Abstract
Caspases have functions particularly in apoptosis and inflammation. Increasing evidence indicates novel roles of these proteases in cell differentiation, including those involved in osteogenesis. This investigation provides a complex screening of osteogenic markers affected by pan caspase inhibition in micromass cultures derived from mouse forelimbs. PCR Array analysis showed significant alterations in expression of 49 osteogenic genes after 7 days of inhibition. The largest change was a decrease in CD36 expression, which was confirmed at organ level by caspase inhibition in cultured mouse ulnae followed by CD36 immunohistochemical analysis. So far, available data point to osteogenic potential of pro-apoptotic caspases. Therefore, the expression of pro-apoptotic caspases (-3, -6, -7, -8, -9) within the growth plate of mouse forelimbs at the stage where the individual zones are clearly apparent was studied. Caspase-9 was reported in the growth plate for the first time as well as caspase-6 and -7 in the resting zone, caspase-7 in the proliferation, and caspase-6 and -8 in the ossification zone. For all caspases, there was a gradient increase in activation toward the ossification zone. The distribution of staining varied significantly from that of apoptotic cells, and thus, the results further support non-apoptotic participation of caspases in osteogenesis.
Keywords: caspases, endochondral ossification, growth plate, immunohistochemistry, micromass cultures, PCR Array analysis
Introduction
Caspases are a family of cysteine proteases with well-established roles in apoptosis and inflammation. Recent work has been uncovering their manifold non-apoptotic functions, including those in differentiation of various cell types, cell cycle control, metabolism, and autophagy.1–3
These non-apoptotic roles of caspases appear to be important also in osteogenesis.4–8 The majority of bones develop via endochondral ossification, where condensing mesenchymal cells differentiate into chondrocytes, which gradually proliferate, mature, and hypertrophy in the forming growth plate.9 Chondrocytes create an extracellular matrix of cartilage that is later invaded by blood vessels, osteoblasts, osteoclasts, and bone marrow cells. Recent evidence shows that some of the chondrocytes undergo apoptosis, some divide but many of them become bone cells.10 The endochondral ossification is controlled and influenced by plenty of factors, molecules, and hormones,9,11,12 including caspases. Caspases are thought to be involved in chondrocyte terminal differentiation and apoptotic death of osteoblasts.13,14 Osteogenic potential of caspases has so far been described for caspase-3,5,8 caspase-7,7 and suggested for caspase-8.4
The objective of this work was to provide a complex screening of the osteogenic potential of caspases related to endochondral ossification. Osteogenic PCR Arrays of 84 candidate genes was performed after pan caspase inhibition in micromass cultures derived from embryonic mouse forelimbs. The greatest change (decrease) in expression in inhibited cultures found for CD36 was corroborated by further immunohistochemical study in caspase-inhibited cultured mouse ulnae. As the so far available data point particularly to osteogenic potential of pro-apoptotic caspases, activation of caspase-8 and -9 as the dominant initiator caspases and caspase-3, -6, -7 as the trio of executioner caspases was investigated across zones of the mouse ulnae growth plate and correlated with distribution of apoptotic cells.
Materials and Methods
Animals, Samples
Mouse (Mus musculus var. alba) forelimbs at embryonic day (E)18 from the mice strain CD1 were collected and decalcified in buffered ethylenediaminetetraacetic acid (EDTA) after immediate fixation in 4% buffered paraformaldehyde. Mice were killed according to the experimental protocol related to the project GACR 16-18430S approved by the University of Veterinary and Pharmaceutical Sciences, Brno, Czech Republic. Decalcified samples were dehydrated in gradient series of ethanol, treated with xylene, and embedded in paraffin. Serial sections (5 µm) were prepared and split over several slides: hematoxylin–eosin (H&E) and Alcian blue staining, cleaved caspases, and TdT-mediated biotin-dUTP nick end labeling (TUNEL) assay.
Micromass Cultures
Micromass cultures (primary mesenchymal cell cultures) were prepared from E12 mouse forelimbs (as, for example, in Mello and Tuan,15 and Chlastakova et al.16). The forelimbs were disintegrated mechanically in Puck’s saline A buffer (PSA; 0.4 g of KCl/l, 8 g of NaCl/l, 0.35 g of NaHCO3/l, 1 g of glucose/l) with 10% fetal bovine serum (FBS; Invitrogen, Karlsruhe, Germany, 9:1) and enzymatically in 10 U/ml of Dispase II (Sigma-Aldrich; Darmstadt, Germany) in 10% FBS/PSA at 37C, 1–1.5 hr, 1000 rpm, and vortexed every 15 min. After the disintegration, differentiating medium (60% F12/40% Dulbecco’s modified Eagle’s medium [DMEM], 10% FBS, 50 µg/ml ascorbic acid, 10 mM β-glycerol phosphate) was added to neutralize the Dispase activity, and the cell suspension was centrifuged at 1000 × g/5 min. Sediment was resuspended in medium, run through a 40 µm cell strainer (Corning Life Sciences; Falcon) and centrifuged. The final concentration of the cell suspension was 2 × 107 cells/ml. Spots (10 µl) were applied on Nunclon 6-well Delta surface culture dishes (Thermo Fisher Scientific; Waltham, MA) and after 1 hr at 37C in a 5% CO2 incubator, 2 ml of medium was added into each plate. Pan caspase inhibitor FMK001 (R&D Systems; Minneapolis, MN) diluted in dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MI) to the final concentration of 100 µM was applied to the micromass medium after stabilization of the micromass cultures per the manufacturer’s recommendation and previous experiments.17 Differentiating medium with DMSO (control group) and pan caspase inhibitor (experimental group) was changed every second day. After 7 days, cells were collected using 350 µl lysis buffer RLT (Qiagen; Hilden, Germany) with β-ME (Sigma-Aldrich) and prepared for RNA isolation.
Limb Organ Cultures
Limb organ cultures were prepared from dissected mouse E18 ulnae. These were placed on Millipore filters above a metal grid and cultured in differentiating medium with DMSO (control group, inhibitor vehicle) and pan caspases inhibitor (experimental group, 100 µM) at 37C in a 5% CO2 incubator; the medium was changed every day. After 7 days, ulnae were fixed in 4% buffered paraformaldehyde overnight and histologically processed for further analysis. They were dehydrated in gradient series of ethanol, treated with xylene, and embedded in paraffin. Serial sections (5 µm) were prepared and split over several slides: H&E and Alcian blue staining, CD36 antigen immunohistochemistry.
