Abstract
Background
The efficacy of novel targeted therapies is often tested at the time of tumor recurrence. However, for glioblastoma (GBM) patients, surgical resections at recurrence are performed only in a minority of patients; therefore, molecular data are predominantly derived from the initial tumor. Molecular data of the initial tumor for patient selection into personalized medicine trials can therefore be used only when the specific genetic change is retained in the recurrent tumor.
Methods
In this study we determined whether EGFR amplification and expression of the most common mutation in GBMs (EGFRvIII) is retained at tumor recurrence. Because retention of genetic changes may be dependent on the initial treatment, we only used a cohort of GBM samples that were uniformly treated according to the current standard of care (ie, chemo-irradiation with temozolomide).
Results
Our data show that, in spite of some quantitative differences, the EGFR amplification status remains stable in the majority (84%) of tumors evaluated. EGFRvIII expression remained similar in 79% of GBMs. However, within the tumors expressing EGFRvIII at initial diagnosis, approximately one-half lose their EGFRvIII expression at tumor recurrence.
Conclusions
The relative stability of EGFR amplification indicates that molecular data obtained in the primary tumor can be used to predict the EGFR status of the recurrent tumor, but care should be taken in extrapolating EGFRvIII expression from the primary tumor, particularly when expressed at first diagnosis.
Keywords: EGFR, EGFRvIII, glioblastoma, recurrent tumors
Gliomas are the most common type of primary brain tumor, of which 60%–70% are diagnosed as glioblastoma multiforme (GBM), the most aggressive variant.1 The current standard of care for GBM patients includes surgical resection followed by chemo-irradiation.2 However, tumors invariably relapse, and treatment options are limited when this occurs. In fact, no standard of care exists for recurrent GBM patients. Nitrosoureas, retreatment with (dose-intense) temozolomide and reirradiation are often employed but have limited activity. Progression-free survival of recurrent GBM is 2–4 months, and post-progression survival is 6–8 months with conventional chemotherapy.3
Current efforts to improve treatment of GBMs are often based on a personalized medicine approach. In this approach, the efficacy of novel agents is tested on those tumors that harbor specific mutations. Personalized medicine trials will generally be performed after the standard of care treatment at the time of tumor recurrence. However, surgical resections at recurrence are performed on a minority of glioma patients. Since marker testing based on circulating tumor DNA is not feasible (<10% detection rate) for glioma patients,4 molecular data can only be derived from analysis of the tumor itself. Therefore, using molecular data of the initial tumor for inclusion into personalized medicine trials requires the specific genetic change to be retained in the recurrent tumor. A recent study on a limited set of low-grade gliomas indicated that only ∼50% of all mutations present in the primary tumor are also present in the recurrent tumor.5 Although this percentage was higher for the known causal cancer genes, this demonstrates the need to obtain more insight into the correlation between molecular changes of the primary and recurrent tumor, especially if this molecular change is the target for treatment at progression. A substantial difference between newly diagnosed and recurrent tumors will indicate that patients require repeat surgery for inclusion into a personalized medicine trial.
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that is frequently amplified and mutated in GBMs.6,7 The most common mutation found in GBM patients, the EGFRvIII mutation, is an in-frame deletion of exons 2–7 that results in the receptor being constitutively active. Because EGFR amplification and EGFRvIII expression contribute to tumor formation, EGFR is a potential target for treatment in GBM patients.8–12 In this study we therefore screened for differences in EGFR status and EGFRvIII expression between tumors at initial diagnosis and at recurrence.
Methods
Samples
GBM samples were collected from 2 hospitals in the Netherlands (Erasmus MC in Rotterdam and MC Haaglanden in The Hague) from patients operated between 1999 and 2013, who had resurgery at first recurrence. Use of patient material was approved by the Institutional Review Board of the respective hospitals. Patients were uniformly treated with chemoradiation with temozolomide.2 All samples were evaluated for tumor content by a central review pathologist (J.M.K.), and samples with insufficient tumor content (<30%) were omitted from the analysis.
Quantitative Reverse Transcriptase Polymerase Chain Reaction
DNA and RNA were isolated using the Allprep DNA/RNA FFPE kit (Qiagen) according to the manufacturers' instructions except for an extension of the prot K incubation step from 15′ to overnight. EGFR amplification status and EGFRvIII expression were determined by quantitative reverse transcriptase (qRT-)PCR using assays from Life Technologies. The assay for EGFR DNA (assay number Hs02501405_cn) was designed ∼1100 bp downstream of exon 1 because few genomic changes occur in this region; genomic breakpoints giving rise to EGFRvIII occur further downstream in this intron.13 Control probes for DNA were RNase P (TaqMan copy number reference assay) and BRAF (HS04949885). EGFR status was determined as the average of the Ct values of control probes minus the average EGFR Ct values. The qPCR assay used correlated with EGFR amplification status as determined by copy number arrays (n = 5, Oncoscan DX, Affymetrix); examples are shown in Supplementary material, Figure S1.
