Abstract
MicroRNAs refer to small RNA molecules that destroy the messenger RNA by binding on them inhibiting the production of protein. However, the role of miR-155 in uveal melanoma metastasis remains largely unknown. In this study, we found that miR-155 was upregulated in both uveal melanoma cells and tissues. Transfection of miR-155 mimic into uveal melanoma cells led to an increase in cell growth and invasion; in contrast, inhibition of miR-155 resulted in opposite effects. Also, we identified Nedd4-family interacting protein 1 as a direct target of miR-155, and the expression of Nedd4-family interacting protein 1 was inhibited by miR-155. Furthermore, ectopic expression of Nedd4-family interacting protein 1 restored the effects of miR-155 on cell proliferation and invasion of uveal melanoma cells. In conclusion, miR-155 acts as a tumor promotor in uveal melanoma through increasing cell proliferation and invasion. Thus, miR-155 might serve as a potential therapeutic target in patients with uveal melanoma.
Keywords: miRNA, uveal melanoma, proliferation, invasion, NDFIP1
Introduction
Uveal melanoma is a rare cancer that is most common in adults. It develops in the anterior (iris) or the ciliary body or choroid, which is commonly referred to as the posterior. However, uvea is the second most common site for primary melanoma after the skin.1–3 Currently, due to inadequate information about the growth and dissemination of uveal tumors, there is no effective therapy available for patients with this disease.4,5 However, recent investigations have revealed novel prognostic factors and clinical therapeutic targets and have helped in understanding this disease.6–8
MicroRNAs (miRNAs) are an abundant class of endogenously expressed, noncoding, short RNAs that regulate gene expression.9–11 Through direct binding to complementary sequences at 3′ untranslated regions (3′UTRs), these small RNA molecules negatively impact the stability and translation of messenger RNAs (mRNAs).12,13 MicroRNAs are increasingly emerging as important regulators of tumorigenesis. miR-155 acts as an oncomir in various human cancers, including liposarcoma,14 lung cancer,15 cervical cancer,16 acute myeloid leukemia,17 hepatocellular carcinoma,18 breast cancer,19 and oral squamous carcinoma.20 However, its expression and potential molecular mechanism in uveal melanoma are not yet clear. Furthermore, the adaptor protein Nedd4-family interacting protein 1 (NDFIP1) plays a key role in the ubiquitination and nuclear translocation of PTEN.21 Nedd4-family interacting protein 1 represses cell proliferation and thus acts as a tumor suppressor.22 However, the mechanism of NDFIP1 downregulation in uveal melanoma is still unknown.
The function of miR-155 in uveal melanoma cells was investigated by this study. First, we illustrated that miR-155 was downregulated in uveal melanoma cells and tissues. Additionally, NDFIP1 was found to be a potential target of miR-155, and miR-155 decreased the NDFIP1 protein level in uveal melanoma cells. Furthermore, ectopic expression of NDFIP1 restored the effects of miR-155 on proliferation and invasion of uveal melanoma cells. In summary, these data suggest that miR-155 acts as a promoter for proliferation and invasion of uveal melanoma cells.
Materials and Methods
Cell Culture and Tumor Specimens
All studies and procedures involving human tissue were approved by Shanxi Ophthalmology Medical Center review board. All enrolled patients provided written informed consent. Tumor samples and their morphologically normal tissues were obtained from 25 patients with uveal melanoma and were immediately frozen in liquid nitrogen. All uveal melanomas in this study were choroidal melanomas. Human uveal melanoma cell lines (OCM-1A, MUM-2C, C918, and MUM-2B) and human melanocyte cell line (D78) were obtained from the Cell Bank of the Chinese Academy of Sciences (Beijing, People’s Republic of China). The OCM-1A, MUM-2C, and D78 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and C918 and MUM-2B in RPMI 1640. miR-155 mimic, inhibitor, and scramble were obtained from Sigma-Aldrich (St Louis, Missouri).
