent reports about mitochondrial dynamics during developmental stages, in specific organs, and in environmental responses, the molecular mechanisms underlying these dynamics, and the relationship to their genome maintenance. Several factors are involved in mitochondrial biogenesis, including gene expression in the nucleus and mitochondria and protein import from the cytosol to the mitochondria. These processes are covered in some excellent recent reviews (Millar et al., 2008; Carrie et al., 2013; Law et al., 2014; Czarna et al., 2016). Here, I focus on recent investigations of the fission and fusion of plant mitochondria and its relatedness to their genome maintenance.
Mitochondria have an endosymbiotic origin in which they are thought to be descendants of an α-proteobacteria. Mitochondria maintain themselves through growth and division but without de novo formation from other organelles. They differ from plastids and peroxisomes in how they control their numbers, shape, and volume. Plastids are generated by growth and division without de novo synthesis, and they rarely undergo fusion (Sakamoto et al., 2008; Osteryoung and Pyke, 2014). Peroxisomes also are generated by growth and division and rarely undergo fusion. However, they also are de novo synthesized from the endoplasmic reticulum (ER; Hu et al., 2012). Interestingly, peroxisomes share some division factors with mitochondria (Pan and Hu, 2011; Arimura and Tsutsumi, 2016).
The pleomorphy and dynamics of mitochondria were observed over 100 years ago (Lewis and Lewis, 1914) and have been further investigated more recently (Bereiter-Hahn, 1990; Logan and Leaver, 2000; Logan, 2006; Jaipargas et al., 2015). Fission and fusion can change the number and shape of mitochondria from a single network to hundreds of particles by shifting their balance without changing their total volume in a cell. Shifting the balance between fission and fusion of mitochondria can have many direct and indirect effects in plants (Logan, 2006, 2010) and in mammals (Liesa and Shirihai, 2013; Mishra and Chan, 2016; see Box 1).
MITOCHONDRIAL FISSION PROTEINS IN YEAST, ANIMALS, AND A RED ALGA
Dnm1, a dynamin-related protein in the yeast Saccharomyces cerevisiae, is the first mitochondrial fission protein to be identified and studied (Bleazard et al., 1999; Sesaki and Jensen, 1999; for review, see Bui and Shaw, 2013; Table I). Dnm1 polymerizes into a helical ring-like structure that wraps around and constricts the site of dividing mitochondria by their GTPase activity. Dnm1 is recruited from the cytosol through interaction with the adaptor protein Mdv1 (and its paralog Caf4), which interact with Dnm1 and Fis1. Fis1 is a small, mitochondrial outer membrane tail-anchored protein. Animal mitochondrial fissions also are implemented by Dnm1 orthologs, Drp1s. Sequences homologous to Fis1 also are found and well conserved in many eukaryote genomes. Orthologs of Mdv1/Caf4, the adaptors between Dnm1 and Fis1, are present in yeast but are rare or absent in other eukaryote genomes. In animals, Fis1 homologs had originally been thought to be required for the localization of Drp1 to the fission sites, as is the case with Fis1 in yeast. However, instead of Fis1, two other mitochondrial outer-membrane proteins (Mff and MiD49/51) were later found to have some redundant but major roles in the relocalization of Drp1 from the cytosol to mitochondrial fission sites (Otera et al., 2010; Palmer et al., 2011, 2013; Zhao et al., 2011).
Table I. Homologous relationships of mitochondrial fission and fusion factors in selected eukaryotes.
The table is reproduced and modified from Arimura and Tsutsumi (2016) with permission. IM, Inner membrane; OM, outer membrane. Dashes indicate that homologs are not present or not reported.
| Process | Organisms |
Features, Functions, and Localization (Estimated) | ||||
|---|---|---|---|---|---|---|
| Yeast S. cerevisiae | Mammal Homo sapiens | Seed Plant Arabidopsis | Liverwort M. polymorpha | Red alga C. merolae | ||
| Fission | Dnm1 | Drp1 | DRP3A, DRP3B | DRP3 | Dnm1 | Cytosolic, forming ring, GTPase |
| Fis1 | Fis1 | AtFIS1A, AtFIS1B | FIS1 | CMQ197C | OM, for localization of DRPs? (through Mdv1 in yeast) | |
| Mdv1, Caf4 | – | – | – | Mda1 | OM outer surface, interact with DRP and Fis1 (in yeast) | |
| – | Mff | – | – | – | OM, for localization of DRP to fission sites | |
| – | MiD49 and MiD51 | – | – | – | OM, for localization of DRP to fission sites | |
| – | – | ELM1, At5g06180 | ELM1 | – | OM outer surface, interact with DRP3 | |
| – | – | PMD1, PMD2 | – | – | OM, independent of DRP3, Rosidae specific | |
| – | – | – | – | FtsZ1 | Matrix, GTPase, polymerizes into ftsZ ring | |
| – | – | – | – | ZED | Matrix, bacterial cell division ZapA homolog, forms ring | |
| Fusion | Fzo1 | Mfn1, Mfn2 | – | – | – | OM, for OM fusion, GTPase |
| Mgm1 | Opa1 | – | – | – | IM and intermembrane space, for IM fusion, GTPase | |
| Ugo1 | – | – | – | – | OM, for OM fusion, interacts with Fzo1 and Mgm1, OM | |
| Clu1 | Clu | FMT | FMT | – | Cytosol and OM, deficiency causes clusters of mitochondria | |
The unicellular red alga Cyanidioschyzon merolae is an excellent experimental model for mitochondrial division because each cell has a single mitochondrion, whose divisions are linked to the host cell division cycle. For studies of mitochondrial division, the cell cycle in C. merolae cells can be synchronized. The proteins that form mitochondrial division rings have been purified and identified (Yoshida et al., 2006; Nishida et al., 2007). C. merolae has two mitochondrial division proteins, an FtsZ homolog (CmFtsZ1) and a ZapA-like protein (ZED), that are similar to bacterial cell division proteins (Takahara et al., 2000; Yoshida et al., 2009). The genes for both of these proteins have been lost in the genomes of yeast, animals, and green plants. ZED and FtsZ1 make rings in the matrix beneath the inner membrane. Then, another protein (Mda1) and a mitochondrial dividing ring, a structure that is observed by electron microscopy, localize to the outer surface and start to constrict the mitochondrion. In the last step, CmDnm1, which is a dynamin-related protein, forms a ring around the constricted site, and the CmDnm1 ring with the mitochondrial dividing ring cuts the mitochondrion into two pieces (Nishida et al., 2003; for review, see Kuroiwa et al., 2008; Kuroiwa, 2010).