EGFRvIII expression was determined with qRT-PCR using a custom-made primers/probe set designed over the exon 1–8 transition. Control qRT-PCR primers were targeted against EGFR wt (HS01076078_m1), RPL30 (Hs00265497_m1), and POP4 (Hs00198357_m1). Samples in which EGFRvIII expression >35 Ct values were scored as negative. EGFRvIII expression was scored as percentage of all EGFR transcripts (EGFRvIII + EGFR wild-type [wt]). In this case, 30% expression of EGFRvIII indicates that EGFRvIII expression is 1 Ct value lower than that of EGFR wt.
All primers showed linear amplification over a wide range of Ct values (DNA content or RNA expression). This finding was observed in 5 independent samples. Slope of the dilution curve was also similar between the 3 primers used, which allows direct comparison between primers used. All qRT-PCR experiments were run in duplicate. The concordance correlation coefficient (Lin, equivalent to intraclass correlation coefficient ICC), was used to assess similarities between EGFR measurements.14
Results and Discussion
EGFR Amplification
A total of 89 cases were identified, of which tissue from 76 cases was available from both resections. EGFR amplification status could be determined in 55 primary-recurrent tumor pairs (Table 1); in remaining patients, we were unable to determine EGFR status in at least one of the 2 samples due to low tissue amounts (n = 7), too low tumor content (n = 1), insufficient DNA quality (n = 10), or no tumor tissue in the block (n = 3). Of these, EGFR amplification, as defined by a ΔCt >3 between controls and EGFR (which corresponds to an approximately 8-fold (23) increase) was present in 40 of 55 (73%) samples at first diagnosis. High-copy EGFR amplification (ie, those tumors having a ΔCt >5, [∼32-fold, 25]) was observed in 23 of 55 (41%) samples. The patient cohort examined in this study therefore had a higher percentage of tumors with EGFR amplification than reported in other studies.6,15 This higher percentage of EGFR-amplified tumors may reflect sample bias or may be caused by differences in sensitivity of the different techniques used. Alternatively, a higher percentage of EGFR-amplified tumors may also be a result of selective enrichment for second surgeries (and retreatment) of EGFR-amplified tumors.
Table 1.
Patient characteristics and molecular data
| Pat ID | Age (y) | Sex | Extent of Resection |
Loc | RT | TMZ | Tumor (%) |
PD (days) | OS (days) | ev | EGFR (dCt) |
EGFRvIII (%) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pr | Rec | Pr | Rec | Pr | Rec | Pr | Rec | |||||||||
| AAA | 54.2 | M | PR | CR | P | 60 | 40% | 222 | 396 | 1 | 0.43 | |||||
| AAB | 68.3 | M | PR | PR | P | 60 | 80% | 80% | 434 | 584 | 1 | 2.93 | 86.5 | 93.7 | ||
| AAC | 68.3 | F | PR | PR | T | 60 | 90% | 60% | 67 | 304 | 1 | 6.28 | 4.08 | 85.1 | ||
| AAD | 64.3 | M | PR | PR | T | 60 | 250 | 451 | 1 | 5.82 | 0.0 | |||||
| AAF | 43.6 | F | PR | PR | F | 60 | 70% | 50% | 139 | 590 | 1 | 3.43 | 1.75 | 0.0 | 0.0 | |
