Abstract
Background:
The proto-oncogene c-MYC, located on chromosome 8q, can be upregulated through gain of 8q, causing alteration in biology of renal cell carcinoma (RCC). The aim of this study was to evaluate the prevalence of c-MYC through chromosome 8q gain and to correlate findings with cancer-specific mortality (CSM), and overall survival (OS).
Methods:
Cytogenetic analysis by conventional or Chromosomal Genomic Microarray Analysis (CMA) was performed on 414 renal tumors. Nonclear and nonpapillary RCC were excluded. Impact of gain in chromosome 8q status on CSM, OS, and its correlation with clinicopathological variables were evaluated. CSM and OS were assessed using log-rank test and the Cox proportional hazards model.
Results:
A total of 297 RCC tumors with cytogenetic analysis were included. Gain of 8q was detected in 18 (6.1%) tumors (9 clear cell and 9 papillary RCC), using conventional method (n = 11) or CMA (n = 7). Gain of 8q was associated with higher T stage (p < 0.001), grade (p < 0.001), nodal involvement (p = 0.005), and distant metastasis (p < 0.001). No association between gain of 8q and age (p = 0.23), sex (p = 0.46), and Charlson comorbidity index (CCI, p = 0.59) were seen. Gain of 8q was associated with an 8.38-fold [95% confidence interval (CI), 3.83–18.34, p < 0.001] and 3.31-fold (95% CI, 1.56–7.04, p = 0.001) increase in CSM and decrease in OS, respectively, at a median follow up of 56 months.
Conclusion:
Chromosome 8q harbors the proto-oncogene c-MYC, which can be upregulated by gain of 8q. Our findings suggest that gain of 8q, can predict aggressive tumor phenotype and inferior survival in RCC.
Keywords: 8q, c-MYC, cytogenetic analysis, prognosis, proto-oncogene, renal cell carcinoma
Introduction
Kidney cancer, predominantly renal cell carcinoma (RCC), with an approximately 22% rate of cancer-specific mortality (CSM), is among the most lethal of urologic malignancies.1 Similar to other solid malignancies, the TNM stage, grade, histology, and clinical indications are most commonly utilized to predict outcomes in patients with RCC. Nevertheless, heterogeneity makes prediction of individual patient’s clinical course unique and matching the biology to appropriate treatments remains challenging.2 Although current preoperative nomograms assist with better prediction of outcome,3,4 identification and incorporation of the molecular modifications, which contribute to fluctuations in RCC behavior, into existing predictive tools should result in improved decision making and more realistic outcome expectation.5,6
Over the last several years, cytogenetic analysis by conventional or Chromosomal Microarray Analysis (CMA) has not only improved classifying RCCs but also in predicting outcomes. The molecular properties of renal neoplasm and genes involved in development and progression of each tumor have been identified. Although the exact mechanism by which each RCC histological subtype develops remains unclear, cytogenetic studies point out that allelic loss on chromosomes 4p, 8p, 9p, 14q and gains of chromosomes 7, 16, 17p are associated with worse survival in clear and papillary type 2 RCC, respectively.7–10
The c-MYC proto-oncogene is located on chromosome 8q, which its amplification influences the expression of a wide range of human genes involved in the progression of cell carcinogenesis.11–14 Although the gain of chromosome 8q and the effect of c-MYC in tumorigenesis of different solid malignancies have been reported,15–18 its effect on RCC still remains poorly characterized.19 The aim of our study is to report on the prevalence of chromosome 8q gain in our renal cancer tumor registry and to evaluate its prognostic significance on CSM and overall survival (OS) in clear cell, papillary, and sarcomatoid variant RCCs.
