Abstract
Discoveries illuminate highly regulated dynamics of mRNA translation, sequestration, and degradation within the cytoplasm of plants.
The export of an mRNA from the nucleus to the cytoplasm begins an odyssey of dynamic regulation that determines the location, longevity, and use of the transcript in the production of polypeptides by ribosomes in plant cells. Recent leveraging of mutants, imaging of fluorescent proteins, and omics of protein and ribosome association with mRNAs have significantly enriched our understanding of the intricate regulation of the fates of transcripts within the cytoplasm of plant cells. This Update explores the connections between translation, decay, and storage of mRNAs that involve three heterogenous mRNA-ribonucleoprotein (mRNP) complexes: polyribosomes (polysomes), processing bodies (PBs), and stress granules (SGs; Box 1; Eulalio et al., 2007; Horvathova et al., 2017; Hubstenberger et al., 2017; Hummel et al., 2015; Merchante et al., 2017; Moore et al., 2016; Roy and von Armin et al., 2013). We begin with a brief overview of cellular surveillance of mRNA integrity during the first (pioneer) round of translation. Next, we consider mRNA decay initiated on translating ribosomes, including cotranslational exonucleolytic decay and microRNA (miRNA)-targeted decay. Then, we detail the general decay process of mRNAs initiated by enzymatic deadenylation of the 3′ polyadenylated (poly[A]) tail and removal of the 5′ 7-methyl guanosine (7mG)-cap. Finally, we discuss the selective sequestration of translationally repressed mRNAs in SGs under severe environmental stress. This dynamic interplay between mRNA translation, stabilization, and turnover fine-tunes the differential regulation of genes to enable developmental plasticity and effective environmental responses.
CONNECTIONS BETWEEN mRNA DECAY AND TRANSLATION
mRNA turnover occurs via multiple selective pathways that serve to modulate the abundance of transcripts and to eliminate those that are dysfunctional. Upon export from the nucleus to the cytoplasm, an mRNA may be bound at its 5′ terminus by the two-subunit nuclear cap-binding complex (CBC) composed of CAP BINDING PROTEIN20 (CBP20) and CBP80 (Fig. 1). It is not well understood how the CBC is exchanged for a cytoplasmic cap-binding complex, composed of either eIF4E and eIF4G or eIFiso4E and eIFiso4G. Both eIF4E and eIFiso4E bind directly to the 7mG-cap structure at the 5′ terminus and are tethered to the scaffold proteins eIF4G and eIFiso4G, respectively, which interact directly with a PABP among those bound to the poly(A) tail at the 3′ terminus of the transcript (Browning and Bailey-Serres, 2015). This physical circularization of the polysome enhances primary initiation or subsequent ribosome-recycling events (Gallie, 2014; Browning and Bailey-Serres, 2015). It also may safeguard the transcript termini from enzymatic end attacks that can commence its degradation.
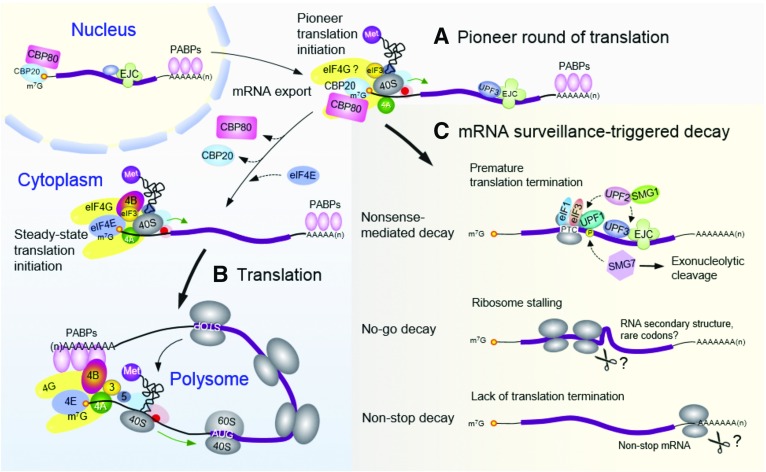
Figure 1.
mRNA surveillance-triggered decay during the pioneer round of translation. A, An mRNA is exported from the nucleus to the cytoplasm as an mRNP with the nuclear CBP20/CBP80 cap-binding complex (CBC) bound to the 5′-cap and poly(A)-binding proteins (PABPs) bound to the 3′ poly(A) tail. Within minutes after export, the mRNA undergoes a pioneer round of translation as a quality-control mechanism. B, After the first round of translation and replacement of CBP20 by the eukaryotic translation initiation factor 4E (eIF4E), only mRNAs that pass the quality-control check can enter into active translation as templates for bulk protein synthesis. C, mRNAs with defects in translation are subjected to degradation via different pathways. mRNAs containing a sequence feature that causes premature translation termination (e.g. a premature termination codon [PTC] upstream of an exon junction complex [EJC]) are targeted for nonsense-mediated decay (NMD). mRNAs with stalled translating ribosomes in the coding region or lacking a termination codon may be targeted to the no-go decay and non-stop decay pathways, respectively. In contrast to NMD, which has been characterized, it is less clear if the no-go decay and non-stop decay pathways function in plants. AUG, Translation initiation codon; STOP, termination codon.
mRNA Surveillance-Triggered Decay
mRNAs exported through the nuclear pore undergo a quality-control round of translation that routes transcripts with aberrant features to the NMD pathway (Fig. 1). Several mRNA qualities that compromise the integrity of translation termination trigger NMD, including a PTC marked by the presence of an exon junction complex 3′ of a stop codon (Reddy et al., 2013; Shaul, 2015). This is often a hallmark of incomplete or alternative splicing. Other mRNA features that trigger NMD include a more than 300-nucleotide-long 3′ untranslated region (UTR), an intron in the 3′ UTR that is 50 or more nucleotides downstream of the stop codon, an upstream open reading frame (ORF) that is out of frame with the start codon of the main (protein-coding) ORF (mORF), a truncated transcript due to premature 3′ polyadenylation, or an inefficient or absent stop codon (Reddy et al., 2013).
The early steps of PTC recognition and NMD complex formation in eukaryotes involve UP FRAMESHIFT1 (UPF1), UPF2, UPF3, and SUPPRESSOR WITH MORPHOLOGICAL EFFECT ON GENITALIAs (SMGs; for review, see Shaul, 2015; Dai et al., 2016). The late steps of NMD involve translational repression and the degradation of target mRNAs. In mammals, the degradation of NMD targets is initiated by SMG6-mediated endonucleolytic cleavage followed by exonucleolytic degradation of the 3′ and 5′ cleavage products by the 5′-3′ exoribonuclease EXORIBONUCLEASE1 (XRN1) and the exosome (see below), respectively (Shaul, 2015). In Arabidopsis (Arabidopsis thaliana), an ortholog of SMG6 has not been identified, but degradation of NMD substrates occurs in PBs and requires UPF1 and SMG7. It was proposed that AtSMG7 recruits the NMD complex to PBs for mRNA degradation through its interaction with phosphorylated AtUPF1. However, the mechanism by which NMD targets are fully degraded to nucleotides is unknown, since AtXRN4, the functional cytoplasmic ortholog of mammalian XRN1, is not required (Kerényi et al., 2013; Mérai et al., 2013).
In addition to restricting the synthesis of nonfunctional proteins from PTC-containing mRNAs, NMD conditionally regulates the abundance and translation of a number of functional transcripts. For example, NMD controls the developmental transition to flowering by down-regulating a functional FLOWERING LOCUS M transcript isoform under elevated temperatures in Arabidopsis (Sureshkumar et al., 2016). It also tempers levels of defense-related transcripts under normal growth conditions (Jeong et al., 2011; Gloggnitzer et al., 2014; Shaul, 2015), contributes to the repression of EIN3-BINDING F BOX PROTEIN2 mRNA translation in response to ethylene (Merchante et al., 2015), and promotes the acclimation response to salt stress in Arabidopsis (Vexler et al., 2016). A general dampening of NMD by stresses could be responsible for the increase in NMD-sensitive splicing variants in response to high salt (Drechsel et al., 2013) and prematurely polyadenylated transcripts under hypoxia (de Lorenzo et al., 2017).
XRN4-Mediated Cotranslational Decay
As polysomes and the decay machinery act antagonistically in controlling the function and fate of cytoplasmic mRNAs, it has been generally thought that turnover takes place on ribosome-free mRNPs following translational repression and removal of the 3′ poly(A) tail. Several new studies demonstrate that, in addition to a deadenylation-dependent mechanism (see below), cytoplasmic mRNA degradation can occur cotranslationally in the 5′ to 3′ direction in a deadenylation-independent manner (Fig. 2). By selective purification of polyadenylated mRNAs with a free 5′-monophosphate, studies of Arabidopsis, rice (Oryza sativa), and soybean (Glycine max) show a global pervasiveness of 5′ decay-intermediate termini that display a three-nucleotide periodicity throughout the mORF, in the same register as the codon periodicity generated by RNase digestion in ribosome footprinting analyses (Hou et al., 2016; Yu et al., 2016; Crisp et al., 2017). This in-frame periodicity in decay intermediates is dependent on XRN4 function, indicating that 5′-3′ mRNA degradation can accompany the codon-by-codon translocation of elongating ribosomes. This cotranslational XRN4 activity most likely follows an inhibition of translational initiation before or as a consequence of removal of the 5′-7mG-cap. Interestingly, cotranslational mRNA degradation is associated with paused or stacked ribosomes on upstream ORFs, within and at the stop codon of certain mORFs, and near to noncleavable miRNA target sites (Hou et al., 2016; Yu et al., 2016). This suggests that cotranslational XRN4-dependent decay may be linked to the kinetics of ribosome translocation, destabilizing mRNAs with sites of rate-limiting translational elongation or termination.