| AAG | 43.6 | F | PR | PR | T | 60 | 2 | 70% | 70% | 108 | 379 | 1 | 0.85 | 0.55 | 0.0 | 0.0 |
| AAI | 57.7 | M | PR | PR | T | 5 | 70% | 60% | 26 | 445 | 1 | 3.64 | 3.51 | 0.0 | ||
| AAJ | 60.9 | F | PR | PR | T | 60 | 2 | 70% | 50% | 143 | 282 | 1 | 0.92 | 0.61 | 0.0 | |
| AAK | 58.0 | F | PR | PR | P | 60 | 4 | 70% | 60% | 182 | 605 | 1 | 3.29 | 3.55 | 0.0 | 0.0 |
| AAL | 60.1 | M | PR | PR | O | 60 | 3 | 70% | 60% | 187 | 373 | 1 | 2.54 | 2.88 | 0.0 | 0.0 |
| AAM | 63.0 | F | PR | PR | T | 60 | 6 | 70% | 60% | 271 | 410 | 1 | 5.82 | 6.32 | 85.2 | 79.7 |
| AAN | 50.3 | F | PR | PR | P | 60 | 70% | 70% | 455 | 527 | 1 | 6.00 | 2.59 | 36.1 | 0.0 | |
| AAS | 37.3 | M | PR | PR | F | 60 | 6 | 80% | 70% | 264 | 508 | 1 | 4.41 | 0.0 | 0.0 | |
| AAT | 62.5 | F | PR | PR | F | 60 | 6 | 80% | 50% | 833 | 1277 | 1 | 6.62 | 4.38 | 68.9 | 0.0 |
| AAU | 52.5 | F | PR | PR | FP | 60 | 6 | 70% | 80% | 647 | 1279 | 1 | 3.84 | 0.0 | 0.0 | |
| AAV | 60.9 | M | PR | PR | T | 60 | 12 | 70% | 60% | 532 | 1412 | 1 | 5.39 | 6.49 | 0.0 | 0.0 |
| AAW | 40.7 | F | PR | CR | F | 60 | 1 | 70% | 60% | 104 | 448 | 1 | 0.54 | 0.51 | 0.0 | 0.0 |
| AAX | 43.0 | F | PR | PR | O | 60 | 1 | 61 | 754 | 1 | 11.29 | 9.41 | 0.2 | |||
| AAY | 69.6 | F | PR | PR | F | 60 | 80% | 80% | 257 | 315 | 1 | 0.36 | 0.84 | 0.0 | 0.0 | |
| ABA | 52.9 | F | PR | PR | T | 60 | 6 | 70% | 80% | 241 | 470 | 1 | 3.52 | 5.97 | 0.0 | 0.0 |
| ACA | 65.3 | M | PR | PR | F | 60 | 6 | 90% | 70% | 147 | 247 | 1 | 8.74 | 3.87 | 0.0 | 0.0 |
| ADA | 55.7 | M | PR | PR | F | 60 | 6 | 363 | 602 | 1 | 6.71 | 4.48 | 0.0 | |||
| AFA | M | PR | PR | F | 60 | 6 | 80% | 80% | 496 | 850 | 1 | 0.26 | 0.20 | 0.0 | 0.0 | |
| AGA | 50.5 | M | CR | PR | T | 70 | 6 | 90% | 70% | 305 | 535 | 1 | 5.04 | 4.70 | 0.0 | 0.0 |
| AHA | 50.8 | M | PR | PR | F | 60 | 4 | 70% | 80% | 195 | 332 | 1 | 4.43 | 7.01 | 0.0 | |
| AIA | 65.2 | M | PR | PR | T | 60 | 6 | 280 | 437 | 1 | 2.28 | 4.05 | 0.0 | |||
| ALA | 50.5 | M | CR | PR | T | 60 | 6 | 90% | 70% | 274 | 0 | 6.97 | 8.86 | 7.5 | 0.0 | |
| AMA | 61.9 | M | PR | CR | P | 60 | 6 | 70% | 70% | 1707 | 1740 | 1 | 4.08 | 3.72 | 91.5 | |
| AOA | 64.5 | F | CR | CR | P | 60 | 12 | 80% | 80% | 434 | 0 | 6.30 | 0.78 | 0.7 | 0.0 | |
| AQA | 75.1 | F | CR | PR | T | 40 | 9 | 60% | 70% | 352 | 0 | 3.67 | 1.10 | 0.0 | 0.0 | |
| ARA | 68.9 | M | PR | CR | F | 60 | 6 | 258 | 0 | 3.06 | 0.38 | 0.0 | ||||
| CAB | 55.8 | M | PR | PR | O | 60 | 4 | 50% | 60% | 214 | 479 | 1 | −0.23 | 3.93 | ||
| CAC | 44.6 | M | PR | CR | T | 60 | 5 | 70% | 30% | 270 | 576 | 1 | 8.45 | 1.93 | 71.5 | 6.5 |
| CAD | 51.6 | M | PR | PR | T | 60 | 5 | 60% | 70% | 252 | 348 | 1 | −0.30 | 3.33 | 0.0 | |
| CAF | 28.4 | M | PR | CR | T | 60 | 6 | 70% | 70% | 276 | 694 | 1 | 4.65 | 6.68 | 54.1 | 27.4 |
| CAK | 45.7 | F | PR | PR | P | 60 | 2 | 40% | 30% | 229 | 395 | 1 | 1.43 | 3.43 | 0.0 | |
| CAM | 47.2 | M | B | PR | T | 60 | 60% | 60% | 388 | 520 | 1 | 7.48 | 8.70 | 0.0 | ||