Methods
Patient selection
The institutional, prospectively-maintained, renal tumor database at the Fox Chase Cancer Center was used to identify patients who underwent radical or partial nephrectomy for suspected RCC between August 2002 and August 2013. Patients who had cytogenetic analysis by conventional or CMA were identified. Nonclear and nonpapillary RCC, and those with incomplete medical records were excluded from the study. All patients had a computed tomography (CT) scan of the abdomen and pelvis, or a CT scan or X-ray of the chest as preoperative staging. If indicated, cranial CT or bone scan were obtained. TNM staging was determined using a collaborative stage approach, combining pathological and clinical findings from patient records, the tumor registry, and the kidney cancer database. Pathologic T (pT) stage was designated pathologically and M stage was assigned mostly clinically (based on cross sectional imaging). If lymphadenectomy was not performed at the time of surgery, N stage was assigned clinically. Collaborative staging was revised according to the cancer staging manual of the American Joint Committee on Cancer, 7th edition.20 The postoperative surveillance was physician-dependent, often using the recommended follow up (www.cancernomograms.com).
Karyotypic and genomic copy number analysis
Cytogenetic analysis was performed on 414 patients who had renal mass extirpation as in our prior report.10 Briefly, immediately after the specimen was diagnosed by uro-oncologic pathologists (TAS and ED), a fresh specimen was cut in half, with one piece used for cytogenetics and the other used for microarray analysis. Each cytogenetic sample was minced mechanically using two scalpels and then treated with 0.2% collagenase for 30–60 min at 37°C. The samples were cultured for 2–14 days in RPMI 1640 medium containing 15% fetal bovine serum. Metaphase slide preparations and G-banding were performed according to our standard method.21 All metaphases (n = 20) were analyzed consistent with the International Standing committee on Human Cytogenetic Nomenclature by cytogeneticists (JRT and JP).22 Tumor tissue used for genomic copy number analysis was macro-dissected to remove obvious necrotic areas, stroma, and adjacent normal tissue. For each tumor, genome-wide copy number analysis was done using Affymetrix Cytogenetics Whole-genome 2.7M or CytoScan HD Array according to the manufacturer’s protocol (www.affymetrix.com). The intensities of probe hybridization were analyzed using Affymetrix software GCOS, and genotyping and copy number analysis were performed using Affymetrix Chromosome Analysis Suite (ChAS) with the default setting.
Assessment of c-MYC expression
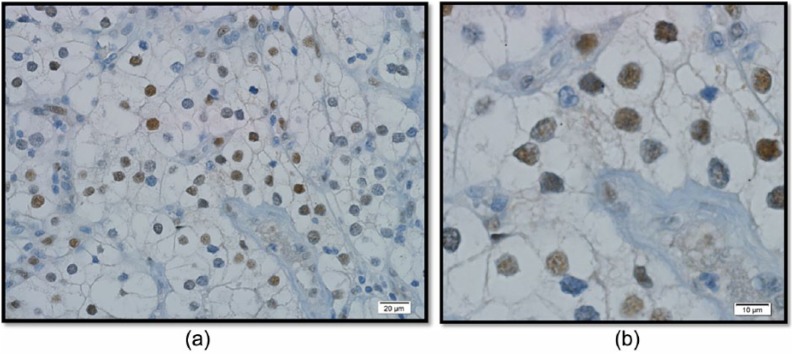
Anti-c-MYC rabbit monoclonal antibody was obtained from Ventana (Tucson, Arizona, USA). Sections were deparaffinized, prepared, and stained on an automated stainer (Figure1).
Figure 1.
Imuunohistochemical stain for clear cell RCC: (a) low power magnification; (b) high power magnification.
Statistical analysis
The association between chromosome 8q and mortality in clear cell renal cell carcinoma and papillary renal cell carcinioma patients was examined. As part of the prospective maintenance of the database, date and cause of death were obtained from the death certificate, patient’s family, or local physician. Length of follow up was calculated from the date of surgery to the date of last follow up or death. OS and CSM were calculated from the date of surgery to the date of death from any cause and date of patients’ cancer-related death, respectively. Kaplan–Meier curves were plotted for OS and CSM for each group of patients showing gain or no gain of chromosome 8q.
Continuous variables were analyzed using the Mann–Whitney test and categorical variables were analyzed using Fisher’s exact test. Univariate log-rank tests and Cox proportional hazards models were used to analyze survival endpoints. All tests were two-sided and used a type I error of 5% to determine statistical significance. The R statistical language and environment were used in the computations.