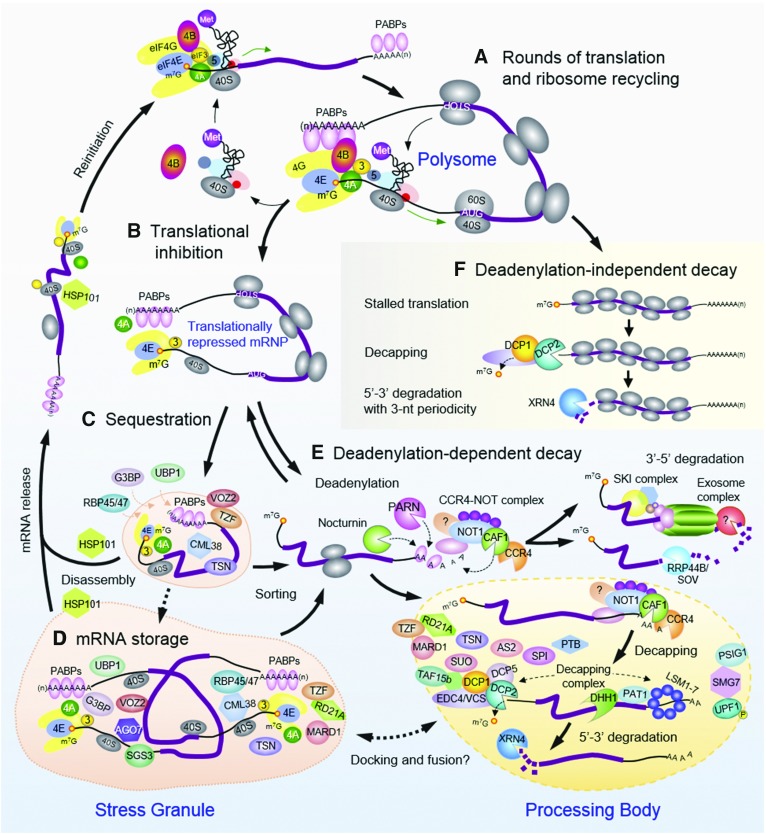
Figure 2.
Overview of cytoplasmic mRNA translation, storage, and decay in plants. A, Cytoplasmic mRNAs undergoing active translation form polysomes and remain in a translationally active state until damaged or translationally repressed. B, Repression can result from multiple causes (i.e. altered abundance or phosphorylation of specific translation factors or RNA-binding proteins and ribosome stalling) that limit translational initiation or ribosome translocation, promoting transition into a translationally repressed mRNP, where the mRNA is either sequestered by RNA-binding proteins (e.g. UBP1, G3BP, and RBP45/47) or degraded via different pathways. C, mRNA sequestration is typically triggered by cellular stress, serving as a sorting nucleation point for mRNP assembly into SGs for storage or PBs for degradation. D, During a stress-recovery period, intact mRNAs stored in SGs can reenter translation via a process facilitated by a chaperone (e.g. HSP101 during heat stress recovery). Some mRNAs released from SGs may be targeted for degradation after stress. E, In general mRNA decay, the 3′ protective poly(A) tail is removed by different classes of deadenylases [Nocturnin, poly(A)-specific RNase (PARN), and the CCR-NOT complex], then degradation proceeds in the 3′ to 5′ direction by the multimeric SKI-exosome complex and/or the RRP44B/SOV exonuclease. After deadenylation, mRNA degradation also can occur from the 5′ to 3′ direction. mRNAs degraded via this mode could be localized in the cytosol and/or PBs, where the multiprotein mRNA-decapping complex removes the protective 5′-cap and, subsequently, the 5′-3′ XRN4 exoribonuclease catalyzes nucleotide hydrolysis. F, In addition to the deadenylation-dependent decay pathway, translationally repressed mRNAs can be degraded directly in the 5′ to 3′ direction while in association with elongating ribosomes via XRN4-mediated cotranslational decay. This mode of degradation bypasses deadenylation but may require mRNA decapping. The progressive 5′-3′ exonucleolytic destruction of the mRNA by XRN4 yields a codon-by-codon three-nucleotide signature that is thought to reflect the movement of the most 5′ ribosome along the transcript. AUG, Translation initiation codon; STOP, termination codon.
Ribosome pausing and cotranslational decay also may serve as an efficient means for rapid transcriptome adjustment in response to environmental stimuli and transcriptome resetting during stress recovery. For example, cotranslational decay appears to control rapid down-regulation of excess-light-activated transcripts during stress recovery in Arabidopsis plants (Crisp et al., 2017). Moreover, within 15 min of pronounced heat stress (38°C), approximately 25% of the Arabidopsis seedling transcriptome is targeted for XRN4-dependent degradation, including both polysome-associated and polysome-released mRNAs (Merret et al., 2013). The heat-induced decay of polysomal mRNAs is dependent on LA RELATED PROTEIN 1A (LARP1A). High heat triggers the accumulation of XRN4 and the decapping proteins DECAPPING1 (DCP1) and DCP2 in polysomes (Merret et al., 2013) and seems to coincide with ribosome pausing in 5′ mRNA regions to elicit XRN4-dependent decay (Merret et al., 2015). It will be of interest to know how heat triggers the prerequisite mRNA decapping and whether the discrimination of mRNAs for heat-regulated turnover is integrated with the recently uncovered role of eIF5B1 in maintaining efficient translation of a subset of mRNAs, notably those that encode low-Mr nonstress proteins, during heat stress in Arabidopsis (Zhang et al., 2017a).
miRNA-Triggered Decay of Translating mRNAs
miRNA-mediated gene silencing regulates key biological processes in plants, including the plasticity of development in the context of the environment (for review, see Ferdous et al., 2015; Fouracre and Poethig, 2016; D’Ario et al., 2017). The fully processed 21- or 22-nucleotide miRNA and its effector ARGONAUTE1 (AGO1) form a core RNA-induced silencing complex (RISC) that controls the fate of specific mRNAs through a mechanism that involves endonucleolytic cleavage and/or translational repression (Rogers and Chen, 2013). Slicing of miRNA targets by AGO1 generates two truncated cleavage products, termed RISC 5′-and 3′-cleaved fragments, which are further degraded by the cellular RNA decay machinery. In Arabidopsis, the RISC 3′-cleaved fragment contains a free 5′-monophosphate, making it a substrate for XRN4 (Souret et al., 2004). RISC 5′-cleaved fragments undergo rapid turnover upon uridylation at their 3′ end by the miRNA nucleotidyl transferase HEN1 SUPPRESSOR1 (Ren et al., 2014). The pathways participating in RISC 5′-cleaved fragment demise could include 5′-3′ decay by XRN4 (Ren et al., 2014), although the mechanism of the prerequisite removal of the 5′ cap from these fragments is largely unknown. Alternatively, the uridylated fragments may be degraded in the 3′ to 5′ direction by the exosome (see below). This is supported by the observation of their increased abundance in genotypes of Arabidopsis deficient in exosome cofactor SUPERKILLER (SKI) proteins (Branscheid et al., 2015). Recently, Arabidopsis RISC-INTERACTING CLEARING 3′-5′ EXORIBONUCLEASE1 (RICE1) and RICE2 were identified as AGO1- and AGO10-binding proteins that degrade uridylated RISC 5′-cleaved fragments and facilitate RISC recycling (Zhang et al., 2017b).
The copurification of AGO1 and several miRNAs with Arabidopsis polysomes was reported some time ago (Lanet et al., 2009), but only recent coupling of mutant analyses, subcellular fractionation, and ribosome footprinting methods has clearly demonstrated their preferential association with membrane-bound polysomes (MBPs), including those at the endoplasmic reticulum (Li et al., 2013, 2016). These studies show that both translational repression and cleavage of miRNA targets occur mainly in these subcellular domains. Notably, MBP association of AtAGO1 is required for miRNA-triggered production of phased secondary small interfering RNAs (phasiRNAs) from select RNAs (e.g. TRANS-ACTING SIRNA [TAS] transcripts; Li et al., 2016). This is corroborated by the presence of two proteins required for phasiRNA biogenesis, SUPPRESSOR OF GENE SILENCING3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE6, in specific granules called siRNA bodies that reside on the cytoplasmic side of membrane compartments (Jouannet et al., 2012). The position of ribosome footprints generated from MBPs suggests that ribosome occupancy contributes to defining the site of phasiRNA biogenesis on these transcripts (Li et al., 2016). Indeed, functional translation of a positionally conserved short ORF of the AtTAS3 transcript is critical to efficient tasiRNA production (Bazin et al., 2017).
The complex interactions between miRNA-targeted mRNAs and the translational and degradation apparatus raise new questions about the subcompartments of the cytoplasm where these processes take place. Although miRNA-mediated translational repression involves MBPs, prior protein colocalization studies suggest that these processes occur in PBs (Brodersen et al., 2008; Yang et al., 2012). The location of XRN4-mediated decay of the RISC 5′- and 3′-cleaved fragments could be in PBs or freely in the cytosol, since XRN4 is present in both locations (Weber et al., 2008).
GENERAL CYTOPLASMIC mRNA DEGRADATION
General cytoplasmic mRNA degradation in eukaryotic cells is initiated by the removal of the 3′ poly(A) tail, a reversible step known as deadenylation, followed by 5′-cap hydrolysis (decapping) and subsequent 5′ to 3′ mRNA decay by XRN1/4 (Siwaszek et al., 2014). Both processes may occur in but are not confined to PBs. Alternatively, deadenylated mRNAs can be degraded in the 3′ to 5′ direction by the exosome. This section summarizes our current understanding of deadenylation-dependent mRNA turnover machinery and mechanisms as well as the significance of their regulation in plants (Fig. 2; Table I).
Table I. Proteins involved in the regulation of bulk cytoplasmic mRNA degradation in plants.