RNA Extraction, Reverse Transcription, and Real-Time Polymerase Chain Reaction
Total RNA from cultured cells was extracted by using the TRIzol reagent (Takara, Japan) in accordance with the manufacturer’s instructions. TaqMan miRNA assays (Applied Biosystems, Foster City, California) were used to test the expression levels of miR-155, and U6 small nuclear RNA was used as an internal control. Three independent experiments were performed to analyze the relative gene expression.
Cell Proliferation Assay
Cell proliferation was determined by the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) assay per manufacturer’s instructions. Absorbance was detected at 450 nm and measured by Quant Universal Microplate Spectrophotometer (BioTek Instruments, Inc, Winooski, USA).
Transwell Invasion Assay
Invasion assay was performed using Matrigel invasion chamber system (BD Biosciences, Bedford, Massachusetts). In brief, 1 × 105 cells in 200 μL of serum-free medium were added to the upper chamber, and 800 μL of DMEM with 10% serum as a chemoattractant was placed in the lower chamber. The plates were incubated for 18 hours at 37°C in 5% CO2. Cells that did not invade through the pores were removed with a cotton swab. Invasion of cells to the lower side of the filters were fixed and stained with crystal violet and were counted. Each experiment was repeated at least 3 times.
Luciferase Reporter Assay
Cells were plated in 24-well plates before transfection. After 36 hours, transfected cells in each well of 12-well plates were harvested with 100 μL of lysis buffer. Ten microliters of the cell extract was used to measure luciferase activity by using the Dual-Luciferase Reporter Assay System (Promega, Madison, Wisconsin). Three independent experiments were performed to analyze the activity.
Western Blot Analysis
Cells were washed with phosphate-buffered saline, collected, and total protein was extracted from the cells using Protein Extraction kit (Key-Gen, Nanjing, China). Protein concentrations were determined by Qubit Protein Assay Kit and Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, USA). Approximately 50 μg of total protein was separated using 10% or 12% SDS-PAGE (Sodium dodecyl sulfate polyacrylamide gel electrophoresis) and then transferred to polyvinylidene difluoride membrane, which was incubated with the antibody for NDFIP1 (ab116124; Abcam, Cambridge, Massachusetts) or GAPDH (Glyceraldehyde-3-phosphate dehydrogenase; Cell Signaling Technology, Beverly, Massachusetts). The membrane was washed and incubated with horseradish peroxidase–conjugated secondary antibody.
Statistical Analysis
Data were presented as mean ± standard deviation. The differences between 2 groups were determined by Student t test and the differences in more than 2 groups were determined by 1-way analysis of variance. All statistical analyses were performed using SPSS 16.0 (SPSS Inc, Chicago, USA). A P < .05 was considered as statistically significant.
Results
miR-155 Expression Is Upregulated in Uveal Melanoma Cells and Specimens
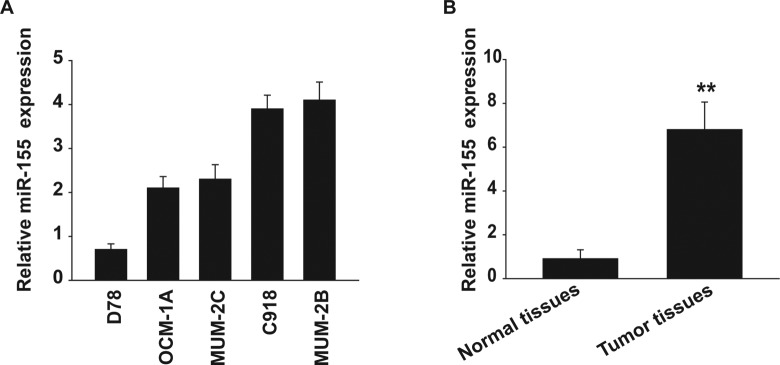
To determine whether miRNA was involved in the regulation of tumorigenesis of uveal melanoma cells, we first compared the expression of miR-155 between normal melanocytes and uveal melanoma cells. Real-time qualitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to detect the expression of miR-155 in 2 poorly invasive human uveal melanoma cell lines, OCM-1A and MUM-2C, 2 highly invasive human uveal melanoma cell lines, C918 and MUM-2B, and the normal uveal melanocyte cell line D78. miR-155 was highly expressed in all the uveal melanoma cell lines examined. In contrast, the expression of miR-155 was low in the melanocytes (Figure 1A).