PCR Arrays
RNeasy Mini Kit (Qiagen) was used for RNA isolation, then mRNA was transcribed into cDNA using SuperScript VILO (Invitrogen; Carlsbad, CA), and PCR Arrays were applied to analyze 84 genes connected with osteogenesis (PAMM-026A-24; SA Biosciences, Frederick, MD). Data were statistically evaluated by PCR Array Data Analysis V4 (SA Biosciences). Statistical significance was determined as p<0.05. The threshold of fold change was established as ±1.5. Housekeeping genes used in this analysis were: Actb, B2m, Gapdh, Gusb and Hsp90ab1. Three independent biological repetitions were performed and analyzed.
Immunohistochemistry
Histological sections were deparaffinized in xylene, rehydrated in alcohol series, and pretreated with citrate buffer (5 min at 97C). Endogenous peroxidase activity was eliminated by 3% hydrogen peroxide in phosphate-buffered saline (PBS; 1 min at room temperature), and nonspecific staining was eliminated by incubation in goat serum (45 min, room temperature, ABC kit; Vectastain, Vector Laboratories). Primary antibodies (Table 1) were used overnight at 4C in a humidified chamber. The next day, biotinylated secondary antibody was applied (30 min, room temperature, ABC kit; Vectastain, Vector Laboratories) followed by streptavidin-peroxidase complex (30 min, room temperature, ABC kit; Vectastain, Vector Laboratories). Chromogen substrate 3,3′-diaminobenzidine tetrachloride (DAB, K3466; Dako, Glostrup, Denmark) was used for visualization of positive cells (brown). Hematoxylin was used for counterstaining to clearly distinguish negative cells in blue. Negative controls were performed by omitting the primary antibodies. Interdigital tissue of embryonic mouse limbs was used as a positive control in the case of caspase-3, -6, -7, -9,18 caspase-8,19 and hypertrophic chondrocytes in the case of CD36.20
Table 1.
Information About Primary Antibodies Used in This Study.
| Primary Antibody | Species/Source and Clonality | Species Cross-Reactivity | Optimized Dilution |
|---|---|---|---|
| CD36, Thermo Fisher Scientific, PA5-27236 | Rabbit, pAb | Human, mouse | 1:100 |
| Cleaved Caspase-3 (Asp175), Cell Signaling, 9664S | Rabbit, mAb | Human, mouse, rat | 1:200 |
| Cleaved Caspase-6 (Asp162), Cell Signaling, 9761S | Rabbit, pAb | Human, mouse, rat | 1:200 |
| Cleaved Caspase-7 (Asp198), Cell Signaling, 9491S | Rabbit, pAb | Human, mouse, rat, monkey | 1:50 |
| Cleaved Caspase-8 (Asp387), Cell Signaling, 8592S | Rabbit, mAb | Mouse | 1:800 |
| Cleaved Caspase-9 (Asp353), Cell Signaling, 9509S | Rabbit, pAb | Mouse | 1:100 |
TUNEL Assay
After rehydration and elimination of endogenous peroxidase activity by 3% hydrogen peroxide in PBS (10 min, room temperature), samples were pretreated with proteinase K (20 µg/ml, 15 min, room temperature; Merck Millipore, Billerica, MA). ApopTag Peroxidase In Situ Apoptosis Detection Kit was used (Merck Millipore), after equilibration buffer (15 min, room temperature), reaction mix was applied for 45 min at 37C in humidified chamber. After anti-digoxigenin-peroxidase reaction (30 min, room temperature), positive cells were visualized by the color reaction using the DAB substrate (K3466; Dako), and slides were counterstained with hematoxylin.
Results
Pan Caspase Inhibition Significantly Impacts Osteogenic Gene Expression Profile in Micromass Cultures
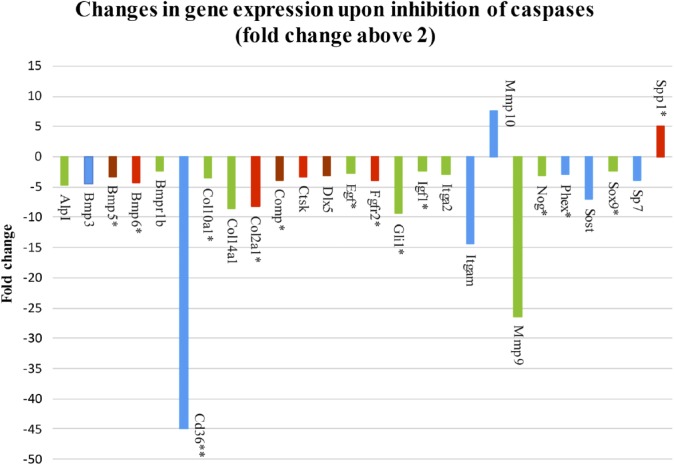
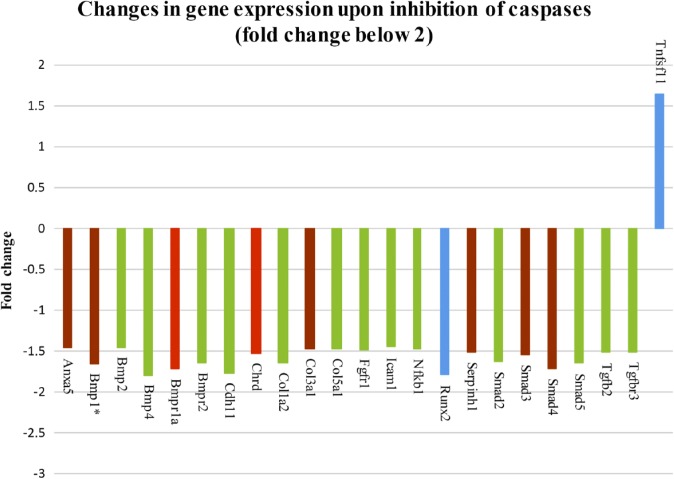
To examine the role of caspases in osteogenic signaling pathways, osteogenic markers were evaluated by PCR Arrays analysis of micromass cultures treated for 1 week with pan caspase inhibitor. Expression of 49 genes was significantly different in the micromass cultures after pan caspase inhibition, compared with non-inhibited controls (treated with inhibitor vehicle DMSO only), 26 of them fold change above 2 (Fig. 1) and 23 of them with fold change between 2 and 1.5 (Fig. 2). Statistical significance was determined using a t-test and distinguished by color in the figures: p<0.05 (blue color), p<0.01 (green color), p<0.001 (brown color), p<0.0001 (red color). Decreased gene expression was seen in the case of Alkaline phosphatase (AlpI), Annexin A5 (Anxa5), Bone morphogenetic proteins (BMPs), and their receptors (Bmp1, Bmp2, Bmp3, Bmp4, Bmp5, Bmp6, Bmpr1a, Bmpr1b, Bmpr2), CD 36 antigen, Cadherin 11 (Cdh11), Chordin (Chrd), Collagens (Col10a1, Col14a1, Col1a2, Col2a1, Col3a1, Col5a1), Cartilage oligomeric matrix protein (Comp), Cathepsin K (Ctsk), Distal-less homeobox 5 (Dlx5), Epidermal growth factor (Egf), Fibroblast growth factor receptors (Fgfr1, Fgfr2), GLI-Kruppel family member (Gli1), Intercellular adhesion molecule 1 (Icam1), Insulin-like growth factor 1 (Igf1), Integrin alpha 2 (Itga2), Integrin alpha M (Itgam), Matrix metalloproteinase 9 (Mmp9), Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (Nfkb1), Noggin (Nog), Phosphate regulating gene with homologies to endopeptidases on the X chromosome (Phex), Runt-related transcription factor 2 (Runx2), Serine (or cysteine) peptidase inhibitor, member 1 (Serpinh1), Smads (Smad2, Smad3, Smad5), Sclerostin (Sost), SRY-box containing gene 9 (Sox9), Sp7 transcription factor 7 (Sp7), Transforming growth factor beta 2 (Tgfb2), and Transforming growth factor beta receptor III (Tgfbr3). On the contrary, increased gene expression was observed in the case of Mmp10, Secreted phosphoprotein 1 (Spp1), and Tumor necrosis factor (ligand) superfamily, member 11 (Tnfsf11).