AtRRP44A, AtRRP6L1, and AtRRP6L2 are components of the exosome complex, but they are not included here because subcellular localization data suggest that they are nuclear proteins and, hence, components of the nuclear exosome (Lange et al., 2008; Zhang et al., 2010). At, Arabidopsis thaliana; Ca, Capsicum annuum; Nb, Nicotiana benthamiana; Os, Oryza sativa; Ta, Triticum aestivum.
| Protein | Molecular Function | Gene Symbol | Gene Identifier | Biological Functions | References |
|---|---|---|---|---|---|
| Deadenylation | |||||
| CCR4-NOT complex | |||||
| Carbon catabolite repressor4 | 3′-5′ exoribonuclease, EEP superfamily | AtCCR4a | AT3G58560 | Suc and starch metabolism | Suzuki et al. (2015) |
| AtCCR4b | AT3G58580 | ||||
| OsCCR4a | Os10g27230 | Unknown | Chou et al. (2017) | ||
| OsCCR4b | Os03g07080 | ||||
| CCR4-associated factor1 | 3′-5′ exoribonuclease, DEDD superfamily | AtCAF1a | AT3G44260 | Defense response to bacterial pathogens; abiotic stress response | Liang et al. (2009); Walley et al. (2010) |
| AtCAF1b | AT5G22250 | ||||
| OsCAF1A | Os08g34170 | Unknown | Chou et al. (2014, 2017) | ||
| OsCAF1B | Os04g58810 | ||||
| OsCAF1G | Os09g24990 | ||||
| OsCAF1H | Os02g55300 | ||||
| CaCAF1 | Development and defense response | Sarowar et al. (2007) | |||
| Negative on TATA1 | CCR4-NOT complex scaffold | OsNOT1 | Os10g40780 | Unknown | Chou et al. (2017) |
| Nocturnin | Poly(A)-specific 3′-5′ exoribonuclease, EEP superfamily | AtHESP | AT1G31500 | Circadian clock regulation | Delis et al. (2016) |
| Poly(A) RNase | Poly(A)-specific 3′-5′ exoribonuclease, DEDD superfamily | AtPARN/AHG2 | AT1G55870 | Embryogenesis; ABA sensitivity; abiotic stress response | Reverdatto et al. (2004); Nishimura et al. (2005) |
| 5′-3′ mRNA decay | |||||
| Decapping complex | |||||
| Decapping1 | Decapping cofactor | AtDCP1 | AT1G08370 | Postembryonic development; dehydration and heat stress response | Xu et al. (2006); Xu and Chua (2012); Motomura et al. (2015) |
| Decapping2 | Decapping enzyme | AtDCP2/TDT | AT5G13570 | Postembryonic development; heat stress response | Xu et al. (2006); Motomura et al. (2015) |
| Decapping5 | Decapping enhancer | AtDCP5 | AT1G26110 | Postembryonic development; osmotic stress response | Xu and Chua (2009, 2012) |
| Enhancer of mRNA decapping4 | Core decapping complex scaffold | AtEDC4/VCS | AT3G13300 | Postembryonic development; osmotic stress response; seed dormancy and germination | Xu et al. (2006); Goeres et al. (2007); Soma et al. (2017) |
| TaEDC4 | Traes_6DL_3FBA5B70E | Plant-fungal pathogen interaction | Petre et al. (2016) | ||
| LSM1-7 complex | Decapping enhancer | AtLSM1a | AT1G19120 | Postembryonic development; heat stress tolerance; ABA-mediated response to cold, drought, and salt tolerance | Perea-Resa et al. (2012, 2016); Golisz et al. (2013); Okamoto et al. (2016) |
| AtLSM1b | AT3G14080 | ||||
| AtLSM2 | AT1G03330 | ||||
| AtLSM3a | AT1G21190 | ||||
| AtLSM3b | AT1G76860 | ||||
| AtLSM4 | AT5G27720 | ||||
| AtLSM5 | AT5G48870 | ||||
| AtLSM6a | AT2G43810 | ||||
| AtLSM6b | AT3G59810 | ||||
| AtLSM7 | AT2G03870 | ||||
| Protein-associated with topoisomerase1 | Decapping enhancer, translation repressor? | AtPAT1 | AT1G79090 | Immune response | Roux et al. (2015) |
| DEAD-box RNA helicase | AtDHH1 | AT3G61240 | Unknown | Xu et al. (2006); Bhullar et al. (2017) | |
| Asymmetric leaf2 | Decapping activator | AtAS2 | AT1G65620 | Plant-virus interaction | Ye et al. (2015) |
| 5′-3′ mRNA decay | |||||
| Exoribonuclease4 | 5′-3′ exoribonuclease | AtXRN4/EIN5 | AT1G54490 | Ethylene response/signaling; heat stress response | Potuschak et al. (2006); Merret et al. (2013) |
| NbXRN4 | gi|242037819 | Defense response to viral pathogen | Peng et al. (2011); Lee et al. (2016) | ||
| LA-related protein1A | Heat-specific cofactor of AtXRN4 | AtLARP1A | AT5G21160 | Heat stress response | Merret et al. (2013) |
| 3′-5′ mRNA decay | |||||
| Cytoplasmic exosome | |||||
| rRNA processing proteins | RNase PH domain-containing proteins; hexameric core ring subunits of exosome | AtRRP41 | AT3G61620 | Female gametogenesis | Chekanova et al. (2007) |
| AtRRP42 | AT3G07750 | Unknown | Chekanova et al. (2007) | ||
| AtRRP43 | AT1G60080 | Unknown | Chekanova et al. (2007) | ||
| AtRRP45A | AT3G12990 | Essential for viability; cuticular wax biosynthesis | Hooker et al. (2007) | ||
| AtRRP45B/CER7 | AT3G60500 | ||||
| AtRRP46 | AT3G46210 | Unknown | Chekanova et al. (2007) | ||
| AtRRP41L/MTR3 | AT4G27490 | Germination and seedling development | Yang et al. (2013) | ||
| S1/KH domain-containing proteins; cap structure subunits of core exosome | AtRRP4 | AT1G03360 | Postzygotic development | Chekanova et al. (2007) | |
| AtRRP40A | AT2G25355 | Unknown | Chekanova et al. (2007) | ||
| AtRRP40B | AT4G32175 | Unknown | Chekanova et al. (2007) | ||
| AtCSL4 | AT5G38885 | Unknown | Chekanova et al. (2007) | ||
| RNase II-type 3′-5′ exoribonuclease | AtRRP44B/SOV | AT1G77680 | Compensation of VCS function in postembryonic development | Zhang et al. (2010); Kumakura et al. (2013) | |
| RNase D-type 3′-5′ exoribonuclease, DEDD superfamily | AtRRP6L3 | AT2G32415 | Unknown | Lange et al. (2008) | |
| Superkiller (SKI) complex | |||||
| Superkiller proteins | Auxiliary factors of the exosome | AtSKI2 | AT3G46960 | Cuticular wax biosynthesis | Zhao and Kunst (2016) |
| AtSKI3 | AT1G76630 | ||||
| AtSKI8/VIP3 | AT4G29830 | ||||
Deadenylation
Progressive cytoplasmic mRNA poly(A) tail shortening displaces PABPs from the mRNA 3′ end, likely disrupting the 5′-cap to 3′-tail (eIF4G-PABP) interaction that promotes translational reinitiation (Gallie, 2014), shifting mRNAs toward a translationally silent state and exposing the 3′ terminus to exonucleases (Łabno et al., 2016). This first, rate-limiting step of mRNA decay is catalyzed by Mg2+-dependent 3′-5′ exoribonucleases called deadenylases that are classified into two superfamilies: EEP (exonuclease-endonuclease-phosphatase) and DEDD (Asp-Glu-Asp-Asp) nucleases (Siwaszek et al., 2014).
In yeast and metazoa, shortening and removal of the mRNA 3′ poly(A) tail is performed by two major deadenylase complexes: poly(A)-nuclease PAN2-PAN3 and CARBON CATABOLITE REPRESSOR4-NEGATIVE ON TATA (CCR4-NOT; Wahle and Winkler, 2013). The PAN2-PAN3 complex is a heterotrimer of the DEDD-type PAN2 deadenylase interacting with a PAN3 homodimer (Siwaszek et al., 2014). Interestingly, the PAN2-PAN3 complex is present across higher eukaryotes, including the green alga Chlamydomonas reinhardtii, but has not been recognized to date in flowering plants. The CCR4-NOT complex contains two catalytic subunits (i.e. CCR4 and CAF1) and at least seven additional proteins (NOT1–NOT5, CAF40, and CAF130), with NOT1 serving as a scaffold (Miller and Reese, 2012). Of these, CCR4, CAF1, and NOT1 orthologs have been identified in plants (Table I). Arabidopsis encodes two CCR4 paralogs, AtCCR4a and AtCCR4b, both of which produce gene products that interact with the decapping complex proteins AtDCP1 and AtDCP2 within and outside of PBs (Suzuki et al., 2015). Mutant analyses have identified some important functions of these functionally redundant EEP deadenylases. For example, they appear to influence Suc and starch metabolism by controlling the poly(A) tail length and steady-state level of GRANULE-BOUND STARCH SYNTHASE1 mRNA (Suzuki et al., 2015). The expansive Arabidopsis CAF1 family includes 11 members. The products of two of these, CAF1a and CAF1b, exhibit 3′-5′ exonucleolytic activity in vitro via their conserved DEDD domain, contribute to effective defense response (Liang et al., 2009), and regulate the poly(A) length of a defined set of stress-related transcripts rather than acting as general deadenylases (Liang et al., 2009; Walley et al., 2010).