Figure 1.
The expression of miR-155 is upregulated in uveal melanoma cells and specimens. A, The expression of miR-155 was measured in uveal melanoma (OCM-1A, MUM-2C, C918, and MUM-2B) and human melanocyte cell line (D78) using qRT-PCR. B, miR-155 was detected in 25 uveal melanoma samples by qRT-PCR. **P < .01. qRT-PCR indicates qualitative reverse transcription polymerase chain reaction.
Specimens from 25 human patients with uveal melanoma were analyzed to determine the expression pattern of miR-155 by using qRT-PCR. Consistent with the results from uveal melanoma cell lines, miR-155 was also increased in human uveal melanoma tissues in comparison with the normal uveal tissues (Figure 1B). These results indicate that the expression of miR-155 is upregulated in human uveal melanoma.
miR-155 Mimic Increases Proliferation and Invasion of Uveal Melanoma Cells
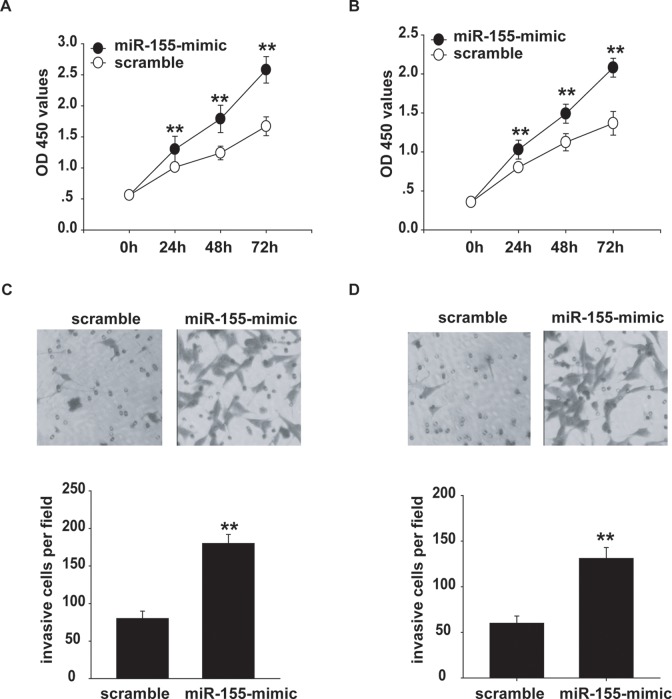
The next step was to determine whether miR-155 had any biological effect on melanoma cells. OCM-1A and MUM-2C cells were transfected with mR-155 mimic or scrambled oligonucleotides. After transfection, CCK-8 assay was carried out to determine the growth ability of the cells. miR-155 caused a dramatic increase in proliferation in the melanoma cells compared with that in the control cells (Figure 2A and B).
Figure 2.
miR-155 mimic increases proliferation and invasion of uveal melanoma cells. A and B, CCK-8 assay was performed to measure the OCM-1A (A) or MUM-2C (B) cell proliferation after transfection with miR-155 mimic or scramble. C and D, Invasion analysis of OCM-1A (C) or MUM-2C (D) cells after treatment with miR-155 mimics or scramble. **P < .01.