Figure 1.
Pan caspase inhibitor altered expression of 49 osteogenic genes, 26 genes by the fold change above 2. Abbreviations: Spp1, Secreted phosphoprotein 1; Sp7, Sp7 transcription factor 7; Sox9, SRY-box containing gene 9; Sost, Sclerostin; Phex, Phosphate regulating gene with homologies to endopeptidases on the X chromosome; Nog, Noggin; MMP9 and 10, Matrix metalloproteinase 9 and 10; Itgam, Integrin alpha M; Itga2, Integrin alpha 2; Igf1, Insulin-like growth factor 1; Gli1, GLI-Kruppel family member; FgFr2, Fibroblast growth factor receptors; Egf, Epidermal growth factor; DIx5, Distal-less homeobox 5; Ctsk, Cathepsin K; Comp, Cartilage oligomeric matrix protein; Col2a1, Col14a1, Col10a1, Collagens; Cd36, CD 36 antigen; Bmpr1b, 6, 5, 3, Bone morphogenetic proteins, and their receptors; AlpI, Alkaline phosphatase. The p value is distinguished by the color: p<0.05 (blue color), p<0.01 (green color), p<0.001 (brown color), p<0.0001 (red color). *Expression of genes impacted by caspase-3, data published in our previous report by Adamova et al.8 **The fold change of CD36 reduced 10 times.
Figure 2.
Pan caspase inhibitor altered expression of 49 osteogenic genes, 23 genes by the fold change between 2 and 1.5. Abbreviations: Tnfsf11, Tumor necrosis factor (ligand) superfamily, member 11; Tgfbr2, Transforming growth factor beta 2; Tgfbr3, Transforming growth factor beta receptor III; Smad5, 4, 3, 2, Smads; Serpinh1, Serine (or cysteine) peptidase inhibitor, member 1; Runx2, Runt-related transcription factor 2; Nfkb1, Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1; Icam1, Intercellular adhesion molecule 1; Fgfr1, Fibroblast growth factor receptors; Col5a1, Col3a1, Col1a2, Collagens; Chrd, Chordin; Cdh11, Cadherin 11; Bmpr2, 1a, 4, Bone morphogenetic proteins, and their receptors; Bmp2, 1, Bone morphogenetic proteins; Anxa5, Annexin A5. The p value is distinguished by the color: p<0.05 (blue color), p<0.01 (green color), p<0.001 (brown color), p<0.0001 (red color). *Expression of genes impacted by caspase-3, data published in our previous report by Adamova et al.8
CD36 Is Expressed Throughout the Prenatal Development of Long Bones and Its Expression Is Significantly Decreased After Caspase Inhibition
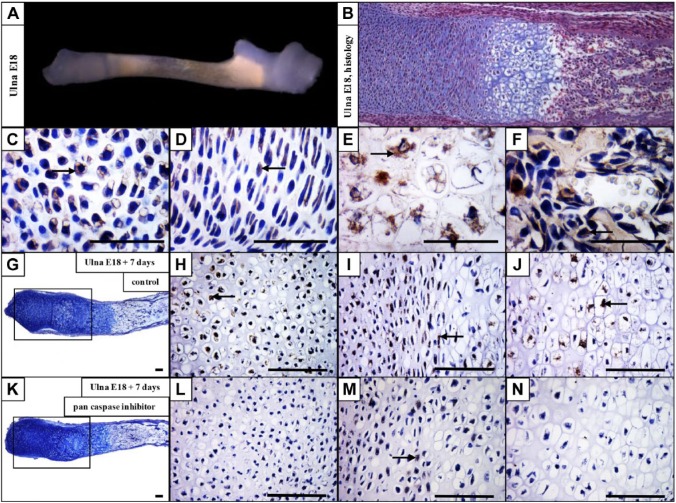
As CD36 expression was the most prominent change after caspase inhibition, its expression on the protein level was examined in developing mouse forelimbs, with a focus on the ulnae. CD36 was detected in the resting chondrocytes, and was particularly abundant in hypertrophic chondrocytes at E15 (data not shown). E18 ulna was used (Fig. 3A) to evaluate CD36 in the growth plate (Fig. 3B). CD36 antigen was present in the resting (Fig. 3C), proliferating (Fig. 3D), hypertrophic (Fig. 3E), and ossification zones (Fig. 3F).
Figure 3.