MITOCHONDRIAL FISSION PROTEINS IN ARABIDOPSIS AND THE LIVERWORT MARCHANTIA POLYMORPHA
Arabidopsis (Arabidopsis thaliana) has well-conserved Dnm1/Drp1 orthologs, DRP3A and DRP3B (formerly known as ADL2A/2B), that are involved in mitochondrial fission (Arimura and Tsutsumi, 2002; Arimura et al., 2004a; Logan et al., 2004; Fujimoto et al., 2009). They localized to the prefission and postfission sites of mitochondria (Fig. 1A). Disruption of these genes causes a decrease in mitochondrial number and the formation of elongated, network-shaped mitochondria. Two homologs of the outer-membrane anchored protein Fis1, AtFis1a (Bigyin) and AtFis1b, also were found, and disruption of their gene expression in Arabidopsis caused the mitochondria to become larger (Scott et al., 2006; Zhang and Hu, 2009). Another factor in mitochondrial fission was identified from forward genetic screening and mapping of Arabidopsis mutants with elongate mitochondria (Arimura et al., 2008). These mutants, called elm1 (elongate mitochondria) mutants, were defective in a gene that we called ELM1. The ELM1 protein is a plant-specific factor that localizes on the surface of the mitochondrial outer membrane, where it is required for the localization of DRP3A (Arimura et al., 2008). In a review (Logan, 2010), elm1 was suggested to be allelic to a previously screened mutant, nmt (network mitochondria; Logan et al., 2003), although the sequence of the responsible gene in nmt was not determined and the two mutants were not tested for allelism.
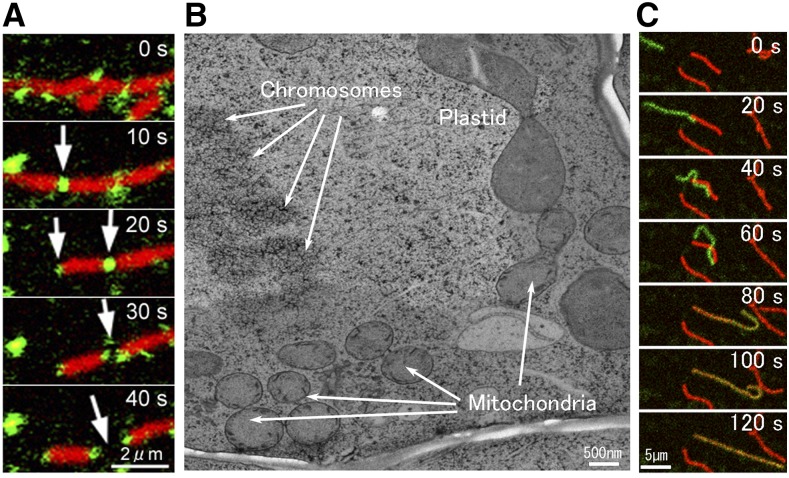
Figure 1.
Plant mitochondrial fission and fusion. A, Mitochondrial fission (arrows) with a dynamin-related protein, DRP3A (in green; GFP-DRP3A), which localizes to predividing and postdividing sites of mitochondria (in red; stained by MitoTracker) in a tobacco cultured BY-2 cell. Images were made by Dr. Masaru Fujimoto. B, Cell cycle-dependent divisions of organelles. The image shows aligned chromosomes, a dividing plastid, three pairs of divided mitochondria, and a dividing mitochondrion in a part of a dividing Arabidopsis root cell in metaphase. The electron micrograph was made by Dr. Mayuko Sato, Mayumi Wakazaki, and Dr. Kiminori Toyooka. The scale bar is approximate. C, Mitochondrial fusion in an onion (Allium cepa) epidermal cell in which mitochondrial fission was disrupted by coexpression of a dominant-negative type of DRP3B(K56A). Mitochondria matrices were labeled with a photoconvertible fluorescent protein, Kaede, causing some mitochondria to be green and others to be red. Fusion results in a yellow color.
DRP3A, FIS1a, and ELM1 have additional homologs (DRP3B, AtFIS1b, and At5g06180, respectively) in the Arabidopsis genome, which complicate their analysis. We recently used another model plant, the liverwort M. polymorpha, whose genome has single copies of DRP3, ELM1, and FIS1 (Nagaoka et al., 2017). Liverworts are considered as one of the earliest diverging distant land plant lineages (Wickett et al., 2014). Therefore, an examination of the characteristics of mitochondria in liverworts could help us to understand the commonalities and differences of mitochondrial fission factors among land plants. M. polymorpha drp3 and elm1 knockout mutants have highly elongated, networked mitochondria, and their growth is severely retarded (Nagaoka et al., 2017). The authors suggested that DRP3 and ELM1 were used for mitochondrial fission before the diversification of these two plants. However, the M. polymorpha fis1 knockout mutant grew almost as well as the wild type, and its mitochondria show little or no elongation. Therefore, FIS1 may have little or no involvement in mitochondrial fission in M. polymorpha epidermal cells, as is the case in mammals (Otera et al., 2010; Palmer et al., 2011, 2013; Zhao et al., 2011). Although Arabidopsis fis1 mutants are reported to have larger mitochondria (Scott et al., 2006; Zhang and Hu, 2009), their phenotypes seemed to be less obvious than those in drp3 and elm1 mutants. The conservation of FIS1 in animals, yeast, and plants raises the possibility that these proteins have other common roles not directly linked to mitochondrial fission with dynamins.
Two other mitochondrial outer-membrane proteins (peroxisomal and mitochondrial division factor1 [PMD1] and PMD2) were shown to be involved in the proliferation of mitochondria (Aung and Hu, 2011). PMD1 has a coiled-coil motif and a C-terminal transmembrane domain. PMD2 localizes specifically to mitochondria. PMD1 and PMD2 act in a nonredundant manner and in a manner independent of DRP3 and FIS1.
Mitochondrial fission is thought to be required for the inheritance and maintenance of mitochondria during the host cell division. Figure 1B shows four synchronized mitochondrial divisions that probably occur in a dividing Arabidopsis root cell in metaphase. In tobacco (Nicotiana tabacum) BY2 suspension cells, AtDRP3A/3B is activated by phosphorylation at metaphase and DRP3A/3B is partially degraded by ubiquitination at interphase (Wang et al., 2012). Overexpression of an outer-membrane-anchored ubiquitin protease, UBP27, changed mitochondrial morphology and reduced the mitochondrial association of DRP3 (Pan et al., 2014b), although it remains unclear whether UBP27 interacts directly with and deubiquitinates DRP3.