The study of rice deadenylases has provided further insight. OsCCR4a and OsCCR4b are located predominantly in visible foci, presumably PBs, that include XRN4. The EEP domain of OsCCR4s is sufficient for in vitro 3′-5′ exonuclease activity on poly(A), poly(U), and poly(C) substrates (Chou et al., 2017). Similarly, recombinant OsCAF1A, OsCAF1B, OsCAF1G, and OsCAF1H are 3′-5′ exonucleases, with at least OsCAF1B associating with XRN4 in PB-like foci (Chou et al., 2014). In yeast and mammals, CAF1 serves as a bridge between CCR4 and NOT1 via interactions between the N-terminal Leu-rich repeat of CCR4 and the MIF4G domain of NOT1. The rice CCR4-NOT complex appears to share this topology, although the N-terminal Leu-rich repeat domain of CCR4s is absent and the interaction with CAF1 is apparently replaced by an N-terminal zf-MYND-like domain of CCR4 (Chou et al., 2017).
Two other conserved deadenylases control the turnover of specific transcripts. The first is the EEP-type deadenylase Nocturnin that is distinguished by its circadian clock-regulated transcript abundance in multicellular eukaryotes (Godwin et al., 2013). Arabidopsis HESPERIN (AtHESP) is closely related to mammalian Nocturnin, with maximal mRNA levels in the evening. Intriguingly, altered AtHESP expression affects the rhythmicity of the core clock oscillator mRNAs TIMING OF CAB EXPRESSION1 and CIRCADIAN CLOCK ASSOCIATED1 (Delis et al., 2016). The second is the homodimeric DEDD-class PARNs found in plants and humans but apparently absent from budding yeast (Saccharomyces cerevisiae) and fruit flies (Drosophila melanogaster; Godwin et al., 2013). Arabidopsis PARN (AtPARN)/ABA HYPERSENSITIVE GERMINATION2 (AHG2) is required for the poly(A) tail shortening of select embryonic transcripts, affecting embryonic development and some stress responses (Chiba et al., 2004; Reverdatto et al., 2004; Nishimura et al., 2005). Altogether, the diversity of factors involved in deadenylation indicates significant control of deadenylation, the first step of general mRNA turnover.
Decapping
Removal of the mRNA 5′-7mG-cap inhibits translation initiation and commits a deadenylated mRNA to complete degradation. mRNA decapping is carried out by DCP2, a conserved bilobed Nudix family enzyme that hydrolyzes the 5′-cap structure in a divalent cation (Mn2+/Mg2+)-dependent manner, releasing 7mGDP and a 5′-monophosphorylated mRNA (Arribas-Layton et al., 2013). DCP2 functions in a multisubunit decapping complex, which includes cofactors that mediate its activity and regulators that coordinate decapping with translational repression, deadenylation, and hydrolysis of the mRNA body (Table I). In Arabidopsis, DCP1 and VARICOSE (VCS)/ENHANCER OF mRNA DECAPPING4 (EDC4) directly modulate DCP2 activity (Xu et al., 2006). All three proteins are considered core decapping complex factors, indispensable for mRNA decapping and essential for postembryonic development (Xu and Chua, 2011). Colocalization of AtDCP1 and AtDCP2 in macromolecular foci detectable by confocal microscopy is dynamically regulated by heat stress (Motomura et al., 2015). Based on mutant analyses, the factor VCS regulates seed dormancy and germination in Arabidopsis (Basbouss-Serhal et al., 2017) and functions in plant-fungal pathogen interactions in Nicotiana benthamiana (Petre et al., 2016). Additionally, AtDCP5, an Sm-like (LSM) domain-containing RNA-binding protein (RBP), interacts with DCP1 and DCP2. Unlike DCP1 and VCS, DCP5 does not directly regulate DCP2 activity; however, it is required for PB formation, mRNA decapping, and translational repression associated with mRNA decay. Attenuation of DCP5 function does not result in lethality but causes developmental defects similar to loss-of-function mutants of core complex proteins, including perturbation of germination and leaf venation (Xu and Chua, 2009), indicating that these developmental processes require tight regulation of mRNAs mediated by the decapping pathway.
Phosphorylation of specific residues of the core decapping proteins constitutes a crucial mechanism for the osmotic stress response in plants. Under this stress, Arabidopsis DCP1 is phosphorylated by MAPK6 (MPK6), leading to enhancement of its association with DCP5, which, in turn, promotes the RNA-binding capacity of DCP5 and the mRNA-decapping activity of DCP2 (Xu and Chua, 2012). By contrast, VCS is phosphorylated by the abscisic acid (ABA)-unresponsive osmotic stress-activated subclass I of SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2s (SnRK2s). Osmotic stress triggers SnRK2 localization in PBs, where they appear to phosphorylate and regulate VCS function (Soma et al., 2017). Mutations that prevent DCP1 or VCS phosphorylation render Arabidopsis plants susceptible to osmotic stress (Xu and Chua, 2012; Soma et al., 2017). These observations firmly suggest that transcriptome adjustments mediated by mRNA decapping are relevant to stress resilience.
In yeast and metazoa, the heptameric LSM1-7 complex, in association with PROTEIN-ASSOCIATED WITH TOPOISOMERASE1 (PAT1), binds to the 3′ end of deadenylated mRNAs and promotes decapping (Tharun, 2009; Haas et al., 2010). Simultaneously, PAT1 interacts with the DEAD-box RNA helicase DHH1/DDX6. Both PAT1 and DDH1 play a role in translational inhibition as well as PB formation (Coller and Parker, 2005; Ozgur et al., 2010; Sharif et al., 2013). This links deadenylation with translational repression and decapping. The LSM1-7 complex has been characterized in Arabidopsis. Genetic analysis of the paralogs AtLSM1A and AtLSM1B revealed their functional redundancy and importance in normal development (Perea-Resa et al., 2012; Golisz et al., 2013). Similar to DCP1, DCP2, and VCS, both LSM1A and LSM1B accumulate in PBs under heat, cold, drought, and salt stress and are required for conditional PB assembly under these conditions (Perea-Resa et al., 2012, 2016). Intriguingly, the Arabidopsis LSM1s differentially regulate tolerance to abiotic stresses by dynamically controlling the decapping and degradation of a select group of stress-responsive transcripts, including those involved in ABA biosynthesis (Perea-Resa et al., 2016). Mutant analysis shows that the LSM5 component of the LSM1-7 complex is essential for heat tolerance in Arabidopsis, targeting both functional and aberrant transcripts for degradation during heat stress (Okamoto et al., 2016). A proteomic analysis revealed that AtLSM1A forms complexes with VCS, PAT1, DHH1/DDX6-like, and NOT1-like proteins (Golisz et al., 2013), so its interaction with the deadenylation machinery or the 3′ deadenylated mRNA remains somewhat unclear.
Arabidopsis PAT1 interacts with LSM1B and regulates the decapping of select transcripts. It is an MPK4 phosphorylation target that accumulates in PBs in response to the bacterial pathogen-associated molecular pattern flagellin22. PAT1 and MPK4 suppress plant autoimmunity, as loss of their function results in the ENHANCED DISEASE SUSCEPTIBILITY1-mediated constitutive immune response through a pathway downstream of the immune receptor SUPPRESSOR OF MKK1 MKK2 2 (Roux et al., 2015). Studies of the Arabidopsis LSM1-7-PAT1 complex extend the contribution of mRNA decapping from abiotic to biotic stress signaling and suggest that both components and architecture of the mRNA-decapping complex of yeast and metazoa are conserved in plants. An Arabidopsis DHH1-like protein has been identified and shown to localize in DCP1 and DCP2 granules (Xu et al., 2006; Bhullar et al., 2017), but whether this protein interacts with PAT1 and functions in translational repression and mRNA decapping has yet to be elucidated.
Arabidopsis also possesses noncanonical, condition-specific decapping activators important for abiotic and biotic stress responses. For example, the BEACH domain-containing protein SPIRRIG (SPI) is a regulator of cellular membrane dynamics that localizes into PBs under salt stress via its interaction with DCP1. SPI function is required for salt stress-triggered PB assembly, and it selectively stabilizes and destabilizes a subset of salt-responsive transcripts by regulating their translocation into PBs (Steffens et al., 2015). ASYMMETRIC LEAF2 (AS2) is a transcription factor playing a role in leaf development. Intriguingly, AS2 colocalizes with PBs as a result of its trafficking there by the geminivirus nuclear shuttle protein BV1 (Ye et al., 2015). This apparently promotes DCP2 decapping activity to accelerate host mRNA decay, leading to the suppression of host antiviral gene silencing. There are numerous examples of viral conscription of the translational apparatus (Browning and Bailey-Serres, 2015); this example illustrates a bizarre means by which a virus manipulates mRNA turnover.
5′-3′ Degradation
In eukaryotes, the hydrolysis of 5′-monophosphorylated (uncapped) transcripts into nucleotides is executed by Mg2+-dependent 5′-3′ exoribonucleases of the XRN family, with XRN4 functioning as the cytoplasmic enzyme in Arabidopsis (Kastenmayer and Green, 2000). Unlike the core decapping proteins, disruption of AtXRN4 function only results in mild morphological defects under standard growth conditions. This may reflect reliance on AtXRN4 for the degradation of specific transcripts (Rymarquis et al., 2011). Nevertheless, XRN4-mediated translational repression and mRNA decay play a role in seed dormancy and germination (Basbouss-Serhal et al., 2017), ethylene signaling (Li et al., 2015; Merchante et al., 2015), heat stress response and tolerance (Merret et al., 2013; Nguyen et al., 2015), and viral defense (Jaag and Nagy, 2009; Peng et al., 2011; Lee et al., 2016). An alternative explanation is that, in the absence of XRN4 function, the 3′-5′ degradation pathways described below may maintain homeostasis.