CD36 is expressed in the growth plate and significantly downregulated after caspase inhibition. Dissected ulna at E18 (A), hematoxylin–eosin and Alcian blue staining of the forelimb at E18 (B). Expression of CD36 in the resting (C), proliferating (D), hypertrophic (E), and ossification (F) zones of the growth plate. E18 ulna + 7 days of cultivation in control medium (G) and expression of CD36 in the ulna cultivated in control medium (H, I, J). E18 ulna + 7 days of cultivation in medium with inhibitor (K) and expression of CD36 in medium with inhibitor (L, M, N). Arrows point to positive cells (brown). Scale bar = 100 µm.
To examine the impact of caspase deficiency on CD36 expression in the intact bone, dissected ulnae were cultured for 7 days under the conditions of caspase inhibition. In non-inhibited control samples (Fig. 3G), CD36 is expressed throughout the growth plate, especially in the resting, proliferation, and hypertrophic zones (Fig. 3H, I, and J). By contrast, in the pan caspase inhibitor treated samples (Fig. 3K), the resting (Fig. 3L), proliferation (Fig. 3M), and hypertrophic (Fig. 3N) zones are mostly CD36 negative.
Pro-Apoptotic Caspases Are Activated in the Growth Plate During Endochondral Ossification, With a Gradient Increase in Activation Toward the Ossification Zone
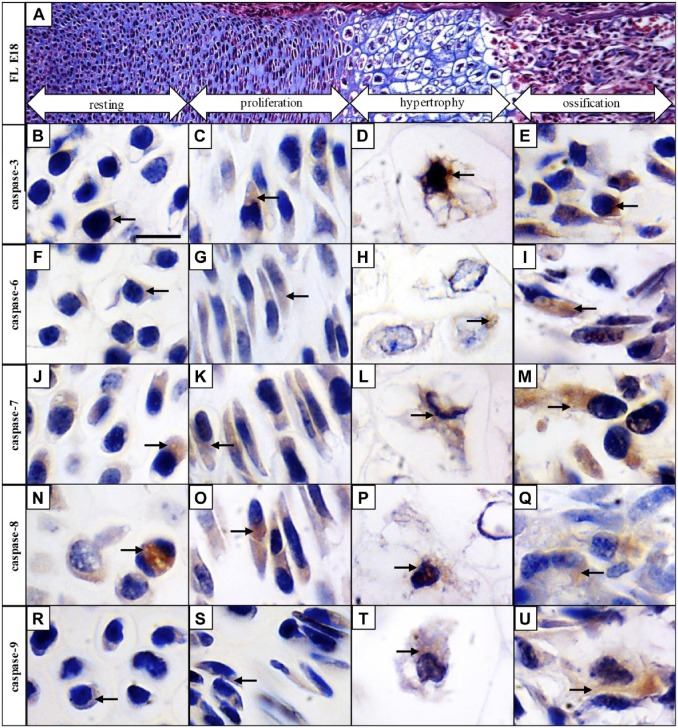
To follow caspase activation in all major zones of the long bone development, mouse forelimbs at E18 were collected for examination. Resting, proliferation, hypertrophic, and ossification zones were clearly distinguishable at this stage. The resting zone of the cartilage has typical round-shaped chondrocytes containing large nuclei compared with the cytoplasm, in the proliferation zone, where round chondrocytes become flattened and create multicellular clusters. Hypertrophic zone is characterized by 5- to 10-fold increased size of chondrocytes and massive secretion of extracellular matrix and in the ossification zone, osteoblasts are differentiated and the matrix is becoming calcified (Fig. 4A).
Figure 4.
Activated pro-apoptotic caspases are expressed throughout the growth plate of developing long bones. Hematoxylin–eosin and Alcian blue staining of the growth plate (A), immunohistochemical detection of individual caspases in resting, proliferation, hypertrophic, and ossification zones of the growth plate: caspase-3 (B, C, D, E), caspase-6 (F, G, H, I), caspase-7 (J, K, L, M), caspase-8 (N, O, P, Q), and caspase-9 (R, S, T, U). Arrows point to positive cells (brown). Scale bar = 50 µm.
Some expression of active caspase-3 was detected in the resting zone of the growth plate (Fig. 4B), and increasing activation was observed in the proliferation (Fig. 4C) and hypertrophic zones (Fig. 4D). Presence of caspase-3 was apparent mostly in the ossification zone (Fig. 4E). Active caspase-6 in the resting (Fig. 4F), proliferating (Fig. 4G), and hypertrophic (Fig. 4H) zones displayed low activation but active caspase-6 was visible particularly in the ossification zone (Fig. 4I). Active caspase-7 was prominent through the growth plate in the resting (Fig. 4J), proliferating (Fig. 4K), hypertrophic (Fig. 4L), as well as ossification zone (Fig. 4M). Similar results were obtained in the case of caspase-8, also activation of this caspase displayed an increasing trend from the resting (Fig. 4N), through proliferating (Fig. 4O), and hypertrophic (Fig. 4P), toward the ossification zone (Fig. 4Q). Some cells positive for caspase-9 were observed in the resting (Fig. 4R), proliferating (Fig. 4S), and hypertrophic zones (Fig. 4T), however, activation of caspase-9 was linked particularly with the ossification zone (Fig. 4U).