OTHER FACTORS, STRUCTURES, AND PHENOMENA RELATED TO MITOCHONDRIAL FISSION
Cardiolipins are phospholipids that are found mainly in the mitochondrial inner membrane. Cardiolipins are required for the localization or function of fission and fusion proteins in yeast and mammalian cells (for review, see Frohman, 2015). In an Arabidopsis cardiolipin synthase mutant, the mitochondria were enlarged and DRP3A/3B did not localize properly to mitochondria (Pan et al., 2014a).
In yeast and mammalian cells, the ER also appears to have a role in mitochondrial fission (Friedman et al., 2011). Mitochondrial fission occurs at the sites where ER tubules contact mitochondria and mediate constriction before the localization of dynamin-related proteins. Recent double-labeling studies of mitochondria and ER in plants have led to speculation that similar processes may occur in Arabidopsis (Jaipargas et al., 2015) and Physcomitrella patens (Mueller and Reski, 2015).
In the adl2a (drp3a) Arabidopsis mutant, matrix-localized fluorescent proteins revealed the presence of thin tubules called matrixules protruding from mitochondria (Logan et al., 2004). Similar tubules called stromules were observed to protrude from plastids (Köhler et al., 1997). We found similar structures in Arabidopsis by double labeling the mitochondrial outer membrane and the matrix (Yamashita et al., 2016b). These structures, which we named mitochondrial outer-membrane protrusions (MOPs), extended several micrometers from the main bodies of mitochondria (Yamashita et al., 2016b). MOPSs, as the name implies, tend to be composed more of the mitochondrial outer membrane than the matrix, whereas matrixules are extensions of the matrix. However, some MOPs have a limited amount of matrix. MOPs also were observed to bridge mitochondria (Yamashita et al., 2016a). The tips of some of the MOPs were pinched off to form smaller pieces that resembled mitochondria-derived vesicles in mammalian cells (for review, see Sugiura et al., 2014). MOPs and mitochondria-derived vesicle-like structures also were seen in the drp3a/3b double mutant (Yamashita et al., 2016b), suggesting that DRP3-dependent mitochondrial division is not required for their formation. These structures were more frequently observed in senescent leaves, suggesting that they were related to the degradation of mitochondria (Yamashita et al., 2016b). During leaf senescence in individually darkened leaves in Arabidopsis, the number of mitochondria decreases (Keech et al., 2007), probably by autophagy (Li et al., 2014, Li and Vierstra, 2014). The remaining mitochondria retain their motility and respiration activity until the last step of leaf senescence (Keech et al., 2007; Keech, 2011; Ruberti et al., 2014; Chrobok et al., 2016). These mitochondria are thought to provide energy and metabolites for degrading the cell components and relocating them to other younger parts of the plant (Keech et al., 2007; Keech, 2011; Ruberti et al., 2014; Chrobok et al., 2016).
Mitochondrial fission could serve not only to divide mitochondria but also to diversify them. Mitochondrial divisions without equal distribution of nucleoids were reported in tobacco cultured cells (Arimura et al., 2004b). Unequal division of mitochondria with different (lower and higher) membrane potentials was observed in mammalian cells (Twig et al., 2008). Mitochondria with low membrane potential (i.e. depolarized mitochondria) would not be able to fuse, resulting in their isolation from the healthy mitochondria that are continually undergoing fission and fusion. The isolated (nonfunctional) mitochondria are thought to be degraded specifically by bulk autophagy or mitophagy (mitochondria-specific autophagy; Twig et al., 2008). The mammalian Parkin and Pink1 genes, the causal genes for familial Parkinson’s diseases, were shown to have a role in mitophagy of degraded mitochondria (Narendra et al., 2008). Because plants have fission-fusion cycles of mitochondria and undergo autophagy, they may have a similar mechanism for mitochondrial quality control.
Different types of mitochondria coexist in germinating seeds (Dai et al., 1998; Logan et al., 2001) and individual senescence-induced leaves (Keech et al., 2007). On shorter time scales (e.g. seconds), individual mitochondria are depolarized spontaneously by an influx of calcium ions and then are immediately repolarized in several seconds (Schwarzländer et al., 2012). These changes, called pulsing, are observed much more frequently under stress conditions (Schwarzländer et al., 2012). At every time point, the mitochondria in each cell seem to have different physiological statuses and components.
MITOCHONDRIAL FUSION
Mitochondrial fusion in animals and yeast is carried out by two kinds of GTPases, Mfn/Fzo1 and Opa1/Mgm1, both of which are distantly related to dynamins (Chan, 2012; Mishra and Chan, 2016; for review, see Willems et al., 2015). Mfn1/Fzo1 and Opa1/Mgm1 are anchored in the outer and inner membranes, respectively, and contribute to the fusions of their respective membranes to those of another mitochondrion. Although mitochondrial fusion clearly occurs in plants (Fig. 1C; Arimura et al., 2004b; Sheahan et al., 2005), functional orthologs of dynamin-related proteins or other proteins involved in mitochondrial fusion have not been found so far in any plant. Arabidopsis has homologs of Mfn1/Fzo1 and Opa1/Mgm1, but the homologs most similar to these dynamin-related proteins are DRP3A/3B, which are apparently mitochondrial division-type dynamins. Another factor structurally similar to Mfn1/Fzo1, named FZL (for FZO-Like), is unrelated to mitochondrial fusion but was shown to localize in chloroplasts and contribute to the organization of thylakoids (Gao et al., 2006). Thus, the factors and molecular mechanisms directly involved in mitochondrial fusion in plants seem to be very different from those in animals and fungi. Forward genetic screenings for mutants of mitochondrial fusion (Logan et al., 2003; Feng et al., 2004) were unsuccessful, possibly because fusion mutants are more lethal than fission mutants, as is the case in yeast (Bleazard et al., 1999; Sesaki and Jensen, 1999). In Arabidopsis, the fmt (friendly mitochondria) mutant has aggregations of portions of mitochondria in each cell (Logan et al., 2003). FMT protein has been proposed to mediate the intermitochondrial association before fusion (El Zawily et al., 2014).