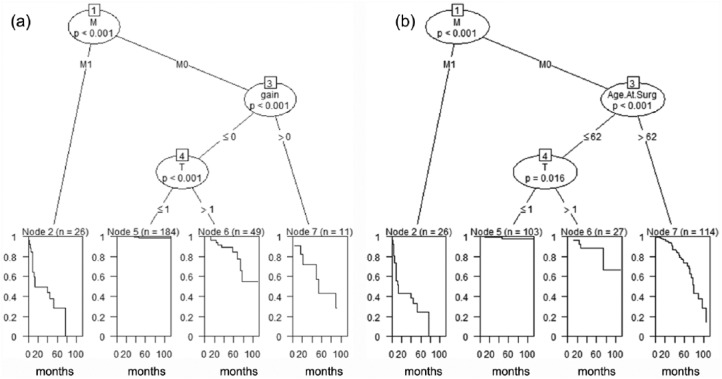
In order to identify clinical and demographic variables related to OS and CSM, using the Classification and Regression Trees (CART), multivariable analyses were performed by constructing Decision Trees (Figure 2). A Decision Tree is a logical model represented as a binary tree that shows how the value of a response variable can be predicted by a set of relevant clinical variables. If the response variable is time to an event, such as OS or CSM, then a regression tree is generated that predicts the probability of OS or CSM. Variables such as gain of chromosome 8q, grade, Charlson comorbidity index (CCI), sex, BMI, T, N and M stages, and age at surgery were used to conduct analysis for OS and CSM. The unified CART framework that embeds recursive binary partitioning into the theory of permutation tests was used in our analyses.23 This approach is well suited for our analysis as it overcomes the problem of over-fitting and selection bias towards clinical variables with many possible splits or missing values. In addition, this approach results in unbiased selection among clinical variables measured at different scales (such as categorical, ordinal or continuous). Significance testing procedures were applied to determine whether no significant association between any of the clinical variables and the response can be stated and the recursion needs to stop. The open-source R package PARTY (www.r-project.org) was used in the computations.
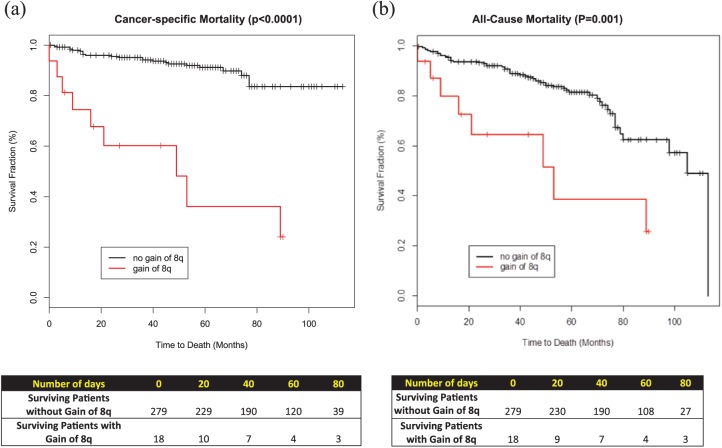
Figure 2.
Kaplan-Meier survival curves: (a) cancer-specific mortality; (b) all-cause mortality.
Results
Clinicopathological characteristics
Patient demographics and clinicopathological variables are summarized in Table 1. A total of 297 patients who had renal mass extirpation and cytogenetic analysis by conventional or CMA met the inclusion criteria. Of those, 231 (78%) had diagnosis of clear cell, 52 (17%) papillary, and 15 patients (5%) had rhabdoid or sarcomatoid variant histology. Radical nephrectomy was performed in 95 patients (32%), whereas 202 (68%) had a nephron-sparing surgery. The median age at the time of surgery was 62 years, 72% were male, and the median CCI was 1. Median body mass index of the cohort was 28.6 kg/m2. Pathologic T3 or higher, positive lymph node, and distant metastasis were present in 22%, 4%, and 13% of patients respectively.
Table 1.