| CAN | 66.0 | M | PR | B | T | 60 | 6 | 80% | 50% | 270 | 494 | 1 | 4.08 | 6.58 | 0.0 | |
| CAO | 50.4 | M | PR | PR | T | 60 | 6 | 70% | 70% | 605 | 940 | 1 | 8.10 | 10.75 | 9.9 | 0.0 |
| CAS | 53.8 | M | CR | B | F | 60 | 80% | 30% | 198 | 560 | 1 | 6.65 | 3.10 | 1.9 | ||
| CAV | 31.4 | F | PR | PR | T | 60 | 6 | 70% | 70% | 451 | 673 | 1 | 3.10 | 0.0 | 0.0 | |
| CAX | 39.8 | M | PR | PR | P | 60 | 2 | 70% | 20% | 162 | 1079 | 1 | 4.18 | 3.40 | 0.0 | |
| CAZ | 43.0 | F | PR | B | T | 60 | 6 | 80% | 50% | 905 | 1240 | 1 | 2.83 | 2.15 | 0.0 | 0.0 |
| CBA | 56.6 | M | PR | PR | F | 60 | 60% | 40% | 109 | 190 | 1 | 2.33 | 0.80 | 0.0 | 0.0 | |
| CBE | 53.9 | F | PR | PR | T | 59 | 6 | 80% | 70% | 297 | 523 | 1 | 4.45 | 6.15 | 0.0 | 0.0 |
| CBF | 59.7 | F | PR | PR | F | 60 | 80% | 70% | 232 | 513 | 1 | 5.98 | 4.33 | 87.3 | 95.9 | |
| CBG | 31.7 | M | PR | PR | F | 648 | 90% | 80% | 389 | 702 | 1 | 1.40 | 0.0 | |||
| CBH | 72.8 | M | PR | PR | O | 40 | 70% | 60% | 120 | 333 | 1 | 1.03 | 0.0 | |||
| CBI | 41.6 | F | PR | PR | O | 60 | 6 | 80% | 90% | 290 | 633 | 1 | 5.35 | 5.55 | 61.2 | 47.0 |
| CBM | 55.3 | M | PR | PR | 60 | 6 | 60% | 60% | 271 | 353 | 1 | 3.03 | 3.58 | 0.0 | 0.0 | |
| CBP | 61.0 | F | PR | PR | P | 60 | 2 | 70% | 80% | 181 | 546 | 1 | −0.05 | 0.10 | 0.0 | 0.0 |
| CBQ | 61.3 | M | PR | PR | P | 60 | 6 | 80% | 70% | 698 | 1283 | 1 | 5.40 | 4.65 | 0.0 | 0.0 |
| CBR | 60.1 | M | PR | T | 60 | 2 | 70% | 90% | 1291 | 343 | 1 | 1.45 | 0.0 | 0.0 | ||
| CBS | 52.7 | F | PR | PR | T | 60 | 2 | 70% | 70% | 170 | 260 | 1 | 4.73 | 2.25 | 0.0 | 0.0 |
| CBT | 52.5 | M | PR | CR | F | 60 | 6 | 60% | 70% | 289 | 681 | 1 | 6.55 | 6.90 | 51.0 | 20.6 |
| CBV | 50.0 | M | PR | PR | F | 60 | 6 | 30% | 308 | 1383 | 1 | 3.75 | ||||
| CBW | 49.3 | M | PR | PR | T | 60 | 8 | 70% | 60% | 1261 | 1903 | 1 | 0.52 | 3.30 | 0.0 | 0.0 |
| CCA | 45.4 | F | CR | PR | P | 60 | 6 | 70% | 70% | 885 | 1488 | 1 | 7.58 | 8.08 | 0.0 | 0.5 |
| CCB | 52.1 | M | PR | B | T | 60 | 5 | 60% | 60% | 202 | 511 | 1 | 4.30 | 3.90 | 12.4 | |
| CCD | 52.5 | F | PR | PR | 60 | 80% | 70% | 283 | 327 | 1 | 4.65 | 4.60 | 0.0 | 0.0 | ||
| CCP | 43.2 | F | PR | PR | F | 59 | 203 | 279 | 1 | 2.10 | −0.95 | 0.0 | ||||
| CCV | 49.2 | M | PR | T | 60 | 3 | 80% | 411 | 413 | 1 | 8.10 | 0.0 | ||||
| CCW | 48.0 | F | PR | PR | T | 60 | 70% | 30% | 191 | 529 | 1 | 7.25 | 6.60 | 52.3 | 0.0 | |
| CCX | 49.9 | M | PR | PR | P | 65 | 30% | 70% | 2069 | 2743 | 1 | 3.48 | 3.50 | 22.7 | 0.0 | |
| CCZ | 51.2 | F | PR | PR | O | 60 | 3 | 80% | 70% | 247 | 277 | 1 | 8.43 | 8.88 | 76.9 | 51.4 |
| CDA | 65.6 | M | PR | PR | T | 60 | 6 | 80% | 50% | 628 | 890 | 1 | 5.88 | 4.10 | 0.0 | |
| CDB | 36.5 | M | PR | PR | T | 60 | 80% | 30% | 109 | 223 | 1 | 3.75 | 0.0 | 83.9 | ||
| CAY | 48.8 | F | PR | PR | O | 60 | 6 | 282 | 336 | 1 | 8.50 | 7.0 | ||||
| CBO | 63.6 | M | PR | B | T | 60 | 5 | 262 | 512 | 1 | 2.20 | 0.0 | ||||