The evidence for mitochondrial fusion in plants was reported indirectly over 30 years ago (Belliard et al., 1979). In cybrids (cytoplasmic hybrids), which are plants regenerated from the fusion of cells of two species, the mitochondrial genomes were found to have new restriction enzyme fragment patterns, suggesting that recombination occurred between the parental mitochondrial genomes (Belliard et al., 1979). On the other hand, genetic recombination was not observed in plastids (i.e. only one of the parental genomes was observed). The mitochondrial genome of a cybrid between two plants in the nightshade family (tobacco and henbane [Hyoscyamus niger]) had more than 35 regions of recombination between homologous sequences, ranging from 100 to 9,000 bp (Sanchez-Puerta et al., 2015). Recombination and coexistence of the mitochondrial genes from different species also were observed in (1) plants generated from the junction of grafted tissues (Fuentes et al., 2014; Gurdon et al., 2016), (2) parasitic plants whose mitochondrial genomes contained some host plant fragments (Davis and Wurdack, 2004; Sanchez-Puerta et al., 2017), and even (3) host plants, in which fragments of parasitic plant mitochondrial genomes were observed in the host mitochondrial genomes (Mower et al., 2004). Furthermore, horizontal gene transfer between nonrelated plants is more frequently observed in mitochondrial genomes than in nuclear and plastid genomes (Bergthorsson et al., 2003). The mitochondrial genome of the flowering shrub Amborella trichopoda includes the entire mitochondrial genome of a moss (Rice et al., 2013; Taylor et al., 2015). At least in some cases, mitochondrial fusion might have contributed to horizontal gene transfer between plant mitochondrial genomes (Rice et al., 2013).
MITOCHONDRIAL DYNAMICS IN DIFFERENT ENVIRONMENTS AND ORGANS
When Arabidopsis plants were kept in the dark for 1 week without Suc, mitochondria in the mid cells of the hypocotyl became longer, but after the mid cells were exposed to light or Suc, their mitochondria fragmented (Jaipargas et al., 2015). Low oxygen also caused mitochondrial elongation and network formation in suspension-cultured tobacco cells (Van Gestel and Verbelen, 2002) and in Arabidopsis leaf mesophyll cells (Ramonell et al., 2001). Recently, transient fragmentation of mitochondria in Arabidopsis leaf epidermal cells was observed after cold treatment (Arimura et al., 2017). Such environmentally induced changes in mitochondria are probably due to changes in the balance between fission and fusion. In mammalian studies, cells with higher energy demand tend to have more elongated mitochondria, and on the other hand, cells with higher energy supply have more fragmented mitochondria (for review, see Liesa and Shirihai, 2013). The findings that plant mitochondria are longer in the dark without Suc (Jaipargas et al., 2015) or in low oxygen (Ramonell et al., 2001; Van Gestel and Verbelen, 2002) and fragmented in the light or with Suc (Jaipargas et al., 2015) seem to be consistent with such mammalian observations (i.e. mitochondrial fragmentation is associated with an increased energy supply).
Mitochondria have a characteristic cup shape or a long, stretched-rod shape, in the egg cells in Pelargonium zonale (Kuroiwa et al., 1996) and Zea mays (Diboll and Larson, 1966), but have relatively numerous small particle or peanut-like shapes in rice (Oryza sativa; Takanashi et al., 2010). Their DNA amounts in mitochondria in egg cells increase compared with those in pollen or leaf cells in species in which mitochondrial DNAs are maternally inherited (for review, see Nagata, 2010). During fertilization, mitochondria in the sperm cells in pollen were found to enter an egg cell and a central cell of a female gamete in Arabidopsis (Matsushima et al., 2008). However, mitochondrial DNA in pollen degraded before fertilization, suggesting that such DNA degradation apparently contributes to the maternal inheritance of mitochondrial genomes (Matsushima et al., 2008; Nagata, 2010; Wang et al., 2010). The DNA was reported to be degraded by an Mg2+-dependent exonuclease, DPD1 (Matsushima et al., 2011). However, the pollen from dpd1 mutants, in which organelle DNAs were clearly detected, could not cause paternal inheritance of mitochondrial DNA to the progeny, suggesting that some other mechanism suppresses the paternal inheritance of organelle DNA, independent of DPD1. Another protein for mitochondrial dynamics, a MIRO1 GTPase, is known to be essential for the proper mitochondrial morphology and distribution in pollen tube and in early embryogenesis, and the knockout of this gene causes lethality (Yamaoka and Leaver, 2008; Yamaoka et al., 2011).
Elongated or networked mitochondria also have been observed in some organs or developmental stages. In germinating Arabidopsis seeds, even just after imbibition, mitochondria first develop an inner-membrane potential and respiration ability, gradually acquire mobility, and then undergo transient elongation and branching (Paszkiewicz et al., 2017). At this latter stage (i.e. at the elongation and branching stage), the percentage of mitochondria with DNA increased. Long and branched tentaculate mitochondria were observed to surround the nucleus in the shoot apical meristems (but not in the root meristems) of Arabidopsis (Seguí-Simarro et al., 2008) and cucurbit plants (watermelon [Citrullis vulgaris] and muskmelon [Cucumis melo]; Bendich and Gauriloff, 1984). In tobacco, Arabidopsis, and Medicago spp. protoplasts before the first cell division, mitochondria underwent a transient massive fusion, resulting in elongated mitochondria (Sheahan et al., 2005). None of these cells with elongate mitochondria (germinating cells, shoot apical meristems, and culture-starting protoplasts) could be considered as somatic cells. Rather, they are meristem-like cells that are the sites of organellar DNA replication just before massive cell proliferation (Rose and McCurdy, 2017). Plant organellar DNA synthesis amplifies DNA copy numbers by 2 or more orders of magnitude during the early few cell divisions, while the nuclei duplicate their genome only once in each cell cycle (Kuroiwa et al., 1992; Suzuki et al., 1992; Fujie et al., 1994). These transient mitochondrial fusions might be related to mitochondrial DNA replication in preparation for the following massive cell proliferation (see below).
MAINTENANCE OF MITOCHONDRIAL DNA WITH DYNAMIC MITOCHONDRIA
In plant somatic cells, the number of mitochondrial genomes is less than the number of mitochondria (Bendich and Gauriloff, 1984; Kuroiwa et al., 1992; Preuten et al., 2010; Wang et al., 2010). During the latter part of leaf development or leaf senescence, the integrity and amount of organellar DNA decrease (for review, see Oldenburg and Bendich, 2015). One of the nucleases identified originally in pollen (DPD1 nuclease) is suggested to contribute to the degradation of organellar DNA during leaf senescence too (Sakamoto and Takami, 2014). Each Arabidopsis mesophyll cell has more than 500 mitochondria (Sheahan et al., 2005) and about 50 copies of mitochondrial genomes (Preuten et al., 2010; Wang et al., 2010). Some mitochondria seem to have no nucleoids (DNA-protein complexes in mitochondria; Arimura et al., 2004b; Takanashi et al., 2006; Wang et al., 2010). Many of the mitochondria with nucleoids have only a part of the mitochondrial genome (Kuroiwa et al., 1992; Satoh et al., 1993; Takanashi et al., 2006; Wang et al., 2010). The shortage of genetic information in the individual mitochondrion is thought to be overcome by constant fission and fusion (Arimura et al., 2004b; Logan, 2006).