3′-5′ Degradation
After deadenylation, the 3′ end of mRNAs can be hydrolyzed directly by an evolutionarily conserved complex of multiple 3′-5′ exoribonucleases called the exosome. In Arabidopsis, as in other eukaryotes, the exosome core complex is composed of nine subunits. Six are RNase PH domain-containing proteins (RIBOSOMAL RNA PROCESSING PROTEIN41 [RRP41], RRP42, RRP43, RRP45, RRP46, and RRP41L/MTR3) that together form a hexameric core ring that is stabilized by the attachment of a cap of three S1/KH-domain proteins (RRP4, RRP40, and CSL4; Lange and Gagliardi, 2010). Whereas all nine core exosome subunits have equal contributions to the structural and functional integrity of the yeast and mammalian exosome, each Arabidopsis exosome subunit appears to play a distinct and, in some cases, dispensable role suggesting target specificity (Table I). For example, AtCSL4 is apparently dispensable and AtRRP4 alone provides the 3′-5′ exoribonuclease activity (Chekanova et al., 2002, 2007), whereas in yeast and mammals, exosome core subunits are catalytically inactive and additionally require RRP6 and/or RRP44 for the 3′-5′ exoribonuclease activity.
The Arabidopsis genome encodes three RRP6 paralogs: AtRRP6L1, AtRRP6L2, and AtRRP6L3. Of these gene products, only RRP6L3 was shown to be cytosolic, but its role in mRNA degradation is unknown (Lange et al., 2008). There are two AtRRP44-like genes: AtRRP44A and AtRRP44B/SUPPRESSOR OF VARICOSE (SOV). RRP44A is an active nucleus-localized RNase that functions in rRNA processing (Kumakura et al., 2016), whereas RRP44B is a cytosolic RNase that forms microscopic PB-like particles. AtRRP44A and RRP44B/SOV fall into distinct phylogenetic clades present across the animal and plant kingdoms (Zhang et al., 2010). The latter lacks the N-terminal PIN domain required for binding to the exosome core. It is suggested that RRP44A is a component of the nuclear exosome whereas RRP44B/SOV is a solo cytoplasmic exoribonuclease (Kumakura et al., 2013). Notably, RRP44B/SOV can partially complement the function of VCS in cytoplasmic mRNA degradation in some Arabidopsis ecotypes (Zhang et al., 2010). This indicates a general functional redundancy of the 3′-5′ decay apparatus, although it seems possible that the different complexes may have some specificity with respect to individual mRNAs, developmental stages, or growth conditions.
The activity of the cytoplasmic exosome is dependent on the SKI complex, a tetrameric assembly of a SKI8 dimer and the RNA helicase SKI2 connected by their mutual interaction with the scaffold protein SKI3, which unwinds and threads RNA substrates into the exosome for degradation (Halbach et al., 2013). Arabidopsis SKI2, SKI3, and SKI8 form a stable complex in vivo, with SKI2 and SKI3 also present in cytoplasmic foci (Dorcey et al., 2012; Zhang et al., 2015; Zhao and Kunst, 2016). Disruption of AtSKI8 function results in the stabilization of mRNAs targeted for degradation by the exosome (Dorcey et al., 2012). Similar to the exosome core components RRP45A and RRP45B, all three SKI proteins are required for the exosomal degradation of the AtECERIFERUM3 transcript during stem wax deposition (Zhao and Kunst, 2016).
mRNA SEQUESTRATION IN SGS: A TRANSIT DEPOT BETWEEN TRANSLATION, STORAGE, AND DECAY?
Polysome disassembly as a consequence of translational repression induces the sequestration of mRNAs into translationally inactive mRNPs. SGs are one type of mRNPs that form in response to stress-induced inhibition of translation initiation, serving as triage centers for mRNA sorting to PBs or protection from degradation (Protter and Parker, 2016; see Box 1). These heterogenous complexes are physically, compositionally, and functionally linked with PBs (Buchan and Parker, 2009). Typically, mRNAs stored in SGs are translationally competent and can reenter the translational pool once released. In plants, this is exemplified by Arabidopsis mRNAs encoding ribosomal proteins that are preferentially stored in SGs under heat shock stress and released and translated during recovery through a mechanism that requires HEAT SHOCK PROTEIN101 (HSP101; Merret et al., 2017).
In yeast and humans, SGs contain a diverse proteome (Jain et al., 2016). Some RBPs are essential for SG formation in these species, such as T-cell-restricted intracellular antigen-1 (TIA-1) and TIA-1-related (TIAR) proteins (Gilks et al., 2004), Ras-GTPase-activating protein SH3-domain-binding proteins (G3BPs; Aulas et al., 2015), and Tudor staphylococcal nucleases (TSNs; Gao et al., 2015). Recent studies have revealed the roles of related proteins in plants (Fig. 2; Table II).
Table II. Proteins associated with SGs and PBs in plants.
CCR4s, CAF1s, XRN4, and components of the mRNA-decapping complex are localized PBs but are only presented in Table I. AtLARP1A, a heat-specific cofactor of XRN4-mediated cotranslational decay, also accumulates in SGs and PBs under heat shock stress and is presented in Table I. All 11 Arabidopsis tandem zinc finger proteins are localized to cytoplasmic granules; however, only those with demonstrated colocalizations with known SG and/or PB components are included. AtSGS3 and AtAGO7 are typically present in distinct siRNA bodies but become positive for SG markers during heat stress. At, Arabidopsis thaliana; Os, Oryza sativa.
| Protein | Symbol | Gene Identifier | mRNP | Biological Functions | References |
|---|---|---|---|---|---|
| Eukaryotic translation initiation factors | AteIF4E | AT4G18040 | SG | Heat stress response | Weber et al. (2008) |
| AteIF3b1 | AT5G27640 | SG | Suzuki et al. (2015) | ||
| Poly(A)-binding proteins | AtPABP2 | AT4G34110 | SG | Hypoxia stress response | Weber et al. (2008); Sorenson and Bailey-Serres (2014) |
| AtPABP8 | AT1G49760 | SG | Heat stress response | ||
| RNA-binding proteins45 and -47 | AtRBP47B | AT1G19130 | SG | Heat stress response | Weber et al. (2008) |
| Oligouridylate binding proteins1 | AtUBP1A | AT1G54080 | SG | Hypoxia stress response | Sorenson and Bailey-Serres (2014) |
| AtUBP1B | AT1G17370 | SG | Salt, osmotic, and heat stress response/tolerance; ABA sensitivity | Weber et al. (2008); Nguyen et al. (2016, 2017) | |
| AtUBP1C | AT3G14100 | SG | Hypoxia stress response/tolerance; Suc-reversible seedling growth arrest | Sorenson and Bailey-Serres (2014) | |
| Ras-GAP SH3 domain-binding protein | AtG3BP-like | AT5G43960 | SG | Plant-virus interaction | Krapp et al. (2017) |
| Tudor staphylococcal nucleases | AtTSN1 | AT5G07350 | SG, PB | GA-mediated growth response under salt stress; heat stress response | Yan et al. (2014); Gutierrez-Beltran et al. (2015) |
| AtTSN2 | AT5G61780 | SG, PB | |||
| Tandem zinc finger proteins | AtTZF1 | AT2G25900 | SG, PB | Sugar and ABA-mediated stress response; GA-dependent growth response | Pomeranz et al. (2010b); Lin et al. (2011) |
| OsTZF1 | Os05s10670 | SG, PB | Salt and drought stress tolerance | Jan et al. (2013) | |
| AtTZF4 | AT1G03790 | SG, PB | ABA-, GA-, and phytochrome-mediated seed germination | Bogamuwa and Jang (2013, 2016) | |
| AtTZF5 | AT5G44260 | SG, PB | |||
| AtTZF6 | AT5G07500 | SG, PB | |||
| AtTZF9 | AT5G58620 | PB | Pathogen-associated molecular pattern-triggered immunity | Maldonado-Bonilla (2014) | |
| Mediator of ABA-regulated dormancy1 | AtMARD1 | AT3G63210 | SG, PB | Interacting partner of AtTZF4, AtTZF5, and AtTZF6 | Bogamuwa and Jang (2016) |
| Responsive to dehydration21A | AtRD21A | AT1G47128 | SG, PB | Interacting partner of AtTZF4, AtTZF5, and AtTZF6 | Bogamuwa and Jang (2016) |
| Calmodulin-like38 | AtCML38 | AT1G76650 | SG | Hypoxia response/tolerance | Lokdarshi et al. (2016) |
| Vascular plant one-zinc-finger protein2 | AtVOZ2 | AT2G42400 | SG, PB | Heat stress response | Koguchi et al. (2017) |
| Heat shock protein101 | AtHSP101 | AT1G74310 | SG | Heat stress response | Merret et al. (2017) |
| Suppressor of gene silencing3 | AtSGS3 | AT5G23570 | SG | Heat stress response | Jouannet et al. (2012) |
| Argonaute7 | AtAGO7 | AT1G69440 | SG | Heat stress response | Jouannet et al. (2012) |
| BEACH domain-containing protein | AtSPIRRIG/AtSPI | AT1G03060 | PB | Salt stress tolerance | Steffens et al. (2015) |
| GW-repeat protein | AtSUO | AT3G48050 | PB | miRNA-mediated translational repression; control of vegetative phase change | Yang et al. (2012) |
| TBP-associated factor1b | AtTAF15b | AT5G58470 | PB | TNL-mediated plant immunity | Dong et al. (2016) |
| Plant SMY2-type ILE-GYF domain-containing protein1 | AtPSIG1 | AT5G42950 | PB | Regulation of cell death during plant-pathogen interaction | Matsui et al. (2017) |
| Polypyrimidine tract-binding proteins | AtPTB1 | AT3G01150 | PB | Pollen germination | Wang and Okamoto (2009); Stauffer et al. (2010) |
| AtPTB2 | AT5G53180 | PB | |||
| Ethylene insensitive2 | AtEIN2 | AT5G03280 | PB | Ethylene response | Li et al. (2015); Merchante et al. (2015) |
| Up frameshift1 | AtUPF1 | AT5G47010 | PB | Nonsense-mediated decay; ethylene response | Kerényi et al. (2013); Merchante et al. (2015) |
| Suppressor with morphological effect on genitalia7 | AtSMG7 | AT5G19400 | PB | Nonsense-mediated decay; TNL-mediated plant immunity | Mérai et al. (2013); Gloggnitzer et al. (2014) |
TIA-1/TIARs are composed of three N-terminal RNA recognition motifs (RRMs) that provide RNA/DNA-binding specificity and a C-terminal prion-related domain that confers self-aggregation, both of which are required for SG formation (Waris et al., 2014). In plants, RBP45/47 and OLIGOURIDYLATE BINDING PROTEIN1 (UBP1) are the triple RRM families most closely related to the animal TIA-1/TIARs. Of the Arabidopsis RBP45/47 family, RBP47B is discernible in the nucleus under normal conditions and relocates to cytoplasmic foci detectable by confocal microscopy in response to heat, salt, and hypoxia stress (Weber et al., 2008; Yan et al., 2014; Gutierrez-Beltran et al., 2015; Lokdarshi et al., 2016). The Arabidopsis UBP1 family is composed of three members, namely AtUBP1A, AtUBP1B, and AtUBP1C. UBP1A and UBP1C reversibly aggregate into cytoplasmic poly(A) mRNA granules upon hypoxic and heat stress (Sorenson and Bailey-Serres, 2014). Similarly, UBP1B reversibly forms SGs under heat stress and plays a crucial role in ABA signaling and heat stress tolerance (Nguyen et al., 2016, 2017). An analysis of RNA decay kinetics suggested that UBP1B complexes (UBP1B-SGs) protect stress-related mRNAs from degradation during heat stress (Nguyen et al., 2016). Likewise, UBP1C is required for plant survival of low-oxygen stress. UBP1C dynamically relocalizes from the nucleus to form cytoplasmic foci that are disassembled upon reoxygenation. Intriguingly, RNA-ribonucleoprotein immunoprecipitation followed by microarray analysis determined that AtUBP1C preferentially associates with mRNAs with U-rich 3′ UTRs under control conditions, but UBP1C-SGs collect mRNAs that are either stored or degraded as opposed to preferentially translated during hypoxia (Sorenson and Bailey-Serres, 2014).