| AEA | 45.0 | F | CR | CR | T | 60 | 6 | 281 | 402 | 1 | 4.08 | 0.0 | ||||
| AAE | 53.0 | F | PR | PR | F | 60 | 6 | 1026 | 1357 | 1 | 4.60 | 0.0 | ||||
| AAH | 46.8 | M | PR | PR | P | 60 | 335 | 545 | 1 | 8.52 | 1.4 | |||||
| ANA | 65.6 | M | CR | PR | T | 60 | 6 | 430 | 0 | 8.95 | 0.0 | |||||
| AAP | 47.9 | F | PR | PR | T | 60 | 4 | 534 | 1802 | 1 | 10.34 | 63.7 | ||||
| AAR | 52.8 | M | CR | PR | P | 60 | 3 | 186 | 393 | 1 | 2.6 | |||||
| AKA | 61.4 | F | CR | PR | F | 60 | 3 | 90% | 70% | 211 | 364 | 1 | 0.94 | 0.0 | ||
Abbreviations: B, biopsy; CR, complete resection; F, female; M; male; Pr, primary tumor; PR, partial resection; Rec, recurrent tumor; TMZ, number of cycles; RT, radiation therapy dose (Gy); Loc, tumor location (F, frontal; O, occipital; P, parietal; T, temporal; FP, posterior fossa).
To test whether EGFR-amplified tumors are more frequently eligible for resurgery, we tested for such a selective enrichment on GBM samples treated within Erasmus MC (between 1989 and 2005) as reported by us.16 For this analysis, we used molecular subtyping based on gene expression data as a surrogate marker for EGFR amplification: EGFR amplification occurs predominantly in one molecular subtype (IGS-18, a subtype similar to “classical” GBMs as defined by The Cancer Genome Atlas [TCGA]).16,17 Of the tumors diagnosed as GBM at initial presentation, 32 were assigned to IGS-18, of which 7 (22%) patients received resurgery. This frequency is ∼2–3-fold lower in tumors assigned to other subtypes (where EGFR amplification is infrequent) including IGS-22 (1/12, 8.3%) or IGS-23 (6/47 12.8%; this subtype shows overlap with the TCGA mesenchymal GBMs). Although this difference is not statistically significant, it does provide some support for the bias towards resurgery of EGFR-amplified tumors found in current. Of note, this potential bias was not observed in the TCGA dataset in which 20 of 39, 2 of 5, and 11 of 18 patients received resurgery (tumors assigned to IGS-18, IGS-22, and IGS-23, respectively).
We also compared clinical data from this study with data from GBMs in a historical cohort (n = 259) to screen for potential sample bias.16 As may be expected, patients in the recurrent GBM cohort had a better performance score compared with the historical cohort (90.1 ± 8.7 vs 81.6 ± 17.0; P < .0001, t test) and were younger in age (51.2 ± 12.7 years vs 55.7 ± 13.6 years; P < .0001, t test). Our cohort also had a significantly lower male-to-female ratio compared with our historic cohort (48:42 vs 175:84; P = .006, Fisher exact test). There were also some differences in tumor location (n = 27, 15, 7, and 36 vs n = 40, 33, 12, and 29 for frontal, parietal, occipital and temporal locations, respectively), although this difference did not reach statistical significance (P = .06, chi-square test). However, re-resection of GBMs will only be performed on tumors that are relatively accessible for surgery, which inevitably results in a location bias.