Pro-Apoptotic Caspases Are Present in Non-Apoptotic Cells of the Growth Plate as Demonstrated in the Ossification Zone
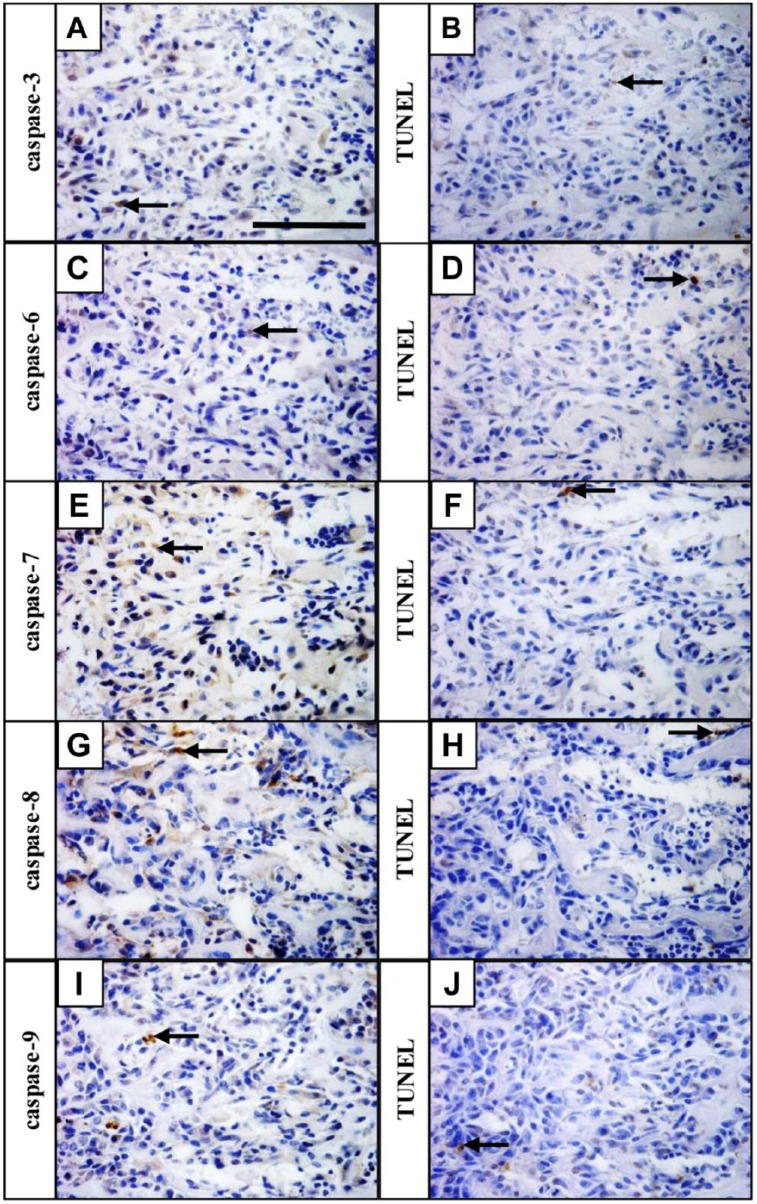
To evaluate the presence of pro-apoptotic caspases in non-apoptotic cells, the pattern of expression of individual active caspases was correlated with localization of apoptotic (TUNEL positive) cells. The ossification zone, where the expression of all caspases was most prominent, was used for examination. The distribution of staining in the ossification zone for individual caspases (Fig. 5A, C, E, G, and I) was different from that for TUNEL (Fig. 5B, D, F, H, and J), showing presence of pro-apoptotic caspases in non-apoptotic cells.
Figure 5.
Pro-apoptotic caspase activation occurs predominantly in non-apoptotic cells. Caspase-3 (A), caspase-6 (C), caspase-7 (E), caspase-8 (G), and caspase-9 (I) expression in the ossification zone, apoptotic cells detected by TUNEL assay on serial sections (B, D, F, H, J). Arrows point to positive cells (brown). Scale bar = 100 µm. Abbreviation: TUNEL, TdT-mediated biotin-dUTP nick end labeling.
Discussion
This work provides novel data related to emerging non-apoptotic roles of caspases in osteogenesis. The general screening of osteogenic impact of caspases related to endochondral ossification identified novel genes whose expression was influenced by active caspases within the osteogenic networks. The evidence about new functions of caspases in endochondral ossification is further supplemented by the activation pattern of caspases-3, -6, -7, -8, and -9 in non-apoptotic cells of the developing mouse growth plate.
In previous studies dealing with caspases and osteogenesis, pan caspase inhibition was performed in limb cultures, and altered expression of Runx2, Alp, Col2a1, and Gdf5 was followed.21 In caspase-inhibited human chondrocytes, apoptosis was prevented and delayed while chondrocytes remained functional.22,23 In our previous article, changes in gene expression (Bmp1, Bmp5, Bmp6, Col10a1, Col2a1, Comp, Egf, Fgfr2, Gli1, Igf1, Nog, Phex, Sox9, and Spp1) following inhibition of caspase-3 were reported, and corresponding alterations of these genes are obvious also after general caspase inhibition.8
The most prominent change in gene expression upon caspase inhibition was observed in the case of the CD36 antigen. The apparent downregulation of CD36 demonstrated by PCR Arrays was confirmed by pan caspase inhibition in cultured ulnae, where the expression of CD36 was maintained in controls but was strongly decreased in treated samples. CD36 functions as a receptor for collagens,24,25 trombospondin,26 and oxidized low-density lipoprotein.27 CD36 is suggested to be involved in bone resorption by activation of c-src signaling in osteoclasts.28,29 CD36 was also presented as a chondrocyte hypertrophy marker,20 and it participates as a key regulator in bone mineralization and osteoblast functions.30,31
After pan caspase inhibition, significantly altered gene expression was also detected in the case of proteins involved in the degradation of the extracellular matrix, the matrix proteinases (Mmp9, Mmp10, and Ctsk). Mmp10 is expressed throughout the growth plate and in osteoclasts.32 Mmp9 is responsible for the invasion of osteoclasts into mineralized cartilage and apoptosis of hypertrophic chondrocytes, and is involved in angiogenesis.33 Ctsk is abundantly expressed by osteoclasts.34 Other extracellular matrix molecules (Itga2, Itgam) were significantly impacted as well, such as integrins participating in cell-cell and cell-extracellular matrix interactions.35
Some BMPs, members of the TGFβ superfamily, were significantly altered by caspase inhibition, and have an important role in cartilage formation (chondrocyte proliferation) during endochondral ossification.36,37 The downregulation of Bmp3 and Bmpr1b receptor following caspase inhibition may indicate a role for caspases in chondrogenesis maintenance, as the upregulation of BMPs in mesenchymal stem cells has osteoinductive activity.38 Moreover, our observations are in agreement with reduction of Alp activation after inhibition of caspase activity in BMP-4-treated cells.4 Alkaline phosphatase has a role in mineralization of the extracellular matrix produced by chondrocytes.39 The BMP antagonist, Sost, is known as a marker of osteocytes40 involved in the inhibition of osteoblast activity and the reduction of differentiation of osteoprogenitors.41
Collagen proteins are the main organic components of bone extracellular matrix.42 The decreased expression of several types of collagens, especially Col14a1 after caspase inhibition indicates changes in the creation of connective tissue in treated micromass cultures. Several transcription factors involved in osteogenesis, particularly Sp7 (Osterix) and Dlx5, were downregulated in the treated cultures. Both molecules are necessary for osteoblast differentiation, and their expression is induced by Bmp2.43
Osteogenic potential has been associated particularly with pro-apoptotic caspases.4,5,7,8 Our temporospatial investigation within the growth plate supplements previous knowledge (Table 2). In our study, the expression of caspase-6, -7, and -9 in the resting and ossification zones, caspase-7 in the proliferation zone, and caspase-9 in the proliferation and hypertrophic zones were demonstrated for the first time. Moreover, comparison of the activation of individual caspases within the ossification zone with the pattern of apoptosis clearly showed presence of these caspases in non-apoptotic cells. In osteogenesis, non-apoptotic activity of pro-apoptotic caspases was earlier demonstrated in the case of caspase-3 in vitro,4–6 caspase-7,7,44 and caspase-8 in vitro.4 Non-apoptotic engagement of these caspases, particularly in proliferation and differentiation, is known also from other organs/tissues/cells, such as skin, brain, muscles, keratinocytes, or immune system cells (reviewed in Shalini et al.3).