G3BPs are phosphorylation-dependent, sequence-specific endoribonucleases that interact with Ras GTPase-activating proteins. In mammalian cells, G3BP1 is a potent SG-nucleating protein that constitutes stable cores of SGs (Jain et al., 2016). As such, G3BP1 is targeted and sequestered by several viral proteins to prevent SG assembly/sequestration of viral RNAs during infection (Reineke and Lloyd, 2013). The first plant G3BP-like protein was isolated from N. benthamiana together with other SG components as targets of the viral nonstructural protein3 of Semliki Forest virus (Krapp et al., 2017). This protein is closely related to the NUCLEAR TRANSPORT FACTOR2 (NTF2) family in Arabidopsis. This G3BP-like NTF2-RRM aggregates into SGs and interacts with the nuclear shuttle protein of the begomovirus Abutilon mosaic virus and Pea necrotic yellow dwarf virus upon heat stress (Krapp et al., 2017). The roles of plant G3BP-like proteins in SG nucleation and mRNA stabilization await further studies.
TSNs are evolutionarily conserved cytoskeleton-associated RBPs characterized by four complete N-terminal staphylococcal nuclease domains followed by a central Tudor domain and a partial SN domain at the C terminus (Gutierrez-Beltran et al., 2016). A plant TSN was first described in rice as an RBP required for the transport of a subgroup of seed storage RNAs to a subdomain of the endosperm endoplasmic reticulum (Tian and Okita, 2014). Arabidopsis encodes functionally redundant TSN1 and TSN2, which play a pivotal role in seed germination and response to high salinity and heat stress (Frei dit Frey et al., 2010; Yan et al., 2014; Gutierrez-Beltran et al., 2015). Root transcriptome and mRNA decay data suggest that TSN1 and TSN2 protect a specific set of transcripts from degradation during salinity stress, including mRNAs encoding GA 20-OXIDASE3, a key enzyme in GA biosynthesis (Yan et al., 2014). In contrast to animal TSNs that seem to localize exclusively to SGs (Gao et al., 2015), the Arabidopsis TSNs are targeted to both SGs and PBs and function in mRNA decapping during heat stress, suggestive of a crucial role in the structural integrity as well as the molecular identity of both SGs and PBs (Gutierrez-Beltran et al., 2015).
Tandem CCCH zinc finger (TZF) proteins constitute another RBP family that associates with SGs and PBs. Plant TZFs are characterized by two zinc-binding CCCH motifs arranged in tandem, with an Arg-rich motif upstream of the TZF motifs (Bogamuwa and Jang, 2014). Arabidopsis encodes 11 TZFs, encoding proteins localized to cytoplasmic granules when expressed heterologously in maize (Zea mays) mesophyll protoplasts (Pomeranz et al., 2010a). AtTZFs play diverse roles in growth, development, and stress responses (Pomeranz et al., 2010b; Lin et al., 2011; Bogamuwa and Jang, 2014). AtTZF1 shuttles between the nucleus and PBs under ambient temperatures but relocalizes into SGs after heat shock (Pomeranz et al., 2010b). Through its Arg-rich and TZF motifs, AtTZF1 binds poly(U) RNAs in the presence of zinc and triggers the decay of AU-rich element-containing mRNAs (Qu et al., 2014). In rice, OsTZF1 is a negative regulator of leaf senescence under stress and also binds poly(U) RNAs. Its colocalization with SG and PB markers was enhanced after ABA and NaCl treatment (Jan et al., 2013). AtTZF4, AtTZF5, and AtTZF6 are expressed specifically in seeds and involved in seed germination (Bogamuwa and Jang, 2013). All three proteins interact physically in both SGs and PBs, along with MEDIATOR OF ABA-REGULATED DORMANCY1 and RESPONSIVE TO DEHYDRATION21A (Bogamuwa and Jang, 2016). The dual localization of these proteins in SGs and PBs hints that they could be involved in the selective sorting of transcripts from a stable state to en masse degradation.
Apart from RBPs, Arabidopsis studies have uncovered proteins with diverse cellular functions as constituents of SGs. These include the purported calcium sensor CALMODULIN-LIKE38 (CML38), which reversibly accumulates into SGs under hypoxic conditions in a Ca2+-dependent manner (Lokdarshi et al., 2016), and the NAM (no apical meristem), ATAF (Arabidopsis transcription activation factor), and CUC (cup-shaped cotyledon) family transcription factor VASCULAR PLANT ONE-ZINC-FINGER2 (VOZ2), which translocates from the cytosol to the nucleus and SGs/PBs during heat stress (Koguchi et al., 2017). AtCML38 function contributes to hypoxia tolerance (Lokdarshi et al., 2016), but it remains to be discovered how AtVOZ2 incorporation into SGs/PBs under heat stress contributes to mRNA metabolism and if this influences heat stress tolerance.
CONCLUSION
The processes of mRNA translation, stabilization, and turnover are intrinsically linked via highly complex and dynamic interactions of the macromolecular complexes that govern the function and fate of each transcript within the cytoplasm of plant cells. Recent studies have uncovered remarkable regulation of these processes, underscored strong conservation of mechanisms across eukaryotes, and illuminated functions that may be limited to the plant lineage. As mRNAs are exported from the nucleus, the pioneer round of translation appears to be the default quality-control pathway that licenses reiterative rounds of translation of a polysomal mRNA. It is now recognized that translation can be linked to mRNA decay or sequestration through cotranslational degradation or a more complex process involving the disassembly of a polysome complex and then advancement to decay or stabilization. Through mutant analyses, confocal microscopy, and omics technologies, the integration of mRNA translation, turnover, and sequestration has become recognized as a highly selective regulatory process of individual mRNAs. Due to space limitations, we did not delve upon how mRNA decay protects the cellular transcriptome from inappropriate posttranscriptional silencing of endogenous genes (for review, see Liu and Chen, 2016; Tsuzuki et al., 2017; Zhang and Guo, 2017). We also did not consider nuclear mechanisms of transcript turnover that can involve CBP80 (Yu et al., 2016). Nor did we consider N6-methyladenosine and other modifications of specific nucleotides (the epitranscriptome; Luo et al., 2014; Vandivier et al., 2015; Zuber et al., 2016; David et al., 2017), which may influence RNA-protein interactions and, hence, the nuclear or cytoplasmic fates of individual transcripts. The wealth of new discoveries of the dynamic cytoplasmic mRNP triumvirate has unearthed new challenging questions (see Outstanding Questions) that may provide insights relevant to other eukaryotes or of value to crop improvement.
Footnotes
This work was supported by the United States National Science Foundation grant no. MCB-1716913 to J.B.-S. and a Royal Thai Government's Development and Promotion of Science and Technology Talents Project scholarship to T.C.
Articles can be viewed without a subscription.