Clinicopathological characteristics and associations of chromosome 8q gain.
| Variable | Total | No gain | Gain | p-value |
|---|---|---|---|---|
| n = 297 | n = 276 | n = 18 | ||
| Age, median (IQR) | 62 (16) | 62 (16) | 63 (11) | 0.234 |
| Sex (%) | ||||
| Male | 216 (73) | 202 (73) | 14 (78) | 0.456 |
| Female | 81 (27) | 77 (27) | 4 (22) | |
| pT stage (%) | ||||
| 1,2 | 231 (78) | 225 (82) | 6 (33) | <0.001 |
| 3,4 | 65 (22) | 53 (18) | 12 (67) | |
| pN stage (%) | ||||
| 0 | 286 (96) | 272 (99) | 14 (78) | 0.005 |
| 1 | 11 (4) | 7 (1) | 4 (22) | |
| M stage (%) | ||||
| 0 | 259 (87) | 249 (89) | 10 (56) | <0.001 |
| 1 | 38 (13) | 30 (11) | 8 (44) | |
| Nuclear grade (%) | ||||
| ⩽2 | 152 (51) | 149 (54) | 3 (17) | <0.001 |
| >2 | 142 (49) | 127 (46) | 15 (83) | |
| CCI, median (IQR) | 1 (2) | 1 (2) | 1 (2) | 0.593 |
| Standard division | 1.69 | 1.71 | 1.47 |
CCI, Charlson comorbidity index; IQR, interquartile range.
Correlation of gain of chromosome 8q with known prognostic factors
Cytogenetic analysis revealed gain of chromosome 8q in 18 (6.1%) tumors by either conventional method (n = 11) or CMA (n = 7), of which 9 were clear cell RCC and 9 were papillary RCC. A total of nine (50%) tumors had sarcomatoid or rhabdoid features. Among tumors with 8q gain, 56% were stage III or higher. Gain of 8q was associated with higher grade (p < 0.001), higher risk of regional lymph node involvement (p = 0.004), and distant metastasis (p < 0.001). No association between gain of 8q and age (p = 0.23), sex (p = 0.46), and CCI (p = 0.59) was seen (Table 1).
c-MYC expression
To correlate with immunohistochemical staining, 11 random cases were stained for c-MYC and 200 nuclei counted in the highest area of expression (supplementary Table 2). All cases expressed c-MYC: mean and median expression of 48% (range 8–90%) and 45%, respectively. As a control, 20 sections from different areas in 5 cases of RCC, with nuclear grade 3 or 4 but without gain of 8q, were stained, of which none of the sections stained positive for c-MYC. Immunohistochemical stains for c-MYC revealed increased expression (>30% of nuclei) in 9 of 11 cases stained (82%).
OS and CSM
To investigate the association between gain of 8q with the clinical outcome, univariate and multivariable analyses for the two end points were performed (Figures 2 and 3, respectively). Within a median follow up of 56 months, gain of chromosome 8q was associated with a hazard ratio (HR) of 8.38 (or 8.38-fold increase in the risk of death) in (95% CI, 3.83–18.34, p < 0.0001). Similarly, gain of chromosome 8q was associated with a 3.31-fold increase in all-cause mortality (HR 3.31, 95% CI: 1.56–7.04, p = 0.001). Median survival for those with gain of 8q was 53 months versus 105 months for those without gain of 8q chromosome.
Figure 3.
The n and p values in this figure show the sample size and statistical significance.
CART (classification and regression trees) for CSM (a) and OS (b): Divides the patients into four groups with different survival profiles. For example, the best survival is exhibited by younger (age at surgery ⩽62) M0 patients with lower T stage (<1). Comparatively, the worst survival is seen in M1 patients.
CSM, cancer-specific mortality; OS, overall survival
Discussion
To our knowledge, this is the first study evaluating the effect of chromosome 8q gain in a combined cohort of clear cell and papillary RCC. As shown by cytogenetic analysis and histoimmunostaining (Figure 1), our findings suggest that proto-oncogene c-MYC is upregulated through gain of 8q in a subset of RCC tumors. On multivariable analysis, gain of 8q, independently, was associated with higher CSM (HR = 8.38) and all-cause mortality (HR = 3.31).
