Abstract
Recent studies advance understanding of the mechanisms, spatial control, and regulation of chloroplast division, but many questions remain.
Chloroplasts descended from a free-living cyanobacterium acquired through endosymbiosis roughly one billion years ago. Three major groups comprising the Archaeplastida, photosynthetic eukaryotes bearing primary plastids, arose subsequently: the glaucophytes, red algae (red lineage), and Viridiplantae, also called Chloroplastida, encompassing green algae and land plants (green lineage; Keeling, 2010; Zimorski et al., 2014). Beyond carrying out photosynthesis, plastids perform many other vital functions, such as fatty acid and amino acid synthesis, and are therefore essential organelles (Pyke, 2009). Plastid division increases chloroplast populations during leaf development, crucial for photosynthetic capacity (Leech and Baker, 1983), and ensures that plastids are faithfully inherited during cytokinesis.
Similar to their free-living ancestors, plastids are propagated through division of preexisting organelles. This process is powered by a macromolecular machine with ring-shaped contractile complexes on both the inner and outer envelope membranes (for review, see Miyagishima et al., 2011; Falconet, 2012; Yoshida et al., 2012; Osteryoung and Pyke, 2014). The division machinery is a mosaic of components of both endosymbiotic and host origin that must cooperate to divide the organelle (Fig. 1B). The stromal components are largely endosymbiont derived, whereas the cytosolic components are strictly eukaryotic. In land plants and many algae, all the plastid division proteins are encoded in the nucleus, but some algae retain a few that are plastid encoded (Onuma et al., 2017). Here, we highlight developments in the division of chloroplasts, particularly in land plants, with an emphasis on findings published since the previous Plant Physiology Update on this topic (Miyagishima, 2011). We refer readers to other reviews for information on the division of other plastid types, including secondary plastids, and for a more evolutionary perspective (for review, see Miyagishima, 2011; Miyagishima et al., 2011, 2014; Pyke, 2016).
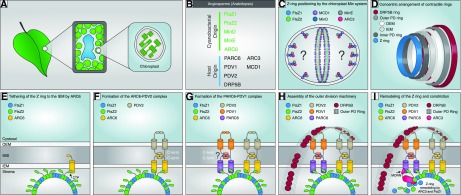
Figure 1.
Working model of the positioning, assembly, and dynamics of the chloroplast division machinery in angiosperms based primarily on studies in Arabidopsis but informed by studies in C. merolae and other organisms. A, Diagram showing chloroplasts within a leaf mesophyll cell. B, Cyanobacterial (endosymbiotic) or host (eukaryotic) origin of chloroplast division components in angiosperms. C, FtsZ1 and FtsZ2 self-assemble as dynamic heteropolymers, possibly with mixed stoichiometry (Olson et al., 2010; Chen et al., 2017); protofilaments may possibly associate laterally to form the Z ring at the division site (Lutkenhaus and Du, 2017). Positioning of the Z ring is confined to the midzone by the chloroplast Min system, comprising ARC3, MCD1, MinD, and MinE, which inhibits Z-ring assembly at nondivision sites. ARC3 acts downstream of MinD and MinE as the direct inhibitor of Z-ring assembly (Zhang et al., 2013). MCD1, a transmembrane IEM protein, recruits MinD to the membrane (Nakanishi et al., 2009), where MinE is also colocalized (Miyagishima et al., 2011). ARC3 interacts with both MinD and MinE (Maple et al., 2007). Thus, we hypothesize that ARC3 forms a complex with MinD and MinE that is tethered to the membrane by MCD1. The exact localization pattern of ARC3 and the inhibitory mechanisms of ARC3 on Z-ring assembly are unclear (indicated by the question marks). The chloroplast Min-system components also localize partly to the division site (Shimada et al., 2004; Nakanishi et al., 2009; Miyagishima et al., 2011), where they may promote Z-ring remodeling during division (Johnson et al., 2015). D, Overview of the four contractile ring structures formed across the two envelope membranes. The composition of the inner PD ring is unknown and is not shown in further panels. The outer PD ring is synthesized by PDR1 (not shown) in the red alga C. merolae (Yoshida et al., 2010). The order of assembly based on studies in C. merolae is Z ring, inner PD ring, outer PD ring, DRP5B ring (Miyagishima et al., 2001, 2003). E to I, Stepwise assembly and dynamics of the division complex at the middle of the chloroplast. E, Tethering of the Z ring to the IEM is achieved mainly through interaction of the conserved FtsZ2 C-terminal peptide (CTP) with ARC6 (Maple et al., 2005), which probably stabilizes the Z ring and facilitates its assembly (Vitha et al., 2003; Johnson et al., 2013). F, ARC6 recruits PDV2 to the division site through direct interaction between their C-terminal IMS regions (Glynn et al., 2008). Dimerization of the cytosolic regions of two PDV2 molecules induces dimerization of two ARC6 molecules (Wang et al., 2017). G, PARC6 acts downstream of ARC6 to localize PDV1 to the division site through direct interaction between their C-terminal IMS regions (Glynn et al., 2009; Zhang et al., 2016). Based on ARC6-PDV2 studies (Wang et al., 2017), PDV1 dimerization might also promote PARC6 dimerization (indicated by question mark). H, In C. merolae, PDR1 (not shown) is recruited from the cytosol to construct the outer PD ring, composed of polyglucan fibrils (Yoshida et al., 2010). In Arabidopsis, PDV1 and PDV2 function together to recruit DRP5B from the cytosol (Miyagishima et al., 2006) to form the DRP5B ring (Gao et al., 2003; Miyagishima et al., 2003; Holtsmark et al., 2013). Coordination of the stromal Z ring and the cytosolic DRP5B ring is established through the ARC6-PDV2 and PARC6-PDV1 complexes. Whether the outer PD ring interacts with the PDV proteins is unclear. I, Remodeling of the Z ring and constriction. We speculate that PARC6 recruits ARC3 to the division site via interaction with the ARC3 MORN domain, enabling ARC3 to interact with FtsZ in the Z ring. The latter interaction may be facilitated by interaction of PARC6 with the FtsZ2 CTP (Zhang et al., 2016). As an FtsZ assembly inhibitor, ARC3 activation at the division site may promote Z-ring remodeling. MinD, MinE, and MCD1 also localize partly to the division site (Nakanishi et al., 2009; Miyagishima et al., 2011; not shown in I). Dynamic remodeling of the Z ring probably also depends on FtsZ1 (TerBush and Osteryoung, 2012; Yoshida et al., 2016; Terbush et al., 2018). OEM, Outer envelope membrane; IMS, intermembrane space; IEM, inner envelope membrane; MORN, membrane occupation and recognition nexus domain of ARC3; PD ring, plastid-dividing ring.
FUNCTION, ASSEMBLY, AND DYNAMICS OF FTSZ
A central and nearly ubiquitous component of the chloroplast division machinery is FtsZ, a tubulin-like cytoskeletal GTPase that descended from the cyanobacterial ancestor of chloroplasts, where it functioned in cell division (Erickson et al., 2010; Miyagishima et al., 2011; TerBush et al., 2013; Osteryoung and Pyke, 2014; Haeusser and Margolin, 2016). In both bacteria and chloroplasts, FtsZ assembles into a contractile “Z ring” inside the cell or organelle that defines the division site (Fig. 2; Box 1; Friedman and Nunnari, 2014; Addinall et al., 1996; Beech et al., 2000; Gilson et al., 2003; Bilsson-Filho et al., 2017; Osawa et al., 2009; Rothfield et al., 2005; Yang et al., 2017; Wagstaff et al., 2017; Strepp et al., 1997; Nishida et al., 2003; Lutkenhaus et al., 1980; Wang et al., 2017). Purified FtsZ undergoes GTP-dependent self-assembly into single-stranded polymers called protofilaments. Because the GTPase active site is formed within the subunit interface by the interaction of two monomers, polymerization catalyzes GTP hydrolysis. Hydrolysis destabilizes the interface, leading to protofilament fragmentation and subunit dissociation (Mukherjee and Lutkenhaus, 1998; Scheffers et al., 2002; Redick et al., 2005; Huecas et al., 2007; Chen and Erickson, 2009; Erickson et al., 2010). Released subunits can exchange nucleotides and recycle back into protofilaments. This dynamic GTPase-dependent turnover was essential for constriction of Z rings reconstituted on tubular liposomes (Osawa et al., 2008; Osawa and Erickson, 2011) and is likely required for Z-ring constriction in vivo.