References
- Arribas-Layton M, Wu D, Lykke-Andersen J, Song H (2013) Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim Biophys Acta 1829: 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Caron G, Gkogkas CG, Mohamed NV, Destroismaisons L, Sonenberg N, Leclerc N, Parker JA, Vande Velde C (2015) G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J Cell Biol 209: 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbouss-Serhal I, Pateyron S, Cochet F, Leymarie J, Bailly C (2017) 5′ to 3′ mRNA decay contributes to the regulation of Arabidopsis seed germination by dormancy. Plant Physiol 173: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J (2017) Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA 114: E10018–E10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar DS, Sheahan MB, Rose RJ (2017) RNA processing body (P-body) dynamics in mesophyll protoplasts re-initiating cell division. Protoplasma 254: 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa S, Jang JC (2013) The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ 36: 1507–1519 [DOI] [PubMed] [Google Scholar]
- Bogamuwa S, Jang JC (2016) Plant tandem CCCH zinc finger proteins interact with ABA, drought, and stress response regulators in processing-bodies and stress granules. PLoS ONE 11: e0151574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa SP, Jang JC (2014) Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol 55: 1367–1375 [DOI] [PubMed] [Google Scholar]
- Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O, Brodersen P (2015) SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res 43: 10975–10988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation. The Arabidopsis Book 13: e0176, doi/10.1199/tab.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Dutko JA, Mian IS, Belostotsky DA (2002) Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3′→5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res 30: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ (2004) AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 328: 95–102 [DOI] [PubMed] [Google Scholar]
- Chou WL, Chung YL, Fang JC, Lu CA (2017) Novel interaction between CCR4 and CAF1 in rice CCR4-NOT deadenylase complex. Plant Mol Biol 93: 79–96 [DOI] [PubMed] [Google Scholar]
- Chou WL, Huang LF, Fang JC, Yeh CH, Hong CY, Wu SJ, Lu CA (2014) Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol Biol 85: 443–458 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R (2005) General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly DR, Smith AB, Murray KD, Estavillo GM, Searle I, Ford E, Bogdanović O, Lister R, Borevitz JO, et al. (2017) Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29: 1836–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Li W, An L (2016) NMD mechanism and the functions of Upf proteins in plant. Plant Cell Rep 35: 5–15 [DOI] [PubMed] [Google Scholar]
- D’Ario M, Griffiths-Jones S, Kim M (2017) Small RNAs: big impact on plant development. Trends Plant Sci 22: 1056–1068 [DOI] [PubMed] [Google Scholar]
- David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR (2017) Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29: 445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis C, Krokida A, Tomatsidou A, Tsikou D, Beta RAA, Tsioumpekou M, Moustaka J, Stravodimos G, Leonidas DD, Balatsos NAA, et al. (2016) AtHESPERIN: a novel regulator of circadian rhythms with poly(A)-degrading activity in plants. RNA Biol 13: 68–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo L, Sorenson R, Bailey-Serres J, Hunt AG (2017) Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell 29: 1262–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong OX, Meteignier LV, Plourde MB, Ahmed B, Wang M, Jensen C, Jin H, Moffett P, Li X, Germain H (2016) Arabidopsis TAF15b localizes to RNA processing bodies and contributes to snc1-mediated autoimmunity. Mol Plant Microbe Interact 29: 247–257 [DOI] [PubMed] [Google Scholar]
- Dorcey E, Rodriguez-Villalon A, Salinas P, Santuari L, Pradervand S, Harshman K, Hardtke CS (2012) Context-dependent dual role of SKI8 homologs in mRNA synthesis and turnover. PLoS Genet 8: e1002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, Rätsch G, Wachter A (2013) Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Ferdous J, Hussain SS, Shi BJ (2015) Role of microRNAs in plant drought tolerance. Plant Biotechnol J 13: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouracre JP, Poethig RS (2016) The role of small RNAs in vegetative shoot development. Curr Opin Plant Biol 29: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei dit Frey N, Muller P, Jammes F, Kizis D, Leung J, Perrot-Rechenmann C, Bianchi MW (2010) The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22: 1575–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. (2014) The role of the poly(A) binding protein in the assembly of the Cap-binding complex during translation initiation in plants. Translation (Austin) 2: e959378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Fu X, Song J, Zhang Y, Cui X, Su C, Ge L, Shao J, Xin L, Saarikettu J, et al. (2015) Poly(A)+ mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics. FEBS J 282: 874–890 [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer J, Akimcheva S, Srinivasan A, Kusenda B, Riehs N, Stampfl H, Bautor J, Dekrout B, Jonak C, Jiménez-Gómez JM, et al. (2014) Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16: 376–390 [DOI] [PubMed] [Google Scholar]
- Godwin AR, Kojima S, Green CB, Wilusz J (2013) Kiss your tail goodbye: the role of PARN, Nocturnin, and Angel deadenylases in mRNA biology. Biochim Biophys Acta 1829: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golisz A, Sikorski PJ, Kruszka K, Kufel J (2013) Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res 41: 6232–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Denisenko TV, Zhivotovsky B, Bozhkov PV (2016) Tudor staphylococcal nuclease: biochemistry and functions. Cell Death Differ 23: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV (2015) Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27: 926–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E (2010) HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 189: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Reichelt P, Rode M, Conti E (2013) The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell 154: 814–826 [DOI] [PubMed] [Google Scholar]
- Hooker TS, Lam P, Zheng H, Kunst L (2007) A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvathova I, Voigt F, Kotrys AV, Zhan Y, Artus-Revel CG, Eglinger J, Stadler MB, Giorgetti L, Chao JA (2017) The dynamics of mRNA turnover revealed by single-molecule imaging in single cells. Mol Cell 68: 615–625 [DOI] [PubMed] [Google Scholar]
- Hou CY, Lee WC, Chou HC, Chen AP, Chou SJ, Chen HM (2016) Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. (2017) P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68: 144–157 [DOI] [PubMed] [Google Scholar]
- Hummel M, Dobrenel T, Cordewener JJ, Davanture M, Meyer C, Smeekens SJ, Bailey-Serres J, America TA, Hanson J (2015) Proteomic LC-MS analysis of Arabidopsis cytosolic ribosomes: identification of ribosomal protein paralogs and re-annotation of the ribosomal protein genes. J Proteomics 128: 436–449 [DOI] [PubMed] [Google Scholar]
- Jaag HM, Nagy PD (2009) Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386: 344–352 [DOI] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161: 1202–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HJ, Kim YJ, Kim SH, Kim YH, Lee IJ, Kim YK, Shin JS (2011) Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol 52: 2147–2156 [DOI] [PubMed] [Google Scholar]
- Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A (2012) Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J 31: 1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer JP, Green PJ (2000) Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA 97: 13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerényi F, Wawer I, Sikorski PJ, Kufel J, Silhavy D (2013) Phosphorylation of the N- and C-terminal UPF1 domains plays a critical role in plant nonsense-mediated mRNA decay. Plant J 76: 836–848 [DOI] [PubMed] [Google Scholar]
- Koguchi M, Yamasaki K, Hirano T, Sato MH (2017) Vascular plant one-zinc-finger protein 2 is localized both to the nucleus and stress granules under heat stress in Arabidopsis. Plant Signal Behav 12: e1295907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp S, Greiner E, Amin B, Sonnewald U, Krenz B (2017) The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res 227: 6–14 [DOI] [PubMed] [Google Scholar]
- Kumakura N, Otsuki H, Ito M, Nomoto M, Tada Y, Ohta K, Watanabe Y (2016) Arabidopsis AtRRP44 has ribonuclease activity that is required to complement the growth defect of yeast rrp44 mutant. Plant Biotechnol 33: 77–85 [Google Scholar]
- Kumakura N, Otsuki H, Tsuzuki M, Takeda A, Watanabe Y (2013) Arabidopsis AtRRP44A is the functional homolog of Rrp44/Dis3, an exosome component, is essential for viability and is required for RNA processing and degradation. PLoS ONE 8: e79219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łabno A, Tomecki R, Dziembowski A (2016) Cytoplasmic RNA decay pathways: enzymes and mechanisms. Biochim Biophys Acta 1863: 3125–3147 [DOI] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crété P, Voinnet O, Robaglia C (2009) Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Gagliardi D (2010) The exosome and 3′-5′ RNA degradation in plants. In Jensen TH, ed, RNA Exosome, Vol. 2. Springer, New York, pp 50–62 [Google Scholar]
- Lange H, Holec S, Cognat V, Pieuchot L, Le Ret M, Canaday J, Gagliardi D (2008) Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana. Mol Cell Biol 28: 3038–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Lin TL, Lin JW, Han YT, Huang YT, Hsu YH, Meng M (2016) Promotion of bamboo mosaic virus accumulation in Nicotiana benthamiana by 5′→3′ exonuclease NbXRN4. Front Microbiol 6: 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Le B, Ma X, Li S, You C, Yu Y, Zhang B, Liu L, Gao L, Shi T, et al. (2016) Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H (2015) EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683 [DOI] [PubMed] [Google Scholar]
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C (2009) The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 19: 307–316 [DOI] [PubMed] [Google Scholar]
- Lin PC, Pomeranz MC, Jikumaru Y, Kang SG, Hah C, Fujioka S, Kamiya Y, Jang JC (2011) The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J 65: 253–268 [DOI] [PubMed] [Google Scholar]
- Liu L, Chen X (2016) RNA quality control as a key to suppressing RNA silencing of endogenous genes in plants. Mol Plant 9: 826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokdarshi A, Conner WC, McClintock C, Li T, Roberts D (2016) Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 170: 1046–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. (2014) Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 5: ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD. (2014) Composition and function of P bodies in Arabidopsis thaliana. Front Plant Sci 5: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Nomura Y, Egusa M, Hamada T, Hyon GS, Kaminaka H, Watanabe Y, Ueda T, Trujillo M, Shirasu K, et al. (2017) The GYF domain protein PSIG1 dampens the induction of cell death during plant-pathogen interactions. PLoS Genet 13: e1007037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérai Z, Benkovics AH, Nyikó T, Debreczeny M, Hiripi L, Kerényi Z, Kondorosi É, Silhavy D (2013) The late steps of plant nonsense-mediated mRNA decay. Plant J 73: 50–62 [DOI] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enríquez P, Binder BM, Heber S, Stepanova AN, Alonso JM (2015) Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697 [DOI] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM (2017) Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J 90: 628–653 [DOI] [PubMed] [Google Scholar]