The human c-MYC oncogene is found on chromosome 8 at position 8q24.11,14 This region of the long arm of chromosome 8 has been previously shown to be involved in tumorigenesis of variety of malignancies and it is known as a prognosticator of cancer progression and poor outcome.15–19,24 The c-MYC acts as a transcription factor and modulates the expression of target genes by binding to specific DNA sequences and impacts the expression of genes which influence cell growth, metabolism, angiogenesis, and apoptosis.11–13 One of the proposed mechanisms that enables this oncogene to change the biology of the tumor is via the mitogen-activated protein kinase pathway (MAPK/ERK).12 Also, it has been shown via regulating the cyclin-dependent kinase activity, MYC-target gene cyclin D1 (CCND1) promotes the G1/S phase transition in the cell cycle progression.25
Several studies have examined the role of c-MYC in RCC. Tang and colleagues13 using functional network analysis of the differentially expressed genes, they investigated the deregulated pathways in clear cell RCC. They identified 37 differentially expressed genes as MYC-target genes, denoting that MYC pathway is activated in clear cell RCC tissues. Furthermore, their findings demonstrated that in clear cell RCC, MYC promotes both cell cycle progression and disruption by inducing expression of CCDN1 and knockdown of MYC, respectively. Using monoclonal antibody (mAb) MYC-1 immunostaining, Kinouchi and colleagues26 reported a direct correlation of c-MYC expression with nuclear pleomorphism in 41 primary and 17 metastatic RCC tumors. Among the primary tumors, positive MYC staining was noted in 12%, 81%, and 100% of tumors with grade 1, 2, and 3, respectively. Moreover, in metastatic tumors, positive staining was seen in 0% of grade 1, 50% of grade 2, and all of tumors with grade 3. Furge and colleagues8 using molecular genetics and computational analysis, were able to show that over-expression of MYC maps to amplification of chromosome 8q24, which correlated with poor OS in patients with papillary RCC. Furthermore, although Klatte and colleagues did not directly correlate the impact of chromosome 8q gain on c-MYC level,19 they identified the gain of 8q, in a subset of clear cell RCCs, as a strong and independent predictor of poor survival. In this current study, for the first time, using both cytogenetic analysis and immunostaining, we are able to show that gain of 8q amplifies c-MYC in clear cell and papillary RCC, in which inversely affects the survival.
In addition to the c-MYC proto-oncogene, other genes located in the 8q24 region may contribute directly to the outcome of patients harboring the genomic rearrangement involving 8q24 amplification. In a 17 gene/probe tissue gene expression model, genes mapping to 8q24 (FAM49B, RAD21, TAF2, C8ORF53, and KIAA0196) were strongly associated with systemic progression in prostate cancer; and 8q24 was one of four highly aberrant chromosomal regions identified by gene expression microarray that selected CYC1, SIAHBP, and SCRIB as potential oncogenes.24,27 Furthermore, existing data implies that alternate MYC/chromosome 8 copy number alterations may be associated with differential response to specific immunomodulatory therapies and eluting that MYC may be a surrogate for other genes located on chromosome 8.18 Ongoing large-scale whole-genome expression profiling of RCC by The Cancer Genome Atlas (CTGA) will provide imperative information regarding the relationship between MYC and other pertinent genes and the benefit of neoadjuvant or adjuvant targeted therapies.28
With a demographic shift towards an aging population with more medical comorbidities, objectifying the risks and benefits of individual treatment options for patients is a crucial undertaking for medical providers who treat renal tumors. Traditional clinical staging has focused solely on tumor factors, and assessment of patients has been mostly qualitative and prone to inaccuracy.6 Clinicians have difficulty identifying patients who are prone to progression and dying from RCC. With further discovery of RCC prognostic biomarkers and their incorporation into quantitative predictive models, physician and patients will be armed with better understanding and a more realistic view of outcomes.29 The current body of evidence suggests that gain of 8q may serve as a useful adjunct to already established RCC prognosticators to help identify patients with clinically significant disease, who may benefit from enrollment into neoadjuvant or adjuvant clinical trials, and provide cues to tailor individual patient postoperative surveillance schedules.30
As is the case in any retrospective study, there are some inherent limitations to our study, such as data selection and analysis which are susceptible to biases. One limitation is that majority of our cohort is made up of pT1–T2 renal tumors. Additionally, the average follow up for the study was relatively short and as a result, other prognostically relevant factors related to later course of the disease could have been underestimated. Although we showed the MYC expression by immunostaining, we did not evaluate the presence of other genes located near chromosome 8q. Even though we have substantial numbers of RCC tumors with cytogenetic analysis, due to the sample size and low prevalence of 8q gain, the power to detect its interaction with clinical and pathologic factors needs to be further studied. Further molecular studies and using larger cohorts (multi-institutional) on the role of chromosome 8q in RCC are necessary to validate our results.