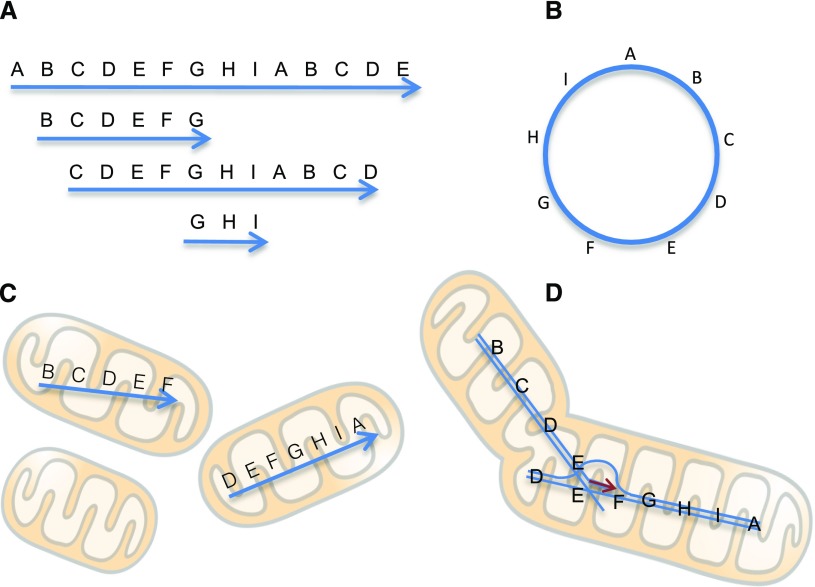
Plant mitochondrial genomes are much larger (from around 200 kb to more than 10 Mb) than the 16.5-kb single circular molecules of human and animal mitochondrial genomes (Kubo and Newton, 2008; Sloan, 2013; Gualberto and Newton, 2017). Two different structures have been proposed for plant mitochondrial genomes. They are usually depicted as circles, called master circles. Homologous sequences in different regions actively recombine, resulting in a multipartite genome structure consisting of many kinds of DNA (Palmer and Shields, 1984; Kubo and Newton, 2008). The circular structure was deduced from the mapping of sequences or restriction fragment length polymorphisms. However, the physical structure of mitochondrial DNA is now thought to be mostly linear with various sizes, some even longer than the genome size (Oldenburg and Bendich, 1996; Backert and Börner, 2000; Sloan, 2013; Cheng et al., 2017). Such linear structures are thought to have random fragments of a long head-to-tail concatemer sequence of the mitochondrial genomes (Fig. 2; Bendich, 1993; for review, see Sloan, 2013). For example, if the circular genome has the sequence ABCDE, the linear molecules would have sequences like ABCDEABCDEABCDEA or a mixture of ABC, DEABCD, BCDEA, and so on. Such structures that are physically linear but with a circularly permutated sequence are similar to those of some viruses and bacteriophages. The replication of plant mitochondrial genomes has been suggested to be similar to one of the replication mechanisms of T4 bacteriophages, called RDR (Backert and Börner, 2000; Maréchal and Brisson, 2010). At first, a T4 bacteriophage starts DNA replication from Ori sequences inside a host cell at the early stage of infection, generating one or a few copies (Kreuzer and Brister, 2010). However, when the copy number reaches about 20, replication depends mainly on RDR. In RDR, DNA synthesis starts from the 3′ end of each linear molecule, with other molecules acting as templates. In other words, both 3′ ends of each linear molecule function as primers for DNA synthesis, resulting in massive amplification of the genome (Broker and Doermann, 1975; Mosig, 1998; Kreuzer, 2000; Kreuzer and Brister, 2010). An advantage of RDR for circularly permutated linear molecules is that the DNAs are able to replicate without telomeres, which are necessary for the replication of the linear DNAs in nuclear chromosomes. Replication origins have not yet been identified in plant mitochondrial genomes. If they do not exist, replication might occur only by RDR.
Figure 2.
Mitochondrial genome structures, mitochondrial fusion, and DNA replication. A, In vivo variable-length linear DNA structures with circularly permutated sequences. B, A master circle of the mitochondrial genome deduced from these sequences, although such circles are probably rare. C, Individual mitochondria having zero or some portions of the genome as linear DNA molecules. Double-strand breaks that form linear DNA fragments might be generated by respiration-induced reactive oxygen species. D, Mitochondrial fusion provides an opportunity for recombination of DNA fragments. DNA replication by recombination-dependent replication (RDR) may start at this stage. Fusion is dominant where organellar DNA synthesis is active, such as in shoot apical meristems, and during germination and regeneration.
Cheng et al. (2017) recently observed the physical structures of DNA molecules isolated from mitochondria during the early germination of mung bean (Vigna radiata) seeds. At first, the DNA was in the form of simple linear molecules with sizes shorter than the genome size. Subsequently, during stratification, imbibition, and germination, the DNAs became more branched and longer, gradually acquiring sizes longer than the genome. Finally, the DNAs reverted to simple short linear molecules. These changes may reflect the gathering and recombination of short linear molecules at the start of RDR during germination. Similar changes in mitochondrial DNA structures have been observed in the growth cycles (but not cell cycles) of cultured tobacco cells in pioneering studies by Oldenburg and Bendich (1996; for review, see Oldenburg and Bendich, 2015) and Chenopodium album cells (Backert and Börner, 2000). Rapid amplification of organellar DNAs has been observed around meristems (Kuroiwa et al., 1992; Fujie et al., 1994) and at specific stages, such as during germination (Cheng et al., 2017) and at the start of cell culture (Satoh et al., 1993). The rapid amplification of organellar DNA independent of the host cell cycle also seems to be like that in phages that use RDR for DNA synthesis.