EGFRvIII Expression
Of the 76 cases with tissue available from the primary and recurrent tumors, EGFRvIII expression could be evaluated in 111 samples from 69 patients (Table 1). Data from both primary and recurrent samples was generated for 42 patients; data from either the primary or recurrent tumor were of insufficient quality in the remaining 27 patients (in most cases, qRT-PCR could detect transcripts, but the Ct values were too high to reliably allow quantification of EGFRvIII expression). EGFRvIII expression was detected in 34 samples and, apart from one recurrent sample, only occurred in samples with a genomic amplification of the EGFR locus (Fig. 1B). For the single sample with EGFRvIII expression without EGFR amplification (patient CAC) it should be noted that high copy EGFR amplification and EGFRvIII expression were detected in the primary tumor, but the recurrent tumor had a much lower tumor content (30%). EGFRvIII expression was detected in 17 of 35 (49%) primary tumors with EGFR amplification (ΔCt >3), which is a similar frequency as previously reported.6
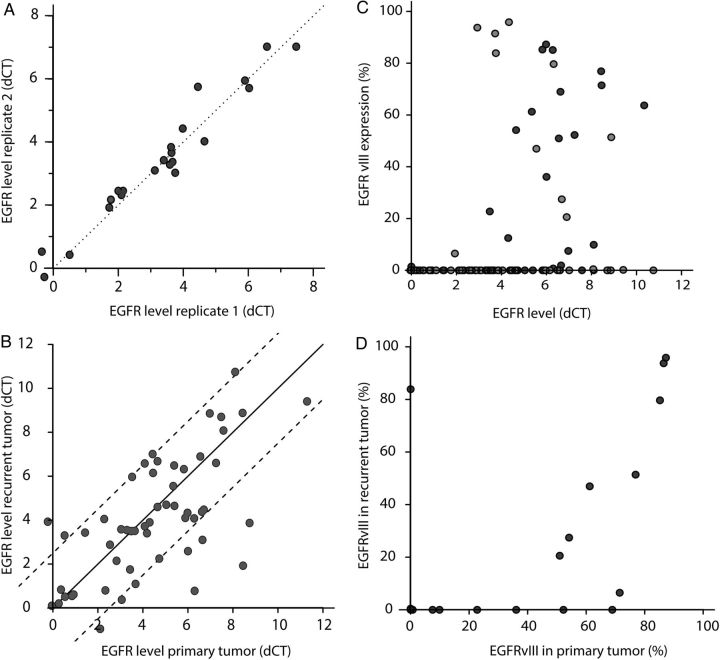
Fig. 1.
(A) Variability of EGFR amplification within biological replicates. As can be seen, the EGFR status between replicates was relatively constant in our samples. (B) EGFR amplification of primary versus recurrent glioblastomas. Although EGFR amplification varied between the primary and recurrent tumor, the difference was generally within 2.5 ΔCt values (dotted lines) of each other. (C) EGFRvIII expression, plotted as a percentage of all EGFR transcripts, is predominantly observed in samples with EGFR amplification (ie, those with dCt >3). Points in dark grey are from initial diagnoses, and light grey is from the recurrent tumor. (D) EGFRvIII expression in primary versus recurrent tumors. As can be seen, the relative expression of EGFRvIII was often lower in recurrent tumors than in primary tumors, with 7 samples showing EGFRvIII expression only in the primary tumor.
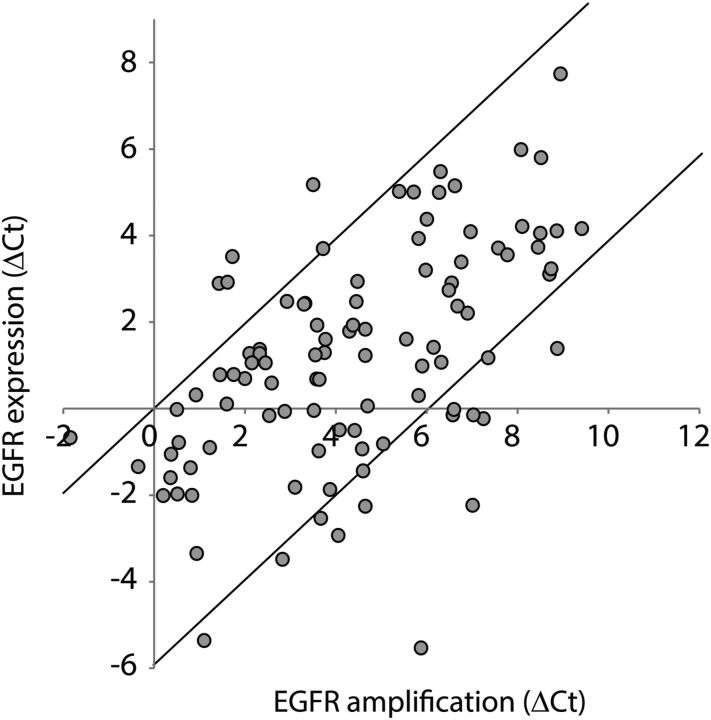
Similar to a report by Hobbs et al,15 our data show that EGFR amplification status was highly variable between tumors. While some tumors showed only modest amplification levels (3–4 ΔCt values), other tumors showed a much stronger amplification (up to 10 ΔCt value difference between EGFR and controls). Although the EGFR amplification status was variable between tumors, the EGFR status was relatively constant within biological replicates (n = 22, Fig. 1A). EGFRvIII expression was also highly variable between different tumors and ranged from <1% up to >90% of all EGFR transcripts being EGFRvIII. EGFR amplification and EGFR gene expression levels were correlated (Fig. 2).