Consequently, we assessed the effect of miR-155 on invasion, a prerequisite for malignant transformation and metastasis, using transwell invasion assay. After transfection of OCM-1A and MUM-2C cells with either miR-155 mimic or scrambled oligonucleotides, invasion assays were performed. As shown in Figure 2C and D, the invasive ability of the miR-155 mimic-transfected cells increased significantly compared with that of the negative control–transfected cells. Thus, the results show that miR-155 promotes proliferation and invasion of uveal melanoma cells.
miR-155 Inhibitor Decreases Proliferation and Invasion of Uveal Melanoma Cells
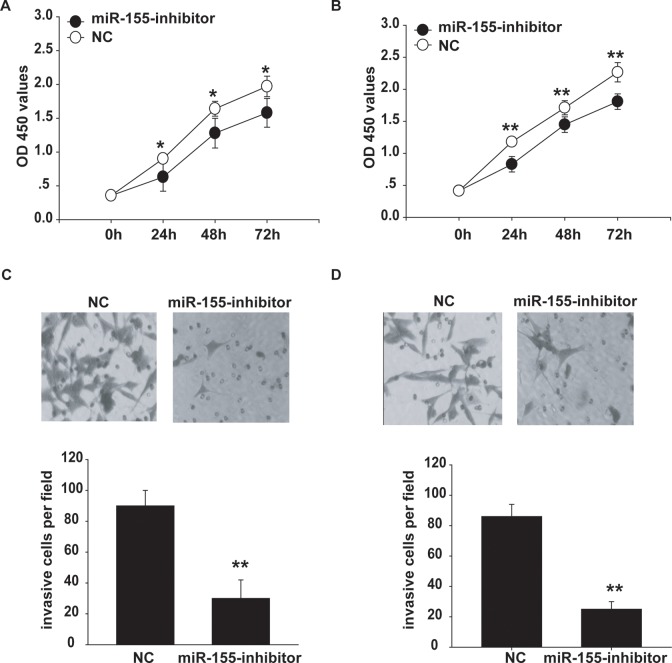
Next, we also determined whether inhibition of miR-155 had an impact on uveal melanoma cells. Two uveal melanoma cell lines, MUM-2B and C918, were transfected with miR-155 inhibitor or negative control. The inhibition of miR-155 markedly reduced proliferation in the 2 uveal melanoma cell lines over a 5-day interval (Figure 3A and B).
Figure 3.
miR-155 inhibitor decreases proliferation and invasion of uveal melanoma cells. A and B, CCK-8 assay was performed to measure the MUM-2B (A) or C918 (B) cell proliferation after transfection with miR-155 inhibitor or negative control. C and D, Invasion analysis of MUM-2B (C) or C918 (D) cells after treatment with miR-155 inhibitor or negative control. *P < .05, **P < .01.
We next performed transwell invasion assays. After transfection of MUM-2B and C918 cells with miR-155 inhibitor or negative control, the invasive capacity of the miR-155 inhibitor-transfected cells was found to be significantly lower than that of the negative control–transfected cells (Figure 3C and D). Thus, these results indicate that miR-155 inhibitor reduces cell proliferation and invasion of uveal melanoma cells.
Nedd4-Family Interacting Protein 1 Is a Target of miR-155
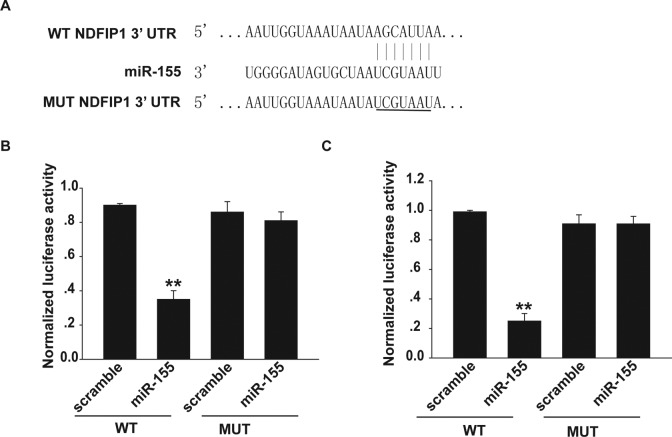
After demonstrating the role of miR-155 in uveal melanoma, we sought to determine the cellular mechanism underlying the miR-155-induced decrease in cell proliferation and invasion. We found that the 3′UTR of NDFIP1 contained highly conserved putative miR-155 binding sites (Figure 4A). As expected, luciferase activity of the wild-type pLuc-NDFIP1 3’UTR construct was significantly inhibited after the introduction of miR-155 into OCM-1A and MUM-2C cells but not after the introduction of the negative control (Figure 4B and C). The mutant pLuc-NDFIP1 3′UTR construct completely abolished the ability of miR-155 to regulate the expression of luciferase (Figure 4B and C). These results demonstrate that NDFIP1 is a potential target of miR-155.