- Merret R, Carpenier MC, Favory JJ, Picart C, Descombin J, Bousquet-Antonelli C, Tillard P, Lejay L, Deragon JM, Charng YY (2017) Heat-shock protein HSP101 affects the release of ribosomal protein mRNAs for recovery after heat shock. Plant Physiol 174: 1216–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R, Descombin J, Juan YT, Favory JJ, Carpentier MC, Chaparro C, Charng YY, Deragon JM, Bousquet-Antonelli C (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep 5: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Merret R, Nagarajan VK, Carpentier MC, Park S, Favory JJ, Descombin J, Picart C, Charng YY, Green PJ, Deragon JM, et al. (2015) Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res 43: 4121–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, Reese JC (2012) Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47: 315–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Gossmann N, Dietz KJ (2016) Redox regulation of cytosolic translation in plants. Trends Plant Sci 21: 388–397 [DOI] [PubMed] [Google Scholar]
- Motomura K, Le QTN, Hamada T, Kutsuna N, Mano S, Nishimura M, Watanabe Y (2015) Diffuse decapping enzyme DCP2 accumulates in DCP1 foci under heat stress in Arabidopsis thaliana. Plant Cell Physiol 56: 107–115 [DOI] [PubMed] [Google Scholar]
- Nguyen AH, Matsui A, Tanaka M, Mizunashi K, Nakaminami K, Hayashi M, Iida K, Toyoda T, Nguyen DV, Seki M (2015) Loss of Arabidopsis 5′-3′ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol 56: 1762–1772 [DOI] [PubMed] [Google Scholar]
- Nguyen CC, Nakaminami K, Matsui A, Kobayashi S, Kurihara Y, Toyooka K, Tanaka M, Seki M (2016) Oligouridylate binding protein 1b plays an integral role in plant heat stress tolerance. Front Plant Sci 7: 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CC, Nakaminami K, Matsui A, Watanabe S, Kanno Y, Seo M, Seki M (2017) Overexpression of oligouridylate binding protein 1b results in ABA hypersensitivity. Plant Signal Behav 12: e1282591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T (2005) Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44: 972–984 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Matsui A, Tanaka M, Morosawa T, Ishida J, Iida K, Mochizuki Y, Toyoda T, Seki M (2016) Sm-like protein-mediated RNA metabolism is required for heat stress tolerance in Arabidopsis. Front Plant Sci 7: 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, Stoecklin G (2010) Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol 30: 4308–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yang J, Yan F, Lu Y, Jiang S, Lin L, Zheng H, Chen H, Chen J (2011) Silencing of NbXrn4 facilitates the systemic infection of Tobacco mosaic virus in Nicotiana benthamiana. Virus Res 158: 268–270 [DOI] [PubMed] [Google Scholar]
- Perea-Resa C, Carrasco-López C, Catalá R, Turečková V, Novak O, Zhang W, Sieburth L, Jiménez-Gómez JM, Salinas J (2016) The LSM1-7 complex differentially regulates Arabidopsis tolerance to abiotic stress conditions by promoting selective mRNA decapping. Plant Cell 28: 505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C, Hernández-Verdeja T, López-Cobollo R, del Mar Castellano M, Salinas J (2012) LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24: 4930–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B, Saunders DGO, Sklenar J, Lorrain C, Krasileva KV, Win J, Duplessis S, Kamoun S (2016) Heterologous expression screens in Nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PLoS ONE 11: e0149035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz M, Lin PC, Finer J, Jang JC (2010a) AtTZF gene family localizes to cytoplasmic foci. Plant Signal Behav 5: 190–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, Jang JC (2010b) The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol 152: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P (2006) The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18: 3047–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Kang SG, Wang W, Musier-Forsyth K, Jang JC (2014) The Arabidopsis thaliana tandem zinc finger 1 (AtTZF1) protein in RNA binding and decay. Plant J 78: 452–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Lloyd RE (2013) Diversion of stress granules and P-bodies during viral infection. Virology 436: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B (2014) Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA 111: 6365–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA (2004) mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 10: 1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux ME, Rasmussen MW, Palma K, Lolle S, Regué ÀM, Bethke G, Glazebrook J, Zhang W, Sieburth L, Larsen MR, et al. (2015) The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J 34: 593–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, von Arnim AG (2013) Translational regulation of cytoplasmic mRNAs. The Arabidopsis Book 11: e0165, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, Green PJ (2011) Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarowar S, Oh HW, Cho HS, Baek KH, Seong ES, Joung YH, Choi GJ, Lee S, Choi D (2007) Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J 51: 792–802 [DOI] [PubMed] [Google Scholar]
- Scarpin MR, Sigaut L, Temprana SG, Boccaccio GL, Pietrasanta LI, Muschietti JP (2017) Two Arabidopsis late pollen transcripts are detected in cytoplasmic granules. Plant Direct (in press) http://doi.org/10.1002/pld3.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H, Ozgur S, Sharma K, Basquin C, Urlaub H, Conti E (2013) Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res 41: 8377–8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O. (2015) Unique aspects of plant nonsense-mediated mRNA decay. Trends Plant Sci 20: 767–779 [DOI] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, Barna M (2017) Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwaszek A, Ukleja M, Dziembowski A (2014) Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol 11: 1122–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma F, Mogami J, Yoshida T, Abekura M, Takahashi F, Kidokoro S, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2017) ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat Plants 3: 16204. [DOI] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 111: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Stauffer E, Westermann A, Wagner G, Wachter A (2010) Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J 64: 243–255 [DOI] [PubMed] [Google Scholar]
- Steffens A, Bräutigam A, Jakoby M, Hülskamp M (2015) The BEACH domain protein SPIRRIG is essential for Arabidopsis salt stress tolerance and functions as a regulator of transcript stabilization and localization. PLoS Biol 13: e1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens A, Jaegle B, Tresch A, Hülskamp M, Jakoby M (2014) Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol 164: 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S (2016) Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat Plants 2: 16055. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Arae T, Green PJ, Yamaguchi J, Chiba Y (2015) AtCCR4a and AtCCR4b are involved in determining the poly(A) length of granule-bound starch synthase 1 transcript and modulating sucrose and starch metabolism in Arabidopsis thaliana. Plant Cell Physiol 56: 863–874 [DOI] [PubMed] [Google Scholar]
- Tharun S. (2009) Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol 6: 228–232 [DOI] [PubMed] [Google Scholar]
- Tian L, Okita TW (2014) mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr Opin Plant Biol 22: 77–85 [DOI] [PubMed] [Google Scholar]
- Tsuzuki M, Motomura K, Kumakura N, Takeda A (2017) Interconnections between mRNA degradation and RDR-dependent siRNA production in mRNA turnover in plants. J Plant Res 130: 211–226 [DOI] [PubMed] [Google Scholar]
- Vandivier LE, Campos R, Kuksa PP, Silverman IM, Wang LS, Gregory BD (2015) Chemical modifications mark alternatively spliced and uncapped messenger RNAs in Arabidopsis. Plant Cell 27: 3024–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vexler K, Cymerman MA, Berezin I, Fridman A, Golani L, Lasnoy M, Saul H, Shaul O (2016) The Arabidopsis NMD factor UPF3 is feedback-regulated at multiple levels and plays a role in plant response to salt stress. Front Plant Sci 7: 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Winkler GS (2013) RNA decay machines: deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim Biophys Acta 1829: 561–570 [DOI] [PubMed] [Google Scholar]
- Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K (2010) Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 152: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Okamoto T (2009) Involvement of polypyrimidine tract-binding protein (PTB)-related proteins in pollen germination in Arabidopsis. Plant Cell Physiol 50: 179–190 [DOI] [PubMed] [Google Scholar]
- Waris S, Wilce MCJ, Wilce JA (2014) RNA recognition and stress granule formation by TIA proteins. Int J Mol Sci 15: 23377–23388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Xu J, Chua NH (2009) Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH (2011) Processing bodies and plant development. Curr Opin Plant Biol 14: 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH (2012) Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J 31: 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Yan Z, Wang Y, Yan X, Han Y (2014) Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Bot 65: 5933–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS (2012) Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc Natl Acad Sci USA 109: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Zhang B, Jia J, Yan C, Habaike A, Han Y (2013) RRP41L, a putative core subunit of the exosome, plays an important role in seed germination and early seedling growth in Arabidopsis. Plant Physiol 161: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Yang J, Sun Y, Zhao P, Gao S, Jung C, Qu J, Fang R, Chua NH (2015) Geminivirus activates ASYMMETRIC LEAVES 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLoS Pathog 11: e1005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Willmann MR, Anderson SJ, Gregory BD (2016) Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mRNA cap-binding complex in co-translational RNA decay in Arabidopsis. Plant Cell 28: 2385–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu X, Gaikwad K, Kou X, Wang F, Tian X, Xin M, Ni Z, Sun Q, Peng H, et al. (2017a) Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. Plant Cell 29: 1952–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Murphy C, Sieburth LE (2010) Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci USA 107: 15981–15985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo H (2017) mRNA decay in plants: both quantity and quality matter. Curr Opin Plant Biol 35: 138–144 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu Y, Liu X, Hong X, Xu Y, Zhu P, Shen Y, Wu H, Ji Y, Wen X, et al. (2015) Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348: 120–123 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hu F, Sung MW, Shu C, Castillo-González C, Koiwa H, Tang G, Dickman M, Li P, Zhang X (2017b) RISC-interacting clearing 3′-5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. eLife 6: e24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kunst L (2016) SUPERKILLER complex components are required for the RNA exosome-mediated control of cuticular wax biosynthesis in Arabidopsis inflorescence stems. Plant Physiol 171: 960–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber H, Scheer H, Ferrier E, Sement FM, Mercier P, Stupfler B, Gagliardi D (2016) Uridylation and PABP cooperate to repair mRNA deadenylated ends in Arabidopsis. Cell Rep 14: 2707–2717 [DOI] [PubMed] [Google Scholar]