Fig. 2.
Correlation between EGFR amplification status (x-axis) and EGFR gene expression levels (y-axis) as determined by quantitative reverse-transcriptase PCR on tumor DNA or RNA.
Most Glioblastomas Retain Their EGFR Amplification Status at Tumor Recurrence
EGFR amplification of the recurrent tumor did differ from the primary tumor, but the difference was generally within 2–2.5 ΔCt values of each other (Fig. 1C). The overall concordance correlation coefficient between primary and recurrent tumors was 0.65. Cases in which the difference between primary and recurrent tumors was <2.5 ΔCt values (n = 42 tumor pairs) were considered to have retained their EGFR amplification status. In 13 tumors, the difference in EGFR amplification between primary and recurrent tumors was >2.5 ΔCt values; only 4 tumors showed a marked (≥4 ΔCt values) difference between the initial tumor at recurrence. More detailed analysis failed to detect any specific characteristics for these tumors with respect to extent of resection, use of steroids, MGMT promoter methylation, and tumor location. Also, while we did observe a slightly higher tumor content in the primary tumor (71% ± 14% vs 63% ± 17%; P<.001, paired t test), this change is unlikely to explain discrepancies in EGFR amplification status between the tumor at initial diagnosis and at recurrence: a 2-fold decrease in tumor content would result in a maximal decrease in Ct value of one (ie, one PCR cycle). The EGFR amplification status would change in 8 of 13 samples showing a change >2.5 ΔCt values between primary and recurrent: 5 from amplified to nonamplified (with 3 from high copy amplification, ie, ΔCt values >5 to nonamplified) and 3 from EGFR not amplified to amplified, all of which resulted in moderate levels of EGFR amplification (ie, ΔCt values >3 but <5). Overall, the EGFR amplification status (dichotomized to either nonamplified or amplified) remained identical in most tumor pairs (46/55; 84%, Table 2).
Table 2.
Summary of EGFR and EGFRvIII data
|
EGFR in Recurrent Tumor |
||||
|---|---|---|---|---|
| Nonamp | Amp | n | ||
| EGFR in primary tumor | Nonamp | 10 | 5a | 15 |
| Amp | 7a | 33 | 40 | |
| n | 17 | 38 | 55 | |
|
EGFRvIII in Recurrent Tumor |
||||
| Absent | Present | n | ||
| EGFRvIII in primary tumor | Absent | 25 | 2 | 27 |
| Present | 7 | 8 | 15 | |
| n | 32 | 10 | 42 | |
Abbreviations: Amp, amplified; Nonamp, nonamplified.
Cutoff value for EGFR amplification is ΔCt>3 between EGFR and control probes.
aOf the samples that changed EGFR status from nonamplified to amplified or from amplified to nonamplified, 9 showed a difference in ΔCt value >2.5 between the primary and recurrent tumor. When considering that a change in EGFR amplification status also requires >2.5 ΔCt values difference between primary and recurrent tumors, 46 of 55 (84%) tumors retained their EGFR status. Only 5 showed a difference in ΔCt value >3 between the primary and recurrent tumor.
Glioblastomas Can Lose EGFRvIII Expression at Tumor Recurrence
The relative expression of EGFRvIII was often lower in recurrent tumors than in the primary tumor. Of the 15 tumors with detectable EGFRvIII expression in the primary tumor, 8 showed a>20% decrease in relative abundance of EGFRvIII transcripts (Fig. 1D). In fact, the EGFRvIII variant was lost at the time of progression in 7 of 15 EGFRvIII-positive tumors at first surgery. These data are in line with data reported in a different study using an unselected patient cohort,18 although intratumoral heterogeneity may also explain part of this variability.19,20
Of the 15 tumors with EGFRvIII expression, corresponding EGFR amplification status was available for 14. The majority of these (9/14) showed a relative increase in EGFR amplification (ΔCt between the tumor at initial diagnosis and at recurrence between 0 and 3), even though EGFRvIII expression decreased (n = 8) or stayed the same (n = 1). In fact only 3 of 14 showed concordant decrease in EGFR amplification status (> 2.0 ΔCt values between initial recurrent tumors) and decrease in EGFRvIII expression.