Figure 4.
The NDFIP1 is a target of miR-155. A, The 3′UTR of NDFIP1 contained the highly conserved putative miR-155 binding sites by using TargetScan prediction software. B, Luciferase reporter analysis demonstrated that overexpression of miR-155 reduced luciferase activity in the NDFIP1 wild-type reporter gene but not the mutant type in OCM-1A cells. C, Luciferase reporter analysis was performed in MUM-2C cells. **P < .01. NDFIP1 indicates nedd4-family interacting protein 1; 3′UTR, 3′ untranslated regions.
miR-155 Downregulates NDFIP1 Expression in Uveal Melanoma Cells
To confirm whether miR-155 was indeed responsible for the downregulation of NDFIP1 in uveal melanoma cells, OCM-1A and MUM-2C cells were transfected with miR-155 mimic or scrambled oligonucleotides. Western blot analysis showed that the expression of NDFIP1 was dramatically reduced when OCM-1A and MUM-2C cells were transfected with miR-155 mimic (Figure 5A and B). In contrast, after the MUM-2B and C918 cells were transfected with miR-155 inhibitor or negative control, the expression of NDFIP1 was dramatically increased due to the inhibition of miR-155 (Figure 5C and D). These results demonstrate that miR-155 downregulates the expression of NDFIP1 in uveal melanoma cells.
Figure 5.
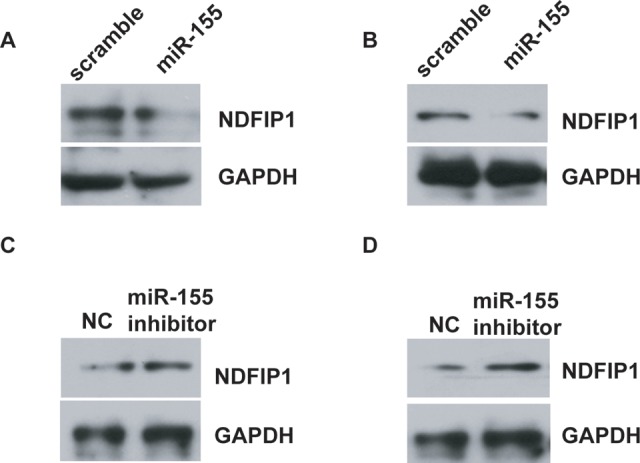
miR-155 downregulates NDFIP1 expression in uveal melanoma cells. A and B, Western blot analysis has shown that miR-155 mimic inhibited the protein expression of NDFIP1 in OCM-1A (A) or MUM-2C (B) cells. GAPDH was also detected as a loading control. C and D, Western blot analysis has shown that miR-155 inhibitor increased the protein expression of NDFIP1 in MUM-2B (C) or C918 (D) cells. GAPDH was also detected as a loading control. NDFIP1 indicates nedd4-family interacting protein 1.