Table 2.
References Reporting About Activated Caspases in the Growth Plate of Long Bones Supplemented by Our Data.
| Caspase | Resting Zone | Proliferation Zone | Hypertrophic Zone | Ossification Zone |
|---|---|---|---|---|
| c3 | Chicken Pucci et al.45 Mouse Blumer et al.46 |
Rat Chrysis et al.47 Chicken Pucci et al.45 |
Rat Chrysis et al.47 Chicken Pucci et al.45 Mouse Blumer et al.46 |
Human Krajewska et al.48 |
| c6 | * | Rat Chrysis et al.47 |
Rat Chrysis et al.47 |
* |
| c7 | * | * | Mouse Svandova et al.7 |
Mouse Svandova et al.7 |
| c8 | Human Trieb et al.49 |
Human Trieb et al.49 |
Human Trieb et al.49 |
* |
| c9 | * | * | * | * |
Individual references are given for published works.
Data investigated and presented here for the first time.
Regarding engagement of examined caspases in apoptosis of hypertrophic chondrocytes, our results are consistent with those of previous research.50,51 A number of possible mechanisms have been posited for apoptotic versus non-apoptotic engagement of caspases, including subcellular localization,6,52,53 co-expressions with inhibitor of apoptosis proteins,54 levels of activation of caspase-activated DNase,55 sublethal concentrations of caspases,56 and substrate specificity of different caspases determined by posttranslational modifications and interactions with other proteins.57–59 Apparently, the complex cellular context determines the consequences of caspase activation.60
This investigation added missing data to pro-apoptotic caspase activation within the growth plate and, thus, completed a comprehensive overview of pro-apoptotic caspase pattern related to apoptotic and non-apoptotic events in the resting, proliferating, hypertrophic, and osteogenic zones of the growth plate. The increasing gradient of caspase-positive cells from resting to ossification zone pointed to non-apoptotic functions of caspases in osteogenic pathways. PCR Array-based analysis of general osteogenic potential of caspases indicated several genes within osteogenic molecular networks whose expression is likely to interfere with active caspases in a non-apoptotic way and, thus, provided a list of candidates for further research or targeted modulations related to chondrogenesis and osteogenesis.
Supplementary Material
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: EJ performed the experiments, analyzed the results, and drafted the manuscript, PB performed the experiments, and EM designed the study and revised the manuscript. All authors have read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Czech Science Foundation (GACR 16-18430S) and P.B. by the IGA UVPS Brno, Czech Republic (117/2017/FVL).
ORCID iD: E Janečková  https://orcid.org/0000-0002-3991-8199
https://orcid.org/0000-0002-3991-8199
Contributor Information
Eva Janečková, Department of Physiology, University of Veterinary and Pharmaceutical Sciences, Brno, Czech Republic; Department of Experimental Biology, Faculty of Science, Masaryk University, Brno, Czech Republic.
Petra Bíliková, Department of Physiology, University of Veterinary and Pharmaceutical Sciences, Brno, Czech Republic.
Eva Matalová, Department of Physiology, University of Veterinary and Pharmaceutical Sciences, Brno, Czech Republic; Institute of Animal Physiology and Genetics CAS, v.v.i., Brno, Czech Republic.
Literature Cited
- 1. Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14(1):44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- 2. Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep. 2012;13(4):322–30. doi: 10.1038/embor.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22(4):526–39. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mogi M, Togari A. Activation of caspase is required for osteoblastic differentiation. J Biol Chem. 2003;278(48):47477–82. doi: 10.1074/jbc.M307055200. [DOI] [PubMed] [Google Scholar]
- 5. Miura M, Chen XD, Allen MR, Bi Y, Gronthos S, Seo BM, Lakhani S, Favell RA, Feng XH, Robey PG, Young M, Shi S. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J Clin Invest. 2004;114(12):1704–13. doi: 10.1172/JCI20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Szymczyk KH, Freeman TA, Adams CS, Srinivas V, Steinbeck MJ. Active caspase-3 is required for osteoclast differentiation. J Cell Physiol. 2006;209(3):836–44. doi: 10.1002/jcp.20770. [DOI] [PubMed] [Google Scholar]
- 7. Svandova E, Lesot H, Vanden Berghe T, Tucker AS, Sharpe PT, Vandenabeele P, Matalová E. Non-apoptotic functions of caspase-7 during osteogenesis. Cell Death Dis. 2014;5:e1366. doi: 10.1038/cddis.2014.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adamova E, Janečková E, Kleparnik K, Matalová E. Caspases and osteogenic markers—in vitro screening of inhibition impact. In Vitro Cell Dev Biol Anim. 2016;52(2):144–8. doi: 10.1007/s11626-015-9964-1. [DOI] [PubMed] [Google Scholar]
- 9. Provot S, Schipani E. Molecular mechanisms of endochondral bone development. Biochem Biophys Res Commun. 2005;328(3):658–65. doi: 10.1016/j.bbrc.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 10. Jing Y, Hinton RJ, Chan KS, Feng JQ. Co-localization of cell lineage markers and the tomato signal. J Vis Exp. 2016;118:e54982. doi: 10.3791/54982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. White A, Wallis G. Endochondral ossification: a delicate balance between growth and mineralisation. Curr Biol. 2001;11(15):R589–91. doi: 10.1016/S0960-9822(01)00359-1. [DOI] [PubMed] [Google Scholar]
- 12. Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40(1):46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 13. Hock JM, Krishnan V, Onyia JE, Bidwell JP, Milas J, Stanislaus D. Osteoblast apoptosis and bone turnover. J Bone Miner Res. 2001;16(6):975–84. doi: 10.1359/jbmr.2001.16.6.975. [DOI] [PubMed] [Google Scholar]