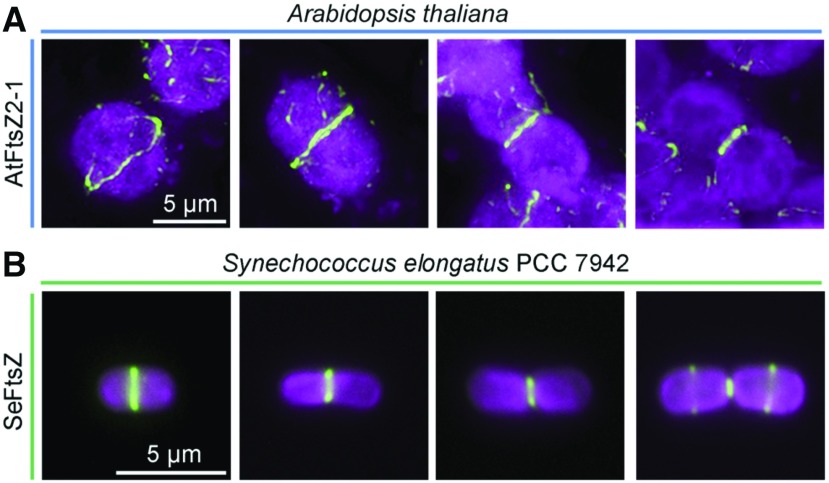
Figure 2.
Immunofluorescence localization of the contractile Z ring in chloroplasts and cyanobacterial cells during division. A, Arabidopsis FtsZ2-1 (AtFtsZ2-1) detected with an anti-AtFtsZ2-1 antibody (McAndrew et al., 2001) in mesophyll cells of a fully expanded leaf obtained from a 3-week-old plant (Col-0). B, FtsZ in the cyanobacterium Synechococcus elongatus PCC 7942 (SeFtsZ) detected with an anti-Anabaena FtsZ antibody (Agrisera). Green, FtsZ; magenta, chlorophyll fluorescence. Bars, 5 μm.
Because most bacteria have only a single FtsZ gene, bacterial Z rings are composed of homopolymers. In contrast, the majority of photosynthetic eukaryotes appear to encode two plastid-targeted, stroma-localized paralogs of FtsZ (Osteryoung and Vierling 1995; McAndrew et al., 2001; Leger et al., 2015; Box 1). These are called FtsZ1 and FtsZ2 in the green lineage, where they appear to be universally conserved, and FtsZA and FtsZB in red algae (Miyagishima et al., 2004; TerBush et al., 2013). FtsZ1 and FtsZ2 colocalize to Z rings in vivo (McAndrew et al., 2001; Vitha et al., 2001), are both required for normal chloroplast division in Arabidopsis (Arabidopsis thaliana; Osteryoung et al., 1998; Yoder et al., 2007; Schmitz et al., 2009), and coassemble in heteropolymers (Olson et al., 2010; TerBush and Osteryoung, 2012; Yoshida et al., 2016; Fig. 1). FtsZ2 and FtsZA are structurally more similar to bacterial FtsZs, in that they both retain a conserved C-terminal peptide (CTP), which in bacteria interacts with membrane proteins to anchor Z rings to the plasma membrane (Ma and Margolin, 1999; Vaughan et al., 2004; Margolin, 2005; Haeusser and Margolin, 2016). Similarly, the Arabidopsis FtsZ2 CTP interacts with two proteins in the chloroplast inner envelope membrane (IEM; Fig. 1, E–G; described below). In contrast, FtsZ1 and FtsZB lack the CTP, and no interaction between FtsZ1 and any membrane protein has been detected, suggesting its presence in the Z ring is a consequence of coassembly with FtsZ2 (Maple et al., 2005; Glynn et al., 2008; Zhang et al., 2016).
Recently, FtsZA and FtsZB from the red alga Galdieria sulphuraria were also shown to copolymerize in vitro (Chen et al., 2017), indicating that FtsZ heteropolymerization is a conserved feature and may be the physiologically relevant state. Studies of FtsZ dynamics in heterologous yeast systems are beginning to provide insight into the functional significance of heteropolymerization. These systems lack FtsZ and any native assembly regulators, allowing the intrinsic assembly and dynamic properties of FtsZ proteins to be investigated (Srinivasan et al., 2008; TerBush et al., 2016; Yoshida et al., 2016). When fluorescent fusions of Arabidopsis FtsZ2 (AtFtsZ2) and AtFtsZ1 were expressed separately in the fission yeast Schizosaccharomyces pombe, each assembled homopolymeric filamentous structures (homofilaments) in the cytosol (TerBush and Osteryoung, 2012), as do purified FtsZ2 and FtsZ1 in vitro (El-Kafafi et al., 2005; Lohse et al., 2006; Olson et al., 2010; Smith et al., 2010). Fluorescence recovery after photobleaching (FRAP) experiments showed that AtFtsZ2 homofilaments exhibited a much lower degree of subunit turnover than AtFtsZ1 homofilaments. When coexpressed, AtFtsZ2 and AtFtsZ1 colocalized in heterofilaments, which were considerably more dynamic than AtFtsZ2 homofilaments. Similar results were obtained for G. sulphuraria FtsZA (GsFtsZA) and GsFtsZB expressed in S. pombe; GsFtsZA homofilaments were less dynamic than GsFtsZB homofilaments, and GsFtsZA/GsFtsZB heterofilaments were more dynamic than GsFtsZA homofilaments (Terbush et al., 2017). In a related study, Yoshida et al. (2016) reconstituted Z rings in the yeast Pichia pastoris by fusing a membrane-tethering sequence (MTS; Osawa et al., 2008) to the C terminus of AtFtsZ2, enabling it to bind directly to the plasma membrane in the yeast cells. MTS-tagged AtFtsZ2 assembled into a well-defined ring, but MTS-tagged AtFtsZ1 did not. When expressed together, MTS-tagged AtFtsZ2 and AtFtsZ1 (without the MTS) coassembled in the membrane-tethered ring, and FRAP showed these rings were more dynamic than AtFtZ2 rings. Further, both types of rings could be induced to constrict, and coassembled rings constricted more rapidly. These findings, along with studies of Arabidopsis ftsZ mutants (Yoder et al., 2007; McAndrew et al., 2008; Schmitz et al., 2009), suggest that the more bacterial-like FtsZ2 and FtsZA proteins establish the structural framework and impart stability to chloroplast Z rings, while FtsZ1 and FtsZB enhance Z-ring dynamics through copolymerization.
The biochemical mechanism by which FtsZ1 and FtsZB increase protofilament turnover is not yet clear, but one possibility is that they introduce lower-affinity FtsZ-FtsZ interfaces into heteropolymers. This is suggested partly by experiments showing that coassembled GsFtsZA/GsFtsZB protofilaments were much more dynamic than GsFtsZA protofilaments in vitro (Chen et al., 2017). Additionally, mutation of a conserved residue required for GTP hydrolysis drastically reduced turnover of AtFtsZ2 and GsFtsZA homofilaments in S. pombe, as expected based on the GTPase-dependent turnover of bacterial FtsZ described above. Surprisingly, however, the equivalent mutations in AtFtsZ1 and GsFtsZB reduced but did not abolish turnover of these homofilaments, suggesting their dynamic behavior is not solely dependent on GTPase activity (TerBush and Osteryoung, 2012; Terbush et al., 2017). Collectively, these studies provide evidence that the duplication, functional divergence, and coassembly of plastid FtsZs imparted a new mechanism for facilitating Z-ring dynamics in red- and green-lineage chloroplasts.
Interestingly, two FtsZ types are also widespread in organisms bearing secondary plastids or that retain mitochondrial FtsZ (Miyagishima et al., 2004; Leger et al., 2015; Box 1). Additionally, a third FtsZ family, FtsZ3, is found in a subset of photosynthetic eukaryotes, where its occurrence may be correlated with the retention of a chloroplast peptidoglycan wall (Box 2; Cassier-Chauvat and Chauvat, 2014; Grosche and Rensing, 2017; Hirano et al., 2016; Kasten et al., 1997; Katayama et al., 2003; Homi et al., 2009; Takano and Takechi, 2010; Martin et al., 2009b, 2009b; Machida et al., 2006; Lin et al., 2017).