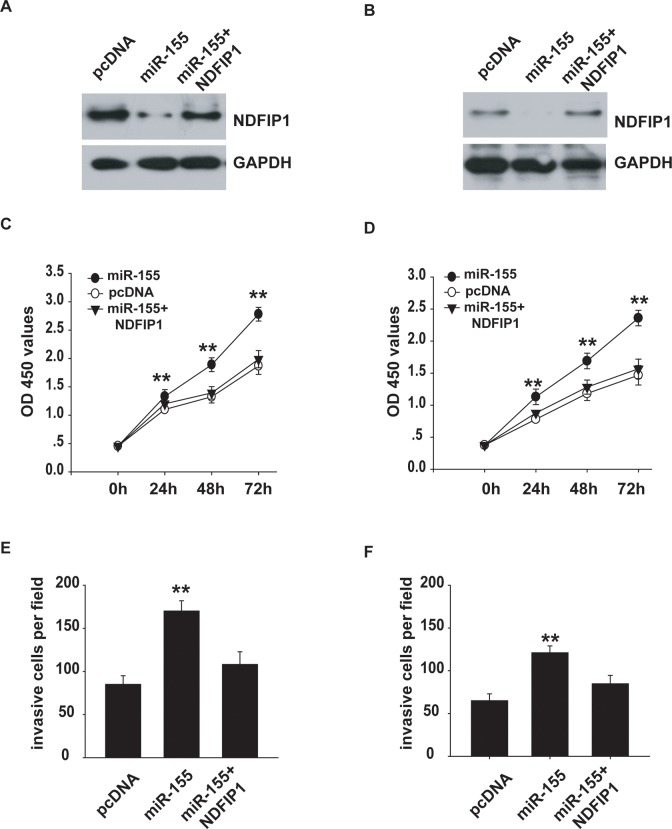
Ectopic Expression of NDFIP1 Restores the Effects of miR-155 on Proliferation and Invasion of Uveal Melanoma Cells
Finally, we studied the functional involvement of NDFIP1 in the regulation of proliferation and invasion of uveal melanoma cells in rescue experiments. We cotransfected OCM-1A and MUM-2C cells with miR-155 and pcDNA3.1-NDFIP1 or empty vector (pcDNA3.1). Western blot analysis was used to confirm the transfection efficiency (Figure 6A and B). Subsequently, a CCK-8 assay was performed with the treated uveal melanoma cells, and our results showed that NDFIP1 overexpression partially reversed the promotion of cell proliferation caused by miR-155 in OCM-1A and MUM-2C cells (Figure 6C and D). Furthermore, NDFIP1 overexpression overcame miR-155-dependent promotion of cell invasion (Figure 6E and F) in OCM-1A and MUM-2C cells. Collectively, our data reveal that NDFIP1 participates in the miR-155-dependent negative regulation of proliferation and invasion of uveal melanoma cells.
Figure 6.
Ectopic expression of NDFIP1 restored the effects of miR-155 on cell proliferation and invasion of uveal melanoma cells. A and B, Western blot analysis has shown that the protein expression of NDFIP1 in OCM-1A (A) or MUM-2C (B) cells. GAPDH was also detected as a loading control. C and D, The CCK-8 assay was done to evaluate the reverse by NDFIP1 of miR-155-induced promotion of cell growth in MUM-2B (C) or C918 (D) cells. E and F, The restorative effect of NDFIP1 on miR-155 upregulated invasion in MUM-2B (E) or C918 (F) cells. **P < .01. CCK-8 indicates Cell Counting Kit-8; NDFIP1, nedd4-family interacting protein 1.
Discussion
According to recent evidence, miRNAs have a regulatory role in the pathogenesis of cancer in humans. This is achieved through the suppression of genes involved in cell proliferation, differentiation, and invasion.23,24 The global suppression of miRNAs leads to increased tumorigenesis and cellular transformation. Recent studies have unveiled a list of potential candidates crucial for human tumors.25,26 Studies have demonstrated that miR-155 promotes or inhibits the proliferation and invasion of many types of cancer cell lines via posttranscriptional silencing of mRNAs.27–29 Thus far, there is no known role of miR-155 in uveal melanoma cells.