- 14. Adams CS, Shapiro IM. The fate of the terminally differentiated chondrocyte: evidence for microenvironmental regulation of chondrocyte apoptosis. Crit Rev Oral Biol Med. 2002;13(6):465–73. [DOI] [PubMed] [Google Scholar]
- 15. Mello MA, Tuan RS. High density micromass cultures of embryonic limb bud mesenchymal cells: an in vitro model of endochondral skeletal development. In Vitro Cell Dev Biol Anim. 1999;35(5):262–9. doi: 10.1007/s11626-999-0070-0. [DOI] [PubMed] [Google Scholar]
- 16. Chlastakova I, Liskova M, Kudelova J, Dubska L, Kleparnik K, Matalová E. Dynamics of caspase-3 activation and inhibition in embryonic micromasses evaluated by a photon-counting chemiluminescence approach. In Vitro Cell Dev Biol Anim. 2012;48(9):545–9. doi: 10.1007/s11626-012-9542-8. [DOI] [PubMed] [Google Scholar]
- 17. Cho JH, Lee PY, Son WC, Chi SW, Park BC, Kim JH, Park SG. Identification of the novel substrates for caspase-6 in apoptosis using proteomic approaches. BMB Rep. 2013;46(12):588–93. doi: 10.5483/BMBRep.2013.46.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zuzarte-Luis V, Berciano MT, Lafarga M, Hurle JM. Caspase redundancy and release of mitochondrial apoptotic factors characterize interdigital apoptosis. Apoptosis. 2006;11(5):701–15. doi: 10.1007/s10495-006-5481-8. [DOI] [PubMed] [Google Scholar]
- 19. Svandova E, Vesela B, Lesot H, Poliard A, Matalová E. Expression of Fas, FasL, caspase-8 and other factors of the extrinsic apoptotic pathway during the onset of interdigital tissue elimination. Histochem Cell Biol. 2017;147(4):497–510. doi: 10.1007/s00418-016-1508-6. [DOI] [PubMed] [Google Scholar]
- 20. Cecil DL, Appleton CT, Polewski MD, Mort JS, Schmidt AM, Bendele A, Beier F, Terkeltaub R. The pattern recognition receptor CD36 is a chondrocyte hypertrophy marker associated with suppression of catabolic responses and promotion of repair responses to inflammatory stimuli. J Immunol. 2009;182(8):5024–31. doi: 10.4049/jimmunol.0803603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Valck D, Luyten FP. Caspase inhibition supports proper gene expression in ex vivo mouse limb cultures. Cell Death Differ. 2001;8(10):985–94. doi: 10.1038/sj.cdd.4400912. [DOI] [PubMed] [Google Scholar]
- 22. Nutall ME, Nadeau DP, Fisher PW, Wang F, Keller PM, DeWolf WE, Jr, Goldring MB, Badger AM, Lee D, Levy MA, Gowen M, Lark MW. Inhibition of caspase-3-like activity prevents apoptosis while retaining functionality of human chondrocytes in vitro. J Orthop Res. 2000;18(3):356–63. doi: 10.1002/jor.1100180306. [DOI] [PubMed] [Google Scholar]
- 23. Gibson G, Lin DL, Wang X, Zhang L. The release and activation of transforming growth factor β2 associated with apoptosis of chick hypertrophic chondrocytes. J Bone Miner Res. 2001;16(12):2330–8. doi: 10.1359/jbmr.2001.16.12.2330. [DOI] [PubMed] [Google Scholar]
- 24. Tandon NN, Kralisz U, Jamieson GA. Identification of glycoprotein IV (CD36) as a primary receptor for platelet-collagen adhesion. J Biol Chem. 1989;264(13):7576–83. [PubMed] [Google Scholar]
- 25. Mercier N, Catimel B, Reck MP, Pellecchia D, McGregor JL. Identification of a functional site on CD36 involved in the interaction between platelets and collagen. Platelets. 1995;6(3):139–45. doi: 10.3109/09537109509013266. [DOI] [PubMed] [Google Scholar]
- 26. Silverstein RL, Baird M, Lo SK, Yesner LM. Sense and antisense cDNA transfection of CD36 (glycoprotein IV) in melanoma cells. J Biol Chem. 1992;267(23):16607–12. [PubMed] [Google Scholar]
- 27. Nicholson AC, Febbraio M, Han J, Silverstein RL, Hajjar DP. CD36 in atherosclerosis. The role of a class B macrophage scavenger receptor. Ann N Y Acad Sci. 2000;902:128–33. doi: 10.1111/j.1749-6632.2001.tb03944.x. [DOI] [PubMed] [Google Scholar]
- 28. Huang MM, Bolen JB, Barnwell JW, Shattil SJ, Brugge JS. Membrane glycoprotein IV (CD36) is physically associated with the Fyn, Lyn, and Yes protein-tyrosine kinases in human platelets. Proc Natl Acad Sci U S A. 1991;88(17):7844–8. doi: 10.1073/pnas.88.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carron JA, Wagstaff SC, Gallagher JA, Bowler WB. A CD36-binding peptide from trombospondin-1 can stimulate resorption by osteoclasts in vitro. Biochem Biophys Res Commun. 2000;270(3):1124–7. doi: 10.1006/bbrc.2000.2574. [DOI] [PubMed] [Google Scholar]
- 30. Kevorkova O, Martineau C, Martin-Falstrault L, Sanchez-Dardon J, Brissette L, Moreau R. Low-bone-mass phenotype of deficient mice for the cluster of differentiation 36 (CD36). PLoS ONE. 2013;8(10):e77701. doi: 10.1371/journal.pone.0077701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staines KA, Zhu D, Farquharson C, MacRae VE. Identification of novel regulators of osteoblast matrix mineralization by time series transcriptional profiling. J Bone Miner Metab. 2014;32(3):240–51. doi: 10.1007/s00774-013-0493-2. [DOI] [PubMed] [Google Scholar]
- 32. Ortega N, Behonick DJ, Werb Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004;14(2):86–93. doi: 10.1016/j.tcb.2003.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Engsig MT, Chen QJ, Vu TH, Pedersen AC, Therkidsen B, Lund LR, Henriksen K, Lenhard T, Foged NT, Werb Z, Delaisse JM. Matrix metalloproteinase 9 and vascular endothelial growth factor are essential for osteoclast recruitment into developing long bones. J Cell Biol. 2000;151(4):879–89. doi: 10.1083/jcb.151.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Littlewood-Evans A, Kokubo T, Ishibashi O, Inaoka T, Wlodarski B, Gallegher JA. Localization of cathepsin K in human osteoclasts by in situ hybridization and immunohistochemistry. Bone. 1997;20(2):81–6. doi: 10.1016/S8756-3282(96)00351-1. [DOI] [PubMed] [Google Scholar]