Conclusion
Chromosome 8q harbors the proto-oncogene c-MYC, which can be over expressed by gain of 8q. Our findings suggest that gain of 8q, shown by cytogenetics, can potentially predict aggressive tumor phenotype and inferior survival in patients with RCC. If cytogenetic studies are not available, c-MYC stains can be helpful in detecting c-MYC over-expression.
Supplementary Material
Footnotes
Funding: This publication was supported in part by grant number P30 CA006927 from the National Cancer Institute. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Additional funds were provided by Fox Chase Cancer via institutional support of the Kidney Cancer Keystone Program.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Supplementary Material: Supplementary Table for this article is available online.
Contributor Information
Reza Mehrazin, Department of Urology and Oncological Science, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Essel Dulaimi, Divisions of Pathology and Cancer Biology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA.
Robert G. Uzzo, Divisions of Urologic Oncology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA
Karthik Devarjan, Divisions of Biostatistics; Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA.
Jianming Pei, Divisions of Pathology and Cancer Biology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA.
Marc C. Smaldone, Divisions of Urologic Oncology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA
Alexander Kutikov, Divisions of Urologic Oncology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA.
Joseph R. Testa, Divisions of Pathology and Cancer Biology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA
Tahseen Al-Saleem, Divisions of Pathology and Cancer Biology, Fox Chase Cancer Center-Temple Health System, Philadelphia, PA, USA.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- 2. Mehrazin R, Smaldone MC, Kutikov A, et al. Growth kinetics and short term outcomes of cT1b and cT2 renal masses under active surveillance. J Urol 2014; 192: 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kutikov A, Egleston BL, Wong YN, et al. Evaluating overall survival and competing risks of death in patients with localized renal cell carcinoma using a comprehensive nomogram. J Clin Oncol 2010; 28: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cindolo L, Chiodini P, Brookman-May S, et al. Assessing the accuracy and generalizability of the preoperative and postoperative Karakiewicz nomograms for renal cell carcinoma: results from a multicentre European and US study. BJU Int 2013; 112: 578–584. [DOI] [PubMed] [Google Scholar]
- 5. Soung Sullivan P, Rao J, Cheng L, et al. Classical pathology versus molecular pathology in renal cell carcinoma. Curr Urol Rep 2007; 8: 5–11. [DOI] [PubMed] [Google Scholar]
- 6. Chen DY, Uzzo RG, Viterbo R. Thinking beyond surgery in the management of renal cell carcinoma: the risk to die from renal cell carcinoma and competing risks of death. World J Urol 2014; 32: 607–613. [DOI] [PubMed] [Google Scholar]
- 7. Cheng L, Williamson SR, Zhang S, et al. Understanding the molecular genetics of renal cell neoplasia: implications for diagnosis, prognosis and therapy. Expert Rev Anticancer Ther 2010; 10: 843–864. [DOI] [PubMed] [Google Scholar]
- 8. Furge KA, Chen J, Koeman J, et al. Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high-grade papillary renal cell carcinoma. Cancer Res 2007; 67: 3171–3176. [DOI] [PubMed] [Google Scholar]
- 9. Klatte T, Rao PN, de Martino M, et al. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J Clin Oncol 2009; 27: 746–753. [DOI] [PubMed] [Google Scholar]