Qualitatively EGFRvIII status (present or absent) remained similar between the primary and recurrent tumor in 33of 42 (79%) samples: EGFRvIII was absent from the primary and recurrent tumor in 25 samples and expressed in both primary and recurrent tumor in 8 samples (Table 2). The loss of EGFRvIII expression may be explained by the hypothesis that EGFRvIII deletions occur after EGFR amplification and that individual cells harbor varying levels of EGFRvIII.5 Loss of EGFRvIII expression at tumor recurrence then represents clonal selection of the tumor. Indeed, gliomas are heterogeneous tumors in which distinct subpopulations of cells exist, each with different genetic makeup.5,21 However, recent evidence also suggests that genomic EGFRvIII deletion is an early event and that EGFRvIII expression is regulated by the tumor.19 In fact, mice experiments demonstrated that the ratio of EGFRwt-to-EGFRvIII expression was similar to the primary tumor at regrowth, even when sorting for EGFRvIII high- or low-expressing tumor cells22 (see also23). Therefore, loss of EGFRvIII expression is a result of epigenetic regulation.
In summary, our data show that, in spite of some quantitative differences, the EGFR amplification status remained stable in the majority (∼84%) of tumors evaluated. EGFRvIII status also remained similar in 79% of GBMs; however, when focusing on EGFRvIII-expressing tumors, only 50% retained EGFRvIII expression at recurrence. The relative stability of EGFR amplification expression therefore indicates that molecular data obtained in the primary tumor can be used to predict the EGFR status of the recurrent tumor. Care should be taken when extrapolating EGFRvIII expression; a repeat biopsy should be considered in trials on recurrent glioblastoma targeting EGFRvIII mutations.
Funding
This work was supported by a grant from AbbVie.
Conflict of interest statement. Consultancy for AbbVie, M. van den Bent.
Supplementary Material
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. 4th ed.Lyon: The World Health Organization; 2007. [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Gorlia T, Stupp R, Brandes AA, et al. New prognostic factors and calculators for outcome prediction in patients with recurrent glioblastoma: a pooled analysis of EORTC Brain Tumour Group phase I and II clinical trials. Eur J Cancer. 2012;48(8):1176–1184. [DOI] [PubMed] [Google Scholar]
- 4. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rich JN, Reardon DA, Peery T, et al. Phase II trial of gefitinib in recurrent glioblastoma. J Clin Oncol. 2004;22(1):133–142. [DOI] [PubMed] [Google Scholar]
- 9. van den Bent MJ, Brandes AA, Rampling R, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27(8):1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vivanco I, Robins HI, Rohle D, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2(5):458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gan HK, Fichtel L, Lassman AB, et al. A phase 1 study evaluating ABT-414 in combination with temozolomide (TMZ) for subjects with recurrent or unresectable glioblastoma (GBM). J Clin Oncol. 2014;32(5S):2021. [Google Scholar]
- 12. Sampson JH, Heimberger AB, Archer GE, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28(31):4722–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frederick L, Eley G, Wang XY, et al. Analysis of genomic rearrangements associated with EGRFvIII expression suggests involvement of Alu repeat elements. Neuro Oncol. 2000;2(3):159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45(1):255–268. [PubMed] [Google Scholar]
- 15. Hobbs J, Nikiforova MN, Fardo DW, et al. Paradoxical relationship between the degree of EGFR amplification and outcome in glioblastomas. Am J Surg Pathol. 2012;36(8):1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. [DOI] [PubMed] [Google Scholar]
- 17. Erdem-Eraslan L, Gravendeel LA, de Rooi J, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol. 2013;31(3):328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montano N, Cenci T, Martini M, et al. Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Neoplasia. 2011;13(12):1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Del Vecchio CA, Giacomini CP, Vogel H, et al. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene. 2013;32(21):2670–2681. [DOI] [PubMed] [Google Scholar]
- 20. Francis JM, Zhang CZ, Maire CL, et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4(8):956–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 22. Nathanson DA, Gini B, Mottahedeh J, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343(6166):72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA. 2012;109(8):3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.