As stated above, fusion-dominant tentacle-shaped mitochondria have been observed in shoot apical meristems, and transient mitochondrial fusions have been observed in germinating cells and culture-starting protoplasts. These are the regions during the organellar DNA amplifications. A model of the relationship between mitochondrial fusion and DNA replication is shown in Figure 2. The mitochondria in plant somatic cells have linear DNA molecules, each containing a portion of the genome. Before the start of consecutive cell divisions (e.g. before the start of culture or the start of germination), mitochondria fuse to bring their DNA molecules together. The ends then undergo homologous recombination to start replication by RDR and to obtain the sequences that their genome lacks.
CONCLUSION
Many questions remain about the molecular factors and mechanisms of mitochondrial fission and fusion in plants (see Outstanding Questions box). Since the discovery of the balance between mitochondrial fission and fusion and the identification of molecular factors in yeast and mammals more than 15 years ago, research in this area has expanded enormously. Some of the new fields of study include mitophagy, mitochondrial quality control, submitochondrial structures, sites of contact with other organelles, cell death, and Parkinson’s and other neurodegenerative diseases. Plant mitochondria have many characteristics that distinguish them from other mitochondria, such as smallness, abundance, fast movement, and roles in photorespiration and cytoplasmic male sterility. These plant-specific characteristics present many challenging and exciting opportunities for future research.
Acknowledgments
I apologize for not being able to cite all of the relevant publications due to space limitations. The unpublished electron micrograph was kindly provided by Dr. Mayuko Sato, Mayumi Wakazaki, and Dr. Kiminori Toyooka (RIKEN Center for Sustainable Resource Science). I thank Dr. James Raymond for critical reading and helpful comments.
Footnotes
This work was supported by a grant from the Japan Society for the Promotion of Science (grant no. 16K14827).
Articles can be viewed without a subscription.
References
- Arimura S, Aida GP, Fujimoto M, Nakazono M, Tsutsumi N (2004a) Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol 45: 236–242 [DOI] [PubMed] [Google Scholar]
- Arimura S, Fujimoto M, Doniwa Y, Kadoya N, Nakazono M, Sakamoto W, Tsutsumi N (2008) Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell 20: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Tsutsumi N (2002) A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc Natl Acad Sci USA 99: 5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Tsutsumi N (2016) Mitochondrial and peroxisomal division. In Rose, DJ, ed, Molecular Cell Biology of the Growth and Differentiation of Plant Cells 15, CRC Press, Boca Raton, FL, pp. 51–65
- Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N (2004b) Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA 101: 7805–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura SI, Kurisu R, Sugaya H, Kadoya N, Tsutsumi N (2017) Cold treatment induces transient mitochondrial fragmentation in Arabidopsis thaliana in a way that requires DRP3A but not ELM1 or an ELM1-like homologue, ELM2. Int J Mol Sci 18: 2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Hu J (2011) The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23: 4446–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Börner T (2000) Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr Genet 37: 304–314 [DOI] [PubMed] [Google Scholar]
- Belliard G, Vedel F, Pelletier G (1979) Mitochondrial recombination in cytoplasmic hybrids of Nicotiana tabacum by protoplast fusion. Nature 281: 401–403 [Google Scholar]
- Bendich AJ. (1993) Reaching for the ring: the study of mitochondrial genome structure. Curr Genet 24: 279–290 [DOI] [PubMed] [Google Scholar]
- Bendich AJ, Gauriloff LP (1984) Morphometric analysis of cucurbit mitochondria: the relationship between chondriome volume and DNA content. Protoplasma 119: 1–7 [Google Scholar]
- Bereiter-Hahn J. (1990) Behavior of mitochondria in the living cell. Int Rev Cytol 122: 1–63 [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Adams KL, Thomason B, Palmer JD (2003) Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424: 197–201 [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1: 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broker TR, Doermann AH (1975) Molecular and genetic recombination of bacteriophage T4. Annu Rev Genet 9: 213–244 [DOI] [PubMed] [Google Scholar]
- Bui HT, Shaw JM (2013) Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol 23: R891–R899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Giraud E, Ng S, Zhang MF, Narsai R, Whelan J (2013) How do plants make mitochondria? Planta 237: 429–439 [DOI] [PubMed] [Google Scholar]
- Chan DC. (2012) Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46: 265–287 [DOI] [PubMed] [Google Scholar]
- Cheng N, Lo YS, Ansari MI, Ho KC, Jeng ST, Lin NS, Dai H (2017) Correlation between mtDNA complexity and mtDNA replication mode in developing cotyledon mitochondria during mung bean seed germination. New Phytol 213: 751–763 [DOI] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, Lindén P, Ziolkowska A, Liebsch D, Narsai R, Szal B, Moritz T, Rouhier N, et al. (2016) Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol 172: 2132–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarna M, Kolodziejczak M, Janska H (2016) Mitochondrial proteome studies in seeds during germination. Proteomes 4: E19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Lo YS, Jane WN, Lee LW, Chiang KS (1998) Population heterogeneity of higher-plant mitochondria in structure and function. Eur J Cell Biol 75: 198–209 [DOI] [PubMed] [Google Scholar]
- Davis CC, Wurdack KJ (2004) Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science 305: 676–678 [DOI] [PubMed] [Google Scholar]
- Diboll AG, Larson DA (1966) An electron microscopic study of the mature megagametophyte in Zea mays. Am J Bot 53: 391–402 [PubMed] [Google Scholar]
- El Zawily AM, Schwarzländer M, Finkemeier I, Johnston IG, Benamar A, Cao Y, Gissot C, Meyer AJ, Wilson K, Datla R, et al. (2014) FRIENDLY regulates mitochondrial distribution, fusion, and quality control in Arabidopsis. Plant Physiol 166: 808–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Arimura S, Hirano HY, Sakamoto W, Tsutsumi N (2004) Isolation of mutants with aberrant mitochondrial morphology from Arabidopsis thaliana. Genes Genet Syst 79: 301–305 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK (2011) ER tubules mark sites of mitochondrial division. Science 334: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA. (2015) Role of mitochondrial lipids in guiding fission and fusion. J Mol Med (Berl) 93: 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes I, Stegemann S, Golczyk H, Karcher D, Bock R (2014) Horizontal genome transfer as an asexual path to the formation of new species. Nature 511: 232–235 [DOI] [PubMed] [Google Scholar]