- 35. Loeser RF. Integrins and chondrocyte-matrix interactions in articular cartilage. Matrix Biol. 2014;39:11–6. doi: 10.1016/j.matbio.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kamiya N, Mishina Y. New insights on the roles of BMP signaling in bone—a review of recent mouse genetic studies. Biofactors. 2011;37(2):75–82. doi: 10.1002/biof.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beederman M, Lamplot JD, Nan G, Wang J, Liu X, Yin L, Li R, Shui W, Zhang H, Kim SH, Zhang W, Zhang J, Kong Y, Denduluri S, Rogers MR, Pratt A, Haydon RC, Luu HH, Angeles J, Shi LL, He TC. BMP signaling in mesenchymal stem cell differentiation and bone formation. J Biomed Sci Eng. 2013;6(8A):32–52. doi: 10.4236/jbise.2013.68A1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22(4):233–41. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 39. Golub EE, Boesze-Battaglia K. The role of alkaline phosphatase in mineralization. Curr Opin Orthop. 2007;18:444–8. doi: 10.1097/BCO.0b013e3282630851. [DOI] [Google Scholar]
- 40. Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, Latham JA. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22(23):6267–76. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sutherland MK, Geoghegan JC, Yu C, Tucott E, Skonier JE, Winkler DG, Latham JA. Sclerostin promotes the apoptosis of human osteoblastic cells: a novel regulation of bone formation. Bone. 2004;35(4):828–35. doi: 10.1016/j.bone.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 42. Gelse K, Poschl E, Aigner T. Collagens—structure, function and biosynthesis. Adv Drug Deliv Rev. 2003;55(12):1531–46. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 43. Ulsamer A, Ortuno MJ, Ruiz S, Susperregui AR, Osses N, Rosa JL, Ventura F. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283(7):3816–26. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 44. Matalová E, Lesot H, Svandova E, Vanden Berghe T, Sharpe PT, Healy C, Vandenabeele P, Tucker AS. Caspase-7 participates in differentiation of cells forming dental hard tissues. Dev Growth Differ. 2013;55(5):615–21. doi: 10.1111/dgd.12066. [DOI] [PubMed] [Google Scholar]
- 45. Pucci B, Adams CS, Fertala J, Snyder BC, Mansfield KD, Tafani M, Freeman T, Shapiro IM. Development of the terminally differentiated state sensitizes epiphyseal chondrocytes to apoptosis through caspase-3 activation. J Cell Physiol. 2007;210(3):609–5. doi: 10.1002/jcp.20857. [DOI] [PubMed] [Google Scholar]
- 46. Blumer MJ, Longato S, Schwarzer C, Fritsch H. Bone development in the femoral epiphysis of mice: the role of cartilage canals and the fate of resting chondrocytes. Dev Dyn. 2007;236(8):2077–88. doi: 10.1002/dvdy.21228. [DOI] [PubMed] [Google Scholar]
- 47. Chrysis D, Nilsson O, Ritzen EM, Savendahl L. Apoptosis is developmentally regulated in rat growth plate. Endocrine. 2002;18(3):271–8. doi: 10.1385/ENDO:18:3:271. [DOI] [PubMed] [Google Scholar]
- 48. Krajewska M, Wang HG, Krajewski S, Zapata JM, Shabaik A, Gascoyne R, Reed JC. Immunohistochemical analysis of in vivo patterns of expression of CPP32 (Caspase-3), a cell death protease. Cancer Res. 1997;57(8):1605–13. [PubMed] [Google Scholar]
- 49. Trieb K, Cetin E, Girsch W, Brand G. Distinct expression of Apo-1 and caspase-8 in human growth plate. Eur Cell Mat. 2003;5(2):57–8. [DOI] [PubMed] [Google Scholar]
- 50. Roach HI, Erenpreisa J, Aigner T. Osteogenic differentiation of hypertrophic chondrocytes involves asymmetric cell divisions and apoptosis. J Cell Biol. 1995;131(2):483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zenmyo M, Komiya S, Kawabata R, Sasaguri Y, Inoue A, Morimatsu M. Morphological and biochemical evidence for apoptosis in the terminal hypertrophic chondrocytes of the growth plate. J Pathol. 1996;180(4):430–3. doi: [DOI] [PubMed] [Google Scholar]
- 52. Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, Wakeham A, Bouchard D, Yeh WC, McGlade JC, Ohashi PS, Hakem R. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev. 2003;17(7):883–95. doi: 10.1101/gad.1063703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Botton S, Sabri S, Daugas E, Zermati Y, Guidotti JE, Hermine O, Kroemer G, Vainchenker W, Debili N. Platelet formation is the consequence of caspase activation within megakaryotes. Blood. 2002;100(4):1310–7. doi: 10.1182/blood-2002-03-0686. [DOI] [PubMed] [Google Scholar]
- 54. Sadowski-Debbing K, Coy JF, Mier W, Hug H, Los M. Caspases—their role in apoptosis and other physiological processes as revealed by knock-out studies. Arch Immunol Ther Exp (Warsz). 2002;50(1):19–34. [PubMed] [Google Scholar]
- 55. Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci U S A. 2010;107(9):4230–5. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Basu S, Rajakaruna S, Menko AS. Insulin-like growth factor receptor-1 and nuclear factor κB are crucial survival signals that regulate caspase-3-mediated lens epithelial cell differentiation initiation. J Biol Chem. 2012;287(11):8384–97. doi: 10.1074/jbc.M112.341586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zermati Y, Garrido C, Amsellem S, Fishelson S, Bouscary D, Valensi F, Varet B, Solary E, Hermine O. Caspase activation is required for terminal erythroid differentiation. J Exp Med. 2001;193(2):247–54. doi: 10.1084/jem.193.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sordet O, Rebe C, Plenchette S, Zermati Y, Hermine O, Vainchenker W, Garrido C, Solary E, Dubrez-Daloz L. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002;100(13):4446–53. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- 59. Nhan TQ, Liles WC, Schwartz SM. Physiological functions of caspases beyond cell death. Am J Pathol. 2006;169(3):729–37. doi: 10.2353/ajpath.2006.060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abraham MC, Shaham S. Death without caspases, caspases without death. Trends Cell Biol. 2004;14(4):184–93. doi: 10.1016/j.tcb.2004.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.