SPATIAL REGULATION OF DIVISION: THE CHLOROPLAST MIN SYSTEM
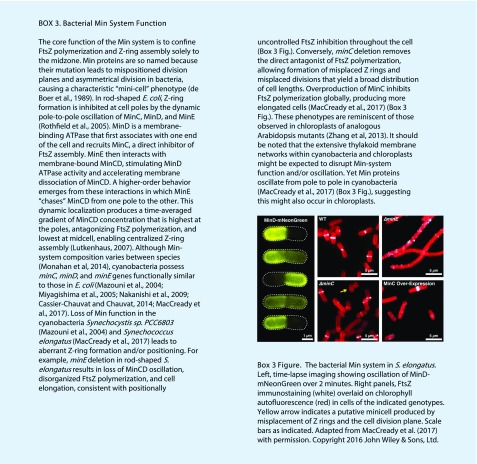
The Z ring is the first structure to assemble at the division site (Miyagishima et al., 2001; Fig. 1), and its placement likely establishes the placement of downstream components. In bacteria, Z-ring positioning is controlled by a negative regulatory system called the Min system that prevents self-assembly of Z rings everywhere but at the division site (Box 3; Miyagishima et al., 2005; Monahan et al., 2014; de Boer et al., 1989; reviewed in Lutkenhaus, 2007; Rowlett and Margolin, 2013). In E. coli and cyanobacteria (MacCready et al., 2017), the Min system concentrates MinC, the direct inhibitor of FtsZ polymerization, near the cell poles through a remarkable oscillatory mechanism driven by MinD and MinE, which function as regulators of MinC localization (Box 3). Homologs of cyanobacterial MinD and MinE acquired through endosymbiosis have been retained in the green lineage and localize to the stroma, where they play roles in the spatial regulation of chloroplast division and Z-ring placement analogous to those in bacteria (Colletti et al., 2000; Itoh et al., 2001; Vitha et al., 2003; Fujiwara et al., 2004; Aldridge and Møller, 2005; Glynn et al., 2007; Fujiwara et al., 2008). However, in many species, MinC has been lost and instead replaced by the stromal protein ARC3 (Accumulation and Replication of Chloroplasts3; Shimada et al., 2004; Maple et al., 2007). A MinC-like role for ARC3 was initially suggested by the phenotypes of Arabidopsis arc3 mutants (Pyke and Leech, 1992), which displayed multiple chloroplast constrictions, multiple Z rings, and mispositioning of chloroplast division sites, leading to heterogeneity in chloroplast size and number (Glynn et al., 2007; Maple et al., 2007; Fig. 3), reminiscent of bacterial minicell phenotypes (de Boer et al., 1990; Yu and Margolin, 1999; Box 3). ARC3 interacts directly with AtFtsZ1 and AtFtsZ2 and inhibits their assembly in S. pombe (Maple et al., 2007; TerBush and Osteryoung, 2012; Zhang et al., 2013). Its overexpression in Arabidopsis produces dose-dependent chloroplast enlargement and fragmented FtsZ filaments (Maple et al., 2007; Zhang et al., 2013), resembling MinC overexpression in E. coli and cyanobacteria (de Boer et al., 1990; MacCready et al., 2017; Box 3). Finally, the large-chloroplast phenotypes in Arabidopsis MinD overexpressors and minE mutants (Colletti et al., 2000; Glynn et al., 2007; Fig. 3) were completely suppressed in the absence of ARC3, consistent with a role for these proteins as ARC3 regulators (Zhang et al., 2013). These findings established ARC3 as a functional replacement for MinC and the proximal inhibitor of Z-ring assembly in Arabidopsis chloroplasts. However, ARC3 is not found in all green-lineage organisms; some, including Physcomitrella patens bear sequences with partial similarity to MinC (Yang et al., 2008; Osteryoung and Pyke, 2014), whose functions have not been tested.
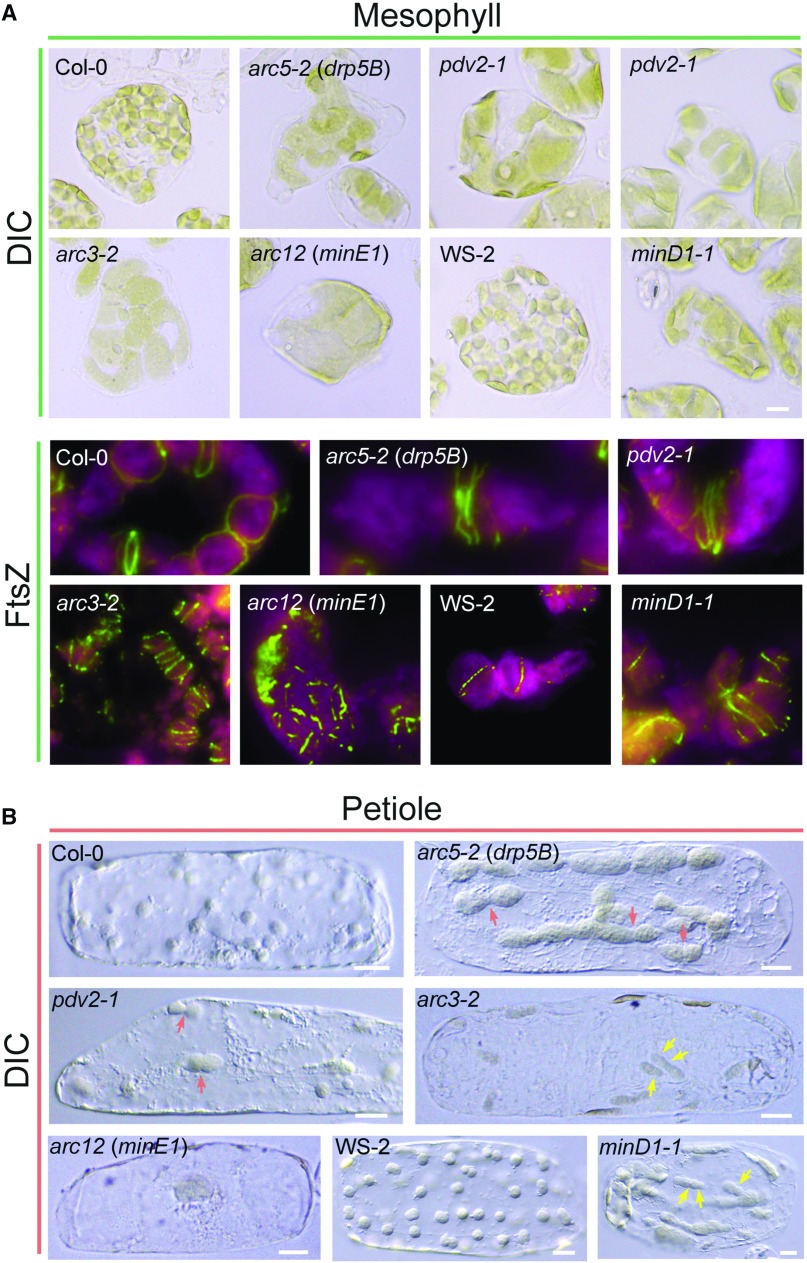
Figure 3.
Chloroplast morphology and Z-ring localization patterns in Arabidopsis wild-type plants and various chloroplast division mutants. Although phenotypes vary, most mutants display reduced numbers of enlarged chloroplasts (Osteryoung and Pyke, 2014). A, Chloroplast morphology observed by differential interference contrast (DIC) microscopy (top) and Z-ring localization detected by immunofluorescence staining (bottom) in mesophyll cells. The minD1-1 mutant (Zhang et al., 2013) is in the WS-2 background; all others are in Col-0. Images of FtsZ localization were adapted from Miyagishima et al. (2006) (Col-0, pdv2-1 and arc5-2) and Zhang et al. (2013) (arc12 and minD1-1) with permission; copyright © 2006 and 2013 by the American Society of Plant Biologists. Green, FtsZ; magenta/red, chlorophyll fluorescence. B, Chloroplast morphology in cells isolated from leaf petioles. Petiole cells contain fewer chloroplasts than mesophyll cells, and the morphology phenotypes are more evident. Red arrows indicate dumbbell-shaped chloroplasts in pdv2-1 and arc5-2 mutants. Yellow arrows denote asymmetric division planes in arc3-2 and minD1-1 mutants. Bars, 10 μm.
ARC3 and MinC share little sequence similarity and may inhibit FtsZ assembly by different mechanisms. MinC acts by promoting protofilament breakage at GDP-bound subunit interfaces, competing with Z-ring anchoring proteins for FtsZ binding and inhibiting protofilament bundling (Shen and Lutkenhaus, 2009, 2010; Lutkenhaus and Du, 2017). Unlike MinC, ARC3 bears an FtsZ-like region, though it lacks conserved residues required for GTP binding and hydrolysis (Shimada et al., 2004). This region may interact with FtsZ (Maple et al., 2007), perhaps sequestering FtsZ subunits and thereby antagonizing polymerization. Like MinC, ARC3 may also have multiple assembly inhibitory activities.
Although it is unknown whether MinD, MinE, and ARC3 oscillate in chloroplasts and the full localization of ARC3 is not entirely clear, the MinC-like role of ARC3 in preventing Z-ring formation at nondivision sites suggests that ARC3 and MinD must localize at least partly to membrane regions away from the midplastid. Such localization is suggested by immunostaining showing that MinD and MinE localize partly to punctate structures dispersed over the envelope membrane (Nakanishi et al., 2009; Miyagishima et al., 2011; Fig. 1C). However, the chloroplast Min system in Arabidopsis differs from bacterial systems in several other respects. In bacteria, MinD binds directly to the membrane toward polar zones, where it recruits MinC (Lutkenhaus, 2007). In Arabidopsis, MinD association with the membrane requires the green-lineage-specific transmembrane protein MCD1 (Nakanishi et al., 2009; Fig. 1C). ARC3 might be recruited to the membrane by an MCD1-MinD complex, but this is not yet known. Additionally, MinC only interacts with MinD, whereas ARC3 interacts with both MinD and MinE (Maple et al., 2007), which colocalize in vivo (Miyagishima et al., 2011). Defining the complex interactions among ARC3, MinD, MinE, MCD1, and FtsZ will be important for understanding how the chloroplast Min system spatially regulates Z-ring formation.