- Fujie M, Kuroiwa H, Kawano S, Mutoh S, Kuroiwa T (1994) Behavior of organelles and their nucleoids in the shoot apical meristem during leaf development in Arabidopsis thaliana L. Planta 194: 395–405 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Mano S, Kondo M, Saito C, Ueda T, Nakazono M, Nakano A, Nishimura M, Tsutsumi N (2009) Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J 58: 388–400 [DOI] [PubMed] [Google Scholar]
- Gao H, Sage TL, Osteryoung KW (2006) FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc Natl Acad Sci USA 103: 6759–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ (2017) Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol 68: 225–252 [DOI] [PubMed] [Google Scholar]
- Gurdon C, Svab Z, Feng Y, Kumar D, Maliga P (2016) Cell-to-cell movement of mitochondria in plants. Proc Natl Acad Sci USA 113: 3395–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaipargas EA, Barton KA, Mathur N, Mathur J (2015) Mitochondrial pleomorphy in plant cells is driven by contiguous ER dynamics. Front Plant Sci 6: 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O. (2011) The conserved mobility of mitochondria during leaf senescence reflects differential regulation of the cytoskeletal components in Arabidopsis thaliana. Plant Signal Behav 6: 147–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardeström P (2007) The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant Cell Environ 30: 1523–1534 [DOI] [PubMed] [Google Scholar]
- Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR (1997) Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042 [DOI] [PubMed] [Google Scholar]
- Kreuzer KN. (2000) Recombination-dependent DNA replication in phage T4. Trends Biochem Sci 25: 165–173 [DOI] [PubMed] [Google Scholar]
- Kreuzer KN, Brister JR (2010) Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J 7: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Newton KJ (2008) Angiosperm mitochondrial genomes and mutations. Mitochondrion 8: 5–14 [DOI] [PubMed] [Google Scholar]
- Kuroiwa H, Ohta T, Kuroiwa T (1996) Studies on the development and three-dimensional reconstruction of giant mitochondria and their nuclei in egg cells of Pelargonium zonale Ait. Protoplasma 192: 235–244 [Google Scholar]
- Kuroiwa T. (2010) Mechanisms of organelle division and inheritance and their implications regarding the origin of eukaryotic cells. Proc Jpn Acad Ser B Phys Biol Sci 86: 455–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T, Fujie M, Kuroiwa H (1992) Studies on the behavior of mitochondrial DNA: synthesis of mitochondrial DNA occurs actively in a specific region just above the quiescent center in the root meristem of Pelargonium zonale. J Cell Sci 101: 483 [Google Scholar]
- Kuroiwa T, Misumi O, Nishida K, Yagisawa F, Yoshida Y, Fujiwara T, Kuroiwa H (2008) Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int Rev Cell Mol Biol 271: 97–152 [DOI] [PubMed] [Google Scholar]
- Law SR, Narsai R, Whelan J (2014) Mitochondrial biogenesis in plants during seed germination. Mitochondrion 19: 214–221 [DOI] [PubMed] [Google Scholar]
- Lewis MR, Lewis WH (1914) Mitochondria in tissue culture. Science 39: 330–333 [DOI] [PubMed] [Google Scholar]
- Li F, Chung T, Vierstra RD (2014) AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2014) Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 10: 1466–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Shirihai OS (2013) Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab 17: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC. (2006) The mitochondrial compartment. J Exp Bot 57: 1225–1243 [DOI] [PubMed] [Google Scholar]
- Logan DC. (2010) Mitochondrial fusion, division and positioning in plants. Biochem Soc Trans 38: 789–795 [DOI] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Logan DC, Millar AH, Sweetlove LJ, Hill SA, Leaver CJ (2001) Mitochondrial biogenesis during germination in maize embryos. Plant Physiol 125: 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DC, Scott I, Tobin AK (2003) The genetic control of plant mitochondrial morphology and dynamics. Plant J 36: 500–509 [DOI] [PubMed] [Google Scholar]
- Logan DC, Scott I, Tobin AK (2004) ADL2a, like ADL2b, is involved in the control of higher plant mitochondrial morphology. J Exp Bot 55: 783–785 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N (2010) Recombination and the maintenance of plant organelle genome stability. New Phytol 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Hamamura Y, Higashiyama T, Arimura S, Sodmergen, Tsutsumi N, Sakamoto W (2008) Mitochondrial dynamics in plant male gametophyte visualized by fluorescent live imaging. Plant Cell Physiol 49: 1074–1083 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Tang LY, Zhang L, Yamada H, Twell D, Sakamoto W (2011) A conserved, Mg2+-dependent exonuclease degrades organelle DNA during Arabidopsis pollen development. Plant Cell 23: 1608–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Small ID, Day DA, Whelan J (2008) Mitochondrial biogenesis and function in Arabidopsis. The Arabidopsis Book 6: e0111, doi/10.1199/tab.0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC (2016) Metabolic regulation of mitochondrial dynamics. J Cell Biol 212: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G. (1998) Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet 32: 379–413 [DOI] [PubMed] [Google Scholar]
- Mower JP, Stefanović S, Young GJ, Palmer JD (2004) Plant genetics: gene transfer from parasitic to host plants. Nature 432: 165–166 [DOI] [PubMed] [Google Scholar]
- Mueller SJ, Reski R (2015) Mitochondrial dynamics and the ER: the plant perspective. Front Cell Dev Biol 3: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka N, Yamashita A, Kurisu R, Watari Y, Ishizuna F, Tsutsumi N, Ishizaki K, Kohchi T, Arimura SI (2017) DRP3 and ELM1 are required for mitochondrial fission in the liverwort Marchantia polymorpha. Sci Rep 7: 4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N. (2010) Mechanisms for independent cytoplasmic inheritance of mitochondria and plastids in angiosperms. J Plant Res 123: 193–199 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, Kuroiwa T (2003) Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA 100: 2146–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Yagisawa F, Kuroiwa H, Yoshida Y, Kuroiwa T (2007) WD40 protein Mda1 is purified with Dnm1 and forms a dividing ring for mitochondria before Dnm1 in Cyanidioschyzon merolae. Proc Natl Acad Sci USA 104: 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ (1996) Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell 8: 447–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ (2015) DNA maintenance in plastids and mitochondria of plants. Front Plant Sci 6: 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Pyke KA (2014) Division and dynamic morphology of plastids. Annu Rev Plant Biol 65: 443–472 [DOI] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT (2013) Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem 288: 27584–27593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep 12: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Shields CR (1984) Tripartite structure of the Brassica campestris mitochondrial genome. Nature 307: 437–440 [Google Scholar]