- 10. Pei J, Feder MM, Al-Saleem T, et al. Combined classical cytogenetics and microarray-based genomic copy number analysis reveal frequent 3;5 rearrangements in clear cell renal cell carcinoma. Genes Chromosomes Cancer 2010; 49: 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neel BG, Jhanwar SC, Chaganti RS, et al. Two human c-onc genes are located on the long arm of chromosome 8. Proc Natl Acad Sci U S A 1982; 79: 7842–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosenwald IB. The role of translation in neoplastic transformation from a pathologist’s point of view. Oncogene 2004; 23: 3230–3247. [DOI] [PubMed] [Google Scholar]
- 13. Tang SW, Chang WH, Su YC, et al. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett 2009; 273: 35–43. [DOI] [PubMed] [Google Scholar]
- 14. Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El Gammal AT, Bruchmann M, Zustin J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res 2010; 16: 56–64. [DOI] [PubMed] [Google Scholar]
- 16. Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 2002; 109: 321–334. [DOI] [PubMed] [Google Scholar]
- 17. Weber RG, Pietsch T, von Schweinitz D, et al. Characterization of genomic alterations in hepatoblastomas. A role for gains on chromosomes 8q and 20 as predictors of poor outcome. Am J Pathol 2000; 157: 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perez EA, Jenkins RB, Dueck AC, et al. C-MYC alterations and association with patient outcome in early-stage HER2-positive breast cancer from the north central cancer treatment group N9831 adjuvant trastuzumab trial. J Clin Oncol 2011; 29: 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klatte T, Kroeger N, Rampersaud EN, et al. Gain of chromosome 8q is associated with metastases and poor survival of patients with clear cell renal cell carcinoma. Cancer 2012: 118: 5777–5782. [DOI] [PubMed] [Google Scholar]
- 20. Edge SB, Byrd DR, Compton CC, et al. AJCC cancer staging manual. 7th ed. New York, NY: Springer-Verlag, 2010. [Google Scholar]
- 21. Miura I, Siegfried JM, Resau J, et al. Chromosome alterations in 21 non-small cell lung carcinomas. Genes Chromosomes Cancer 1990; 2: 328–338. [DOI] [PubMed] [Google Scholar]
- 22. An international system for human cytogenetic nomenclature (1985) ISCN 1985. Report of the Standing Committee on Human Cytogenetic Nomenclature. Birth Defects Orig Artic Ser 2013; 21: 1–137. [PubMed] [Google Scholar]
- 23. Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat 2006: 15: 651–674. [Google Scholar]
- 24. Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One 2008; 3: e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Q, Hopp T, Fuqua SA, et al. Cyclin D1 in breast premalignancy and early breast cancer: implications for prevention and treatment. Cancer Lett 2001; 162: 3–17. [DOI] [PubMed] [Google Scholar]
- 26. Kinouchi T, Saiki S, Naoe T, et al. Correlation of c-MYC expression with nuclear pleomorphism in human renal cell carcinoma. Cancer Res 1989; 49: 3627–3630. [PubMed] [Google Scholar]
- 27. Buness A, Kuner R, Ruschhaupt M, et al. Identification of aberrant chromosomal regions from gene expression microarray studies applied to human breast cancer. Bioinformatics 2007; 23: 2273–2280. [DOI] [PubMed] [Google Scholar]
- 28. Hakimi AA, Mano R, Ciriello G, et al. Impact of recurrent copy number alterations and cancer gene mutations on the predictive accuracy of prognostic models in clear cell renal cell carcinoma. J Urol 2014; 192: 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mehrazin R, Uzzo RG, Kutikov A, et al. Lymphopenia is an independent predictor of inferior outcome in papillary renal cell carcinoma. Urol Oncol. Epub ahead of print 11 July 2014. DOI: 10.1016/j.urolonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mehrazin R, Galsky MD. Kidney cancer: Systemic therapy–differentiating the achievable from the achieved. Nat Rev Urol 2015; 12: 128–129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.