ARC3 also colocalizes with FtsZ to the midplastid (Shimada et al., 2004), likely through interaction with the IEM transmembrane protein PARALOG OF ARC6 (PARC6; also called CDP1; Fig. 1, C and I). Chloroplasts in Arabidopsis parc6 mutants displayed multiple and misplaced constrictions, multiple Z rings (though less well defined than in arc3 and minD mutants; Fig. 3) and excessively long FtsZ filaments. Further, PARC6 overexpression produced enlarged chloroplasts with small FtsZ fragments, similar to ARC3 overexpression. These phenotypes implicated PARC6 as an additional negative regulator of FtsZ assembly and Z-ring positioning factor (Glynn et al., 2009; Zhang et al., 2009). The stromal region of PARC6 interacts with both FtsZ2 and ARC3 (Zhang et al., 2016; Fig. 1I), and in vivo FRAP data suggest ARC3 may accelerate Z-ring remodeling (Johnson et al., 2015). PARC6-ARC3 interaction is mediated by a C-terminal region of ARC3 called the Membrane Occupation and Recognition Nexus (MORN) domain (Glynn et al., 2009; Zhang et al., 2016; Fig. 1I). However, the MORN domain also inhibits ARC3 interaction with FtsZ (Maple et al., 2007; Zhang et al., 2013). These and related findings suggested a model in which PARC6 may bring ARC3 and FtsZ into close proximity at the division site, where PARC6-ARC3 interaction would sequester the MORN domain, enabling ARC3 to interact with FtsZ (Zhang et al., 2016). The negative effect of ARC3 on FtsZ assembly would facilitate Z-ring remodeling during constriction. This effect would presumably be less pronounced than at non-division sites, where Z-ring formation is fully inhibited (Fig. 1C). MinD, MinE, and MCD1 may also contribute to ARC3-mediated Z-ring remodeling because they also localize partly to the midplastid (Nakanishi et al., 2009; Miyagishima et al., 2011; Fig. 1C).
PARC6 has also been detected near plastid poles, which may indicate its retention at newly formed poles immediately following division (Glynn et al., 2009). If ARC3 is also retained or concentrated at these positions following division, this could potentially prevent premature Z-ring assembly and misplacement prior to reestablishment of a new division site (Osteryoung and Pyke, 2014; Zhang et al., 2016). These ideas are speculative but suggest avenues for future research.
OUTER DIVISION COMPONENTS
The outer chloroplast division machinery comprises two key contractile rings, the DYNAMIN-RELATED PROTEIN5B (DRP5B) ring, and the outer plastid-dividing (PD) ring (Fig. 1, H and I). DRP5B, also called ACCUMULATION AND REPLICATION OF CHLOROPLASTS5 (ARC5; Gao et al., 2003), is a plant-specific subfamily of the dynamin GTPases (Gao et al., 2003; Miyagishima et al., 2003), which mediate constriction of many organelle types (Purkanti and Thattai, 2015). Arabidopsis arc5 mutants have fewer chloroplasts than wild type that display an enlarged dumbbell shape, suggesting DRP5B is required for sustaining and/or completing constriction (Pyke and Leech, 1994). DRP5B is recruited from cytosolic patches to the external surface of the chloroplast by the OEM proteins PDV1 and PDV2 (Gao et al., 2003; Miyagishima et al., 2006; Fig. 1H), which may regulate its GTPase activity (Holtsmark et al., 2013). DRP5B also participates in the division of peroxisomes and mitochondria, indicating possible cross-talk in the replication of these organelles (Zhang and Hu, 2010; Aung and Hu, 2012; Kao et al., 2018; Arimura, 2018).
The outer PD ring (Fig. 1, D, H, and I) is an electron-dense contractile structure observed in algae and land plants and characterized most extensively in the unicellular red alga Cyanidioschyzon merolae (Kuroiwa et al., 2008; Miyagishima et al., 2011; Yoshida et al., 2012), where it is composed primarily of fine polyglucan fibrils. These are probably synthesized by Plastid-Dividing Ring1 (PDR1), a putative glycosyltransferase that relocalizes from the cytosol to a ring at the chloroplast-division site and disperses following division (Yoshida et al., 2010). DRP5B is recruited to the chloroplast after appearance of the outer PD ring and was speculated to provide the motive force for sliding of PD-ring filaments during constriction (Yoshida et al., 2006; Yoshida et al., 2010). To date, neither the functions of PDR1 homologs nor outer PD-ring composition have been investigated in other organisms.
COORDINATION OF DIVISION COMPLEXES ACROSS THE ENVELOPE MEMBRANES
Chloroplast division requires the coordinated formation and constriction of the stromal and cytosolic contractile machineries across the two envelope membranes. The stromal machinery appears to include an additional contractile structure termed the inner PD ring (Fig. 1D) of unknown composition (Kuroiwa et al., 2002, 2008; Hashimoto, 2005). At present, no data exist on how coordination of the PD rings is achieved, but in Arabidopsis, coordination between the Z ring and DRP5B ring is governed by two sets of paralogous proteins: ARC6 and PARC6 in the IEM, and PDV1 and PDV2 in the OEM (Fig. 1, E–H).
ARC6 descended from the cyanobacterial cell division protein Ftn2 (also called ZipN) and is conserved throughout the green lineage (Koksharova and Wolk, 2002; Vitha et al., 2003; Mazouni et al., 2004; Marbouty et al., 2009). arc6 mutants (Pyke et al., 1994) possess one or two giant chloroplasts with fragmented FtsZ filaments, while ARC6 overexpressors, though still bearing enlarged chloroplasts, exhibit exceptionally long FtsZ filaments. These phenotypes indicated that ARC6 promotes FtsZ assembly and probably stabilizes the Z ring (Pyke et al., 1994; Vitha et al., 2003). ARC6 spans the IEM with its large N terminus exposed to the stroma and its smaller C terminus protruding into the intermembrane space (IMS; Fig. 1E). The stromal region of ARC6 binds specifically to FtsZ2 via the CTP and probably acts as the primary membrane tether for the Z ring (Johnson et al., 2013; Fig. 1E). The IMS region interacts with the C-terminal IMS region of the land-plant-specific OEM protein PDV2 (Fig. 1F), and this interaction is required for PDV2 localization to the division site (Glynn et al., 2008). A similar relationship exists between PARC6 and PDV1 (Fig. 1G), which arose by duplication and divergence of ARC6 and PDV2, respectively, and may both be confined to vascular plants (Miyagishima et al., 2006; Glynn et al., 2009). In turn, PDV2 and PDV1 recruit DRP5B. Thus, ARC6 and PARC6 convey positional information from the stromal Z ring to the outside of the chloroplast through PDV2 and PDV1 to localize the DRP5B ring.
The dumbbell-shaped appearance and presence of multiple, laterally associated Z rings near the division site in the enlarged chloroplasts of Arabidopsis pdv1 and pdv2 mutants (Fig. 3) indicates that division is initiated but not completed in these mutants (Miyagishima et al., 2006) and suggests that information is also relayed from the external to internal division complexes during normal chloroplast division. A recent study combining x-ray crystallography, interaction assays, and genetic analysis in Arabidopsis provides evidence for this hypothesis (Wang et al., 2017). The authors demonstrated that part of the IMS region of PDV2 (PDV2IMS) inserts into a pocket formed by a highly conserved region of the ARC6IMS (Kumar et al., 2016) and that a second PDV2 molecule induces ARC6IMS dimerization, resulting in the formation of a heterotetramer (Fig. 1F). ARC6IMS dimerization depended on interaction between the cytosolic regions of the two PDV2 molecules. A dimerization-deficient mutant of PDV2 produced multiple, uncondensed ARC6 rings in vivo instead of the single ring observed in wild type (Wang et al., 2017). Thus, PDV2 dimerization on the cytosolic surface transmits information inside the chloroplast, resulting in the formation of a single ARC6 ring. Though not specifically tested in this study, PDV2-induced ARC6 dimerization may also contribute, along with the chloroplast Min system, to the formation or maintenance of a single, condensed Z ring in the stroma, though by an unknown mechanism (Wang et al., 2017).
There are many additional questions regarding coordination of inner and outer complexes. One concerns the role of DRP5B in the division process. DRP5B recruitment to the chloroplast probably involves direct interaction with PDV proteins (Miyagishima et al., 2006; Holtsmark et al., 2013). Therefore, DRP5B might facilitate the dimerization of PDV2, leading to condensed ARC6 and Z rings. This possibility is suggested by the phenotype of arc5, a DRP5B mutant (Robertson et al., 1996; Gao et al., 2003), that also exhibits multiple, uncondensed Z rings near the middle of its dumbbell-shaped chloroplasts (Miyagishima et al., 2006; Fig. 3). Another question is whether PDV1-PARC6 interaction (Fig. 1G) might also relay information from outside to inside the chloroplast. Assuming so, the effect might be to destabilize rather than stabilize Z rings, since PARC6 negatively regulates Z-ring formation, probably at least partly through its interaction with ARC3 (Glynn et al., 2009; Zhang et al., 2009, 2016; Fig. 1I).