In the present study, we found that the function of miR-155 is associated with NDFIP1. We compared the expression of miR-155 between melanoma tissues and normal tissues. miR-155 overexpression promoted proliferation and invasion of uveal melanoma cells; inversely, the inhibition of miR-155 resulted in reverse effects. Furthermore, we identified that NDFIP1 is a direct target of miR-155. Our results demonstrated that ectopic expression of NDFIP1 restored the effects of miR-155 on proliferation and invasion of uveal melanoma cells.
To explore the molecular mechanism of miR-155, we first determined that the 3′UTR of NDFIP1 contained highly putative miR-155 binding sites by using TargetScan prediction software. Next, we revealed that ectopic expression of miR-155 was linked to the suppression of luciferase activity. Additionally, we observed that miR-155 overexpression decreased the level of NDFIP1 protein and the inhibition of miR-155 increased its level.
PTEN has been demonstrated to be a tumor suppressor and a critical factor in the development and progression of various cancers, including uveal melanoma.30–32 Nedd4-family interacting protein 1 is required for both the ubiquitination and nuclear translocation of PTEN on endosomes.21 Previous studies also identified that NDFIP1 inhibits cell proliferation by regulating localization and signaling specificity of PTEN.22 The ability of miR-155 to target NDFIP1 may be one such mechanism of posttranscriptional control of NDFIP1.
In conclusion, we demonstrated that miR-155 is frequently overexpressed in uveal melanoma tissues and cell lines. miR-155 plays a major role in the malignant progression of uveal melanoma cells by directly regulating the expression of NDFIP1. According to the above findings, it is possible that miR-155 can be used as a therapeutic target to treat patients with this disease.
Abbreviations
- CCK-8
Cell Counting Kit-8
- DMEM
Dulbecco’s modified Eagle’s medium
- miRNAs
microRNAs
- mRNAs
messenger RNAs
- NDFIP1
Nedd4-family interacting protein 1
- qRT-PCR
qualitative reverse transcription polymerase chain reaction
- 3′UTRs
3′ untranslated regions.
Footnotes
Author Contributions: J.P. and H.L. contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by key project of Shanxi Health and Family Planning Commission (2016D041 and 2014D50).
References
- 1. Oliva M, Rullan AJ, Piulats JM. Uveal melanoma as a target for immune-therapy. Ann Transl Med. 2016;4(9):172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nichols EE, Richmond A, Daniels AB. Tumor characteristics, genetics, management, and the risk of metastasis in uveal melanoma. Semin Ophthalmol. 2016;31(4):304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chattopadhyay C, Kim DW, Gombos DS, et al. Uveal melanoma: from diagnosis to treatment and the science in between. Cancer. 2016;122(15):2299–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tarlan B, Kıratlı H. Uveal melanoma: current trends in diagnosis and management. Turk J Ophthalmol. 2016;46(3):123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carvajal RD, Schwartz GK, Tezel T, Marr B, Francis JH, Nathan PD. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol. 2017;101(1):38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caltabiano R, Puzzo L, Barresi V, et al. ADAM 10 expression in primary uveal melanoma as prognostic factor for risk of metastasis. Pathol Res Pract. 2016;212(11):980–987 [DOI] [PubMed] [Google Scholar]
- 7. Field MG, Durante MA, Decatur CL, et al. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in class 1 and class 2 uveal melanomas. Oncotarget. 2016;7(37):59209–59219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pan H, Ni H, Zhang L, et al. P2RX7-V3 is a novel oncogene that promotes tumorigenesis in uveal melanoma. Tumour Biol. 2016;37(10):13533–13543 [DOI] [PubMed] [Google Scholar]