- Pan R, Hu J (2011) The conserved fission complex on peroxisomes and mitochondria. Plant Signal Behav 6: 870–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Jones AD, Hu J (2014a) Cardiolipin-mediated mitochondrial dynamics and stress response in Arabidopsis. Plant Cell 26: 391–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Kaur N, Hu J (2014b) The Arabidopsis mitochondrial membrane-bound ubiquitin protease UBP27 contributes to mitochondrial morphogenesis. Plant J 78: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Paszkiewicz G, Gualberto JM, Benamar A, Macherel D, Logan DC (2017) Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell 29: 109–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T, Cincu E, Fuchs J, Zoschke R, Liere K, Börner T (2010) Fewer genes than organelles: extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J 64: 948–959 [DOI] [PubMed] [Google Scholar]
- Ramonell KM, Kuang A, Porterfield DM, Crispi ML, Xiao Y, McClure G, Musgrave ME (2001) Influence of atmospheric oxygen on leaf structure and starch deposition in Arabidopsis thaliana. Plant Cell Environ 24: 419–428 [DOI] [PubMed] [Google Scholar]
- Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, et al. (2013) Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342: 1468–1473 [DOI] [PubMed] [Google Scholar]
- Rose RJ, McCurdy DW (2017) New beginnings: mitochondrial renewal by massive mitochondrial fusion. Trends Plant Sci 22: 641–643 [DOI] [PubMed] [Google Scholar]
- Ruberti C, Barizza E, Bodner M, La Rocca N, De Michele R, Carimi F, Lo Schiavo F, Zottini M (2014) Mitochondria change dynamics and morphology during grapevine leaf senescence. PLoS ONE 9: e102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Miyagishima SY, Jarvis P (2008) Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. The Arabidopsis Book 6: e0110, doi/10.1199/tab.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Takami T (2014) Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J Exp Bot 65: 3835–3843 [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, García LE, Wohlfeiler J, Ceriotti LF (2017) Unparalleled replacement of native mitochondrial genes by foreign homologs in a holoparasitic plant. New Phytol 214: 376–387 [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Zubko MK, Palmer JD (2015) Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol 206: 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Nemoto Y, Kawano S, Nagata T, Hirokawa H, Kuroiwa T (1993) Organization of heterogeneous mitochondrial DNA molecules in mitochondrial nuclei of cultured tobacco cells. Protoplasma 175: 112–120 [Google Scholar]
- Schwarzländer M, Logan DC, Johnston IG, Jones NS, Meyer AJ, Fricker MD, Sweetlove LJ (2012) Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell 24: 1188–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I, Tobin AK, Logan DC (2006) BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J Exp Bot 57: 1275–1280 [DOI] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Coronado MJ, Staehelin LA (2008) The mitochondrial cycle of Arabidopsis shoot apical meristem and leaf primordium meristematic cells is defined by a perinuclear tentaculate/cage-like mitochondrion. Plant Physiol 148: 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol 147: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, McCurdy DW, Rose RJ (2005) Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J 44: 744–755 [DOI] [PubMed] [Google Scholar]
- Sloan DB. (2013) One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol 200: 978–985 [DOI] [PubMed] [Google Scholar]
- Sugiura A, McLelland GL, Fon EA, McBride HM (2014) A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J 33: 2142–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kawano S, Sakai A, Fujie M, Kuroiwa H, Nakamura H, Kuroiwa T (1992) Preferential mitochondrial and plastid DNA synthesis before multiple cell divisions in Nicotiana tabacum. J Cell Sci 103: 831–837 [Google Scholar]
- Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Takano H, Sakai A, Kawano S, Kuroiwa T (2000) A putative mitochondrial ftsZ gene is present in the unicellular primitive red alga Cyanidioschyzon merolae. Mol Gen Genet 264: 452–460 [DOI] [PubMed] [Google Scholar]
- Takanashi H, Arimura S, Sakamoto W, Tsutsumi N (2006) Different amounts of DNA in each mitochondrion in rice root. Genes Genet Syst 81: 215–218 [DOI] [PubMed] [Google Scholar]
- Takanashi H, Ohnishi T, Mogi M, Okamoto T, Arimura S, Tsutsumi N (2010) Studies of mitochondrial morphology and DNA amount in the rice egg cell. Curr Genet 56: 33–41 [DOI] [PubMed] [Google Scholar]
- Taylor ZN, Rice DW, Palmer JD (2015) The complete moss mitochondrial genome in the angiosperm Amborella is a chimera derived from two moss whole-genome transfers. PLoS ONE 10: e0137532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gestel K, Verbelen JP (2002) Giant mitochondria are a response to low oxygen pressure in cells of tobacco (Nicotiana tabacum L.). J Exp Bot 53: 1215–1218 [DOI] [PubMed] [Google Scholar]
- Wang DY, Zhang Q, Liu Y, Lin ZF, Zhang SX, Sun MX, Sodmergen (2010) The levels of male gametic mitochondrial DNA are highly regulated in angiosperms with regard to mitochondrial inheritance. Plant Cell 22: 2402–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu P, Zhang Q, Zhu J, Chen T, Arimura S, Tsutsumi N, Lin J (2012) Phosphorylation and ubiquitination of dynamin-related proteins (AtDRP3A/3B) synergically regulate mitochondrial proliferation during mitosis. Plant J 72: 43–56 [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111: E4859–E4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ (2015) Redox homeostasis and mitochondrial dynamics. Cell Metab 22: 207–218 [DOI] [PubMed] [Google Scholar]
- Yamaoka S, Leaver CJ (2008) EMB2473/MIRO1, an Arabidopsis Miro GTPase, is required for embryogenesis and influences mitochondrial morphology in pollen. Plant Cell 20: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Nakajima M, Fujimoto M, Tsutsumi N (2011) MIRO1 influences the morphology and intracellular distribution of mitochondria during embryonic cell division in Arabidopsis. Plant Cell Rep 30: 239–244 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Fujimoto M, Katayama K, Tsutsumi N, Arimura S (2016a) Mitochondrial outer membrane forms bridge between two mitochondria in Arabidopsis thaliana. Plant Signal Behav 11: e1167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Fujimoto M, Katayama K, Yamaoka S, Tsutsumi N, Arimura S (2016b) Formation of mitochondrial outer membrane derived protrusions and vesicles in Arabidopsis thaliana. PLoS ONE 11: e0146717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Hirooka S, Fujiwara T, Ohnuma M, Yoshida M, Misumi O, Kawano S, Kuroiwa T (2009) The bacterial ZapA-like protein ZED is required for mitochondrial division. Curr Biol 19: 1491–1497 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, Nanamiya H, Kawamura F, Kuroiwa T (2006) Isolated chloroplast division machinery can actively constrict after stretching. Science 313: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Zhang X, Hu J (2009) Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J 57: 146–159 [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlén P, Tomilin N, Shupliakov O, Lendahl U, Nistér M (2011) Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J 30: 2762–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]