OTHER CHLOROPLAST DIVISION PROTEINS
A few other proteins with less well-defined functions also contribute to chloroplast division (Basak and Møller, 2013; Osteryoung and Pyke, 2014). Final separation of chloroplasts appears to involve two proteins: CLUMPED CHLOROPLASTS1, a cystolic protein localized partly near the plasma membrane and partly on the chloroplast (Yang et al., 2011), and CRUMPLED LEAF, located in the OEM (Asano et al., 2004; Chen et al., 2009; Sugita et al., 2012). Both proteins were speculated to mediate chloroplast attachment to the cytoskeleton. GIANT CHLOROPLAST1 is a stromal protein associated with the IEM that bears some similarity to the bacterial cell-division inhibitor SulA and may play an indirect role in division-site placement (Maple et al., 2004; Raynaud et al., 2004).
REGULATION OF CHLOROPLAST DIVISION
Knowledge on how chloroplast division is regulated is still rudimentary, particularly in land plants with multiple chloroplasts per cell whose division is not tightly coordinated with cell division (Miyagishima, 2011; Pedroza-Garcia et al., 2016). Here, we highlight recent work on selected aspects of division regulation.
To achieve a permanent endosymbiotic relationship and ensure faithful organelle inheritance, the eukaryotic host needed to establish synchrony between endosymbiont division and host cell cycles (Pedroza-Garcia et al., 2016). Massive gene transfer to the nucleus and loss from the chloroplast genome following endosymbiosis solved part of this problem by bringing most genes under host control (Keeling, 2010). Miyagishima et al. (2012) used synchronized cultures of several unicellular organisms representing the major lineages of primary plastid-bearing algae to investigate the expression patterns of key chloroplast division genes and proteins during the cell cycle. They found that all the nuclear genes were expressed during S phase, when Z-ring formation and chloroplast division were initiated, except FtsZ from the glaucophyte Cyanophora paradoxa, which was constitutively expressed (Miyagishima et al., 2012). Nevertheless, Z-ring formation in C. paradoxa was still confined to the S phase. In contrast, minD and other chloroplast division genes in the plastid genomes of the green algae Chlorella vulgaris and Mesostgma viride were expressed constitutively, but chloroplast division still began during S phase. The results suggested that expression of endosymbiont-derived plastid-division genes that reside in the nucleus is under tighter host control than expression of genes retained in the plastid genome and that nuclear-gene expression governs the timing of chloroplast division (Miyagishima et al., 2012). The molecular mechanisms controlling the phase-specific timing of gene expression and chloroplast division remain unclear.
Accumulating evidence indicates that retrograde signals from the chloroplast also exert control over the host cell cycle (Garton et al., 2007; Kobayashi et al., 2009, 2011; Pedroza-Garcia et al., 2016). A recent study in C. merolae (Sumiya et al., 2016) revealed that cell-cycle progression was arrested in prophase when chloroplast division, which is sensitive to FtsZ level (Vitha et al., 2001), was disrupted by FtsZ overexpression prior to assembly of the mature division machinery. Two indicators of the G2-to-M transition, increased cyclin B expression and relocalization of cyclin-dependent kinase B, were blocked in the arrested cells. However, once the assembled chloroplast division complex began to constrict, disruption of chloroplast division no longer impeded cell-cycle progression. These results revealed that insufficient assembly of the chloroplast division machinery imposes a checkpoint on the cell cycle. Further analysis suggested the checkpoint is sensed between Z-ring formation and DRP5B recruitment (Sumiya et al., 2016). How this retrograde signal is conveyed to the nucleus is unknown.
Cytokinin has been implicated in the control of chloroplast division in P. patens (Abel et al., 1989; Reutter et al., 1998), and recent work indicates this is partly through regulation of PDV gene expression. Overexpression of the PDV proteins results in increased chloroplast number and decreased chloroplast size in Arabidopsis and P. patens, indicating that PDV1 and PDV2 levels influence the frequency of chloroplast division (Okazaki et al., 2009; Chang et al., 2017). Overexpression of Cytokinin-Responsive Transcription Factor2 or exogenous cytokinin treatment produced a similar phenotype in Arabidopsis, and these plants had elevated levels of PDV1 and PDV2 but not other division proteins. Similarly, treatment of moss with cytokinin specifically increased PDV2 transcript levels and chloroplast division (Okazaki et al., 2009). Recently, Vercruyssen et al. (2015) reported that overexpression of another transcription factor, GROWTH REGULATING FACTOR5 (GRF5), stimulates chloroplast and cell division. Because cytokinin has similar effects, they proposed that GRF5 and cytokinin cooperate in the control of chloroplast division. However, PDV2 expression was not increased in GRF5 overexpressors, suggesting GRF5 promotes chloroplast division by a different mechanism (Vercruyssen et al., 2015).
Another study showed that chloroplast division is impaired in mutants deficient in gibberllins (GA) (Jiang et al., 2012). FtsZ2, ARC6, DRP5B, and PDV transcript levels were dramatically decreased in the mutant. Exogenous GA treatment restored wild-type chloroplast division and transcript levels. Based on additional mutant studies, the authors proposed that GA might indirectly stimulate chloroplast division through promoting the degradation of DELLA protein family members, which negatively regulate chloroplast division (Jiang et al., 2012).
A mutant screen in Arabidopsis identified the transcription factor FHY3/CPD45, a key regulator of far-red light signaling, and its homolog FRS4/CPD25, as coactivators of ARC5 (DRP5B) expression and chloroplast division (Gao et al., 2013). fhy3/cpd45 and frs4/cpd25 mutants had enlarged dumbbell-shaped chloroplasts, similar to arc5 (Fig. 3). Expression of a downstream target of FHY3/CPD45 or of ARC5 in fhy3/cpd45 rescued only the far-red light signaling or chloroplast division defect, respectively, suggesting that FHY3/CPD45 activates ARC5 expression and far-red light signaling through independent pathways (Chang et al., 2015).
Recently, Okazaki et al. (2015) reported that the phosphoinositide phosphatidylinositol 4-phosphate (PI4P) negatively regulates chloroplast division in Arabidopsis. Inhibition of phosphatidylinositol 4-kinase to reduce PI4P levels in chloroplast membranes accelerated chloroplast division, producing a larger population of smaller chloroplasts, similar to overexpression of PDV proteins (Okazaki et al., 2009). However, this effect was due primarily to increased recruitment of DRP5B from the cytosol to the chloroplast surface and not to increased PDV protein levels. PDV1 and PDV2 both bound to PI4P, but the effect of PI4P depletion on chloroplast division was largely abolished in pdv1 but not pdv2 mutants. Based on these and other results, the authors proposed that PDV1 interaction with PI4P in the chloroplast envelope alters PDV1 affinity for DRP5B, which may alter the rate of chloroplast division. These findings implicate phosphoinositide signaling in the regulation of chloroplast division (Okazaki et al., 2015).