- 9. Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 2013;5(12):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leal JA, Lleonart ME. MicroRNAs and cancer stem cells: therapeutic approaches and future perspectives. Cancer Lett. 2013;338(1):174–183. [DOI] [PubMed] [Google Scholar]
- 11. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6(11):8474–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kapodistrias N, Mavridis K, Batistatou A, et al. Assessing the clinical value of microRNAs in formalin-fixed paraffin-embedded liposarcoma tissues: overexpressed miR-155 is an indicator of poor prognosis. Oncotarget. 2017;8(4):6896–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xue X, Liu Y, Wang Y, et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget. 2016;7(51):84508–84519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang H, Shuang D, Yi Z, Sheng H, Liu Y. Up-regulated microRNA-155 expression is associated with poor prognosis in cervical cancer patients. Biomed Pharmacother. 2016;83:64–69. [DOI] [PubMed] [Google Scholar]
- 17. Ramamurthy R, Hughes M, Morris V, et al. miR-155 expression and correlation with clinical outcome in pediatric AML: a report from Children’s Oncology Group. Pediatr Blood Cancer. 2016;63(12):2096–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ji J, Xu M, Tu J, et al. MiR-155 and its functional variant rs767649 contribute to the susceptibility and survival of hepatocellular carcinoma. Oncotarget. 2016;7(37):60303–60309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tiago DM, Conceição N, Caiado H, Laizé V, Cancela ML. Matrix Gla protein repression by miR-155 promotes oncogenic signals in breast cancer MCF-7 cells. FEBS Lett. 2016;590(8):1234–1241 [DOI] [PubMed] [Google Scholar]
- 20. Zeng Q, Tao X, Huang F, et al. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int J Mol Med. 2016;37(5):1274–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howitt J, Lackovic J, Low LH, et al. Ndfip1 regulates nuclear Pten import in vivo to promote neuronal survival following cerebral ischemia. J Cell Biol. 2012;196(1):29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Howitt J, Low LH, Putz U, et al. Ndfip1 represses cell proliferation by controlling Pten localization and signaling specificity. J Mol Cell Biol. 2015;7(2):119–131 [DOI] [PubMed] [Google Scholar]
- 23. Behbahani GD, Ghahhari NM, Javidi MA, Molan AF, Feizi N, Babashah S. MicroRNA-mediated post-transcriptional regulation of epithelial to mesenchymal transition in cancer. Pathol Oncol Res. 2017;23(1):1–12. [DOI] [PubMed] [Google Scholar]
- 24. Momtazi AA, Shahabipour F, Khatibi S, Johnston TP, Pirro M, Sahebkar A. Curcumin as a MicroRNA regulator in cancer: a review. Rev Physiol Biochem Pharmacol. 2016;171:1–38. [DOI] [PubMed] [Google Scholar]
- 25. Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kinoshita T, Yip KW, Spence T, Liu FF. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J Hum Genet. 2017;62(1):67–74. [DOI] [PubMed] [Google Scholar]
- 27. Liu WJ, Zhao YP, Zhang TP, et al. MLH1 as a direct target of MiR-155 and a potential predictor of favorable prognosis in pancreatic cancer. J Gastrointest Surg. 2013;17(8):1399–1405. [DOI] [PubMed] [Google Scholar]
- 28. Kopp KL, Ralfkiaer U, Gjerdrum LM, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013;12(12):1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu F, Jia X, Du F, et al. miR-155-deficient bone marrow promotes tumor metastasis. Mol Cancer Res. 2013;11(8):923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okumura N, Yoshida H, Kitagishi Y, Nishimura Y, Matsuda S. Alternative splicings on p53, BRCA1 and PTEN genes involved in breast cancer. Biochem Biophys Res Commun. 2011;413(3):395–399. [DOI] [PubMed] [Google Scholar]
- 31. Georgescu MM. PTEN tumor suppressor network in PI3K-akt pathway control. Genes Cancer. 2010;1(12):1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abdel-Rahman MH, Yang Y, Zhou XP, Craig EL, Davidorf FH, Eng C. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J Clin Oncol. 2006;24(2):288–295. [DOI] [PubMed] [Google Scholar]