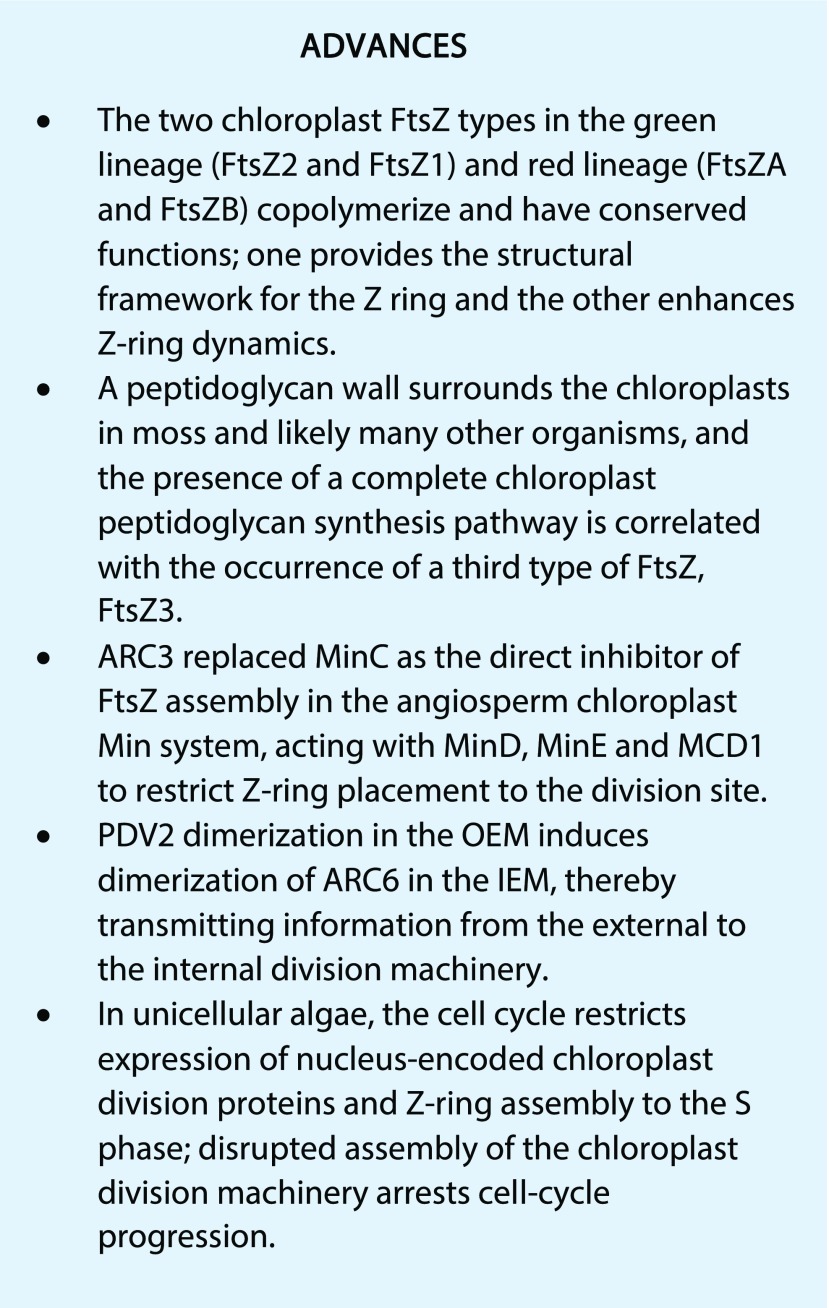
CONCLUDING REMARKS
While many of the players in chloroplast division have been identified, particularly in Arabidopsis, we still lack detailed mechanistic understanding of many of their biochemical activities and functional interactions as components of a dynamic molecular machine. Moreover, phylogenomic and functional studies indicate significant evolutionary diversity in the composition of the division machinery (Miyagishima et al., 2011). For example, although ARC6 descended from a closely related cyanobacterial cell division gene (Koksharova and Wolk, 2002; Vitha et al., 2003), no ARC6 homolog or other membrane-tethering protein for the Z ring has been reported in red algae. We still know absolutely nothing about how thylakoids divide; ultrastructural evidence suggests it may occur independently from division of the envelope membranes (Whatley, 1988). We have little information about the division of plastid types other than chloroplasts or about other potential modes of plastid replication, such as a reported budding mechanism (Forth and Pyke, 2006; Pyke, 2016). The mechanisms regulating the control of chloroplast compartment size remain mostly unidentified (Pyke, 1999; Larkin et al., 2016). The extent to which division is coordinated with lipid biosynthesis (Wu and Xue, 2010; Fan and Xu, 2011; Nobusawa and Umeda, 2012) and other metabolic processes is unknown. Finally, how plastid division is regulated and integrated with cell division and expansion, chloroplast biogenesis, and plant growth and development remain poorly understood (Jarvis and López-Juez, 2013; Pedroza-Garcia et al., 2016; see “Outstanding Questions”). Clearly, it is still early days in the study of plastid division—many rich avenues remain to be explored.
Acknowledgments
We thank Dr. Min Zhang for contributing images and current and former members of the Osteryoung laboratory for research and insightful discussions.
Footnotes
This work was supported by US Department of Energy grant number DE-FG02-06ER15808 to K.W.O. and National Science Foundation grant numbers 1121943 and 1719376 to K.W.O. and 1517241 to K.W.O and D.C.D.
Articles can be viewed without a subscription.
References
- Abel WO, Knebel W, Koop H-U, Marienfeld JR, Quader H, Reski R, Schnepf E, Spörlein B (1989) A cytokinin-sensitive mutant of the moss, Physcomitrella patens, defective in chloroplast division. Protoplasma 152: 1–13 [Google Scholar]
- Addinall SG, Bi E, Lutkenhaus J (1996) FtsZ ring formation in fts mutants. J Bacteriol 178: 3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge C, Møller SG (2005) The plastid division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J Biol Chem 280: 31673–31678 [DOI] [PubMed] [Google Scholar]
- Arimura S. (2018) Fission and fusion of plant mitochondria, and genome maintenance. Plant Physiol 176: 152-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Yoshioka Y, Kurei S, Sakamoto W, Sodmergen, Machida Y (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J 38: 448–459 [DOI] [PubMed] [Google Scholar]
- Aung K, Hu J (2012) Differential roles of Arabidopsis dynamin-related proteins DRP3A, DRP3B, and DRP5B in organelle division. J Integr Plant Biol 54: 921–931 [DOI] [PubMed] [Google Scholar]
- Basak I, Møller SG (2013) Emerging facets of plastid division regulation. Planta 237: 389–398 [DOI] [PubMed] [Google Scholar]
- Beech PL, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson PR, McFadden GI (2000) Mitochondrial FtsZ in a chromophyte alga. Science 287: 1276–1279 [DOI] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, et al. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355: 739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassier-Chauvat C, Chauvat F (2014) Cell division in cyanobacteria. In Flores E, Herrero A, eds, The Cell Biology of Cyanobacteria. Caister Academic Press, Poole, UK, pp 7–27 [Google Scholar]
- Chang N, Gao Y, Zhao L, Liu X, Gao H (2015) Arabidopsis FHY3/CPD45 regulates far-red light signaling and chloroplast division in parallel. Sci Rep 5: 9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun Q, Li Y, Mu Y, Hu J, Feng Y, Liu X, Gao H (2017) PDV2 has a dosage effect on chloroplast division in Arabidopsis. Plant Cell Rep 36: 471–480 [DOI] [PubMed] [Google Scholar]
- Chen Y, Asano T, Fujiwara MT, Yoshida S, Machida Y, Yoshioka Y (2009) Plant cells without detectable plastids are generated in the crumpled leaf mutant of Arabidopsis thaliana. Plant Cell Physiol 50: 956–969 [DOI] [PubMed] [Google Scholar]
- Chen Y, Erickson HP (2009) FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry 48: 6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Porter K, Osawa M, Augustus AM, Milam SL, Joshi C, Osteryoung KW, Erickson HP (2017) The chloroplast tubulin homologs FtsZA and FtsZB from the red alga Galdieria sulphuraria co-assemble into dynamic filaments. J Biol Chem 292: 5207–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol 10: 507–516 [DOI] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56: 641–649 [DOI] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI (1990) Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA 87: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kafafi S, Mukherjee S, El-Shami M, Putaux JL, Block MA, Pignot-Paintrand I, Lerbs-Mache S, Falconet D (2005) The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. Biochem J 387: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M (2010) FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74: 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconet D. (2012) Origin, evolution and division of plastids. In Eaton-Rye JJ, Tripathy BC, Sharkey TD, eds, Photosynthesis-Plastid Biology, Energy Conversion and Carbon Assimilation. Springer, Dordrecht, the Netherlands, pp 35–61 [Google Scholar]
- Fan J, Xu C (2011) Genetic analysis of Arabidopsis mutants impaired in plastid lipid import reveals a role of membrane lipids in chloroplast division. Plant Signal Behav 6: 458–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth D, Pyke KA (2006) The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J Exp Bot 57: 1971–1979 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Nunnari J (2014) Mitochondrial form and function. Nature 505: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara MT, Hashimoto H, Kazama Y, Abe T, Yoshida S, Sato N, Itoh RD (2008) The assembly of the FtsZ ring at the mid-chloroplast division site depends on a balance between the activities of AtMinE1 and ARC11/AtMinD1. Plant Cell Physiol 49: 345–361 [DOI] [PubMed] [Google Scholar]
- Fujiwara MT, Nakamura A, Itoh R, Shimada Y, Yoshida S, Møller SG (2004) Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J Cell Sci 117: 2399–2410 [DOI] [PubMed] [Google Scholar]
- Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA 100: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liu H, An C, Shi Y, Liu X, Yuan W, Zhang B, Yang J, Yu C, Gao H (2013) Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J 75: 795–807 [DOI] [PubMed] [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ (2007) crinkled leaves 8—a mutation in the large subunit of ribonucleotide reductase--leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. Plant J 50: 118–127 [DOI] [PubMed] [Google Scholar]
- Gilson PR, Yu XC, Hereld D, Barth C, Savage A, Kiefel BR, Lay S, Fisher PR, Margolin W, Beech PL (2003) Two Dictyostelium orthologs of the prokaryotic cell division protein FtsZ localize to mitochondria and are required for the maintenance of normal mitochondrial morphology. Eukaryot Cell 2: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Froehlich JE, Osteryoung KW (2008) Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Miyagishima SY, Yoder DW, Osteryoung KW, Vitha S (2007) Chloroplast division. Traffic 8: 451–461 [DOI] [PubMed] [Google Scholar]
- Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, Osteryoung KW (2009) PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J 59: 700–711 [DOI] [PubMed] [Google Scholar]
- Grosche C, Rensing SA (2017) Three rings for the evolution of plastid shape: A tale of land plant FtsZ. Protoplasma 254: 1879–1885 [DOI] [PubMed] [Google Scholar]
- Haeusser DP, Margolin W (2016) Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14: 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H. (2005) The ultrastructural features and division of secondary plastids. J Plant Res 118: 163–172 [DOI] [PubMed] [Google Scholar]
- Hirano T, Tanidokoro K, Shimizu Y, Kawarabayasi Y, Ohshima T, Sato M, Tadano S, Ishikawa H, Takio S, Takechi K, et al. (2016) Moss chloroplasts are surrounded by a peptidoglycan wall containing d-amino acids. Plant Cell 28: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtsmark I, Lee S, Lunde KA, Auestad K, Maple-Grødem J, Møller SG (2013) Plastid division control: The PDV proteins regulate DRP5B dynamin activity. Plant Mol Biol 82: 255–266 [DOI] [PubMed] [Google Scholar]
- Homi S, Takechi K, Tanidokoro K, Sato H, Takio S, Takano H (2009) The peptidoglycan biosynthesis genes MurA and MraY are related to chloroplast division in the moss Physcomitrella patens. Plant Cell Physiol 50: 2047–2056 [DOI] [PubMed] [Google Scholar]
- Huecas S, Schaffner-Barbero C, García W, Yébenes H, Palacios JM, Díaz JF, Menéndez M, Andreu JM (2007) The interactions of cell division protein FtsZ with guanine nucleotides. J Biol Chem 282: 37515–37528 [DOI] [PubMed] [Google Scholar]
- Itoh R, Fujiwara M, Nagata N, Yoshida S (2001) A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol 127: 1644–1655 [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jiang X, Li H, Wang T, Peng C, Wang H, Wu H, Wang X (2012) Gibberellin indirectly promotes chloroplast biogenesis as a means to maintain the chloroplast population of expanded cells. Plant J 72: 768–780 [DOI] [PubMed] [Google Scholar]
- Johnson CB, Shaik R, Abdallah R, Vitha S, Holzenburg A (2015) FtsZ1/FtsZ2 turnover in chloroplasts and the role of ARC3. Microsc Microanal 21: 313–323 [DOI] [PubMed] [Google Scholar]
- Johnson CB, Tang LK, Smith AG, Ravichandran A, Luo Z, Vitha S, Holzenburg A (2013) Single particle tracking analysis of the chloroplast division protein FtsZ anchoring to the inner envelope membrane. Microsc Microanal 19: 507–512 [DOI] [PubMed] [Google Scholar]
- Kao YT, Gonzalez KL, Bartel B (2018) Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol 176: 162–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten B, Buck F, Nuske J, Reski R (1997) Cytokinin affects nuclear- and plastome-encoded energy-converting plastid enzymes. Planta 201: 261–272 [DOI] [PubMed] [Google Scholar]
- Katayama N, Takano H, Sugiyama M, Takio S, Sakai A, Tanaka K, Kuroiwa H, Ono K (2003) Effects of antibiotics that inhibit the bacterial peptidoglycan synthesis pathway on moss chloroplast division. Plant Cell Physiol 44: 776–781 [DOI] [PubMed] [Google Scholar]
- Keeling PJ. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Imamura S, Hanaoka M, Tanaka K (2011) A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat Cell Biol 13: 483–487 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kanesaki Y, Tanaka A, Kuroiwa H, Kuroiwa T, Tanaka K (2009) Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc Natl Acad Sci USA 106: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksharova OA, Wolk CP (2002) A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J Bacteriol 184: 5524–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Radhakrishnan A, Su CC, Osteryoung KW, Yu EW (2016) Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6. Protein Sci 25: 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T, Misumi O, Nishida K, Yagisawa F, Yoshida Y, Fujiwara T, Kuroiwa H (2008) Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int Rev Cell Mol Biol 271: 97–152 [DOI] [PubMed] [Google Scholar]
- Kuroiwa H, Mori T, Takahara M, Miyagishima SY, Kuroiwa T (2002) Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215: 185–190 [DOI] [PubMed] [Google Scholar]
- Larkin RM, Stefano G, Ruckle ME, Stavoe AK, Sinkler CA, Brandizzi F, Malmstrom CM, Osteryoung KW (2016) REDUCED CHLOROPLAST COVERAGE genes from Arabidopsis thaliana help to establish the size of the chloroplast compartment. Proc Natl Acad Sci USA 113: E1116–E1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech R, Baker NR (1983) The development of photosynthetic capacity in leaves. In Dale JE, Milthorpe FL, eds, The Growth and Functioning of Leaves. Cambridge University Press, Cambridge, pp 271–307 [Google Scholar]
- Leger MM, Petrů M, Žárský V, Eme L, Vlček Č, Harding T, Lang BF, Eliáš M, Doležal P, Roger AJ (2015) An ancestral bacterial division system is widespread in eukaryotic mitochondria. Proc Natl Acad Sci USA 112: 10239–10246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Li N, Kudo H, Zhang Z, Li J, Wang L, Zhang W, Takechi K, Takano H (2017) Genes sufficient for synthesizing peptidoglycan are retained in gymnosperm genomes, and MurE from Larix gmelinii can rescue the albino phenotype of Arabidopsis MurE mutation. Plant Cell Physiol 58: 587–597 [DOI] [PubMed] [Google Scholar]
- Lohse S, Hause B, Hause G, Fester T (2006) FtsZ characterization and immunolocalization in the two phases of plastid reorganization in arbuscular mycorrhizal roots of Medicago truncatula. Plant Cell Physiol 47: 1124–1134 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. (2007) Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76: 539–562 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Du S (2017) E. coli cell cycle machinery. In Löwe J, Amos L, eds, Prokaryotic Cytoskeletons. Springer International Publishing, Cham, Switzerland, pp 27–65 [Google Scholar]
- Lutkenhaus JF, Wolf-Watz H, Donachie WD (1980) Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol 142: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Margolin W (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol 181: 7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCready JS, Schossau J, Osteryoung KW, Ducat DC (2017) Robust Min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol Microbiol 103: 483–503 [DOI] [PubMed] [Google Scholar]
- Machida M, Takechi K, Sato H, Chung SJ, Kuroiwa H, Takio S, Seki M, Shinozaki K, Fujita T, Hasebe M, et al. (2006) Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc Natl Acad Sci USA 103: 6753–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J, Aldridge C, Møller SG (2005) Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J 43: 811–823 [DOI] [PubMed] [Google Scholar]
- Maple J, Fujiwara MT, Kitahata N, Lawson T, Baker NR, Yoshida S, Møller SG (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr Biol 14: 776–781 [DOI] [PubMed] [Google Scholar]
- Maple J, Vojta L, Soll J, Møller SG (2007) ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep 8: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Saguez C, Cassier-Chauvat C, Chauvat F (2009) ZipN, an FtsA-like orchestrator of divisome assembly in the model cyanobacterium Synechocystis PCC6803. Mol Microbiol 74: 409–420 [DOI] [PubMed] [Google Scholar]
- Margolin W. (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Lang D, Hanke ST, Mueller SJX, Sarnighausen E, Vervliet-Scheebaum M, Reski R (2009a) Targeted gene knockouts reveal overlapping functions of the five Physcomitrella patens FtsZ isoforms in chloroplast division, chloroplast shaping, cell patterning, plant development, and gravity sensing. Mol Plant 2: 1359–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Lang D, Heckmann J, Zimmer AD, Vervliet-Scheebaum M, Reski R (2009b) A uniquely high number of ftsZ genes in the moss Physcomitrella patens. Plant Biol 11: 744–750 [DOI] [PubMed] [Google Scholar]
- Mazouni K, Domain F, Cassier-Chauvat C, Chauvat F (2004) Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol Microbiol 52: 1145–1158 [DOI] [PubMed] [Google Scholar]
- McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol 127: 1656–1666 [PMC free article] [PubMed] [Google Scholar]
- McAndrew RS, Olson BJSC, Kadirjan-Kalbach DK, Chi-Ham CL, Vitha S, Froehlich JE, Osteryoung KW (2008) In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem J 412: 367–378 [DOI] [PubMed] [Google Scholar]
- Miyagishima SY. (2011) Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol 155: 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Froehlich JE, Osteryoung KW (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Nakamura M, Uzuka A, Era A (2014) FtsZ-less prokaryotic cell division as well as FtsZ- and dynamin-less chloroplast and non-photosynthetic plastid division. Front Plant Sci 5: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Nakanishi H, Kabeya Y (2011) Structure, regulation, and evolution of the plastid division machinery. Int Rev Cell Mol Biol 291: 115–153 [DOI] [PubMed] [Google Scholar]
- Miyagishima SY, Nishida K, Mori T, Matsuzaki M, Higashiyama T, Kuroiwa H, Kuroiwa T (2003) A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Nozaki H, Nishida K, Nishida K, Matsuzaki M, Kuroiwa T (2004) Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: the duplication of FtsZ is implicated in endosymbiosis. J Mol Evol 58: 291–303 [DOI] [PubMed] [Google Scholar]
- Miyagishima SY, Suzuki K, Okazaki K, Kabeya Y (2012) Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol Biol Evol 29: 2957–2970 [DOI] [PubMed] [Google Scholar]
- Miyagishima SY, Takahara M, Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T (2001) Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13: 2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima SY, Wolk CP, Osteryoung KW (2005) Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol 56: 126–143 [DOI] [PubMed] [Google Scholar]
- Monahan LG, Liew AT, Bottomley AL, Harry EJ (2014) Division site positioning in bacteria: One size does not fit all. Front Microbiol 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J 17: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Suzuki K, Kabeya Y, Miyagishima SY (2009) Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr Biol 19: 151–156 [DOI] [PubMed] [Google Scholar]
- Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, Kuroiwa T (2003) Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA 100: 2146–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa T, Umeda M (2012) Very-long-chain fatty acids have an essential role in plastid division by controlling Z-ring formation in Arabidopsis thaliana. Genes Cells 17: 709–719 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, Nakanishi H, Miyagishima SY (2009) The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Miyagishima SY, Wada H (2015) Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson BJ, Wang Q, Osteryoung KW (2010) GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J Biol Chem 285: 20634–20643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma R, Mishra N, Miyagishima SY (2017) Regulation of chloroplast and nucleomorph replication by the cell cycle in the cryptophyte Guillardia theta. Sci Rep 7: 2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP (2008) Reconstitution of contractile FtsZ rings in liposomes. Science 320: 792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP (2009) Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J 28: 3476–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Erickson HP (2011) Inside-out Z rings--constriction with and without GTP hydrolysis. Mol Microbiol 81: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Pyke KA (2014) Division and dynamic morphology of plastids. Annu Rev Plant Biol 65: 443–472 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Vierling E (1995) Conserved cell and organelle division. Nature 376: 473–474 [DOI] [PubMed] [Google Scholar]
- Pedroza-Garcia JA, Domenichini S, Bergounioux C, Benhamed M, Raynaud C (2016) Chloroplasts around the plant cell cycle. Curr Opin Plant Biol 34: 107–113 [DOI] [PubMed] [Google Scholar]
- Purkanti R, Thattai M (2015) Ancient dynamin segments capture early stages of host-mitochondrial integration. Proc Natl Acad Sci USA 112: 2800–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA. (1999) Plastid division and development. Plant Cell 11: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA. (2009) Plastid Biology. Cambridge University Press, Cambridge [Google Scholar]
- Pyke KA. (2016) Plastid division. In Rose RJ, ed., Molecular Cell Biology of the Growth and Differentiation of Plant Cells. CRC Press, Boca Raton, Florida, pp 37–50 [Google Scholar]
- Pyke KA, Leech RM (1992) Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol 99: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1994) A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol 104: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Rutherford SM, Robertson EJ, Leech RM (1994) arc6, a fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol 106: 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Cassier-Chauvat C, Perennes C, Bergounioux C (2004) An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16: 1801–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redick SD, Stricker J, Briscoe G, Erickson HP (2005) Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J Bacteriol 187: 2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reutter K, Atzorn R, Hadeler B, Schmülling T, Reski R (1998) Expression of the bacterial ipt gene in Physcomitrella rescues mutations in budding and in plastid division. Planta 206: 196–203 [Google Scholar]
- Robertson EJ, Rutherford SM, Leech RM (1996) Characterization of chloroplast division using the Arabidopsis mutant arc5. Plant Physiol 112: 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL (2005) Spatial control of bacterial division-site placement. Nat Rev Microbiol 3: 959–968 [DOI] [PubMed] [Google Scholar]
- Rowlett VW, Margolin W (2013) The bacterial Min system. Curr Biol 23: R553–R556 [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJ (2002) GTP hydrolysis of cell division protein FtsZ: Evidence that the active site is formed by the association of monomers. Biochemistry 41: 521–529 [DOI] [PubMed] [Google Scholar]
- Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW (2009) Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant 2: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J (2009) The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol 72: 410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J (2010) Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol Microbiol 75: 1285–1298 [DOI] [PubMed] [Google Scholar]
- Shimada H, Koizumi M, Kuroki K, Mochizuki M, Fujimoto H, Ohta H, Masuda T, Takamiya K (2004) ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol 45: 960–967 [DOI] [PubMed] [Google Scholar]
- Smith AG, Johnson CB, Vitha S, Holzenburg A (2010) Plant FtsZ1 and FtsZ2 expressed in a eukaryotic host: GTPase activity and self-assembly. FEBS Lett 584: 166–172 [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Mishra M, Wu L, Yin Z, Balasubramanian MK (2008) The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev 22: 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95: 4368–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita C, Kato Y, Yoshioka Y, Tsurumi N, Iida Y, Machida Y, Sugita M (2012) CRUMPLED LEAF (CRL) homologs of Physcomitrella patens are involved in the complete separation of dividing plastids. Plant Cell Physiol 53: 1124–1133 [DOI] [PubMed] [Google Scholar]
- Sumiya N, Fujiwara T, Era A, Miyagishima SY (2016) Chloroplast division checkpoint in eukaryotic algae. Proc Natl Acad Sci USA 113: E7629–E7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano H, Takechi K (2010) Plastid peptidoglycan. Biochim Biophys Acta 1800: 144–151 [DOI] [PubMed] [Google Scholar]
- Terbush AD, MacCready JS, Chen C, Ducat DC, Osteryoung KW (2018) Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant Physiol 176: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, Osteryoung KW (2012) Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J Cell Biol 199: 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, Porzondek CA, Osteryoung KW (2016) Functional analysis of the chloroplast division complex using Schizosaccharomyces pombe as a heterologous expression system. Microsc Microanal 22: 275–289 [DOI] [PubMed] [Google Scholar]
- TerBush AD, Yoshida Y, Osteryoung KW (2013) FtsZ in chloroplast division: Structure, function and evolution. Curr Opin Cell Biol 25: 461–470 [DOI] [PubMed] [Google Scholar]
- Vaughan S, Wickstead B, Gull K, Addinall SG (2004) Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol 58: 19–29 [DOI] [PubMed] [Google Scholar]
- Vercruyssen L, Tognetti VB, Gonzalez N, Van Dingenen J, De Milde L, Bielach A, De Rycke R, Van Breusegem F, Inzé D (2015) GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol 167: 817–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff JM, Tsim M, Oliva MA, García-Sanchez A, Kureisaite-Ciziene D, Andreu JM, Löwe J (2017) A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. MBio 8: e00254–e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li J, Sun Q, Yu X, Zhang W, Jia N, An C, Li Y, Dong Y, Han F, et al. (2017) Structural insights into the coordination of plastid division by the ARC6-PDV2 complex. Nat Plants 3: 17011. [DOI] [PubMed] [Google Scholar]
- Whatley JM. (1988) Mechanisms and morphology of plastid division. In Boffey SA, Lloyd D, eds, Division and Segregation of Organelles. Cambridge University Press, Cambridge, pp 63–84 [Google Scholar]
- Wu GZ, Xue HW (2010) Arabidopsis β-ketoacyl-[acyl carrier protein] synthase i is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 22: 3726–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Glynn JM, Olson BJ, Schmitz AJ, Osteryoung KW (2008) Plastid division: across time and space. Curr Opin Plant Biol 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355: 744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sage TL, Liu Y, Ahmad TR, Marshall WF, Shiu SH, Froehlich JE, Imre KM, Osteryoung KW (2011) CLUMPED CHLOROPLASTS 1 is required for plastid separation in Arabidopsis. Proc Natl Acad Sci USA 108: 18530–18535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder DW, Kadirjan-Kalbach D, Olson BJ, Miyagishima SY, Deblasio SL, Hangarter RP, Osteryoung KW (2007) Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol 48: 775–791 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, Nanamiya H, Kawamura F, Kuroiwa T (2006) Isolated chloroplast division machinery can actively constrict after stretching. Science 313: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Kuroiwa H, Misumi O, Yoshida M, Ohnuma M, Fujiwara T, Yagisawa F, Hirooka S, Imoto Y, Matsushita K, et al. (2010) Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 329: 949–953 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Miyagishima SY, Kuroiwa H, Kuroiwa T (2012) The plastid-dividing machinery: Formation, constriction and fission. Curr Opin Plant Biol 15: 714–721 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Mogi Y, TerBush AD, Osteryoung KW (2016) Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat Plants 2: 16095. [DOI] [PubMed] [Google Scholar]
- Yu XC, Margolin W (1999) FtsZ ring clusters in min and partition mutants: Role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Zhang M, Chen C, Froehlich JE, TerBush AD, Osteryoung KW (2016) Roles of Arabidopsis PARC6 in coordination of the chloroplast division complex and negative regulation of FtsZ assembly. Plant Physiol 170: 250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hu J (2010) The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Hu Y, Jia J, Li D, Zhang R, Gao H, He Y (2009) CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res 19: 877–886 [DOI] [PubMed] [Google Scholar]
- Zhang M, Schmitz AJ, Kadirjan-Kalbach DK, Terbush AD, Osteryoung KW (2013) Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in Arabidopsis through interaction with FtsZ2. Plant Cell 25: 1787–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB (2014) Endosymbiotic theory for organelle origins. Curr Opin Microbiol 22: 38–48 [DOI] [PubMed] [Google Scholar]